Abstract
In a recent report by the Centers for Disease Control and Prevention (CDC), multidrug resistant (MDR) Acinetobacter baumannii is a pathogen described as an “urgent threat.” Infection with this bacterium manifests as different diseases such as community and nosocomial pneumonia, bloodstream infections, endocarditis, infections of the urinary tract, wound infections, burn infections, skin and soft tissue infections, and meningitis. In particular, nosocomial meningitis, an unwelcome complication of neurosurgery caused by extensively-drug resistant (XDR) A. baumannii, is extremely challenging to manage. Therefore, understanding how A. baumannii adapts to different host environments, such as cerebrospinal fluid (CSF) that may trigger changes in expression of virulence factors that are associated with the successful establishment and progress of this infection is necessary. The present in-vitro work describes, the genetic changes that occur during A. baumannii infiltration into CSF and displays A. baumannii’s expansive versatility to persist in a nutrient limited environment while enhancing several virulence factors to survive and persist. While a hypervirulent A. baumannii strain did not show changes in its transcriptome when incubated in the presence of CSF, a low-virulence isolate showed significant differences in gene expression and phenotypic traits. Exposure to 4% CSF caused increased expression of virulence factors such as fimbriae, pilins, and iron chelators, and other virulence determinants that was confirmed in various model systems. Furthermore, although CSF's presence did not enhance bacterial growth, an increase of expression of genes encoding transcription, translation, and the ATP synthesis machinery was observed. This work also explores A. baumannii’s response to an essential component, human serum albumin (HSA), within CSF to trigger the differential expression of genes associated with its pathoadaptibility in this environment.
Subject terms: Microbiology, Pathogenesis
Introduction
Acinetobacter baumannii has emerged as an important pathogen due to its ability to resist multiple antibiotics, persist in hospital settings, and cause a wide variety of infections such as pneumonia, bacteremia, urinary tract infections, skin and soft-tissue infections, and meningitis, showing its capacity to infect diverse host environments (lung, blood, CSF, etc.)1–4. The acquisition of resistance to carbapenems by certain strains (carbapenem-resistant Acinetobacter baumannii, CRAB) increased the problematic nature of this pathogen5,6, which has been qualified as an “urgent” threat in a recent report by the Centers for Disease Control and Prevention7.
Bacterial meningitis, which is considered a medical emergency, is a serious infection that can cause permanent disabilities (brain damage, hearing loss, and learning problems) or death if untreated8–10. Post neurosurgical A. baumannii meningitis is reported between ~ 10–17% of hospitalized inpatients11–13, can cause death or leave permanent sequelae, and is usually associated with high mortality rates reaching up to 40 to 70%14,15. Illustrative of these infections' dangerous nature is the recent reported case of the A. baumannii infection of a 39-year-old man treated with external ventricular drainage of cerebrospinal fluid (CSF)16. Although the strain was susceptible to colistin at the time of detection, it quickly acquired resistance while maintaining virulence16. This genetic plasticity, a consequence of its ability to acquire and integrate foreign DNA, gives A. baumannii a tremendous metabolic versatility that permits the bacterium to adapt and persist in harsh conditions2,17–21. A. baumannii's success in causing numerous infections, where it gets in contact with different body components and fluids, must be the result of its capabilities to not only capture adequate genetic determinants, but also regulate expression of the proper cell components2,6,17–19,22–24. Previously, we demonstrated that human serum albumin (HSA) and pleural fluid (HSA-containing fluid) affect A. baumannii behavior, triggering an adaptive response that modulates DNA uptake, cytotoxicity, immune evasion, stress responses and metabolism22,24–26.
Understanding the virulence determinants of this bacterium requires a thorough comprehension of the general genotypic and phenotypic responses when it is exposed to different human body fluids. As part of our studies on A. baumannii pathogenicity in relation to meningitis, we assessed in these experiments how gene expression changes when in contact with CSF. Our goal was to enhance our understanding of why this disease and pathogen are so problematic with the intent to develop better therapies for this fatal infection.
Results and discussion
CSF enhances the expression of genes involved in transcription and translation machineries, ATP production, and specific metabolic pathways without increasing growth rate
To uncover the impact of specific host environments at the transcriptional level, two different A. baumannii strains, A118 (low pathogenicity and high antibiotic susceptibility) and AB5075 (hypervirulent and multi-drug resistant), were exposed to CSF27–29. Transcriptomic analysis of A. baumannii A118, using a fold-change cutoff of log2 > 1 (with adjusted P-value < 0.05), showed 275 differentially expressed-genes (DEGs), 7.76% of the total genes in the A. baumannii A118 reference genome. However, statistically significant changes were not observed when A. baumannii AB5075, a hypervirulent and highly resistant strain, was exposed to CSF under the conditions tested. Previous observations have shown that A. baumannii’s response to different stimuli correlates with each particular strain’s degree of pathogenicity. Less pathogenic strains induced more changes in their phenotypic behavior to overcome the stressful environment and persist30.
The analysis of A. baumannii A118 DEGs revealed an increase in the expression of many genes involved in gene expression processes and energy production machineries (Table 1 and Supplementary Table S1). Notably, a large proportion of ribosomal protein genes are overexpressed upon exposure to CSF. Among the ribosomal protein associated genes, 47 out of 55 displayed a significant increase of expression of twofold or more. Coincidently, key translation genes such as those encoding elongation factors (EF) EF-G, EF-F and EF-P were also up-regulated. Concurrently, the main genes of the transcriptional machinery (RNA polymerase) were similarly overexpressed. The rpoB and rpoC genes, which code for the beta and beta' subunits of RNA polymerase (core of the transcription machinery), were overexpressed with a log2fold just below 1. However, the gene encoding the alpha subunit was also upregulated with a log2fold change of 1.48 (Table 1 and Supplementary Table S1).
Table 1.
CSF regulated genes in A. baumannii A118.
