Abstract
The canonical DNA glycosylase role is global base damage repair but includes functions in epigenetic gene regulation, immune response modulation, replication, and transcription. In this issue, Eckenroth et al., 2020 present the NEIL2 glycosylase structure. Its catalytic domain flexibility differentiates it from most other glycosylases and suggests novel regulatory mechanisms.
Critical for genome maintenance, DNA repair pathways face two major mechanistic challenges. First, DNA repair intermediates can be more toxic than the DNA damage itself. Thus, many eukaryotic DNA repair proteins remain bound to their product and act as chaperones to prevent diversion off pathway. Commonly interacting through intrinsically disordered regions at the N- and C-termini, downstream proteins then promote product release (Liu and Roy, 2002). Second, DNA repair proteins must distinguish uncommon occurrences of DNA damage from undamaged DNA, whether within double stranded (ds) or single stranded (ss) DNA in replication forks or transcription. For recognition, DNA repair proteins test for their specific DNA damage through distortion. The first enzymes in the base excision repair (BER), of which NEIL2 is a member, often recognize their target, DNA base damage, by destabilized base stacking. NEIL2 acts on 5-hydroxyuracil (5-hU), abasic sites, 5-guanidinohydantoin (Gh), Spiroiminodihydantoin (Sp), and 4,6-diamino-5-formamidopyrimidine (FapyA) in preferentially ssDNA over dsDNA. Such base damages are predicted to disrupt base stacking by disturbing the base structure or mispairing. To find their target damage substrate, glycosylases bend and/or flip nucleotides out from the double helix, prior to specific recognition of the base damage.
In this issue, Eckenroth and coworkers present the first NEIL2 structure. Although it is DNA-free and the exact structural mechanisms underlying its glycosylase activity was not revealed, it is clear from crystal structure and the Small Angle X-ray Scattering (SAXS) data that NEIL2 is unlike most BER glycosylases. Typically, the BER enzyme’s catalytic core acts as an inflexible mold, testing for the characteristics of the DNA to be distorted. Intrinsically disordered regions are typically in the termini. BER crystal structures of both monofunctional (Uracil N-Glycosylase/UNG, Alkylation DNA glycosylase/AlkA) and bifunctional glycosylases (8-Oxoguanine DNA Glycosylase /OGG1, endonuclease III/NTH, Nei Like DNA Glycosylase 1/NEIL1, and NEIL3) show little change between DNA-free and DNA-bound structures (Fig. 1) (Duclos et al., 2012; Faucher et al., 2009; Faucher et al., 2010; Fromme and Verdine, 2003; Golan et al., 2005; Hollis et al., 2000; Parikh et al., 1998; Sarre et al., 2015; Slupphaug et al., 1996; Teale et al., 2002; Zhu et al., 2016). Local shifts in loops and closing to pinch the damaged strand and bend DNA occur, but the protein core fold remains mostly unchanged with root mean square deviation (RMSD) of less than 3 Å.
Fig. 1. NEIL2 catalytic domain flexibility breaks the rule from most DNA repair glycosylases.
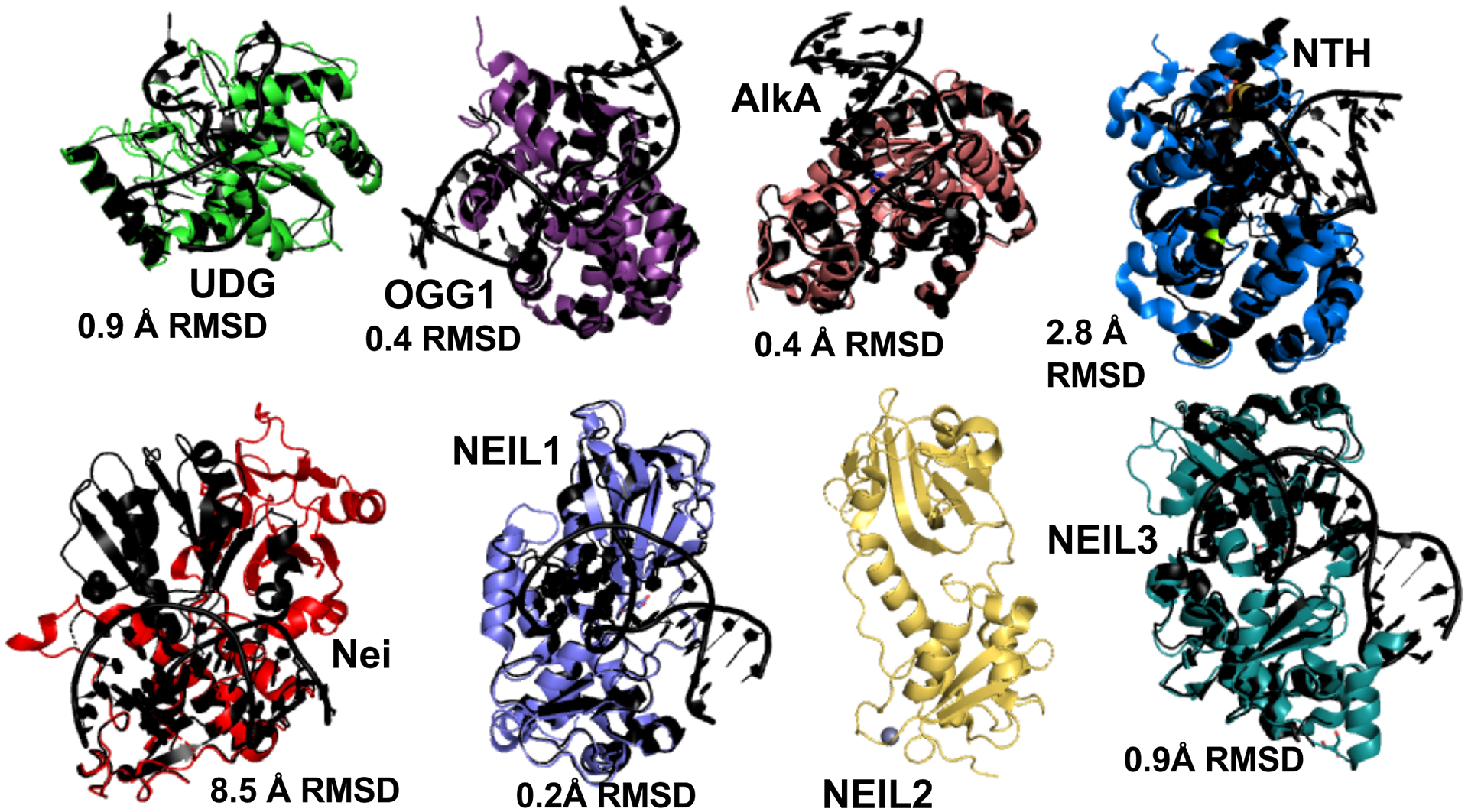
