Abstract
Despite intensive research, brain tumors are amongst the malignancies with the worst prognosis; therefore, a prompt diagnosis and thoughtful assessment of the disease is required. The resistance of brain tumors to most forms of conventional therapy has led researchers to explore the underlying biology in search of new vulnerabilities and biomarkers. The unique metabolism of brain tumors represents one potential vulnerability and the basis for a system of classification. Profiling this aberrant metabolism requires a method to accurately measure and report differences in metabolite concentrations. Magnetic resonance-based techniques provide a framework for examining tumor tissue and the evolution of disease. Nuclear Magnetic Resonance (NMR) analysis of biofluids collected from patients suffering from brain cancer can provide biological information about disease status. In particular, urine and plasma can serve to monitor the evolution of disease through the changes observed in the metabolic profiles. Moreover, cerebrospinal fluid can be utilized as a direct reporter of cerebral activity since it carries the chemicals exchanged with the brain tissue and the tumor mass. Metabolic reprogramming has recently been included as one of the hallmarks of cancer. Accordingly, the metabolic rewiring experienced by these tumors to sustain rapid growth and proliferation can also serve as a potential therapeutic target. The combination of 13C tracing approaches with the utilization of different NMR spectral modalities has allowed investigations of the upregulation of glycolysis in the aggressive forms of brain tumors, including glioblastomas, and the discovery of the utilization of acetate as an alternative cellular fuel in brain metastasis and gliomas. One of the major contributions of magnetic resonance to the assessment of brain tumors has been the non-invasive determination of 2-hydroxyglutarate (2HG) in tumors harboring a mutation in isocitrate dehydrogenase 1 (IDH1). The mutational status of this enzyme already serves as a key feature in the clinical classification of brain neoplasia in routine clinical practice and pilot studies have established the use of in vivo magnetic resonance spectroscopy (MRS) for monitoring disease progression and treatment response in IDH mutant gliomas. However, the development of bespoke methods for 2HG detection by MRS has been required, and this has prevented the wider implementation of MRS methodology into the clinic. One of the main challenges for improving the management of the disease is to obtain an accurate insight into the response to treatment, so that the patient can be promptly diverted into a new therapy if resistant or maintained on the original therapy if responsive. The implementation of 13C hyperpolarized magnetic resonance spectroscopic imaging (MRSI) has allowed detection of changes in tumor metabolism associated with a treatment, and as such has been revealed as a remarkable tool for monitoring response to therapeutic strategies. In summary, the application of magnetic resonance-based methodologies to the diagnosis and management of brain tumor patients, in addition to its utilization in the investigation of its tumor-associated metabolic rewiring, is helping to unravel the biological basis of malignancies of the central nervous system.
Keywords: Brain tumors, Metabolomics, 13C-tracing, MRS, Hyperpolarization
1. Introduction
1.1. Brain malignancies
Central nervous system (CNS) tumors account for about 1.5% of all new cases of cancer and for 3% of related deaths [1]. CNS tumors come in many types, but the majority of malignant brain tumors originate in the glial cells (gliomas), which are further subdivided in the latest World Health Organization (WHO) classification (2016) by the specific cell type into astrocytic and oligodendroglial tumors, other astrocytic tumors, ependymal tumors and neuronal and mixed neuronal-glial tumors [2,3]. Secondary CNS tumors also frequently occur from metastasis into the brain from another tumor location and are the most common group of intracranial malignancies (10 times higher than primary brain tumors) [4]. Almost 30% of cancer patients suffer from secondary CNS tumors [5], with lung and breast adenocarcinomas and melanoma being the main primary sources. Once diagnosed, brain metastasis is always fatal, with survival rates ranging from 2 months for untreated to 1 year for intensively treated cases. The brain metastatic process involves the migration of tumor cells from the primary tumor, followed by intravasation of the surrounding vessels, dissemination over the body throughout the circulatory system, and finally, extravasation and crossing of the blood-brain-barrier, after which the cells settle and proliferate within the brain.