| Gene ID | Gene name | Log2fold change | p-adj | Gene associated function |
|---|---|---|---|---|
| AbA118F_3259 | bauB | 3.55 | 1.4 E-21 | Iron compound ABC uptake transporter substrate-binding protein |
| AbA118F_1017 | fimA | 3.09 | 1.2 E-160 | Fimbrial protein |
| AbA118F_3260 | bauE | 2.84 | 1.1 E-06 | putative iron compound ABC uptake transporter, ATP-binding protein |
| AbA118F_0516 | 2.81 | 3.4 E-126 | Exporter protein, RND family | |
| AbA118F_0517 | 2.79 | 9.5 E-142 | Polyketide synthase module | |
| AbA118F_0746 | 2.77 | 9.1 E-36 | hypothetical protein | |
| AbA118F_0519 | 2.58 | 2.2 E-94 | Acyl-CoA dehydrogenase | |
| AbA118F_0523 | 2.54 | 1.1 E-63 | Autoinducer synthesis protein SolI | |
| AbA118F_3262 | bauD | 2.52 | 2.9 E-02 | Iron transport protein |
| AbA118F_0514 | 2.50 | 1.2 E-117 | hypothetical protein | |
| AbA118F_0518 | 2.45 | 2.2 E-66 | Acyl carrier protein | |
| AbA118F_0515 | 2.39 | 2.4 E-65 | hypothetical protein | |
| AbA118F_0745 | 2.37 | 1.9 E-12 | Hypothetical protein | |
| AbA118F_2656 | 2.34 | 9.0 E-3 | Urease beta subunit | |
| AbA118F_3134 | fimB | 2.32 | 4.7 E-18 | P pilus assembly protein, chaperone PapD |
| AbA118F_1015 | 2.30 | 1.1 E-124 | outer membrane usher protein precursor | |
| AbA118F_2504 | 2.16 | 1.9 E-102 | Biotin carboxylase of acetyl-CoA carboxylase | |
| AbA118F_0483 | 2.14 | 1.2 E-107 | ATP synthase delta chain | |
| AbA118F_1447 | 2.12 | 6.7 E-08 | hypothetical protein | |
| AbA118F_2933 | rpmC | 2.12 | 4.5 E-85 | LSU ribosomal protein L29p (L35e)-rpmC |
| AbA118F_1014 | 2.10 | 1.5 E-78 | fimbrial adhesin precursor | |
| AbA118F_2505 | 2.09 | 1.4 E-58 | Biotin carboxyl carrier protein of acetyl-CoA carboxylase | |
| AbA118F_3136 | 2.09 | 8.8 E-19 | Fimbrial adhesin | |
| AbA118F_0481 | 2.08 | 7.4 E-108 | ATP synthase gamma chain | |
| AbA118F_2932 | 2.08 | 3.0 E-94 | SSU ribosomal protein S17p (S11e) | |
| AbA118F_0041 | 2.03 | 1.2 E-90 | 33–36 kDa outer membrane protein | |
| AbA118F_2934 | 1.99 | 1.3 E-92 | LSU ribosomal protein L16p (L10e) | |
| AbA118F_3202 | 1.97 | 1.3 E-96 | Translation elongation factor Ts | |
| AbA118F_1819 | 1.97 | 1.3 E-104 | LSU ribosomal protein L10p (P0) | |
| AbA118F_2936 | 1.96 | 1.7 E-92 | LSU ribosomal protein L22p (L17e) | |
| AbA118F_2935 | 1.96 | 2.1 E-93 | SSU ribosomal protein S3p (S3e) | |
| AbA118F_1818 | 1.93 | 5.6 E-95 | LSU ribosomal protein L7p/L12p (P1/P2) | |
| AbA118F_0484 | atpF | 1.92 | 2.7 E-90 | ATP synthase F0 sector subunit b |
| AbA118F_0113 | 1.91 | 7.9 E-58 | hypothetical protein | |
| AbA118F_3133 | 1.91 | 9.3 E-43 | Fimbrial protein precursor | |
| AbA118F_2937 | 1.90 | 1.5 E-88 | SSU ribosomal protein S19p (S15e) | |
| AbA118F_3135 | 1.89 | 3.5 E-08 | type 1 fimbriae anchoring protein FimD | |
| AbA118F_0482 | atpA | 1.88 | 7.0 E-97 | ATP synthase alpha chain |
| AbA118F_2359 | 1.87 | 1.5 E-05 | hypothetical protein | |
| AbA118F_3256 | basD | 1.86 | 2.1 E-08 | Non-ribosomal peptide synthetase modules, siderophore biosynthesis |
| AbA118F_1925 | 1.85 | 4.2 E-55 | Succinyl-CoA ligase [ADP-forming] beta chain | |
| AbA118F_0480 | atpD | 1.83 | 9.1 E-76 | ATP synthase beta chain |
| AbA118F_1016 | 1.80 | 2.1 E-67 | P pilus assembly protein, chaperone PapD | |
| AbA118F_2749 | 1.77 | 1.5 E-08 | Homocysteine S-methyltransferase-like protein | |
| AbA118F_0513 | 1.76 | 4.2 E-11 | 4′-phosphopantetheinyl transferase | |
| AbA118F_3345 | 1.76 | 1.8 E-20 | Hypothetical protein | |
| AbA118F_1919 | 1.76 | 2.5 E-14 | tRNA-Thr | |
| AbA118F_3265 | basB | 1.76 | 3.8 E-08 | Non-ribosomal peptide synthetase |
| AbA118F_2938 | 1.75 | 2.9 E-79 | LSU ribosomal protein L2p (L8e) | |
| AbA118F_2939 | 1.75 | 1.1 E-73 | LSU ribosomal protein L23p (L23Ae) | |
| AbA118F_0485 | 1.73 | 3.2 E-66 | ATP synthase F0 sector subunit c | |
| AbA118F_2311 | 1,72 | 2.3 E-49 | SSU ribosomal protein S18p : SSU ribosomal protein S18p, zinc-independent | |
| AbA118F_0479 | atpC | 1.72 | 3.8 E-64 | ATP synthase epsilon chain |
| AbA118F_3254 | basF/entF | 1.72 | 1.6 E-04 | Isochorismatase of siderophore biosynthesis |
| AbA118F_2312 | 1.71 | 1.7 E-66 | SSU ribosomal protein S6p | |
| AbA118F_1530 | 1.69 | 2.0 E-30 | hypothetical protein | |
| AbA118F_3044 | 1.68 | 5.0 E-62 | 3-oxoacyl-[acyl-carrier protein] reductase | |
| AbA118F_0520 | 1.68 | 3.7 E-41 | Polyketide synthase module | |
| AbA118F_2927 | 1.67 | 6.2 E-71 | SSU ribosomal protein S8p (S15Ae) | |
| AbA118F_3258 | bauA | 1.67 | 1.9 E-11 | Ferrichrome-iron receptor |
| AbA118F_3481 | 1.67 | 3.4 E-14 | Oxidoreductase, short-chain dehydrogenase/reductase family | |
| AbA118F_3201 | 1.66 | 2.9 E-02 | hypothetical protein | |
| AbA118F_1820 | 1.66 | 9.5E-72 | LSU ribosomal protein L1p (L10Ae) | |
| AbA118F_1821 | 1.64 | 2.3 E-69 | LSU ribosomal protein L11p (L12e) | |
| AbA118F_1340 | 1.64 | 1.5 E-30 | Ferrichrome-iron receptor | |
| AbA118F_0638 | tssL | 1,63 | 1.2 E-39 | Putative transmembrane protein |
| AbA118F_2322 | pgaB | 1.63 | 1.5 E-19 | Biofilm PGA synthesis deacetylase PgaB |
| AbA118F_2321 | pgaA | 1.63 | 2.1 E-26 | Biofilm PGA outer membrane secretin PgaA |
| AbA118F_0387 | 1.61 | 1.3 E-54 | Hemolysin | |
| AbA118F_2915 | 1.60 | 2.9 E-53 | LSU ribosomal protein L17p | |
| AbA118F_2024 | 1.59 | 9.3 E-43 | Sodium-alanine symporter family protein | |
| AbA118F_2923 | 1.59 | 4.0 E-42 | LSU ribosomal protein L30p (L7e) | |
| AbA118F_1509 | 1.59 | 1.8 E-63 | Translation elongation factor Tu | |
| AbA118F_0149 | 1.58 | 8.3 E-26 | Imidazolonepropionase | |
| AbA118F_2323 | pgaC | 1.56 | 2.8 E-13 | Biofilm PGA synthesis N-glycosyltransferase PgaC |
| AbA118F_2920 | 1.56 | 6.5 E-44 | LSU ribosomal protein L36p : LSU ribosomal protein L36p, zinc-dependent | |
| AbA118F_1621 | 1.55 | 4.6 E-54 | Glyceraldehyde-3-phosphate dehydrogenase, putative | |
| AbA118F_1623 | 1.55 | 2.0 E-43 | Gluconate transporter family protein | |
| AbA118F_3243 | 1.54 | 3.1 E-52 | hypothetical protein | |
| AbA118F_0791 | 1.54 | 1.0 E-50 | Phosphoglycerate kinase | |
| AbA118F_2940 | 1.53 | 8.5 E-60 | LSU ribosomal protein L4p (L1e) | |
| AbA118F_2921 | 1.53 | 4.0 E-65 | Protein translocase subunit SecY | |
| AbA118F_3255 | entE/basE | 1.52 | 2.5 E-06 | 2,3-dihydroxybenzoate-AMP ligase of siderophore biosynthesis |
| AbA118F_0114 | 1.52 | 5.7 E-29 | hypothetical protein | |
| AbA118F_0148 | 1.51 | 2.6 E-11 | Formiminoglutamase | |
| AbA118F_2924 | 1.51 | 1.3 E-48 | SSU ribosomal protein S5p (S2e) | |
| AbA118F_1954 | 1.50 | 7.5 E-53 | LSU ribosomal protein L21p | |
| AbA118F_3316 | -1.50 | 1.59 E-29 | Phosphate ABC transporter, periplasmic phosphate-binding protein PstS | |
| AbA118F_1136 | -1.52 | 2.8 E-04 | putative zinc-type alcohol dehydrogenase-like protein YbdR | |
| AbA118F_1643 | -1.56 | 5.3 E-04 | Twin-arginine translocation protein TatB | |
| AbA118F_0751 | -1.57 | 1.6 E-04 | TPR-repeat-containing protein | |
| AbA118F_2679 | -1.69 | 1.5 E-02 | hypothetical protein | |
| AbA118F_1644 | -1.82 | 5.8 E-05 | Twin-arginine translocation protein TatC | |
| AbA118F_2820 | -1.86 | 3.9 E-02 | Allantoin racemase | |
| AbA118F_2477 | -1.87 | 4.6 E-10 | Mg(2 +)-transport-ATPase-associated protein MgtC | |
| AbA118F_0711 | -1.89 | 5.4 E-16 | Acyl-CoA dehydrogenase | |
| AbA118F_0704 | -1.99 | 1.4 E-02 | 2-aminoethylphosphonate ABC transporter substrate-binding protein | |
| AbA118F_2199 | -2.06 | 2.4 E-04 | Protein co-occuring with molybdenum cofactor biosynthesis protein B | |
| AbA118F_0907 | -2.14 | 5.2 E-20 | outer membrane porin, putative | |
| AbA118F_2367 | -2.30 | 4.0 E-02 | Phage tail/DNA circulation protein | |
| AbA118F_1645 | -2.39 | 1.2 E-27 | Alkaline phosphatase | |
| AbA118F_1190 | -2.62 | 4.9 E-02 | Lysozyme (N-acetylmuramidase) family | |
| AbA118F_1376 | -2.93 | 3.4 E-02 | Lysozyme (N-acetylmuramidase) family | |
| AbA118F_3137 | -3.07 | 2.8 E-03 | hypothetical protein | |
| AbA118F_2475 | -3.31 | 2.0 E-08 | hypothetical protein | |
| AbA118F_2476 | -3.83 | 2.7 E-11 | Mg(2 +) transport ATPase, P-type |
The table lists all A. baumanii A118 ORFs that are regulated by CSF with a Log2FC ≥|1.5|, p < 0.05.
