Abstract
Purpose
We conducted the study to elucidate how LncRNA LINC00941 affects colon cancer progression and its possible regulatory mechanism.
Methods
The expression level of LINC00941 in colon cancer tissues and cells was detected by qRT-PCR. The function of LINC00941 on colon cancer cell proliferation, migration, and invasion was detected by CCK-8 and Transwell assay respectively. The target interactions among LINC00941, miR-205-5p, and MYC were further confirmed by dual-luciferase reporter gene assays and RNA pull-down experiments. Meanwhile, in vivo experiments were carried out to study the role of LINC00941 in the xenotransplantation model.
Results
LINC00941 expression level was elevated in colon cancer tissues and cells. LINC00941 overexpression accelerated proliferation, migration, and invasion of colon cancer cells, while the LINC00941 knockdown showed the opposite results. In addition, LINC00941 regulated the expression of MYC by sponging miR-205-5p as a competitive endogenous RNA, and miR-205-5p knockdown reversed the tumor inhibition of LINC00941 knockdown on colon cancer cells. Xenograft model assay confirmed that LINC00941 silencing could inhibit colon cancer cell growth and metastasis.
Conclusion
LINC00941 may markedly promote colon cancer progression by acting on the miR-205-5p/MYC axis as a ceRNA, which offers novel clues for lncRNA to guide the treatment and prognosis of colon cancer.
Keywords: colon cancer, LINC00941, miR-205-5p, MYC, ceRNA
Introduction
Colon cancer is one of the most common gastrointestinal malignant tumors clinically, with rising morbidity and mortality. Its occurrence is related to many factors such as environment, genetics, diet, and lifestyle. With the development of molecular biology and clinical diagnosis technology in recent years, the diagnosis and treatment of colon cancer in its early stage have made some progress, but the overall survival of colon cancer patients has not been significantly rising.1,2 About 20–25% of the diagnosed colon cancer patients have distant metastasis, and the prognosis is poor.3–5 In addition, the potential molecular mechanism in colon cancer is not entirely known.6,7 Therefore, it has become a focus of current research to study the potential mechanism of colon cancer in its occurrence and development, and to search the pivotal molecules participating in tumor growth and metastasis, for the purpose of enhancing the therapeutic efficacy and prognosis of colon cancer.
Long non-coding RNAs (LncRNAs) are usually located in the nucleus or cytoplasm with a length of more than 200 nucleotides generally. They are a kind of non-coding RNA transcript that has aroused considerable research interest in recent years.8 LncRNAs have extensive functions, for instance, regulating the organism development and differentiation.9–11 They are involved in the occurrence of many diseases. Recently, a growing number of research has shown that the abnormality of lncRNAs expression profile and the occurrence of various malignant tumors are going hand in hand.12–16 LncRNAs, whose change can result in aberrant gene expression, are important regulatory factors in tumor formation and development. The potential mechanism of LncRNAs in tumorigenesis is very intricate. lncRNAs serving as support in the nuclei promote proteins to form ribonucleoprotein complexes that guide chromatin-modifying complexes recruited to target genes.17–19 LncRNAs can competitively combine with miRNAs functioning as CeRNAs in the cytoplasm, and thus regulate miRNAs target expression.20,21
LINC00941, also known as the MSC upregulation factor, is a new type of LncRNA located in the 12p11.21 region of the human genome. Some studies have shown that it can promote the growth and metastasis of many tumors. In oral squamous cell carcinoma, the Wnt/β-catenin signal transduction pathway was activated by highly expressed LINC00941;22 the up-regulation of LINC00941 can accelerate the development of hepatocellular carcinoma by promoting epithelial-mesenchymal transition (EMT) and malignancy.23 LINC00941 was highly expressed in gastric cancer, which was related to its biological processes.24,25 In lung adenocarcinoma, LINC00941 participated in regulating the phosphorylation of the PI3K-AKT signaling pathway.26 However, the biological effect and potential molecular mechanism of LINC00941 in colon cancer are still uncertain, so it is necessary to discuss the characteristics of LINC00941 in colon cancer.
We conducted the experiments in vitro and in vivo to explore LINC00941 expression, biological effects, and possible mechanisms in colon cancer. The findings showed that LINC00941 had the function of miRNA sponge to regulate miR-205-5p expression, positively regulated MYC expression, and promoted colon cancer progression. Our results demonstrated that the LINC00941-miR-205-5p-MYC axis may provide new clues to guide the treatment and prognosis of colon cancer.
Materials and Methods
Tissue Specimen Collection
We collected the paired cancer tissues and paracancerous tissues of 42 patients with colon cancer who underwent surgery in the Affiliated Cancer Hospital of Zhengzhou University from 2012 to 2014. All specimens were confirmed by histopathology and the informed written consents were collected from every patient prior to tissue collection. All patients did not receive any preoperative treatment. All tissue samples were frozen and stored in liquid nitrogen at −80 °C. The clinical features of 42 colon cancer patients are listed in Table 1. The study had gained the approval of the Ethics Committee of Affiliated Cancer Hospital of Zhengzhou University.
Table 1.
Correlation Between LINC00941 Expression and Clinical Features
| Group | Number | LINC00941 Expression | p-value | |
|---|---|---|---|---|
| (n=42) | High (n=31) | Low (n=11) | ||
| Age | ||||
| >60 | 25 | 17 | 8 | 0.299 |
| ≤60 | 17 | 14 | 3 | |
| Gender | ||||
| Male | 22 | 16 | 6 | 0.867 |
| Female | 20 | 15 | 5 | |
| Location | ||||
| Left | 18 | 12 | 6 | 0.362 |
| Right | 24 | 19 | 5 | |
| Tumor stage | ||||
| T1+T2 | 22 | 13 | 9 | 0.023 |
| T3+T4 | 20 | 18 | 2 | |
| Lymph node metastasis | ||||
| Absent | 16 | 8 | 8 | 0.006 |
| Present | 26 | 23 | 3 | |
| Distant metastasis | ||||
| Absent | 19 | 10 | 9 | 0.005 |
| Present | 23 | 21 | 2 | |
Cell Culture
Colorectal cancer cells and normal colon epithelial cells were purchased from the Shanghai Cell Bank of the Chinese Academy of Sciences, and all cells were cultured in incubators at 37 °C, 90%RH, and 5% CO2. Modified RPMI-1640 medium (Thermo-Fisher) was served for incubation, and replaced regularly every 48–72 hours.
Cell Transfection
LINC00941 cDNA was cloned into the expression vector pcDNA3.1(+) (Invitrogen) for the overexpression of this lncRNA in colon cancer cells. Two siRNA targeting LINC00941 (siLINC00941-1, 2), negative control siRNA (siNC), miR-205-5p mimics, NC mimics, miR-205-5p inhibitor and NC inhibitor were all purchased from Genepharma (Suzhou, Jiangsu, China). All the sequences are listed in Table 2. 1x106 cells were transfected with 50 pg/μL siRNA, miRNA mimics, miRNA inhibitor, or corresponding NCs using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) and harvested after 48–72 h.
