Abstract
Natural products and natural product-derived compounds have been widely used for pharmaceuticals for many years, and the search for new natural products that may have interesting activity is on going. Abyssomicins are natural product molecules that have antibiotic activity via inhibition of the folate synthesis pathway in microbiota. These compounds also appear to undergo a required [4+2] cycloaddition in their biosynthetic pathway. Here we report the structure of an FAD-dependent reductase, AbsH3, from the biosynthetic gene cluster of novel abyssomicins found in Streptomyces sp. LC-6-2.
Keywords: Natural Product, Oxidoreductase, X-ray Crystallography, Antibiotics, Ice Rings
Introduction:
Abyssomicins are novel microbial natural products produced by Verrucosispora and Streptomyces species (1-15). Abyssomicin C is a polycyclic polyketide-type antibiotic that was initially detected in a screen for inhibition of p-aminobenzoate (pABA) synthesis, which is a key cofactor in the tetrahydrofolate (THF) biosynthesis pathway. Unique to microorganisms, the THF pathway is an attractive drug target for antibiotics (16). Abyssomicin C is the first natural product inhibitor of this pathway, and it inhibits the aminodeoxychorismate synthase enzyme (ADCS), which converts chorismate and glutamine into 4-amino-4-deoxychorismate and glutamate. Other inhibitors of this pathway include sulfonamides and trimethoprim, which inhibit much further along the pathway at dihydropteroate synthase (sulfonamides) or dihydrofolate reductase (trimethoprim) (17-19). Abyssomicin C inhibits ADCS via covalent modification of the ADCS (20). In this reaction, abyssomicin C functions as a Michael acceptor, with the Cys263 sulfur acting as the nucleophile. After this reaction abyssomicin C is thought to rearrange into an abyssomicin D derivative, thus irreversibly inhibiting ADCS. Other interesting activities have been ascribed to abyssomicin compounds separate from their ability to inhibit the folate pathway in gram-positive bacteria, such as antitumor properties and activation of latent HIV replication (10, 21).
As exemplified by abyssomicin, versipelostatin and pyrroindomycin biosynthetic studies (22-24), novel Diels-Alderase-catalyzed stereoselective [4 + 2] cycloaddition reactions are central to forming the cyclic abyssomicin core architecture. Recent structural and mechanistic studies of AbyA5 (25), an esterase implicated in the formation of the key Diels-Alderase substrate, and biochemical characterization of AbmV (26), a P450 oxidase central to bridged ether formation, further extend understanding of abyssomicin core construction. However, biochemical and/or structural studies of the tailoring enzymes involved in abyssomicin core scaffold maturation remain sparse. Here we report the crystal structure of AbsH3 (GenBank Accession number: ARE67860, Uniprot: A0A1V0QH64), a putative FAD-dependent reductase encoded by the abyssomicin biosynthetic gene cluster found in the coal mine fire isolate Streptomyces sp. LC-6-2 (GenBank accession number KY432814) (14). AbsH3 is homologous to a number of mono-oxygenases, including TetX2 (PDB ID: 3P9U, 3V3N) and PhzS (PDB ID: 2RGJ) (30% and 33% identity, respectively) (27, 28). Unique Streptomyces sp. LC-6-2 abyssomicin metabolic features to which enzyme-catalyzed oxidation may contribute include the production of novel “enantiomeric”-like abyssomicin metabolites (abyssomicins M–X), unprecedented core cyclization patterns (e.g., the abyssomicin W 8/6/6/6 tetracyclic core) and linear spirotetronates (e.g., abyssomicin X).
Materials and Methods:
Cloning, Expression, and Purification.
Genomic DNA of Streptomyces sp. LC-6-2 was extracted using GenElute Bacterial Genomic DNA Kit (Sigma-Aldrich) and stored in Tris-EDTA buffer. The gene absH3 was amplified by PCR using Phusion Hot Start II DNA Polymerase (Thermo Scientific) using genomic DNA as template. The DNA fragments of the expected sizes were separated by agarose electrophoresis and purified by gel extraction kit (Zymo Research) and annealed with T4 DNA ligase into pET30 vector (Novagen) as previously described (29). Cloned genes were verified by DNA sequencing. The primers used for cloning were as follows: absH3-pET30-fw 5’-GGTATTGAGGGTCGCATGAACACGACCGAC-3’/ absH3-pET30-rv 5’-AGAGGAGAGTTAGAGCCTCAGCTCGCCGCTGT-3’.
