Abstract
Objective
To compare two clinically available MR volumetry software, NeuroQuant® (NQ) and Inbrain® (IB), and examine the inter-method reliabilities and differences between them.
Materials and Methods
This study included 172 subjects (age range, 55–88 years; mean age, 71.2 years), comprising 45 normal healthy subjects, 85 patients with mild cognitive impairment, and 42 patients with Alzheimer's disease. Magnetic resonance imaging scans were analyzed with IB and NQ. Mean differences were compared with the paired t test. Inter-method reliability was evaluated with Pearson's correlation coefficients and intraclass correlation coefficients (ICCs). Effect sizes were also obtained to document the standardized mean differences.
Results
The paired t test showed significant volume differences in most regions except for the amygdala between the two methods. Nevertheless, inter-method measurements between IB and NQ showed good to excellent reliability (0.72 < r < 0.96, 0.83 < ICC < 0.98) except for the pallidum, which showed poor reliability (left: r = 0.03, ICC = 0.06; right: r = −0.05, ICC = −0.09). For the measurements of effect size, volume differences were large in most regions (0.05 < r < 6.15). The effect size was the largest in the pallidum and smallest in the cerebellum.
Conclusion
Comparisons between IB and NQ showed significantly different volume measurements with large effect sizes. However, they showed good to excellent inter-method reliability in volumetric measurements for all brain regions, with the exception of the pallidum. Clinicians using these commercial software should take into consideration that different volume measurements could be obtained depending on the software used.
Keywords: MRI, Alzheimer's disease, Softwares, Brain volumetry, NeuroQuant®, Reliability
INTRODUCTION
Volumetric measurements of brain atrophy have demonstrated close correlations with actual atrophy, neuropathological changes, and cognitive impairment in various neurodegenerative diseases (1,2,3,4). Hippocampal and/or medial temporal lobar atrophy has been already integrated into the diagnostic framework of Alzheimer's disease (AD) (5). Although visual assessment of brain atrophy has been commonly performed in clinical practice, it suffers from high inter-observer variability and low sensitivity (6,7). In contrast, the quantitative volumetric measurement method is an objective method with good repeatability and reliability (8,9). Thus, volumetric measurement of brain atrophy could be used as an imaging marker for clinical differential diagnosis and prediction of disease progression.
There are several freely available software packages for brain volume measurements: FSL (10), Voxel-Based Morphometry (11), FreeSurfer (12), and Statistical Parametric Mapping (13). However, the labor-intensive nature of these research software has limited generalization to routine clinical practice until the introduction of clinically available software (3).
Currently, the FDA has approved several commercially available software for volume measurements: NeuroQuant (14,15,16), Neuroreader (7), and MSmetrix (17,18). Among these, NeuroQuant® (NQ, CorTechs Labs) is the most widely used software because of its fast processing time and the provision of information regarding the cortices of both hemispheres and white matter volume. Moreover, it provides normalized information of patients' data considering the intracranial volume (ICV) and relative atrophy report compared with age-matched normal data (8,9,15).
The most recently introduced clinically available software is Inbrain® (IB, MIDAS Information Technology Co., Ltd.) which is a Korean FDA-cleared software based on the FreeSurfer platform enhanced with its own deep learning algorithm (19,20). While NQ provides only volume measurements of brain structures, IB provides not only volume measurements but also cortical thicknesses. In a previous study using IB, IB was able to classify the disease status and predict the progression into AD using cortical thickness in patients with mild cognitive impairment (MCI) (20). However, it has yet to be validated in terms of reliability with established software such as NQ or FreeSurfer. Therefore, in this study, we aimed to evaluate the inter-method reliability of the two commercially available software, IB and NQ, for brain volumetry in normal healthy subjects as well as in subjects with MCI and AD.
MATERIALS AND METHODS
This retrospective study received Institutional Review Board approval, and the requirement for written informed consent was waived in accordance with the requirements of a retrospective study.
Subjects
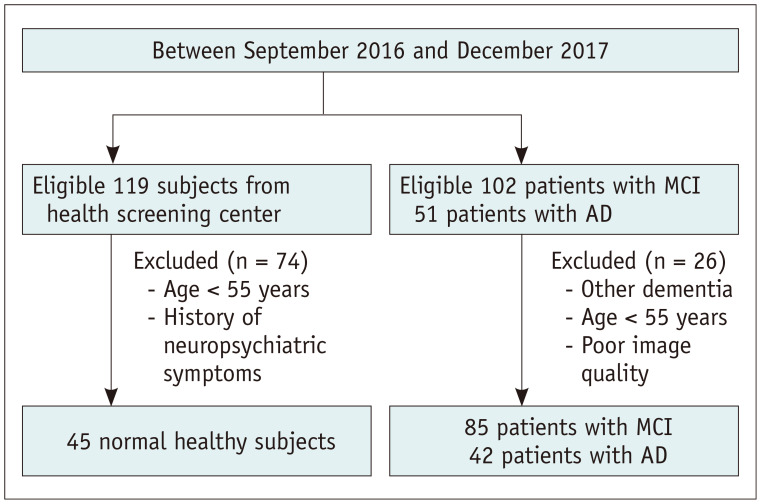
Table 1 shows the demographic data of the study population. A flowchart detailing the recruitment of subjects is shown in Figure 1. As part of a clinical practice guideline development research initiative by the Korean Society of Neuroradiology, this study used the imaging database of 102 and 51 patients with MCI and AD, respectively, who underwent brain magnetic resonance imaging (MRI) and visited the memory clinics between September 2016 and December 2017. The diagnosis of MCI and AD was confirmed by two dementia specialists (one neurologist and one psychiatrist), based on the criteria of the Diagnostic and Statistical Manual of Mental Disorders (4th edition), the National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer's Disease and Related Disorders Association, McKhann et al. (21), and Petersen et al. (22). After excluding patients who had other forms of dementia, those younger than 55 years of age, patients with poor image quality, 85 patients with MCI (33 male and 52 female; age range, 57–85 years; mean age, 71.76 years), and 42 patients with AD (14 male and 28 female; age range, 67–88 years; mean age, 79.00 years) were finally included. For comparison, we searched the imaging database of 119 normal healthy subjects who underwent brain MRI in the health screening center during the same time period. The inclusion criteria for healthy controls were as follows: over 55 years of age, no clinical evidence of neurological or psychiatric symptoms as evaluated by a physician. Finally, 45 normal healthy subjects (23 male, 22 female; age range, 55–74 years; mean age, 62.76 years) were included. Patients with MCI and AD were diagnosed using neuropsychiatric evaluations such as the Mini-Mental State Examination, Clinical Dementia Rating, Seoul Neuropsychological Screening Battery, or Consortium to Establish a Registry for Alzheimer's Disease.
