Abstract
Background and Objectives
Targeting cancer-associated fibroblast (CAF) is being explored as an approach to improve cancer therapies. The roles of CAF remain unclarified in malignant transformation of papillary thyroid cancer (PTC) into dedifferentiated thyroid cancer (DDTC). This study aimed to investigate correlations of CAF with dedifferentiation and clinicopathological characteristics of thyroid cancer.
Materials and Methods
We applied three different mRNA-based CAF gene signatures to quantify CAF in our cohort, the Gene Expression Omnibus (GEO) cohort and The Cancer Genome Atlas (TCGA) cohort, and analyzed expression of α-SMA by immunohistochemistry in thyroid cancer. The CAF score was analyzed for its associations with clinicopathological characteristics, genetic mutations, tumor-associated signaling pathways and immune landscape.
Results
The CAF score increased significantly in DDTCs compared with normal thyroid tissues and PTCs, and the α-SMA-positive CAFs were found enriched in DDTCs. The high CAF score showed a significant correlation with the anaplastic phenotype in DDTC and low thyroid differentiation score in PTC. Patients with a high CAF score remarkably increased the risk of aggressive outcomes in both DDTC and PTC. Furthermore, the CAF score was positively correlated with genetic mutations, oncogenic signaling pathways, the immune score and increased expression of tumor microenvironment (TME) target markers.
Conclusion
Our findings suggest CAFs positively correlate with dedifferentiation and aggressive outcomes of thyroid cancer, and targeting CAFs as a therapeutic approach may benefit DDTC patients.
Keywords: CAF, dedifferentiation, DDTC, PTC
Introduction
A series of studies support tumoral progression from well-differentiated thyroid cancer to dedifferentiated thyroid cancer (DDTC), including poorly differentiated thyroid cancer (PDTC) and anaplastic thyroid cancer (ATC).1–4 Dedifferentiation of papillary thyroid cancer (PTC) into DDTC contributes to aggressive and even lethal outcomes of thyroid cancer patients as mentioned in our previous study.5 It is well known that accumulation of genetic alterations is responsible for the malignant transformation of dedifferentiation.1,2
Tumor microenvironment (TME) is the key driving factor of tumor progression.6 Within the TME, cells of the innate (eg, macrophages, mast cells, neutrophils) and the adaptive (eg, lymphocytes) immune responses are interconnected with epithelial cancer cells, fibroblasts, and endothelial cells via cytokines, chemokines, and adipocytokines, of which the molecular pattern has a key role and could explain the involvement of the immune system in tumor initiation and progression.7 Cancer-associated fibroblasts (CAFs), as a type of activated fibroblasts, are implicated to be a critical modulator within TME, which crosstalk with both cancer cells and TME to sustain cancer cell survival, to promote invasion and metastasis, to induce angiogenesis, and to attenuate chemotherapeutic sensitivity.8–10 CAF’s clinical implications have been shown adequate to serve as a biomarker and a potential target in some cancer.10,11 Several studies have revealed that presence of CAF is associated with invasion of thyroid cancer.12–15 However, on one aspect, there is no study investigating the roles of CAF in dedifferentiation of thyroid cancer, and on the other aspect, CAF was commonly identified based on the expression of one or two CAF markers, such as alpha-smooth muscle actin (α-SMA) or fibroblast activation protein alpha (FAP) in the previous studies,12,13,15 which are not representative of the overall CAF population.
The present study aimed to use three different CAF gene signatures to investigate the associations of CAF with differentiation status and clinicopathological outcomes in thyroid cancer patients from Fudan University Shanghai Cancer Center (FUSCC), the Gene Expression Omnibus (GEO) database and The Cancer Genome Atlas (TCGA) database. The CAF signature score was found to significantly increase in DDTC, and the patients with a high CAF score showed aggressive outcomes. Additionally, we further analyzed molecular abbreviations of cancer cell and TME associated with CAF to uncover possible mechanism underlying CAF-modulating progression.
Materials and Methods
CAF Gene Signatures
Considering the existence of different CAF subpopulations in cancer stroma, the usage of one or two positive markers for CAF selection has been demonstrated to be heterogeneous. Thus, we applied three different mRNA-based CAF gene signatures to quantify CAF in thyroid cancer according to the previous studies from Wu,16 Nurmik17 and Calon.18 The gene sets of the above CAF signatures were provided in Table-S1. A summary score of CAF is defined in each sample as the median of the absolute expression value of the genes in the signature, as described in the previous study.19
RNA Sequencing Data from FUSCC
We previously performed high-throughput RNA sequencing in six normal thyroid (NT) tissues, five PTC tissues, and five DDTC tissues from PTC cases undergoing dedifferentiation, and the RNA sequencing methods and the clinicopathological parameters of these cases were described in the previous study.20 The RNA sequencing data of the five NTs, five PTCs, and five DDTCs were used for analyses in this study due to the bias of the sequencing outcomes from a normal sample, of which gene expression profiles of the three CAF signatures were shown in Table-S2. Our study was approved by the Medical Ethics Committee of the FUSCC, and all patients provided written informed consents for their specimens and information to be used for research and stored in the hospital database. All procedures performed in this study were in accordance with the ethical standards of our institutional research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Immunohistochemistry (IHC)
We obtained the surgical samples of PTC cases undergoing dedifferentiation from the Department of Pathology at the FUSCC, which were fixed in formalin and embedded in paraffin. Hematoxylin- and eosin-stained (H&E) sections were used to identify regions of PTC and dedifferentiated carcinoma. IHC staining was performed to detect α-SMA (Clone 1A4, M0851, 1:500, Dako) expression on a Roche BenchMark ULTRA system according to the manufacturer’s instructions. The staining extent was scored as 0 (<10%) and 1 (≥10%) based on the percentage of α-SMA positive-staining fibroblasts in cancer stroma according to the previous study.15,21 The intensity score from 0–2 was given for each sample according to the staining intensity (0, negative; 1, low intensity; 2, high intensity). Scores for staining intensity and positivity percentage were then multiplied to generate the immunoreactivity score for each case.