To identify damaged bases, DNA repair glycosylases typically provide rigid frames to sculpt DNA into a distorted conformation, often with damage nucleotides flipped out. DNA-free NEIL2, in contrast, would require a major rearrangement of its catalytic domains to be catalytically competent. Shown are DNA-free glycosylase structures (color) overlaid on DNA-bound structures (black). UNG (PDB:1AKZ/4SKN), 3-methyladenine DNA glycosylase AlkA (PDB:1PVS, 1DIZ), OGG1 (PDB:3FHF, 3KNT), NTH (PDB:4UNF,1P59), Nei (PDB:1Q3b, 2EA0), NEIL1(PDB:1TDH, 5ITR), NEIL2 (6VJI), NEIL3 (3TWK,3TWM).
NEIL2 breaks this rigidity rule. In Eckenroth et al, (2020), the crystal structure and SAXS of the DNA-free enzyme indicate that the two lobes of its catalytic core are not in a catalytically competent orientation, as compared to its paralogs, NEIL1 and NEIL3 structures. This flexibility is distinct from more common disordered termini that tether partner proteins. By breaking the catalytic core, NEIL2 is by default inactive until the catalytic domains come together in the proper orientation. Thus, NEIL2 would require a rare conformational change within the catalytic core, to achieve catalytic competency. Its bacterial ortholog Endonuclease VIII (Nei), as noted by Eckenroth and coworkers (2020), also shows a requirement for significant reorganization.
Why NEIL2 requires such a regulatory mechanism remains enigmatic. The NEIL protein family is exceptional to other repair enzymes in its links to non-repair-related functions in epigenetic gene activation, modulation of immune response, replication, and transcription), so perhaps NEIL2 needs greater regulation. One potential reason for flexibility is to mechanistically recognize its diverse damaged DNA substrates. Only when its targeted DNA substrate is bound will the enzyme transform into a catalytically competent conformation. NEIL2 works on damage preferentially in ssDNA and bubble DNA, and the energetic cost of nucleotide flipping of undamaged DNA versus damaged DNA is not as severe as in paired ds DNA, thus setting the need for a clever substrate recognition mechanism. FEN1 nuclease uses both DNA-induced protein conformational changes and protein-induced DNA sculpting to validate its flap substrate and license incision (Tsutakawa et al., 2011). NEIL3 also acts on damage in ssDNA, but its catalytic core is intact. Eckenroth and co-workers reason that catalytic core flexibility allows tethering of partner proteins, while still allowing specific DNA binding. Perhaps these proteins license the glycosylase activity to occur at specific locations as exemplified by the interaction and stimulation of NEIL2 activity by YB-1 transcription factor (Das et al., 2007). Yet, NEIL2 has similar activity to NEIL1 and NEIL3 in vitro in the absence of partner proteins, suggesting that the DNA substrate is sufficient to reorient the catalytic domains, putting partner protein interactions as a means to raise or lower glycosylase activity, but not for absolute activation. NEIL2 may have additional non-enzymatic roles where glycosylase activity is purposefully disengaged, as observed for XPG nuclease (Sarker et al., 2005). This NEIL2 structure, breaking the rules for a rigid and preformed catalytic core, suggests that exploration of NEIL2 regulation will be interesting in the years to come.