Table 1. Demographic Data of Study Population.
| Characteristics | NL | MCI | AD | P | |
|---|---|---|---|---|---|
| No. | 45 | 85 | 42 | ||
| Age (yr) | 62.8 ± 5.3 | 71.8 ± 6.9 | 79 ± 4.7 | < 0.001 | |
| Sex | 0.22 | ||||
| Female | 23 | 52 | 14 | ||
| Male | 22 | 33 | 28 | ||
| Mini-Mental State Examination score | NA | 23.99 ± 4.26 | 17.98 ± 4.53 | < 0.001 | |
| CDR | NA | 0.58 ± 0.46 | 0.93 ± 0.43 | < 0.001 | |
Datas are mean ± SD. AD = Alzheimer's disease, CDR = Clinical Dementia Rating, MCI = mild cognitive impairment, NA = not applicable, NL = normal, SD = standard deviation
Fig. 1. Flow chart of the study population.
AD = Alzheimer's disease, MCI = mild cognitive impairment
Image Acquisition
All patients underwent MRI in a 3T unit (Discovery MR750; GE Healthcare). Routine brain MRI with additional T1-volume images was obtained in all subjects. During the time period, all subjects with MCI and AD were scanned for T1 volume images with a slice thickness of either 1 or 1.2 mm according to the preference of the referring physician. However, all normal healthy subjects were scanned for T1 volume images with a slice thickness of 1 mm. The preferred use of 1.2 mm was based on a recommendation from the NQ developers, while 1 mm was preferred based on the assumption that it provides a higher spatial resolution. The parameters of sagittal T1-weighted volumetric fast spoiled gradient-recalled echo were as follows: repetition time/echo time (TR/TE), 8.224/3.192; section thickness, 1 mm; matrix, 256 × 256; flip angle, 12°; field of view (FOV), 250 × 250 mm or TR/TE, 5.692/2.36; section thickness, 1.2 mm; matrix, 192 × 192; flip angle, 8°; FOV, 240 × 240 mm. Overall, three-dimensional (3D) T1 images with a slice thicknesses of 1 and 1.2 mm were obtained in 96 and 76 subjects, respectively. All of the normal healthy subjects were scanned with a slice thickness of 1 mm, and the 85 patients with MCI were scanned with slice thicknesses of 1 and 1.2 mm in 42 and 43 patients, respectively. Patients with AD were scanned with slice thicknesses of 1 and 1.2 mm in 9 and 33 patients, respectively.
Magnetic Resonance Volumetry
Sagittal T1-weighted volumetric images were used for analysis with the automated segmentation methods. The brain MRI data for each subject were uploaded on the tool's server.
The processing in NQ was as follows: removal of the scalp, skull, and meninges; inflation of the brain to a spherical shape; mapping of the spherical brain to a common spherical space shared with the Talairach atlas coordinates; identification of segmented brain regions; and deflation of the brain to its original shape. Each brain region volume was corrected for head size differences by normalizing to the ICV, and the resulting output was expressed as a percentage. The result was compared with the data from the healthy controls, which were saved in the NQ database. The subject's brain region was classified as abnormally small if it fell below the fifth normative percentile. In addition, the automated tool provided an age-related atrophy report, which contained absolute volume and relative volume as a percentage of the ICV of the hippocampi, lateral ventricles, and inferior lateral ventricles. The processing time was 10–15 minutes.
IB (https://www.inbrain.co.kr/index.html) is similar to the segmentation method of FreeSurfer, which is based on the volumetric- and surface-based segmentation and uses a template-driven approach (12,23). The processing in IB was as follows: analysis failure prediction, intensity normalization, brain extraction, registration into the volume and surface atlas, white matter segmentation, white matter surface smoothing, topology correction, pial and white matter surface optimization, comparisons between output results and database, and analysis quality management. Finally, the volume of the regional brain structures and of cortical thickness were obtained. A deep learning algorithm was applied to the multiple steps, including analysis failure prediction, brain extraction, white matter segmentation, and analysis quality management to enhance the quality of the segmentation results. The processing took about 4 hour.
Statistical Analysis
The paired t test was used to compare the mean of volume measures between IB and NQ and Pearson's correlation was used to assess the relationships between the two methods. The inter-method agreement between the two software was assessed using the intraclass correlation coefficient (ICC). Effect sizes were obtained for the evaluation of the standardized mean difference between the two software via the following equation: effect size = mean difference/pooled standard deviation (15,24). Effect sizes were defined as follows: small, 0.2; medium, 0.5; and large, 0.8 (15,24). Further, comparisons were performed separately for the normal healthy subjects, and the MCI and AD subgroups. We performed the subgroup analysis based on the slice thickness. Statistical analyses were performed using commercially available software (SPSS, version 24 for Windows; IBM Corp.).
