Abstract
Objective
The mitotic count of gastrointestinal stromal tumors (GIST) is closely associated with the risk of planting and metastasis. The purpose of this study was to develop a predictive model for the mitotic index of local primary GIST, based on deep learning algorithm.
Materials and Methods
Abdominal contrast-enhanced CT images of 148 pathologically confirmed GIST cases were retrospectively collected for the development of a deep learning classification algorithm. The areas of GIST masses on the CT images were retrospectively labelled by an experienced radiologist. The postoperative pathological mitotic count was considered as the gold standard (high mitotic count, > 5/50 high-power fields [HPFs]; low mitotic count, ≤ 5/50 HPFs). A binary classification model was trained on the basis of the VGG16 convolutional neural network, using the CT images with the training set (n = 108), validation set (n = 20), and the test set (n = 20). The sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated at both, the image level and the patient level. The receiver operating characteristic curves were generated on the basis of the model prediction results and the area under curves (AUCs) were calculated. The risk categories of the tumors were predicted according to the Armed Forces Institute of Pathology criteria.
Results
At the image level, the classification prediction results of the mitotic counts in the test cohort were as follows: sensitivity 85.7% (95% confidence interval [CI]: 0.834–0.877), specificity 67.5% (95% CI: 0.636–0.712), PPV 82.1% (95% CI: 0.797–0.843), NPV 73.0% (95% CI: 0.691–0.766), and AUC 0.771 (95% CI: 0.750–0.791). At the patient level, the classification prediction results in the test cohort were as follows: sensitivity 90.0% (95% CI: 0.541–0.995), specificity 70.0% (95% CI: 0.354–0.919), PPV 75.0% (95% CI: 0.428–0.933), NPV 87.5% (95% CI: 0.467–0.993), and AUC 0.800 (95% CI: 0.563–0.943).
Conclusion
We developed and preliminarily verified the GIST mitotic count binary prediction model, based on the VGG convolutional neural network. The model displayed a good predictive performance.
Keywords: Gastrointestinal stromal tumors, Mitotic index, Deep learning Multidetector computed tomography
INTRODUCTION
Gastrointestinal stromal tumors (GIST) are the most common mesenchymal tumors of the gastrointestinal system. These tumors originate from Cajal stromal cells, with 95% staining positive for CD117 (c-KIT), and 70% staining positive for CD34 (1,2). Previously, unless a characteristic expression of c-KIT and CD34 antigens was identified, the histopathological classification of tumors was challenging (1). As a result, these tumors have been variably referred to as leiomyomas, leiomyosarcomas, and leiomyoblastomas. GIST have different degrees of tumor differentiation and malignant potential. Moreover, the degree of malignancy is closely related to the risk of tumor implantation and metastasis (1,2,3). In recent years, numerous risk prediction standards have been established, including the National Institutes of Health (NIH) modified criteria (4), and the Armed Forces Institute of Pathology (AFIP) criteria (5). The most commonly used risk grading standard is the NIH modified criteria, that classifies the risk of GIST according to the location, size, mitotic count, and tumor rupture, and ranges from very low to high risk (4). Among the various risk grading standards, the mitotic count is a major evaluation criteria for tumor differentiation. The preoperative pathological examination is not routinely applied to GIST (4,5,6,7,8). Percutaneous biopsy has the risk of causing tumor hemorrhage and intra-abdominal tumor spread (9). On the other hand, it is difficult to perform Endoscopic Ultrasound-Guided Fine Needle Aspiration on some GIST growth sites. Therefore, it may not be easy to obtain the preoperative biopsy data of patients diagnosed with (7,9,10). Preoperative abdominal CT enhancement examination can make a more accurate observation of the size, location, and rupture of the tumor (11,12). However, there are limited studies investigating whether it can predict the mitotic count of GIST.
Artificial intelligence (AI) has developed rapidly in the medical field. Especially in medical imaging, computer vision technology is used to rapidly read and intelligently diagnose disease (13,14,15). As one of the power algorithms of AI, deep learning can simplify the procedure by learning predictive features directly. Moreover, compared to classical machine learning, it can automatically extract more high-level and abstract features, and strongly support the translation from AI into clinical application (16). Existing studies of various diseases such as lung cancer have demonstrated that deep learning can be used to comprehensively assess the malignancy potential of a tumor. It may potentially improve the accuracy of diagnosis, prognosis, and prediction (17,18,19,20). The purpose of this study was to develop a predictive model for the mitotic index of local primary GIST, based on a deep learning algorithm.
MATERIALS AND METHODS
Ethics
Ethics Committee of Peking University First Hospital approved this retrospective study (2017-1382). This study was exempt from patient informed consent requirements.
Study Population
This retrospective study included 237 patients, that underwent GIST surgery between December 31, 2018, and January 1, 2010. Sixteen patients were excluded for the following reasons: ten patients had incomplete imaging data, two patients had received neoadjuvant therapy before surgery, and for four patients, the pathological mitotic counts were not available. Thus, the total number of patients enrolled was 221 (105 males and 116 females: mean age 60.9 ± 11.3 years, range 31–82 years). All included patients met the following criteria: 1) patient underwent radical surgery in our hospital; 2) pathological examination of postoperative specimens, combined with immunohistochemistry and morphological features, confirmed that the patient had GIST; 3) contrast-enhanced CT was performed within 15 days prior to surgery; 4) detailed clinical and pathological data was available; 5) patient did not receive imatinib or other tyrosine kinase inhibitors as neoadjuvant therapy prior to surgery.