Transcriptional Profiles from the GEO Database and the TCGA Database
We obtained raw microarray cell intensity (CEL) files of thyroid cancer including GSE29265, GSE33630,22,23 GSE53157,24 GSE6514425 and GSE760393 from the GEO database (http://www.ncbi.nlm.nih.gov/geo/).26,27 The files were background-adjusted and normalized using Robust Multichip Average.28 The same chip platform (Affymetrix Human Genome U133 Plus 2.0 Array) was used by the files of the GEO cohort. The ComBat method was used to remove the batch effects. All probes were mapped to the latest version of the NetAffx Annotation File. If a gene symbol was recorded with multiple probes, the average value was used as its expression level. The combined GEO cohort included the GSE29265 cohort (20 NTs, 20 PTCs and 9 ATCs), the GSE33630 cohort (45 NTs, 49 PTCs and 11 ATCs), the GSE53157 cohort (3 NTs, 15 PTCs and 5 PDTCs), the GSE65144 cohort (13 NTs and 12 ATCs) and the GSE76039 cohort (17 PDTCs and 20 ATCs). TCGA cohort data were available at the website of DATA SETS of University of California Santa Cruz (UCSC) Xena (THCA_22 datasets: https://xenabrowser.net/datapages/; https://www. biorxiv.org/content/early/2018/08/28/326470) and The cBioPortal for Cancer Genomics (http://www.cbioportal.org/). The data from the GEO cohort and the TCGA cohort were described in detail in our previous study,20 Table-S3 and Table-S4.
Gene Set Enrichment Analysis (GSEA)
GSEA was performed using GSEA software, Version 2.0, which was obtained from the Broad Institute (http://www.broad.mit.edu/gsea), as previously described.29,30 Enrichment Map was used for visualization of the GSEA results. Normalized enrichment score (NES) and false discovery rate (FDR) were used to sort the Hallmark Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enriched in each phenotype after gene set permutations were performed 1000 times for each analysis.
Enrichment Analyses of Immune Cell Types
As mentioned in the previous studies,31,32 we used the xCell tool (http://xcell.ucsf.edu/), a gene-signature based method for inferring cell types from tissue transcriptome profiles, to evaluate the heterogeneous cellular landscape of TME. The xCell tool inferred enrichment scores of immune cell types in TME by using the transcriptome data of the 74 DDTC cases from the GEO database and the 391 PTC cases from the TCGA database.
Statistical Analysis
Continuous variables were, respectively, expressed as the mean±standard deviation (SD) in analyses of associations between clinical parameters and molecular variables, and categorical data were summarized with frequencies and percentages. Student’s t-test and one-way analysis of variance were performed to evaluate the differences of continuous variables between two groups and among more than two groups, respectively. Associations between continuous variables and categorical variables were evaluated using Mann–Whitney U-tests for two groups and Kruskal–Wallis tests for more than two groups. χ2 and Fisher’s exact test were used for categorical variables. To analyze the associations between the CAF score and clinicopathological parameters, patients were divided into two subgroups (Low-CAF score and high-CAF score) according to the median value of the CAF score. Based on the median value of thyroid differentiation score (TDS), we divided the PTC patients from the TCGA cohort into a low-differentiated group and a high-differentiated group. Nonparametric receiver operating characteristic (ROC) analyses were performed to calculate area under curve (AUC) for the signature that would be predictive of low differentiation status. Pearson’s/Spearman correlation was performed to analyse the associations of the CAF score with the TDS, BRAF-RAS score, enrichment scores of immune cell types and expression value of immune microenvironment markers. Furthermore, the CAF score was tested for overall survival (OS) analyses in thyroid cancer patients using the GSE76039 cohort.3 The Kaplan-Meier method was used to construct OS curves, and the univariate survival difference was determined by the Log rank test. A p value <0.05 was considered significant. Data preparation and statistical analyses were performed using the SPSS for Windows (version 22.0; IBM Corp., Armonk, NY), the R software (version 3.5.1; R Foundation for Statistical Computing, Vienna, Austria) and the GraphPad Prism (version 6.01; GraphPad Software Inc., La Jolla, Calif).
Results
CAFs Enriched in DDTC
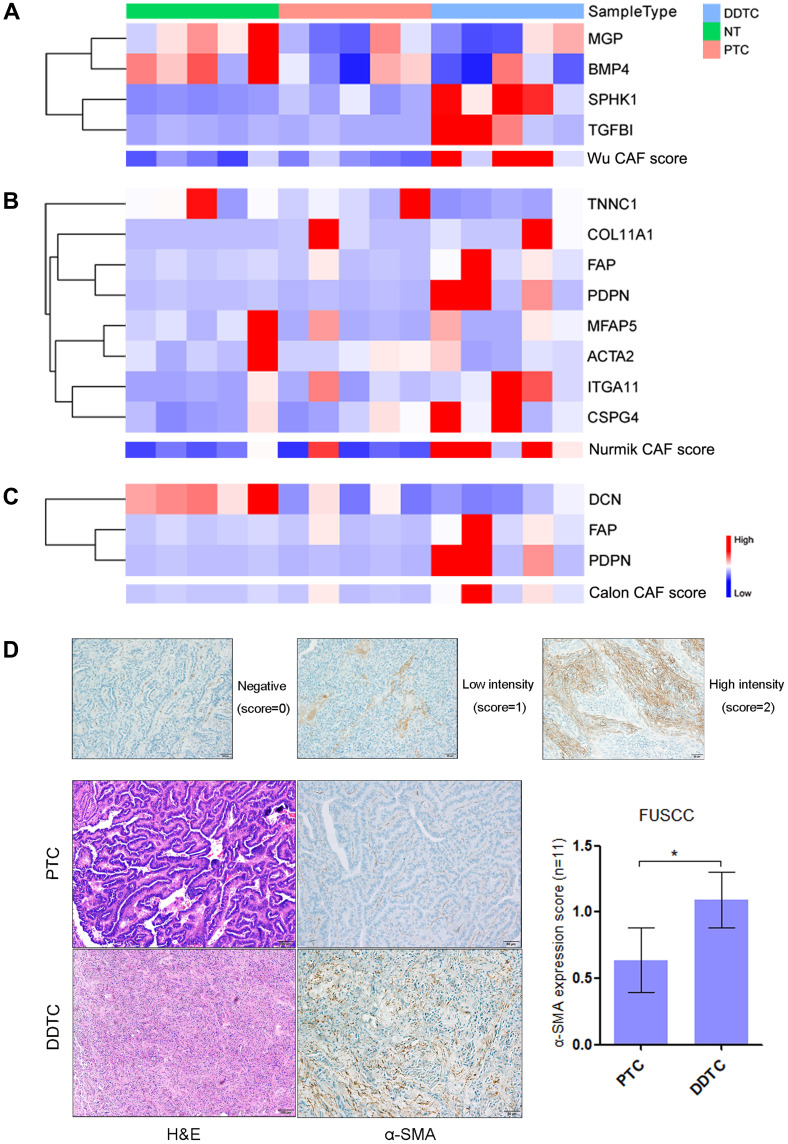
We initially analyze the mRNA-based CAF signatures from Wu, Nurmik and Calon in the five DDTCs, five PTCs and five NTs from dedifferentiated patients at FUSCC. A consistent increase of the three CAF signature scores was found in DDTC compared with PTC and NT though statistical significance was observed in the Wu and Nurmik CAF signatures but not in the Calon signature (Figure 1A–C and Figure-S1A-1C). To validate alteration of CAF presence in the process of thyroid cancer dedifferentiation, we further detected the common marker of CAF, α-SMA, by IHC in eleven dedifferentiated PTC patients from FUSCC, and found that α-SMA significantly increased in stroma area of DDTC compared with the matched PTC stroma area (Figure 1D, p=0.02).