RESULTS
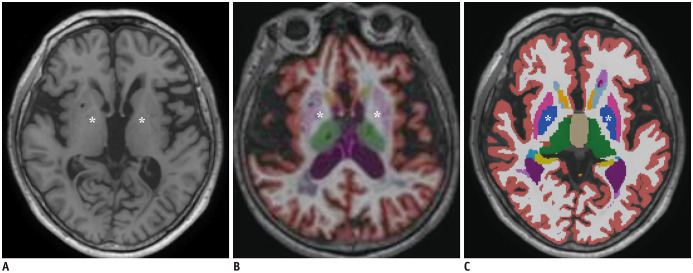
The statistical results are shown in Table 2. Between IB and NQ, there were significant mean differences for most regions. The mean volume in cortical gray matter, cerebral gray matter, cerebral white matter, putamen, and thalamus in NQ were larger than those in IB. The mean volume of the caudate, pallidum, hippocampus, and cerebellum in IB were larger than those in NQ. Especially, there were significant mean differences in the volume of the putamen and pallidum (p < 0.001). The volume of the putamen in NQ was larger than that in IB (5.50 ± 0.79 mL vs. 3.90 ± 0.62 mL in the left hemisphere, 5.26 ± 0.72 mL vs. 3.99 ± 0.62 mL in the right hemisphere). The pallidum volume in NQ was smaller than that in IB (0.50 ± 0.17 mL vs. 1.81 ± 0.25 mL in the left hemisphere, 0.47 ± 0.17 mL vs. 1.82 ± 0.26 mL in the right hemisphere). Figure 2 shows the color-coded images of NQ and IB. In these representative images, the pallidum in NQ appears smaller than that in IB, while the putamen in NQ appears larger than that in IB.
Table 2. Comparisons of Volume Obtained from NeuroQuant® and Inbrain® in All Subjects and Each Subgroup.
| Left Hemisphere | Right Hemisphere | ||||||
|---|---|---|---|---|---|---|---|
| NeuroQuant® | Inbrain® | P | NeuroQuant® | Inbrain® | P | ||
| Mean ± SD | Mean ± SD | Mean ± SD | Mean ± SD | ||||
| Cortical gray matter | 215.69 ± 27.21 | 200.09 ± 25.04 | < 0.001 | 217.99 ± 27.25 | 200.11 ± 24.79 | < 0.001 | |
| NL | 238.116 ± 26.13 | 218.64 ± 21.59 | < 0.001 | 241.73 ± 25.82 | 218.05 ± 21.38 | < 0.001 | |
| MCI | 213.79 ± 22.21 | 201.12 ± 20.27 | < 0.001 | 215.78 ± 22.18 | 201.50 ± 20.15 | < 0.001 | |
| AD | 195.51 ± 19.18 | 177.80 ± 19.58 | < 0.001 | 197.04 ± 17.17 | 178.06 ± 19.60 | < 0.001 | |
| Caudate | 2.80 ± 0.70 | 3.28 ± 0.54 | < 0.001 | 2.83 ± 0.70 | 3.29 ± 0.53 | < 0.001 | |
| NL | 2.62 ± 0.49 | 3.31 ± 0.38 | < 0.001 | 2.82 ± 0.55 | 3.38 ± 0.42 | < 0.001 | |
| MCI | 2.84 ± 0.70 | 3.34 ± 0.60 | < 0.001 | 2.87 ± 0.77 | 3.33 ± 0.56 | < 0.001 | |
| AD | 2.90 ± 0.85 | 3.13 ± 0.57 | < 0.001 | 2.73 ± 0.72 | 3.09 ± 0.52 | < 0.001 | |
| Putamen | 5.50 ± 0.79 | 3.90 ± 0.62 | < 0.001 | 5.26 ± 0.72 | 3.99 ± 0.62 | < 0.001 | |
| NL | 5.88 ± 0.56 | 4.31 ± 0.50 | < 0.001 | 5.68 ± 0.62 | 4.46 ± 0.43 | < 0.001 | |
| MCI | 5.55 ± 0.76 | 3.93 ± 0.50 | < 0.001 | 5.25 ± 0.66 | 3.98 ± 0.52 | < 0.001 | |
| AD | 5.01 ± 0.82 | 3.39 ± 0.60 | < 0.001 | 4.83 ± 0.69 | 3.48 ± 0.60 | < 0.001 | |
| Pallidum | 0.50 ± 0.17 | 1.81 ± 0.25 | < 0.001 | 0.47 ± 0.17 | 1.82 ± 0.26 | < 0.001 | |
| NL | 0.66 ± 0.14 | 1.89 ± 0.25 | < 0.001 | 0.62 ± 0.12 | 1.90 ± 0.23 | < 0.001 | |
| MCI | 0.48 ± 0.14 | 1.80 ± 0.24 | < 0.001 | 0.44 ± 0.14 | 1.79 ± 0.25 | < 0.001 | |
| AD | 0.35 ± 0.11 | 1.74 ± 0.24 | < 0.001 | 0.35 ± 0.12 | 1.79 ± 0.29 | < 0.001 | |
| Thalamus | 7.05 ± 0.79 | 6.33 ± 0.75 | < 0.001 | 7.05 ± 0.84 | 6.10 ± 0.76 | < 0.001 | |
| NL | 7.31 ± 0.92 | 6.78 ± 0.75 | < 0.001 | 7.27 ± 0.96 | 6.44 ± 0.74 | < 0.001 | |
| MCI | 7.04 ± 0.69 | 6.26 ± 0.61 | < 0.001 | 7.05 ± 0.78 | 6.05 ± 0.61 | < 0.001 | |
| AD | 6.80 ± 0.75 | 5.98 ± 0.79 | < 0.001 | 6.80 ± 0.79 | 5.81 ± 0.91 | < 0.001 | |
| Amygdala | 1.44 ± 0.28 | 1.20 ± 0.25 | < 0.001 | 1.37 ± 0.26 | 1.39 ± 0.29 | 0.06† | |
| NL | 1.66 ± 0.25 | 1.40 ± 0.20 | < 0.001 | 1.55 ± 0.23 | 1.62 ± 0.24 | < 0.001 | |
| MCI | 1.43 ± 0.25 | 1.20 ± 0.22 | < 0.001 | 1.38 ± 0.22 | 1.39 ± 0.25 | 0.33† | |
| AD | 1.22 ± 0.19 | 0.98 ± 0.19 | < 0.001 | 1.17 ± 0.22 | 1.14 ± 0.22 | 0.14† | |
| Hippocampus | 3.31 ± 0.75 | 3.55 ± 0.58 | < 0.001 | 3.35 ± 0.74 | 3.60 ± 0.62 | < 0.001 | |
| NL | 4.09 ± 0.56 | 4.07 ± 0.47 | 0.65† | 4.10 ± 0.52 | 4.17 ± 0.50 | 0.02 | |