Diagnosis of a CNS cancer typically involves the acquisition of an MRI (Magnetic Resonance Imaging) scan, usually including gadolinium as contrast agent. Different modalities of MR-based imaging methods can be applied to inform about the location, cellularity, and vascularity of the tumor [6], which are essential features in determining treatment and monitoring the disease. Once diagnosed, radiotherapy, chemotherapy including temozolomide, and surgical resection are the most common management approaches, generally in combination. Prognosis is highly variable among CNS malignancies. Glioblastomas (GBM) present the lowest survival rates (ca. 5% of diagnosed cases survive beyond 5 years) compared to the more favorable prognosis of pilocytic astrocytoma of > 90% 10-year survival rates, which is even higher than for isocitrate dehydrogenase 1 (IDH1) mutant diffuse gliomas that present intermediate survival rates ranging from 20% (anaplastic astrocytoma, WHO III) to 65% (oligodendroglioma, WHO II).
While anatomical MRI is often sufficient for the diagnosis of a CNS malignancy, prognosis depends on the underlying molecular characteristics of the tumor, which are probed at best indirectly by anatomical imaging. Gliomas, for instance, which account for the 75% of total CNS malignancies, are further stratified based on molecular markers; thus IDH1 mutational status is the first node for grades II-IV gliomas [7]. It is now well-established that tumors experience a metabolic rewiring to maintain high levels of synthesis in a challenging environment [8,9]. The metabolic reprogramming undergone by tumors is an important element of the microenvironment in which the treatment response occurs and a shift in the metabolic profile may even emerge as an active adaptive response to treatment [10]. Beyond serving as a marker of treatment response [11], knowledge of metabolism can also inform treatment planning. For example, high rates of glycolysis strongly correlate with radiation resistance in multiple cancer models [12,13]. As more treatment modalities become available, more precise molecular profiling is expected to occupy a more central role in designing an efficient therapeutic strategy. Magnetic resonance-based analysis of biofluids, with its high reproducibility [14–16] and relative insensitivity to sample environmental factors [17], can form a quantitative basis for the profiling of this metabolic shift. Characteristic NMR-based metabolic profiles can also be obtained from samples collected from diseased individuals as well as from diseased tissue in vivo.
1.2. High resolution NMR spectroscopy
NMR studies of tissue and biofluids are commonly focused on 1H and 31P in the field of metabolomics, since 1H and 31P are both sensitive in comparison to other biologically relevant nuclei and present at or near 100% natural abundance (Table 1). While 1H is present in almost all biomolecules, 31P NMR is useful for studies of cellular energy states in vivo and ex vivo, but the overlapping of 31P signals from many phosphorylated compounds is a limitation that must be overcome. Other nuclei of interest include 13C, 15N and 2H, for which isotopic enrichment is usually required.
Table 1.
Properties of NMR-active nuclei frequently utilized in NMR-based metabolic investigations: Sensitivity is given normalized to that of 1H.
| Nucleus | Natural abundance (%) | Spin (I) | Relative Sensitivity |
|---|---|---|---|
| 1H | 99.99 | ½ | 1.0000 |
| 2H | 0.01 | 1 | 0.0035 |
| 13C | 1.11 | ½ | 0.0159 |
| 15N | 0.36 | ½ | 0.0010 |
| 31P | 100 | ½ | 0.0665 |
High-Resolution NMR (HR-NMR) allows the identification of metabolites in complex mixtures based on their chemical structure. Nuclei in different chemical environments (e.g. the CH3 and CH2 protons of ethanol) experience different local fields, and hence give rise to signals of different frequencies, because of the shielding effects of the local electrons. These frequency differences are normally expressed in terms of the chemical shift, which has dimensionless units of parts per million (ppm), and they can be used to identify different molecules within a sample. Additional analytical information is provided by the phenomenon known as spin-spin coupling, whereby resonances are split into multiplets as a result of through-bond interactions between nuclear spins. The frequency difference between the components of a given multiplet is known as the spin-spin coupling constant, J.
