Abstract
The genetic architecture of each individual is comprised of common and rare variants acting alone and in combination to confer risk for disease. The cell-type-specific and/or context-dependent functional consequences of the risk variants linked to brain disease needs to be resolved. Coupling human induced pluripotent stem cell (hiPSC)-based technology with CRISPR-based genome engineering facilitates precise isogenic comparisons of variants across genetic backgrounds. Although functional validation studies are still typically performed of one variant in isolation, and in one cell type at a time, complex genetic diseases require multiplexed gene perturbations to interrogate combinations of genes and resolve physiologically relevant disease biology. Our aim is to discuss advances at the intersection of genomics, hiPSCs and CRISPR. A better understanding of the molecular mechanisms underlying disease risk will improve genetic diagnosis, drive phenotypic drug discovery, and pave the way towards precision medicine.
Keywords: Human induced pluripotent stem cells, neurons, genetics, psychiatric disease, neurodegenerative disease, CRISPR-Cas, disease modeling
INTRODUCTION
Although genetic studies have identified hundreds of common and rare variants significantly associated with neurodegenerative and psychiatric diseases (reviewed 1), large-scale genetic approaches have often failed to deliver actionable results or new guidance for clinical or translational research 2. Genetic, epigenetic and transcriptomic datasets can inform the search for putative causal variants, but novel experimental paradigms are urgently needed to explore the phenotypic impact of hundreds of disease associated loci (reviewed in 1). Moreover, because risk loci for brain diseases are enriched at defined developmental windows 3 and in specific cell types 4,5, genetic variants must be evaluated in the relevant neural cell types at the appropriate stage of maturity.
Human induced pluripotent stem cells (hiPSCs)-based models represent patient-specific platforms with which to study neurodegenerative and psychiatric disease. First, we discuss how hiPSCs can now be used to generate all of the major cell types in the brain, which fortuitously, generally resemble fetal brain tissue6-10, making them particularly well-suited for the functional evaluation of genetic variants linked to psychiatric disease risk (as opposed to the study of processes observed late in disease-state). Second, we consider how combining hiPSC-based models with advanced applications of Clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9-based systems for (epi)genome and transcriptome engineering makes possible the study of combinatorial disease-relevant perturbations in cell-type specific isogenic systems (as highlighted by 11). Third, we consider how genotype-based diagnosis and treatment makes possible a future whereby patients are identified and treated prior to symptom onset, dramatically expanding the therapeutic window of intervention, and making possible the prevention rather than treatment of disease 12,13.
hiPSC-BASED PLATFORMS TO EVALUATE GENOMIC HYPOTHESES
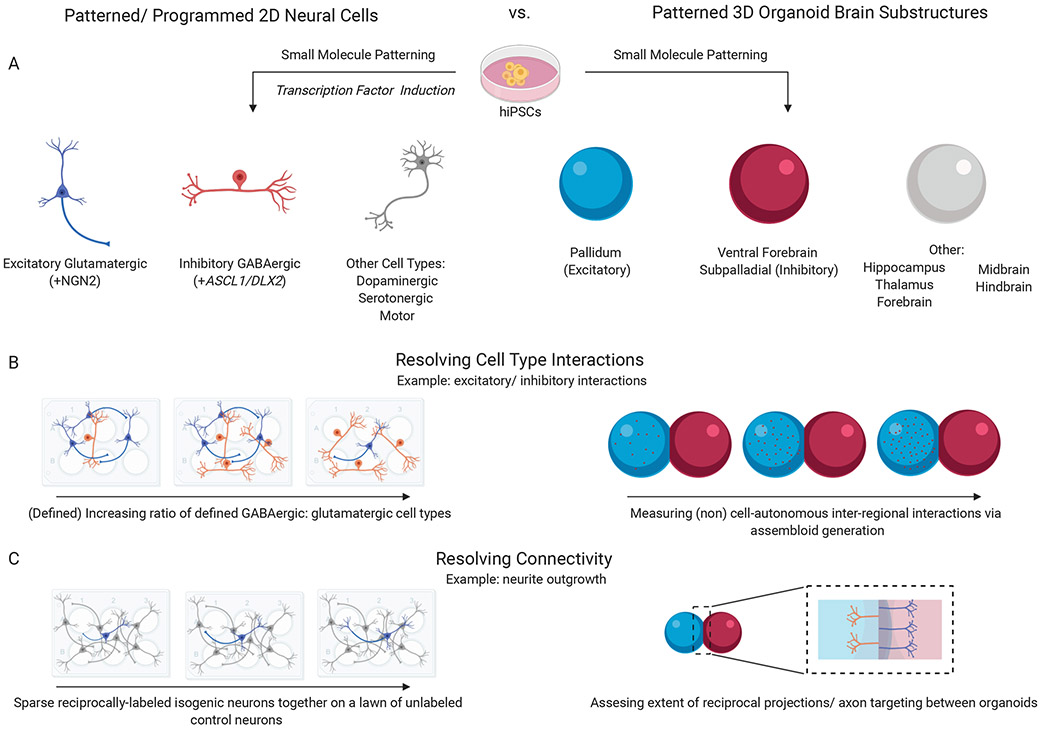
Today, hiPSC studies can be applied to defined and scalable two-dimensional (2D) cultures and/or more complex and physiologically relevant three-dimensional (3D) organoid systems (Fig. 1). While 2D neuron cultures require weeks to achieve electrophysiological maturity, organoids typically require months to demonstrate comparable properties8. In either case, the immaturity of hiPSC-derived neurons and glia relative to the human brain, generally resembling fetal brain tissue6-10, makes them particularly well-suited for the functional evaluation of genetic variants linked to psychiatric disease risk. The strengths and limitations of both approaches, discussed below, are well-counter balanced. We see great utility in applying these approaches together to screen, discover and validate the physiological relevance of disease-specific risk variants.