| MCI | 3.28 ± 0.54 | 3.53 ± 0.44 | < 0.001 | 3.31 ± 0.57 | 3.57 ± 0.49 | < 0.001 | |
| AD | 2.55 ± 0.34 | 3.02 ± 0.40 | < 0.001 | 2.61 ± 0.37 | 3.02 ± 0.37 | < 0.001 | |
| Cerebellum | 61.41 ± 6.16 | 61.70 ± 6.23 | 0.04 | 59.70 ± 5.70 | 60.43 ± 6.14 | < 0.001 | |
| NL | 64.67 ± 6.96 | 64.46 ± 6.91 | 0.46† | 62.18 ± 6.31 | 62.88 ± 6.85 | 0.02 | |
| MCI | 60.82 ± 5.70 | 61.13 ± 6.04 | 0.14† | 59.22 ± 5.25 | 60.00 ± 5.73 | < 0.001 | |
| AD | 59.09 ± 4.66 | 59.90 ± 4.85 | 0.003 | 58.00 ± 5.09 | 58.65 ± 5.40 | 0.03 | |
| Cerebral gray matter* | 431.48 ± 62.85 | 400.11 ± 49.62 | < 0.001 | ||||
| NL | 480.04 ± 51.63 | 436.69 ± 42.85 | < 0.001 | ||||
| MCI | 429.72 ± 44.02 | 402.61 ± 40.14 | < 0.001 | ||||
| AD | 383.03 ± 68.14 | 355.86 ± 38.83 | 0.003 | ||||
| Cerebral white matter* | 447.96 ± 54.46 | 438.53 ± 67.79 | 0.008 | ||||
| NL | 466.59 ± 53.23 | 447.59 ± 48.86 | < 0.001 | ||||
| MCI | 450.32 ± 53.97 | 432.90 ± 59.37 | < 0.001 | ||||
| AD | 423.21 ± 48.38 | 440.23 ± 96.04 | 0.12† | ||||
The units are mL. *Cerebral gray matter and white matter mean volume measured in both hemisphere, †Statistically not significant.
Fig. 2. Representative images of the color-coded images of NQ and IB.
An axial T1-weighted image (A) is shown at basal ganglia level with color-coded images of NQ (B) and IB (C). In these representative images, the pallidum in NQ appears smaller (B) compared to that in IB (C), while the putamen in NQ appears larger (B) than that in IB (C). The pallidum is indicated with asterisks. IB = Inbrain®, NQ = NeuroQuant®
Pearson's correlation analysis between IB and NQ showed a significantly strong linear correlation (0.72 < r < 0.96), except for the pallidum (Table 3). ICC also showed significantly good to excellent correlations between IB and NQ (0.83 < ICC < 0.98) (Table 3), except for the pallidum. There was no significant correlation between the two software in the pallidum (r = 0.03, p = 0.67 in the left and r = −0.05, p = 0.52 in the right, ICC = 0.06, p = 0.34 in the left and ICC = −0.09, p = 0.72 in the right).
Table 3. Results of Pearson's Correlation, ICC and Effect Size in All Subjects and Each Subgroup.
| Left Hemisphere | Right Hemisphere | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| r | P | ICC | P | Effect Size | r | P | ICC | P | Effect Size | ||
| Cortical gray matter | 0.89 | < 0.001 | 0.94 | < 0.001 | 0.6 | 0.9 | < 0.001 | 0.95 | < 0.001 | 0.69 | |
| NL | 0.92 | < 0.001 | 0.95 | < 0.001 | 0.81 | 0.91 | < 0.001 | 0.94 | < 0.001 | 0.99 | |
| MCI | 0.83 | < 0.001 | 0.91 | < 0.001 | 0.6 | 0.87 | < 0.001 | 0.93 | < 0.001 | 0.67 | |
| AD | 0.8 | < 0.001 | 0.89 | < 0.001 | 0.91 | 0.78 | < 0.001 | 0.87 | < 0.001 | 1.03 | |
| Caudate | 0.77 | < 0.001 | 0.85 | < 0.001 | 0.77 | 0.81 | < 0.001 | 0.87 | < 0.001 | 0.74 | |
| NL | 0.78 | < 0.001 | 0.86 | < 0.001 | 1.57 | 0.75 | < 0.001 | 0.84 | < 0.001 | 1.14 | |
| MCI | 0.83 | < 0.001 | 0.9 | < 0.001 | 77 | 0.82 | < 0.001 | 0.88 | < 0.001 | 0.68 | |
| AD | 0.76 | < 0.001 | 0.82 | < 0.001 | 0.32 | 0.83 | < 0.001 | 0.88 | < 0.001 | 0.57 | |
| Putamen | 0.72 | < 0.001 | 0.83 | < 0.001 | 2.25 | 0.77 | < 0.001 | 0.87 | < 0.001 | 1.89 | |
| NL | 0.64 | < 0.001 | 0.78 | < 0.001 | 2.96 | 0.71 | < 0.001 | 0.8 | < 0.001 | 2.29 | |
| MCI | 0.65 | < 0.001 | 0.75 | < 0.001 | 2.52 | 0.72 | < 0.001 | 0.82 | < 0.001 | 2.14 | |
| AD | 0.7 | < 0.001 | 0.8 | < 0.001 | 2.25 | 0.7 | < 0.001 | 0.82 | < 0.001 | 2.09 | |
| Pallidum | 0.03 | 0.67* | 0.06 | 0.34* | 6.13 | -0.05 | 0.52* | -0.09 | 0.72* | 6.15 | |
| NL | 0.03 | 0.85* | 0.05 | 0.43* | 6.07 | 0.03 | 0.83* | 0.06 | 0.43* | 6.98 | |
| MCI | -0.28 | 0.01 | -0.66 | 0.99* | 6.72 | -0.25 | 0.02 | -0.56 | 0.98* | 6.66 | |