E. coli BL21 DE3 was transformed with pET30-absH3 and seed culture was started from a single colony in LB medium supplemented with 50 μg/mL kanamycin. A sample of 2 mL seed culture was transferred to 1 L of LB medium supplemented with 50 μg/mL kanamycin. Cultures were grown at 37 °C until the cell density reached an OD600 of 0.4–0.6, when protein expression was induced with 0.1 mM IPTG. After growing at 18°C for 12 hours, bacterial cells were harvested at 4,000 rpm and lysed by sonication. The resulting cell debris was removed by centrifugation at 21,000 rpm. Supernatant was loaded on HisPur Ni-NTA resin (ThermoFisher) for affinity purification. The resin was first washed with wash buffer (25 mM HEPES, 30 mM imidazole, 200 mM NaCl, 5 mM β-mercapto ethanol (BME), pH 7.5). Proteins were eluted from the resin using elution buffer (25 mM HEPES, 300 mM imidazole, 200 mM NaCl, 5 mM BME, pH 7.5). Fractions containing desired protein (visualized on an SDS-PAGE gel) were concentrated. The imidazole-containing buffer in the concentrated protein was removed using a desalting column (GE PD-10) and exchanged with storage buffer (25 mM HEPES, 200 mM NaCl, pH 7.5). The purity of protein was determined by SDS-PAGE. Purified proteins were promptly flash frozen in sealed microcentrifuge tubes using liquid N2.
Crystallization and Structure Determination.
A Mosquito crystallization robot was used for initial screening of AbsH3 with the Hampton® crystal screens HR2-086, HR2-130, and HR2-134. Screens were conducted via the sitting drop method using 0.2 μL of protein solution and 0.1 μL of screening solution or, alternatively, 0.1 μL of protein and 0.2 μL of reservoir. Starting protein solution concentration was 10mg/mL. Initial crystals of AbsH3 observed in HR2-134 well D8: 100 mM HEPES pH 7.5 and 25% w/v PEG 3350. The final crystal condition after optimization was 22% PEG3350, 20 mM magnesium chloride, 100 mM HEPES, pH 7.4, with drops composed of of 0.2 μL protein and 0.2 μL reservoir solution.
X-Ray diffraction data were collected at the LRL-CAT beam line at the Advanced Photon Source at Argonne National Laboratory at a wavelength of 0.9793 A. Diffraction patterns were processed via De-ice in order to remove ice rings prior to indexing (30). De-Ice is a program that removes ice rings via modeling the ice and subtracting the average intensity working out from the beam stop radially on the raw diffraction image. By subtracting the radial average of intensity in the ice ring, ice ring diffraction can be at least partially accounted for without losing Bragg peaks at the resolution of the ice rings. Indexing and scaling was done in XDS (31). Molecular replacement was used to solve the structure, using PDB ID 3v3n (32) split via SCEDS (33) into multiple search models in Phaser (34). Real-space refinement was carried out manually in Coot (35) with final refinement via Phenix.refine (36). Analysis of the structure was aided using a 3D semi-immersive collaborative graphics system (37).
Docking Experiments.
The X-ray crystal structure was used to explore how each abyssomicin analogue may interact with the protein in any possible binding modes. For this purpose, each ligand molecule was docked into the protein by using the Autdock Vina program (38). The molecular docking was carried out by using the default optimization parameters of the Autdock Vina program for the pose sampling, with a large grid map (74 Å × 80 Å × 64 Å) to cover the entire protein. The docking score was calculated by using the Lamarckian algorithm (39). Finally, the docked protein-ligand binding structures were energy-minimized by using the Amber12 program (40).
Results and Discussion:
Diffraction data was collected at LRL-CAT beamline at the Advanced Photon Source. The highest resolution (1.99 Å) data set exhibited ice rings and attempts to recollect data with cryo-protection (20% ethylene glycol, 10-20% glycerol, saturated sucrose solution) instead of looping from reservoir solution resulted in lessened diffracting power. Therefore, as a workaround to preserve resolution, data were preprocessed with De-Ice (30).. Since the structure of AbsH3 was solved, a new global background model has been developed in DIALS for better estimation of Bragg peak intensities within ice ring data (41). Using this pipeline yields similar statistical results to De-Iced data and requires less intervention on the part of the crystallographer.