Table 2.
The Sequences of the Vectors or Mimics
| Sort | The Sequence |
|---|---|
| siLINC00941-1 | 5ʹ-ACGCGTTGCATAACCTGA-3’ |
| siLINC00941-2 | 5′- GAGACAGTTGATAGCCAAA-3′ |
| siNC | 5ʹ-UUCUCCGAACGUGUCACGUTT-3’ |
| miR-205-5p mimics | 5ʹ-UCCUUCAUUCCACCGGAGUCUG-3’ |
| miR-205-5p inhibitor | 5ʹ-UCCUUCAUUCCACCGGAGUCUG-3’ |
| NC mimics | 5ʹ-UUCUCCGAACGUGUCACGUTT-3’ |
| NC inhibitor | 5ʹ-UCUACUCUUUCUAGGAGGUUGUGA-3’ |
CCK8 Assay
The transfected cell viability was measured according to the CCK-8 assay kit instructions (Beyotime, China). LoVo and HCT116 were inoculated into a 96-well plate (100μL/well), and separately incubated for 24, 48, 72, and 96 h. 10 μ l CCK8 solution was added into each well. The absorbance at 450 nm (OD450) was measured by a micro board reader, to evaluate the cell viability.
Transwell Assay
We adopted the Transwell assay to detect the invasion capability of LoVo and HCT116 cells in vitro. The cells (1 × 105) eluted and re-suspended using the serum-free medium were seeded into Transwell’s parietal chamber which was coated with Matrigel. The culture medium containing 10% fetal bovine serum was placed in the lower chamber. After 24 hours, cells in the lower chamber were removed, fixed, and stained with methanol and 0.1% crystal violet respectively. Five random regions were selected to photograph and count the cells. Said experiment was carried out 3 times.
qRT-PCR
The total RNA of tissues and cells was obtained through the use of Trizol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The reverse transcription for cDNA synthesis was performed by PrimeScript RT kit (Toyobo Life Science). Quantitative analysis was carried out referring to Fast SYBR® Green Master Mix (Thermo Fisher Scientific, Inc.). The internal references for miR-205-5p and LINC00941 were U6 and GAPDH, respectively. Relative RNA expression level was analyzed by the 2−ΔΔct method. All primer sequences are listed in Table 3.
Table 3.
Primer Sequences for qRT-PCR
| Gene | Primer Sequence |
|---|---|
| LINC00941 | |
| Forward: | 5ʹ-GACCTTTTCAGGCCAGCATT-3ʹ; |
| Reverse: | 5ʹ-ACAATCTGGATAGAGGGCTCA-3’ |
| miR-205-5p | |
| Forward: | 5ʹ-TCCTTCATTCCACCGGAGTCTG-3’ |
| Forward: | 5ʹ-GCGAGCACAGAATTAATACGAC-3’ |
| U6 | |
| Forward: | 5′-ATTGGAACGATACAGAGAAGATT-3′ |
| Reverse: | 5′-GGAACGCTTCACGAATTTG-3′ |
| MYC | |
| Forward: | 5′-GGATTCCCGCCTCAGAATAAC-3′ |
| Reverse: | 5′-GTGGGTGTGGGTTGTTCAGG-3′ |
| GAPDH | |
| Forward: | 5ʹ-TTGGTATCGTGGAAGGACTCA-3’ |
| Reverse: | 5ʹ-TGTCATCATATTTGGCAGGTT-3’ |
Fluorescence in situ Hybridization Analysis
RNA-FISH experiment was carried out to assess the subcellular localization of LINC00941. In short, cells were fixed for 15 min using 4% PFA, and permeabilized on ice for 5 min using 0.5% Triton X-100, followed by the incubation between cells and RNA-FISH probes in hybridization buffer overnight at 37°C. RNA-FISH probes were obtained from Sevivebio (Wuhan, China). The cell nucleus was stained with DAPI.
Dual-Luciferase Reporter Gene Assay
Based on the binding sites between LINC00941 and miR-205-5p, MYC and miR-205-5p, the binding and mutant sequences of LINC00941 and MYC were respectively inserted into pmirGLO vector (Promega) to construct LINC00941-wt (wild-type), LINC00941-mut (mutant-type), MYC-wt (wild-type), and MYC-mut (mutant-type). LINC00941-wt or LINC00941-mut was co-transfected with miR-205-5p mimics or NC mimics into colon cancer cells, and with miR-205-5p inhibitor or NC inhibitor into colon cells. Similarly, MYC-wt or MYC-mut was co-transfected with miR-205-5p mimics or NC mimics. 36 hours after the transfection, the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) was applied to test the relative luciferase activity, which was standardized according to Renilla luciferase activity.
RNA Immunoprecipitation (RIP) Assay
RIP assay was performed using Magna rip RNA binding protein immunoprecipitation Kit (Millipore, Billerica, Ma, USA). Simply, the transfected colon cancer cells were collected, and lysed in lysis buffer. Then the cell lysate and RIP buffer containing magnetic-AgO2 complex (the magnetic beads were pre-incubated with AgO2 antibody at room temperature) were incubated at 4 °C. Magnetic beads were coupled with anti-IgG as a negative control. Thereafter, immunoprecipitated RNA was extracted and the AgO2-bound RNA expression was measured by qRT-PCR.
Nude Mice Xenograft Models
BALB/c mice (6–8 weeks old, male) were purchased from Shanghai SLAC Laboratory Animal Co. Ltd., and raised under pathogen-free conditions. Nude mice were randomly divided into two groups: the shNC group and the shLINC00941 group. For in vivo tumor growth model, HCT116 cells resuspended in RPMI-1640 medium (1x108 cells/mL) were subcutaneously injected into the posterior side of BALB/c mice (n=5 per group). The size of the tumor was measured every 5 days, and based on this, the growth curve of the tumor was drawn. The tumor volume was calculated as follows: volume = length × width2 × 0.5. After 30 days, the nude mice were killed, and their tumor weights were measured for further experiments. For in vivo lung-colonization assays, HCT116 cells (2x106) or PBS were injected into each mouse from the caudal vein. 5 weeks later, the mice were euthanized in accordance with Institutional Animal Care and Use Committee protocols (n=5 per group). The lungs were taken out to count the number of tumor nodules. Hematoxylin-eosin staining was utilized for the histological examination of tumor tissues. Animal studies were approved by the Ethics Committee of Affiliated Cancer Hospital of Zhengzhou University in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals.
Western Blot Analysis
Colon cancer cells were lysed in the RIPA protein extraction solution supplemented by protease inhibitors. An equal amount of protein was run on 10%SDS-PAGE and then shifted to the PVDF membrane. The membrane was sealed using 5% skimmed milk at 37 °C for 1 hour and then cultured overnight at 4 °C using primary antibodies. The antibodies involved were: Anti-MYC (1:1,000; cat. no. 9402; Cell Signaling Technology, Inc.) and GAPDH (1:4,000; cat. no. GB11002; Wuhan Servicebio Technology Co., Ltd.). After, the membrane was washed in PBST 3 times, incubated with the corresponding secondary antibody at room temperature for 2 hours, and finally washed again in PBST. The gray value of each protein band was obtained by the use of ImageJ software, and GAPDH was used as an internal reference.