| AD | -0.07 | 0.67* | -0.11 | 0.63* | 7.45 | -0.27 | 0.09* | -0.47 | 0.89* | 6.49 | |
| Thalamus | 0.77 | < 0.001 | 0.87 | < 0.001 | 0.93 | 0.82 | < 0.001 | 0.9 | < 0.001 | 1.19 | |
| NL | 0.8 | < 0.001 | 0.88 | < 0.001 | 0.63 | 0.89 | < 0.001 | 0.92 | < 0.001 | 0.97 | |
| MCI | 0.73 | < 0.001 | 0.84 | < 0.001 | 1.2 | 0.82 | < 0.001 | 0.89 | < 0.001 | 1.43 | |
| AD | 0.75 | < 0.001 | 0.86 | < 0.001 | 1.06 | 0.77 | < 0.001 | 0.86 | < 0.001 | 1.16 | |
| Amygdala | 0.87 | < 0.001 | 0.93 | < 0.001 | 0.9 | 0.91 | < 0.001 | 0.95 | < 0.001 | 0.07 | |
| NL | 0.78 | < 0.001 | 0.86 | < 0.001 | 1.15 | 0.9 | < 0.001 | 0.95 | < 0.001 | 0.3 | |
| MCI | 0.85 | < 0.001 | 0.92 | < 0.001 | 0.98 | 0.87 | < 0.001 | 0.93 | < 0.001 | 0.04 | |
| AD | 0.76 | < 0.001 | 0.86 | < 0.001 | 1.26 | 0.89 | < 0.001 | 0.94 | < 0.001 | 0.14 | |
| Hippocampus | 0.89 | < 0.001 | 0.93 | < 0.001 | 0.36 | 0.94 | < 0.001 | 0.96 | < 0.001 | 0.37 | |
| NL | 0.85 | < 0.001 | 0.91 | < 0.001 | 0.04 | 0.92 | < 0.001 | 0.96 | < 0.001 | 0.14 | |
| MCI | 0.84 | < 0.001 | 0.9 | < 0.001 | 0.51 | 0.91 | < 0.001 | 0.95 | < 0.001 | 0.49 | |
| AD | 0.62 | < 0.001 | 0.76 | < 0.001 | 1.27 | 0.82 | < 0.001 | 0.9 | < 0.001 | 1.11 | |
| Cerebellum | 0.96 | < 0.001 | 0.98 | < 0.001 | 0.05 | 0.95 | < 0.001 | 0.97 | < 0.001 | 0.12 | |
| NL | 0.96 | < 0.001 | 0.98 | < 0.001 | 0.03 | 0.96 | < 0.001 | 0.98 | < 0.001 | 0.11 | |
| MCI | 0.95 | < 0.001 | 0.97 | < 0.001 | 0.05 | 0.94 | < 0.001 | 0.97 | < 0.001 | 0.14 | |
| AD | 0.94 | < 0.001 | 0.97 | < 0.001 | 0.17 | 0.94 | < 0.001 | 0.97 | < 0.001 | 0.12 | |
| Cerebral gray matter | 0.84 | < 0.001 | 0.9 | < 0.001 | 0.55 | ||||||
| NL | 0.92 | < 0.001 | 0.95 | < 0.001 | 0.91 | ||||||
| MCI | 0.86 | < 0.001 | 0.92 | < 0.001 | 0.64 | ||||||
| AD | 0.59 | < 0.001 | 0.67 | < 0.001 | 0.49 | ||||||
| Cerebral white matter | 0.74 | < 0.001 | 0.84 | < 0.001 | 0.15 | ||||||
| NL | 0.93 | < 0.001 | 0.96 | < 0.001 | 0.37 | ||||||
| MCI | 0.79 | < 0.001 | 0.88 | < 0.001 | 0.31 | ||||||
| AD | 0.73 | < 0.001 | 0.74 | < 0.001 | 0.22 | ||||||
*Statistically not significant. ICC = intraclass correlation coefficient
With regard to the effect size, the putamen and pallidum showed the largest effect sizes among the brain regions: the effect sizes of the putamen were 2.25 and 1.89 in the left and right hemispheres, respectively, and that of the pallidum were 6.13 and 6.15 in the left and right hemispheres, respectively.
When subjects were divided into the normal control, MCI, and AD groups, similar results were demonstrated in each group (Tables 2, 3). For the normal control group, most regions showed significant mean differences between the two methods (p < 0.001). There were no significant differences in the hippocampus and cerebellum (p = 0.65 and 0.46). For the MCI group, the cerebellum (p = 0.14) and amygdala (p = 0.33) did not show significant differences. For the AD group, the amygdala (p = 0.14) and cerebral white matter (p = 0.12) did not show significant mean differences. The Pearson's correlation analysis showed significantly moderate to strong linear correlations in each subgroup, except for the pallidum: 0.64 < r < 0.96 in the normal control group, 0.65 < r < 0.95 in the MCI group, and 0.59 < r < 0.94 in the AD group. The ICC was also good to excellent: 0.78 < ICC < 0.98 in the normal control group, 0.75 < ICC < 0.97 in the MCI group, and 0.67 < ICC < 0.97 in the AD group. Effect sizes were within a wide range, from 0.03 in the cerebellum to 6.98 in the pallidum in the normal control group, and from 0.05 in the cerebellum to 6.72 in the pallidum in the MCI group. Likewise, the effect size was the smallest with 0.12 in the cerebellum and the largest with 7.45 in the pallidum in the patients with AD.
