Abstract
The widespread use of drugs has exacerbated the resistance of Eimeria tenalla to anti-coccidial drugs. Using RNA-seq, we previously found the ATPase ASNA1 homolog of E. tenella (EtASNA1) was differentially expressed in resistant strains and drug sensitive (DS) strain. In our study, we used western blotting and quantitative real-time PCR (qRT-PCR) to analyze the translational and transcriptional levels of EtASNA1 in a diclazuril-resistant (DZR) strain, maduramicin-resistant (MRR) strain, salinomycin-resistant (SMR) strain, and DS strain and found EtASNA1 was highly expressed in three drug-resistant strains. The qRT-PCR and western blotting results also showed that the expression levels of EtASNA1 increased with increasing drug concentration, and the transcription levels of the DZR strains isolated from the field were higher than those of the DS strain. In addition, we used in vivo and in vitro tests to analyze the changes of EtASNA1 expression after DZR, MRR, and DS strain infections in chickens, and in vitro inoculation of DF-1 cells in the presence of drugs. The addition of drugs caused expression to be upregulated. The results of qRT-PCR and western blotting also showed that the expression levels of EtASNA1 in second-generation merozoites (SM) and unsporulated oocysts (UO) were significantly higher than those in the other two developmental stages. The immunofluorescence localization of EtASNA1 indicated that the protein was distributed throughout the sporozoites (SZ) and SM, except for the refractile bodies of SZ. In vitro inhibition experiments showed that anti-EtASNA1 antibody incubation significantly inhibited SZ invasion of DF-1 cells. The above results showed that EtASNA1 may be related to host cell invasion of E. tenella and may be involved in the development of E. tenella resistance to some drugs.
Keywords: Eimeria tenella, EtASNA1, Resistant strains
Graphical abstract
In vitro and in vivo culture to study the effect of drugs on transcript levels of EtASNA1. DS+, drug-sensitive strains + diclazuril or maduramicin; DS-, drug-sensitive strains, drug-free; DZR+, diclazuril-resistant strains + diclazuril; DZR-, diclazuril-resistant strains, drug-free; MRR+, maduramicin-resistant strains + maduramicin; MRR-, maduramicin-resistant strains, drug-free.

Highlights
-
•
ATPase ASNA1 homolog was first studied in E. tenella.
•EtASNA1 was highly expressed in different drug-resistant strains (diclazuril-resistant strains, maduramicin-resistant strains) compare to drug sensitive strain of E. tenella, and it might be related to drug resistance of E. tenella.
•The high expression observed in second-generation merozoites may be associated with the propagation and development of E. tenella.
•EtASNA1 may be involved in sporozoites invasion.
1. Introduction
Coccidiosis is a major intestinal parasitic disease of poultry caused by Eimeria spp., which has caused worldwide economic losses to the poultry industry (McDonald et al., 2009; Blake et al., 2014). It has a high morbidity and a mortality rate of 50%, so the losses are significant (Karaer et al., 2012; Györke et al., 2013). Eimeria spp., which infect the intestines of chickens, mainly cause intestinal lesions, leading to a decline in poultry growth and feed utilization (Arabkhazaeli et al., 2013). Among several Eimeria spp., Eimeria tenella is the most common and harmful species.
Presently, the prevention and treatment of coccidiosis mainly depends on vaccination and anti-coccidial drugs. Nevertheless, due to its weak antiviral activity, environmental pollution, and high production costs, the use of vaccines is limited (Vermeulen 1998; Sharman et al., 2010). Therefore, anti-coccidial drugs are still used widely to control Eimeria infection (Peek et al., 2011). The anti-coccidial drugs currently in use mainly include synthetic drugs and ionophores (Min et al., 2004). Among them, diclazuril and maduramicin are representative drugs. Diclazuril can affect the synthesis of nucleic acids of coccidia, which leads to reproductive failure by arresting the further nuclear differentiation of normal growth and division in schizonts and microgametocytes (Vanparijs et al., 1989; McDougald et al., 1990). Studies have suggested that maduramicin kills coccidia by interrupting their normal intracellular ion balance and by influencing Na + -K + -ATPase activity (Chapman et al., 2010). They also affect merozoites by destroying the cell border and internal organelles (Mehlhorn et al., 1983).
However, as with the widespread use of other antimicrobials, the widespread use of anti-coccidial drugs has inevitably resulted in the emergence and development of resistance (Chapman 1997; Blake et al., 2014). The emergence of drug resistance has greatly shortened the service life of anti-coccidial drugs, and the control effect has gradually decreased or even failed, especially in intensive chicken farms (Suo et al., 2001). Studies have investigated the anti-coccidial drug resistance of E. tenella and found its prevalence and severity (Jiang et al., 2005; Thabet et al., 2017a). Since the discovery of drug resistance in Eimeria spp., investigators have studied the mechanism of drug resistance from many aspects, hoping to solve the fundamental problems of drug resistance. However, we still do not fully understand the underlying molecular mechanism of resistance. Therefore, it is important to identify the fundamental mechanism of avian coccidian resistance, and to develop rapid detection methods for drug resistance, to establish a rational treatment strategy.
To further investigate the mechanism of drug resistance in the early stages, the drug concentration increasing method was used to induce the diclazuril-resistant (DZR) strain and maduramicin-resistant (MRR) strain from drug-sensitive (DS) strain of E. tenella in our laboratory. The acquired MRR strain was fully resistant to 7.0 ppm maduramicin, while the DZR strain was only fully resistant to 1.2 ppm diclazuril (Han et al., 2004). Transcriptome sequencing (RNA-seq) was then used to sequence and analyze the DS strain and the two resistant (DZR and MRR) strains of E. tenella, to screen for differentially expressed genes. The results showed that the differential expression of the ATPase ASNA1 homolog of E. tenella (EtASNA1) was upregulated in DZR and MRR strains (Xie et al., 2020). Transcriptome sequencing showed that the log2 ratio of DZR/DS reached 2.45 and the log2 ratio of MRR/DS reached 2.27.
Arsenite-stimulated ATPase (ASNA1) is an arsenic exporter homologous to the ATPase ArsA in Escherichia coli. Studies have shown that the homologues of ArsA in nature are widely distributed in all three domains of eucarya, archaea, and prokarya (Bhattacharjee et al., 1999). ASNA1 (also known as TRC40) is found in the human nucleolus, perinuclear region, and cytosol (Kurdi-Haidar et al., 1998). Constitutively expressed ArsA has been found to be a detoxification pump for arsenic, and this pump actively removes arsenicals and antimonials from cells, reducing the intracellular concentration of the metalloids to subtoxic levels, resulting in resistance (Hsu et al., 1989). Previous studies have also shown that downregulation of ASNA1 in Caenorhabditis elegans leads to increased arsenite sensitivity and growth stagnation (Tseng et al., 2007; Kao et al., 2007), embryonic lethality in mice (Mukhopadhyay et al., 2006), and increased apoptosis and retarded growth in humans (Hemmingsson et al., 2008). However, there have been no reports of ASNA1-related studies in protozoan parasites, especially in Eimeria spp.
In this study, we cloned and successfully expressed the EtASNA1 gene of E. tenella, and studied some of its biological functions. We evaluated the differential expressions of EtASNA1 in drug-resistant and DS strains. In addition, we analyzed the changes in transcription levels of EtASNA1 during drug stimulation. Overall, this study found there may be some relationship between EtASNA1 and drug resistance, which may be provided the basis for further studies on the mechanism of drug resistance.
2. Materials and methods
2.1. Animals, parasites, and cells
The chickens used in this study were supplied by a farm in Fengxian District, Shanghai, China. The Jiagan Biology Company (Shanghai, China) provided us with New Zealand white rabbits. All experimental animals were raised in a coccidia-free environment.
The DS strain of E. tenella was provided by the Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences (Resource Number CAAS21111601) (Huang et al., 1993), and was maintained and propagated by passage through coccidia-free 2-week-old chickens as previously described (Tomley et al., 1997). The DS strain was sensitive to the anticoccidial drugs diclazuril, maduramicin, and salinomycin. Our laboratory induced drug-resistant strains from low to high concentrations in the DS strain by the concentration gradient method. The DZR strain was completely resistant to 1.2 ppm diclazuril and obtained from 0.05 ppm to 1.2 ppm by inducing DS strains after 18 passages. The MRR strain was completely resistant to 7.0 ppm maduramicin and obtained from low concentrations of 2.0 ppm to high concentrations of 7.0 ppm by inducing DS strains after 20 passages (Han et al., 2004). The salinomycin-resistant (SMR) strain was completely resistant to 60 ppm diclazuril and obtained from 15 ppm to 60 ppm by inducing DS strains after 10 passages (Wang et al., 2019). Chickens were fed with a mixture of maduramicin, diclazuril, or salinomycin in the feed from 2 days before inoculation of parasites. The chickens inoculated with DS strain were not given any drugs.