Figure 1: 2D vs. 3D culture systems to resolve cell type effects and interactions.
A, Schematic of types of 2D cell cultures and 3D substructures via small molecule patterning or transcription factor induction (2D only). B, Example assays to resolve cell type interactions. 2D-defined generation of glutamatergic and GABAergic cultures and 3D fusion of excitatory and inhibitory organoids (assembloid). C. Example assays to resolve connectivity in 2D sparsely seeded isogenic neurons on unlabeled control cells, or 3D organoid projection/axon targeting assessment. All figures were created using Biorender.com.
Today, it is possible to rapidly generate highly pure populations of glutamatergic 14, GABAergic 15, dopaminergic 16, serotonergic 17 and motor neurons 18, as well as astrocytes 19 and oligodendrocytes 20 through transcription factor-based induction methods. Defined “co-cultures” of two or more cell types further permits the analysis of neuron-glia 21 and neuron-neuron 22 interactions, and so can better recapitulate a more physiologically relevant context. While the advantages of 2D platforms include scalability and ease of manipulation, limitations reflect technical constraints on the length of time that cells can adhere to plastic, which limit extended time courses of maturation, coupled with unnatural physical restrictions on cellular dynamics (e.g., migration of neural progenitor cells) 23. Conversely, organoids improve the physiological relevance of hiPSC experiments by making possible complex 3D interactions between neurons, astrocytes 24, oligodendrocytes 25, vascular 26,27 and microglia 28. Furthermore, directed differentiation results in region-specific patterned organoids (e.g. excitatory 29-31, inhibitory29-31, and dorsal thalamic32) that can be fused to yield “assembloids” for the study of cell-cell interactions and non-cell autonomous effects in a more physiologically relevant context 31-33. While early (undirected) 3D approaches yielded highly variable populations within and between organoids, differentiations, and donors 34, new methods that apply patterning signals to achieve improved reproducibility, particular across extended differentiation timelines 35,36, make possible controlled differentiation for disease modeling.
CRISPR-MEDIATED FUNCTIONAL VALIDATION OF DISEASE RISK
By precisely targeting a growing diversity of CRISPR proteins and effectors to defined genomic loci via a synthetically delivered guide RNA (gRNA), CRISPR-based technologies have transformed our ability to reengineer the human genome, epigenome and transcriptome (reviewed in 37). Today, the integration of CRISPR technology with patient-specific hiPSC-based studies makes possible the functional validation of putative causal variants and genes in a cell-type-specific and donor-dependent manner.38 Of course, complex genetic diseases do not arise from unidirectional differences in gene network expression (e.g. some common variants are predicted to up-regulate cis-genes, others to down-regulate), and so we also discuss future strategies to engineer large-scale bidirectional gene network perturbations via combinatorial effectors and systems.
Prioritization of Risk Variants for CRISPR-based Perturbation
Most loci identified in genome-wide association studies (GWAS) are poor candidates for CRISPR editing. Fine mapping analysis 39 only rarely identifies one single-nucleotide polymorphism (SNP) that is an excellent candidate for CRISPR editing, particularly if this SNP is expected to overlap with putative promoters or enhancers. More commonly, post-mortem expression and GWAS data are integrated to test for colocalization of expression quantitative trait loci (eQTL) and GWAS associations (e.g. COLOC40,41) and to calculate predicted differential expression in the brain (e.g. prediXcan42). Such multi-SNP approaches infer the magnitude and directionality of gene expression perturbations with tissue-level precision, and so instead prioritize genes for CRISPRa- or CRISPRi-based studies.