Acknowledgements.
We thank John Tainer for feedback on the preview. Dr. Tsutakawa is supported by NCI P01 CA092584 and NIH R01GM110387, and Altaf Sarker by Grant 26IR-0017 UC-TRDRP.
References:
- Bacolla A, Sengupta S, Ye Z, Yang C, Mitra J, De-Paula RB, Hegde ML, Ahmed Z, Mort M, Cooper DN, et al. (2020). Heritable pattern of oxidized DNA base repair coincides with pre-targeting of repair complexes to open chromatin. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty A, Wakamiya M, Venkova-Canova T, Pandita RK, Aguilera-Aguirre L, Sarker AH, Singh DK, Hosoki K, Wood TG, Sharma G, et al. (2015). Neil2-null Mice Accumulate Oxidized DNA Bases in the Transcriptionally Active Sequences of the Genome and Are Susceptible to Innate Inflammation. J Biol Chem 290, 24636–24648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Chattopadhyay R, Bhakat KK, Boldogh I, Kohno K, Prasad R, Wilson SH, and Hazra TK (2007). Stimulation of NEIL2-mediated oxidized base excision repair via YB-1 interaction during oxidative stress. J Biol Chem 282, 28474–28484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duclos S, Aller P, Jaruga P, Dizdaroglu M, Wallace SS, and Doublie S (2012). Structural and biochemical studies of a plant formamidopyrimidine-DNA glycosylase reveal why eukaryotic Fpg glycosylases do not excise 8-oxoguanine. DNA Repair (Amst) 11, 714–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucher F, Duclos S, Bandaru V, Wallace SS, and Doublie S (2009). Crystal structures of two archaeal 8-oxoguanine DNA glycosylases provide structural insight into guanine/8-oxoguanine distinction. Structure 17, 703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucher F, Wallace SS, and Doublie S (2010). The C-terminal lysine of Ogg2 DNA glycosylases is a major molecular determinant for guanine/8-oxoguanine distinction. J Mol Biol 397, 46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromme JC, and Verdine GL (2003). Structure of a trapped endonuclease III-DNA covalent intermediate. EMBO J 22, 3461–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golan G, Zharkov DO, Feinberg H, Fernandes AS, Zaika EI, Kycia JH, Grollman AP, and Shoham G (2005). Structure of the uncomplexed DNA repair enzyme endonuclease VIII indicates significant interdomain flexibility. Nucleic Acids Res 33, 5006–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, and Mitra S (2002). Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc Natl Acad Sci U S A 99, 3523–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollis T, Ichikawa Y, and Ellenberger T (2000). DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA. EMBO J 19, 758–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, and Roy R (2002). Truncation of amino-terminal tail stimulates activity of human endonuclease III (hNTH1). J Mol Biol 321, 265–276. [DOI] [PubMed] [Google Scholar]
- Parikh SS, Mol CD, Slupphaug G, Bharati S, Krokan HE, and Tainer JA (1998). Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J 17, 5214–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker AH, Tsutakawa SE, Kostek S, Ng C, Shin DS, Peris M, Campeau E, Tainer JA, Nogales E, and Cooper PK (2005). Recognition of RNA polymerase II and transcription bubbles by XPG, CSB, and TFIIH: insights for transcription-coupled repair and Cockayne Syndrome. Mol Cell 20, 187–198. [DOI] [PubMed] [Google Scholar]
- Sarre A, Okvist M, Klar T, Hall DR, Smalas AO, McSweeney S, Timmins J, and Moe E (2015). Structural and functional characterization of two unusual endonuclease III enzymes from Deinococcus radiodurans. J Struct Biol 191, 87–99. [DOI] [PubMed] [Google Scholar]
- Schomacher L, Han D, Musheev MU, Arab K, Kienhofer S, von Seggern A, and Niehrs C (2016). Neil DNA glycosylases promote substrate turnover by Tdg during DNA demethylation. Nat Struct Mol Biol 23, 116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupphaug G, Mol CD, Kavli B, Arvai AS, Krokan HE, and Tainer JA (1996). A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature 384, 87–92. [DOI] [PubMed] [Google Scholar]
- Teale M, Symersky J, and DeLucas L (2002). 3-methyladenine-DNA glycosylase II: the crystal structure of an AlkA-hypoxanthine complex suggests the possibility of product inhibition. Bioconjug Chem 13, 403–407. [DOI] [PubMed] [Google Scholar]
- Tsutakawa SE, Classen S, Chapados BR, Arvai AS, Finger LD, Guenther G, Tomlinson CG, Thompson P, Sarker AH, Shen B, et al. (2011). Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell 145, 198–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Lu L, Zhang J, Yue Z, Song J, Zong S, Liu M, Stovicek O, Gao YQ, and Yi C (2016). Tautomerization-dependent recognition and excision of oxidation damage in base-excision DNA repair. Proc Natl Acad Sci U S A 113, 7792–7797. [DOI] [PMC free article] [PubMed] [Google Scholar]