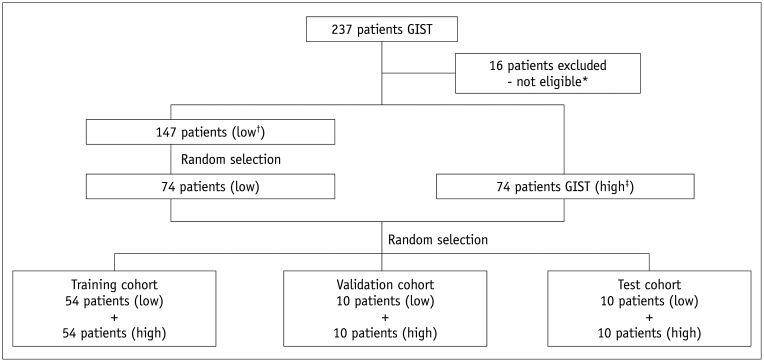
According to the postoperative pathological results and the mitotic count classification criteria of the National Comprehensive Cancer Network (NCCN) prognosis prediction guidelines (7), 147 cases were classified into a low mitotic count group (mitotic count ≤ 5/50 HPFs), and 74 cases were classified into a high mitotic count group (mitotic count > 5/50 HPFs). A detailed flow diagram of the study patients is described in Figure 1. The results included a training set of 108 cases, a validation set of 20 cases, and a test set of 20 cases.
Fig. 1. Flow diagram of the study patients.
*Sixteen patients were excluded for the following reasons: 10 patients had incomplete imaging data, two patients received neoadjuvant therapy before surgery, and four patients had missing pathological mitotic counts. †Low mitotic count (≤ 5/50 HPFs), ‡High mitotic count (> 5/50 HPFs). GIST = gastrointestinal stromal tumors, HPFs = high-power fields
Image Acquisition and Selection
Prior to surgical resection, abdominal enhanced CT examinations of all enrolled patients were performed, using one of the following CT equipment: Siemens Somatom Definition Flash (Siemens Medical Solutions); Philips Brilliance iCT (Philips Medical Systems); Philips Brilliance 64 (Philips Medical Systems); GE Discovery CT750 (GE Healthcare); and GE LightSpeed VCT (GE Healthcare). The other acquisition parameters were as follows: 120 kV tube voltage, 150–350 mAs tube current, 0.5–0.8 seconds tube rotation time, 64 × 0.625 mm detector collimation, 350 × 350 mm field of view, 512 × 512 matrix, 1 mm section thickness, and 1 mm reconstruction interval. Prior to CT examinations, the patients were required to fast for at least for six hours. For CT examinations, 80 mL to 100 mL of iodinated contrast medium was injected intravenously at a rate of 3.0 mL/s. The portal venous phase images (obtained 60 seconds after injection of the contrast agent) were selected for this study.
Segmentation of Region of Interests
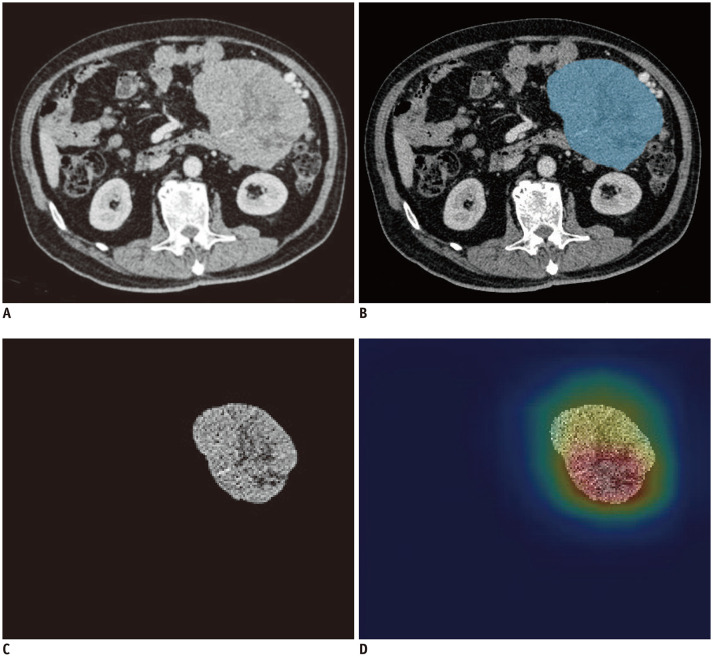
As shown in Figure 2B, the tumor area was manually labelled as a region of interest (ROI) on the workstation (IntelliSpace Discovery). A radiologist with four years of work experience used the software to volumetrically depict tumors in thin-slice contrast-enhanced CT (CE-CT) images in the portal venous phase and recorded the tumor sites. The data obtained were validated and modified by a radiologist with 20 years of experience in abdominal imaging diagnosis.
Fig. 2. A 72-year-old man with primary GIST.