The subgroup analysis based on the slice thickness of 3D T1 images was performed in patients with MCI and AD, because 3D T1 images with a slice thickness of 1 mm were obtained from all healthy subjects. The results based on the slice thickness of 3D T1 images were similar (Supplementary Tables 1, 2). There were significant mean differences between the two software in most regions (p < 0.001), except in the right amygdala, cerebellum, and cerebral white matter. The volumes obtained from images with a slice thickness of 1 mm were different from those obtained with a slice thickness of 1.2 mm in each software. For the cortical gray matter, caudate, hippocampus, and cerebral gray matter, images with a slice thickness of 1 mm resulted in a larger volume than those with a slice thickness of 1.2 mm in both NQ and IB. For the thalamus and cerebral white matter, images with a slice thickness of 1.2 mm resulted in a larger volume than those with a slice thickness of 1 mm in both NQ and IB. Inter-method reliability for images with a slice thickness of 1 mm showed better correlations than images with a slice thickness of 1.2 mm in most regions.
DISCUSSION
In this validation study of inter-method reliability, we found good to excellent correlations and reliability between IB and NQ for most brain regions. However, we found that there were significant differences in volume between IB and NQ. The measurements of cortical gray matter volume resulted in a significant mean difference between the two methods with medium effect sizes. Furthermore, the differences observed for some deep gray matter structures, especially the pallidum, were not negligible, which can be a potential obstacle in the clinical application of volumetry.
Since the introduction of NQ in 2009 (8), many studies have investigated the clinical use of NQ (6,7,8,15,17,25,26). Several studies have compared NQ with FreeSurfer, MSmetrix, or Neuroreader (7,15,16,25,26). The volumetric results of NQ were comparable to those of FreeSurfer, a reference standard of volumetry (15,26). Although the segmentation method of NQ is reportedly similar to FreeSurfer, NQ uses a different atlas, independent code base, and separate methods for intensity normalization and gradient distortion correction to accommodate scanner-specific acquisition-level differences (15). Instead of providing each gyral thickness as in FreeSurfer, NQ gives only the volume of the cortex, white matter, and deep gray matter, thereby achieving a faster processing time.
Since the introduction of IB in 2017 (19), there have only been a few clinical studies on IB (20,27). In addition, a validation study in terms of reliability has not been conducted yet. In contrast to NQ, IB uses the same registration atlas as that of FreeSurfer, and the segmentation method of IB is almost identical to that of FreeSurfer (20). IB has added several steps into the process in FreeSurfer, such as analysis failure prediction, brain extraction, white matter segmentation, and analysis quality management by applying the deep learning technique to reduce the error rates.
In this study, we found that the volume measurements could be different depending on the software used. There were significant mean differences between the two methods in most regions, except the amygdala. Moreover, subcortical gray matter regions showed large effect sizes. The pallidum showed the largest effect size. In this study, the volume of the pallidum in NQ was smaller than that in IB, and the volume of the putamen in NQ was larger than that in IB. Given that the IB uses the FreeSurfer platform, our finding is broadly in line with the previous observation on the difference in volume measurements of the pallidum between NQ and FreeSurfer (15). It has been reported that the difference in the volumes of the pallidum appears to arise from the fundamental problem of similar intensities of the pallidum and white matter in T1-weighted images, which makes it difficult to segment the pallidum from white matter accurately (15,28). Besides that, we speculated that the different results between the two software are mainly attributable to the different pipeline, including the registration atlas. The atlas is the basis for segmentation: NQ uses a different probabilistic atlas from that of FreeSurfer (14), and IB uses the same atlas as that of FreeSurfer. The potential effect of the type of atlas on volumetric results has been demonstrated in a study by using different atlases for hippocampus segmentation, which resulted in differences in accuracy depending on the atlas used (29). Our findings suggest that at least some deep gray matter structures such as the putamen and pallidum are still susceptible to the use of different atlases despite overall good reliability.
Previous studies demonstrated that the patients with AD showed cortical atrophy of the medial temporal, temporoparietal, posterior cingulate, and precuneus regions (30,31); however, no study has focused on volume measurements of the basal ganglia. However, decreases in the volume of subcortical gray matter including the putamen and pallidum have been reported in patients with AD in previous studies (32,33). This decrease in the volume of the basal ganglia could be explained by the neuronal loss caused by amyloid deposition and neurofibrillary tangles (33). In addition, because iron deposition (34) and tau pathology (35) might influence the basal ganglia in patients with AD, changes in the volume of the basal ganglia could not be neglected. Accordingly, the software users should be aware of the fact that the volume results of the basal ganglia could be markedly different depending on the software used.
Atrophy of the hippocampus has been regarded as an imaging marker of AD (36). The volume of the hippocampus was significantly larger with IB than with NQ in patients with MCI and AD. The correlation between IB and NQ tended to be lower in patients with AD compared to normal controls and patients with MCI. For the hippocampal volume, there was a larger difference between patients with MCI and AD in NQ than in IB.
In this study, we used the effect sizes in statistical analysis. Effect sizes are defined as standardized measurements of the size of the mean difference among the study groups (24). Effect size could be obtained with the mean difference between two groups divided by the standard deviation. Therefore, when a result shows the same mean difference, the standard deviation determines the effect size. The paired t test showed significant differences in the amygdala, cerebellum, and cerebral white matter; however, they showed small effect sizes. This meant that the standardized mean difference between the two methods was small, even though they showed statistically significant differences. Furthermore, other deep gray matter structures such as the pallidum, putamen, and thalamus showed large effect sizes. This effect size result implies that the results between the two software were not identical. Thus, we believe that the results for these smaller structures should be carefully interpreted because the interpretation could differ depending on the software used for volume measurements.