Statistical Analysis
Data were expressed as the mean ± SEM. An unpaired two-tailed t-test was used for comparison between two groups. ANOVA or repeated ANOVA followed by Bonferroni’s post hoc test was used to perform multiple comparisons through GraphPad Prism® software (version 6.0; GraphPad Software, Inc.). Pearson χ2 test was used to assess the difference of clinical characteristic variables between the two groups. P<0.05 was considered to be statistically significant.
Results
LINC00941 is Upregulated in Colon Cancer Tissues and Cell Lines, Associated with Clinicopathological Features
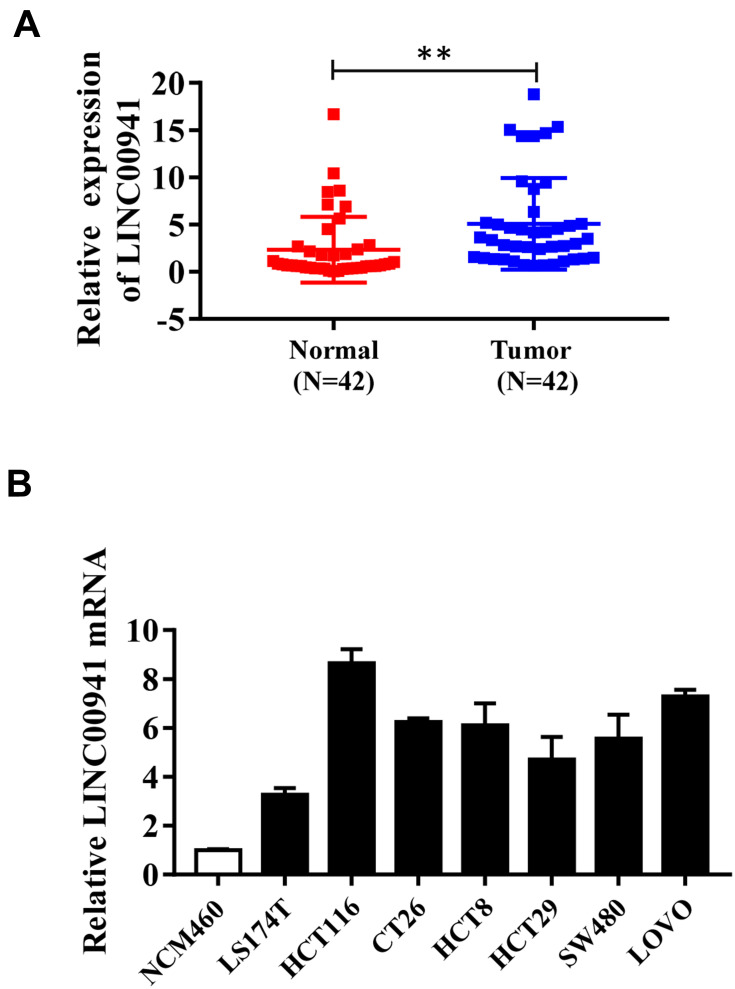
LINC00941 expression level in tissue specimens was measured using qRT-PCR. The results displayed that in contrast with adjacent normal tissues, the expression level of LINC00941 in colon cancer tissues was remarkably elevated (Figure 1A). To explore the clinical significance of LINC00941, we also investigated the relationship between its expression level and clinical characteristics. LINC00941 expression was related to the tumor invasive depth, lymph node metastasis, and distant metastasis, but irrelevant to the sex, age, and tumor location (Table 1). Furthermore, LINC00941 expression in colon epithelial cells and colon cancer cell lines was measured respectively. We found that LINC00941 expression showed a significant increase in colon cancer cell lines, especially in HCT116 and LoVo cell lines (Figure 1B), so the two cell lines were selected for further experiments. Taken together, all the results indicated that LINC00941 may promote tumor formation and development in colon.
Figure 1.
LINC00941 expression is increased in colon cancer tissues and cell lines. (A) LINC00941 expression in colon cancer and adjacent normal tissues was detected by RT-qPCR. (B) LINC00941 expression in colon cancer cell lines LS174T,HCT116,CT26, HCT8, HCT29, SW480 and LoVo compared colonic epithelial cells NCM460 was detected by RT-qPCR. **P < 0.01.
LINC00941 Overexpression Promotes Proliferation, Migration, and Invasion of Colon Cancer Cells
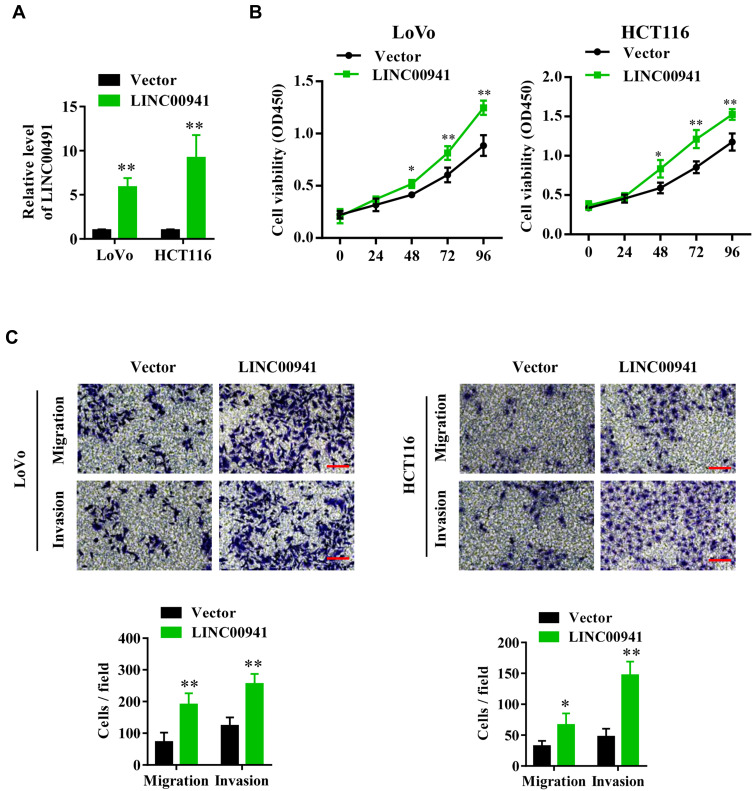
To investigate how LINC00941 acts on the cell biological behavior, pcDNA-LINC00941 plasmid conducted to overexpress LINC00941 was transfected into HCT116 and LoVo cell lines with pcDNA-vector as the negative control. CCK8 and Transwell tests were carried out to determine the proliferation, migration, and invasion of colon cancer cells with LINC00941 overexpression. The results reflected that HCT116 and LoVo cells with overexpressed LINC00941 showed a higher proliferation and invasion ability than the control group (Figure 2A–C).
