DYBLUP is a blue light sensor protein that binds to ciliary dynein motors and regulates light acclimation.
Abstract
Light-responsive regulation of ciliary motility is known to be conducted through modulation of dyneins, but the mechanism is not fully understood. Here, we report a novel subunit of the two-headed f/I1 inner arm dynein, named DYBLUP, in animal spermatozoa and a unicellular green alga. This subunit contains a BLUF (sensors of blue light using FAD) domain that appears to directly modulate dynein activity in response to light. DYBLUP (dynein-associated BLUF protein) mediates the connection between the f/I1 motor domain and the tether complex that links the motor to the doublet microtubule. Chlamydomonas lacking the DYBLUP ortholog shows both positive and negative phototaxis but becomes acclimated and attracted to high-intensity blue light. These results suggest a mechanism to avoid toxic strong light via direct photoregulation of dyneins.
INTRODUCTION
Eukaryotic cilia and flagella are oscillatory bending machinery that drives motility of a single cell and generates fluid flow in epithelial cells (1). The internal structures of these organelles include well-conserved microtubule-based 9+2 arrays, called axonemes, in which molecular motor dyneins are responsible for microtubule sliding and propagation of successive waves. Axonemal dyneins are composed of heavy chains (HCs), intermediate chains (ICs), and light chains (LCs). HCs are the motor subunits that generate force for microtubule sliding. ICs and LCs are associated subunits that assemble and regulate the motor subunits. Axonemal dyneins are grouped into outer- and inner-arm dyneins (OADs and IADs, respectively) according to their position on a doublet microtubule (2–4). Recent cryo–electron tomography (cryo-ET) studies have revealed other structures that are associated with dyneins or connect distinct types of dynein (5–7). Although these structures may play important roles in regulation of ciliary and flagellar motility, how they regulate dynein activity remains unknown.
Ciliary and flagellar motility is modulated by changes in the cellular environment. Extracellular stimuli affect cytoplasmic signaling molecules, such as Ca2+ and adenosine 3′,5′-cyclic monophosphate (cAMP), across the plasma membrane and alter the motor activity of dynein (8). In animal cilia and flagella, these signaling molecules act on the ICs and LCs of OADs through direct binding or protein phosphorylation and alter the motor activity of dynein HCs (4, 9). The molecular composition and roles of IADs are less well characterized in animals, although the molecular composition and regulation of an IAD called f/I1 dynein have been well studied in Chlamydomonas reinhardtii, a unicellular green alga (10–12).
RESULTS
Identification of a novel BLUF protein associated with f/I1 dynein of animal sperm
We started this work by biochemically isolating IADs from the sperm of the sea squirt Ciona intestinalis (phylum Chordata, subphylum Tunicata), a marine invertebrate. After extraction of OADs from the axoneme of Ciona sperm flagella, subsequent extraction with KCl/adenosine 5′-triphosphate (ATP) (fig. S1A) (13) solubilized a two-headed dynein that sedimented at 20S in sucrose density gradient centrifugation (Fig. 1A). Negative-stain electron microscopy showed that this dynein had an asymmetric shape different from that of OAD but quite similar to that of Chlamydomonas f/I1 dynein, a two-headed subspecies of IAD (Fig. 1B) (14).
Fig. 1. Characterization of DYBLUP from sperm flagella of the ascidian C. intestinalis.
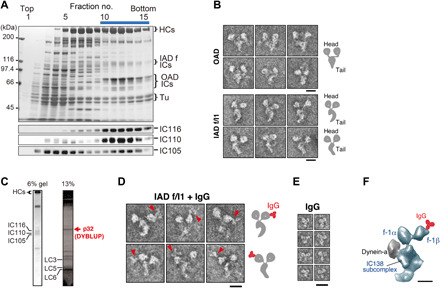
(A) Sucrose density gradient centrifugation of high salt/ATP extract from flagellar axonemes. Proteins separated in 6% SDS gels and corresponding Western blots using antibodies against orthologs of Chlamydomonas IC140 (IC116), IC138 (IC110), and IC97 (IC105) are shown. The bar represents the fractions subjected to Uno Q anion-exchange column chromatography (fig. S1B). (B) Electron micrographs of the negatively stained OAD and IAD f/I1 dynein. Both show two-headed structures but differ in the shape of their tails. Schematic drawings are shown on the right. Scale bars, 20 nm. (C) Subunit composition of Ciona f/I1 dynein. The 32-kDa protein DYBLUP is indicated in red. (D) Labeling of f/I1 dynein with anti-DYBLUP IgG. The images in the top and bottom rows correspond to two opposite orientations when the molecules are adsorbed to the carbon support film. The labeled motor domain is located on the convex side of the tail surface. Schematic drawings are shown on the right. Scale bar, 20 nm. (E) Images of IgG molecule. Scale bar, 20 nm. (F) Localization of DYBLUP on the 1β motor domain. The f/I1 dynein image extracted from the cryo-ET structure of an axoneme [EMD-5330; (18)] is oriented as in the top row of (D). Comparison with the IgG-labeled negative-stain images specifies the position of the label on the motor domain. Scale bar, 10 nm.
Uno Q anion-exchange column chromatography further separated this dynein from the fraction of OAD that was mixed with it in the sucrose density gradient fraction (fig. S1B). Mass spectrometry (MS) analysis of the components in this dynein identified two dynein HCs orthologous to Chlamydomonas 1α and 1β HCs and three ICs, namely, IC116, IC110, and IC105, which are orthologs of Chlamydomonas f/I1 dynein IC140, IC138, and IC97, respectively (Fig. 1C, fig. S1C, and table S1; numbers on the gel in fig. S1C correspond to those in table S1). We detected two HCs of OAD, and homologs of the OAD β HC, which were likely from OAD included in the f/I1 dynein fraction (table S1). These findings indicate that the architecture of f/I1 dynein molecules is conserved between Chlamydomonas and Ciona flagella.
We next analyzed the lower molecular mass components (the LCs) of Ciona f/I1 dynein. We found that the Ciona f/I1 dynein contained LCs that were reported in Chlamydomonas f/I1 dynein (table S1), including orthologs of Chlamydomonas LC9 (Tctex1), LC7b (roadblock), and LC8. In the order of their molecular masses, these LCs were termed Ciona LC3, LC5, and LC6 (Fig. 1C, fig. S1C, and table S1).
We found that an additional protein with molecular mass 32 kDa was contained in the Ciona f/I1 dynein fraction as a major component (Fig. 1C). This protein also comigrated in the subsequent separation on the Uno Q column (fig. S1B). We identified this protein by MS (table S1) as a Ciona protein that consisted of 247 amino acid residues and showed homology to the product of a gene on human chromosome 7, open reading frame 62 (C7orf62), and mouse gene testis-expressed 47 (Tex47). This protein has a BLUF (sensors of blue light using FAD) domain (residues 37 to 140) that functions as a sensor of blue light, which induces structural changes in the protein (15, 16) (fig. S2A); thus, we named the protein “DYBLUP” for dynein-associated BLUF protein.
DYBLUP is distributed in both animals and a green alga
Orthologs of DYBLUP are widely distributed in metazoans, except arthropods, as well as in other groups of eukaryotes, such as chytrid fungi and Chlamydomonas (fig. S2A). The BLUF domain of DYBLUP shares high similarity with homologs in animals and Chlamydomonas, as well as non-DYBLUP photoreceptor proteins in other organisms, including a photosynthetic flagellate, Euglena gracilis, and a photoheterotrophic bacteria, Rhodobacter sphaeroides. Structural modeling of the BLUF domain of DYBLUP showed high conservation with the BLUF domains of the Chlamydomonas DYBLUP ortholog MOT7 and the Rhodobacter photoprotein AppA (fig. S2B).
We could not find clear orthologs of DYBLUP with BLUF domains in nonanimal species except Chlamydomonas, Batrachochytrium (chytrid fungi), and Paramecium (ciliates) (fig. S2C). Leishmania, a genus of the supergroup Excavata, has a homolog of DYBLUP, LAX28 (17); however, it appears to lack the BLUF domain and shares low homology with the DYBLUP orthologs of animals and Chlamydomonas MOT7. Among other Excavata, Trypanosoma have a DYBLUP homolog with no BLUF domain (17), and Euglena show no clear ortholog of DYBLUP (fig. S2C). Like Chlamydomonas, haptophytes, cryptophytes, and the gametes of brown algae have motile flagella and show phototaxis. We found that DYBLUP homologs were present in these organisms but with no clear BLUF domain (fig. S2C).
DYBLUP binds flavin nucleotides
Thin-layer chromatography (TLC) and reversed-phase high-performance liquid chromatography (HPLC) were used to detect flavin adenine dinucleotide (FAD) or flavin mononucleotide (FMN) in the axoneme and purified f/I1 dynein fraction from Ciona sperm (fig. S2, D and E). FAD was detected in the dynein fraction by HPLC (fig. S2E); however, neither FAD nor FMN was detected by TLC or LC–electrospray ionization (ESI)–MS. Instead, a fast-migrating substance was detected in TLC (fig. S2D). Molecular species with masses of 365.2, 603.4, and 707.4 in positive ion mode and 579.3 and 683.3 in negative ion mode were detected by LC-ESI-MS (fig. S2F). Considering possible paired ion types of a common substance, three of these ions were consistent with a species with molecular mass of 684 (M) as follows: mass/charge ratio (m/z) 365.2: (M + 2Na)2+, m/z 707.4: (M + Na)+, and m/z 683.3: (M − H)−.