We collected and purified unsporulated oocysts (UO) and sporulated oocysts (SO) by using standard procedures (Han et al., 2010). Sporozoites (SZ) were recovered from cleaned SO by in vitro excystation and purified (Miska et al., 2004). Second generation merozoites (SM) were collected and purified from the caecal mucosa of chickens at 112 h post-inoculation (Shirley et al., 1995).
We isolated four field diclazuril-resistant strains (D2, D4, D5, and D8) of E. tenella using the single oocyst method (Khalafalla et al., 2010) and demonstrated the resistance to 1.0 ppm diclazuril by drug sensitivity experiments in chickens (unpublished). The SO of these strains were propagated, obtained, and purified for future use.
The chicken embryo fibroblast DF-1 cell line (ATCC CRL-12203) was cultured in our laboratory in Dulbecco's Modified Eagle's Medium (DMEM) (Gibco BRL, Paisley, UK) for immunofluorescence tests and invasion experiments.
2.2. Total RNA extraction and complementary DNA synthesis
According to the manufacturer's protocol, TRIzol reagent (TaKaRa, Tokyo, Japan) used to extract total RNA was from E. tenella SO. The quantity and quality of total RNA were assessed using a NanoDrop 2000C (Thermo Fisher Scientific, Waltham, MA, USA) and electrophoresis under denaturing conditions in 1% agarose gels. Complementary DNA (cDNA) was synthesized from the total RNA using an M-MLV Reverse Transcriptase kit (Invitrogen, Carlsbad, CA, USA). The cDNA was then used as a template for further studies.
2.3. Amplification, cloning, and sequencing of EtASNA1
The polymerase chain reaction (PCR) was used to amplify the complete coding region of the EtASNA1 gene by using a pair of primers designed based on the reported sequences of the EtASNA1 open reading frame (Gene symbol: ETH_00036550). The primer sequences were as follows: 5ʹ- GCGGAATTCATGACAGACGACGAGTTTCCCCTTG-3ʹ (forward primer); 5ʹ- GCGGTCGACTGCATCAATTCGGCGAAAGAAAGGG-3ʹ (reverse primer), including EcoRI and SalI restriction sites. The SO cDNA obtained in the previous step was used as a template for PCR amplification in a 20 μL reaction. The amplification conditions were 95 °C for 3 min; 32 cycles of 95 °C for 45 s, 62 °C for 45 s, 72 °C for 2 min, and 10 min at 72 °C. Then, 1% agarose gel electrophoresis was used to analyze the PCR products, and target bands were gel purified (Qiagen, Duesseldorf, Germany) and subcloned into the pGEM-T-easy vector (Promega, Madison, WI, USA), and positive recombinant plasmid clones were verified by Sangon Biotech (Shanghai, China).
2.4. Sequence analysis of EtASNA1
The sequence of the full-length cDNA of the EtASNA1 gene was used in BLAST searches of GenBank (http://www.ncbi.nlm.nih.gov/BLAST/) and the E. tenella genome database (https://toxodb.org/toxo/). The molecular mass and theoretical isoelectric point were obtained using ProtParam tools (http://www.expasy.org/tools/protparam.html). Signal peptide sequences, protein motifs, and transmembrane regions of the putative EtASNA1 protein were predicted using SignalP (http://www.cbs.dtu.dk/services/SignalP/), TMHMM (http://www.cbs.dtu.dk/services/TMHMM-2.0/), and Motifscan (http://hits.isb-sib.ch/cgi-bin/motif_scan), respectively.
2.5. Expression and purification of recombinant EtASNA1 protein
The identified recombinant plasmid pGEM-T-EtASNA1 was digested with EcoRI and SalI. The target fragment was purified and cloned into a frame of digested expression vector pGEX-6P-1 (Novagen, Darmstadt, Germany) and the recombinant plasmid was identified by sequencing. The correct recombinant plasmid was transferred into E. coli BL21 (DE3) (Tiangen, Beijing, China) and the expression of recombinant protein in E. coli was induced by adding 1.0 mM isopropyl β-D-1-thiogalactopyranoside (Sigma-Aldrich, St. Louis, MO, USA) at an optical density of 0.6 at 600 nm. Induced bacterial cells were incubated for 6 h post-induction and collected by centrifugation. Cell lysates were prepared using sonication and analyzed by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to confirm the subcellular distribution of recombinant protein. Recombinant EtASNA1 (rEtASNA1) protein was purified from SDS-PAGE gel bands (Richard et al., 2009). The purity of the protein was detected by 12% SDS-PAGE, and its concentration was determined by the BCA protein assay kit (Beyotime, Haimen, China). The purified protein was stored in aliquots at −20 °C.
2.6. Polyclonal sera against recombinant EtASNA1 protein
The rEtASNA1 protein was used to subcutaneously immunize 2-month-old rabbits with 200 μg protein emulsified in Freund's complete adjuvant (Sigma-Aldrich) in a 1:1 mixture. The rabbits were boosted with the same protein emulsified in Freund's incomplete adjuvant (Sigma-Aldrich) after 2 weeks. Immunization was carried out once every 7 days for a total of five immunizations. One week after the final immunization, antiserum against rEtASNA1 was collected. Serum collected before protein injection was used as a negative control.
2.7. Quantitative real-time PCR (qRT-PCR)
The mRNA expression profiles of EtASNA1 at different developmental stages (UO, SO, SZ, and SM) of the E. tenella sensitive strain were examined using quantitative real-time PCR (qRT-PCR). The qRT-PCR was performed using the SYBR1 Green I dye method. Total RNA was isolated from four developmental stages using TRIzol reagent (Invitrogen) and treated with DNase I (Invitrogen) to remove all DNA contamination. Synthesis of cDNA was from 2 μg of total RNA using SuperScript II reverse transcriptase (Invitrogen) and random primers (Invitrogen). The 18S rRNA housekeeping gene (GenBank accession number: EF122251) from E. tenella was used as an internal control (Jiang et al., 2012). The qRT-PCR primers for EtASNA1 were 5′-GGCGTCGGCAAGACAACCAC-3′ (sense) and 5′-GTGGACAGCAGCAGCACTGATTC-3′ (antisense). The primers for 18S rRNA were 5′-TGTAGTGGAGTCTTGGTGATTC-3′ (sense) and 5′-CCTGCTGCCTTCCTTAGATG-3′ (antisense). Each reaction was performed in triplicate, and the experiment was repeated three times. The 2−ΔΔCt method (Livak and Schmittgen, 2001) was used to measure relative changes in gene expression.
We also detected the EtASNA1 transcription levels of EtASNA1 in SO of DS, DZR, MRR, and SMR strains using qRT-PCR. The EtASNA1 mRNA transcripts in field isolated DZR strains of E. tenella (D2, D4, D5, and D8) were also detected using the same method. In addition, EtASNA1 mRNA transcripts were compared between the SO of DZR strains resistant to diclazuril at different concentrations (0.2 ppm, 0.5 ppm, 0.8 ppm, and 1.0 ppm) and MRR strains resistant to maduramicin at different concentrations (3 ppm and 5 ppm) with the DS strain.
2.8. SDS-PAGE and western blot analysis
Protein lysates were prepared from four different life-cycle stages of E. tenella DS strains (UO, SO, SZ, and SM) using cell lysis buffer. The concentration of protein in each lysate was determined using a BCA protein assay kit (Beyotime). Then, the parasite protein lysate and rEtASNA1 proteins were resolved by 14% SDS-PAGE and transferred to polyvinylidene difluoride membranes (Millipore, Burlington, MA, USA) for western blotting. The membranes were blocked with 5% (w/v) skimmed milk in phosphate-buffered saline (PBS) for 2 h at 37 °C, and washed five times with PBS containing 0.5% Tween 20 (PBST). The membranes were then incubated for 2 h at 37 °C in rEtASNA1-immunized rabbit sera (1:100 dilution) or SZ-immunized rabbit sera (1:100 dilution), which were previously obtained (Han et al., 2015). A 1:1,000 dilution of anti-GST monoclonal antibody or 1:100 dilution of naive rabbit sera was used as a control, and a 1:2,000 dilution of mouse monoclonal anti-α-tubulin (Beyotime) was used as an internal reference for each group. After five washes in PBST, IRDye® 680RD donkey anti-mouse IgG or IRDye® 680CW goat anti-rabbit IgG (1:10,000 dilution) (LI-COR Biosciences, Lincoln, NE, USA) was used as secondary antibody. After five washes in PBST, the bands were detected with an Odyssey® Infrared Imaging System (LI-COR Biosciences).