Figure 2.
LINC00941 promotes the proliferation, migration and invasion of colon cancer cells. (A) LINC00941 expression level in LoVo and HCT116 cells transfected with LINC00941 overexpression plasmid was detected by RT-qPCR. (B) Cell viability of LoVo and HCT116 cells transfected with LINC00941 overexpression plasmid was detected by CCK8 assay. (C) The invasion and migration of LoVo and HCT116 cells transfected with LINC00941 overexpression plasmid was detected by Transwell assay.Scale bars,200μm. *P < 0.05, **P < 0.01.
LINC00941 Knockdown Impairs Proliferation, Migration, and Invasion of Colon Cancer Cells in vitro
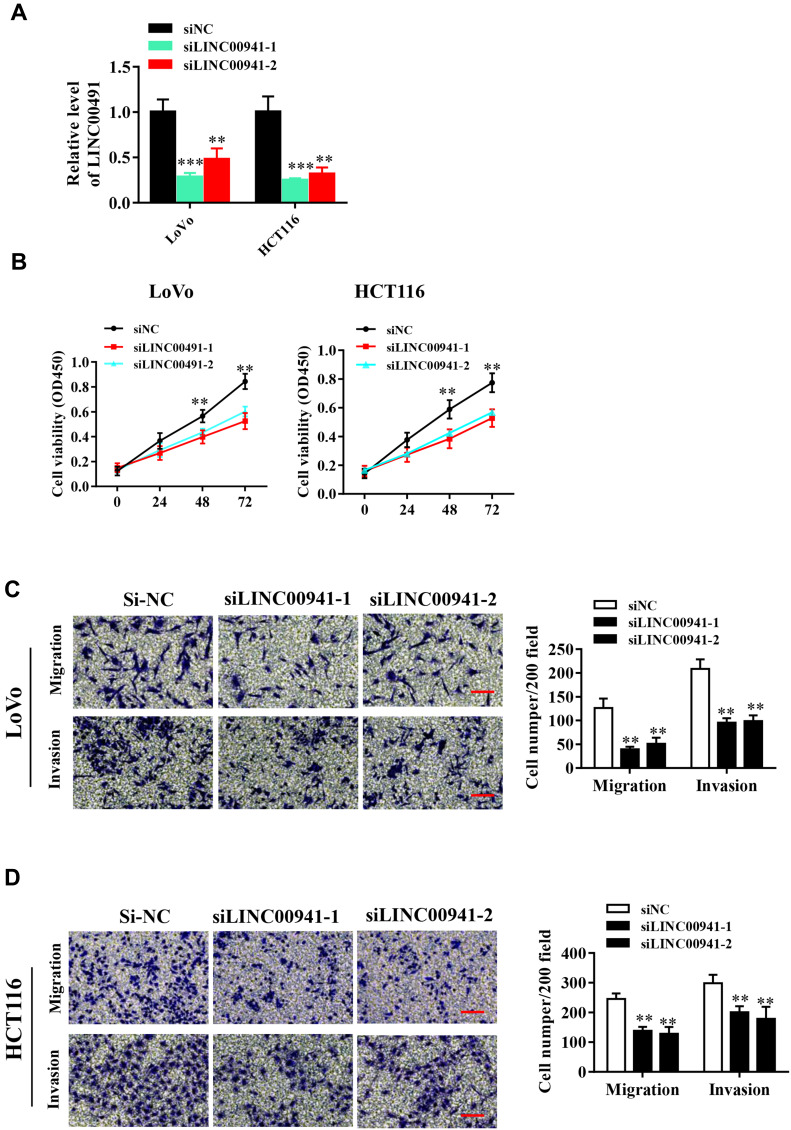
LINC00941 expression in HCT116 and LoVo cells was decreased by two specific siRNA (siLINC00941-1 and siLINC00941-2). The down-regulation of LINC00941 was confirmed by qRT-PCR analysis (Figure 3A). CCK8 assay demonstrated that when LINC00941 decreased, the HCT116 and LoVo cell proliferation was impaired significantly (Figure 3B). Similarly, Transwell analysis showed that knockdown of LINC0094 prevented cancer cells from migrating and invading (Figure 3C and D).
Figure 3.
LINC00941 knockdown impairs proliferation,migration and invasion of colon cancer cells in vitro. (A) LINC00941 expression level in LoVo and HCT116 cells transfected with siLINC00941 was detected by RT-qPCR. (B) Cell viability of LoVo and HCT116 cells transfected with siLINC00941 was detected by CCK8 assay. (C and D) The invasion and migration of LoVo and HCT116 cells transfected with siLINC00941 was detected by Transwell assay.Scale bars,200μm. **P < 0.01, ***P<0.001.
LINC00941 Knockdown Impairs Colon Tumor Growth and Metastasis in vivo
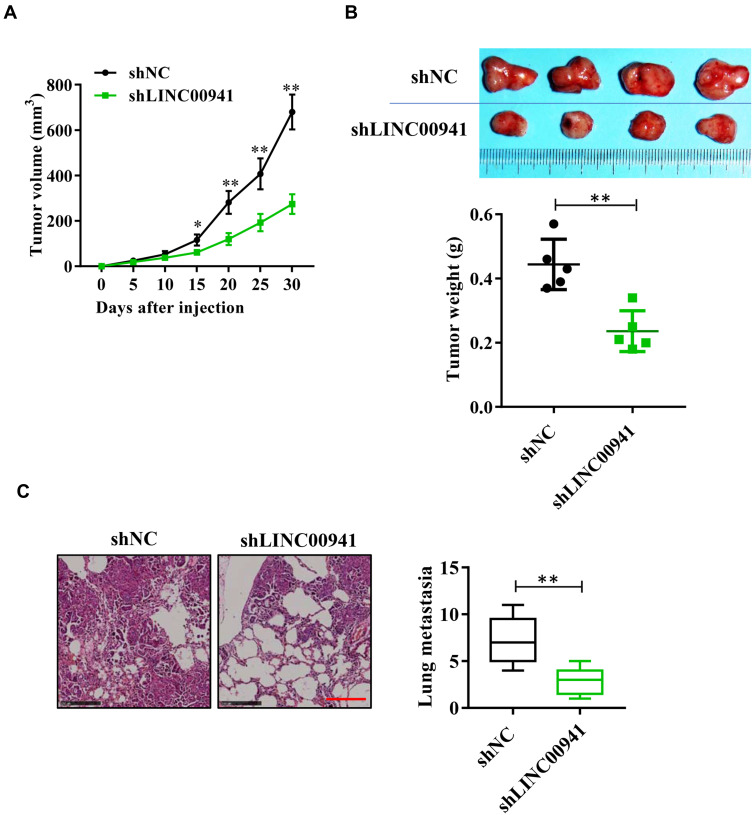
In vivo, the effect of LINC00941 on tumor growth and metastasis was verified using a naked mouse tumorigenesis experiment. As can be seen from Figure 4A and B, the tumor volume in nude mice of the shLINC00941 group was significantly smaller than that of the shNC group, and tumor weight showed similar performance. The tumor cells with low expression of linc00941 were injected into the caudal vein to detect the metastasis of colon cancer cells in vivo. Results showed that knocking down LINC00941 can significantly inhibit the formation of metastatic pulmonary nodules. A histological examination of the lung by hematoxylin-eosin staining further confirmed this difference. (Figure 4C)
Figure 4.