Postoperative pathology showed a mitotic count of 8/50 HPFs. (A–C) showed the original axial CT image, segmented image, and pre-processed image of the tumor respectively. We used image labeling software to segment the tumor area in the CT image as the region of interest, which is the blue area in (B). Next, image preprocessing was performed on the basis of image segmentation and got the preprocessed image (C). The developed model predicted the image mitotic index classification and output the corresponding class activation heatmap (D).
Image Preprocessing
Image data format was converted from Nifty to PNG. The final PNG format was used as the training input data. To achieve this, the images were pre-processed as follows. Finally, as depicted in Figure 2C, the pre-processed images were obtained.
1) The CT images were set to a window width 300 Hounsfield unit (HU) and window level 30 HU, which are the usual settings required for observing a CT image of the abdomen.
2) The images were cropped based on the labelled areas to obtain images with only the ROIs.
3) Images that had an area of less than 1 cm2 after cropping were removed.
4) The cropped images were resized to a 224 × 224 matrix.
Transfer Learning Based on the VGG16 Convolutional Neural Network
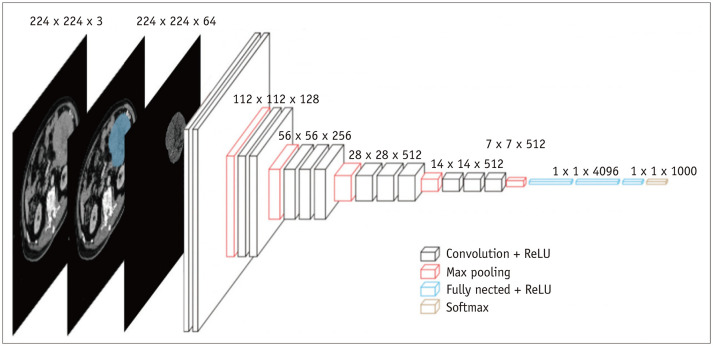
VGG16 was used for transfer learning of the binary classification model. The VGG16 architecture (Fig. 3) consisted of 13 convolutional layers, 5 maximum pooling layers, 3 fully-connected layers, and finally, a 1000-way softmax classifier. Max-pooling was performed over a 2 × 2 pixel window, with stride 2 (21). In this study, we modified the VGG16 model. The details of the model modifications are provided in the Supplementary Materials.
Fig. 3. VGG16 neural network architecture.
Model Training Parameters and Details
The parameters and details of our model training are described below: software environment: Python 3.6; hardware environment: NVIDIA Tesla P100 16 G; memory: 256 G. The initial learning rate was set to 0.00001. The batch size was set to 3 and the original image size was 224 × 224. As soon as the epoch reached 60, the loss value of the validation set was dropped to a stable level. We then halted the training. The final model had the lowest validation loss.
Output of Prediction Results and Model Performance Evaluation Indicators
The prediction results were studied for each image from each patient, at both, the image-level and the patient level. The image-level prediction results were integrated into the prediction results for each patient. At the patient level, if more than half of the images predicted a high mitotic count, the patient was considered to be in the high mitotic count group, and vice versa. Sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of the model were evaluated at these two levels. Furthermore, the model performance, at both, the image level and patient level, was assessed with ROC curves.
Further, according to the AFIP criteria (5), the predicted mitotic count group of each patient was combined with the tumor site and size (according to the volume of interest, the long diameter of the tumor was the output value for the model) to predict the risk category of each the tumor.
Statistical Analysis
Statistical analysis was performed using SPSS version 20.0 (IBM Corp.). T tests and chi-square tests were applied for the continuous variables and the categorical variables, respectively. ROC analysis was used to evaluate the efficacy of the model at the image level and the patient level. A two-sided p value < 0.05 was considered as statistically significant.
RESULTS
A total of 148 contrast-enhanced abdominal CT images from GIST patients were studied. Patient characteristics in the training, validation, and test datasets are presented in Table 1.
Table 1. Clinical Characteristics of Patients in Different Cohorts.