Using rEtASNA1-immunized rabbit sera as a primary antibody, the differences in EtASNA1 translation between DS, DZR, MRR, and SMR strains were determined using western blotting. Using the same method, we compared the translation levels of EtASNA1 in SO at different concentrations of DZR (0.2 ppm, 0.5 ppm, 0.8 ppm, and 1.0 ppm) and MRR (3 ppm and 5 ppm) strains.
2.9. Analysis of the DNA sequence of EtASNA1 in the DS, DZR, and MRR strains
According to the instructions for the TIANamp Genomic DNA Kit (Tiangen), we extracted genomic DNA from the SO of DS, DZR, and MRR strains of E. tenella. The DNA sequence of EtASNA1 (ID: ETH_00036550) was then amplified with the following primers: 5ʹ- ATGACAGACGACGAGTTTCCCCTTG-3ʹ (forward primer) and 5ʹ- CTACTGCATCAATTCGGCGAAAGAA-3ʹ (reverse primer). Then, we cloned the amplified DNA sequences (1,270 bp) into the pGEM-Teasy cloning vector (Promega) and sent it to Sangon Biotech (Shanghai, China) for sequencing. BLAST was used to compare the DNA sequences of the three strains. All samples were sequenced and compared in triplicate.
2.10. Immunofluorescence for EtASNA1 localization
Purified SZ and SM were transferred to glass slides and air-dried as previously described (Peroval et al., 2006; Jiang et al., 2012). SZ previously incubated in complete Dulbecco's Modified Eagle's Medium (DMEM) for 2 h at 41 °C were used to invade DF-1 cells. The DF-1 cells (3 × 105 cells per well) were inoculated into the 6-well plates (Corning, Corning, NY, USA) with precoated glass coverslips, and added to purified SZ (9 × 105 cells per well) to invade cells. At various times post-inoculation, the DF-1 cells were collected and washed. SZ and cells were fixed in 2% paraformaldehyde in PBS for 15 min, followed by 1% Triton X-100 infiltration in PBS for 15 min, and then overnight in PBS containing 2% (w/v) bovine serum albumin at 4 °C. SZ and cells were incubated with rabbit anti-rEtASNA1 (1:100 dilution) for 2 h at 37 °C, and then goat anti-rabbit IgG fluorescein isothiocyanate-conjugated antibody (1:500 dilution; Sigma-Aldrich) was incubated for 1 h at 37 °C. Finally, the nuclei were incubated in 200 μL of 10 μg/mL of 4, 6-diamidino-2-phenylindole (DAPI) (Beyotime) for 20 min at 37 °C for nuclear staining. After each step described above, the slides were washed three times with PBST. The slides were then treated with 60 μL Fluoromount Aqueous Mounting Medium (Sigma-Aldrich) and observed with a fluorescence microscope (Olympus, Tokyo, Japan) and a laser scanning confocal microscope (Zeiss, Oberkochen, Germany).
2.11. SZ invasion inhibition assays
DF-1 cells (2 × 105 cells per well) were seeded in 24-well plates and cultured in DMEM with 10% fetal bovine serum at 37 °C and 5% CO2 for 24 h. Freshly isolated SZ were counted and irreversibly labeled with carboxyfluorescein diacetate succinimidyl ester (Beyotime), and incubated at 37 °C with 50, 100, 200, 300, or 400 μg/mL purified IgG against rEtASNA1 for 2 h. All antibodies in the serum were purified using protein A + G agarose (Beyotime), according to the manufacturer's instructions. At the same time, the same concentration of rabbit IgG (Sigma-Aldrich) was used as a negative control, and SZ incubated without antibody were used as the positive control. DF-1 cells were then infected with SZ (6 × 105 parasites per well) in 24-well plates and cultured for 6 h at 41 °C and 5% CO2, and then washed, trypsinized and collected for analysis using a Cytomics FC500 flow cytometer (Beckman Coulter, Brea, CA, USA). All assays were performed in triplicate, and the positive control and the invasion rate were used to calculate the inhibition rates.
2.12. In vitro effects of drugs on the transcriptional levels during reproduction
Based on Thabet et al., (2017b) and our previous qRT-PCR results, we designed the following experiment to examine the effects of drugs on transcription levels during development of the parasite. DF-1 cells (3 × 105 cells per well) were cultivated in complete medium in 6-well plates for 12 h at 37 °C and 5% CO2. The purified SZ (6 × 105 cells per well) of DS, DZR, or MRR strains were added to the cells. After 24 h of incubation, cultures were washed four times with PBS to remove extracellular SZ. After further incubation for 24 h, diclazuril (10 μg/mL) or maduramicin (10 μg/mL) was added to the DS SZ-infected group, diclazuril (10 μg/mL) to the DZR SZ-infected group, and maduramicin to the MRR (10 μg/mL) to SZ-infected group, followed by continued culturing for 48 h. The drug was added at 48–96 h after the SZ infection of cells, and then the parasites were collected for qRT-PCR. In addition, each group included a blank control without drugs.
The mRNA expression profiles of EtASNA1 at different groups were examined using qRT-PCR as previously described. Total RNA was isolated using TRIzol reagent (Invitrogen) and reverse transcribed it into cDNA as a template. A fragment of the 18S rRNA gene was used as a control.
2.13. In vivo effects of drugs on transcription levels during reproduction
We also studied the effect of drug feeding on the transcription level of SM during in vivo development. The experiment was divided into the DS-infected, non-medicated control group (DS); the DZR-infected, non-medicated control group (DZR-); the DZR-infected, fed diclazuril (1 ppm) group (DZR+); the MRR-infected, non-medicated control group (MRR-); and the MRR-infected, fed maduramicin (5 ppm) group (MRR+). In addition, we did not collect SM after adding drugs to the oocyst-infected group of DS strains. Two-week-old chickens were inoculated with oocysts of sensitive or resistant strains and were fed with or without the corresponding drugs during their growth. SM were collected and purified from the caecal mucosa of chickens at 112 h post-inoculation, followed by qRT-PCR detection.
2.14. Statistical analysis
Statistical analysis was performed using SPSS statistical software for Windows version 22 (SPSS, Chicago, IL, USA). Differences among groups were tested by the one-way analysis of variance using the Duncan test. Values of 0.01 < p < 0.05 were considered significant (denoted by *); 0.001 < p < 0.01 was considered very significant (denoted by **); and p < 0.001 was considered highly significant (denoted by ***). In the figures, significant differences (p < 0.05) between groups are represented by different letters such as a, b, c, and d.
3. Results
3.1. Cloning and sequence analysis of EtASNA1
EtASNA1 was amplified by PCR, and a 408-bp product was obtained. Nucleotide sequence analysis showed that the 408-bp open reading frame (ORF) sequence encoded a polypeptide of 135 amino acid residues with a predicted molecular mass of approximately 14.6 kDa. The sequence displayed 100% homology with E. tenella ATPase ASNA1 homolog partial mRNA (GenBank: XM013379956.1) and 98% with homology Eimeria necatrix arsenical pump-driving ATPase, putative partial mRNA (GenBank: XM_013577295.1), which proved that the EtASNA1 gene of the E. tenella Shanghai strain was successfully cloned. We submitted the sequence to NCBI GenBank (GenBank accession number: MT274691).
Sequence analyses showed that the theoretical isoelectric point of the protein was 4.67. SignalP program analysis revealed that EtASNA1 had no signal peptide or transmembrane domains. The structural module and conservative structure indicated that EtASNA1 contained two N-glycosylation sites (residues 62–65 and 102–105), an ATP/GTP-binding site motif A (residue 28–35), a cAMP and cGMP dependent protein kinase phosphorylation site (residue 48–51), three casein kinase II phosphorylation sites (residues 2–5, 11–14, and 129–132), two protein kinase C phosphorylation sites (residues 20–22 and 68–70), two N-myristoylation sites (residues 27–32 and 123–128), a FtsK domain profile (residue 5–135), a NACHT-NTPase domain profile (residue 22–47), a NIFH_FRXC family profile (residue 21–135), a protein prenyltransferases alpha subunit repeat profile (residue123–135), an anion-transporting ATPase (residue 21–134), a CobQ/CobB/MinD/ParA nucleotide binding (residue 23–68), a NifH/frxC family site (residue 22–59), and a NB-ARC domain (residue 8–41).
3.2. Expression and purification of recombinant EtASNA1
We ligated the ORF of EtASNA1 to the prokaryotic expression vector pGEX-6p-1, and successfully constructed the prokaryotic recombinant expression plasmid pGEX-6p-EtASNA1. The rEtASNA1 was expressed in E. coli BL21 as a GST-tagged fusion protein. The recombinant protein was found mainly in the inclusion bodies. After purification, by excising and extracting SDS-PAGE protein bands, the molecular mass of the rEtASNA1 fused to the GST-tag (26 KDa) was found to be approximately 40.6 kDa, as expected (Fig. 1A).