In addition, genes important for energy production in the cell were also upregulated upon CSF exposure. The atpIBEFHAGCD locus, an operon encoding the FoF1-ATP synthase (the main ATP generator in the bacterial cell) displayed a threefold transcriptional increase in expression (Table 1 and Supplementary Table S1). These transcriptional responses suggest that when CSF is present in the environment, there is an increase in expression of transcription- and translation-related genes, as well as of FoF1-ATP synthase, the main ATP generator. Also, CSF exposure induces the transcription of specific metabolic routes in A. baumannii. In particular, several dehydrogenases of the tricarboxylic acid cycle intermediates as well as a citrate symporter were significantly overexpressed, together with two proline symporters (Supplementary Table S1). The transcriptomic data showed that type I glutamine synthetase (AbA118F_3228) and the proline symporter putP genes are upregulated by a log2fold change of 0.58 and 0.87 respectively.
Studies on Salmonella typhimurium showed that putP codes for a proline permease, an integral membrane protein, that is the primary transport protein when this amino acid is the only carbon or nitrogen source31. The transcriptomic analysis showed upregulation (log2fold change 0.62) of AbA118F_2664, a gene that encodes a CitMHS citrate-magnesium hydrogen complex symporter (Supplementary Table S1). Proteins of the CitMHS family transport citrate-Mg2+ complex coupled with one proton per complex molecule32. Interestingly, increased citrate levels help survival of A. baumannii in certain conditions26. Thus, the net effect of CSF might be an increase in expression of this transporter, which would lead to higher citrate intracellular concentrations that may result in increased rate of growth.
While most of the DEGs were upregulated, only 66 showed a lower transcription level. Among the most downregulation genes we found those coding for transporters (e.g. Mg2 + transporters, AbA118F_2476 and AbA118F_2477) and catabolic proteins (such as AbA118F_1645 Alkaline Phosphatase and AbA118F_0711 Acyl-CoA dehydrogenase) (Table 1 and Supplementary Table S1).
Gene ontology (GO) analysis was next undertaken to identify molecular functions and biological pathways associated to A. baumannii’s adaptive responses to CSF. Consistent with the above mentioned DEGs, GO enrichment analysis revealed a statistically significant overrepresentation of the GO categories ATP synthesis coupled proton transport, translation, tricarboxylic acid cycle, and aerobic respiration by 13.8-, 8.8-, 4.9- and 4.3-fold, respectively (adj. P-value < 0.05). Other studies found that exposure of A. baumannii strains to amikacin, imipenem, and meropenem was associated with increased expression of genes involved in the tricarboxylic acid cycle, biosynthesis of amino acids, purines, and pyrimidines, as well as the operons related to ATP, RNA, and protein synthesis33. Taken together, these results suggest that various stressful environments (low nutrient availability and antibiotic treatment) induce expression of genes associated to energy production, protein synthesis, and metabolism in A. baumannii. As a consequence of these transcriptomic changes, the bacterial cell adapts to survive and accelerate metabolism rive in these hostile conditions.
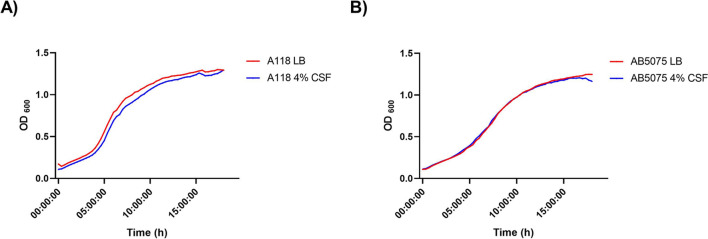
The CSF-mediated upregulation of genes coding for the elements necessary for transcription, translation, expression and ATP synthesis was not accompanied by a decrease in generation time (Fig. 1). These results suggest that cells respond to CSF enhancing the expression of pathways that produce specific effects rather than increasing growth capacity.
Figure 1.
A118 and AB5075 growth curves in LB or LB plus 4% CSF. Strains (A) A118 and (B) AB5075 were grown in LB or LB supplemented with 4% CSF. Growth curves were conducted in independent experimental triplicates.
There are numerous reports supporting the hypothesis that increasing the expression of enzymes involved in transcription, translation, and synthesis of ATP is correlated with faster growth rate28–32. However, these growth differences were not evident in either of the A. baumannii strains in the presence of CSF. An attractive notion to explain this observation is that the increase in gene expression capabilities is channeled toward the synthesis of cell components necessary for survival in the human body, e.g., adhesins and pilins.
The data described in this set of investigations indicate that certain modifications in the A. baumannii metabolism are uncoupled from growth rate. This is not anticipated because bacterial cells are characterized by allocating resources to maximize growth according to the needs for each environmental condition34. Our data also suggest that in depleted medium such as CSF, A. baumannii may be allocating all possible resources towards metabolism using an uncoupled metabolism to optimize its survival.
CSF affects the expression of A. baumannii virulence genes
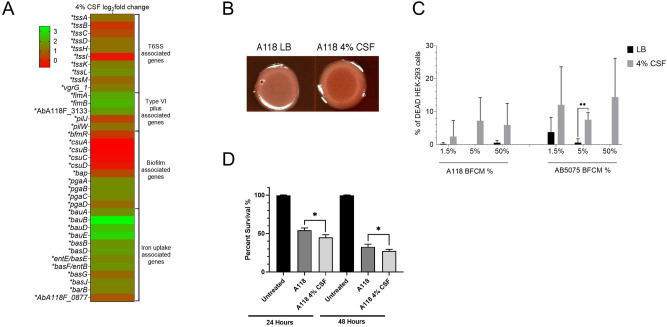
We next observed that the addition of CSF to A. baumannii A118 cultures induced an increase in the expression of a set of genes that code for virulence-associated functions such as type IV pili, iron uptake systems, the type VI secretion system (T6SS), and poly-N-acetylglucosamine (PNAG) production.
Type IV pili participate in microbial adherence as well as motility (gliding or twitching). While A. baumannii lacks flagellum-mediated motility, twitching and surface-associated motility was demonstrated in several strains35,36. Numerous studies on twitching and surface-associated motility in A. baumannii A118 showed dependence on changes in light and temperature27 as well as on the components of the growth media. In particular, addition of HSA resulted in increased motility and concomitantly upregulation of the cognate genes26.