CRISPR Engineering the Genome and Transcriptome
The Cas9 nuclease achieves genomic editing via double stranded DNA breaks to induce insertions and deletions at gRNA targeted sites 43, with its greatest limitations being the efficiency of editing achieved and the frequency of off-target effects. Even as new CRISPR systems are identified and applied to genome editing, including Cpf1 44 with increased activity and targeting ranges 45, and CasX 46 with increased editing specificities, research and clinical applications of genome editors remain constrained by the occurrence of off-target effects 47. “Prime editing”, which relies upon the fusion of Cas9 to an engineered reverse transcriptase, enables efficient genome editing without double stranded DNA breaks, dramatically reducing off-target effects and promising wider applications of genome base editing 48.
Alternative applications of CRISPR platforms function via an enzymatically dead nuclease (dCas9) fused to a variety of effectors, which can be targeted to specific regions of the genome or transcriptome49. For example, CRISPRi/a use fusions of dCas9 to a Krüppel-associated box (KRAB) repressor domain for inactivation 50, or tripartite activator VP64-p65-Rta (VPR) 51 for activation, among others (Table 1). While the ability to edit single genes with relative ease is useful, the power of CRISPR systems is greatly enhanced by the ability to manipulate large numbers of genes simultaneously using pooled or multiplexable platforms.
Table 1:
A brief list of CRISPR-based technologies with potential applicability to hiPSC-based experiments. More plasmids available at: https://www.addgene.org/crispr/.
| CRISPR Technology | Mammalian Cell Applications | hiPSC/hESC Applications |
Addgene No(s). |
|
|---|---|---|---|---|
| Genome Engineering | Cas9 | HEK293T, HUES975 | hiPSCs76 | 62988 |
| Cpf1 | HEK293T44 | hiPSC77 | 69982 | |
| CasX | HEK293T46 | |||
| Transcriptional Regulation | dCas9-VPR (Gene Activation) | HEK293T51 | hiPSCs51 hiPSC-derived NPCs, neurons and astrocytes78 |
63798 99373 |
| dCas9-KRAB (Gene Repression) | HEK293T50 | hiPSC-derived NPCs, neurons and astrocytes78 | 71237 99372 |
|
| dCas9-SunTag (Gene Activation) | U20S79 | hiPSCs80 | 60904 | |
| SAM (Gene Activation) | HEK293T81 | 75112 | ||
| dCpf1-VPR (Gene Activation) | HEK293T82 | 104567 | ||
| Epigenetic Regulation | dCas9-p300 (Histone Acetylation) | HEK293T83 | 83889 | |
| dCas9-LSD1 (Histone Demethylation) | Chicken Embryos84 | 92362 | ||
| dCas9-DNMT3A or dCas9-MQ1 (Cytosine methylation) | Mouse Embryonic Stem Cells85 | hESCs86 | 84569 | |
| dCas9-Tet1 (Cytosine demethylation) | Mouse Embryonic Stem Cells85 | hiPSC-derived neurons87 | 84475 | |
| Transcriptome Engineering | Cas13a (RNA Targeting) | HEK293T88 | 91902 | |
| Cas13b (RNA Editing) | HEK293T89 | 103854 | ||
| CasRx (Cas13d) | HEK293T54 | hiPSCs54 | 109049 |
Large-Scale Network Engineering
We recently applied combinatorial perturbation of four schizophrenia (SZ)-associated risk genes 52. Our results suggested that the downstream effects of combinatorial perturbation exceeded what would be expected from the additive effect of individually perturbed genes. Observed synergistic genes converged on synaptic function, and linked rare and common variant genes implicated in psychiatric disease risk, emphasizing the importance of considering the polygenic nature of SZ and other neuropsychiatric disorders. Future studies must investigate the impact of combinatorial perturbations of dozens to hundreds of risk variants, each in the appropriate disease-relevant direction (some up, others down).