Next, we examined the binding of flavin nucleotides to DYBLUP. Because recombinant Ciona DYBLUP showed low solubility, we recombinantly prepared the Chlamydomonas ortholog of DYBLUP, MOT7. Gel filtration of mixtures of recombinant MOT7 and flavin nucleotides indicated that MOT7 could weakly bind FAD or FMN (fig. S2G). These results suggest that FAD is first bound to DYBLUP and then converted to a smaller substance with a molecular mass of 684 as a ligand of DYBLUP; this conversion may be catalyzed by an enzyme in the flagella.
DYBLUP interacts with the 1β motor domain of f/I1 dynein
We prepared a specific antibody against Ciona DYBLUP, which revealed that DYBLUP was exclusively localized along sperm flagella (fig. S3). Immunoprecipitation of the dynein fraction obtained from sucrose density gradient centrifugation showed that all three ICs of f/I1 dynein coprecipitated with DYBLUP, indicating that DYBLUP is associated with f/I1 (fig. S1D). Negative-stain electron microscopy of f/I1 dynein labeled with anti-DYBLUP immunoglobulin G (IgG) showed that DYBLUP was located on one of the two motor domains (Fig. 1, D and E). The labeled motor domain was always located on the convex side of the dynein tail surface (Fig. 1D). Comparison with a cryo-ET structure of f/I1 dynein in the Chlamydomonas axoneme (18) indicated that the labeled motor domain is that of the 1β HC (Fig. 1F). Biochemical analysis using chemical cross-linking and ultraviolet (UV)–mediated cleavage of dynein HCs further confirmed binding of DYBLUP to the motor domain of f/I1 dynein (fig. S1E).
To determine whether the association of DYBLUP with f/I1 dynein is evolutionally conserved, we analyzed the ortholog of DYBLUP, MOT7, in C. reinhardtii. We prepared flagellar axonemes from wild-type (WT) and several mutant strains lacking axonemal components. MOT7 was detected in the WT strain and in mutants lacking f/I1 dynein (ida1) or lacking other axonemal structures (Fig. 2A). We obtained a mutant lacking MOT7 from the Chlamydomonas Library Project (CLiP) (19) and verified disruption of the MOT7 gene using both polymerase chain reaction (PCR) and Western blotting (fig. S4, A and B). Western blots of WT and mot7 axonemes showed that the mutant lacked MOT7 but retained components of OAD and IADs (Fig. 2B). A rescued strain expressing MOT7–BCCP (biotin carboxyl carrier protein)–3HA (hemagglutinin) showed clear localization of MOT7 along flagella (Fig. 2C). We extracted dyneins from the WT and mutant axonemes and separated them by Mono Q anion exchange column chromatography. MOT7 was detected not only in the f/I1 dynein fraction of WT cells and the OAD-less mutant (oda6) but also in a mutant lacking f/I1dynein (ida1), although ida1 completely lacked IC138 (Fig. 2D). This suggests that DYBLUP/MOT7 is associated with f/I1 dynein but is not an integral component.
Fig. 2. Characterization of the DYBLUP ortholog MOT7 from flagella of the green alga C. reinhardtii.
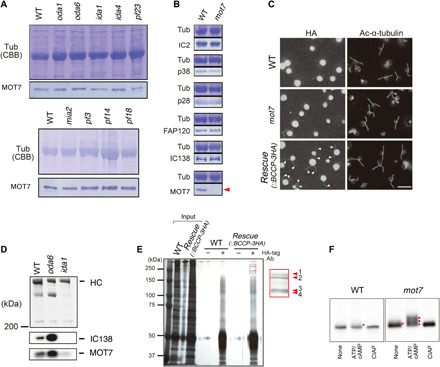
(A) Western blot analysis of MOT7 in the axonemes of the WT and several mutants. MOT7 is present in mutant axonemes with a defect in OAD (oda1 and oda6), two-headed IAD f/I1 (ida1), several single-headed IADs (ida4), most IADs (pf23), the modifier of inner arms (MIA) complex (mia2), the nexin-dynein regulatory complex (N-DRC) complex (pf3), radial spokes (pf14), and the central-pair complex (pf18). Loadings (tubulin bands, Coomassie Brilliant Blue) and the Western blot with anti-MOT7 are shown. (B) Western blot analysis of dynein subunits in axonemes of the WT and mot7 mutant. The mot7 mutant has no apparent axonemal defect in IC2 (OAD), p38 (IAD d), p28 (IADs a, c, d), FAP120, or IC138 (IAD f/I1) but lacks MOT7 (red arrowhead). (C) Immunofluorescence image showing MOT7 along the flagella. Images of staining with anti-HA antibody (left) and anti–acetylated α-tubulin antibody (right) are shown. The flagella of the mot7-rescue mutant expressing MOT7-BCCP-3HA are recognized with anti-HA antibody. The strong fluorescence in the cell body is autofluorescence. Scale bar, 20 μm. (D) Immunoblots of f/I1 dynein purified with a Mono Q column showing the presence of MOT7 in the WT strain and the oda6 and ida1 mutants. (E) Immunoprecipitation of axonemal extract with anti–HA-tag antibody. Four bands (red arrowheads) are specifically immunoprecipitated with antibody from the mot7-rescue mutant expressing MOT7-BCCP-3HA. Two of these (numbers 2 and 4) were identified as FAP44, a tether complex protein associated with the f/I1 motor domain (see table S2). (F) IC138 is hyperphosphorylated in the mot7 mutant. Western blots of axonemes from the WT strain and mot7 mutant using anti-IC138 antibody are shown. Phosphorylation was induced by addition of ATP and cAMP to the isolated axonemes. Phosphorylated IC138 bands are indicated by red dots. CIAP, calf intestine alkaline phosphatase.
MOT7 is associated with the IAD f/I1 tether complex and regulates IC138 phosphorylation
We further examined the roles of DYBLUP/MOT7 in f/I1 dynein through two biochemical experiments. First, axonemal extract from Chlamydomonas expressing BCCP-3HA–tagged recombinant MOT7 was immunoprecipitated with anti-HA antibody (Fig. 2E and fig. S4C). Four high–molecular mass bands (~250 kDa) were observed in immunoprecipitation. Two of them were identified by matrix-assisted laser desorption/ionization–time-of-flight (mass spectrometry) (MALDI-TOF/MS) as FAP44, a recently identified component of the tether complex, which anchors the motor domains of f/I1 dynein to the A-tubule of a doublet microtubule (20, 21); the remaining two bands showed no significant hit to the Chlamydomonas protein database (Fig. 2E and table S2). The band corresponding to FAP44 with lower molecular mass was likely a degradation product or modified form of this protein. The amount of MOT7 was significantly decreased in a Chlamydomonas fap44 mutant (20), and recent work on a homolog of MOT7 in Leishmania, LAX28, showed that it was linked to f/I1 dynein and the tether complex (17). We could not find FAP44 in the Ciona f/I1 dynein fraction, possibly because of differences in the interactions among the tether, DYBLUP, and dynein motor domain in Chlamydomonas and Ciona.
Second, IC138 was hyperphosphorylated in the mot7 strain (Fig. 2F), suggesting a significant role of DYBLUP/MOT7 in the regulation of f/I1dynein through IC138 phosphorylation. Thin-section electron microscopy showed no significant difference in the dynein arms or axonemal structure between the WT Chlamydomonas and a mot7 strain (fig. S4E). To determine the molecular interactions of MOT7 in the axoneme, as partially described above, we introduced a BCCP-3HA tag to the C terminus of MOT7 (22). The BCCP-labeled recombinant MOT7 construct was properly incorporated into the axoneme in the mot7 mutant (fig. S4C). The axoneme with MOT7-BCCP-3HA was decorated with streptavidin and biotinylated cytochrome c to visualize the position of MOT7 (fig. S4F). Cryo-ET and subtomogram averaging of WT axonemes showed that both the 1α and 1β motor domains were associated with the tether complex, which connects f/I1 dynein to the A-tubule of a doublet microtubule (Fig. 3, A and B) (20, 21). Enhanced BCCP was clearly observed between the tether complex and the 1β motor domain of f/I1 dynein (Fig. 3, A and B). Isosurface renderings from a different orientation revealed that a link between the tether and the motor domain was retained in the 1α motor but lost in the 1β motor in the mot7 mutant. The density of tether complex appeared significantly decreased in the mot7 mutant (Fig. 3C), most likely due to structural fluctuations caused by loss of MOT7 protein. The BCCP tag was located on a link from the tether at the opposite side of the stalk of 1β (18, 20, 21). Thus, it is likely that the BCCP tag was located in the vicinity of the 1β AAA1 domain.
Fig. 3. Cryo-ET showing the localization of MOT7 in the link structure between the tether complex and f/I1-1β motor domain.