Figure 1.
Cancer-associated fibroblasts (CAFs) enriched in dedifferentiated papillary thyroid cancer (PTC) cases in the Fudan University Shanghai Cancer Center (FUSCC) cohort. (A–C) Heatmaps of expression of the Wu (A), Nurmik (B) and Calon (C) CAF signature genes and the CAF scores in normal thyroid (NT) tissues, PTCs and dedifferentiated thyroid cancers (DDTCs) from dedifferentiated PTC cases. (D) Expression of alpha-smooth muscle actin (α-SMA) detected by immunohistochemistry in matched PTCs and DDTCs from 11 dedifferentiated PTC cases. *p<0.05.
The CAF Score Was Significantly Associated with Differentiation Status and Poor Clinical Outcomes of Thyroid Cancer
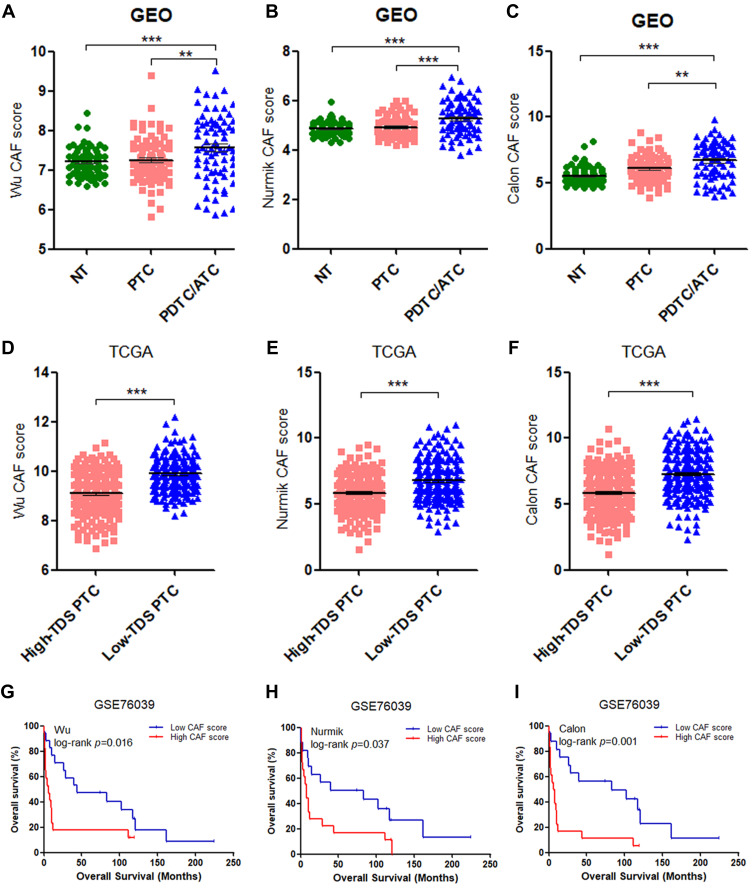
The clinicopathological associations of the CAF score were analyzed in thyroid cancer using transcriptional profiles of the combined GEO cohort and the TCGA cohort to validate the primary findings in our cohort. In the combined GEO cohort, the CAF score showed a significant increase in 74 DDTCs compared with 84 PTCs and 81 NTs in all of the three signatures (Figure 2A–C). Moreover, we confirmed that the low-TDS PTC patients had remarkably higher CAF scores than the high-TDS PTC patients (Figure 2D–F). The CAF score was negatively correlated with the TDS of PTC, and the Calon signature showed a higher prediction for low differentiation status with the smallest number of the signature genes (Figure-S1D-1L).
Figure 2.
Validation of correlations of the CAF score with differentiation status and overall survival (OS). (A–C) Analyses of the CAF scores of the Wu (A), Nurmik (B) and Calon (C) CAF signatures in poorly differentiated thyroid cancer (PDTC)/anaplastic thyroid cancer (ATC) compared with normal thyroid (NT) tissues and papillary thyroid cancer (PTC). (D–F) Analyses of the CAF scores of the Wu (D), Nurmik (E) and Calon (F) CAF signatures in patients with low thyroid differentiation score (TDS) compared with high-TDS patients in PTC. (G–I) The correlation of OS outcomes with the CAF scores of the Wu (G), Nurmik (E) and Calon (F) CAF signatures in PDTC/ATC patients from the GSE76039 cohort. **p<0.01, ***p<0.001.
For the DDTC patients (19 PDTC and 18 ATC patients) from the GSE76039 cohort, the three signatures consistently showed the significant correlation of the high CAF score with the decreased overall survival (OS) (Figure 2G–I), the ATC phenotype (Table 1) and lymph node metastasis (LNM) (Table 1). Furthermore, the CAF score was closely associated with aggressiveness of PTC such as the high-risk histological subtype, extrathyroidal extension (ETE), LNM and T3/T4 stage in the TCGA cohort (Table 2).
Table 1.