| Mitotic Index | Training Cohort (n = 108) | Validation Cohort (n = 20) | Test Cohort (n = 20) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ≤ 5/50 HFPs | > 5/50 HFPs | P | ≤ 5/50 HFPs | > 5/50 HFPs | P | ≤ 5/50 HFPs | > 5/50 HFPs | P | |
| Gender | 0.700 | 0.350 | 1.000 | ||||||
| Male | 28 | 25 | 8 | 5 | 5 | 4 | |||
| Female | 26 | 29 | 2 | 5 | 5 | 6 | |||
| Age (years) | 60.8 ± 10.2 | 60.2 ± 11.2 | 0.768 | 63.8 ± 10.9 | 62.7 ± 12.4 | 0.835 | 58.9 ± 11.3 | 63.3 ± 11.1 | 0.392 |
| Tumor site | 0.321 | 1.000 | 1.000 | ||||||
| Gastric | 36 | 31 | 6 | 7 | 5 | 6 | |||
| Non-gastric | 18 | 23 | 4 | 3 | 5 | 4 | |||
| Tumor size (cm) | 5.4 ± 4.1 | 7.7 ± 4.7 | 0.007 | 4.5 ± 1.6 | 9.5 ± 5.6 | 0.014 | 7.2 ± 4.2 | 13.2 ± 9.8 | 0.089 |
| Risk categories | |||||||||
| None | 8 | 4 | 1 | 0 | 0 | 0 | |||
| Very low | 19 | 0 | 3 | 0 | 3 | 0 | |||
| Low | 16 | 0 | 5 | 0 | 2 | 0 | |||
| Moderate | 6 | 13 | 0 | 2 | 2 | 0 | |||
| High | 5 | 37 | 1 | 0 | 3 | 10 | |||
Data are presented as mean ± SD or n. Independent samples t test was applied in continuous variables. Chi-Squared test was applied in categorical variables. The risk stratification adopted the Armed Forces Institute of Pathology criteria (Miettinen's criteria). HPF = high-power field
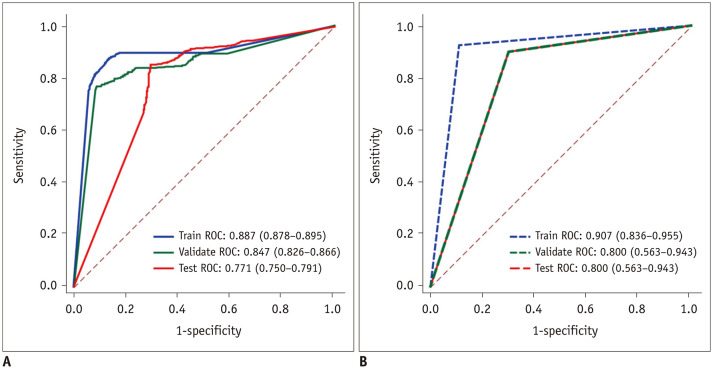
The statistical results at the image level are represented in Table 2. Three ROCs were plotted, and the area under the curves (AUCs) were 0.887 (95% confidence interval [CI]: 0.878–0.895), 0.847 (95% CI: 0.826–0.866), and 0.771 (95% CI: 0.750–0.791), respectively (Fig. 4A). In the test dataset, the sensitivity was 85.7% (95% CI: 0.834–0.877) and the specificity was 67.5% (95% CI: 0.636–0.712). The ROCs at the patient level are represented in Figure 4B. The AUCs in the training, validation, and test sets were 0.907 (95% CI: 0.836–0.955), 0.800 (95% CI: 0.563–0.943), and 0.800 (95% CI: 0.563–0.943), respectively. The sensitivity of the test dataset was 90.0% (95% CI: 0.541–0.995), the specificity was 70.0% (95% CI: 0.354–0.919), the PPV was 75.0% (95% CI: 0.428–0.933), and the NPV was 87.5% (95% CI: 0.467–0.993) (Table 3). In addition, a class activation heatmap was also obtained (Fig. 2D).
Table 2. The Statistical Results at the Image Level (Number of Pictures).
| Mitotic Index | Training Cohort (n = 108) | Validation Cohort (n = 20) | Test Cohort (n = 20) | |||
|---|---|---|---|---|---|---|
| ≤ 5/50 HFPs | > 5/50 HFPs | ≤ 5/50 HFPs | > 5/50 HFPs | ≤ 5/50 HFPs | > 5/50 HFPs | |
| True | 2044 | 3130 | 331 | 717 | 411 | 910 |
| False | 453 | 354 | 104 | 139 | 198 | 152 |
| Total | 2497 | 3484 | 435 | 856 | 609 | 1062 |
| Accuracy (95% CI), % | 86.5 (0.856–0.874) | 81.2 (0.790–0.833) | 79.1 (0.771–0.810) | |||
| Sensitivity (95% CI), % | 89.8 (0.888–0.908) | 83.8 (0.811–0.861) | 85.7 (0.834–0.877) | |||
| Specificity (95% CI), % | 81.9 (0.803–0.833) | 76.1 (0.717–0.800) | 67.5 (0.636–0.712) | |||
| Positive predictive value (95% CI), % | 87.4 (0.862–0.884) | 87.3 (0.848–0.895) | 82.1 (0.797–0.843) | |||
| Negative predictive value (95% CI), % | 85.2 (0.837–0.866) | 70.4 (0.660–0.744) | 73.0 (0.691–0.766) | |||
CI: confidence interval
Fig. 4. ROC curves of different data set.
A. Image-level ROC. B. Patient-level ROC. ROC = region of interest
Table 3. The Statistical Results at the Patient Level (Number of Patients).