Exposure of A. baumannii A118 to CSF produced an increase in the expression of pilW (log2fold change 1.22), pilJ (log2fold change 0.43), fimA (log2fold change 3.09), fimB (log2fold change 2.32), and the fimbrial protein precursor AbA118F_3133 (log2fold change 1.91) (Fig. 2A). All of the type IV fimbriae genes have been experimentally shown to be associated with motility, cell adhesion, and biofilm formation37,38. In addition, our transcriptomic data showed reduced expression in the biofilm associated genes csuABCD, the two-component system response regulator bfmR, and the bap ortholog (biofilm-associated protein, see Supplementary Table S1). Significant differences were not observed in biofilm formation or motility in the presence of CSF (Fig. S1). This result is in contrast to our previous studies with 4% pleural fluid. The absence of changes in biofilm formation and motility can be explained by the different compositions of each fluid. Pleural fluid is clearly inducing more changes than CSF when comparing the number of genes affected by both fluids (1120 vs 275, respectively). Components such as neutrophils, lymphocytes, monocytes, proteins, reactive oxygen species, and neutralization agents are found in pleural fluid and could contribute to the different effect22.
Figure 2.
Exposure to CSF can affect multiple virulence factors in A. baumannii. (A) Heat map of multiple virulence factor associated genes that were differentially expressed in A. baumannii strain A118. Asterisks represent a P-value of < 0.05. (B) Poly-N-acetylglucosamine (PNAG) assays were conducted with strains A118 in LB or LB supplemented with 4% CSF. (C) Percentage viability of HEK-293 cells under exposure to various concentrations of A. baumannii strains A118 or AB5075 supplemented with or without 4% CSF. (D) Percentage survival of Galleria mellonella when inoculated with A. baumannii strain A118 with or without 4% CSF.
The presence of CSF was also correlated with higher expression of twelve genes associated with the acinetobactin iron uptake system (Fig. 2A and Table S1). These genes are part of the ferric-acinetobactin receptor-translocation machinery (bauABDE, bauA log2fold change 1.67), the acinetobactin biosynthesis (basBDFGJ, basD log2fold change 1.86) and export (barB)39,40 (Fig. 2A and Table S1). Besides their direct role in iron uptake in the iron starvation conditions found in the human host, the products of basD and basA are needed for A. baumannii to persist and cause apoptosis of human alveolar epithelial cells41. Bacterial iron uptake systems that are virulence factors are usually highly regulated and are induced under conditions of iron starvation. The transcriptomic data showed that, genes that code for functions in siderophore biosynthesis, export, and import are upregulated of in the presence of CSF. This finding adds another regulatory signal that enhances expression of acinetobactin iron uptake system. This increase in expression could be directly related to growth in the host or to biofilm formation, which is dependent on efficient iron uptake42.
The structures of bacterial biofilms are usually dependent on polysaccharides such as poly-β-1,6-N-acetyl-d-glucosamine (PGA) or cellulose. Previous studies showed that functional production of PGA in Escherichia coli depends on the products of four genes, pgaABCD. pgaC and pgaD are essential for biosynthesis, and pgaB, which specifies a N-deacetylase, together with pgaA are needed to export the polysaccharide from the periplasm to the extracellular milieu43. All four homologs were significantly upregulated when A. baumannii A118 was cultured in the presence of CSF (log2fold change of 1.63, 1.63, 1.56 and 1.08 for pgaA, pgaB, pgaC and pgaD, respectively) (Fig. 2A and Table S1). As expected, Congo red staining showed that A. baumannii A118 cells cultured in the presence of CSF produced higher levels of PGA (Fig. 2B).
Another system known to be involved in A. baumannii’s virulence is the T6SS44. The transcriptomic data showed that nine out of the 14 A. baumannii A118 T6SS genes were significantly upregulated in the presence of CSF. These genes tssABCDHIKLM, code for essential components of the T6SS (Fig. 2A and Table S1).
Other genes of interest related with resistance and pathogenesis of A. baumannii were analyzed. We analyzed the genes involved with antibiotic resistance, quorum sensing, osmotic stress, DNA damage, outer membrane vesicle production, and capsule formation. While many were not found to be differentially expressed, a non-statistically significant upregulation of carO*, which is involved in selective uptake of basic amino acids and also found to be related with carbapenem resistance, was observed45. A non-statistically decrease in expression of blaOXA-69, β-lactamase found in A118 genome, was also observed. Only abaI, which codifies for the autoinducer synthesis protein46, was differentially expressed among the quorum sensing genes, while the rest where downregulated with the exception of fadD (See Fig. S2). In addition, we observed that the osmotic stress regulators, bet (a high-affinity choline uptake protein) and betI (a transcriptional regulator) were differentially upregulated by a log2fold change of -1.20 and -1.40, respectively. For genes associated with capsule formation (K-locus), pgm which encodes a phosphomannomutase was upregulated by 1.08 log2fold (P-value = 2.16 E-21). Regarding the SOS response associated genes, any of them was differentially expressed (See Fig. S2. This result differs from our previous work with pleural fluid exposure, where we found an overrepresentation of the expression of SOS response associated genes difference that can be explain by the presence of reactive oxygen species in pleural fluid.
Lastly, among the genes related with outer-membrane vesicle (OMV) production, ompA was the only differentially expressed (Fig. S2). OmpA is known to have a cytotoxic effect and is considered a key virulence factor associated with bacterial biofilm formation, eukaryotic cell infection, resistance to antibiotics and also poses immunomodulatory effects22,47.
CSF enhances the release of A. baumannii’s cytotoxic agents
An initial assessment of the effect of CSF on A. baumannii virulence was determined using cytotoxicity assays. Bacteria-free conditioned medium (BFCM) obtained from A. baumannii A118 and AB5075 cultured in LB with or without CSF was added to human embryonic kidney cells (HEK-293), and the cells were inspected after 1 h.
Figure 2C shows that BFCM samples obtained from CSF-containing A. baumannii AB5075 cultures were significantly more cytotoxic than BFCM from cultures that lacked CSF. This increase in cytotoxicity was observed at all tested concentrations, 1.5% BFCM (P-value = 0.006), 5% BFCM (P-value = 0.001), and 50% BFCM (P-value < 0.0001). Conversely, BFCM obtained from A. baumannii A118 cultures containing CSF showed an increased cytotoxic effect only at the highest concentration tested (50%) (P-value = 0.002). The results of these assays show that CSF induces the release of one or more cytotoxic substances by A. baumannii (Fig. 2C).
CSF-treatment increases A. baumannii virulence
The effect of CSF on A. baumannii’s virulence was tested using the Galleria mellonella model41,48. Infection with A. baumannii A118 cultured in LB plus 4% CSF resulted in increased mortality compared to the infection with cells cultured in LB (Fig. 2D). These results were consistent with the transcriptional changes in expression of virulence genes observed in vitro.
HSA contribution in A. baumannii pathoadaptation when exposed to CSF
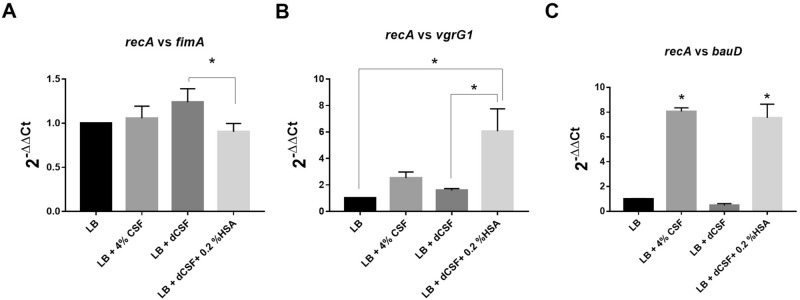
Previous work showed that the presence of pleural fluid (PF) is correlated with modifications of the expression of more than 1100 A. baumannii genes including many virulence factors involved in motility, biofilm formation, efflux, T6SS, fibrinolytic activity and capsule genes22, and with an increase in cytotoxicity and immune evasion30. The experiments shown in previous sections indicate that CSF produces effects similar to those observed with PF such as increase in cytotoxicity and changes in expression of virulence genes. Both fluids, PF and CSF, share as a component, HSA. Thus, we posited that this component may be responsible for the effects produced by both human fluids. To test this hypothesis, we compared levels of expression of the Type 1 fimbrial protein FimA, the iron transport protein BauD, and the Type IV secretion protein VgrG in A. baumannii A118 cells cultured in LB or LB supplemented with either CSF or HSA.