LINC00941 knockdown impairs colon cancer cell growth and metastasis in vivo. (A) Tumor volumes in nude mice transfected with shLINC00941 dramatically smaller than shNC. (B) Compared to shNC, tumor size smaller and tumor weights dramatically lighter in nude mice transfected with shLINC00941. (C) Number of pulmonary nodules in nude mice transfected with shLINC00941 less dramatically than shNC.Scale bars,250μm. *P < 0.05, **P < 0.01.
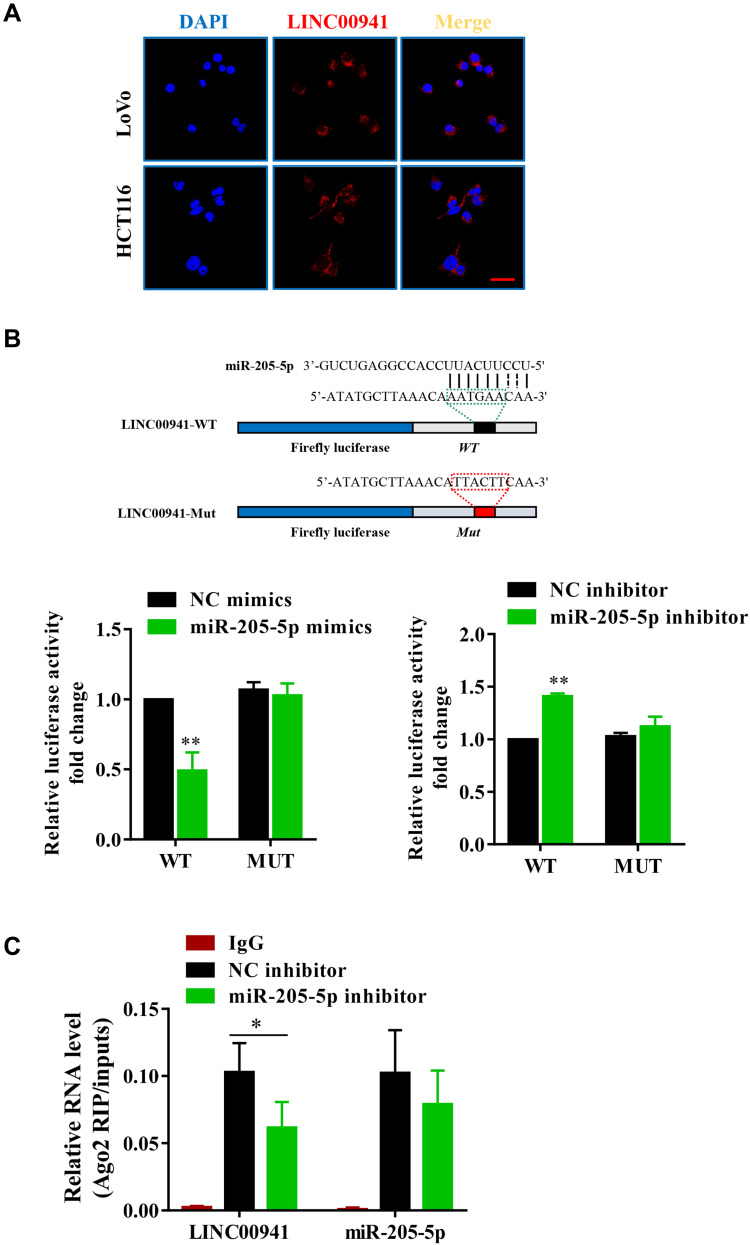
LINC00941 Exists Mainly in the Cytoplasm and Targets miR-205-5p
Further experiments were implemented for exploring the specific mechanism of LINC00941 in regulating colon cancer. Because the activity and function of lncRNAs are related to subcellular distribution, LINC00941 localization in colon cancer cells was detected by FISH. Results confirmed that LINC00941 was mainly localized in the cytoplasm, which hinted that LINC00941 may exert a regulatory function via the ceRNA network (Figure 5A). The potential downstream target of LINC00941 was forecasted according to StarBase v2.0 (http://starbase.sysu.edu.cn/), and it was discovered that LINC00941 may target miR-205-5p. To study the interaction between lncRNA and miRNA, full-length LINC00941 and LINC00941 containing mutation sites binding to miR-205-5p were respectively inserted into double luciferase reporter vector, as LINC00941-wt and LINC00941-mut (Figure 5B). Then the plasmids were transfected into colon cancer cells together with miR-205-5p inhibitor or mimics. As shown in Figure 5B, the luciferase activity of LINC00941-wt declined when miR-205-5p was overexpressed, while increased when the miR-205-5p expression was knocked down. At the same time, the luciferase activity of LINC00941-mut had no significant change. To further confirm the said result, RIP experiments were carried out, and results revealed that LINC00941 and miR-205-5p were all enriched in immunoprecipitated AgO2,but LINC00941 expressed less enrichment in miR-205-5p knock-down cells (Figure 5C). In short, LINC00941 may be a competing endogenous RNA that is effective in regulating miR-205-5p expression.
Figure 5.
LINC00941 targets miR-205-5p. (A) Sublocalization of LINC00941 in LoVo and HCT116 cells detected by FISH analysis. (B) LINC00941 can target miR-205-5p was proved by luciferase reporter assays. (C) LINC00941 and miR-205-5p were enriched in Ago2 immunoprecipitates relative to control IgG immunoprecipitates.Scale bars,25μm. *P < 0.05, **P < 0.01.