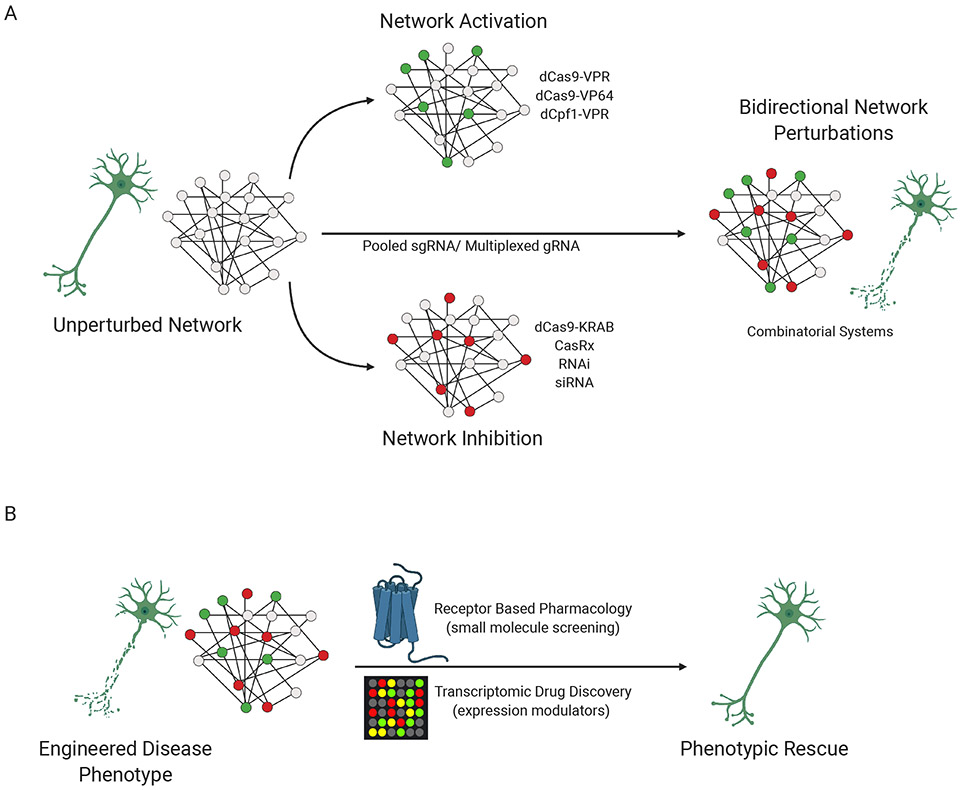
Although dCas9-based CRISPRa and CRISPRi cannot be used in conjunction due to their common Cas protein, a combination of non-complementary CRISPR systems from different bacterial classes would theoretically enable simultaneous, bidirectional manipulation of gene networks (Fig. 2a). Not only do Cpf1 53 and CasRx 54 represent compatible Cas proteins for such bidirectional perturbations, but both also possess the ability to self-process pre-gRNAs, simplifying multiplexed genome engineering through the expression of a single CRISPR array.
Figure 2: Unidirectional and bidirectional network perturbations.
A, The workflow schematic represents a concept of using modulatory and combinatorial CRISPR systems to perturb gene networks unidirectionally, and potentially bidirectionally. B, Phenotypic rescue based drug screens originating from CRISPR-engineered disease phenotypes.
The functional genomic approaches we have described so far are ‘genotype-to-phenotype’ approaches, applying prior knowledge in a hypothesis-driven manner to test the causal role of specific genes. By contrast, forward genetic screens are ‘phenotype-to-genotype’ approaches, broadly manipulating many genes and then characterizing the mutations that resulted in selected phenotypic changes. Recent advances in CRISPR-based approaches have opened new opportunities to conduct forward genetic screens in an unbiased manner 55-57. Notably, approaches such as Perturb-seq58, CRISPR-seq59, CROP-seq60 and ECCITE-seq61 are promising new avenues to conduct large scale genetic screens that couple CRISPR-based perturbations to single-cell RNA sequencing for analytical readouts. Reciprocally, massively parallel reporter assays (MPRAs) can test the activity of regulatory sequences specifically in neuronal cell types 62, functionally validating the impact of non-coding variants at a massive scale. Such large-scale screens may also be extended into high-throughput phenotypic drug screens (Fig. 2b). Thus, the systematic targeting of multiple loci via combinatorial systems can provide a powerful platform to model complex genetic disease risk, through the interrogation of entire gene networks within genetic architectures, and subsequent evaluation of functional deficits resulting from these combined biological processes.
Combinatorial perturbations still face certain limitations: (i) the directional expression of some causal genetic variants are yet unknown, (ii) the magnitude of individual and combinatorial CRISPR-based perturbation may not recapitulate physiologically relevant effect sizes, and (iii) combinatorial perturbations do not recapitulate the entirety of an individual’s genetic architecture. More broadly, CRISPR-based perturbations still cannot mimic megabase-sized copy number variations (CNVs) found in rare genetic disorders. Overall, as large-scale applications of CRISPR-based methods are increasingly feasible, such strategies will help to functionally elucidate the impact of causal genetic variants linked to neurodegenerative and psychiatric disease. Ultimately, the combination of hiPSC- and CRISPR-based platforms will help elucidate downstream phenotypic and functional deficits that genomic approaches alone cannot resolve (Table 1).