| Mitotic Index | Training Cohort (n = 108) | Validation Cohort (n = 20) | Test Cohort (n = 20) | |||
|---|---|---|---|---|---|---|
| ≤ 5/50 HFPs | > 5/50 HFPs | ≤ 5/50 HFPs | > 5/50 HFPs | ≤ 5/50 HFPs | > 5/50 HFPs | |
| True | 48 | 50 | 7 | 9 | 7 | 9 |
| False | 6 | 4 | 3 | 1 | 3 | 1 |
| Total | 54 | 54 | 10 | 10 | 10 | 10 |
| Accuracy (95% CI), % | 90.7 (0.852–0.963) | 80.0 (0.608–0.992) | 80.0 (0.608–0.992) | |||
| Sensitivity (95% CI), % | 92.6 (0.813–0.976) | 90.0 (0.541–0.995) | 90.0 (0.541–0.995) | |||
| Specificity (95% CI), % | 88.9 (0.767–0.954) | 70.0 (0.354–0.919) | 70.0 (0.354–0.919) | |||
| Positive predictive value (95% CI), % | 88.9 (0.767–0.954) | 75.0 (0.428–0.933) | 75.0 (0.428–0.933) | |||
| Negative predictive value (95% CI), % | 92.3 (0.806–0.975) | 87.5 (0.467–0.993) | 87.5 (0.467–0.993) | |||
Furthermore, given the differences between the anatomic sites (i.e., stomach, non-stomach), we tentatively explored diagnostic performance by location. For all gastric GIST cases (67) in the training set, the prediction model performed with an accuracy of 91.0% (95% CI: 0.840–0.981), a sensitivity of 87.1% (95% CI: 0.692–0.958), a specificity of 94.4% (95% CI: 0.800–0.990), a PPV of 93.1% (95% CI: 0.758–0.988), and an NPV of 89.5% (95% CI: 0.743–0.966). In all non-gastric cases (41), the prediction model performed with an accuracy of 90.2% (95% CI: 0.808–0.997), a sensitivity of 100.0% (95% CI: 0.822–1.000), a specificity of 77.8% (95% CI: 0.519–0.926), a PPV of 85.2% (95% CI: 0.654–0.951), and an NPV of 100.0% (95% CI: 0.732–1.000).
The predicted tumor risk category results are represented in Table 4. The diagnostic performance indices for the test set were as follows: sensitivity of 100.0% (95% CI: 0.656–1.000), specificity of 80% (95% CI: 0.442–0.965), PPV of 83.3% (95% CI: 0.509–0.971), and NPV of 100.0% (95% CI: 0.598–1.000).
Table 4. The Predicted Tumor Risk Category Results (Number of Patients).
| Mitotic Index | Training Cohort (n = 108) | Validation Cohort (n = 20) | Test Cohort (n = 20) | |||
|---|---|---|---|---|---|---|
| Low-Mitotic | High-Mitotic | Low-Mitotic | High-Mitotic | Low-Mitotic | High-Mitotic | |
| True | 50 | 50 | 7 | 9 | 8 | 10 |
| False | 4 | 4 | 3 | 1 | 2 | 0 |
| Total | 54 | 54 | 10 | 10 | 10 | 10 |
| Accuracy (95% CI), % | 92.6 (0.876–0.976) | 80.0 (0.608–0.992) | 90.0 (0.756–1.000) | |||
| Sensitivity (95% CI), % | 92.6 (0.813–0.986) | 90.0 (0.541–0.995) | 100.0 (0.656–1.000) | |||
| Specificity (95% CI), % | 92.6 (0.813–0.986) | 70.0 (0.354–0.919) | 80.0 (0.442–0.965) | |||
| Positive predictive value (95% CI), % | 92.6 (0.813–0.986) | 75.0 (0.428–0.933) | 83.3 (0.509–0.971) | |||
| Negative predictive value (95% CI), % | 92.6 (0.813–0.986) | 87.5 (0.467–0.993) | 100.0 (0.598–1.000) | |||
DISCUSSION
On the basis of the VGG16 convolutional neural network, we trained a binary classification prediction model for the mitotic index of GIST. This prediction model has a potential clinical value for patient management (22,23). GISTs are potentially malignant tumors. Preoperative evaluation of the degree of malignancy of each tumor is of great significance for the choice of surgery. With the development of neoadjuvant therapy, studies suggest that, for higher risk GISTs, surgical resection following neoadjuvant therapy may improve patient prognosis (24,25,26). The mitotic count of a GIST could be used as a prediction target. Several studies have demonstrated the applicability and clinical value of the mitotic count (27,28).
In previous studies, radiologists have reviewed findings from CT imaging, and compared them with the findings from tumor pathology. Some studies indicate that abdominal CT enhancement examination, which is a routine examination for GIST, reveals certain imaging characteristics that are closely related to the degree of malignancy of the tumor (29,30). Additionally, the presence of necrosis or cystic degeneration, tumor contour, tumor margin, and pattern of enhancement are also associated with risk stratification (31). However, the available research is influenced by the experience of the researcher and the factors of subjectivity. Moreover, the extracted information is limited. Thus, it is difficult to conduct quantitative research. We used a deep learning algorithm to extract additional image information. This approach can yield more information than traditional methods and can overcome the influence of observer subjectivity. A few studies demonstrate that radiomics can capture intra-tumoral heterogeneity, exploit high-level features, assess tumor biological behavior, and improve diagnostic accuracy and disease prognosis (32,33,34,35). Wang et al. (27) built radiomic prediction models using CE-CT images to preoperatively predict mitotic count of GIST (sensitivity = 63.3%; specificity = 91.3%; accuracy = 85.4%). However, only the largest cross-section of the tumor was chosen as the ROI (27). In our present study, we built a CT-based deep learning model with better sensitivity (90.0%) and a similar prediction performance, with an accuracy of 80.0% in the test cohort. As is consistent with the conventional situation, the prediction performance of our model on the validation set and test set is slightly lower than that of the training set (36). Radiomics is based on the analysis of hand-crafted features, that depends on arbitrary decisions to apply statistical analysis to a particular image, as a form of feature engineering.