The intensities, or areas, of the signals are directly proportional to the number of nuclei that give rise to them, though they are dependent in some circumstances on other factors too, including the pulse sequence that is used for spectral acquisition.
2. Ex vivo NMR metabolomics in body fluids and tissue in brain malignancies
2.1. NMR data acquisition and processing
Both urine and blood can be employed to seek biomarkers of disease or to monitor treatment response in a non-invasive fashion by ex vivo NMR (Fig. 1). One of the main advantages of both these biofluids is that the samples can be collected non-invasively and the spectra can be acquired from the neat sample in the case of urine, or directly after blood fractionation for plasma, without any extraction procedure, making them ideal for large scale screening. As the samples are simple fluids, conventional solution NMR techniques can be employed for detection [18,19]. Typically, a 1D NOESY sequence is used for the analysis of biological samples [20], with presaturation for water suppression [21] rather than WET [22], excitation sculpting [23] or WATERGATE [24].
Fig. 1.
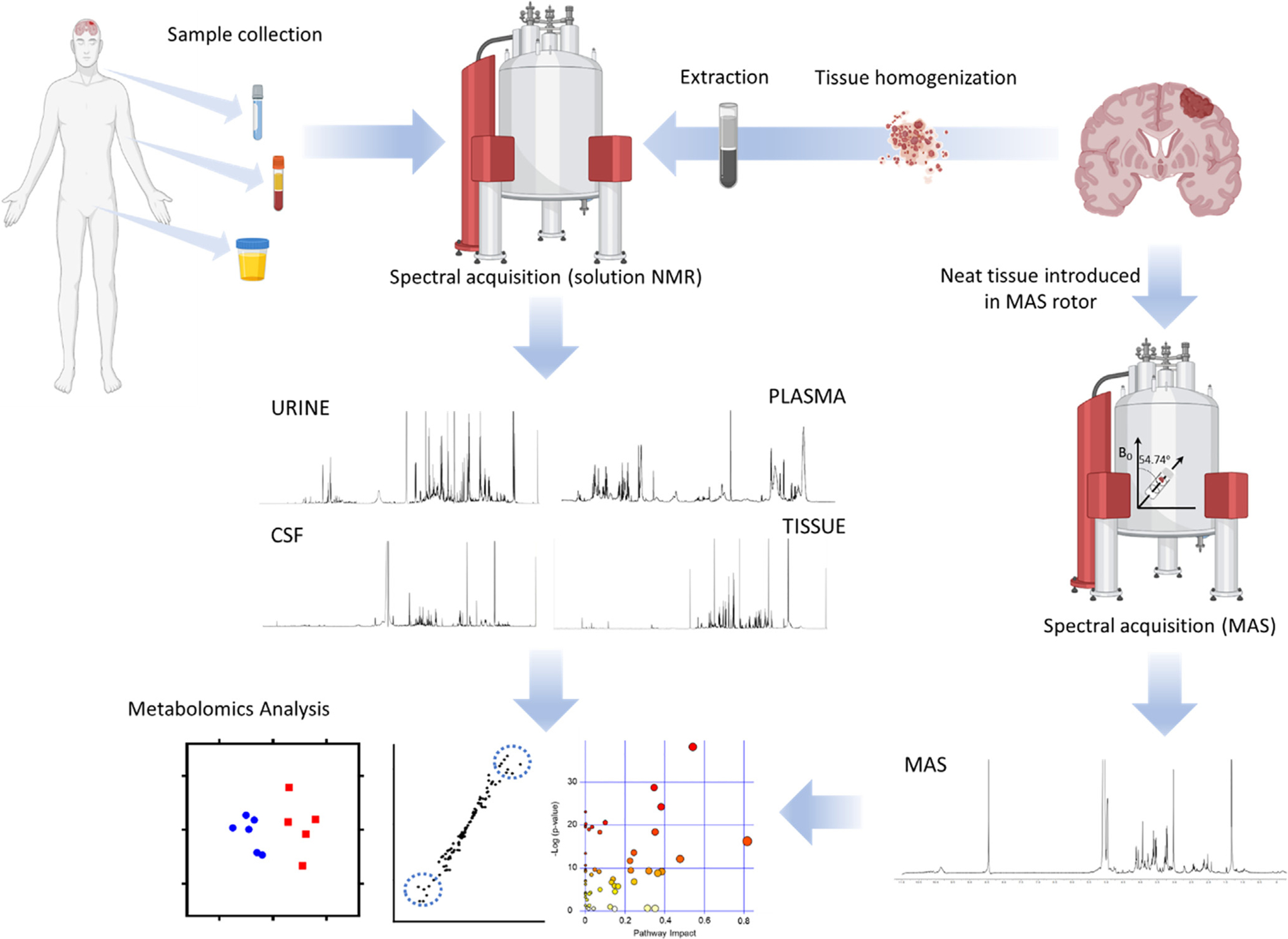
Workflow for metabolomics investigations of brain tumors. After sample (CSF, blood, urine) collection from patients or animal models the preparation steps are minimal, involving mixing the sample with D2O and a buffer, prior to transfer the sample to an NMR tube and the acquisition of the spectra. Analysis of tissue requires the mechanical homogenization of the specimen and subsequent extraction utilizing organic solvents to create a biphasic preparation after centrifugation. Both polar (upper phase) and organic (bottom phase) fractions are dried and reconstituted in a deuterated solvent. Alternatively, tissue can be directly inserted into the MAS rotor for spectral acquisition (MAS spectrum courtesy of Dr. Madhu Basetti, University of Cambridge). Spectra are then analyzed by quantifying the assigned metabolites or by bucketing of the spectra to generate datasets that can be employed for multivariate analysis. This approach usually involves the clustering of samples according to disease status or treatment based on their metabolic profiles, the search of dysregulated metabolites that can be utilized as biomarkers and the pathway analysis to assign the changes in metabolite levels to a specific metabolic route.