A PATHWAY TO PRECISION MEDICINE
Perhaps the greatest promise of hiPSC-based disease models is the potential to discover drugs capable of ameliorating observed in vitro phenotypes, with the hope that these drugs might represent novel clinical therapeutics that could be tailored to patients with greater precision. This potential is based on three critical biological premises: i) patient-specific drug response is predictable based either on patient genotype or the in vitro response of patient-specific hiPSCs-derived neural cells, ii) drug responsiveness is life-time stable, iii) clinical drug responsiveness is correlated to target engagement in disease relevant cell types, rather than arising as an indirect result of side effect tolerance of a potentially limitless number of non-disease-relevant cell types. Today, numerous hiPSC-based studies have shown that established and novel drugs can ameliorate key cellular and molecular disease phenotypes 63.
Given the complex polygenic nature of neurodegenerative and psychiatric disease, it seems unreasonable to expect that the effectiveness of any new drug might hold true for all patients. Although not true for all patients, defined subsets of SZ cases show aberrant pathway, transcriptomic or functional deficits downstream of GWAS-identified variants (e.g. C4A 64, miR-9 65). Moving forward, it is imperative that clinical treatment strategies include better ways to stratify patients, perhaps through a combination of genetics and in vitro testing, matching them to the most appropriate drug. Integrating hiPSC-based models with CRISPR engineering could yield a platform to conduct drug screens and predict clinical response.
Molecular and cellular phenotypes observed in patient hiPSC-derived neurons have been ameliorated by pharmacological treatment across a variety of disease models. For example, loxapine improved neuronal connectivity deficits in SZ hiPSC-neurons 66, insulin growth factor-1 rescued deficits in neural activity in autism spectrum disorder (ASD) hiPSC-neurons 67, and β- and γ-secretase inhibitors reduced amyloid and tau pathology in hiPSC-neurons from Alzheimer’s disease (AD) patients 68. By comparing the transcriptional responses of hiPSC-derived neural progenitor cells derived from SZ cases and controls, we reported differential regulation of neuropsychiatric disease-associated genes in a diagnosis-dependent manner 69, demonstrating the potential value of patient-specific platforms in drug discovery. It is critical to move towards predicting clinical response, either through genetic and/or hiPSC-based strategies. Neurons derived from lithium-responsive, but not non-responsive, bipolar disorder (BD) patients showed ameliorated hyperexcitability following lithium treatment 70. A follow-up study applied to a second cohort trained a naïve Bayes classifier capable of predicting with more than 92% accuracy whether a new patient would show clinical response to lithium 71. An important next step will be CRISPR-based functional validation of the genetic variant(s) linked to lithium responsiveness 72. Towards this, a recent study demonstrated genotype-dependent changes in mRNA expression following treatment of hiPSC-neurons with clinically relevant dosages of valproic acid 73, indicating that precision medicine drug screening approaches might be possible. Although these studies indicate that hiPSC-based models may represent a new drug screening strategy, they fall short of demonstrating that hiPSC-based models can be used to guide precision patient-based medicine in the clinic. Improvements in the efficiency and complexity of cell-based drug screening assays are necessary to accelerate phenotypic drug discovery applications 74.