The structure was solved with molecular replacement in Phaser, utilizing PDB ID 3V3N for the search model split into two “domains” with the SCEDS program. The SCEDS program separated the TetX2 structure 3v3n into two domains which involved multiple cut sites. Fragment A contained residues 14-54,70-79,122-189,287-317, and 330-366. Fragment B contained residues 55-69, 80-121, 190-246, 249-286,318-329, and 367-382. The crystal structure of AbsH3 was solved to 1.99 Å resolution in space group P212121. The final refined structure had R-work and R-free scores of 0.183 and 0.236 respectively. Table 1 provides diffraction and modeling statistics. Data collection and merging statistics are for data after Deice processing.
Table 1.
Data collection and refinement statistics.
| PDB ID | 6N04 |
|---|---|
| Wavelength | 0.9793 |
| Resolution range | 35.62 - 1.998 (2.07 - 1.998) |
| Space group | P 21 21 21 |
| Unit cell | 53.316 109.491 143.637 90 90 90 |
| Total reflections | 379890 (37404) |
| Unique reflections | 53291 (5578) |
| Multiplicity | 7.1 (6.7) |
| Completeness (%) | 91.96 (97.22) |
| Mean I/sigma(I) | 12.06 (1.97) |
| Wilson B-factor | 29.53 |
| R-merge | 0.1237 (1.015) |
| R-meas | 0.1336 (1.101) |
| R-pim | 0.04993 (0.4195) |
| CC1/2 | 0.997 (0.71) |
| CC* | 0.999 (0.911) |
| Reflections used in refinement | 53221 (5560) |
| Reflections used for R-free | 2023 (211) |
| R-work | 0.1828 (0.2713) |
| R-free | 0.2365 (0.2986) |
| CC(work) | 0.964 (0.835) |
| CC(free) | 0.945 (0.731) |
| Number of non-hydrogen atoms | 6573 |
| macromolecules | 5559 |
| ligands | 108 |
| solvent | 906 |
| Protein residues | 731 |
| RMS(bonds) | 0.005 |
| RMS(angles) | 0.77 |
| Ramachandran favored (%) | 98.19 |
| Ramachandran allowed (%) | 1.81 |
| Ramachandran outliers (%) | 0.00 |
| Rotamer outliers (%) | 1.46 |
| Clashscore | 3.64 |
| Average B-factor | 34.84 |
| macromolecules | 33.65 |
| ligands | 25.70 |
| solvent | 43.21 |
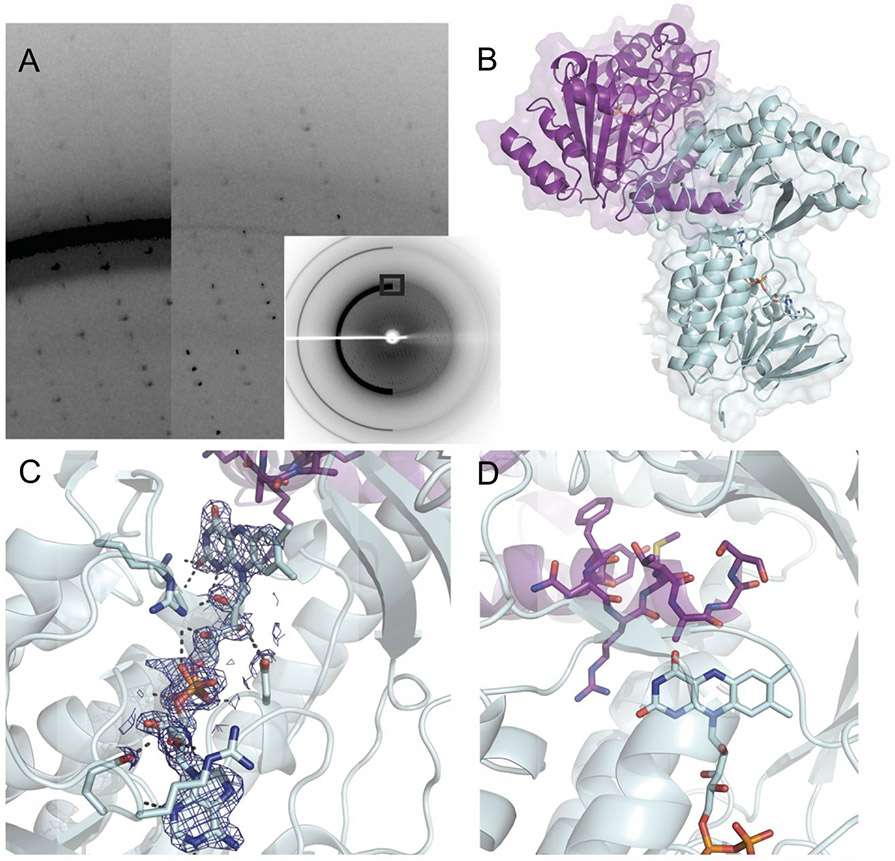
Even though AbsH3 behaves as a monomer in solution, there are two molecules in the asymmetric unit, which have an interesting interaction. The two molecules are aligned in an anti-parallel fashion where the lid domain β-sheets hydrogen bond to each other at the interface of the two monomers, maintaining the secondary structure motif. This feature is accomplished by aligning the lid domains of both monomers, as shown in Figure 1. In addition, in this packing arrangement an AbsH3 monomers obstructs their neighbor’s active site with a C-terminal tail that maintains helical secondary structure. This capping of the active site appears to have stabilized the apo structure and locked the lid domain into place. This hints at possible self-inactivation at very high protein concentration, but it is unclear at this time if this phenomenon is in any way biologically relevant.
Figure 1.