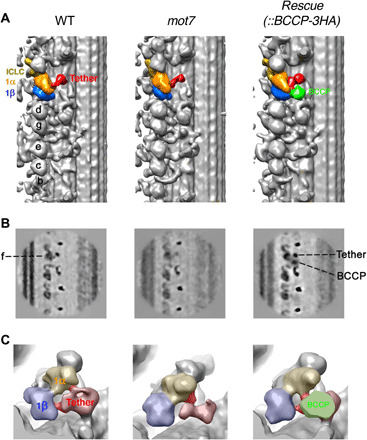
(A) Isosurface rendering longitudinal images of the axonemes from the WT C. reinhardtii strain (left), mot7 mutant (middle), and the mutant rescued with MOT7-BCCP-3HA (right). The 1α and 1β motor domains of f/I1dynein (58), the tether complex, and the enhanced biotin carboxyl carrier protein (BCCP) tag are indicated in orange, blue, red, and green, respectively. BCCP is associated with both the 1β domain and the tether complex. A part of the tether structure is absent in the mot7 mutant. (B) Tomographic slices of the averaged longitudinal view in axonemes from the WT strain (left), mot7 mutant (middle), and the mutant rescued with MOT7-BCCP-3HA (right). The density of BCCP is clearly seen between f/I1 dynein and the tether complex. The amount of tether complex is significantly decreased in the mot7 mutant, likely due to structural fluctuation. (C) Detailed comparison of isosurface rendering images. The WT strain (left), mot7 mutant (middle), and the mutant rescued with MOT7-BCCP-3HA (right) are shown. Two links between the f/I1 motor domain and the tether complex are colored in dark pink. The link from the tether to the 1β domain is lost in the mot7 mutant, but that from the tether to the 1α domain is retained.
MOT7 regulates Chlamydomonas motility for avoidance of blue light
We next investigated the difference in motility between WT Chlamydomonas and the mot7 mutant. Swimming velocity, flagellar bending, and photoshock response showed no notable difference between the WT and the mot7 mutant (fig. S5, A to C). Because MOT7 was shown to be bound to f/I1 dynein and to affect phosphorylation of IC138, we expected some defects in phototactic behavior in the mutant. However, both the WT and the mot7 mutant showed positive and negative phototaxis (swimming toward and away from a light source) against weak (~1 μmol photons m−2 s−1) and strong (~1000 μmol photons m−2 s−1) blue light, respectively (Fig. 4, A and B). However, during the dish assay of negative phototaxis, we observed that the mot7 mutant once showed negative phototaxis but started to move toward the blue light source within 30 min (Fig. 4B and fig. S6A), suggesting that the mutant lacked the ability to sustainably stay away from strong light. This “acclimation” was not observed in the WT and occurred at a slower rate in the mutant rescued with MOT7-3HA (fig. S6A); notably, the expression level of HA-tagged MOT7 in the rescue strain was a little lower than that of WT MOT7, and the exogeneous MOT7 only partially rescued the phenotype. The acclimation was observed in reducing conditions, but not in oxidizing conditions, in which all the strains showed positive phototaxis (fig. S6, B and C), suggesting that MOT7 is involved in regulation of flagellar motility in reducing conditions.
Fig. 4. MOT7-deficient C. reinhardtii mutant shows acclimation to blue light.
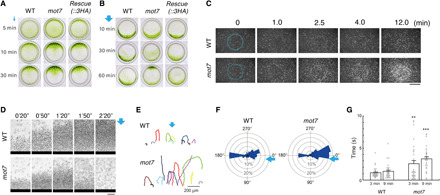
(A) Positive phototactic behavior induced by weak blue light is seen for both the WT strain and the mot7 mutant. (B) Negative phototaxis induced by strong blue light is also seen for both the WT strain and the mot7 mutant. However, the mot7 mutant soon acclimates and turns to the blue light (see also fig. S6A). This acclimation is significantly delayed in the mutant rescued with MOT7-3HA. (C) Irradiation with strong blue light in the center of a microscopic field under red light illumination over the full field. The area of blue light irradiation is shown with a blue line. mot7 mutants become accumulated to the blue light after a time, but the WT strain avoided the area. Scale bar, 50 μm. (D) Accumulation of Chlamydomonas at the opposite end (black bar) of a chamber to avoid strong blue light. WT cells stay and accumulate at the opposite end from the light, whereas mot7 mutant cells initially accumulate at that end but soon turn around toward the blue light, resulting in dispersion of the cell mass. Scale bar, 200 μm. (E) Swimming trajectories of WT cells and the mot7 mutant around the edge of the chamber, recorded for 20 s at 9 min after irradiation. (F) Circular histogram showing the increase in the number of mot7 mutants that swim toward the blue light. Swimming directions 9 min after irradiation are indicated in 18 sections with the bin of 20°; n = 421 WT and 397 mutant cells. Arrows show the direction of blue light. (G) Duration of swimming in a straight path toward the blue light 3 and 9 min after irradiation; n = 30 WT and 31 mutant cells. **P < 0.01 and ***P < 0.001 versus WT.
We confirmed the MOT7-mediated blue light acclimation of Chlamydomonas by using a strain mutated in a tether component, FAP44 (20, 21), and a different mot7 mutant generated by CRISPR/Cas9 gene editing (mot7CRISPR) (fig. S7). The fap44 mutant showed positive or negative phototaxis in oxidizing or reducing conditions, respectively (fig. S7A). However, the mutant never showed acclimation to strong blue light even in reducing conditions (fig. S7, A and B). The mot7CRISPR mutant (fig. S7, C to E) showed acclimation to strong blue light, as well as positive and negative phototaxis. The acclimation became clearer in reducing conditions (fig. S7F).
We further verified the acclimation using two assay systems. First, we illuminated the entire microscopic field with red light-emitting diode (LED) light and provided spot illumination of its center with strong blue light (Fig. 4C). WT Chlamydomonas showed photoshock around the edge of the blue light and never accumulated in the field of blue light (Fig. 4C, top, and movie S1). In contrast, although the mot7 mutant exhibited photoshock and escaped from the strong blue light a short time after illumination, the cells eventually accumulated in the blue light (Fig. 4C, bottom, and movie S2). Turning off the blue light after the reaction induced a rapid dispersion of the cells from the center of the illuminated field, indicating that the accumulation was not due to attachment on the glass or to damage of the cells (movie S2).
Second, to describe the behavior of Chlamydomonas on exposure to strong blue light, we illuminated one side of a chamber on a glass slide with strong blue light and observed the edge of the other side. Control (WT) cells were stacked at the edge away from the light source at the start of the illumination. Then, they briefly tended to move away from the edge (in the “reverse direction”), but soon turned back, resulting in substantial accumulation of cells near the edge (Fig. 4D and movie S3). These movement patterns were observed in the mutant shortly after illumination, but the frequency of direction change and the duration of movement in the reverse direction were soon increased, and the cells began to disperse away from the edge, i.e., toward the blue light (Fig. 4, E and F, and movie S4). Analysis of flagellar waveforms showed that the WT strain transiently moved toward the strong light with a breaststroke-like beating but soon changed the balance of beating of both flagella so as to turn around and move back to the edge of the chamber (fig. S5, D to F, and movies S5 and S6). The mot7 mutant showed asymmetric beating, resulting in increased breaststroke-like movement toward the strong light with time (Fig. 4G).
DISCUSSION
This work demonstrates that DYBLUP/MOT7 is a ciliary photoreceptor protein in distantly related eukaryotes, Ciona and Chlamydomonas. DYBLUP is directly associated with both the 1β motor domain of f/I1 dynein and the tether complex (Figs. 2 and 3). The binding of DYBLUP to the 1β motor domain is likely not strong and could be light sensitive, because DYBLUP is partially dissociated during f/I1 dynein purification in Ciona (fig. S1A) and loss of MOT7 does not affect the assembly of f/I1 dynein in Chlamydomonas (Fig. 2B). Instead, MOT7 is likely to be tightly associated with the tether complex (Figs. 2E and 3) (20), which mechanically stabilizes f/I1 dynein in the axoneme (20, 21). This differs from findings on a homolog of DYBLUP in Leishmania, LAX28, loss of which from the axoneme causes failure in the assembly of both f/I1 dynein and the tether complex (17). Thus, the dependency of assembly of f/I1 dynein on DYBLUP orthologs varies among organisms.
The regulation of IC138 via phosphorylation is thought to be important for regulation of f/I1 dynein (10, 11). IC138 is thought to be phosphorylated and dephosphorylated by casein kinase 1 (CK1) and protein phosphatase 1/2A (PP1/PP2A), respectively (23). Because IC138 is hyperphosphorylated in the mot7 (Fig. 2F) and fap44 Chlamydomonas mutants (21), loss of the tether structure is thought to make the CK1 activity dominant over PP1/PP2A activity in these mutants, resulting in hyperphosphorylation of IC138 (Fig. 5). The mot7 and fap44 mutants exhibit both positive and negative phototaxis, although the responses of the fap44 mutant are weaker than those of the WT strain or the mot7 mutant (fig. S7, A and B). Therefore, the tether structure or IC138 phosphorylation might not be critical for determination of the phototactic sign but might accelerate or maintain phototaxis.
Fig. 5. Schematic model of MOT7-mediated avoidance of blue light acclimation.