Clinicopathological and Molecular Correlations of the CAF Score in DDTC Patients from the GSE76039 Cohort
| Variables | N | Wu CAF Score | p value | Nurmik CAF Score | p value | Calon CAF Score | p value | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Low-Risk | High-Risk | Low-Risk | High-Risk | Low-Risk | High-Risk | |||||
| Age (years) Mean±SD |
37 | 64.16±15.39 | 66.94±12.04 | 0.545 | 65.42±14.71 | 65.61±13.06 | 0.967 | 64.00±14.94 | 67.11±12.58 | 0.499 |
| Gender | 1.000 | 0.242 | 1.000 | |||||||
| Male | 3 | 2 (66.7%) | 1 (33.3%) | 1 (33.3%) | 2 (66.7%) | 2 (66.7%) | 1 (33.3%) | |||
| Female | 12 | 7 (58.3%) | 5 (41.7%) | 9 (75.0%) | 3 (25.0%) | 8 (66.7%) | 4 (33.3%) | |||
| Histology | 0.009 | 0.049 | 0.009 | |||||||
| PDTC | 19 | 14 (73.7%) | 5 (26.3%) | 13 (68.4%) | 6 (31.6%) | 14 (73.7%) | 5 (26.3%) | |||
| ATC | 18 | 5 (27.8%) | 13 (72.2%) | 6 (33.3%) | 12 (66.7%) | 5 (27.8%) | 13 (72.2%) | |||
| T Stage | 0.192 | 1.000 | 1.000 | |||||||
| T1-T2 | 2 | 2 (100.0%) | 0 (0.0%) | 1 (50.0%) | 1 (50.0%) | 1 (50.0%) | 1 (50.0%) | |||
| T3-T4 | 27 | 11 (40.7%) | 16 (59.3%) | 13 (48.1%) | 14 (51.9%) | 11 (40.7%) | 16 (59.3%) | |||
| LNM | 0.007 | 0.046 | 0.007 | |||||||
| N0 | 10 | 8 (80.0%) | 2 (20.0%) | 8 (80.0%) | 2 (20.0%) | 8 (80.0%) | 2 (20.0%) | |||
| N1 | 17 | 4 (23.5%) | 13 (76.5%) | 6 (35.3%) | 11 (64.7%) | 4 (23.5%) | 13 (76.5%) | |||
| Distant metastasis | 0.660 | 1.000 | 0.640 | |||||||
| M0 | 6 | 4 (66.7%) | 2 (33.3%) | 4 (66.7%) | 2 (33.3%) | 4 (66.7%) | 2 (33.3%) | |||
| M1 | 17 | 9 (52.9%) | 8 (47.1%) | 10 (58.8%) | 7 (41.2%) | 8 (47.1%) | 9 (52.9%) | |||
| Mutation count | 35 | 6.59±4.82 | 9.11±8.96 | 0.311 | 9.71±8.94 | 6.17±4.89 | 0.152 | 8.00±8.23 | 7.78±6.46 | 0.930 |
| BRAF V600E | 0.091 | <0.001 | 0.002 | |||||||
| Mutation | 13 | 4 (30.8%) | 9 (69.2%) | 1 (7.7%) | 12 (92.3%) | 2 (6.7%) | 11 (84.6%) | |||
| Wild-type | 24 | 15 (62.5%) | 9 (37.5%) | 18 (75.0%) | 6 (25.0%) | 17 (70.8%) | 7 (29.2%) | |||
| RAS | 0.295 | 0.013 | 0.295 | |||||||
| Mutation | 12 | 8 (66.7%) | 4 (33.3%) | 10 (83.3%) | 2 (16.7%) | 8 (66.7%) | 4 (33.3%) | |||
| Wild-type | 25 | 11 (44.0%) | 14 (56.0%) | 9 (36.0%) | 16 (64.0%) | 11 (44.0%) | 14 (56.0%) | |||
| EIF1AX | 1.000 | 0.046 | 0.340 | |||||||
| Mutation | 5 | 3 (60.0%) | 2 (40.0%) | 5 (100.0%) | 0 (0.0%) | 4 (80.0%) | 1 (20.0%) | |||
| Wild-type | 32 | 16 (50.0%) | 16 (50.0%) | 14 (43.8%) | 18 (56.3%) | 15 (46.9%) | 17 (53.1%) | |||
| TERT promoter | 0.515 | 0.049 | 0.049 | |||||||
| Mutation | 20 | 9 (45.0%) | 11 (55.0%) | 7 (35.0%) | 13 (65.0%) | 7 (35.0%) | 13 (65.0%) | |||
| Wild-type | 17 | 10 (58.8%) | 7 (41.2%) | 12 (70.6%) | 5 (29.4%) | 12 (70.6%) | 5 (29.4%) | |||
| TP53 | 0.049 | 0.515 | 0.049 | |||||||
| Mutation | 16 | 5 (31.3%) | 11 (68.8%) | 7 (43.8%) | 9 (56.3%) | 5 (31.3%) | 11 (68.8%) | |||
| Wild-type | 21 | 14 (66.7%) | 7 (33.3%) | 12 (57.1%) | 9 (42.9%) | 14 (66.7%) | 7 (33.3%) | |||
| PI3KCA/AKT/mTOR | 0.062 | 0.269 | 0.062 | |||||||
| Mutation | 9 | 2 (22.2%) | 7 (77.8%) | 3 (33.3%) | 6 (66.7%) | 2 (22.2%) | 7 (77.8%) | |||
| Wild-type | 28 | 17 (60.7%) | 11 (39.3%) | 16 (57.1%) | 12 (42.9%) | 17 (60.7%) | 11 (39.3%) | |||
Note: Italic and bold type indicates statistical significance.
Abbreviations: CAF, cancer-associated fibroblast; SD, standard deviation; NA, not available; PDTC, poorly differentiated thyroid cancer; ATC, anaplastic thyroid cancer; LNM, lymph node metastasis.
Table 2.