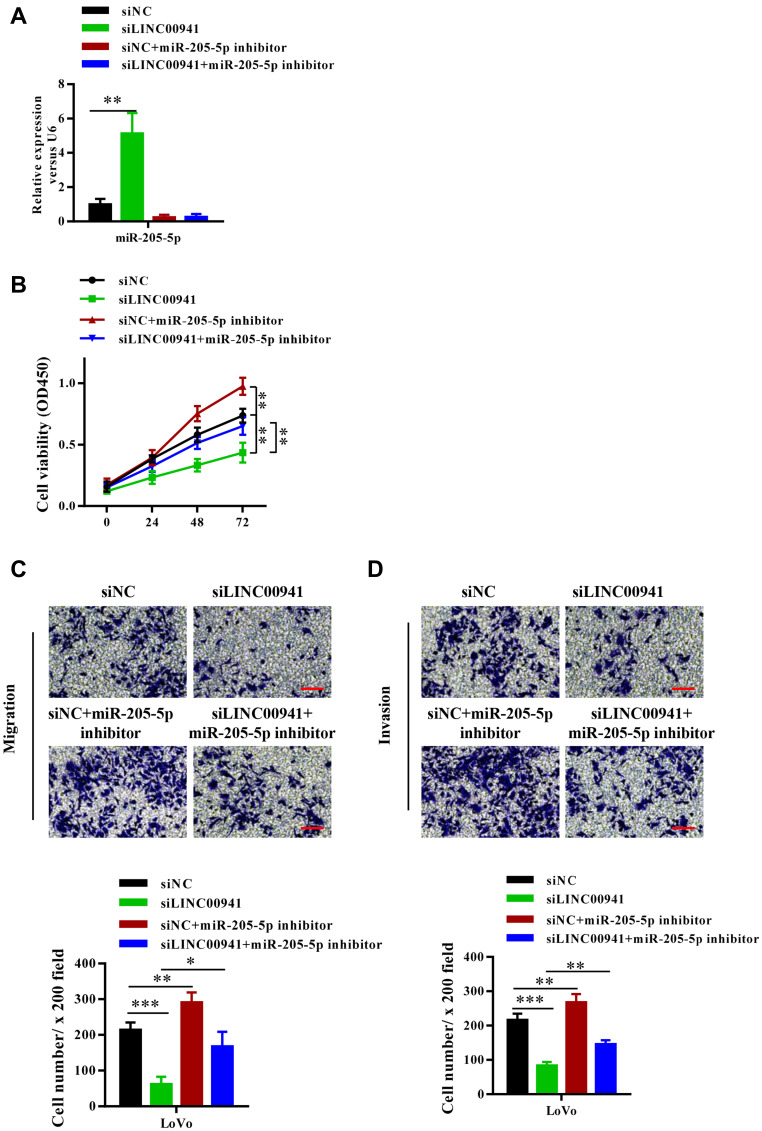
Silencing miR-205-5p Reverses the Effect of LINC00941 Knockdown on Colon Cancer Progression
Based on the above findings, we further researched the interaction between LINC00941 and miR-205-5p. LoVo cells were served to be transfected with SiNC, SiLINC00941, SiNC+miR-205-5p inhibitor, and SiLINC00941+miR-205-5p inhibitor respectively. It can be seen from Figure 6A that miR-205-5p expression of colon cancer cells with siLINC00941 distinctly increased, which was reversed by adding miR-205-5p inhibitor. The results of CCK8 assay and Transwell assay demonstrated that silencing LINC00941 significantly prevented the proliferation, migration, and invasion of colon cancer cells, however partial reversal of the inhibition was achieved by silencing miR-205-5p (Figure 6B–D). These findings manifested that LINC00941 participated in colon cancer development by targeting miR-205-5p.
Figure 6.
Knockdown of miR-205-5p reverses the effects of LINC00941 knockdown on colon cancer progression. (A) MiR-205-5p expression level of LoVo cells in four groups was detected by RT-qPCR. (B) Cell viability of transfected LoVo cells in four groups was detected by CCK8 assay. (C and D) The invasion and migration of transfected LoVo cells in four groups was detected by Transwell assay.Scale bars,200μm. *P < 0.05, **P < 0.01, ***P<0.001.
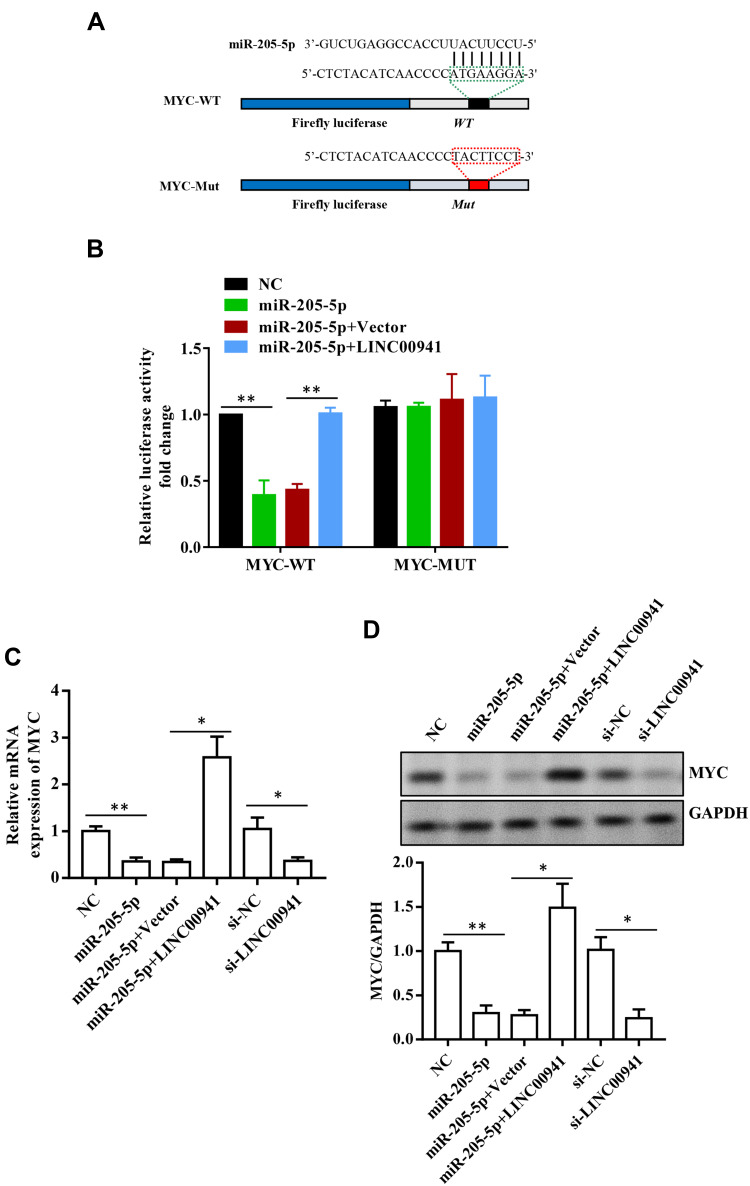
miR-205-5p Targets MYC
The binding site between MYC and miR-205-5p was predicted by TargetScan (http://www.targetscan.org/) (Figure 7A). Double luciferase report gene experiment verified miR-205-5p and MYC had a targeting correlation. Luciferase activity was significantly inhibited in the cells co-transfected with MYC-wt and miR-205-5p mimics, but not in the cells co-transfected with MYC-mut and miR-205-5p mimics (Figure 7B). In addition, the qRT-PCR experiment and Western blot analysis showed that the up-regulation of miR-205-5p significantly decreased MYC expression level, and LINC00941 expression was positively correlated with MYC (Figure 7C and D). The findings illustrated that MYC might be the downstream target of LINC00941 and miR-205-5p.
Figure 7.
MiR-205-5p targets MYC. (A and B) MiR-205-5p can target MYC was proved by luciferase reporter assays. (C and D) MYC expression level in each group of transfected LoVo cells was detected by RT-qPCR and Western blot analysis. *P < 0.05, **P < 0.01.