Crystal Structure of AbsH3 (PDB ID: 6N04) A. Comparison of frames before and after De-Ice processing. On left, frame as collected at LRL-CAT, compared to after processing, right. Inset shows entire frame from which zoomed in imaged is pulled. B. Crystal packing of AbsH3. The crystal packing in the structure shows the C-terminal tail of the purple monomer plunged into the active site of the other aqua AbsH3 monomer. C. FAD density and polar contacts in the core domain of AbsH3. 2Fo-Fc map of FAD at 2.0σ level showing high quality density of this co-factor. Polar contacts with sidechains are also shown. D. C-terminal tail occludes the active site of adjacent monomer in the crystal and may be a self-inactivation function at high AbsH3 concentration.
Similar oxidoreductases to AbsH3 are reliant on FAD as a prosthetic group to supply electrons for their chemical processes, and AbsH3 is no exception. In our structure, FAD is clearly bound, and, in fact, has lower average B-factors than the overall structure. It sits down in the core domain, with the electron-donating ring system extending into the bottom of the active site. This arrangement is similar to homologous reductases including TetX2, which hold their respective FAD molecules in similar fashions (27). PhzS, even though it has a similar overall fold and utilizes a FAD cofactor for oxidoreductase activity, appears to have different substrate binding morphology due to different FAD prosthetic group orientation in the active site, as well as a more open lid domain in AbsH3 that should be accessible from both sides.
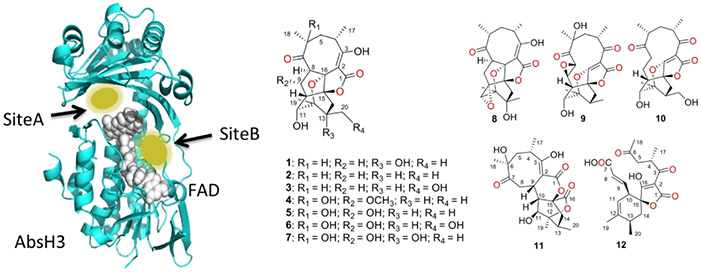
In order to better understand possible substrate binding modes and help determine a likely substrate, docking studies with novel abyssomicin compounds discovered by Wang et al. (14) from the Streptomyces strain that AbsH3 was cloned from were performed. These docking studies showed a number of potentially important residues for binding in the active site. See Supplemental Table 1 and Figure 2 for docking energy table and structures, respectively, for representative docked structures, see Supplemental Figure 1. In site A, the putative active site, Arg89, Tyr162, Gln164, and Arg206 potentially use electrostatic interactions or hydrogen bonds to guide the abyssomicin analogues toward the quinone of the FAD cofactor. The rest of the binding pocket stabilizes the large multi-cyclic ring system via hydrophobic interactions. Leu114, Leu241, Met239, and Leu247 appear to be especially important for stabilizing the compounds in the hydrophobic pocket. Tyr162, Met187, and Arg206 seem to gate the compounds into this pocket and lock them in orientations that allow exposed oxygen moieties to be reduced by the FAD cofactor. An additional binding site, site B, in the FAD binding groove allows for the possibility of diffusion across the surface of the AbsH3 molecule into the active site. Interestingly, compounds 8-12 show tighter binding in this second site rather than the putative active site. Future work unraveling this Abyssomycin biosynthetic pathway will yield new insights into how this fascinating natural product is produced.