Clinicopathological and Molecular Correlations of the CAF Score in PTC Patients from the TCGA Cohort
| Variables | N | Wu CAF Score | p value | Nurmik CAF Score | p value | Calon CAF Score | p value | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Low-Risk Score | High-Risk Score | Low-Risk Score | High-Risk Score | Low-Risk Score | High-Risk Score | |||||
| Age (years) | 0.375 | 0.163 | 0.526 | |||||||
| < 55 | 336 | 163 (48.5%) | 173 (51.5%) | 175 (45.5%) | 161 (47.9%) | 171 (50.9%) | 165 (49.1%) | |||
| ≥ 55 | 165 | 87 (52.7%) | 78 (47.3%) | 75 (45.5%) | 90 (54.5%) | 79 (47.9%) | 86 (52.1%) | |||
| Gender | 0.783 | 0.037 | 0.783 | |||||||
| Male | 135 | 66 (48.9%) | 69 (51.1%) | 57 (42.2%) | 78 (57.8%) | 66 (48.9%) | 69 (51.1%) | |||
| Female | 366 | 184 (50.3%) | 182 (49.7%) | 193 (52.7%) | 173 (47.3%) | 184 (50.3%) | 182 (49.7%) | |||
| Multifocality | 0.801 | 0.215 | 0.955 | |||||||
| Unifocal | 265 | 132 (49.8%) | 133 (50.2%) | 138 (52.1%) | 127 (47.9%) | 132 (49.8%) | 133 (50.2%) | |||
| Multifocal | 226 | 110 (48.7%) | 116 (51.3%) | 105 (46.5%) | 121 (53.5%) | 112 (49.6%) | 114 (50.4%) | |||
| Coexistent HT | 0.861 | 0.528 | 0.581 | |||||||
| Yes | 65 | 34 (52.3%) | 31 (47.7%) | 35 (53.8%) | 30 (46.2%) | 31 (47.7%) | 34 (52.3%) | |||
| No | 397 | 203 (51.1%) | 194 (48.9%) | 197 (49.6%) | 200 (50.4%) | 204 (51.4%) | 193 (48.6%) | |||
| Histological subtype | <0.001 | <0.001 | <0.001 | |||||||
| Low risk | 459 | 242 (52.7%) | 217 (47.3%) | 244 (53.2%) | 215 (46.8%) | 244 (53.2%) | 215 (46.8%) | |||
| High risk | 42 | 8 (19.0%) | 34 (81.0%) | 6 (14.3%) | 36 (85.7%) | 6 (14.3%) | 36 (85.7%) | |||
| T Stage | <0.001 | 0.001 | 0.001 | |||||||
| T1-T2 | 308 | 176 (57.1%) | 132 (42.9%) | 172 (55.8%) | 136 (54.8%) | 172 (55.8%) | 136 (44.2%) | |||
| T3-T4 | 190 | 74 (38.9%) | 116 (61.1%) | 78 (41.1%) | 112 (58.9%) | 78 (41.1%) | 112 (58.9%) | |||
| ETE | <0.001 | <0.001 | <0.001 | |||||||
| Yes | 152 | 48 (31.6%) | 104 (68.4%) | 52 (34.2%) | 100 (65.8%) | 49 (32.2%) | 103 (67.8%) | |||
| No | 331 | 190 (57.4%) | 141 (42.6%) | 189 (57.1%) | 142 (42.9%) | 191 (57.7%) | 140 (42.3%) | |||
| LNM | <0.001 | 0.004 | <0.001 | |||||||
| N0 | 227 | 130 (57.3%) | 97 (42.7%) | 126 (55.5%) | 101 (44.5%) | 129 (56.8%) | 98 (43.2%) | |||
| N1 | 224 | 88 (39.3%) | 136 (60.7%) | 94 (42.0%) | 130 (58.0%) | 87 (38.8%) | 137 (61.2%) | |||
| Distant metastasis | 0.738 | 0.183 | 0.189 | |||||||
| M0 | 279 | 119 (42.7%) | 160 (57.3%) | 119 (42.7%) | 160 (57.3%) | 121 (43.4%) | 158 (56.6%) | |||
| M1 | 9 | 3 (33.3%) | 6 (66.7%) | 6 (66.7%) | 3 (33.3%) | 6 (66.7%) | 3 (33.3%) | |||
| BRAF V600E | <0.001 | <0.001 | <0.001 | |||||||
| Mutation | 239 | 82 (34.3%) | 157 (65.7%) | 89 (37.2%) | 150 (62.8%) | 82 (34.3%) | 157 (65.7%) | |||
| Wild-type | 254 | 166 (65.4%) | 88 (34.6%) | 159 (62.6%) | 95 (37.4%) | 166 (65.4%) | 88 (34.6%) | |||
| TERT promoter | 0.098 | 0.135 | 0.026 | |||||||
| Mutation | 36 | 14 (38.9%) | 22 (61.1%) | 14 (38.9%) | 22 (61.1%) | 12 (33.3%) | 24 (66.7%) | |||
| Wild-type | 360 | 192 (53.3%) | 168 (46.7%) | 187 (51.9%) | 173 (48.1%) | 190 (52.8%) | 170 (47.2%) | |||
| TERT exon | 1.000 | 0.246 | 0.246 | |||||||
| Mutation | 2 | 1 (50.0%) | 1 (50.0%) | 0 (0.0%) | 2 (100.0%) | 0 (0.0%) | 2 (100.0%) | |||
| Wild-type | 491 | 247 (50.3%) | 244 (49.7%) | 248 (50.5%) | 243 (49.5%) | 248 (50.5%) | 243 (49.5%) | |||
| RAS | <0.001 | <0.001 | <0.001 | |||||||
| Mutation | 51 | 45 (88.2%) | 6 (11.8%) | 42 (82.4%) | 9 (17.6%) | 45 (88.2%) | 6 (11.8%) | |||
| Wild-type | 442 | 203 (45.9%) | 239 (54.1%) | 206 (46.6%) | 236 (53.4%) | 203 (45.9%) | 239 (54.1%) | |||
| EIF1AX | 0.216 | 0.216 | 0.686 | |||||||
| Mutation | 6 | 5 (83.3%) | 1 (16.7%) | 5 (83.3%) | 1 (16.7%) | 4 (66.7%) | 2 (33.3%) | |||
| Wild-type | 487 | 243 (49.9%) | 244 (50.1%) | 243 (49.9%) | 244 (50.1%) | 244 (50.1%) | 243 (49.9%) | |||
| ETV6_NTRK3 | 0.372 | 0.214 | 1.000 | |||||||
| Fusion | 5 | 4 (80.0%) | 1 (20.0%) | 1 (20.0%) | 4 (80.0%) | 3 (60.0%) | 2 (40.0%) | |||
| Wild-type | 488 | 244 (50.0%) | 244 (50.0%) | 247 (50.6%) | 241 (49.4%) | 245 (50.2%) | 243 (49.8%) | |||
| CCDC6_RET | 0.112 | 0.253 | 0.112 | |||||||
| Fusion | 21 | 7 (33.3%) | 14 (66.7%) | 8 (38.1%) | 13 (61.9%) | 7 (33.3%) | 14 (66.7%) | |||
| Wild-type | 472 | 241 (51.1%) | 231 (48.9%) | 240 (50.8%) | 232 (49.2%) | 241 (51.1%) | 231 (48.9%) | |||
| PAX8_PPARG | 0.060 | 1.000 | 0.124 | |||||||
| Fusion | 4 | 0 (0.0%) | 4 (100.0%) | 2 (50.0%) | 2 (50.0%) | 4 (100.0%) | 0 (0.0%) | |||
| Wild-type | 489 | 248 (50.7%) | 241 (49.3%) | 246 (50.3%) | 243 (49.7%) | 244 (49.9%) | 245 (50.1%0 | |||
Note: Italic and bold type indicates statistical significance.
Abbreviations: CAF, cancer-associated fibroblast; TCGA, The Cancer Genomics Atlas; HT, Hashimoto’s thyroiditis; ETE, extrathyroidal extension; LNM, lymph node metastasis.