Discussion
More and more evidence points out that lncRNAs participate in the regulation of tumor formation and development and have emerged as a tumor biomarker.27,28 As a novel lncRNA, LINC00941 is elevated in several tumors and plays its numerous functions to affect tumor progression.24–26,29 Liu et al reported that LINC00941 facilitated the initiation of gastric cancer.24 Because of its obvious carcinogenic effect in gastric cancer, we speculated that LINC00941 may also have the same effect on colon cancer. We believed that the research on the biological effect and potential mechanism of LINC00941 in colon cancer is worth studying.
Our report discovered that LINC00941 expression showed a distinct increase in colon cancer tissues and was associated with clinical features. Elevated LINC00941 can accelerate multiplication, migration, and invasion of colon cancer cells in vivo and in vitro. As to the mechanism, LINC00941 participated in miR-205-5p/MYC regulation as a competitive endogenous RNA, thus promoting the progression of colon cancer. Specifically, LINC00941 expression in tumor tissues was higher than that in adjacent normal tissues. The up-regulation of LINC00941 was also related to invasive depth, lymph node, and distant metastasis in colon cancer. These results suggested that LINC00941 shows a high potential in predicting the poor prognosis of patients suffering from colon cancer. Previous investigations have shown that silencing LINC00941 can significantly inhibit gastric cancer cell proliferation, migration, and invasion. Similarly, our functional analysis found that LINC00941 overexpression improved proliferative, migratory, and invasive capacity of colon cancer cells, while siRNA-mediated LINC00941 knockdown showed the opposite effect on multiplication, migration, and invasion of colon cancer cells. The Xenograft in nude mice also exhibited the same trend, which indicated that LINC00941 knockdown could repress the growth and metastasis of transplanted tumors in nude mice. It is suggested that LINC00941 may be a new target for anti-tumor therapy.
Emerging research reveals that lncRNAs play a more extensive role than previously recognized.30 LncRNAs exist in both the nucleus and cytoplasm,31,32 and regulating biological processes and gene expression may be related to their subcellular distribution.33 LncRNAs play a pivotal role in gene modification, transcription, post-transcription, and translation, besides, the series network is multi-level and complex. Our RNA-FISH experiments evaluated that LINC00941 was relatively abundant in the cytoplasm of HCT116 and LoVo cells, suggesting that LINC00941 may play a part at the post-transcriptional level. Lately, a new regulatory mechanism at the post-transcriptional level has been discovered, that is, lncRNAs can adsorb miRNAs as a natural sponge to disturb miRNAs pathway, further regulating the expression of miRNAs target.21,34 Given the structural characteristics and subcellular localization, LINC00941 may also perform the same function in colon cancer. In our experiments, luciferase reported that there was a physical relationship between LINC00941 and miR-205-5p, and RIP analysis further proved the combination of LINC00941 and miR-205-5p. Also, subsequent experiments manifested that miR-205-5p silencing reversed the anti-tumor effect of colon cancer cells with LINC00941 gene knockdown. These results suggested that LINC00941 can directly interact with miR-205-5p,and then regulate the progression of colon cancer via targeting miR-205-5p.
Many discussions have reported that miRNAs play a vital role in the origination and development of tumors by regulating particular genes.35–37 MYC is a crucial oncogene situated on chromosome 8q24, which participates in controlling cell multiplication, differentiation, and apoptosis.38 MYC gene translocation mutates into a proto-oncogene with high transcriptional activity, which can promote malignant transformation of cells and have a hand in the formation and progression of multiple tumors.39 We discovered that miR-205-5p can directly inhibit MYC expression in colon cancer at the post-transcript level by targeting MYC 3ʹ- untranslated region (UTR), which confirmed that MYC was the target of miR-205-5p. To sum up, we demonstrated that LINC00941 can control the MYC expression level at the post-transcript level by acting as a miR-205-5p sponge in the colon cancer cell line. They formed a competitive network of endogenous RNAs in colon cancer.
All in all, this study creatively verified the binding relationship between LINC00941 and miR-205-5p as well as the targeting relationship between miR-205-5p and MYC, which had not been discussed previously. These data contribute to the progress of research on the molecular mechanism of colon cancer, and these findings may provide new hopeful targets for colorectal cancer treatment. However, it is unclear whether other major regulatory factors are involved in this process. Therefore, the functions of LINC00941, miR-205-5p and MYC, and more potential mechanisms in the initiation and progression of colon cancer remain to be further clarified.
Disclosure
The authors report no conflicts of interest in this study.
References
- 1.Chen HY, Lin YM, Chung HC, et al. MiR-103/107 promote metastasis of colorectal cancer by targeting the metastasis suppressors DAPK and KLF4. Cancer Res. 2012;72(14):3631–3641. doi: 10.1158/0008-5472.CAN-12-0667 [DOI] [PubMed] [Google Scholar]
- 2.Kato T, Yasui K, Hirai T, et al. Therapeutic results for hepatic metastasis of colorectal cancer with special reference to effectiveness of hepatectomy - Analysis of prognostic factors for 763 cases recorded at 18 institutions. Dis Colon Rectum. 2003;46(10):22–31. [DOI] [PubMed] [Google Scholar]
- 3.Eadens MJ, Grothey A. Curable metastatic colorectal cancer. Curr Oncol Rep. 2011;13(3):168–176. doi: 10.1007/s11912-011-0157-0 [DOI] [PubMed] [Google Scholar]
- 4.Van Cutsem E, Nordlinger B, Adam R, et al. Towards a pan-European consensus on the treatment of patients with colorectal liver metastases. Eur J Cancer. 2006;42(14):2212–2221. doi: 10.1016/j.ejca.2006.04.012 [DOI] [PubMed] [Google Scholar]
- 5.Alberts SR, Horvath WL, Sternfeld WC, et al. Oxaliplatin, fluorouracil, and leucovorin for patients with unresectable liver-only metastases from colorectal cancer: a north central cancer treatment group phase II study. J Clin Oncol. 2005;23(26):9243–9249. doi: 10.1200/JCO.2005.07.740 [DOI] [PubMed] [Google Scholar]