Figure 2.
Binding sites from docking studies and abyssomicin analogs used in the docking calculations. At left, FAD model showing the two sites where abyssomicin analogues bound in the docking study. Site A is the putative active site, while Site B is in the FAD binding groove. The presence of this second binding site may allow substrates to diffuse across the enzyme surface to the active site, even when not recruited directly to Site A. At right, chemical structures of the abyssomicin analogues used in the docking computations. Interestingly, compounds 8-12 preferred Site B to Site A, which is the active site.
Supplementary Material
Acknowledgements:
This work was supported by the Protein Structure Initiative project, Enzyme Discovery for Natural Product Biosynthesis (NATPRO) with NIH/NIGMS Grant number:U01GM098248, as well as NIH grants R01 CA217255 and R01 GM115261. This work was partially supported by a training fellowship for Jonathan Clinger from the Keck Center of the Gulf Coast Consortia, as part of the Houston Area Molecular Biophysics Program, National Institute of General Medical Sciences (NIGMS) T32GM008280.
This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Use of the Lilly Research Laboratories Collaborative Access Team (LRL-CAT) beamline at Sector 31 of the Advanced Photon Source was provided by Eli Lilly Company, which operates the facility.
References
- 1.Bister B, Bischoff D, Strobele M, Riedlinger J, Reicke A, Wolter F, Bull AT, Zahner H, Fiedler HP, Süssmuth RD. Abyssomicin C—A polycyclic antibiotic from a marine Verrucosispora Strain as an inhibitor of the p-Aminobenzoic acid/Tetrahydrofolate biosynthesis pathway. Angew. Chem. Int. Ed 2004, 43, 2574–2576. [DOI] [PubMed] [Google Scholar]
- 2.Keller-Schierlein W, Muntwyle R, Pache W, Zähner H. Metabolic products of micro-organisms. Chlorothricin and deschlorothricin. Helv. Chim. Acta 1969, 52, 127–142. [Google Scholar]
- 3.Ding WD, Williams DR, Northcote P, Siegel MM, Tsao R, Ashcroft J, Morton GO, Alluri M, Abbanat D, Maiese WM, Ellestad GA. Pyrroindomycins, novel antibiotics produced by Streptomyces rugosporus sp. LL-42D005. J. Antibiot 1994, 47, 1250–1257. [DOI] [PubMed] [Google Scholar]
- 4.Kellerjuslen C, King HD, Kuhn M, Loosli HR, Pache W, Petcher TJ, Weber HP, Vonwartburg A. Tetronomycin, a novel polyether of unusual structure. J. Antibiot 1982, 35, 142–150. [DOI] [PubMed] [Google Scholar]
- 5.Park HR, Furihata K, Hayakawa Y, Shin-ya K. Versipelostatin, a novel GRP78/Bip molecular chaperone down-regulator of microbial origin. Tetrahedron Lett. 2002, 43, 6941–6945. [Google Scholar]
- 6.Lacoske MH, Theodorakis EA. Spirotetronate polyketides as leads in drug discovery. J. Nat. Prod 2015, 78, 562–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vieweg L, Reichau S, Schobert R, Leadlay PF, Süssmuth RD. Recent advances in the field of bioactive tetronates. Nat. Prod. Rep 2014, 31, 1554–1584. [DOI] [PubMed] [Google Scholar]
- 8.Niu XM, Li SH, Gorls H, Schollmeyer D, Hilliger M, Grabley S, Sattler. Abyssomicin E, a Highly Functionalized Polycyclic Metabolite from Streptomyces Species. Org. Lett 2007, 9, 2437–2440. [DOI] [PubMed] [Google Scholar]
- 9.Keller S, Nicholson G, Drahl C, Sorensen E, Fiedler HP, Süssmuth RD. Abyssomicins G and H and atrop-Abyssomicin C from the Marine Verrucosispora Strain AB-18-032. J. Antibiot 2007, 60, 391–394. [DOI] [PubMed] [Google Scholar]
- 10.Igarashi Y, Yu LK, Miyanaga S, Fukuda T, Saitoh N, Sakurai H, Saiki I, Alonso-Vega P, Trujillo ME. Abyssomicin I, a Modified Polycyclic Polyketide from Streptomyces sp. CHI39. J. Nat. Prod 2010, 73, 1943–1946. [DOI] [PubMed] [Google Scholar]