RNA-seq showed that bauD and vgrG, were up-regulated in both, CSF (log2fold-change = 2.5247 and 1.3611 respectively) and HSA (log2fold-change = 0.8046 and 1.1063 respectively)26. On the other hand, the fimA gene was up-regulated with a log2 fold change of 3.0938 in the presence of LB supplemented with CSF but surprisingly, it was slightly down-regulated in medium supplemented with HSA (log2 fold change of 1.6434) (non-statically significant considering a P-value < 0.05)26. While the results obtained with bauD and vgrG seemed to uphold the hypothesis, those produced when assessing expression of fimA did not seem to support that HSA is responsible for CSF-induced upregulation.
To confirm the results obtained by RNA-seq, another set of experiments was carried out measuring levels of expression by quantitative PCR using total RNA from A. baumannii A118 cells cultured in LB or LB supplemented with either CSF, HSA-depleted CSF (dCSF), or dCSF + 0.2% HSA (Fig. 3A-C). Expression of vgrG1 was slightly up-regulated in presence of 4% CSF (2.527-fold; P-value 0.4113) and dCSF (1.586-fold; P-value 0.9040). However, supplementation of LB with dCSF + 0.2% HSA, resulted in a robust increase of 6.065-fold (P-value 0.0152) and 3.825-fold (P-value 0.0234) with respect to LB or LB supplemented with dCSF, respectively (Fig. 3B). Expression of bauD was increased by 8.064-fold (P-value 0.0009) and 7.544-fold (P-value 0.0012) in cells growing in LB supplemented with CSF or dCSF + 0.2% HSA, respectively. Moreover, a 15.847-fold increase (P-value 0.0009) was observed in levels of expression of bauD in cells growing in LB supplemented with dCSF + 0.2% HSA with respect to those in cells growing in LB supplemented with dCSF (Fig. 3C). Conversely, while a slight increase in fimA expression was noted in cells growing in LB supplemented with CSF (1.057-fold, P-value 0.9190) or dCSF (1.238-fold, P-value 0.1349), there was a decrease of 1.107-fold (P-value 0.6526) when the medium was LB supplemented with dCSF + 0.2% HSA. A comparison between levels of expression in cells growing in LB supplemented with dCSF + 0.2% HSA or dCSF showed that the presence of HSA resulted in 1.370-fold decrease (P-value 0.0384) (Fig. 3A). These results confirmed that while HSA may be responsible for increasing expression of certain genes, it is not for others like fimA. In fact, it seems to induce a weak but consistent inhibition of expression of fimA. Future studies will need to carry out to determine if other components of CSF produce an increase in fimA expression.
Figure 3.
HSA is an essential component for the differential expression of genes in A. baumannii. A. baumannii A118 cells were cultured in LB or LB supplemented with one of the following: 4% CSF, HSA-depleted CSF (dCSF), or dCSF + 0.2% HSA and its cDNA was synthesized. RT-qPCR was conducted with three genes (A) fimA encoding gene, (B) vgrG1, and (C) bauD. Shown are the means of the results obtained from three independent experiments. The y axis refers to the fold difference of each gene to the threshold cycle (CT) values corresponding to recA; the standard deviation SD is shown. Asterisks indicate significant differences among treatments, as determined by ANOVA followed by Tukey’s multiple comparison test (P < 0.05).
Bacterial cells express genes that code for factors that allow growth in the hostile environments they encounter upon invading the human body. The results shown in this section indicate that HSA is one of the signals that triggers the expression of several A. baumannii genes when the bacterial cells are in contact with HSA-containing fluids, PF or CSF. Furthermore, our previous studies showed that HSA also enhances DNA acquisition through modulation of natural competence-related gene expression and affects the expression of genes related to motility, efflux pumps, pathogenicity and antibiotic resistance, among others23,26. These characteristics are not unique to A. baumannii, various bacterial pathogens and protozoa49–52 modify gene expression to adapt and thrive within the host utilizing HSA as one of the signals. For example, in Bordetella pertussis, the causative agent of the whooping cough, albumin combined with calcium induces an increase in production and release of the major toxin, adenylate cyclase toxin (ACT)52. Another example is the case of Pseudomonas aeruginosa, in which the presence of albumin is correlated with increased expression of iron-controlled genes (pvdS and regA)53. In summary, our observations, together with the evidence available from studies with other bacteria, suggest an important role of HSA as signal for expression of genes and systems essential for survival within the human body.
Conclusion
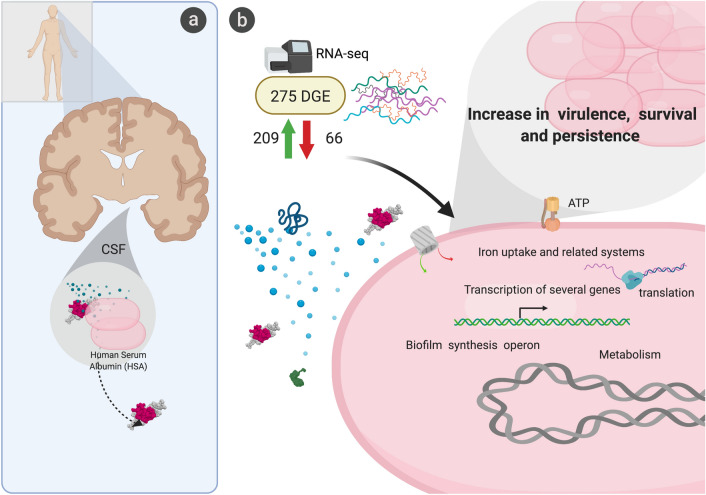
A. baumannii is one of many causative agents of nosocomial bacterial meningitis, an infection associated with high morbidity and mortality rates. During the infection, bacteria can be found in CSF. In fact, of the several methods available for diagnosis of bacterial meningitis, CSF culture is the most favored54. This study describes changes in expression of numerous genes when A. baumannii is exposed to CSF (Fig. 4). These genes code for a variety of proteins that participate in the gene expression machinery, energy production, motility, metabolism, survival, and virulence factors among others. Our results in combination with previous work suggest that HSA may be a contributor in signaling these transcriptomic responses (Fig. 4). HSA is one of the main components of CSF and is also present in blood and PF, all body fluids that trigger similar responses in bacteria. Utilizing HSA as the signal for gene expression of elements that facilitate progression of the infection is an intelligent strategy that permits bacteria to sense the presence of human environments. However, not all strains respond equally when HSA is present, slight differences were identified when comparing A. baumannii strains A118 and AB5075. These changes are correlated with differences in levels of pathogenicity and probably the kind of infections that are more commonly caused by each variant.
Figure 4.
Graphical representation of A. baumannii’s transcriptomic and behavioral response to CSF. (A) General representation showing possible association of colonized A. baumannii in CSF towards HSA. (B) Hypothetical schematic showing possible role of HSA in inducing differential gene expression in A. baumannii. The differential expression of genes associated with metabolism, iron uptake systems, and translation machinery, among other, were found in the 275 DEGs. Created with BioRender.com.
Another remarkable effect of HSA on A. baumannii is the augmentation of natural competency26,55. Traglia et al. proposed that a random coil stretch in the structure of HSA is responsible for increasing the ability of A. baumannii to take up DNA. Identifying factors that are similar between human host environments may be potential therapeutic target candidates. Considering the pleiotropic effects caused by the presence of HSA on A. baumannii, an alternative path to design therapeutic agents against this infection could be to identify compounds that interfere with the ability of A. baumannii to detect HSA. Compounds that interact with the HSA regions that are detected by A. baumannii could mask the presence of the protein impeding expression of the necessary systems for survival and progression of the infection.
Material and methods
Bacterial strains and human fluids
Two A. baumannii strains already used in previous studies27–29, exhibiting different degree of susceptibility and virulence were used. A. baumannii A118 strain is known to be susceptible to variety of antibiotics28,56 and A. baumannii AB5075 possesses increased virulence and is resistant to many antibiotics29.