Correlations of CAFs with Genetic Mutations and Oncogenic Signaling Pathways in Thyroid Cancer
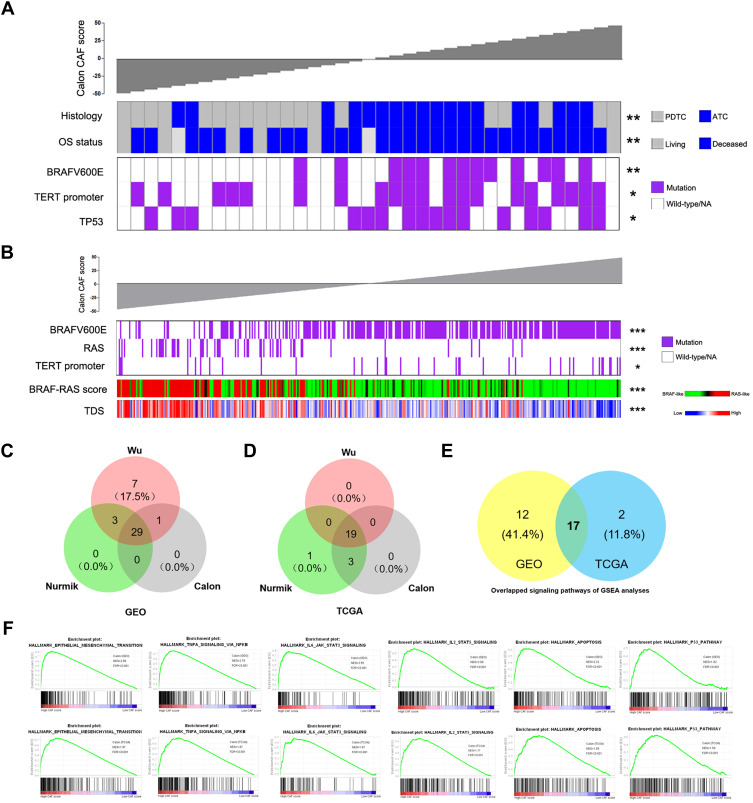
The genetic mutation data of the GSE76039 cohort available were used to investigate correlations of the CAF score with genomic changes in DDTC. The CAF score was significantly associated with BRAFV600E, TERT and TP53 mutations in the Calon signature (Table 1 and Figure 3A), which was also confirmed in the Wu signature or the Nurmik signature (Table 1, Figure-S2A and Figure-S3A). For PTC patients from the TCGA cohort, the Calon signature score showed positive correlations with BRAFV600E and TERT mutations, and exhibited a negative correlation with RAS mutation (Table 2 and Figure 3B), whereas no significant association was observed between the CAF score and EIF1AX mutation, ETV6_NTRK3 fusion, CCDC6_RET fusion and PAX8_PPARG fusion (Table 2). As shown in Table 2, Figure-S2B and Figure-S3B, the CAF score revealed a positive correlation with BRAFV600E mutation and a negative correlation with RAS mutation for both the Wu signature and the Nurmik signature as well. Additionally, the CAF score was confirmed to be remarkably associated with the BRAF-RAS score and the TDS score (Figure 3B, Figure-S2B and Figure-S3B).
Figure 3.
Correlations of the CAF score with genetic mutations and oncogenic signaling pathways based on the Calon CAF signature in thyroid cancer. (A) The DDTC patients from the GSE76039 cohort were ranked by histological subtypes, OS status, BRAFV600E mutation, TERT promoter mutation and TP53 mutation based on the CAF score. (B) The PTC patients from the TCGA cohort were ranked by BRAFV600E mutation, RAS mutation, TERT promoter mutation, BRAF-RAS score and thyroid differentiation score (TDS) based on the CAF score. (C–D) Venn diagrams showing overlapped analyses of tumor-associated signaling pathways in the high-CAF score patients of the three CAF signatures in DDTC patients from the combined GEO cohort (C) and PTC patients from the TCGA cohort (D). (E) Overlapped analyses of tumor-associated signaling pathways in (C) and (D). (F) Oncogenic signaling pathways associated with CAFs based on the Calon signature in the GEO DDTC cohort (up) and the TCGA PTC cohort (down). *p<0.05, **p<0.01, ***p<0.001.
We next performed GSEA using RNA-sequencing data of DDTC cases from the GEO cohort and PTC cases from the TCGA cohort to identify oncogenic signaling pathways associated with CAF. The high score of the CAF signature was used as the phenotype label. Among all the predefined Hallmark KEGG pathways, there were 29 signaling pathways enriched in the DDTC patients with a high-CAF score from the GEO cohort (Figure 3C and Table-S5) and 19 signaling pathways enriched in PTC patients with a high-CAF score from the TCGA cohort (Figure 3D and Table-S6). The three signatures shared a high rate of overlapped signaling pathways, and the number of enriching signaling pathways was higher in DDTC than in PTC. The further overlapped analyses revealed that CAFs enriched a total of 17 signaling pathways (Figure 3E), including a series of pathways mediating malignant transformation, such as epithelial mesenchymal transition, TNF-α signaling via NF-κB, IL6/JAK/STAT3 signaling pathway, IL2/STAT5 signaling pathway, apoptosis and TP53 signaling pathway in both DDTC and PTC patients (Figure 3F and Table-S7).