Fig. 1.
A: Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of purified rEtASNA1. B: Western blotting analysis of rEtASNA1.
Monoclonal anti-GST or rabbit antiserum against E. tenella sporozoites antibody was used as the primary antibody. Lane 1, rEtASNA1 probed with naive rabbit serum. Lane 2, rEtASNA1 probed with monoclonal anti-GST antibody. Lane 3, rEtASNA1 probed with anti-sporozoite rabbit serum.
Western blotting analyses indicated that the recombinant protein could be recognized by anti-GST monoclonal antibody and rabbit serum against SZ. Naive rabbit sera failed to detect rEtASNA1 (Fig. 1B). Overall, these results indicated that rEtASNA1 had good reactivity and was specifically recognized by anti-GST monoclonal antibody or rabbit sera against SZ.
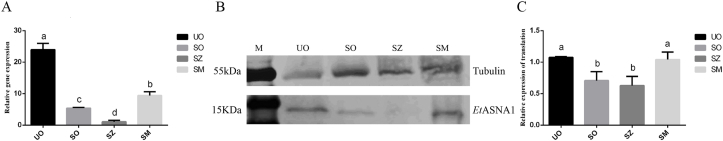
3.3. The transcription and translation levels of EtASNA1 at different developmental stages of the DS strain
Using 18S rRNA as an internal reference, we analyzed the mRNA transcription levels of EtASNA1 at UO, SO, SZ, and SM of E. tenella using qRT-PCR. The results showed that EtASNA1 transcripts were most abundant in UO and SM, and lowest in SZ (Fig. 2A).
Fig. 2.
Quantitative real-time PCR and western blotting analysis of EtSASNA1 expression at four developmental stages of the DS strain.
UO, unsporulated oocysts; SO, sporulated oocysts; SZ, sporozoites; SM, second-generation merozoites. Anti-α-tubulin antibody was used as a control. EtASNA1 was recognized by rabbit anti-rEtASNA1. A: Transcription levels of EtASNA1. B and C: Translation levels of EtASNA1. The data represent the mean ± S.D. of triplicate determinations and are representative of three independent experiments. a, b, c, and d indicate significant differences (p < 0.05) between groups.
Using mouse anti-α-tubulin monoclonal antibody as a control, western blotting was performed to analyze the translation levels of EtASNA1 in UO, SO, SZ, and SM. The levels of EtASNA1 in the UO and SM were much higher than those in the SO and SZ (Fig. 2B), which was consistent with the qRT-PCR results.
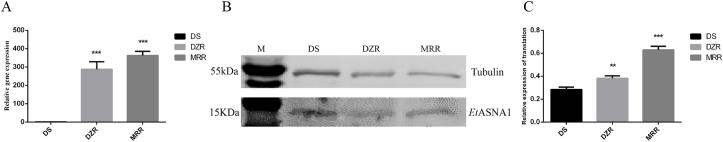
3.4. Differences in transcription and translation levels of EtASNA1 between sensitive and resistant strains
The mRNA levels of EtASNA1 in the SO stage of DS, DZR, and MRR strains were determined by qRT-PCR. Compared with the DS strain, the mRNA transcription in the DZR and MRR strains were highly significant (Fig. 3A), which were consistent with the RNA-seq results (Xie et al., 2020).
Fig. 3.
Transcription and translation levels of EtASNA1 in resistant strains in SO.
DS, drug-sensitive strain; DZR, diclazuril-resistant strain; MRR, maduramicin-resistant strain. Anti-α-tubulin antibody was used as a control. EtASNA1 was recognized by rabbit anti-rEtASNA1. A: Transcription levels of EtASNA1. B and C: Translation levels of EtASNA1. The data represent the mean ± S.D. of triplicate determinations and are representative of three independent experiments. **p < 0.01; ***p < 0.001.
Using the mouse α-tubulin monoclonal antibody as a control, we also compared the translation levels of EtASNA1 in DS, DZR, and MRR strains (SO stage) with rabbit anti-rEtASNA1 serum by western blotting. Analysis of the results showed that compared with the DS strain, the expression levels of EtASNA1 in the DZR and MRR strains were significantly upregulated (Fig. 3B), which agreed with the qRT-PCR and RNA-seq results.
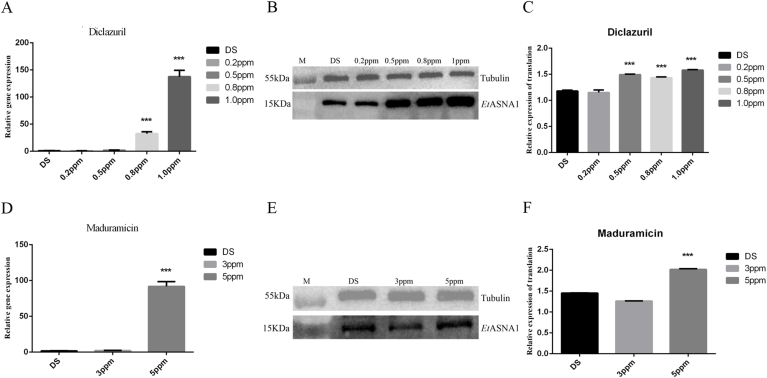
The presence of EtASNA1 mRNA in the SO of 0.2 ppm, 0.5 ppm, 0.8 ppm, and 1.0 ppm DZR strains and 3 ppm and 5 ppm MRR strains were analyzed using qRT-PCR. The expression levels increased with increases of drug concentration, and compared with the DS strain, the transcription levels in the 0.8 ppm, 1.0 ppm DZR strains, and 5 ppm MRR strain were significantly upregulated (Fig. 4A and D). Using α-tubulin as a control, we studied translation levels using western blotting. Consistent with the results of qRT-PCR, the translation levels of the 0.5 ppm, 0.8 ppm, 1.0 ppm DZR and 5 ppm MRR strains were significantly upregulated compared with the DS strain (Fig. 4B and C and 4E-F).
Fig. 4.
Transcription and translation levels of EtASNA1 at different concentrations of diclazuril-resistant (DZR) and maduramicin-resistant (MRR) strains in SO. Anti-α-tubulin antibody was used as a control. EtASNA1 was recognized by rabbit anti-rEtASNA1. A: Transcription levels of EtASNA1 at different concentrations of DZR strains. B and C: Translation levels of EtASNA1 at different concentrations of DZR strains. D: Transcription levels of EtASNA1 at different concentrations of MRR strains. E and F: Translation levels of EtASNA1 at different concentrations of MRR strains. The data represent the mean ± S.D. of triplicate determinations and are representative of three independent experiments. ***p < 0.001.
Similarly, we used qRT-PCR to analyze the transcription levels of EtASNA1 in different field isolated DZR strains (SO stage) obtained from the field from single oocysts, which showed significant upregulations in D2, D4, D5, and D8, when compared with the DS strain (Fig. 5).
Fig. 5.
Transcription levels of EtASNA1 at field diclazuril-resistant strains (SO stage). DS, drug-sensitive strain; D2-D8, four field diclazuril-resistant strains; DZR, diclazuril-resistant strain. The data represent the mean ± S.D. of triplicate determinations and are representative of three independent experiments. ***p < 0.001.
Using qRT-PCR, we also compared the transcription levels of EtASNA1 in the SO of DS and SMR strains. It was found that the transcription level of EtASNA1 in the SMR strain was significantly upregulated compared with the DS strain (Fig. 6A). Using mouse α-tubulin monoclonal antibody as a control and rabbit anti-rEtASNA1 serum as the primary antibody, we compared translation levels of EtASNA1 by western blotting. The results showed that the transcription level of EtASNA1 in the SMR strain was significantly upregulated compared with the DS strain, which was consistent with the qRT-PCR (Fig. 6B and C).
Fig. 6.
Transcription and translation levels of EtASNA1 of the salamycin-resistant (SMR) strain in SO. DS, drug-sensitive strain; SMR, salamycin-resistant strain. Anti-α-tubulin antibody was used as a control. EtASNA1 was recognized by rabbit anti-rEtASNA1. A: Transcription levels of EtASNA1 at SMR strain. B and C: Translation levels of EtASNA1 at SMR strain. The data represent the mean ± S.D. of triplicate determinations and are representative of three independent experiments. **p < 0.01; ***p < 0.001.
3.5. EtASNA1 DNA sequence analysis of the DS, DZR, and MRR strains
We obtained the 1270-bp full-length DNA sequences of EtASNA1 (Gene ID: ETH_00036550) from three strains (DS, DZR, and MRR) of E. tenella. Comparisons of sequencing and BLAST sequences revealed that the DNA sequences of DZR and MRR strains had no mutations compared with the DS strain.