- 6.Lin J, Tan X, Qiu L, et al. Long Noncoding RNA BC032913 as a novel therapeutic target for colorectal cancer that suppresses metastasis by upregulating TIMP3. Mol Ther Nucleic Acids. 2017;8:469–481. doi: 10.1016/j.omtn.2017.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rychahou PG, Kang J, Gulhati P, et al. Akt2 overexpression plays a critical role in the establishment of colorectal cancer metastasis. Proc Natl Acad Sci USA. 2008;105(51):20315–20320. doi: 10.1073/pnas.0810715105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu J, Zhou MY, Yu XP, et al. Long noncoding RNA OR3A4 promotes the migration and invasion of melanoma through the PI3K/AKT signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23(16):6991–6996. doi: 10.26355/eurrev_201908_18739 [DOI] [PubMed] [Google Scholar]
- 9.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. CELL. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006 [DOI] [PubMed] [Google Scholar]
- 10.Lopez-Pajares V. Long non-coding RNA regulation of gene expression during differentiation. Pflugers Arch. 2016;468(6):971–981. doi: 10.1007/s00424-016-1809-6 [DOI] [PubMed] [Google Scholar]
- 11.Blythe AJ, Fox AH, Bond CS. The ins and outs of lncRNA structure: how, why and what comes next? Biochim Biophys Acta. 2016;1859(1):46–58. doi: 10.1016/j.bbagrm.2015.08.009 [DOI] [PubMed] [Google Scholar]
- 12.Dhamija S, Diederichs S. From junk to master regulators of invasion: lncRNA functions in migration,EMT and metastasis. Int J Cancer. 2016;139(2):269–280. doi: 10.1002/ijc.30039 [DOI] [PubMed] [Google Scholar]
- 13.Xin Y, Li Z, Shen J, Chan MT, Wu WK. CCAT1: a pivotal oncogenic long non-coding RNA in human cancers. Cell Prolif. 2016;49(3):255–260. doi: 10.1111/cpr.12252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartonicek N, Maag JL, Dinger ME. Long non-coding RNAs in cancer: mechanisms of action and technological advancements. Mol Cancer. 2016;15(1):43. doi: 10.1186/s12943-016-0530-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, Song Z, Feng C, et al. The long non-coding RNA SUMO1P3 facilitates breast cancer progression by negatively regulating miR320a. Am J Transl Res. 2017;9(12):5594–5602. [PMC free article] [PubMed] [Google Scholar]
- 16.Baratieh Z, Khalaj Z, Honardoost MA, et al. Aberrant expression of PlncRNA-1 and TUG1: potential biomarkers for gastric cancer diagnosis and clinically monitoring cancer progression. Biomark Med. 2017;11(12):1077–1090. doi: 10.2217/bmm-2017-0090 [DOI] [PubMed] [Google Scholar]
- 17.Engreitz JM, Pandya-Jones A, McDonel P, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341(6147):1237973. doi: 10.1126/science.1237973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. doi: 10.1016/j.cell.2010.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simon MD, Pinter SF, Fang R, et al. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 2013;504(7480):465–469. doi: 10.1038/nature12719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465(7301):1033–1038. doi: 10.1038/nature09144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ai Y, Wu S, Zou C, Wei H. LINC00941 promotes oral squamous cell carcinoma progression via activating CAPRIN2 and canonical WNT/β-catenin signaling pathway. J Cell Mol Med. 2020;24(18):10512–10514. doi: 10.1111/jcmm.15667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan X, Zhang D, Wu W, et al. Mesenchymal stem cells promote hepatocarcinogenesis via lncRNA-MUF interaction with ANXA2 and miR-34a. Cancer Res. 2017;77(23):6704–6716. doi: 10.1158/0008-5472.CAN-17-1915 [DOI] [PubMed] [Google Scholar]
- 24.Luo C, Tao Y, Zhang Y, et al. Regulatory network analysis of high expressed long non-coding RNA LINC00941 in gastric cancer. Gene. 2018;662:103–109. doi: 10.1016/j.gene.2018.04.023 [DOI] [PubMed] [Google Scholar]
- 25.Liu H, Wu N, Zhang Z, et al. Long non-coding RNA LINC00941 as a potential biomarker promotes the proliferation and metastasis of gastric cancer. Front Genet. 2019;10:5. doi: 10.3389/fgene.2019.00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Zhao H, Xu Y, et al. Systematic identification of lincRNA-based prognostic biomarkers by integrating lincRNA expression and copy number variation in lung adenocarcinoma. Int J Cancer. 2019;144(7):1723–1734. doi: 10.1002/ijc.31865 [DOI] [PubMed] [Google Scholar]
- 27.Zhang S, Ma H, Zhang D, et al. LncRNA KCNQ1OT1 regulates proliferation and cisplatin resistance in tongue cancer via miR-211-5p mediated Ezrin/Fak/Src signaling. Cell Death Dis. 2018;9(7):742. doi: 10.1038/s41419-018-0793-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S, Ke H, Zhang H, et al. LncRNA MIR100HG promotes cell proliferation in triple-negative breast cancer through triplex formation with p27 loci. Cell Death Dis. 2018;9(8):805. doi: 10.1038/s41419-018-0869-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gugnon M, Manicardi V, Torricelli F, et al. Linc00941 is a novel transforming growth factor β target that primes papillary thyroid cancer metastatic behavior by regulating the expression of cadherin 6. Thyroid. 2020. doi: 10.1089/thy.2020.0001 [DOI] [PubMed] [Google Scholar]
- 30.Troy A, Sharpless NE. Genetic “lnc”-age of noncoding RNAs to human disease. J Clin Invest. 2012;122(11):3837–3840. doi: 10.1172/JCI66645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fort A, Hashimoto K, Yamada D, et al. Deep transcriptome profiling of mammalian stem cells supports a regulatory role for retrotransposons in pluripotency maintenance. Nat Genet. 2014;46(6):558–566. doi: 10.1038/ng.2965 [DOI] [PubMed] [Google Scholar]
- 33.Hu G, Tang Q, Sharma S, et al. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat Immunol. 2013;14(11):1190–1198. doi: 10.1038/ni.2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lv J, Fan HX, Zhao XP, et al. Long non-coding RNA Unigene56159 promotes epithelial-mesenchymal transition by acting as a ceRNA of miR-140-5p in hepatocellular carcinoma cells. Cancer Lett. 2016;382(2):166–175. doi: 10.1016/j.canlet.2016.08.029 [DOI] [PubMed] [Google Scholar]
- 35.Zeng CY, Zhan YS, Huang J, Chen YX. MicroRNA7 suppresses human colon cancer invasion and proliferation by targeting the expression of focal adhesion kinase. Mol Med Rep. 2016;13(2):1297–1303. doi: 10.3892/mmr.2015.4643 [DOI] [PubMed] [Google Scholar]
- 36.Koo KH, Kwon H. MicroRNA miR-4779 suppresses tumor growth by inducing apoptosis and cell cycle arrest through direct targeting of PAK2 and CCND3. Cell Death Dis. 2018;9(2):77. doi: 10.1038/s41419-017-0100-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chandramouli A, Onyeagucha BC, Mercado-Pimentel ME, et al. MicroRNA-101 (miR-101) post-transcriptionally regulates the expression of EP4 receptor in colon cancers. Cancer Biol Ther. 2012;13(3):175–183. doi: 10.4161/cbt.13.3.18874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, Croce CM. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci U S A. 1982;79(24):7824–7827. doi: 10.1073/pnas.79.24.7824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Green TM, Nielsen O, de Stricker K, Xu-Monette ZY, Young KH, Møller MB. High levels of nuclear MYC protein predict the presence of MYC rearrangement in diffuse large B-cell lymphoma. Am J Surg Pathol. 2012;36(4):612–619. doi: 10.1097/PAS.0b013e318244e2ba [DOI] [PubMed] [Google Scholar]