Our study used the deep learning approach. As a further development of machine learning, deep learning can be simplified by direct learning prediction and can extract learning features from images that are not affected by subjective extraction features. Deep learning automatically extracts more advanced and abstract features than manual extraction features (37). The VGG16 convolutional neural network was selected as the basis of the model. This network emerged from the VGG at Oxford University (21). In their eponymously named VGG network, it is easy to implement repeated structures in code in any modern deep learning framework with the use of loops and subroutines. This network is characterized by the repeated use of 3 × 3 convolution kernels and 2 × 2 convolution kernels, that increase the depth of the network (21). In this research, we adopted the transfer learning method. We replaced the fully connected layer of the VGG16 to achieve our multi-classification task. Furthermore, we selected multiple layers of tumor images that better reflected the imaging features of the tumor. The results obtained from comprehensive multi-slice CT images are more likely to reflect the overall biological behavior and mitotic rate of the tumor, as compared to hose obtained from a single layer of images (38).
Current research suggests that the malignant risk and the probability of metastasis of GISTs located in the small intestine and rectum are higher than that for gastric cases (39,40). Our study explored the diagnostic performance of the model by location. In the training set, the performance of the model in the gastric group, including the specificity and PPV, was better than that of the non-gastric group. On the other hand, the sensitivity and NPV were worse than the non-gastric group. However, the differences between the two groups were not statistically significant. In other words, the model is applicable to both gastric GISTs and non-gastric GISTs, and there is no significant difference in diagnostic efficacy. However, given our limited number of cases, future studies must have an increased sample size. Further research should also explore the impact of the tumor site on the model's diagnostic efficacy, or establish benign and malignant prediction models for different tumor sites.
The pattern classification for GIST is of great significance for its clinical management, as it can provide information for preoperative evaluation of tumors. According to the AFIP criteria (5), the predicted mitotic count group of each patient was combined with the tumor site and size to predict the tumor risk category. The risk was classified as very low risk, low risk, moderate risk, and high risk. In our study, the results of the tumor risk category were based on the predictive model results, the tumor site, and the tumor size. Moreover, as compared with the gold standard, the results were of satisfactory accuracy. The tumor risk category predicted by the model further enhanced the diagnostic efficiency of tumor malignancy potential. In addition, the predictive model for mitotic count grouping allows prediction of the biological behavior of GISTs according to the NCCN guidelines. This enables the description of a specific range of metastasis rates (7). This is important for clinical decision-making.
Additionally, contrary to previous group studies, the predictive model is also able output a class activation heatmap of the corresponding image. The meaning of the current class activation heatmap is still unclear. However, we observed that, for the high mitotic count group, the region in the image displaying the higher color temperature (red) corresponds to the heterogeneous region, or the periphery of the necrotic region. There was no obvious high color temperature region in the class activation heatmap of the low mitotic count group. This may indicate that the necrotic and heterogeneous areas are image features of high-risk GISTs (31,41).
There are some notable limitations of our study. First, in most cases, the mitotic count reported by pathological findings can reflect the overall state of tissue proliferation, and the malignant potential of the tumor. However, we are currently unable to match CT images to pathology sections layer by layer. This issue will be addressed in future studies. Second, at present, due to the variability in tumor sites and the complexity introduced due to adjacent cavity organs, we have been unable to achieve automatic segmentation of GISTs. In the future, we will further research the automatic segmentation of tumors to achieve segmentation and sequential classification of GISTs. Furthermore, in order to provide more valuable image reports for clinical use, the prediction results should be implanted into structural reports.
In this study, we developed a GIST nuclear split phase two-category prediction model based on the VGG convolutional neural network. We were able to obtain satisfactory prediction results. The model can excavate abdominal CT imaging information. Moreover, it has reference value for preoperative prediction of mitotic index, that can help guide the clinical management of GIST.
Footnotes
Conflicts of Interest: The authors have no potential conflicts of interest to disclose.
Supplementary Materials
The Data Supplement is available with this article at https://doi.org/10.3348/kjr.2020.0851.