The Pooled Human Cerebrospinal Fluid (CSF) sample was acquired from Innovative Research, MI, which is a certified vendor that obtains human samples from Food and Drug Administration (FDA)-approved facilities. The samples were collected from normal healthy individual donors and pooled. Lysogeny Broth, LB, supplemented with 4% CSF was used for all CSF conditions. The following concentration was used since 4% of other human fluids has been used in previously studies22,30,57.
RNA extraction and sequencing
A. baumannii colonies (A118 and AB5075) were suspended in LB with or without 4% CSF and incubated with agitation for 18 h at 37 °C. Overnight cultures were then diluted 1:10 in fresh LB broth and incubated with agitation for 7 h at 37 °C. RNA was extracted from each strain using the TRI REAGENT Kit (Molecular Research Center, Inc., Cincinnati, Ohio, USA) as previously described26. Total RNA extractions were performed in two biological replicates for each condition.
RNA sequencing was outsourced to Novogene (Novogene Corporation, CA) for mRNA-seq analysis, which includes rRNA depletion, library preparation following the protocols of the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (New England Biolabs) (New England Biolabs) and HiSeq 2500 paired-end 150 bp sequencing.
RNA–seq data analysis
RNA-seq reads (GEO accession No GSE153967) corresponding to A. baumannii strain A118 and AB5075 exposed to LB or LB plus 4% CSF were analyzed as follows. Trimming of low-quality bases at the ends of the reads to a minimum length of 100 bp and removal of Illumina adaptor sequences was performed using Trimmomatic58. FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used to assess the quality of the reads before and after trimming. Burrows-Wheeler Alignment software (BWA) was used to align the RNA-seq reads to sequences of the whole genome shotgun sequencing project of strain Acinetobacter baumannii A118F (DDBJ/ENA/GenBank accession VCCO01000000). FeatureCounts was used to calculate the read counts per gene, and differential expression analysis was performed using DEseq259,60. Principal component analysis (PCA) and gene expression heat map with clustering dendrograms of the RNA-seq data analysis of LB and CSF treatments are shown in Supplementary Fig. S3. Features exhibiting FDR < 0.05 and log2fold change > 1 were considered statistically significant.
Gene ontology (GO) analysis
GO terms were retrieved from UniProt for the best BLASTx hits to A. baumannii A118F genes. Using GO.db Bioconductor annotation data package in R language, GO terms and ancestor terms were assigned for all DEGs from this study. GO enrichment analysis was performed using custom-made scripts as described previously61. The enrichment factor was estimated as the ratio between the proportions of genes associated with a particular GO category present in the dataset under analysis, relative to the proportion of the number of genes in this category in the whole genome. P-values were calculated using the Fisher Exact Test and adjusted by the Benjamini–Hochberg method.
Growth curves
Growth curves of both strains, A118 and AB5075, were conducted on 96-well plates. Overnight cultures were subcultured 1:50 in LB or LB + 4% CSF and incubated for 18 h at 37 °C with medium shaking. OD600 nm was measured every 20 min using a Synergy 2 multi-mode plate reader (BioTek, Winooski, VT, USA) and Gen5 microplate reader software (BioTek). To study the effect of different carbon sources, both strains were culture overnight under different condition (LB broth and LB broth + 4% CSF).
Motility and biofilm assays
Motility and biofilms assays were performed as previously described26. A118 and AB5075 cells were cultured in LB broth with or without 4% CSF. Bacterial cells were incubated with agitation for 18 h at 37 °C. Experiments were performed in triplicate, with at least three technical replicates per biological replicate.
Cytotoxicity assays
In a Nunclon Delta Surface opaque 96-well microplate (ThermoScientific), we added colorless DMEM, 4% CSF, and A118 or AB5075 BFCM diluted in LB broth to make 50 μL of BFCM at final concentrations of 1.5%, 5%, and 50%. An additional 50 μL of ATCC HEK-293 cells at a concentration of 1 × 106 cells/mL in colorless DMEM were suspended in the well and intoxicated for 1 h at 37 °C, 5% CO2. CellTiter-Glo Reagent (100 μl) was added to each experimental and standard curve well and then placed on an orbital shaker for 2 min. Following mixing, plates were incubated at room temperature for 10 min to stabilize the luminescent signal. The viability of HEK cells was measured at room temperature using the “all” luminescence function of SpectraMax M3.
Galleria mellonella infection model
To assess the virulence of A. baumannii with and without CSF in vivo, the G. mellonella insect model of infection was used62. Larvae weighing between 200 and 400 mg were maintained on wood chips in the dark at 4 °C. A. baumannii A118 was grown overnight in either LB or LB with 4% CSF. An equivalent of 1.0 OD600 unit of overnight culture was pelleted and resuspended in 1 mL of cold sterile 20 mM phosphate buffered saline, pH 7.4 (PBS). The cells were further diluted 1:10 in sterile PBS and used for injections. A Hamilton syringe was used to inject 5 μL of the diluted bacterial suspension via the left proleg of each larva. A control group of untreated larvae was used to assess overall larval viability for the duration of the assay. One hundred G. mellonella larvae were used in each condition and incubated at (37 °C) in a sterile Petri dish for 24 h intervals for 48 h total. Larvae viability was monitored by observing response to gentle prodding with a glass rod; those with no response were considered dead. Four replicates with 100 larvae per Petri dish were performed for each condition.
PNAG assays
To study extracellular matrix (ECM) production, microcolony biofilm was used as model system. 5 μl of overnight cultures of A118 cultured in LB broth and LB broth + 4% CSF were inoculated on LB agar and supplemented with Congo red as previously described63. Plates were incubated at 28 °C in static incubator for up to 48hs. Results were recorded at 24 h with a Plugable USB 2.0 Digital Microscope.
HSA depletion
HSA was depleted from CSF by placing 1 mL of CSF into a 30 kDa Amicon Ultra Centrifugal Filter (Millipore, Temecula, CA, United States) and the solution was centrifuged at 20,000 × g for 10 min. To identify HSA was successfully depleted, an SDS-PAGE was conducted that contained 4% CSF, depleted CSF (dCSF), and dCSF plus 0.2% HSA (Fig. S4).
RT-qPCR
Previously extracted and DNase-treated RNA from A. baumannii strain A118 grown in LB, 4% CSF, 4% depleted CSF and 4% depleted CSF + 0.2% HSA, were synthesized to cDNA using the manufacturer protocol provided within the iScript Reverse Transcription Supermix for qPCR (Bio-Rad, Hercules, CA, United States). The cDNA concentrations were measured with a DeNovix DS-11 + spectrophotometer; each sample was then diluted to a concentration of 50 ng/μl. qPCR was conducted using the iQ SYBR Green Supermix through the manufacturer’s instructions. At least three biological replicates of cDNA were used and were run in quadruplet. All samples were then run on the CFX96 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, CA, United States).
The transcript levels of each sample were normalized to the recA rRNA transcript levels for each cDNA sample. The relative quantification of gene expression was performed using the comparative threshold method 2−ΔΔCt. The ratios obtained after normalization were expressed as folds of change compared with cDNA samples isolated from bacteria cultures on LB. Asterisks indicate significant differences as determined by ANOVA followed by Tukey’s multiple comparison test (P < 0.05), using GraphPad Prism (GraphPad software, San Diego, CA, United States).
Statistical analysis
All experiments were performed at least in technical and biological triplicate. Data was expressed as means ± standard deviation. Statistical analysis using Mann–Whitney test or ANOVA followed by Tukey’s multiple comparison test were performed using GraphPad Prism (GraphPad software, San Diego, CA, USA), and a P-value < 0.05 was considered statistically significant.
All procedures performed in this study were in accordance with the CSUF Institutional Biosafety Committee Approval plan (DBH117-01) and are in compliance with the NIH, CDC, OSHA and other environmental and occupational regulations.