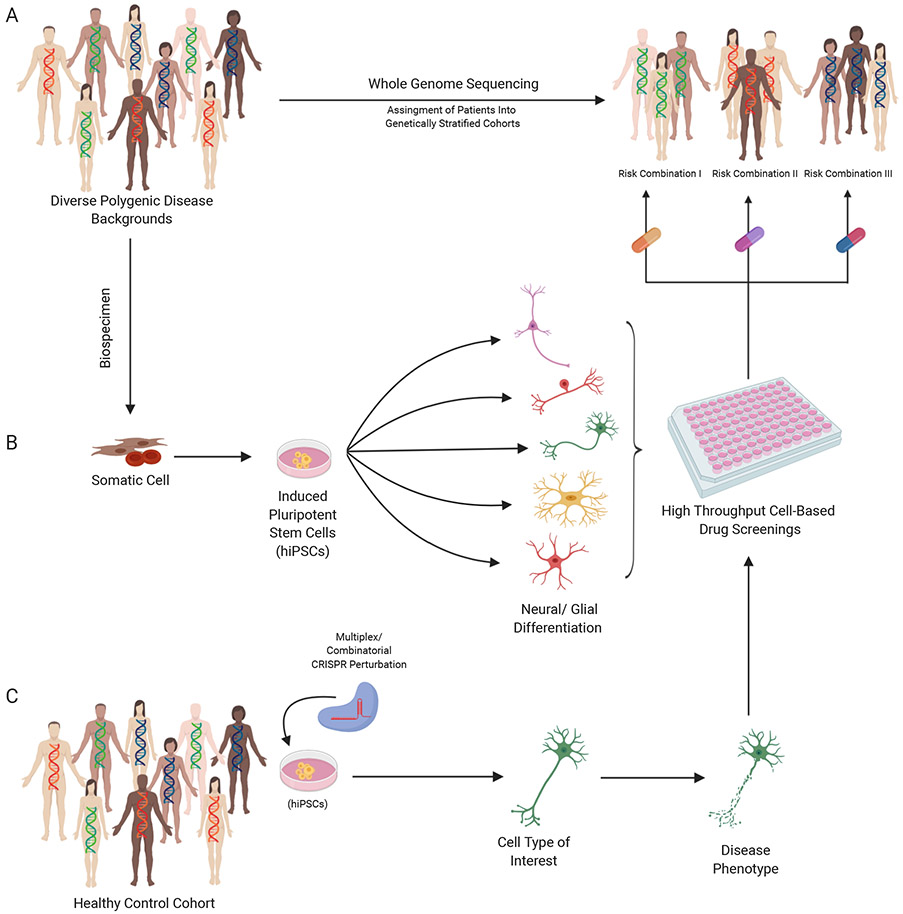
We anticipate that future genomic approaches will provide the means to stratify patients with overlapping risk combinations into “genetically defined” cohorts. This would yield specific hypotheses and drug targets to be evaluated by integrating patient-derived hiPSCs and CRISPR-engineered isogenic models. Convergent genes and gene networks significantly associated with disease risk enriched with rare and common variant disease risk represent promising targets for drug screens (Fig. 3).
FIGURE 3: Coupling hiPSC and CRISPR platforms to accelerate functional validations of brain disease risk loci.
The rising reality of precision medicine via the integration of A, whole-genome sequencing to stratify patients with overlapping risk combinations into genetically defined cohorts, B, hiPSCs platforms to obtain patient-specific cell types and screened for phenotypic rescue, C, CRISPR-mediated network perturbations for engineered risk combinations and systematic cell-based drug screening, ultimately for genetically targeted therapeutics.
CONCLUDING REMARKS AND FUTURE PERSPECTIVES
Given the growing number of identified variants linked to disease risk and the extensive clinical heterogeneity between patients, neurodegenerative and psychiatric diseases are notoriously challenging to treat effectively. Nonetheless, proof-of-principle hiPSC-based models have demonstrated the efficacy of patient-derived neurons and glia to recapitulate transcriptomic and cellular features of brain disease. Today, CRISPR-based isogenic experiments in patient and control hiPSC-derived neural cells make possible more precise interrogation of variants, genes, and gene networks relevant to disease biology. However, there still is a vital need to further develop and improve these models, especially in recapitulating the complex phenotypic characterizations readily available in animal models. Our hope is that the integration of whole-genome sequencing, hiPSC-based disease modeling, CRISPR-mediated functional validation and phenotypic drug discovery in neural-cell-based screens will enable genotype-based diagnoses and drug treatment predictions, making possible a future for precision medicine.
ACKNOWLEDGEMENTS
This work was partially supported by National Institute of Health (NIH) grants R56 MH101454 (K.J.B), R01 MH106056 (K.J.B.) and R01 MH109897 (K.J.B.).
Footnotes
COMPETING FINANCIAL INTEREST STATEMENT
The authors declare no conflicts of interest.