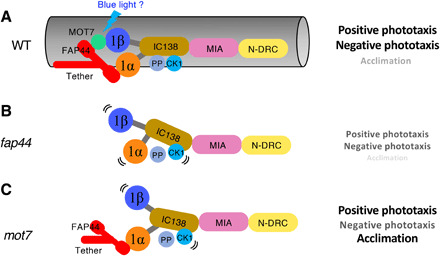
(A) In WT Chlamydomonas flagella, the motor activity of IAD f/I1 is regulated by the proper arrangement of motors and regulatory components, such as the tether complex, protein kinase CK1, and protein phosphatase PP1/2A. The molecular interactions among these components are essential for positive and negative phototaxis and suppression of light acclimation. MOT7 links the 1β motor domain and the tether component FAP44, regulating the motor activity for avoidance of acclimation to strong blue light. (B) The fap44 mutant lacks the entire tether structure but shows both positive and negative phototaxis, although these responses are weak compared with the WT strain. The lack of the tether complex decreases the binding of CK1 to the doublet microtubule, causing hyperphosphorylation of IC138. (C) In the mot7 mutant, the link between the 1β motor and the tether complex is missing, although that between the 1α motor and the tether complex is present. IC138 is constitutively hyperphosphorylated due to decreased binding of CK1 to the microtubule. The mutant shows both positive and negative phototaxis but becomes acclimated to strong light. The extent of three photoresponses is indicated on the right. The motility regulation of the two flagella during positive phototaxis is associated with anchoring of the 1α motor, but not 1β motor, to the tether complex and not related to dephosphorylation of IC138. Negative phototaxis does not require the tether-f/I1 connection or regulation of IAD f/I1 activity by IC138 dephosphorylation. Regarding the acclimation to strong blue light, MOT7-mediated binding of 1β motor to the tether complex is required for unbalanced flagellar beating, which prevents the cells from swimming toward the light source. The anchoring of 1α by the tether structure is thought to promote light acclimation.
We found that the acclimation of the mot7 mutant to strong blue light is due to loss of turning movement induced by unbalanced beating of the two flagella (fig. S5D). The unbalanced beating is accompanied by a decrease in beat frequency in one of the two flagella (fig. S5F). The 1β motor of f/I1 dynein is effective in increasing sliding velocity, whereas the 1α motor loads local restraint for sliding (24). Therefore, we suppose that DYBLUP/MOT7 inhibits the 1β motor function in a light-dependent manner, decreases the flagellar beat frequency, and induces unbalanced beating to avoid blue light. In addition, the fap44 mutant, which lacks most of the axonemal MOT7 and the entire tether complex (20, 21), shows no acclimation to blue light. The mot7 mutant is devoid of the link between 1β and the tether but retains the link between 1α and the tether (Figs. 3 and 5). Thus, we suggest that the anchoring of 1α by the tether structure is a prerequisite for light acclimation.
Strong blue light induces the production of excess reactive oxygen species and damages organisms. Land plants prevent this light toxicity by enhanced thermal dissipation (25), which is driven by phototropin, a blue light receptor protein with flavin-binding, light, oxygen, or voltage (LOV) domains (26). LOV and BLUF are two blue light reception domains known to function as intracellular signal transducers that use light. These domains are present in prokaryotes and eukaryotes. In Euglena, photoactivated adenylyl cyclase (PAC), a BLUF protein, regulates flagellar motility through a light-induced increase in intracellular cAMP (16). Similarly, the brown algae Ectocarpus have an LOV protein that responds to blue light and regulates signaling in flagellar motility (27). Phototropin is localized along flagella and is suggested to regulate cAMP-dependent mating in Chlamydomonas in a light-dependent manner (28). These photoreceptor proteins are localized in flagella, but in flagellar accessary structures (16, 27) or axonemal structures other than dynein arms, central apparatus, or radial spokes (28), and thus are not likely to directly alter dynein activity but rather to mediate signaling for flagellar motility via cyclic nucleotides (29, 30).
We showed that flavin nucleotides could weakly interact with the BLUF domains of MOT7 but are unlikely to be an intrinsic ligand (fig. S2, D to G). It is possible that a small substance, likely to be a derivative of FAD/FMN, acts as a ligand of the BLUF domains and regulates the function of DYBLUP/MOT7 to mediate the binding of the tether complex to the motor domain of f/I1 dynein. Nonetheless, the BLUF domains are conserved in distantly related organisms with different requirements for light, i.e., Chlamydomonas, ciliates, and animals (fig. S2, A and C). This makes it difficult to correlate the function of DYBLUP/MOT7 with photodependent responses. The prevention of light acclimation via MOT7 depends on the redox state (figs. S6B and S7, A and F), suggesting that the regulation of f/I1 motor activity is redox-mediated downstream of light reception by MOT7. Considering the properties of FAD in both sensing blue light and as a redox molecule (31), DYBLUP might have conserved the function of transducing redox signaling for regulation of dynein activity in ciliates and animals.
In 1980, there was a very curious report on the stoppage of live sea urchin sperm by blue light (32). We have not obtained any evidence showing how blue light changes the structure of DYBLUP and regulates dynein or sperm motility in animals. Nevertheless, DYBLUP is a protein of great interest with regard to its function and molecular evolution. Greater understanding of this protein might also shed light on optogenetic technology to enable direct manipulation of molecular motors.
MATERIALS AND METHODS
Isolation of f/I1 dynein from sperm flagella of C. intestinalis
Sperm flagella and axonemes from C. intestinalis were prepared as described previously (33). Sperm were collected from adult sperm ducts. Flagella were isolated via homogenization of sperm in artificial Ca2+-free seawater [462 mM NaCl, 9.39 mM KCl, 59.08 mM MgCl2, 5 mM EGTA, and 10 mM Hepes-NaOH (pH 8.0)], followed by pooling of the supernatant after centrifugation at 1500g for 5 min. The isolated flagella were pelleted by centrifugation at 9100g for 5 min. The axonemes were prepared by demembranation of flagella with 0.05% Triton X-100 in an axoneme buffer [AXB; 150 mM KCl, 1 mM MgSO4, 0.5 mM EGTA, 0.2 mM dithiothreitol, and 20 mM tris-HCl (pH 8.0)], followed by centrifugation.
Isolation of the f/I1 dynein from axonemes was performed following the methods of previous papers (13, 33, 34) with slight modifications. The axonemes were treated with 0.6 M KCl in AXB for 30 min on ice and centrifuged at 17,360g for 15 min. This treatment was repeated twice to remove as much of the OAD as possible. The axonemes were further treated with 0.7 M KCl and 5 mM ATP in AXB for 30 min on ice to extract the f/I1 dynein and centrifuged at 17,360g for 15 min to recover the extract. The extraction of f/I1 dynein was repeated twice. The extracts were subjected to ultracentrifugation at 452,000g for 15 min to remove aggregates before loading onto a sucrose density gradient.
A linear sucrose gradient (5 to 20%) with J-buffer [5 mM MgSO4, 2 mM EGTA, 0.5 mM dithiothreitol, and 20 mM Hepes (pH 7.8)] containing 200 mM potassium acetate was prepared using Gradient Station (BioComp Instruments, Fredericton, NB, Canada). The extracts were loaded onto the linear sucrose gradient and centrifuged in a Himac CP65β centrifuge with a P28S2 rotor (Hitachi, Japan) at 121,930g for 16 hours at 4°C. Fractions of 650 μl were collected. The protein concentrations and the adenosine triphosphatase (ATPase) activity of each fraction were measured using the Bradford method (35) and Taussky and Schorr method (36), respectively, to determine the position of dynein. The fractions containing f/I1 dynein were determined by Western blot analysis using anti–Ci-IC116, anti–Ci-IC110, and anti–Ci-IC105 antibodies.
The sucrose density gradient fractions containing f/I1 dynein were separated on a Uno Q anion exchange column (Bio-Rad, Hercules, CA, USA) using an ÄKTA purifier HPLC system (Amersham Pharmacia Biotech, USA). The column was equilibrated with J-buffer, and the proteins were eluted with a linear gradient of 150 to 450 mM KCl in J-buffer. The fractions containing f/I1 dynein were determined on the basis of ATPase activity and Western blotting results using anti–Ci-IC116, anti–Ci-IC110, and anti–Ci-IC105 antibodies.
Ligand analysis
Axonemes, KCl extracts, and dynein fractions were dialyzed against 10 mM ammonium bicarbonate (pH 7.7) and 0.5 mM MgSO4, lyophilized, dissolved in 70% ethanol, and heated at 95°C for 3 min. After centrifugation, the supernatant was applied to a silica gel plate for TLC; plates were developed in a solvent containing 1-butanol, acetic acid, and water (4:1:5, v/v). The spots were visualized with a Pharos FX molecular imager (Bio-Rad; excitation, 488 nm; emission, 605 nm).
Fluorescent HPLC analysis was carried out with a Shimadzu RF-20AXS fluorescence detector using a Develosil ODS-HG-5 column (1.5 mm × 150 mm). Flavins were eluted in a 10 to 40% methanol gradient in 10 mM NaH2PO4 and detected by fluorescence at 530 nm (excitation, 445 nm). For ESI-LC-MS, samples were dissolved in acetonitrile and 0.1% HCOOH and analyzed with a Shimadzu LCMS-2020 system.
Gel filtration was carried out in a dark room using a Sephadex G-50 column (0.5 mm × 50 mm) equilibrated with 10 mM Hepes-NaOH (pH 8.0). Bacterially expressed Chlamydomonas MOT7 or bovine serum albumin (BSA) was mixed with 1.5 μM FAD or FMN and incubated on ice in a dark box for 2 hours. Samples were applied to the column, eluted with the same buffer, and collected in 100-μl fractions.