Whole blood can be separated by centrifugation to obtain plasma and serum components. Both plasma and serum contain macromolecules which give rise to broad resonances that obscure the underlying signals. The broad macromolecule signals can be attenuated by the Carr-Purcell-Meiboom-Gill (CPMG) pulse block [25,26], which works as a T2 filter to attenuate signals from macromolecules with short T2 relaxation times. However, the filter is not perfectly efficient, and substantial sample heterogeneity can exist from both the pulse imperfections and paramagnetic effects from the bound heme. For this reason, proteins are normally removed by either precipitation or ultrafiltration to enhance accurate quantification. In contrast to plasma, serum contains red blood cells which, once extracted, give rise to an additional set of metabolites [27].
Although 1D acquisition protocols for routine samples are standardized [28] and relatively insensitive to radiofrequency pulse imperfections except in the region near the water peak [29], analysis of the spectra is not straightforward. Several alternatives exist for the quantification of spectral peaks. The first is to simply measure the peak intensities or areas. While this method is frequently used for the targeted analysis of individual well-resolved peaks, it often poses a problem in congested regions of the spectrum where overlapping peaks are not easily resolved. Alternatively, the spectra can be approximated to a linear combination of the spectra of reference metabolites [30–32], using least squares [33] or Bayesian methods of analysis [34]. While this approach avoids the need for manual assignment, it has the drawback of being highly dependent on exact referencing and alignment [35]. Most 1H chemical shifts are reproducible to within 0.03 ppm [36]. A subset of peaks, however, is particularly sensitive to environmental effects and can vary widely with sample conditions [37]. Weak acids and bases such as citrate and histidine are particularly prone to this effect, due to acid/base equilibrium and possible chelation of metal ions by the base [37]. Careful sample handling can minimize variability [38], but the residual error can still complicate analysis. The alignment problem can be mitigated by binning the spectra over a narrow spectral region. The optimal binning procedure is an area of active research; proposed methods including genetic algorithms and [39,40] probabilistic graphical models [41]. Analysis itself can proceed either by univariate (each metabolite is treated independently) or multivariate methods. Multivariate analysis is often preferred as a natural correlation exists between metabolites within the same pathway, and different strategies have been implemented in metabolomics investigations [42,43].