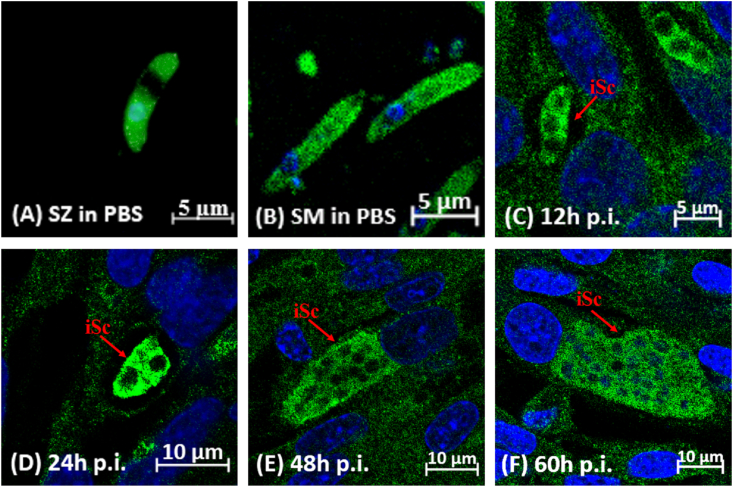
3.6. Immunofluorescence localization of EtASNA1 in parasites
Using antibody against rEtASNA1, the localization of EtASNA1 in SZ, SM, and first generation schizont was investigated in vitro by immunofluorescence. The EtASNA1 protein was located in most areas of the SZ with the exception of the refractile body and most regions of SM (Fig. 7A and B). When SZ were added to DF-1 cells for 12–60 h, EtASNA1 protein was still located in most areas of the parasites during the development from SZ to schizonts (Fig. 7C–F), but the fluorescence intensity was significantly increased when the SZ were newly developed into immature merozoites (Fig. 7D).
Fig. 7.
Indirect immunofluorescence localization of EtASNA1 at different developmental stages of E. tenella.
The rEtASNA1-immunized rabbit serum was used as primary antibody. A: Sporozoites (SZ) in phosphate buffered saline (PBS). B: Second-generation merozoites (SM) in PBS. Infected DF-1 cells were collected at the indicated times post infection (p.i.). C: Immature schizont (iSc) at 12 h p.i. D: iSc at 24 h p.i. E: iSc at 48 h p.i. F: iSc at 60 h p.i.
3.7. Inhibition of E. tenella SZ invasion by antibodies against rEtASNA1
To evaluate the role of EtASNA1 in E. tenella SZ invasion of DF-1 cells, the invasion inhibition experiment was used to test the ability of rabbit anti-rEtASNA1 antibody to inhibit the invasion of DF-1 cells by E. tenella SZ. Fig. 8 shows that compared with the same dose of naive rabbit serum IgG (used as a negative control), pretreatment with 300 and 400 μg/mL anti-rEtASNA1 IgG significantly decreased the invasion ability of E. tenella SZ, reaching an inhibition rate of 20%.
Fig. 8.
Inhibition of sporozoite invasion in vitro by antibodies against rEtASNA1. Anti-rEtASNA1, rabbit antiserum against recombinant EtASNA1 protein; IgG, naive rabbit serum. The data represent the mean ± S.D. of triplicate determinations and are representative of three independent experiments. **p < 0.01.
3.8. In vitro effects of drugs on transcriptional levels during reproduction
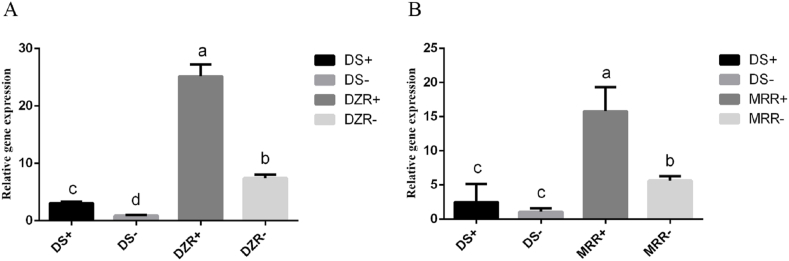
We investigated the effect of the addition of drugs on the transcription levels of EtASNA1 during the in vitro propagation of SZ of the DS, DZR, and MRR strains. We used “+” to indicate that drugs were added during reproduction, and “-” to indicate that no drugs were added. Fig. 9A shows that the transcription levels of EtASNA1 in DZR strains were usually higher than those in DS strains. We also found that in the DS or DZR strains, when compared with the non-medicated control groups, the addition of diclazuril caused an increase of EtASNA1 transcription levels. Fig. 9B shows the expressions of the DS and MRR strains with or without maduramicin, indicating that the transcription levels of the MRR strains were higher than those of DS strain, while in the MRR strains, the addition of drugs caused the transcription levels to increase.
Fig. 9.
In vitro culturing to study the effect of drugs on transcript levels of EtASNA1.
A: Treatment with diclazuril. DS+, drug-sensitive strains + diclazuril; DS-, drug-sensitive strains, drug-free; DZR+, diclazuril-resistant strains + diclazuril; DZR-, diclazuril-resistant strains, drug-free. B: Treatment with maduramicin. DS+, drug-sensitive strains + maduramicin; DS-, drug-sensitive strains, drug-free; MRR+, maduramicin-resistant strains + maduramicin; MRR-, maduramicin-resistant strains, drug-free. The data represent the mean ± S.D. of triplicate determinations and are representative of three independent experiments. a, b, c, and d indicate significant differences (p < 0.05) between groups.
3.9. In vivo effects of drugs on transcription levels during reproduction
In addition, we studied the transcriptional levels of EtASNA1 of DS, DZR, and MRR strains, which were used to infect chickens with or without drugs during development. Overall, the transcription level of EtASNA1 of drug-resistant strains was higher than that of the DS strain (p < 0.01), which was consistent with our previous qRT-PCR and western blotting results. Whether in the DZR group or the MRR group, the addition of the drugs caused a significant increase in its transcription levels (p < 0.01) (Fig. 10).
Fig. 10.
In vivo culture to study the effect of drugs on transcript levels of EtASNA1. DS, drug-sensitive strains; DZR+, diclazuril-resistant strains + diclazuril; DZR-, diclazuril-resistant strains, drug-free; MRR+, maduramicin-resistant strains + maduramicin; MRR-, maduramicin-resistant strains, drug-free. The data represent the mean ± S.D. of triplicate determinations and are representative of three independent experiments. a and b indicate significant differences (p < 0.05) between groups. **p < 0.01; ***p < 0.001.
4. Discussion
The emergence of drug resistance of Eimeria spp to anti-coccidial drugs has caused huge economic damage to the chicken industry and is currently the main problem in the prevention and control of chicken coccidiosis. In our previous study, we found that EtASNA1 was highly expressed in two drug-resistant strains (DZR and MRR), which we hypothesized might be related to the resistance of E. tenella to maduramicin and diclazuril (Xie et al., 2020). Other studies have also shown that ASNA1 plays an important role in regulating the sensitivity of melanoma cells to arsenic, antimony, and platinum, and promoting the discharge of metal ions (Hemmingsson et al., 2008). They speculated that ASNA1 is a key protein in a mechanism of RASP-phenotype mediating drug resistance and that downregulation of ASNA1 results in a specific phenotype.
In the present study, the ATPase ASNA1 homolog gene of E. tenella was cloned and analyzed. The results showed that the ORF sequence encoded a polypeptide of 135 amino acid residues. The amino acid sequence shared 96% (135/140) identity with E. necatrix putative arsenical pump-driving ATPase (XP_013432749.1) which has 140 amino acids, and 81% (109/135) identity with Eimeria praecox putative arsenical pump-driving ATPase (CDI86691.1), which has 135 amino acids and 80% (108/135) identity with Cyclospora cayetanensis ATPase ASNA1 homolog (XP_026191661.1), which has 135 amino acids. Blast searches also found that the amino acid sequence of EtASNA1 shared some identity with typical arsenite-activated ATPase family proteins of other protozoans which have approximately 400 amino acids, for example, Toxoplasm gondii and Besnoitia besnoiti. Although, the polypeptide size of EtASNA1 was much smaller than these typical ASNA1 proteins, EtASNA1 contained the same important functional sites, when compared to the typical ASNA1 proteins, so we propose that EtASNA1 also has some typical ASNA1 protein functions.