REFERENCES
- 1.Sullivan PF & Geschwind DH Defining the Genetic, Genomic, Cellular, and Diagnostic Architectures of Psychiatric Disorders. Cell 177, 162–183 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flint J & Ideker T The great hairball gambit. PLoS Genet 15, e1008519 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finucane HK et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat Genet 50, 621–629 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skene NG et al. Genetic identification of brain cell types underlying schizophrenia. Nat Genet 50, 825–833 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sims R et al. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer's disease. Nat Genet 49, 1373–1384 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brennand K et al. Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Mol Psychiatry 20, 361–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mariani J et al. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc Natl Acad Sci U S A 109, 12770–5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pasca AM et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods 12, 671–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qian X et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell 165, 1238–54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicholas CR et al. Functional maturation of hPSC-derived forebrain interneurons requires an extended timeline and mimics human neural development. Cell Stem Cell 12, 573–86 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajarajan P, Flaherty E, Akbarian S & Brennand KJ CRISPR-based functional evaluation of schizophrenia risk variants. Schizophr Res (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brennand KJ Personalized medicine in a dish: the growing possibility of neuropsychiatric disease drug discovery tailored to patient genetic variants using stem cells. Stem Cell Investig 4, 91 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haggarty SJ, Silva MC, Cross A, Brandon NJ & Perlis RH Advancing drug discovery for neuropsychiatric disorders using patient-specific stem cell models. Mol Cell Neurosci 73, 104–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78, 785–98 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang N et al. Generation of pure GABAergic neurons by transcription factor programming. Nat Methods 14, 621–628 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Theka I et al. Rapid generation of functional dopaminergic neurons from human induced pluripotent stem cells through a single-step procedure using cell lineage transcription factors. Stem Cells Transl Med 2, 473–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vadodaria KC et al. Generation of functional human serotonergic neurons from fibroblasts. Mol Psychiatry 21, 49–61 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Goto K et al. Simple Derivation of Spinal Motor Neurons from ESCs/iPSCs Using Sendai Virus Vectors. Molecular Therapy - Methods & Clinical Development 4, 115–125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Canals I et al. Rapid and efficient induction of functional astrocytes from human pluripotent stem cells. Nat Methods 15, 693–696 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Ehrlich M et al. Rapid and efficient generation of oligodendrocytes from human induced pluripotent stem cells using transcription factors. Proc Natl Acad Sci U S A 114, E2243–E2252 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuijlaars J et al. Sustained synchronized neuronal network activity in a human astrocyte co-culture system. Sci Rep 6, 36529 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marro SG et al. Neuroligin-4 Regulates Excitatory Synaptic Transmission in Human Neurons. Neuron 103, 617–626 e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawaguchi K, Kageyama R & Sano M Topological defects control collective dynamics in neural progenitor cell cultures. Nature 545, 327–331 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Dezonne RS et al. Derivation of Functional Human Astrocytes from Cerebral Organoids. Sci Rep 7, 45091 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marton RM et al. Differentiation and maturation of oligodendrocytes in human three-dimensional neural cultures. Nat Neurosci 22, 484–491 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mansour AA et al. An in vivo model of functional and vascularized human brain organoids. Nat Biotechnol 36, 432–441 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cakir B et al. Engineering of human brain organoids with a functional vascular-like system. Nat Methods 16, 1169–1175 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abud EM et al. iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases. Neuron 94, 278–293 e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Birey F et al. Assembly of functionally integrated human forebrain spheroids. Nature 545, 54–59 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiang Y et al. Fusion of Regionally Specified hPSC-Derived Organoids Models Human Brain Development and Interneuron Migration. Cell Stem Cell 21, 383–398.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bagley JA, Reumann D, Bian S, Levi-Strauss J & Knoblich JA Fused cerebral organoids model interactions between brain regions. Nat Methods 14, 743–751 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiang Y et al. hESC-Derived Thalamic Organoids Form Reciprocal Projections When Fused with Cortical Organoids. Cell Stem Cell 24, 487–497 e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paşca SP Assembling human brain organoids. Science 363, 126 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Quadrato G et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature 545, 48–53 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Velasco S et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature 570, 523–527 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoon SJ et al. Reliability of human cortical organoid generation. Nat Methods 16, 75–78 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adli M The CRISPR tool kit for genome editing and beyond. Nat Commun 9, 1911 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajarajan P, Gil SE, Brennand KJ & Akbarian S Spatial genome organization and cognition. Nat Rev Neurosci 17, 681–691 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benner C et al. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics 32, 1493–501 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giambartolomei C et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet 10, e1004383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dobbyn A et al. Co-localization of Conditional eQTL and GWAS Signatures in Schizophrenia. bioRxiv (2017). [Google Scholar]