SDS–polyacrylamide gel electrophoresis and Western blotting
SDS–polyacrylamide gel electrophoresis (SDS-PAGE) was performed following a modification of the procedure of Laemmli (37). The gels were stained with Coomassie Brilliant Blue R-250, silver, or SYPRO Ruby (Invitrogen, Carlsbad, CA, USA). The molecular weight standards used were from Bio-Rad and Nacalai Tesque (Kyoto, Japan). For Western blotting, proteins separated by SDS-PAGE were transferred to polyvinylidene difluoride membranes (Merck Millipore, Billerica, MA, USA). The membranes were blocked with 5 to 7.5% skim milk in PBS-T (50 mM NaCl, 2.8 mM NaH2PO4·2H2O, 7.2 mM Na2HPO4, 0.05% Tween 20, or 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 0.05% Tween 20) and incubated with various primary antibodies. After three washes with PBS-T, the membranes were incubated with goat anti-rabbit or anti-mouse IgG (H+L) secondary antibody and horseradish peroxidase conjugate (Invitrogen) and washed with PBS-T three times. The immunoreactive signals were detected using a tetramethylbenzidine (TMB) substrate kit (Vector Laboratories Inc.) or enhanced chemiluminescence (ECL) prime Western blotting detection reagents (GE Healthcare) and a Las4000 mini luminescence image analyzer with a cooled charge-coupled device (CCD) camera (Fujifilm, Tokyo, Japan). The procedure for Western blotting was performed at 4°C or room temperature (RT). The antibodies used in this study are listed in table S3.
Antibodies
Production of polyclonal antibodies against Ci-IC110, Ci-IC105, DYBLUP, and MOT7 was performed by following the methods reported in a previous paper (38) with slight modifications. PCR was performed to amplify protein-coding regions using complementary DNAs (cDNAs) for these proteins as the templates. The PCR primers used were 5′-GCGCGTCGACATGTCTAAGGTGGTTAAG-3′ (sense) and 5′-GCGCCTCGAGCAAAGGACTTTTAGGAGT-3′ (antisense) for Ci-IC110, 5′-GCGCGGATCCATGCCTCCCAAATCCCCG-3′ and 5′-GCGCGAATTCCTTCCTCCACAACTGACT-3′ for Ci-IC105, 5′-GCGCGGATCCATGGGTTCTATACATGAAA-3′ and 5′-GCGCCTCGAGCTATTTTAAATTGACTGC-3′ for DYBLUP, and 5′-GCGCGGATCCATGGCTGACACGCTCCGAA-3′ and 5′-GCGCGAATTCTTAGTATTTGAGCGGCGG-3′ for MOT7. The PCR products were subcloned into vector pET28a(+) (Novagen, Merck Group, Madison, WI, USA) and used to transfect Escherichia coli BL21 (DE3) (Novagen). Protein expression was induced by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside overnight at RT. Recombinant proteins containing a His6-tag were extracted from E. coli with lysis buffer [6 M urea, 0.5% Triton X-100, 1% CHAPS, 5 mM dithiothreitol, 5 mM imidazole, 500 mM NaCl, and 20 mM tris-HCl (pH 8.0)] and purified on a Ni Sepharose High Performance column (GE Healthcare, Buckinghamshire, UK). The purified proteins were dialyzed against phosphate-buffered saline (PBS) at 4°C for 4 hours three times and used as antigens to raise polyclonal antibodies. Antigens were emulsified with complete Freund’s adjuvant (Difco, Detroit, MI, USA), and Balb/c mice were given four subcutaneous injections at intervals of 10 to 14 days. Crude serum was used for experiments unless otherwise noted.
LC-MS/MS and MALDI-TOF/MS analyses
Before LC/ESI-MS/MS (LC-MS/MS) analysis, each Uno Q fraction containing f/I1 dynein or OAD was concentrated by lyophilization. The lyophilized samples were solubilized with SDS sample buffer and incubated at 95°C for 2 min. The samples were separated by SDS-PAGE. For separation of higher molecular weight (~500 kDa) and lower molecular weight (~10 kDa) proteins, 6 and 13% gels were used, respectively. The 6 and 13% gels were stained with Coomassie Brilliant Blue and SYPRO Ruby, respectively. Selected bands were cut out, trimmed into small pieces, and subjected to in-gel trypsin digestion. Peptides were analyzed by LC-MS/MS as described in a previous paper (39). For some proteins, gel pieces were subjected to peptide mass fingerprinting using MALDI-TOF/MS. The database for MS analysis was constructed from the Ciona KH gene model (40) and the CIPRO model (41).
Isolation of rescue strains of the C. reinhardtii mot7 mutant
The C. reinhardtii strains used in this study are listed in table S4. Chlamydomonas was maintained and harvested using normal procedures and tris-acetate phosphate (TAP) medium (42).
For isolation of the rescue strain expressing MOT7-BCCP-3HA, the synthesized WT MOT7 cDNA sequence (GenScript Japan) was cloned into the Nde I/Bam HI sites of the Chlamydomonas expression vector pIC2L-MCS-BCCP-3HA-Hyg (a gift from H. Yanagisawa, The University of Tokyo) (43). For isolation of the rescue strain expressing MOT7-3HA, the WT MOT7 cDNA sequence was cloned into the Nde I/Bam HI sites of the Chlamydomonas expression vector pGend (44), which includes DNA encoding a 3HA-tag. In addition, a hygromycin or paromomycin resistance cassette (AphVII/AphVIII) was incorporated into the pGend vector so that transformants would have hygromycin or paromomycin resistance. The mot7 mutant was transformed with the constructed vectors using electroporation (45), and screening of the rescued cells was performed by hygromycin B or paromomycin resistance selection and Western blotting using anti-HA antibodies.
Chlamydomonas MOT7 gene knockout by CRISPR/Cas9-based genome editing
Gene knockout was achieved using the CRISPR/Cas9 system carried out following the method of Greiner et al. (46), in which ribonucleoprotein (RNP) composed of guide RNAs and Cas9 protein was introduced into WT cells by electroporation together with HDR (homology-directed repair) DNA and a pHyg3 vector (47). In brief, CRISPR RNA (crRNA), trans-activating crRNA (tracrRNA), and Cas9 protein were purchased from Integrated DNA Technologies (Alt-R CRISPR/Cas9 system). Two sites in the second exon of the MOT7 gene were selected as crRNA targets using Benchling (https://benchling.com/): 5′-ACGTGGCACGACTAGCGCAAAGG-3′ as the first site, and 5′-CACCACCCGGATTGTCTACGTGG-3′ as the second site, where protospacer adjacent motif (PAM) sequences are underlined. HDR DNAs were designed as follows and purchased with their complementary sequences to prepare double-stranded DNAs (Eurofins Genomics): 5′-C*A*C*CCGGATTGTCTACGTGGCACGACTAGCGTAATGACCACGACATCGACTACAAGGACCAAAGGCAGAACAGCTCGGATAGCGTAA*G*A*G-3′ for the first site, and 5′-C*G*C*AGACCGTCCTCACCACCCGGATTGTCTTATAATGACCACGACATCGACTACAAGGACACGTGGCACGACTAGCGCAAAGGCAGA*A*C*A-3′ for the second site (bold letters, termination codons; underline, FLAG-tag sequences for PCR-based genotyping; *, S-oligo). RNP was prepared by mixing crRNA, tracrRNA, and Cas9 protein following the method of Greiner et al. (46). WT cells were suspended in 40 μl of MAX Efficiency transformation buffer for algae (A24229, Thermo Fisher Scientific, Waltham, MA, USA) with 40 mM sucrose containing 15 pmol RNP, 15 pmol HDR, and ~0.2 μg of linearized pHyg3 vector and subjected to electroporation (1 × 108 cells/ml). Transformed cells were grown on a TAP-agar plate containing hygromycin B, and colonies were genotyped by PCR using the following primers to select MOT7-KO strains (mot7CRISPR): 5′-CTTCGAGTTCCCGTAAGCAG-3′ and 5′-TCCGCCAAGTACATGAAACA-3′ as MOT7-specific primers, and a FLAG-tag sequence–specific primer (46).
Motility analysis of Chlamydomonas
Chlamydomonas strains were cultured in TAP medium (42) at 20°C with aeration under a 14-hour:10-hour light:dark cycle. Cells were usually recovered and washed with a wash buffer [1 mM KCl, 0.3 mM CaCl2, 0.2 mM EGTA, 5 mM Hepes-KOH (pH 7.4)] 1 hour before onset of the light period. They were kept under red LED light before illumination with blue light. For the dish assay, washed Chlamydomonas were transferred to a sterile plastic culture dish (35 mm diameter) and illuminated with weak (~1 μmol photons m−2 s−1) or strong (~1000 μmol photons m−2 s−1) blue light (465 to 475 nm) from one side of the dish using a midget LED lamp (5mm Round LED, OptoSupply, Hong Kong). For illumination on a microscopic stage, an inverted microscope (IX71, Olympus, Japan) was used. The whole field was illuminated with a laboratory-made red stroboscopic LED lamp (620 to 630 nm, Power LED, Edison, Taiwan). The center area was illuminated with blue light (440 nm; Spectra X, Lumencor Inc., OR, USA) using an iris diaphragm through the objective (~220 μmol photons m−2 s−1). In another experiment, a midget LED lamp was used to illuminate one side of a glass slide edge with blue light (~350 μmol photons m−2 s−1). Images were recorded with a high-speed camera (HAS-220 or HAS-U2, DITECT, Japan) at 50, 100, or 500 frames per second for trajectory or waveform analysis. Photon flux density was measured with an MQ-200 quantum meter (Apogee Instruments Inc., UT, USA). The trajectories of cells or flagellar waveforms were analyzed using Bohboh software (Bohboh Soft, Tokyo, Japan). For manipulating the redox state of solution, we used tert-butyl hydroperoxide (0.2 mM) and 3-(3′,4′-dichlorophenyl)-l,1-dimethylthiourea (75 mM) for oxidative and reductive conditions, respectively.