Supplementary information
Author contributions
J.M., P.S., A.S.B., K.M.P.W, R.S., S.A.B, and M.S.R conceived the study and designed the experiments. J.M., C.R.G., C.L., R.C., C.P., C.L., S.E.F., M.R.T., P.S., K.P., V.J., S.A.B, K.M.P.W., A.S.B., R.S., and M.S.R. performed the experiments and genomics and bioinformatics analyses. J.M., R.C., S.E.F, M.R.T., K.M.P.W., A.S.B., R.S., and M.S.R. analyzed the data and interpreted the results. P.S., V.J., M.E.T., K.M.P.W., R.A.B., R.S., and M.S.R. contributed reagents/materials/analysis tools. J.M., C.R.G.., C.L., R.C., C.P., S.E.F., A.J.V., P.S., V.J., M.E.T., K.M.P.W., R.A.B., A.S.B., R.S., and M.S.R. wrote and revised the manuscript. All authors read and approved the final manuscript.
Funding
The authors’ work was supported by NIH SC3GM125556 to MSR, 2R15AI047115 to MET, R01AI100560 to RAB and AJV, R01AI063517, R01AI072219 to RAB, and Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT) to AJV. This study was supported in part by funds and/or facilities provided by the Cleveland Department of Veterans Affairs, VA Merit Review Award Numbers 1I01BX001974 to RAB and 1I01 BX002872 to KMPW from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development, and the Geriatric Research Education and Clinical Center VISN 10 to RAB. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Department of Veterans Affairs. A.J.V., AS-B and RS are staff members from CONICET.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-81714-6.
References
- 1.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 2008;21:538–582. doi: 10.1128/cmr.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roca I, Espinal P, Vila-Farres X, Vila J. The Acinetobacter baumannii oxymoron: commensal hospital dweller turned pan-drug-resistant menace. Front. Microbiol. 2012;3:148. doi: 10.3389/fmicb.2012.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong D, et al. Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin. Microbiol. Rev. 2017;30:409–447. doi: 10.1128/CMR.00058-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hartstein AI, et al. Multiple intensive care unit outbreak of Acinetobacter calcoaceticus subspecies anitratus respiratory infection and colonization associated with contaminated, reusable ventilator circuits and resuscitation bags. Am. J. Med. 1988;85:624–631. doi: 10.1016/s0002-9343(88)80233-x. [DOI] [PubMed] [Google Scholar]
- 5.Ramirez MS, Bonomo RA, Tolmasky ME. Carbapenemases: transforming Acinetobacter baumannii into a yet more dangerous menace. Biomolecules. 2020 doi: 10.3390/biom10050720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin DL, et al. Functional analysis of the Acinetobacter baumannii XerC and XerD site-specific recombinases: potential role in dissemination of resistance genes. Antibiotics (Basel) 2020 doi: 10.3390/antibiotics9070405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.CDC. Antibiotic Resistance Threats in the United States. Atlanta, GA: U.S. Department of Health and Human Services, CDC; 2019 (2019).
- 8.Erdem I, et al. Clinical features, laboratory data, management and the risk factors that affect the mortality in patients with postoperative meningitis. Neurol India. 2008;56:433–437. doi: 10.4103/0028-3886.44629. [DOI] [PubMed] [Google Scholar]
- 9.Sacar S, et al. A retrospective study of central nervous system shunt infections diagnosed in a university hospital during a 4-year period. BMC Infect. Dis. 2006;6:43. doi: 10.1186/1471-2334-6-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tuon FF, Penteado-Filho SR, Amarante D, Andrade MA, Borba LA. Mortality rate in patients with nosocomial Acinetobacter meningitis from a Brazilian hospital. J Brazilian J. Infect. Dis.. 2010;14:437–440. doi: 10.1016/S1413-8670(10)70090-8. [DOI] [PubMed] [Google Scholar]
- 11.Alvarez-Vega M, et al. Multidrug-resistant Acinetobacter meningitis in neurosurgical patients with intraventricular catheters: assessment of different treatments-authors' response. J. Antimicrob. Chemother. 2020;75:783–784. doi: 10.1093/jac/dkz541. [DOI] [PubMed] [Google Scholar]
- 12.assessment of different treatments. Rodriguez Guardado A, et al. Multidrug-resistant Acinetobacter meningitis in neurosurgical patients with intraventricular catheters. J. Antimicrob. Chemother. 2008;61:908–913. doi: 10.1093/jac/dkn018. [DOI] [PubMed] [Google Scholar]
- 13.Chang JB, et al. Prevalence and antibiotic resistance of bacteria isolated from the cerebrospinal fluid of neurosurgical patients at Peking union medical college hospital. Antimicrob. Resist. Infect. Control. 2018;7:41. doi: 10.1186/s13756-018-0323-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huttova M, et al. Postsurgical meningitis caused by Acinetobacter baumannii associated with high mortality. Neuro. Endocrinol. Lett. 2007;28(Suppl 2):15–16. [PubMed] [Google Scholar]
- 15.Metan G, Alp E, Aygen B, Sumerkan B. Acinetobacter baumannii meningitis in post-neurosurgical patients: clinical outcome and impact of carbapenem resistance. J. Antimicrob. Chemother. 2007;60:197–199. doi: 10.1093/jac/dkm181. [DOI] [PubMed] [Google Scholar]
- 16.López-Rojas R, Jiménez-Mejías ME, Lepe JA, Pachón J. Acinetobacter baumannii resistant to colistin alters its antibiotic resistance profile: a case report from Spain. J. Infect. Dis. 2011;204:1147–1148. doi: 10.1093/infdis/jir476. [DOI] [PubMed] [Google Scholar]
- 17.Jawad A, Seifert H, Snelling AM, Heritage J, Hawkey PM. Survival of Acinetobacter baumannii on dry surfaces: comparison of outbreak and sporadic isolates. J. Clin. Microbiol. 1998;36:1938–1941. doi: 10.1128/JCM.36.7.1938-1941.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juttukonda LJ, Chazin WJ, Skaar EP. Acinetobacter baumannii coordinates urea metabolism with metal import to resist host-mediated metal limitation. MBio. 2016 doi: 10.1128/mBio.01475-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mortensen BL, Skaar EP. The contribution of nutrient metal acquisition and metabolism to Acinetobacter baumannii survival within the host. Front. Cell Infect. Microbiol. 2013;3:95. doi: 10.3389/fcimb.2013.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imperi F, et al. The genomics of Acinetobacter baumannii: Insights into genome plasticity, antimicrobial resistance and pathogenicity. IUBMB Life. 2011;63:1068–1074. doi: 10.1002/iub.531. [DOI] [PubMed] [Google Scholar]
- 21.Snitkin ES, et al. Genome-wide recombination drives diversification of epidemic strains of Acinetobacter baumannii. Proc. Natl. Acad. Sci. USA. 2011;108:13758–13763. doi: 10.1073/pnas.1104404108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez J, et al. Human pleural fluid triggers global changes in the transcriptional landscape of Acinetobacter baumannii as an adaptive response to stress. Sci. Rep. 2019;9:17251. doi: 10.1038/s41598-019-53847-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quinn B, et al. Effect of host human products on natural transformation in Acinetobacter baumannii. Curr. Microbiol. 2018 doi: 10.1007/s00284-017-1417-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodman N, et al. Human pleural fluid elicits pyruvate and phenylalanine metabolism in acinetobacter baumannii to enhance cytotoxicity and immune evasion. Front Microbiol. 2019 doi: 10.3389/fmicb.2019.01581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobs AC, et al. Characterization of the Acinetobacter baumannii growth phase-dependent and serum responsive transcriptomes. FEMS Immunol. Med. Microbiol. 2012;64:403–412. doi: 10.1111/j.1574-695X.2011.00926.x. [DOI] [PubMed] [Google Scholar]
- 26.Quinn B, et al. Human serum albumin alters specific genes that can play a role in survival and persistence in Acinetobacter baumannii. Sci. Rep. 2018;8:14741. doi: 10.1038/s41598-018-33072-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramirez MS, et al. Identification of potential virulence factors in the model strain Acinetobacter baumannii A118. Front. Microbiol. 2019;10:1599. doi: 10.3389/fmicb.2019.01599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramirez MS, et al. Naturally competent Acinetobacter baumannii clinical isolate as a convenient model for genetic studies. J. Clin. Microbiol. 2010;48:1488–1490. doi: 10.1128/JCM.01264-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobs AC, et al. AB5075, a highly virulent isolate of Acinetobacter baumannii, as a model strain for the evaluation of pathogenesis and antimicrobial treatments. MBio. 2014;5:e01076–e1014. doi: 10.1128/mBio.01076-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.30Rodman Nyah, M. J., Fung Sammie, Nakanouchi Jun, Myers Amber L., Harris Caitlin M., Dang Emily, Fernandez Jennifer S., Liu Christine, Mendoza Anthony M., Jimenez Veronica, Nikolaidis Nikolas, Brennan Catherine A., Bonomo Robert A., Sieira Rodrigo, Ramirez Maria Soledad. Human Pleural Fluid Elicits Pyruvate and Phenylalanine Metabolism in Acinetobacter baumannii to Enhance Cytotoxicity and Immune Evasion Frontiers in microbiology10, 1581. 10.3389/fmicb.2019.01581 (2019). [DOI] [PMC free article] [PubMed]