- 42.Huckins LM et al. Gene expression imputation across multiple brain regions provides insights into schizophrenia risk. Nat Genet 51, 659–674 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cong L et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zetsche B et al. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat Biotechnol 35, 31–34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kleinstiver BP et al. Engineered CRISPR–Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nature Biotechnology 37, 276–282 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J-J et al. CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature 566, 218–223 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grunewald J et al. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature 569, 433–437 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anzalone AV et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garneau JE et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Thakore PI et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nature Methods 12, 1143 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chavez A et al. Highly efficient Cas9-mediated transcriptional programming. Nature Methods 12, 326 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schrode N et al. Synergistic effects of common schizophrenia risk variants. Nat Genet 51, 1475–1485 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang X et al. Multiplex gene regulation by CRISPR-ddCpf1. Cell Discovery 3, 17018 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Konermann S et al. Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell 173, 665–676 e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu C et al. Overexpression of NEUROG2 and NEUROG1 in human embryonic stem cells produces a network of excitatory and inhibitory neurons. FASEB J 33, 5287–5299 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Y et al. CRISPR Activation Screens Systematically Identify Factors that Drive Neuronal Fate and Reprogramming. Cell Stem Cell (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tian R et al. CRISPR Interference-Based Platform for Multimodal Genetic Screens in Human iPSC-Derived Neurons. Neuron (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dixit A et al. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell 167, 1853–1866 e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adamson B et al. A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell 167, 1867–1882 e21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Datlinger P et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat Methods 14, 297–301 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mimitou EP et al. Multiplexed detection of proteins, transcriptomes, clonotypes and CRISPR perturbations in single cells. Nat Methods 16, 409–412 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Inoue F, Kreimer A, Ashuach T, Ahituv N & Yosef N Identification and Massively Parallel Characterization of Regulatory Elements Driving Neural Induction. Cell Stem Cell 25, 713–727 e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rowe RG & Daley GQ Induced pluripotent stem cells in disease modelling and drug discovery. Nat Rev Genet 20, 377–388 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sekar A et al. Schizophrenia risk from complex variation of complement component 4. Nature 530, 177–83 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Topol A et al. Dysregulation of miRNA-9 in a Subset of Schizophrenia Patient-Derived Neural Progenitor Cells. Cell Rep 15, 1024–36 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brennand KJ et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature 473, 221–5 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marchetto MC et al. Altered proliferation and networks in neural cells derived from idiopathic autistic individuals. Mol Psychiatry 22, 820–835 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Choi SH et al. A three-dimensional human neural cell culture model of Alzheimer's disease. Nature 515, 274–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Readhead B et al. Expression-based drug screening of neural progenitor cells from individuals with schizophrenia. Nat Commun 9, 4412 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mertens J et al. Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature 527, 95–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stern S, Linker S, Vadodaria KC, Marchetto MC & Gage FH Prediction of response to drug therapy in psychiatric disorders. Open Biol 8(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hou L et al. Genetic variants associated with response to lithium treatment in bipolar disorder: a genome-wide association study. Lancet 387, 1085–1093 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jiang X et al. Sodium valproate rescues expression of TRANK1 in iPSC-derived neural cells that carry a genetic variant associated with serious mental illness. Mol Psychiatry 24, 613–624 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moffat JG, Vincent F, Lee JA, Eder J & Prunotto M Opportunities and challenges in phenotypic drug discovery: an industry perspective. Nat Rev Drug Discov 16, 531–543 (2017). [DOI] [PubMed] [Google Scholar]
- 75.Ran FA et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Poon A et al. Modeling neurodegenerative diseases with patient-derived induced pluripotent cells: Possibilities and challenges. N Biotechnol 39, 190–198 (2017). [DOI] [PubMed] [Google Scholar]
- 77.Zhou M et al. Seamless Genetic Conversion of SMN2 to SMN1 via CRISPR/Cpf1 and Single-Stranded Oligodeoxynucleotides in Spinal Muscular Atrophy Patient-Specific Induced Pluripotent Stem Cells. Hum Gene Ther 29, 1252–1263 (2018). [DOI] [PubMed] [Google Scholar]
- 78.Ho SM et al. Evaluating Synthetic Activation and Repression of Neuropsychiatric-Related Genes in hiPSC-Derived NPCs, Neurons, and Astrocytes. Stem Cell Reports 9, 615–628 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS & Vale RD A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–46 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nihongaki Y et al. CRISPR-Cas9-based photoactivatable transcription systems to induce neuronal differentiation. Nat Methods 14, 963–966 (2017). [DOI] [PubMed] [Google Scholar]
- 81.Konermann S et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tak YE et al. Inducible and multiplex gene regulation using CRISPR–Cpf1-based transcription factors. Nature Methods 14, 1163 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Klann TS et al. CRISPR-Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nat Biotechnol 35, 561–568 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Williams RM et al. Genome and epigenome engineering CRISPR toolkit for in vivo modulation of cis-regulatory interactions and gene expression in the chicken embryo. Development 145(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu XS et al. Editing DNA Methylation in the Mammalian Genome. Cell 167, 233–247 e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ziller MJ et al. Dissecting the Functional Consequences of De Novo DNA Methylation Dynamics in Human Motor Neuron Differentiation and Physiology. Cell Stem Cell 22, 559–574 e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu XS et al. Rescue of Fragile X Syndrome Neurons by DNA Methylation Editing of the FMR1 Gene. Cell 172, 979–992 e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Abudayyeh OO et al. RNA targeting with CRISPR-Cas13. Nature 550, 280–284 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cox DBT et al. RNA editing with CRISPR-Cas13. Science 358, 1019–1027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]