Immune Implications of CAFs in Thyroid Cancer
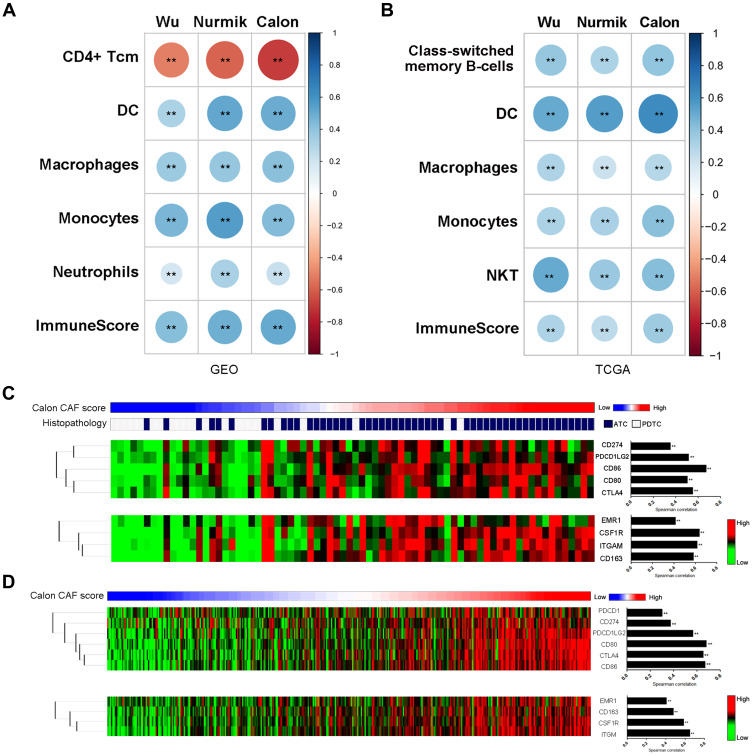
To evaluate the associations of CAFs with immune cell types in TME of thyroid cancer, we performed linear correlation analyses between CAF scores and enrichment scores of immune cell types. In both DDTC and PTC, the CAF score showed a positive correlation with the immune score and the enrichment scores of dendritic cells (DC), macrophages and monocytes (Figure 4A and B). The CAF score was negatively correlated with the enrichment score of CD4+ Tcm and positively correlated with the enrichment score of neutrophils in DDTC (Figure 4A) while class-switched memory B cells and NKT cells enriched in PTC with a high-CAF score (Figure 4B). In the further analyses, the CAF score was found to display significant associations with the key immune microenvironment markers. The patients with a high CAF score showed significantly increased expression of immune checkpoint markers including CD274, PDCD1LG2, CD86, CD80 and CTLA4 and activated tumor-associated macrophage markers including EMR1, CSF1R, CD163 and ITGM in DDTC and PTC (Figure 4C and D, Figure-S2C-2D and Supplementary Figure-S3C-3D).
Figure 4.
Impact of CAFs on immune microenvironment. (A–B) Enrichment analyses of CAF-associated immune cell types in DDTC patients from the combined GEO cohort (A) and PTC patients from the TCGA cohort (B). (C) The DDTC patients were ranked by the Calon signature score, histopathological subtypes, immune checkpoint markers and activated tumor-associated macrophage markers. (D) The PTC patients were ranked by the Calon signature score, immune checkpoint markers and activated tumor-associated macrophage markers. Spearman correlation was performed to analyse the linear correlation of the CAF score with expression value of immune microenvironment markers. **p<0.01.
Discussion
In the past decade, a number of studies have focused on CAF, and we get more deep insight into functional roles and molecular mechanism of CAF in the process of tumor progression.9 Targeting CAF is being explored as a novel therapeutic approach to improve cancer therapies. This study investigated alterations of CAFs in malignant transformation of PTC into DDTC, and further analyzed clinicopathological characteristics, genetic mutations, oncogenic signaling pathways and alterations of immune microenvironment associated with CAF in DDTC and PTC patients.
Initially, we compared the CAF score among NTs, PTCs and DDTCs from the dedifferentiated PTC cases using three CAF gene signatures, and also analyzed α-SMA expression in these cases by IHC. We further compared the CAF score among NTs, PTCs and DDTCs from the combined GEO cohort and between the low-TDS group and the high-TDS group from the TCGA cohort to validate the association of CAF with differentiation status of thyroid cancer. To our knowledge, it is the first time that CAFs have been verified to be positively correlated with dedifferentiation status of thyroid cancer.
Moreover, we analyzed the associations between the CAF score and clinicopathological characteristics in DDTC from the GSE76039 cohort and PTC from the TCGA cohort. As described in the previous studies,13,15 CAFs were confirmed to show a positive correlation with invasive behaviors of thyroid cancer such as LNM and ETE as well in our study. It was also found that the CAF score was closely related to the high-risk histological subtypes and tumor stage, and patients with a high CAF score increased the risk of decreased OS in DDTC. These results revealed that enriched presence of CAF was indicative of poor clinical outcomes in thyroid cancer.
Furthermore, we described molecular abbreviations in cancer cells and TME based on CAFs. It is found that BRAFV600E mutations significantly increase in high-CAF score patients while RAS mutations rise in low-CAF score patients in the TCGA cohort, which is consistent with the close relationship between the CAF score and the BRAF-RAS score. The positive correlations of the CAF score with the BRAFV600E, TP53 and TERT promoter mutations were obvious in DDTC, although they were not completely significant for all of the tree signatures in statistics due to the limited number of the GSE76039 cohort. It is well known that crosstalk of CAFs with tumor cells remarkably exerted impact on hallmarks of cancer.8,9 As described in our study, the high-CAF score group enriched a number of oncogenic signaling pathways compared with the low-CAF score group, suggesting that CAFs enhance aggressive behaviors of thyroid cancer through directly affecting hallmarks of cancer cells. Recently, there are a series of studies indicating that CAFs contribute to tumor progression through crosstalk with the immune microenvironment.33–36 Our study reveals that the CAFs show positive correlations with immune score and high expression of immune checkpoint markers and activated tumor-associated macrophage activated markers. These findings implicate that CAF can indirectly influence biological behavior of thyroid cancer cell by modulating immune cells in stroma.
Finally, we have to mention that some limitations exist in this study. Because the limited number of thyroid cancer patients are tested for α-SMA expression and the CAF gene signatures in the FUSCC cohort, it is necessary to validate the roles of the CAF markers and the CAF signatures in a large cohort of thyroid cancer patients with long-term follow up in our cohort. Moreover, it is not clarified concerning the functional roles and molecular mechanism of CAFs in thyroid cancer dedifferentiation and immune crosstalk. In the next-step study, we will perform experiments of thyroid cancer cells and fibroblasts in vivo and vitro to investigate biological roles.
In summary, our study found CAFs enriched in DDTC, and the CAF score was significantly associated with differentiation status and poor clinical outcomes of thyroid cancer. A close correlation was also confirmed between the CAF score and tumor driver mutations, oncogenic signaling pathways and alterations of immune microenvironment in thyroid cancer. Our findings reveal CAFs are critical markers and mediators associated with tumor progression in thyroid cancer.
Acknowledgments
We thank The UCSC Xena Platform for cancer genomics data sets, the cBioPortal for Cancer Genomics and the GEO platform of NCBI.
Funding Statement
The study was supported by the National Natural Science Foundation of China (81772854 to Qinghai Ji), the Science and Technology Commission of Shanghai Municipality (19411966600 to Yu Wang) and the Shanghai Anticancer Association (SACA-AX106 to Yu Wang and SACA-CY19B01 to Ben Ma).
Ethical Approval
Our study was approved by the Medical Ethics Committee of the FUSCC, and all patients provided written informed consents for their specimens and information to be used for research and stored in the hospital database. All procedures performed in this study were in accordance with the ethical standards of our institutional research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interests.