To further analyze the function of EtASNA1, qRT-PCR and western blotting were used to detect the mRNA transcription and protein translation levels of EtASNA1 at different developmental stages of E. tenella. All the results showed that EtASNA1 expression levels were higher in UO and SM stages. We speculate that the change in expression from UO to SO may be caused by changes in the environment. Some studies have shown that changes in the environment, such as heat, cold, chemical factors and nutrition, may cause changes in protein expression (Montero et al., 2008; Perez-Morales et al., 2009; Liu et al., 2012; Sun et al., 2014). The high expression observed in SM and UO may be associated with the propagation of E. tenella in the host cells and evading the host's immune response. These two stages (UO and SM) developed in the intestine of chickens and needed high metabolism to obtain nutrients from the host cells, as well as to evade the host's immune response. Studies on ASNA1 revealed that ASNA1 mRNA is highly expressed in human testis and other reproductive tissues (Bhattacharjee et al., 2001). Norlin et al. (2018) reported that inactivation of ASNA1 in pancreatic progenitor cells led to cell apoptosis, and disrupted the growth and differentiation of developing cells, which ultimately resulted in severe agenesis. In addition, the metabolic changes caused by hypoxic conditions in chickens may also be responsible for the high expression of EtASNA1 during the UO and SM stages. UO and SM were produced in chickens with insufficient oxygen, SO was obtained from UO after sufficient oxidation, and SZ was collected from SO. Under hypoxic conditions, the body regulates a variety of cellular activities to maintain homeostasis (Pugh et al., 2017). The Nakayama et al. (2019) studies in humans showed that hypoxia induces changes in transcriptional and post-transcriptional regulations of gene expression. Matthew et al. (2018) also showed that the activation of another Na + -K + -ATPase was involved in some human body reactions during hypoxia at rest.
Using indirect immunofluorescence, we found EtASNA1 was distributed throughout the parasites, whether in SZ or SM, and during the development of SZ into schizonts in DF-1 cells. ATPase is an important class of enzymes for living organisms. It exists widely and plays an important role in many biological activities. When the SZ-invaded cells began to develop, the fluorescence intensity was significantly enhanced. The previous qRT-PCR and western blotting studies also found that the protein expression level in the SM was significantly higher than that of SZ, which indicates that EtASNA1 may participate in the reproductive development of E. tenella in host cells. Some studies have reported that ASNA1 was mandatory for the growth in C. elegans (Kao et al., 2007). Other studies reported that downregulated ASNA1 expression was associated with retarded growth of ovarian cancer cells (Hemmingsson et al., 2008). Moreover, we speculate that this protein may affect the invasion of host cells, because invasion inhibition assays showed that pretreatment with the anti-rEtASNA1 polyclonal antibody can inhibit the ability of SZ to invade DF-1 cells.
Previous studies indicated that the expressions of some proteins have changed in some drug-resistant strains of parasites compared with sensitive strains. In a study of resistance to Plasmodium, it was found that Plasmodium berghei treated with dihydroartemisinin significantly reduced the expression of ribosomal protein genes in trophozoites (Shaw et al., 2015). A study of protozoan drug resistance found that Leishmania donovani resistant to antimony gluconate had high expression of cysteine-rich protein (Das et al., 2015). In our previous study, we found high expression of EtASNA1 in drug resistant strains of E. tenella.
At present, the drug resistance of Eimeria to anti-coccidial drugs is widespread. Therefore, we urgently need a method that can quickly diagnose parasite resistance using in vitro tests. In this study, we used qRT-PCR and western blotting to compare the expressions of the EtASNA1 gene between DZR, MRR, and DS strains. Analysis of the results showed that compared with the DS strain, the expression of EtASNA1 was upregulated in two drug-resistant strains. These results agree with previous findings (Xie et al., 2020). In addition, we found that the expression of DZR and MRR strains increased with the increase of drug concentrations. We hypothesized that the long-term effects of the drugs on the parasites may cause an increase in EtASNA1 expression. The study of Chen et al. (2016) on leukemia showed that expression levels of ASNA1 were negatively correlated with cell sensitivities to arsenic trioxide, and ASNA1 expression was higher in resistant cells than parental sensitive cells. Hemmingsson et al. showed that decreased ASNA1 expression was related to increased sensitivity to cisplatin and arsenite (Hemmingsson et al., 2008, 2009). In the present study, we found that compared with drug-sensitive strains, the expression levels of EtASNA1 in drug-resistant strains were all upregulated. We hypothesize that these results may also be related to the outflow of drugs from cells to reduce the effective intracellular drug concentrations. Studies have shown that the well-characterized ArsAB pump actively extrudes arsenicals and antimonials out of the cell, thereby reducing the intracellular concentration of these toxic metalloids and producing resistance (Hsu et al., 1989).
To verify the upregulated expression of EtASNA1 in different drug-resistant strains, we detected expression levels in several resistant strains isolated from the field using qRT-PCR, and found that compared with the DS strain, the expression of EtASNA1 in these strains was also significantly upregulated. However, the expression level of EtASNA1 in the field drug-resistant strains was still significantly different when compared with the laboratory-induced DZR strain, which was completely resistant to diclazuril. This might be because the wild strains we obtained showed a certain resistance to diclazuril, so they were not completely resistant to high concentrations of diclazuril. It was possible that their sensitivities to drugs were different, resulting in different levels of expression. Compared with the DS strain, the transcription and translation levels of EtASNA1 were also significantly upregulated in SMR strains induced in our laboratory. The results of our in vitro and in vivo tests also showed that the drug stimulation of parasites caused an increase in their expression levels to a certain extent, both for DS and resistant strains. In E. coli, the ArsA operates in concert with the ArsB, a transmembrane channel, to efflux arsenite. Therefore, we speculate that EtASNA1 may be related to the excretion of drugs and could participate in the metabolism of parasites, but this requires further research.
Studies have shown that mutations in drug-resistant Plasmodium genes cause increased expression (Kasturi et al., 2018). Therefore, we investigated whether the upregulated expression of EtASNA1 in drug-resistant strains and the development of resistance in drug-resistant strains were related to base mutations. We compared the EtASNA1 genomic DNA sequences of the DS, DZR, and MRR strains. However, no base mutation was found in the sequence alignments. Thus, the emergence of drug resistance in our study may be caused by other mechanisms, such as mutations in other genes encoding proteins that interact with EtASNA1. Studies have shown that mutations at certain sites of the pfkelch13 gene caused a conformational change, which reduced its binding to phosphatidylinositol-3 kinase (PfPI3K), further leading to reduced polyubiquitination of PfPI3K. This change was caused by an increase in the levels of PfPI3K and phosphatidylinositol-3-phosphate (PI3P), and induced the development of artemisinin resistance (Mbengue et al., 2015, Mbengue et al., 2015). Other studies have found that mutations in Plasmodium, Trypanosoma cruzi and T. gondii resistant strains caused amino acid substitutions and gene transporters, and further led to changes in gene expressions (Adjalley et al., 2015; Pulcini et al., 2015; García-Huertas et al., 2017; Montazeri et al., 2018). However, the reasons for the high expressions of EtASNA1 in drug-resistant strains need further study.
The high expression of EtASNA1 in resistant strains suggested that there may be some relationship between EtASNA1 and drug resistance, and it may play an important role in drug resistance and participate in many physiological and biochemical reactions. However, the process of coccidian drug resistance is very complicated, so the exact relationship between EtASNA1 and the drug resistance mechanism still needs further research.
5. Conclusions
The characteristics and partial functions of EtASNA1, an upregulated gene expressed in drug-resistant strains, were investigated in this study. It was found that the protein may be involved in the reproduction and invasion of E. tenella in host cells. The high expressions of EtASNA1 in DZR, MRR, and SMR strains may reflect its relationships with drug resistance and its involvement in many physiological and biochemical reactions.
Funding
The funding agencies played no role in the design or implementation of the study, analysis or interpretation of the data, or the preparation and submission of the manuscript.
Declaration of competing interest
The authors declare no conflict of interests.
Acknowledgments
This work was supported by the National Key R&D Program of China (2018YFD0500302), the National Natural Science Foundation of China (Grant Nos. 31970420 and 31672551), and the National Sharing Service Platform for Parasite Resources (No. TDRC-2019-194-30). We would like to thank all organizations that funded this work and all those who provided technical assistance.