Preparation of axonemes and dyneins from Chlamydomonas
Isolation of Chlamydomonas flagella was achieved through dibucaine treatment as described in a previous paper (48). After collection via centrifugation, flagella were demembraned with 0.2% Igepal in HMDS solution [10 mM Hepes (pH 7.4), 5 mM MgSO4, 1 mM dithiothreitol, and 4% sucrose]. The axonemes were precipitated by centrifugation and washed with HMDEK solution [30 mM Hepes, 5 mM MgSO4, 1 mM dithiothreitol, 1 mM EGTA, and 50 mM potassium acetate (pH 7.4)] to remove Igepal. Preparation of dyneins from Chlamydomonas was performed using an HPLC system with a Mono Q anion exchange column as described previously (49). The axonemes were treated with 0.6 M KCl in HMDEK on ice for 10 to 30 min to extract crude dyneins. The suspension was centrifuged, and the supernatant containing crude dyneins was pooled. The supernatant was diluted 10-fold by addition of HMDEK solution. The diluted supernatant was centrifuged to remove aggregates and loaded onto a Mono Q column. The Mono Q fractions were analyzed by SDS-PAGE using 3 to 5% acrylamide with a 3 to 8 M urea gradient to determine the positions of dynein species.
Immunofluorescence microscopy
Immunofluorescence microscopy of Ciona sperm was performed as described previously (50). Sperm were diluted 1:50 in artificial seawater and activated by addition of sperm-activating and -attracting factors (SAAFs). The activated sperm were dropped onto glass slides coated with poly-l-lysine (1 mg/ml). After incubation for 5 min, the sperm were fixed with 4% paraformaldehyde in 100 mM Mops and 0.5 M NaCl (pH 8.0) at RT for 60 min and permeabilized by incubation in methanol at −20°C for 10 min. After removal of methanol by air blowing, the glass slides were rehydrated with PBS for 5 min and transferred to a moist chamber. The samples were incubated in blocking buffer (10% goat serum in PBS containing 0.05% Triton X-100) at RT for 60 min. The samples were incubated with various primary antibodies in blocking buffer at RT for 60 min and washed with PBS containing 0.05% Triton X-100 for 5 min four times. As primary antibodies, anti-DYBLUP from mouse and anti–acetylated α-tubulin from rabbit (Cell Signaling Technology, Danvers, MA, USA) were used at 1:100 and 1:1000 dilution, respectively, in blocking buffer. Subsequently, the samples were incubated with Alexa 546–conjugated goat anti-mouse IgG or Alexa 488–conjugated goat anti-rabbit IgG (Molecular Probes, Eugene, OR, USA) at 1:1000 dilution in blocking buffer for 60 min at RT. After two 5-min washes with PBS, the samples were treated with 1 μM 4,6-diamidino-phenylindole (DAPI) for 5 min and mounted in 50% glycerol. Immunofluorescence images were observed using a fluorescence microscope BX50 (Olympus) with a 100×/1.35 NA UplanApo oil objective lens (Olympus). The images were recorded on a DP controller (Olympus) using a DP70 digital camera (Olympus).
For Chlamydomonas, cells were treated with autolysin for 1 hour at RT. The cells were attached to glass slides and fixed with 2% paraformaldehyde for 10 min at RT, followed by cold methanol for 2 min at −30°C. Fixed cells were incubated in blocking buffer [10 mM sodium phosphate, 5% normal goat serum, 5% glycerol, 1% cold fish gelatin, and 0.004% sodium azide (pH 7.2)] for 30 min at 37°C and then with an anti–HA-tag antibody (1:200, code no. 561; MBL, Japan) or anti–acetylated α-tubulin antibody (1:1000, clone no. 6-11B-1; Sigma-Aldrich, USA) for 1 hour at 37°C. The cells were washed with PBS containing 0.05% Triton X-100 and stained with Alexa Flour 488–conjugated goat anti-mouse (1:1000) or Alexa Flour 568–conjugated goat anti-rabbit (1:1000) secondary antibody for 1 hour at 37°C. Immunofluorescence images were observed under an IX71 fluorescence microscope (Olympus) and recorded using a DP72 digital camera (Olympus).
Immunoprecipitation
In Ciona, immunoprecipitation was performed as described previously (51) with slight modifications. Anti-DYBLUP antiserum was diluted 1:25 with a binding buffer containing 1.5 M glycine and 3 M NaCl (pH 8.9). The diluted antiserum was added to Protein A Sepharose beads (GE Healthcare) and mixed on a rotator for 4 hours at 4°C. After mixing, the beads were washed three times with binding buffer and then washed twice with IP buffer [200 mM KCl, 5 mM MgSO4, 2 mM EGTA, 0.5 mM dithiothreitol, and 20 mM Hepes (pH 7.8)] containing 3% BSA. The dynein fraction from sucrose density gradients was dialyzed against IP buffer overnight at 4°C and diluted 1:2 with IP buffer containing 3% BSA. The dynein fraction was added to the beads and mixed on a rotator for 1 hour at 4°C. The beads were washed three times with IP buffer without BSA, suspended in SDS sample buffer, and incubated at 95°C for 2 min. The SDS-treated samples were subjected to SDS-PAGE. Immunoprecipitates were detected by Western blotting using anti–Ci-IC116, anti–Ci-IC110, anti–Ci-IC105, and anti-DYBLUP antibodies. Nonimmune serum from mice was used as a negative control.
In Chlamydomonas, axonemal proteins from WT and mutant cells were extracted with 0.6 M KCl in HMDEK buffer containing EDTA-free Protease Inhibitor Cocktail (Roche Diagnostics, Tokyo, Japan). The extracts were diluted 10-fold with IP buffer containing 2% polyvinylpyrrolidone (molecular weight 40,000, PVP40; Sigma-Aldrich), 0.05% Triton X-100, and HMDEK and added to rProteinG Sepharose beads (GE Healthcare, Chicago, IL, USA) conjugated with anti–HA-tag polyclonal antibody and incubated for 1 hour at 4°C in the IP buffer. The beads were washed three times with IP buffer without PVP40 and incubated with SDS sample buffer for 5 min at 95°C. The precipitants were separated by SDS-PAGE and stained with silver, and selected bands were identified by peptide mass fingerprinting analysis by MALDI-TOF/MS (BioGARAGE, Leave a Nest Co. Ltd., Japan).
Cross-linking and vanadate-mediated photocleavage
Before cross-linking, the dynein fractions from sucrose density gradients were dialyzed against J-buffer containing 200 mM potassium acetate overnight at 4°C. BS3 cross-linker (Thermo Fisher Scientific) was added to aliquots of the dialyzed dynein fractions from sucrose density gradients and incubated for 30 min at 25°C. The cross-linking reactions were terminated by addition of 20 mM glycine (pH 7.8). The mixtures were dialyzed against a UV buffer [450 mM sodium acetate, 0.5 mM EDTA, 1 mM dithiothreitol, 2 mM MgCl2, 10 mM Hepes (pH 7.4)] for 5 hours at 4°C. To perform vanadate-mediated photocleavage of the dynein HCs (52), 100 μM sodium metavanadate and 1 mM ATP were added to the dialyzed samples. The samples were placed in a 24-well plate on ice and irradiated with 365-nm light for 30 min. For SDS-PAGE, 4% polyacrylamide/4 M urea gels were used as the separating gel. The cross-linked products were detected by Western blotting using anti-DYBLUP antibody.
Transmission electron microscopy
Ciona axonemes were fixed with 2.5% glutaraldehyde in 100 mM sodium cacodylate buffer (pH 7.4) for 2 hours at RT, washed with 100 mM sodium cacodylate (pH 7.4), and postfixed with 1% OsO4 in the cacodylate buffer on ice for 1 hour. After dehydration in a graded ethanol series, samples were substituted in propylene oxide and embedded in Quetol 812 (Nisshin EM, Tokyo, Japan) or Agar Low Viscosity Resin (Agar Scientific, Stansted, Essex, UK). The resin was cut into sections with an average thickness of 70 nm. Sections were stained with uranyl acetate and observed under a 1200EX electron microscope (JEOL, Tokyo, Japan) at 80 kV.
For negative-stain transmission electron microscopy (TEM), f/I1 dynein or OAD fractions separated by sucrose density gradient were diluted to approximately 3 nM with MME buffer [30 mM Mops, 5 mM MgSO4, 1 mM EGTA, 0.1 mM dithiothreitol (pH 7.4)] containing 100 mM KCl. The samples were applied onto carbon-coated copper grids. After 30 to 60 s, the samples on the grid were stained with 1 to 2% (w/v) uranyl acetate. For analyzing localization of DYBLUP in the f/I1 dynein molecule, the dynein fraction (~120 nM) was mixed 1:0.75 (v/v) with purified anti-DYBLUP IgG and incubated on ice for 1 min. Electron microscopy was performed using a Tecnai F20 electron microscope (FEI, Hillsboro, OR, USA).