- 31.Dila DK, Maloy SR. Proline transport in Salmonella typhimurium: putP permease mutants with altered substrate specificity. J. Bacteriol. 1986;168:590–594. doi: 10.1128/jb.168.2.590-594.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boorsma A, van der Rest ME, Lolkema JS, Konings WN. Secondary transporters for citrate and the Mg(2+)-citrate complex in Bacillus subtilis are homologous proteins. J. Bacteriol. 1996;178:6216–6222. doi: 10.1128/jb.178.21.6216-6222.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qin H, et al. Comparative transcriptomics of multidrug-resistant Acinetobacter baumannii in response to antibiotic treatments. Sci. Rep. 2018;8:3515. doi: 10.1038/s41598-018-21841-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim J, Darlington A, Salvador M, Utrilla J, Jimenez JI. Trade-offs between gene expression, growth and phenotypic diversity in microbial populations. Curr. Opin. Biotechnol. 2019;62:29–37. doi: 10.1016/j.copbio.2019.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harding CM, et al. Acinetobacter baumanniistrain M2 produces type IV pili which play a role in natural transformation and twitching motility but not surface-associated motility. MBio. 2013 doi: 10.1128/mBio.00360-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wood CR, Ohneck EJ, Edelmann RE, Actis LA. a light-regulated type I pilus contributes to Acinetobacter baumannii biofilm, motility, and virulence functions. Infect. Immun. 2018 doi: 10.1128/IAI.00442-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vijayakumar S, et al. Biofilm formation and motility depend on the nature of the Acinetobacter baumannii clinical isolates. Front. Public Health. 2016;4:105. doi: 10.3389/fpubh.2016.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peleg AY, et al. The success of Acinetobacter species; genetic, metabolic and virulence attributes. PLoS ONE. 2012;7:e46984. doi: 10.1371/journal.pone.0046984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eijkelkamp BA, Hassan KA, Paulsen IT, Brown MH. Investigation of the human pathogen Acinetobacter baumannii under iron limiting conditions. BMC Genom. 2011;12:126. doi: 10.1186/1471-2164-12-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Penwell WF, Arivett BA, Actis LA. The Acinetobacter baumannii entA gene located outside the acinetobactin cluster is critical for siderophore production, iron acquisition and virulence. PLoS ONE. 2012;7:e36493. doi: 10.1371/journal.pone.0036493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaddy JA, et al. Role of acinetobactin-mediated iron acquisition functions in the interaction of Acinetobacter baumannii strain ATCC 19606T with human lung epithelial cells, Galleria mellonella caterpillars, and mice. Infect. Immun. 2012;80:1015–1024. doi: 10.1128/IAI.06279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahmed E, Holmstrom SJ. Siderophores in environmental research: roles and applications. Microb. Biotechnol. 2014;7:196–208. doi: 10.1111/1751-7915.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Itoh Y, et al. Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-beta-1,6-N-acetyl-D-glucosamine. J. Bacteriol. 2008;190:3670–3680. doi: 10.1128/JB.01920-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coulthurst SJ. The type VI secretion system - a widespread and versatile cell targeting system. Res. Microbiol. 2013;164:640–654. doi: 10.1016/j.resmic.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 45.Mussi MA, Relling VM, Limansky AS, Viale AM. CarO, an Acinetobacter baumannii outer membrane protein involved in carbapenem resistance, is essential for L-ornithine uptake. FEBS Lett. 2007;581:5573–5578. doi: 10.1016/j.febslet.2007.10.063. [DOI] [PubMed] [Google Scholar]
- 46.Niu C, Clemmer KM, Bonomo RA, Rather PN. Isolation and characterization of an autoinducer synthase from Acinetobacter baumannii. J. Bacteriol. 2008;190:3386–3392. doi: 10.1128/jb.01929-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ansari H, Tahmasebi-Birgani M, Bijanzadeh M, Doosti A, Kargar M. Study of the immunogenicity of outer membrane protein A (ompA) gene from Acinetobacter baumannii as DNA vaccine candidate in vivo. Iran. J. Basic Med. Sci. 2019;22:669–675. doi: 10.22038/ijbms.2019.30799.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peleg AY, et al. Galleria mellonella as a model system to study Acinetobacter baumannii pathogenesis and therapeutics. Antimicrob. Agents Chemother. 2009;53:2605–2609. doi: 10.1128/AAC.01533-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown KM, Lourido S, Sibley LD. Serum albumin stimulates protein kinase G-dependent microneme secretion in toxoplasma gondii. J. Biol. Chem. 2016;291:9554–9565. doi: 10.1074/jbc.M115.700518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Chateau M, Holst E, Bjorck L. Protein PAB, an albumin-binding bacterial surface protein promoting growth and virulence. J Biol Chem. 1996;271:26609–26615. doi: 10.1074/jbc.271.43.26609. [DOI] [PubMed] [Google Scholar]
- 51.Egesten A, Frick IM, Morgelin M, Olin AI, Bjorck L. Binding of albumin promotes bacterial survival at the epithelial surface. J. Biol. Chem. 2011;286:2469–2476. doi: 10.1074/jbc.M110.148171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gonyar LA, Gray MC, Christianson GJ, Mehrad B, Hewlett EL. Albumin, in the presence of calcium, elicits a massive increase in extracellular bordetella adenylate cyclase toxin. Infect. Immun. 2017 doi: 10.1128/iai.00198-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kruczek C, et al. Serum albumin alters the expression of iron-controlled genes in Pseudomonas aeruginosa. Microbiology. 2012;158:353–367. doi: 10.1099/mic.0.053371-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu HM, et al. Accuracy of real-time PCR, gram stain and culture for Streptococcus pneumoniae, Neisseria meningitidis and Haemophilus influenzae meningitis diagnosis. BMC Infect. Dis. 2013;13:26. doi: 10.1186/1471-2334-13-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Traglia GM, Quinn B, Schramm ST, Soler-Bistue A, Ramirez MS. Serum albumin and Ca2+ are natural competence inducers in the human pathogen Acinetobacter baumannii. Antimicrob. Agents Chemother. 2016;60:4920–4929. doi: 10.1128/AAC.00529-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Traglia GM, Chua K, Centron D, Tolmasky ME, Ramirez MS. Whole-genome sequence analysis of the naturally competent Acinetobacter baumannii clinical isolate A118. Genome Biol. Evol. 2014;6:2235–2239. doi: 10.1093/gbe/evu176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martinez J, et al. Human fluids alter DNA-acquisition in Acinetobacter baumannii. Diagn. Microbiol. Infect. Dis. 2018 doi: 10.1016/j.diagmicrobio.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 60.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bonomi HR, et al. Xanthomonas campestris attenuates virulence by sensing light through a bacteriophytochrome photoreceptor. EMBO Rep. 2016;17:1565–1577. doi: 10.15252/embr.201541691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Papp-Wallace KM, et al. Overcoming an extremely drug resistant (XDR) pathogen: avibactam restores susceptibility to ceftazidime for Burkholderia cepacia complex isolates from cystic fibrosis patients. ACS Infect. Dis. 2017;3:502–511. doi: 10.1021/acsinfecdis.7b00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wermser C, Lopez D. Identification of Staphylococcus aureus genes involved in the formation of structured macrocolonies. Microbiology. 2018;164:801–815. doi: 10.1099/mic.0.000660. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.