References
- 1.Xu B, Ghossein R. Genomic landscape of poorly differentiated and anaplastic thyroid carcinoma. Endocrine Pathol. 2016;27(3):205–212. doi: 10.1007/s12022-016-9445-4 [DOI] [PubMed] [Google Scholar]
- 2.Pozdeyev N, Gay LM, Sokol ES, et al. Genetic analysis of 779 advanced differentiated and anaplastic thyroid cancers. Clin Cancer Res. 2018;24(13):3059–3068. doi: 10.1158/1078-0432.CCR-18-0373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Landa I, Ibrahimpasic T, Boucai L, et al. Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers.. J Clin Invest. 2016;126(3):1052–1066. doi: 10.1172/JCI85271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ibrahimpasic T, Xu B, Landa I, et al. Genomic alterations in fatal forms of non-anaplastic thyroid cancer: identification of MED12 and RBM10 as novel thyroid cancer genes associated with tumor virulence. Clin Cancer Res. 2017;23(19):5970–5980. doi: 10.1158/1078-0432.CCR-17-1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma B, Xu W, Wei W, et al. Clinicopathological and survival outcomes of well-differentiated thyroid carcinoma undergoing dedifferentiation: a retrospective study from FUSCC. Int J Endocrinol. 2018;(2018):2383715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. [DOI] [PubMed] [Google Scholar]
- 7.Ferrari SM, Fallahi P, Galdiero MR, et al. Immune and inflammatory cells in thyroid cancer microenvironment. Int J Mol Sci. 2019;20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun Q, Zhang B, Hu Q, et al. The impact of cancer-associated fibroblasts on major hallmarks of pancreatic cancer. Theranostics. 2018;8:5072–5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sahai E, Astsaturov I, Cukierman E, et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer. 2020;20:174–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi H, Enomoto A, Woods SL, Burt AD, Takahashi M, Worthley DL. Cancer-associated fibroblasts in gastrointestinal cancer. Nat Rev Gastroenterol Hepatol. 2019;16:282–295. [DOI] [PubMed] [Google Scholar]
- 11.Hosein AN, Brekken RA, Maitra A. Pancreatic cancer stroma: an update on therapeutic targeting strategies. Nat Rev Gastroenterol Hepatol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Wang Y, Li D, Jing S. Notch and TGF-beta/Smad3 pathways are involved in the interaction between cancer cells and cancer-associated fibroblasts in papillary thyroid carcinoma. Tumour Biol. 2014;35:379–385. [DOI] [PubMed] [Google Scholar]
- 13.Minna E, Brich S, Todoerti K, et al. Cancer associated fibroblasts and senescent thyroid cells in the invasive front of thyroid carcinoma. Cancers. 2020;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fozzatti L, Alamino VA, Park S, et al. Interplay of fibroblasts with anaplastic tumor cells promotes follicular thyroid cancer progression. Sci Rep. 2019;9:8028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho JG, Byeon HK, Oh KH, et al. Clinicopathological significance of cancer-associated fibroblasts in papillary thyroid carcinoma: a predictive marker of cervical lymph node metastasis. Eur Arch Otorhinolaryngol. 2018;275:2355–2361. [DOI] [PubMed] [Google Scholar]
- 16.Wu X, Ruan L, Yang Y, Mei Q. Analysis of gene expression changes associated with human carcinoma-associated fibroblasts in non-small cell lung carcinoma. Biol Res. 2017;50:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nurmik M, Ullmann P, Rodriguez F, Haan S, Letellier E. In search of definitions: cancer-associated fibroblasts and their markers. Int J Cancer. 2020;146:895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calon A, Lonardo E, Berenguer-Llergo A, et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet. 2015;47:320–329. [DOI] [PubMed] [Google Scholar]
- 19.Buffa FM, Harris AL, West CM, Miller CJ. Large meta-analysis of multiple cancers reveals a common, compact and highly prognostic hypoxia metagene. Br J Cancer. 2010;102:428–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma B, Jiang H, Wen D, et al. Transcriptome analyses identify a metabolic gene signature indicative of dedifferentiation of papillary thyroid cancer. J Clin Endocrinol Metab. 2019;104:3713–3725. [DOI] [PubMed] [Google Scholar]
- 21.Kawase A, Ishii G, Nagai K, et al. Podoplanin expression by cancer associated fibroblasts predicts poor prognosis of lung adenocarcinoma. Int J Cancer. 2008;123:1053–1059. [DOI] [PubMed] [Google Scholar]
- 22.Dom G, Tarabichi M, Unger K, et al. A gene expression signature distinguishes normal tissues of sporadic and radiation-induced papillary thyroid carcinomas. Br J Cancer. 2012;107:994–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomas G, Tarabichi M, Gacquer D, et al. A general method to derive robust organ-specific gene expression-based differentiation indices: application to thyroid cancer diagnostic. Oncogene. 2012;31:4490–4498. [DOI] [PubMed] [Google Scholar]
- 24.Pita JM, Banito A, Cavaco BM, Leite V. Gene expression profiling associated with the progression to poorly differentiated thyroid carcinomas. Br J Cancer. 1782;101:2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Roemeling CA, Marlow LA, Pinkerton AB, et al. Aberrant lipid metabolism in anaplastic thyroid carcinoma reveals stearoyl CoA desaturase 1 as a novel therapeutic target. J Clin Endocrinol Metab. 2015;100:E697–E709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41:D991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. [DOI] [PubMed] [Google Scholar]
- 29.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. [DOI] [PubMed] [Google Scholar]
- 31.Choi H, Na KJ. Integrative analysis of imaging and transcriptomic data of the immune landscape associated with tumor metabolism in lung adenocarcinoma: clinical and prognostic implications. Theranostics. 2018;8:1956–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiang H, Ramil CP, Hai J, et al. cancer-associated fibroblasts promote immunosuppression by inducing ros-generating monocytic MDSCS in lung squamous cell carcinoma. Cancer Immunol Res. 2020;8:436–450. [DOI] [PubMed] [Google Scholar]
- 34.Barrett R, Pure E. Cancer-associated fibroblasts: key determinants of tumor immunity and immunotherapy. Curr Opin Immunol. 2020;64:80–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ersek B, Sillo P, Cakir U, et al. Melanoma-associated fibroblasts impair CD8+ T cell function and modify expression of immune checkpoint regulators via increased arginase activity. Cell Mol Life Sci. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ford K, Hanley CJ, Mellone M, et al. NOX4 inhibition potentiates immunotherapy by overcoming cancer-associated fibroblast-mediated CD8 t-cell exclusion from tumors. Cancer Res. 2020;80:1846–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]