References
- Adjalley S.H., Scanfeld D., Kozlowski E., Llinás M., Fidock D.A. Genome-wide transcriptome profiling reveals functional networks involving the Plasmodium falciparum drug resistance transporters PfCRT and PfMDR1. BMC Genom. 2015;16(1):1090. doi: 10.1186/s12864-015-2320-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabkhazaeli F., Modrisanei M., Nabian S., Mansoori B., Madani A. Evaluating the resistance of Eimeria spp. field isolates to anticoccidial drugs using three different indices. Iran. J. Parasitol. 2013;8:234–241. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3724148/ [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee H., Ghosh M., Mukhopadhyay R., Rosen B.P. In: Broome-Smith J.K., Baumberg S., Sterling C.J., Ward Y., editors. vol. 58. Society for General Microbiology; Leeds: 1999. pp. 58–79. (Transport of Molecules across Microbial Membranes). [Google Scholar]
- Bhattacharjee H., Ho Y.S., Rosen B.P. Genomic organization and chromosomal localization of the Asna1 gene, a mouse homologue of a bacterial arsenic-translocating ATPase gene. Gene. 2001;272(1–2):291–299. doi: 10.1016/s0378-1119(01)00522-4. [DOI] [PubMed] [Google Scholar]
- Blake D.P., Tomley F.M. Securing poultry production from the ever-present Eimeria challenge. Trends Parasitol. 2014;30(1):12–19. doi: 10.1016/j.pt.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Chapman H.D. Biochemical, genetic and applied aspects of drug resistance in Eimeria parasites of the fowl. Avian Pathol. 1997;26:221–244. doi: 10.1080/03079459708419208. [DOI] [PubMed] [Google Scholar]
- Chapman H.D., Jeffers T.K., Williams R.B. Forty years of monensin for the control of coccidiosis in poultry. Poultry Sci. 2010;89(9):1788–1801. doi: 10.3382/ps.2010-00931. [DOI] [PubMed] [Google Scholar]
- Chen J., Cheng J., Yi J., Xie B., Lin L., Liu Z., Zhao H.S., Wang B., Ai Z.Y., Yang Y., Wei H.L. Differential expression and response to arsenic stress of MRPs and ASAN1 determine sensitivity of classical multidrug-resistant leukemia cells to arsenic trioxide. Leuk. Res. 2016;50:116–122. doi: 10.1016/j.leukres.2016.10.003. [DOI] [PubMed] [Google Scholar]
- Das S., Shah P., Tandon R., Yadav N.K., Sahasrabuddhe A.A., Sundar S., Siddiqi M.M., Dube A. Over-expression of cysteine leucine rich protein is related to sag resistance in clinical isolates of Leishmania donovani. PLoS Neglected Trop. Dis. 2015;9(8) doi: 10.1371/journal.pntd.0003992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Huertas P., Mejía-Jaramillo A.M., González L., Triana-Chávez O. Transcriptome and functional genomics reveal the participation of adenine phosphoribosyltransferase in Trypanosoma cruzi resistance to benznidazole. J. Cell. Biochem. 2017;118(7):1936–1945. doi: 10.1002/jcb.25978. [DOI] [PubMed] [Google Scholar]
- Györke A., Pop L., Cozma V. Prevalence and distribution of Eimeria species in broiler chicken farms of different capacities. Parasite. 2013;20:50. doi: 10.1051/parasite/2013052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H.Y., Kong C.L., Dong H., Zhu S.H., Zhao Q.P., Zhai Q., Liang S.T., Li S., Yang S.H., Huang B. Molecular characterization and functional analysis of subunit 7 of eukaryotic initiation factor 3 from Eimeria tenella. Exp. Parasitol. 2015;154:118–126. doi: 10.1016/j.exppara.2015.04.002. [DOI] [PubMed] [Google Scholar]
- Han H.Y., Lin J.J., Zhao Q.P., Dong H., Jiang L.L., Xu M.Q., Zhu S.H., Huang B. Identification of differentially expressed genes in early stages of Eimeria tenella by suppression subtractive hybridization and cDNA microarray. J. Parasitol. 2010;96(1):95–102. doi: 10.1645/ge-2221.1. [DOI] [PubMed] [Google Scholar]
- Han H.Y., Zhao Q.P., Chen Z.G., Huang B. Induction of diclazuril resistant strains and madumycin resistant strains of Eimeria tenella in the laboratory. Chin. J. Vet. Sci. 2004;24(2):138–140. [Google Scholar]
- Hemmingsson O., Nöjd M., Kao G., Naredi P. Increased sensitivity to platinating agents and arsenite in human ovarian cancer by downregulation of ASNA1. Oncol. Rep. 2009;22(4):869–875. doi: 10.3892/or_00000511. [DOI] [PubMed] [Google Scholar]
- Hemmingsson O., Zhang Y., Still M., Naredi P. ASNA1, an ATPase targeting tail-anchored proteins, regulates melanoma cell growth and sensitivity to cisplatin and arsenite. Canc. Chemother. Pharmacol. 2008;63(3):491–499. doi: 10.1007/s00280-008-0762-2. [DOI] [PubMed] [Google Scholar]
- Hsu C.M., Rosen B.P. Characterization of the catalytic subunit of an anion pump. J. Biol. Chem. 1989;264:17349–17354. https://www.jbc.org/content/264/29/17349.long [PubMed] [Google Scholar]
- Huang B., Wu X.Z., Shi T.W., Chen Z.G. Study on the identification and pathogenicity of the pure species of Eimeria tenella. Chin. J. Vet. Parasitol. 1993;1(4):18–20. [Google Scholar]
- Jiang L.L., Huang B., Han H.Y., Zhao Q.P., Dong H., Chen Z.G. Comparison of the proteome of the sporutated oocysts of Eimeria tenella diclazuril sensitive strain with diclazuril resistant strain. Chin. J. Biotechnol. 2005;21(3):435–439. https://pubmed.ncbi.nlm.nih.gov/16108370/ [PubMed] [Google Scholar]
- Jiang L.L., Lin J.J., Han H.Y., Zhao Q.P., Dong H., Zhu S.H., Huang B. Identification and partial characterization of a serine protease inhibitor (serpin) of Eimeria tenella. Parasitol. Res. 2012;110(2):865–874. doi: 10.1007/s00436-011-2568-0. [DOI] [PubMed] [Google Scholar]
- Kao G., Nordenson C., Still M., Ronnlund A., Tuck S., Naredi P. ASNA-1 positively regulates insulin secretion in c. elegans and mammalian cells. Cell. 2007;128:577–587. doi: 10.1016/j.cell.2006.12.031. [DOI] [PubMed] [Google Scholar]
- Karaer Z., Guven E., Akcay A., Kar S., Nalbantoglu S., Cakmak A. Prevalence of subclinical coccidiosis in broiler farms in Turkey. Trop. Anim. Health Prod. 2012;44:589–594. doi: 10.1007/s11250-011-9940-z. [DOI] [PubMed] [Google Scholar]
- Kasturi H., Souvik B., Innocent S. Drug resistance in Plasmodium. Nat. Rev. Microbiol. 2018;16(3):156–170. doi: 10.1038/nrmicro.2017.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalafalla R.E., Daugschies A. Single oocyst infection: a simple method for isolation of Eimeria spp. from the mixed field samples. Parasitol. Res. 2010;107(1):187–188. doi: 10.1007/s00436-010-1840-z. [DOI] [PubMed] [Google Scholar]
- Kurdi-Haidar B., Hom D.K., Flittner D.E., Heath D., Fink L., Naredi P., Howell S.B. Dual cytoplasmic and nuclear distribution of the novel arsenite-stimulated human ATPase (hASNA-I) J. Cell. Biochem. 1998;71(1):1–10. https://pubmed.ncbi.nlm.nih.gov/9736449/ [PubMed] [Google Scholar]
- Liu Z., Xi D., Kang M., Guo X., Xu B. Molecular cloning and characterization of Hsp27.6: the first reported small heat shock protein from Apis cerana cerana. Cell Stress Chaperones. 2012;17(5):539–551. doi: 10.1007/s12192-012-0330-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Med. Sci. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Matthew L., Anne R.C., Gary J.L., Dennis G.L., Frank A.D. Inhibition of Na +/K + -ATPase and K IR channels abolishes hypoxic hyperaemia in resting but not contracting skeletal muscle of humans. J. Physiol. 2018;596(15):3371–3389. doi: 10.1113/jp275913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbengue A., Bhattacharjee S., Pandharkar T., Liu H.T., Estiu G., Stahelin R.V., Rizk S.S., Njimoh D.L., Ryan Y., Chotivanich K., Nguon C., Ghorbal M., Lopez-Rubio J., Pfrender M., Emrich S., Mohandas N., Dondorp A.M., Wiest O., Haldar K. A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature. 2015;520(7549):683–687. doi: 10.1038/nature14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbengue A., Bhattacharjee S., Pandharkar T., Liu H., Estiu G., Stahelin R.V., Rizk S.S., Njimoh D.L., Ryan Y., Chotivanich K., Nguon C., Ghorbal M., Lopez-Rubio J.J., Pfrender M., Emrich S., Mohandas N., Dondorp A.M., Wiest O., Haldar K. A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature. 2015;520(7549):683–687. doi: 10.1038/nature14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald V., Shirley M.W. Past and future: vaccination against Eimeria. Parasitology. 2009;136(12):1477–1489. doi: 10.1017/s0031182009006349. [DOI] [PubMed] [Google Scholar]