- 11.Wang Q, Song FH, Xiao X, Huang P, Li L, Monte A, Abdel-Mageed WM, Wang J, Guo H, He WN, Xie F, Dai HQ, Liu MM, Chen CX, Xu H, Liu M, Piggott AM, Liu XT, Capon RJ, Zhang LX. Abyssomicins from the South China Sea Deep-Sea Sediment Verrucosispora sp.: Natural Thioether Michael Addition Adducts as Antitubercular Prodrugs. Angew. Chem., Int. Ed 2013, 52, 1231–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leon B, Navarro G, Dickey BJ, Stepan G, Tsai A, Jones GS, Morales ME, Barnes T, Ahmadyar S, Tsiang M, Geleziunas R, Cihlar T, Pagratis N, Tian Y, Yu H, Linington RG. Abyssomicin 2 reactivates latent HIV-1 by a PKC-and HDAC-independent mechanism. Org. Lett 2015, 17, 262–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abdalla MA, Yadav PP, Dittrich B, Schuffler A, Laatsch H. ent-Homoabyssomicins A and B, Two New Spirotetronate Metabolites from Streptomyces sp. Ank 210. Org. Lett 2011, 13, 2156–2159. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Elshahawi SI, Cai W, Zhang Y, Ponomareva LV, Chen X, Copley GC, Hower JC, Zhan C-G, Paekin S, Rohr S, Van Lanen SG, Shaaban KA, Thorson JS. Bi- and Tetracyclic Spirotetronates from the Coal Mine Fire Isolate Streptomyces sp. LC-6-2. J. Nat. Prod 2017, 80, 1141–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang H, Song Y, Li X, Wang X, Ling C, Qin X, Zhou Z, Li Q, Wei X, Ju J. Abyssomicin Monomers and Dimers from the Marine-Derived Streptomyces koyangensis SCSIO 5802. J. Nat. Prod 2018, 81, 1892–1898. [DOI] [PubMed] [Google Scholar]
- 16.Brown GM. The biosynthesis of folic acid. J. Biol. Chem 1962, 237, 536–540. [PubMed] [Google Scholar]
- 17.Yun MK, Wu Y, Li Z, Zhao Y, Waddell MB, Ferreira AM, Lee RE, Bashford D, White SW. Catalysis and sulfa drug resistance in dihydropteroate synthase. Science 2012, 335, 1110–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li R, Sirawaraporn R, Chitnumsub P, Sirawaraporn W, Wooden J, Athappilly F, Turley S, Hol WG. Three-dimensional structure of M. tuberculosis dihydrofolate reductase reveals opportunities for the design of novel tuberculosis drugs. J. Mol. Biol 2000, 295, 307–323. [DOI] [PubMed] [Google Scholar]
- 19.Sadaka C, Ellsworth E, Hansen PR, Ewin R, Damborg P, Watts JL. Review on abyssomicins: Inhibitors of the chorismate pathway and folate biosynthesis. Molecules 2018, 23, 1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keller S, Schadt HS, Ortel I, Süssmuth RD. Action of atrop-Abyssomicin C as an Inhibitor of 4-Amino-4-deoxychorismate Synthase PabB. Agnew. Chem. Int. Ed 2007, 46, 8284–8286. [DOI] [PubMed] [Google Scholar]
- 21.Song Y, Li Q, Qin F, Sun C, Hao L, Wei X, Wong N-K, Ye L, Zhang Y, Shao M, Ju J. Neoabyssomicins A–C, polycyclic macrolactones from the deep-sea derived Streptomyces koyangensis SCSIO 5802. Tetrahedron 2017, 73, 5366–5372. [Google Scholar]
- 22.Byrne MJ, Lees NR, Han LC, van der Kamp MW, Mulholland AJ, Stach JE, Willis CL, Race PR. The catalytic mechanism of a natural Diels–Alderase revealed in molecular detail. J. Am. Chem. Soc 2016, 138, 6095–6098. [DOI] [PubMed] [Google Scholar]
- 23.Hashimoto T, Hashimoto J, Teruya K, Hirano T, Shin-Ya K, Ikeda H, Liu HW, Nishiyama M, Kuzuyama T. Biosynthesis of versipelostatin: identification of an enzyme-catalyzed [4+ 2]-cycloaddition required for macrocyclization of spirotetronate-containing polyketides. J. Am. Chem. Soc 2015, 137, 572–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian Z, Sun P, Yan Y, Wu Z, Zheng Q, Zhou S, Zhang H, Yu F, Jia X, Chen D, Mandi A, Kurtan T, Liu W. An enzymatic [4+ 2] cyclization cascade creates the pentacyclic core of pyrroindomycins. Nat. Chem. Biol 2015, 11, 259–265. [DOI] [PubMed] [Google Scholar]