References
- 1.Zhao X, Yue C. Gastrointestinal stromal tumor. J Gastrointest Oncol. 2012;3:189–208. doi: 10.3978/j.issn.2078-6891.2012.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miettinen M, Lasota J. Gastrointestinal stromal tumors: review on morphology, molecular pathology, prognosis, and differential diagnosis. Arch Pathol Lab Med. 2006;130:1466–1478. doi: 10.5858/2006-130-1466-GSTROM. [DOI] [PubMed] [Google Scholar]
- 3.Rammohan A, Sathyanesan J, Rajendran K, Pitchaimuthu A, Perumal SK, Srinivasan U, et al. A gist of gastrointestinal stromal tumors: a review. World J Gastrointest Oncol. 2013;5:102–112. doi: 10.4251/wjgo.v5.i6.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joensuu H. Risk stratification of patients diagnosed with gastrointestinal stromal tumor. Hum Pathol. 2008;39:1411–1419. doi: 10.1016/j.humpath.2008.06.025. [DOI] [PubMed] [Google Scholar]
- 5.Miettinen M, Lasota J. Gastrointestinal stromal tumors: pathology and prognosis at different sites. Semin Diagn Pathol. 2006;23:70–83. doi: 10.1053/j.semdp.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 6.Poveda A, García Del Muro X, López-Guerrero JA, Cubedo R, Martínez V, Romero I, et al. GEIS guidelines for gastrointestinal sarcomas (GIST) Cancer Treat Rev. 2017;55:107–119. doi: 10.1016/j.ctrv.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 7.von Mehren M, Randall RL, Benjamin RS, Boles S, Bui MM, Ganjoo KN, et al. Soft tissue sarcoma, version 2.2018, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2018;16:536–563. doi: 10.6004/jnccn.2018.0025. [DOI] [PubMed] [Google Scholar]
- 8.Sepe PS, Brugge WR. A guide for the diagnosis and management of gastrointestinal stromal cell tumors. Nat Rev Gastroenterol Hepatol. 2009;6:363–371. doi: 10.1038/nrgastro.2009.43. [DOI] [PubMed] [Google Scholar]
- 9.Demetri GD, von Mehren M, Antonescu CR, DeMatteo RP, Ganjoo KN, Maki RG, et al. NCCN task force report: update on the management of patients with gastrointestinal stromal tumors. J Natl Compr Canc Netw. 2010;8 Suppl 2:S1–S41. doi: 10.6004/jnccn.2010.0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ha CY, Shah R, Chen J, Azar RR, Edmundowicz SA, Early DS. Diagnosis and management of GI stromal tumors by EUS-FNA: a survey of opinions and practices of endosonographers. Gastrointest Endosc. 2009;69:1039–1044.e1. doi: 10.1016/j.gie.2008.07.041. [DOI] [PubMed] [Google Scholar]
- 11.Vernuccio F, Taibbi A, Picone D, LA Grutta L, Midiri M, Lagalla R, et al. Imaging of Gastrointestinal Stromal Tumors: From Diagnosis to Evaluation of Therapeutic Response. Anticancer Res. 2016;36:2639–2648. [PubMed] [Google Scholar]
- 12.Lee MW, Kim SH, Kim YJ, Lee JM, Lee JY, Park EA, et al. Gastrointestinal stromal tumor of the stomach: preliminary results of preoperative evaluation with CT gastrography. Abdom Imaging. 2008;33:255–261. doi: 10.1007/s00261-007-9253-x. [DOI] [PubMed] [Google Scholar]
- 13.Greenfield D, Wilson S. Artificial intelligence in medicine: applications, implications, and limitations. [Accessed September 24, 2019]. Availabe at: http://sitn.hms.harvard.edu/flash/2019/artificial-intelligence-in-medicine-applications-implications-and-limitations/
- 14.Beam AL, Kohane IS. Translating artificial intelligence into clinical care. JAMA. 2016;316:2368–2369. doi: 10.1001/jama.2016.17217. [DOI] [PubMed] [Google Scholar]
- 15.Aerts HJWL. Data science in radiology: a path forward. Clin Cancer Res. 2018;24:532–534. doi: 10.1158/1078-0432.CCR-17-2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seif G. Deep learning vs classical machine learning. [Accessed September 25, 2019]. Available at: https://towardsdatascience.com/deep-learning-vs-classical-machine-learning-9a42c6d48aa.
- 17.Vial A, Stirling D, Field M, Ros M, Ritz C, Carolan M, et al. The role of deep learning and radiomic feature extraction in cancer-specific predictive modelling: a review. Transl Cancer Res. 2018;7:803–816. [Google Scholar]
- 18.Wang S, Shi J, Ye Z, Dong D, Yu D, Zhou M, et al. Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur Respir J. 2019;53:1800986. doi: 10.1183/13993003.00986-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jung H, Kim B, Lee I, Lee J, Kang J. Classification of lung nodules in CT scans using three-dimensional deep convolutional neural networks with a checkpoint ensemble method. BMC Med Imaging. 2018;18:48. doi: 10.1186/s12880-018-0286-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song Q, Zhao L, Luo X, Dou X. Using deep learning for classification of lung nodules on computed tomography images. J Healthc Eng. 2017;2017:8314740. doi: 10.1155/2017/8314740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition; Proceedings of the ICLR; 2015 May 7-9; San Diego, CA, USA. [Google Scholar]
- 22.Dematteo RP, Gold JS, Saran L, Gönen M, Liau KH, Maki RG, et al. Tumor mitotic rate, size, and location independently predict recurrence after resection of primary gastrointestinal stromal tumor (GIST) Cancer. 2008;112:608–615. doi: 10.1002/cncr.23199. [DOI] [PubMed] [Google Scholar]