- McDougald L.R., Mathis G.F., Seibert B.P. Anticoccidial efficacy of diclazuril against recent field isolates of Eimeria from commercial poultry farms. Avian Dis. 1990;34(4):911–915. https://pubmed.ncbi.nlm.nih.gov/2282020/ [PubMed] [Google Scholar]
- Mehlhorn H., Pooch H., Raether W. The action of polyether ionophorous antibiotics (monensin, salinomycin, lasalocid) on developmental stages of Eimeria tenella (Coccidia, Sporozoa) in vivo and in vitro: study by light and electron microscopy. Z. Parasitenkd. 1983;69(4):457–471. doi: 10.1007/bf00927702. [DOI] [PubMed] [Google Scholar]
- Min W., Dalloul R.A., Lillehoj H.S. Application of biotechnological tools for coccidia vaccine development. J. Vet. Sci. 2004;5(4):279–288. https://pubmed.ncbi.nlm.nih.gov/15613810/ [PubMed] [Google Scholar]
- Miska K.B., Fetterer R.H., Barfield R.C. Analysis of transcripts expressed by Eimeria tenella oocysts using subtractive hybridization methods. J. Parasitol. 2004;90(6):1245–1252. doi: 10.1645/ge-309r. [DOI] [PubMed] [Google Scholar]
- Montazeri M., Mehrzadi S., Sharif M., Sarvi S., Tanzifi A., Aghayan S.A., Daryani A. Drug resistance in toxoplasma gondii. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.02587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero E., Rodriguez M., Gonzalez L.M., Lobo C.A. Babesia divergens: identification and characterization of BdHSP-20, a small heat shock protein. Exp. Parasitol. 2008;119(2):238–245. doi: 10.1016/j.exppara.2008.01.020. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay R., Ho Y.S., Swiatek P.J., Rosen B.P., Bhattacharjee H. Targeted disruption of the mouse Asna1 gene results in embryonic lethality. FEBS Lett. 2006;580:3889–3894. doi: 10.1016/j.febslet.2006.06.017. [DOI] [PubMed] [Google Scholar]
- Nakayama K., Kataoka N. Regulation of gene expression under hypoxic conditions. Int. J. Mol. Sci. 2019;20(13):3278. doi: 10.3390/ijms20133278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norlin S., Parekh V., Edlund H. The ATPase activity of Asna1/TRC40 is required for pancreatic progenitor cell survival. The Company of Biologists Ltd. 2018;145(1):1–12. doi: 10.1242/dev.154468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peek H.W., Landman W.J. Coccidiosis in poultry: anticoccidial products, vaccines and other prevention strategies. Vet. Q. 2011;31(3):143–161. doi: 10.1080/01652176.2011.605247. [DOI] [PubMed] [Google Scholar]
- Perez-Morales D., Ostoa-Saloma P., Espinoza B. Trypanosoma cruzi SHSP16: characterization of an alpha-crystallin small heat shock protein. Exp. Parasitol. 2009;123(2):182–189. doi: 10.1016/j.exppara.2009.06.019. [DOI] [PubMed] [Google Scholar]
- Peroval M., Pery P., Labbe M. The heat shock protein 90 of Eimeria tenella is essential for invasion of host cell and schizont growth. Int. J. Parasitol. 2006;36:1205e1215. doi: 10.1016/j.ijpara.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Pugh C.W., Ratcliffe P.J. New horizons in hypoxia signaling pathways. Exp. Cell Res. 2017;356(2):116–121. doi: 10.1016/j.yexcr.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulcini S., Staines H.M., Lee A.H., Shafik S.H., Bouyer G., Moore C.M., Daley D.A., Hoke M.J., Altenhofen L.M., Painter H.J., Mu J.B., Ferguson D.J.P., Llinás M., Martin R.E., Fidock D.A., Cooper R.A., Krishna S. Mutations in the Plasmodium falciparum chloroquine resistance transporter, PfCRT, enlarge the parasite's food vacuole and alter drug sensitivities. Sci. Rep. 2015;5:14552. doi: 10.1038/srep14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard R.B. Chapter 32 elution of proteins from gels. Methods Enzymol. 2009;463:565–572. doi: 10.1016/S0076-6879(09)63032-9. [DOI] [PubMed] [Google Scholar]
- Sharman P.A., Smith N.C., Wallach M.G., Katrib M. Chasing the golden egg: vaccination against poultry coccidiosis. Parasite Immunol. 2010;32:590–598. doi: 10.1111/j.1365-3024.2010.01209.x. [DOI] [PubMed] [Google Scholar]
- Shaw P.J., Chaotheing S., Kaewprommal P., Piriyapongsa J., Wongsombat C., Suwannakitti N., Koonyosying P., Uthaipibull C., Yuthavong Y., Kamchonwongpaisan S. Plasmodium parasites mount an arrest response to dihydroartemisinin, as revealed by whole transcriptome shotgun sequencing (RNA-seq) and microarray study. BMC Genom. 2015;16:830. doi: 10.1186/s12864-015-2040-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley M.W., Coudert P., Eckert J., Braun R. Biotechnology—Guidelines on Techniques in Coccidiosis Research. The European Commission DGXII; Luxembourg City, Luxembourg: 1995. Eimeria species and strains of chickens; p. 24. [Google Scholar]
- Sun H., Wang L., Wang T., Zhang J., Liu Q., Chen P., Chen Z., Wang F., Li H., Xiao Y., Zhao X. Display of Eimeria tenella EtMic2 protein on the surface of Saccharomyces cerevisiae as a potential oral vaccine against chicken coccidiosis. Vaccine. 2014;32(16):1869–1876. doi: 10.1016/j.vaccine.2014.01.068. [DOI] [PubMed] [Google Scholar]
- Suo X., Wang M., Wu W.X., Pan B.L. Efficacy of einerivac plus, a coccidiosis vaccine, against coccidiosis in broilers under field conditions. J. Anim. Sci. Vet. Med. 2001:265–369. 03. [Google Scholar]
- Thabet A., Honscha W., Daugschies A., Bangoura B. Quantitative proteomic studies in resistance mechanisms of Eimeria tenella against polyether ionophores. Parasitol. Res. 2017;116(5):1553–1559. doi: 10.1007/s00436-017-5432-z. [DOI] [PubMed] [Google Scholar]
- Thabet A., Zhang R., Alnassan A.A., Daugschies A., Bangoura B. Anticoccidial efficacy testing: in vitro Eimeria tenella assays as replacement for animal experiments. Vet. Parasitol. 2017;233:86–96. doi: 10.1016/j.vetpar.2016.12.005. [DOI] [PubMed] [Google Scholar]
- Tomley F.M. Techniques for isolation and characterization of apical organelles from Eimeria tenella sporozoites. Methods Enzymol. 1997;13:171–176. doi: 10.1006/meth.1997.0509. [DOI] [PubMed] [Google Scholar]
- Tseng Y.Y., Yu C.W., Liao V.H. Caenorhabditis elegans expresses a functional ArsA. FEBS J. 2007;274:2566–2572. doi: 10.1111/j.1742-4658.2007.05791.x. [DOI] [PubMed] [Google Scholar]
- Vanparijs O., Marsboom R., Desplenter L. Diclazuril, a new broad spectrum anticoccidial drug in chickens. 1. Dose titration studies and pilot floor pen trials. Poultry Sci. 1989;68(4):489–495. doi: 10.3382/ps.0680489. [DOI] [PubMed] [Google Scholar]
- Vermeulen A.N. Progress in recombinant vaccine development against coccidiosis. A review and prospects into the next millennium. Int. J. Parasitol. 1998;28:1121–1130. doi: 10.1016/s0020-7519(98)00080-0. [DOI] [PubMed] [Google Scholar]
- Wang H.X., Zhao Q.P., Zhu S.H., Huang B., Dong H., Wang Q.J., Yu S.L., Yu Y., Liang S.S., Han H.Y. Laboratory induction of salamycin resistant strains of Eimeria tenella. Chin. J. Anim. Infect. Dis. 2019;1–9 http://kns.cnki.net/kcms/detail/31.2031.s.20190917.1154.025.html [Google Scholar]
- Xie Y.X., Huang B., Xu L.Y., Zhao Q.P., Zhu S.H., Zhao H.Z., Dong H., Han H.Y. Comparative transcriptome analyses of drug-sensitive and drug-resistant strains of Eimeria tenella by RNA-sequencing. J. Eukaryot. Microbiol. 2020;67(1):1–11. doi: 10.1111/jeu.12790. [DOI] [PubMed] [Google Scholar]