The main limitation of this study was that we used two different magnetic resonance (MR) sequences for volumetric measurements. We did not consider the repeatability in the same scanner in terms of the different MR sequences that were applied. Actually, the volume of the cerebral white matter in patients with AD was greater than those of patients with MCI in IB. This is difficult to explain; however, it might be related to the scan protocol, where a slice thickness of 1.2 mm was more frequently used in the AD group than in the other 2 groups. Because the cerebral white matter tended to show greater volumes in scans with a slice thickness of 1.2 mm compared to those with a slice thickness of 1 mm in both NQ and IB, different MR scanning parameters might affect the volume measurements in a different way. Second, we did not investigate the reproducibility in a different MR scanner. The results of the volume measurements could be different in a different MR scanner because brain volumetry is usually influenced by several technical factors including MRI field strength and scanner model, as well as post-processing-related issues (23). Further studies are warranted in the future for complete methodological validation. Finally, we did not compare the result from NQ and IB with that of FreeSurfer or manual segmentation, which is the reference standard. Therefore, we could not determine which software could produce results that are similar to those of FreeSurfer or manual segmentation.
In conclusion, we compared two commercial software for automated volume measurements of brain regions. Overall, they showed good to excellent correlation. However, they showed significant mean differences and large effect sizes. Therefore, clinicians and researchers should take the type of software used into consideration when interpreting the results of volume measurements obtained using commercial software.
Footnotes
This work was supported by Konkuk University Medical Center Research Grant 2017.
Conflicts of Interest: The authors have no potential conflicts of interest to disclose.
Supplementary Materials
The Data Supplement is available with this article at https://doi.org/10.3348/kjr.2020.0518.
Comparisons of Volume Obtained from NeuroQuant® and Inbrain®, Based on the Slice Thickness in Patients with MCI and AD
Results of Pearson's Correlation, ICC and Effect Size, Based on the Slice Thickness in Patients with MCI and AD
References
- 1.Moon WJ, Kim HJ, Roh HG, Han SH. Atrophy measurement of the anterior commissure and substantia innominata with 3T high-resolution MR imaging: does the measurement differ for patients with frontotemporal lobar degeneration and Alzheimer disease and for healthy subjects? AJNR Am J Neuroradiol. 2008;29:1308–1313. doi: 10.3174/ajnr.A1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moon Y, Moon WJ, Kim H, Han SH. Regional atrophy of the insular cortex is associated with neuropsychiatric symptoms in Alzheimer's disease patients. Eur Neurol. 2014;71:223–229. doi: 10.1159/000356343. [DOI] [PubMed] [Google Scholar]
- 3.Park M, Moon WJ. Structural MR imaging in the diagnosis of Alzheimer's disease and other neurodegenerative dementia: current imaging approach and future perspectives. Korean J Radiol. 2016;17:827–845. doi: 10.3348/kjr.2016.17.6.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whitwell JL, Josephs KA, Murray ME, Kantarci K, Przybelski SA, Weigand SD, et al. MRI correlates of neurofibrillary tangle pathology at autopsy: a voxel-based morphometry study. Neurology. 2008;71:743–749. doi: 10.1212/01.wnl.0000324924.91351.7d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jack CR, Jr, Bennett DA, Blennow K, Carrillo MC, Dunn B, Haeberlein SB, et al. NIA-AA Research Framework: toward a biological definition of Alzheimer's disease. Alzheimers Dement. 2018;14:535–562. doi: 10.1016/j.jalz.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ross DE, Ochs AL, DeSmit ME, Seabaugh JM, Havranek MD Alzheimer's Disease Neuroimaging Initiative. Man versus machine part 2: comparison of radiologists' interpretations and NeuroQuant Measures of brain asymmetry and progressive atrophy in patients with traumatic brain injury. J Neuropsychiatry Clin Neurosci. 2015;27:147–152. doi: 10.1176/appi.neuropsych.13040088. [DOI] [PubMed] [Google Scholar]
- 7.Tanpitukpongse TP, Mazurowski MA, Ikhena J, Petrella JR Alzheimer's Disease Neuroimaging Initiative. Predictive utility of marketed volumetric software tools in subjects at risk for Alzheimer disease: do regions outside the hippocampus matter? AJNR Am J Neuroradiol. 2017;38:546–552. doi: 10.3174/ajnr.A5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brewer JB. Fully-automated volumetric MRI with normative ranges: translation to clinical practice. Behav Neurol. 2009;21:21–28. doi: 10.3233/BEN-2009-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Min J, Moon WJ, Jeon JY, Choi JW, Moon YS, Han SH. Diagnostic efficacy of structural MRI in patients with mild-to-moderate Alzheimer disease: automated volumetric assessment versus visual assessment. AJR Am J Roentgenol. 2017;208:617–623. doi: 10.2214/AJR.16.16894. [DOI] [PubMed] [Google Scholar]
- 10.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. FSL. Neuroimage. 2012;62:782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 11.Ashburner J, Friston KJ. Voxel-based morphometry--the methods. Neuroimage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- 12.Fischl B. FreeSurfer. Neuroimage. 2012;62:774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ashburner J, Friston KJ. Unified segmentation. Neuroimage. 2005;26:839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 14.Brewer JB, Magda S, Airriess C, Smith ME. Fully-automated quantification of regional brain volumes for improved detection of focal atrophy in Alzheimer disease. AJNR Am J Neuroradiol. 2009;30:578–580. doi: 10.3174/ajnr.A1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ochs AL, Ross DE, Zannoni MD, Abildskov TJ, Bigler ED Alzheimer's Disease Neuroimaging Initiative. Comparison of automated brain volume measures obtained with NeuroQuant and FreeSurfer. J Neuroimaging. 2015;25:721–727. doi: 10.1111/jon.12229. [DOI] [PubMed] [Google Scholar]