- 25.Lees NR, Han LC, Byrne MJ, Davies JA, Parnell AE, Moreland PEJ, Stach JEM, van der Kamp MW, Willis CL, Race PR. Three-in-One Oxygen Vacancies: Whole Visible-Spectrum Absorption, Efficient Charge Separation, and Surface Site Activation for Robust CO2 Photoreduction. Angew Chem. Int. Ed. Engl 2019, 58, 1–6. [DOI] [PubMed] [Google Scholar]
- 26.Li Q, Ding W, Yao Z, Tu J, Wang L, Huang H, Li S, Ju J. AbmV Catalyzes Tandem Ether Installation and Hydroxylation during Neoabyssomicin/Abyssomicin Biosynthesis. Org. Lett 2018. 20, 4854–4857. [DOI] [PubMed] [Google Scholar]
- 27.Walkiewicz K, Davlieva M, Wu G, Shamoo Y. Crystal structure of Bacteroides thetaiotaomicron TetX2, a tetracycline degrading monooxygenase at 2.8 Å resolution. Proteins 2011, 79, 2335–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenhagen BT, Shi K, Robinson H, Gamage S, Bera AK, Ladner JE, Parsons JF. Crystal structure of the pyocyanin biosynthetic protein PhzS. Biochemistry, 2008, 47, 5281–5289. [DOI] [PubMed] [Google Scholar]
- 29.Cui Z, Liu X, Overbay J, Cai W, Wang X, Lemke A, Wiegmann D, Niro G, Thorson JS, Ducho C, van Lanen SG. Enzymatic Synthesis of the Ribosylated Glycyl-Uridine Disaccharide Core of Peptidyl Nucleoside Antibiotics. J. Org. Chem 2018, 83, 7239–7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chapman MS, Somasundaram T. De-icing: recovery of diffraction intensities in the presence of ice rings. Acta Cryst. D 2010, 66, 741–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta. Cryst. D: Biol. Cryst 2010, 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walkiewicz K, Shammoo Y. PDB ID 3V3N 2013 Rice University. [Google Scholar]
- 33.McCoy AJ, Nicholls RA, Schneider TR. SCEDS: protein fragments for molecular replacement in Phaser. Acta Cryst. D 2013, 69, 2216–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Solving structures of protein complexes by molecular replacement with Phaser. J. Appl. Cryst 2007, 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta. Cryst. D: Biol. Cryst 2010, 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Gorsse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta. Cryst. D: Biol. Cryst 2010, 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yennamali R, Arangarasan R, Bryden A, Gleicher M, Phillips GN Jr.. Using a commodity high-definition television for collaborative structural biology. J. Appl. Cryst 2014, 47, 1153–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput Chem 2010. 31, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 1998. 19, 1639–1662. [Google Scholar]
- 40.Case DA, Darden TA, Cheatham TE III, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts B, Hayik S, Roitberg A, Seabra G, Swails J, Götz AW, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wolf RM, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hseih M-J, Cui G, Roe DR, Mathews DH, Seetin MG, Salomon-Ferrer R, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA. 2012 AMBER 12, University of California, San Francisco. [Google Scholar]
- 41.Parkhurst JM, Thorn A, Vollmar M, Winter G, Waterman DG, Fuentes-Montero L, Gildea RG, Murshudov GN, Evans G. Background modelling of diffraction data in the presence of ice rings. IUCrJ 2017, 4, 626–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.