- 23.Miettinen M, Lasota J, Sobin LH. Gastrointestinal stromal tumors of the stomach in children and young adults: a clinicopathologic, immunohistochemical, and molecular genetic study of 44 cases with long-term follow-up and review of the literature. Am J Surg Pathol. 2005;29:1373–1381. doi: 10.1097/01.pas.0000172190.79552.8b. [DOI] [PubMed] [Google Scholar]
- 24.Ishikawa T, Kanda T, Kameyama H, Wakai T. Neoadjuvant therapy for gastrointestinal stromal tumor. Transl Gastroenterol Hepatol. 2018;3:3. doi: 10.21037/tgh.2018.01.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Mehren M. The role of adjuvant and neoadjuvant therapy in gastrointestinal stromal tumors. Curr Opin Oncol. 2008;20:428–432. doi: 10.1097/CCO.0b013e328302ed82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doyon C, Sidéris L, Leblanc G, Leclerc YE, Boudreau D, Dubé P. Prolonged therapy with imatinib mesylate before surgery for advanced gastrointestinal stromal tumor results of a phase II trial. Int J Surg Oncol. 2012;2012:761576. doi: 10.1155/2012/761576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang C, Li H, Jiaerken Y, Huang P, Sun L, Dong F, et al. Building CT radiomics-based models for preoperatively predicting malignant potential and mitotic count of gastrointestinal stromal tumors. Transl Oncol. 2019;12:1229–1236. doi: 10.1016/j.tranon.2019.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pelandré GL, Djahjah MC, Gasparetto EL, Nacif MS, Marchiori E, Mello EL. Tomographic findings of gastric gastrointestinal stromal tumor and correlation with the mitotic index. Arq Gastroenterol. 2013;50:244–250. doi: 10.1590/S0004-28032013000400002. [DOI] [PubMed] [Google Scholar]
- 29.Iannicelli E, Carbonetti F, Federici GF, Martini I, Caterino S, Pilozzi E, et al. Evaluation of the relationships between computed tomography features, Pathological findings, and prognostic risk assessment in gastrointestinal stromal tumors. J Comput Assist Tomogr. 2017;41:271–278. doi: 10.1097/RCT.0000000000000499. [DOI] [PubMed] [Google Scholar]
- 30.Zhou C, Duan X, Zhang X, Hu H, Wang D, Shen J. Predictive features of CT for risk stratifications in patients with primary gastrointestinal stromal tumour. Eur Radiol. 2016;26:3086–3093. doi: 10.1007/s00330-015-4172-7. [DOI] [PubMed] [Google Scholar]
- 31.Li H, Ren G, Cai R, Chen J, Wu X, Zhao J. A correlation research of Ki67 index, CT features, and risk stratification in gastrointestinal stromal tumor. Cancer Med. 2018;7:4467–4474. doi: 10.1002/cam4.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhuo T, Li X, Zhou H. Combining radiomics and CNNs to classify benign and malignant GIST; Proceedings of the 2018 International Conference on Network, Communication, Computer Engineering (NCCE 2018); 2018 May 26-27; Chongqing, China. Atlantis Press; 2018. [Google Scholar]
- 33.Ning Z, Luo J, Li Y, Han S, Feng Q, Xu Y, et al. Pattern classification for gastrointestinal stromal tumors by integration of radiomics and deep convolutional features. IEEE J Biomed Health Inform. 2019;23:1181–1191. doi: 10.1109/JBHI.2018.2841992. [DOI] [PubMed] [Google Scholar]
- 34.Chen T, Ning Z, Xu L, Feng X, Han S, Roth HR, et al. Radiomics nomogram for predicting the malignant potential of gastrointestinal stromal tumours preoperatively. Eur Radiol. 2019;29:1074–1082. doi: 10.1007/s00330-018-5629-2. [DOI] [PubMed] [Google Scholar]
- 35.Feng C, Lu F, Shen Y, Li A, Yu H, Tang H, et al. Tumor heterogeneity in gastrointestinal stromal tumors of the small bowel: volumetric CT texture analysis as a potential biomarker for risk stratification. Cancer Imaging. 2018;18:46. doi: 10.1186/s40644-018-0182-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maier A, Syben C, Lasser T, Riess C. A gentle introduction to deep learning in medical image processing. Z Med Phys. 2019;29:86–101. doi: 10.1016/j.zemedi.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 37.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 38.Watabe T, Tatsumi M, Watabe H, Isohashi K, Kato H, Yanagawa M, et al. Intratumoral heterogeneity of F-18 FDG uptake differentiates between gastrointestinal stromal tumors and abdominal malignant lymphomas on PET/CT. Ann Nucl Med. 2012;26:222–227. doi: 10.1007/s12149-011-0562-3. [DOI] [PubMed] [Google Scholar]
- 39.Miettinen M, Makhlouf H, Sobin LH, Lasota J. Gastrointestinal stromal tumors of the jejunum and ileum: a clinicopathologic, immunohistochemical, and molecular genetic study of 906 cases before imatinib with long-term follow-up. Am J Surg Pathol. 2006;30:477–489. doi: 10.1097/00000478-200604000-00008. [DOI] [PubMed] [Google Scholar]
- 40.Parab TM, DeRogatis MJ, Boaz AM, Grasso SA, Issack PS, Duarte DA, et al. Gastrointestinal stromal tumors: a comprehensive review. J Gastrointest Oncol. 2019;10:144–154. doi: 10.21037/jgo.2018.08.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pinaikul S, Woodtichartpreecha P, Kanngurn S, Leelakiatpaiboon S. Gastrointestinal stromal tumor (GIST): computed tomographic features and correlation of CT findings with histologic grade. J Med Assoc Thai. 2014;97:1189–1198. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.