- 16.Ross DE, Seabaugh J, Cooper L, Seabaugh J. NeuroQuant® and NeuroGage® reveal effects of traumatic brain injury on brain volume. Brain Inj. 2018;32:1437–1441. doi: 10.1080/02699052.2018.1489980. [DOI] [PubMed] [Google Scholar]
- 17.Steenwijk MD, Amiri H, Schoonheim MM, de Sitter A, Barkhof F, Pouwels PJW, et al. Agreement of MSmetrix with established methods for measuring cross-sectional and longitudinal brain atrophy. Neuroimage Clin. 2017;15:843–853. doi: 10.1016/j.nicl.2017.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Storelli L, Rocca MA, Pagani E, Van Hecke W, Horsfield MA, De Stefano N, et al. Measurement of whole-brain and gray matter atrophy in multiple sclerosis: assessment with MR imaging. Radiology. 2018;288:554–564. doi: 10.1148/radiol.2018172468. [DOI] [PubMed] [Google Scholar]
- 19.Cho Y, Seong JK, Jeong Y, Shin SY Alzheimer's Disease Neuroimaging Initiative. Individual subject classification for Alzheimer's disease based on incremental learning using a spatial frequency representation of cortical thickness data. Neuroimage. 2012;59:2217–2230. doi: 10.1016/j.neuroimage.2011.09.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JS, Kim C, Shin JH, Cho H, Shin DS, Kim N, et al. Machine learning-based individual assessment of cortical atrophy pattern in Alzheimer's disease spectrum: development of the classifier and longitudinal evaluation. Sci Rep. 2018;8:4161. doi: 10.1038/s41598-018-22277-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR, Jr, Kawas CH, et al. The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petersen RC, Smith GE, Waring SC, Ivnik RJ, Tangalos EG, Kokmen E. Mild cognitive impairment: clinical characterization and outcome. Arch Neurol. 1999;56:303–308. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- 23.Guo C, Ferreira D, Fink K, Westman E, Granberg T. Repeatability and reproducibility of FreeSurfer, FSL-SIENAX and SPM brain volumetric measurements and the effect of lesion filling in multiple sclerosis. Eur Radiol. 2019;29:1355–1364. doi: 10.1007/s00330-018-5710-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olejnik S, Algina J. Measures of effect size for comparative studies: applications, interpretations, and limitations. Contemp Educ Psychol. 2000;25:241–286. doi: 10.1006/ceps.2000.1040. [DOI] [PubMed] [Google Scholar]
- 25.Reid MW, Hannemann NP, York GE, Ritter JL, Kini JA, Lewis JD, et al. Comparing two processing pipelines to measure subcortical and cortical volumes in patients with and without mild traumatic brain injury. J Neuroimaging. 2017;27:365–371. doi: 10.1111/jon.12431. [DOI] [PubMed] [Google Scholar]
- 26.Ross DE, Ochs AL, Tate DF, Tokac U, Seabaugh J, Abildskov TJ, et al. High correlations between MRI brain volume measurements based on NeuroQuant® and FreeSurfer. Psychiatry Res Neuroimaging. 2018;278:69–76. doi: 10.1016/j.pscychresns.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 27.Kim HJ, Park JY, Seo SW, Jung YH, Kim Y, Jang H, et al. Cortical atrophy pattern-based subtyping predicts prognosis of amnestic MCI: an individual-level analysis. Neurobiol Aging. 2019;74:38–45. doi: 10.1016/j.neurobiolaging.2018.10.010. [DOI] [PubMed] [Google Scholar]
- 28.Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- 29.Nestor SM, Gibson E, Gao FQ, Kiss A, Black SE Alzheimer's Disease Neuroimaging Initiative. A direct morphometric comparison of five labeling protocols for multi-atlas driven automatic segmentation of the hippocampus in Alzheimer's disease. Neuroimage. 2013;66:50–70. doi: 10.1016/j.neuroimage.2012.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du AT, Schuff N, Kramer JH, Rosen HJ, Gorno-Tempini ML, Rankin K, et al. Different regional patterns of cortical thinning in Alzheimer's disease and frontotemporal dementia. Brain. 2007;130:1159–1166. doi: 10.1093/brain/awm016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh V, Chertkow H, Lerch JP, Evans AC, Dorr AE, Kabani NJ. Spatial patterns of cortical thinning in mild cognitive impairment and Alzheimer's disease. Brain. 2006;129:2885–2893. doi: 10.1093/brain/awl256. [DOI] [PubMed] [Google Scholar]
- 32.de Jong LW, van der Hiele K, Veer IM, Houwing JJ, Westendorp RG, Bollen EL, et al. Strongly reduced volumes of putamen and thalamus in Alzheimer's disease: an MRI study. Brain. 2008;131:3277–3285. doi: 10.1093/brain/awn278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li YD, He HJ, Dong HB, Feng XY, Xie GM, Zhang LJ. Discriminative analysis of early-stage Alzheimer's disease and normal aging with automatic segmentation technique in subcortical gray matter structures: a multicenter in vivo MRI volumetric and DTI study. Acta Radiol. 2013;54:1191–1200. doi: 10.1177/0284185113492971. [DOI] [PubMed] [Google Scholar]
- 34.Moon Y, Han SH, Moon WJ. Patterns of brain iron accumulation in Vascular dementia and Alzheimer's dementia using quantitative susceptibility mapping imaging. J Alzheimers Dis. 2016;51:737–745. doi: 10.3233/JAD-151037. [DOI] [PubMed] [Google Scholar]
- 35.Hamasaki H, Honda H, Suzuki SO, Shijo M, Ohara T, Hatabe Y, et al. Tauopathy in basal ganglia involvement is exacerbated in a subset of patients with Alzheimer's disease: The Hisayama study. Alzheimers Dement (Amst) 2019;11:415–423. doi: 10.1016/j.dadm.2019.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jack CR., Jr Alzheimer disease: new concepts on its neurobiology and the clinical role imaging will play. Radiology. 2012;263:344–361. doi: 10.1148/radiol.12110433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparisons of Volume Obtained from NeuroQuant® and Inbrain®, Based on the Slice Thickness in Patients with MCI and AD
Results of Pearson's Correlation, ICC and Effect Size, Based on the Slice Thickness in Patients with MCI and AD