Cryo-ET of Chlamydomonas axonemes
Flagella were isolated from WT Chlamydomonas, the mot7 mutant, and the rescue strain expressing MOT7-BCCP-3HA using the dibucaine method (48). Flagella from the rescue strain were labeled with streptavidin (53). The samples were plunge-frozen as described previously (54) with slight modifications. We used an R3.5/1 holey grid (Quantifoil, Thuringia, Germany) and Cryoplunge-3 (Gatan, USA). We screened cryo-grids with axonemes in proper appearance using a JEM2200FS TEM (JEOL, Japan) [as described in (54)] and acquired the final data using a Titan Krios TEM (Thermo Fisher Scientific, USA) operated at 300-kV accelerating voltage and equipped with a GIF Quantum energy filter and a K2 direct electron detector. Tomographic acquisition of micrographs was carried out at a nominal magnification of 30,000, corresponding to 4.2 Å per pixel, using the SerialEM program (55). Tomograms were analyzed using the IMOD package (56), and then subtomograms were extracted with IMOD and analyzed using our in-house programs using pseudo-ninefold symmetry and 96-nm periodicity as described previously (54). Averaged subtomograms were visualized using IMOD and UCSF Chimera (57).
Computational methods
The PCR primers for production of anti–Ci-IC110, anti–Ci-IC105, anti-DYBLUP, and anti- MOT7 antibodies were designed using GENETYX (GENETYX Corp., Tokyo, Japan). Identification of axonemal proteins from mass spectra was performed using Mascot (Matrix Science, Boston, MA, USA). To search for DYBLUP homologs, homologs were defined by an E value of <10−3 by BLASTP (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The BLUF domain was predicted using SMART (http://smart.embl-heidelberg.de/). Multiple sequence alignment of the BLUF domain in DYBLUP, DYBLUP homologs, PAC from E. gracilis, and AppA from the purple bacterium R. sphaeroides was produced using Clustal W (http://clustalw.ddbj.nig.ac.jp/) and is displayed using Boxshade (https://embnet.vital-it.ch/software/BOX_form.html). BLUF domain structure homology modeling for DYBLUP/MOT7 from Ciona and Chlamydomonas was performed using the fully automated protein structure homology-modeling server SWISS-MODEL (http://swissmodel.expasy.org/) with AppA as the template structure.
Data accession
DYBLUP from C. intestinalis and MOT7 from C. reinhardtii are registered in DDBJ/EMBL/GenBank with accession numbers LC475460 and LC475459, respectively. The accession numbers for orthologs of Chlamydomonas IC140, IC138, and IC97 in C. intestinalis, i.e., IC116, IC110, and IC105, are LC557018, LC554900, and AB083181, respectively. Data for cryo-ET are available at wwPDB for C. reinhardtii axonemes of WT, mot7 mutant, and the mutant rescued with MOT7-BCCP-3HA as EMD-12162, EMD-12160 and EMD-12161, respectively.
Statistical analysis
Data are expressed as means ± SEM. Statistical significance was calculated using Student’s t test; P < 0.05 was considered significant.
Acknowledgments
We thank the Chlamydomonas Library Project for the mot7 mutant, K. Yan (National Institute of Advanced Industrial Science and Technology) for technical assistance in negative-stain electron microscopy, and S. Takeda (University of Yamanashi) for valuable comments. We thank the EM Facility at Paul Scherrer Institute and ScopeM at ETH Zurich for technical support with cryo-EM. We thank T. Oda and T. Kubo (University of Yamanashi) for providing the fap44 mutant. We thank all staff members of the Education and Research Center of Marine Bio-Resources, Tohoku University, International Coastal Research Center, The Atmosphere and Ocean Research Institute (The University of Tokyo), and the National BioResource Project for supplying C. intestinalis. We thank J. Allen, DPhil, from Edanz Group (https://en-author-services.edanz.com/ac), for editing a draft of this manuscript. Funding: This work was supported, in part, by Nanotechnology Platform Program (JPMXP09A-20-OS-0015) of the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan, and by the Cooperative Study Program (no. 136 in year 2018) of the National Institute for Physiological Sciences. This work was also supported, in part, by Grants-in-Aid for Scientific Research (A) (17H01440) from the Japan Society for the Promotion of Science, Japan (JSPS) to K.I., T.K., K.H., and K.-i.W.; Grants-in-Aid for Scientific Research (B) (22370023), and challenging Exploratory Research (15 K14566) from the Japan Society for the Promotion of Science, Japan (JSPS) to K.I.; a Grant-in-Aid for Innovative Areas (15H01201) from the Ministry of Education, Culture, Sports, Science and Technology, Japan to K.I.; Grants-in-Aid for Scientific Research (B) (JP26291034 and JP17H03665) to T.K.; a grant from the Swiss National Science Foundation and Novartis Biomedical Research Foundation to T.I. (NF310030_192644); a Grant-in-Aid for Young Scientists (B) (16 K18584), Scientific Research on Innovative Areas, Platforms for Advanced Technologies and Research Resources “Advanced Bioimaging Support” from JSPS (16H06280), and University of Tsukuba Basic Research Support Program Type A to O.K.; and a JSPS Research Fellowship for Young Scientists (PD), JSPS Grant-in-Aid for Young Scientists (B) (JP17K15117), The Uehara Memorial Foundation, and The Ito Chubei Foundation to R.Y. Author contributions: K.I. designed the outline of the study. O.K., R.Y., K.H., K.Miz., H.I., T.I., K.-i.W., T.K., and K.I. designed the experimental strategy. O.K., R.Y., K.Miz., and K.I. conducted the main experiments. R.Y., Y.N., Ma.Na., Y.T., Y.Y., Y.A., and K.-i.W. verified Chlamydomonas mutants. K.H., D.S., M.S., and Mami.No. conducted electron microscopy. A.No., J.M.O., N.Z., T.I., C.S., K.Mu., K.Mit., H.I., and T.K. conducted cryo-ET. M.S., K.S., Y.N., Ma.Na., K.-i.W., and K.I. conducted motility analysis. K.Miz., Mi.Na., A.Na., Mamo.No., L.Y., S.N., and H.S. conducted protein MS. M.K. and H.K. conducted HPLC and ESI-MS for flavins. K.I. wrote the manuscript. O.K. and R.Y. edited the manuscript. All authors revised and approved the final version of the manuscript. Competing interests: The authors declare that they have no competing financial interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/9/eabf3621/DC1
REFERENCES AND NOTES
- 1.Gibbons I. R., Cilia and flagella of eukaryotes. J. Cell Biol. 91, 107s–124s (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.King S. M., The dynein microtubule motor. Biochim. Biophys. Acta 1496, 60–75 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Kamiya R., Functional diversity of axonemal dyneins as studied in Chlamydomonas mutants. Int. Rev. Cytol. 219, 115–155 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Inaba K., Sperm flagella: Comparative and phylogenetic perspectives of protein components. Mol. Hum. Reprod. 17, 524–538 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Heuser T., Raytchev M., Krell J., Porter M. E., Nicastro D., The dynein regulatory complex is the nexin link and a major regulatory node in cilia and flagella. J. Cell Biol. 187, 921–933 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bui K. H., Sakakibara H., Movassagh T., Oiwa K., Ishikawa T., Asymmetry of inner dynein arms and inter-doublet links in Chlamydomonas flagella. J. Cell Biol. 186, 437–446 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamamoto R., Song K., Yanagisawa H., Fox L., Yagi T., Wirschell M., Hirono M., Kamiya R., Nicastro D., Sale W. S., The MIA complex is a conserved and novel dynein regulator essential for normal ciliary motility. J. Cell Biol. 201, 263–278 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inaba K., Molecular architecture of the sperm flagella: Molecules for motility and signaling. Zoolog. Sci. 20, 1043–1056 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Inaba K., Calcium sensors of ciliary outer arm dynein: Functions and phylogenetic considerations for eukaryotic evolution. Cilia 4, 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Habermacher G., Sale W. S., Regulation of flagellar dynein by phosphorylation of a 138-kD inner arm dynein intermediate chain. J. Cell Biol. 136, 167–176 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King S. J., Dutcher S. K., Phosphoregulation of an inner dynein arm complex in Chlamydomonas reinhardtii is altered in phototactic mutant strains. J. Cell Biol. 136, 177–191 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wirschell M., Hendrickson T., Sale W. S., Keeping an eye on I1: I1 dynein as a model for flagellar dynein assembly and regulation. Cell Motil. Cytoskeleton 64, 569–579 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Yokota E., Mabuchi I., Isolation and characterization of a novel dynein that contains C and A heavy chains from sea urchin sperm flagellar axonemes. J. Cell Sci. 107, 345–351 (1994). [DOI] [PubMed] [Google Scholar]
- 14.Kotani N., Sakakibara H., Burgess S. A., Kojima H., Oiwa K., Mechanical properties of inner-arm dynein-f (dynein I1) studied with in vitro motility assays. Biophys. J. 93, 886–894 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masuda S., Bauer C. E., AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell 110, 613–623 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Iseki M., Matsunaga S., Murakami A., Ohno K., Shiga K., Yoshida K., Sugai M., Takahashi T., Hori T., Watanabe M., A blue-light-activated adenylyl cyclase mediates photoavoidance in Euglena gracilis. Nature 415, 1047–1051 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Beneke T., Banecki K., Fochler S., Gluenz E., LAX28 is required for the stable assembly of the inner dynein arm f complex, and the tether and tether head complex in Leishmania flagella. J. Cell Sci. 133, jcs239855 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heuser T., Barber C. F., Lin J., Krell J., Rebesco M., Porter M. E., Nicastro D., Cryoelectron tomography reveals doublet-specific structures and unique interactions in the I1 dynein. Proc. Natl. Acad. Sci. U.S.A. 109, E2067–E2076 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X., Zhang R., Patena W., Gang S. S., Blum S. R., Ivanova N., Yue R., Robertson J. M., Lefebvre P. A., Fitz-Gibbon S. T., Grossman A. R., Jonikas M. C., An indexed, mapped mutant library enables reverse genetics studies of biological processes in Chlamydomonas reinhardtii. Plant Cell 28, 367–387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kubo T., Hou Y., Cochran D. A., Witman G. B., Oda T., A microtubule-dynein tethering complex regulates the axonemal inner dynein f (I1). Mol. Biol. Cell 29, 1060–1074 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu G., Wang Q., Phan N., Urbanska P., Joachimiak E., Lin J., Wloga D., Nicastro D., The I1 dynein-associated tether and tether head complex is a conserved regulator of ciliary motility. Mol. Biol. Cell 29, 1048–1059 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oda T., Yanagisawa H., Kamiya R., Kikkawa M., A molecular ruler determines the repeat length in eukaryotic cilia and flagella. Science 346, 857–860 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Gokhale A., Wirschell M., Sale W. S., Regulation of dynein-driven microtubule sliding by the axonemal protein kinase CK1 in Chlamydomonas flagella. J. Cell Biol. 186, 817–824 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toba S., Fox L. A., Sakakibara H., Porter M. E., Oiwa K., Sale W. S., Distinct roles of 1α and 1β heavy chains of the inner arm dynein I1 of Chlamydomonas flagella. Mol. Biol. Cell 22, 342–353 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petroutsos D., Tokutsu R., Maruyama S., Flori S., Greiner A., Magneschi L., Cusant L., Kottke T., Mittag M., Hegemann P., Finazzi G., Minagawa J., A blue-light photoreceptor mediates the feedback regulation of photosynthesis. Nature 537, 563–566 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Christie J. M., Phototropin blue-light receptors. Annu. Rev. Plant Biol. 58, 21–45 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Fu G., Nagasato C., Oka S., Cock J. M., Motomura T., Proteomics analysis of heterogeneous flagella in brown algae (stramenopiles). Protist 165, 662–675 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Huang K., Kunkel T., Beck C. F., Localization of the blue-light receptor phototropin to the flagella of the green alga Chlamydomonas reinhardtii. Mol. Biol. Cell 15, 3605–3614 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pasquale S. M., Goodenough U. W., Cyclic AMP functions as a primary sexual signal in gametes of Chlamydomonas reinhardtii. J. Cell Biol. 105, 2279–2292 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan J., Snell W. J., Signal transduction during fertilization in the unicellular green alga, Chlamydomonas. Curr. Opin. Microbiol. 3, 596–602 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Gomelsky M., Klug G., BLUF: A novel FAD-binding domain involved in sensory transduction in microorganisms. Trends Biochem. Sci. 27, 497–500 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Gibbons B. H., Intermittent swimming in live sea urchin sperm. J. Cell Biol. 84, 1–12 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Padma P., Hozumi A., Ogawa K., Inaba K., Molecular cloning and characterization of a thioredoxin/nucleoside diphosphate kinase related dynein intermediate chain from the ascidian, Ciona intestinalis. Gene 275, 177–183 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Inaba K., Mizuno K., Purification of dyneins from sperm flagella. Methods Cell Biol. 92, 49–63 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Bradford M. M., A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976). [DOI] [PubMed] [Google Scholar]
- 36.Taussky H. H., Shorr E., A microcolorimetric method for the determination of inorganic phosphorus. J. Biol. Chem. 202, 675–685 (1953). [PubMed] [Google Scholar]
- 37.Laemmli U. K., Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 (1970). [DOI] [PubMed] [Google Scholar]
- 38.Mizuno K., Padma P., Konno A., Satouh Y., Ogawa K., Inaba K., A novel neuronal calcium sensor family protein, calaxin, is a potential Ca2+-dependent regulator for the outer arm dynein of metazoan cilia and flagella. Biol. Cell 101, 91–103 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Yamada L., Saito T., Taniguchi H., Sawada H., Harada Y., Comprehensive egg coat proteome of the ascidian Ciona intestinalis reveals gamete recognition molecules involved in self-sterility. J. Biol. Chem. 284, 9402–9410 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Satou Y., Mineta K., Ogasawara M., Sasakura Y., Shoguchi E., Ueno K., Yamada L., Matsumoto J., Wasserscheid J., Dewar K., Wiley G. B., Macmil S. L., Roe B. A., Zeller R. W., Hastings K. E. M., Lemaire P., Lindquist E., Endo T., Hotta K., Inaba K., Improved genome assembly and evidence-based global gene model set for the chordate Ciona intestinalis: New insight into intron and operon populations. Genome Biol. 9, R152 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Endo T., Ueno K., Yonezawa K., Mineta K., Hotta K., Satou Y., Yamada L., Ogasawara M., Takahashi H., Nakajima A., Nakachi M., Nomura M., Yaguchi J., Sasakura Y., Yamasaki C., Sera M., Yoshizawa A. C., Imanishi T., Taniguchi H., Inaba K., CIPRO 2.5: Ciona intestinalis protein database, a unique integrated repository of large-scale omics data, bioinformatic analyses and curated annotation, with user rating and reviewing functionality. Nucleic Acids Res. 39, D807–D814 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.E. H. Harris, The Chlamydomonas Sourcebook. A Comprehensive Guide to Biology and Laboratory Uses (Academic Press, 1989), p. 780. [DOI] [PubMed] [Google Scholar]
- 43.Oda T., Yanagisawa H., Kikkawa M., Detailed structural and biochemical characterization of the nexin-dynein regulatory complex. Mol. Biol. Cell 26, 294–304 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fischer N., Rochaix J. D., The flanking regions of PsaD drive efficient gene expression in the nucleus of the green alga Chlamydomonas reinhardtii. Mol. Genet. Genomics 265, 888–894 (2001). [DOI] [PubMed] [Google Scholar]
- 45.Shimogawara K., Fujiwara S., Grossman A., Usuda H., High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 148, 1821–1828 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greiner A., Kelterborn S., Evers H., Kreimer G., Sizova I., Hegemann P., Targeting of photoreceptor genes in Chlamydomonas reinhardtii via zinc-finger nucleases and CRISPR/Cas9. Plant Cell 29, 2498–2518 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berthold P., Schmitt R., Mages W., An engineered streptomyces hygroscopicus aph 7″ gene mediates dominant resistance against hygromycin B in Chlamydomonas reinhardtii. Protist 153, 401–412 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Witman G. B., Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 134, 280–290 (1986). [DOI] [PubMed] [Google Scholar]
- 49.Kagami O., Kamiya R., Translocation and rotation of microtubules caused by multiple species of Chlamydomonas inner-arm dynein. J. Cell Sci. 103, 653–664 (1992). [Google Scholar]
- 50.Hozumi A., Padma P., Toda T., Ide H., Inaba K., Molecular characterization of axonemal proteins and signaling molecules responsible for chemoattractant-induced sperm activation in Ciona intestinalis. Cell Motil. Cytoskeleton 65, 249–267 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Yamamoto R., Yanagisawa H., Yagi T., Kamiya R., Novel 44-kilodalton subunit of axonemal dynein conserved from Chlamydomonas to mammals. Eukaryot. Cell 7, 154–161 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee-Eiford A., Ow R. A., Gibbons I. R., Specific cleavage of dynein heavy chains by ultraviolet irradiation in the presence of ATP and vanadate. J. Biol. Chem. 261, 2337–2342 (1986). [PubMed] [Google Scholar]
- 53.Oda T., Kikkawa M., Novel structural labeling method using cryo-electron tomography and biotin-streptavidin system. J. Struct. Biol. 183, 305–311 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Bui K. H., Ishikawa T., 3D structural analysis of flagella/cilia by cryo-electron tomography. Methods Enzymol. 524, 305–323 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Mastronarde D. N., Held S. R., Automated tilt series alignment and tomographic reconstruction in IMOD. J. Struct. Biol. 197, 102–113 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Bui K. H., Yagi T., Yamamoto R., Kamiya R., Ishikawa T., Polarity and asymmetry in the arrangement of dynein and related structures in the Chlamydomonas axoneme. J. Cell Biol. 198, 913–925 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hendrickson T. W., Perrone C. A., Griffin P., Wuichet K., Mueller J., Yang P., Porter M. E., Sale W. S., IC138 is a WD-repeat dynein intermediate chain required for light chain assembly and regulation of flagellar bending. Mol. Biol. Cell 15, 5431–5442 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.King S. M., Otter T., Witman G. B., Characterization of monoclonal antibodies against Chlamydomonas flagellar dyneins by high-resolution protein blotting. Proc. Natl. Acad. Sci. U.S.A. 82, 4717–4721 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ikeda K., Yamamoto R., Wirschell M., Yagi T., Bower R., Porter M. E., Sale W. S., Kamiya R., A novel ankyrin-repeat protein interacts with the regulatory proteins of inner arm dynein f (I1) of Chlamydomonas reinhardtii. Cell Motil. Cytoskeleton 66, 448–456 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamamoto R., Yanagisawa H., Yagi T., Kamiya R., A novel subunit of axonemal dynein conserved among lower and higher eukaryotes. FEBS Lett. 580, 6357–6360 (2006). [DOI] [PubMed] [Google Scholar]
- 63.LeDizet M., Piperno G., The light chain p28 associates with a subset of inner dynein arm heavy chains in Chlamydomonas axonemes. Mol. Biol. Cell 6, 697–711 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/9/eabf3621/DC1