NMR, LC- (Liquid Chromatography) and GC-MS (Gas Chromatography-Mass Spectrometry) are the more common analytical techniques employed for metabolomics investigations. While MS-based approaches cover a wider spectrum of the metabolome and have a higher sensitivity (10–100 times higher than NMR), NMR spectroscopy can be used to assess unique classes of metabolites such as lipoproteins, which cannot be done via LC/GC-MS. Sample treatment for GC/LC-MS usually involves more steps, including derivatization sometimes, and a prior chromatographic separation is required. Also, quantification of metabolites can be easily done in an NMR spectrum since the intensity of the signal is directly proportional to the metabolite concentrations and number of nuclei in the molecule; contrarily, the intensity of the peak in mass spectrometry is usually affected by the degree of ionization of the compound.
2.2. Urine and blood
In an early NMR-based metabolomics investigation of plasma from brain tumor patients, using a 300 MHz system for spectral acquisition [44], methyl signals arising from plasma lipoproteins were utilized to classify patients according to disease status; additionally, the authors performed 13C NMR analysis on plasma to support their findings about the correlation of the lipoprotein content in plasma and the presence of a brain neoplasia. However, the accurate quantification of the different classes of lipoproteins by NMR has not been resolved until recently [45], which may have prevented the utilization of less specific classes of lipoproteins as biomarkers. Indeed, NMR-based investigation of plasma from brain tumor patients has been limited, with the most extensive work conducted recently, including the plasma analyses of 70 glioma patients and 70 healthy subjects [46]. These authors found 20 metabolites with differential levels which were mostly assigned to tricarboxylic acid (TCA) cycle intermediates and other energy metabolites [46]; these results were in accordance with other reports [47] highlighting citrate, pyruvate, glucose, formate and creatine as key features for discriminating plasma samples collected from healthy volunteers from those obtained from brain tumor patients. Furthermore, Baranovicova et al. [47] were able to discriminate between each individual type of brain malignancy and the controls based on the plasma metabolite profile. Differences among the plasma profiles of patients with different brain tumors and healthy controls were also reported through the application of 31P and 1H NMR to the serum lipid fraction. This work revealed higher quantities of phospholipids and total cholesterol compared to normal individuals. Additionally, a comparative analysis among tumor types revealed that the ratio of total phospholipids to phosphatidylcholine (computed by 31P) was significantly different for medulloblastoma patients and normal subjects [48]. The authors attribute those increased levels to their higher utilization in brain tumor tissue, leading to enhanced synthesis in the liver, and hence, translated to elevated levels in blood.
Despite the high incidence of brain metastasis in cancer patients [49,50], predictors of this fatal process are not yet fully established; thus, metabolomics studies focused on the profiling of biofluids to search for disease biomarkers may provide a way forward. Accordingly, Larkin et al. investigated the 1H NMR urinary profiles of mice harboring the 4 T1-GFP brain metastasis cell line by collecting urine samples at different time points and applying multivariate analysis strategies such as O-PLS-DA (Orthogonal Projections to Latent Structures Discriminant Analysis) in order to find markers of disease [51]. Key metabolites for clustering samples according to sampling time were allantoin, citrate, trimethylamine, trimethylamine-N-oxide, 2-oxoglutarate, creatinine, taurine and creatine + phosphocreatine.
2.3. Cerebrospinal fluid
Cerebrospinal fluid (CSF) plays a key role in the CNS circulation of nutrients filtered from the blood in addition to cleansing the system of metabolic by-products. Therefore, it can be utilized to investigate the metabolic alterations associated with a CNS disease, such as a brain malignancy.
The first investigations involving NMR analysis of CSF were conducted in the 1980s [52] in which resonances in the 1D 1H spectra were assigned to 17 metabolites on the basis of 2D J-resolved and COSY/TOCSY spectra [53,54]. This original metabolic profiling of CSF was further expanded to a more complete characterization of CSF by 1H NMR years later [55], and the creation of a database for the CSF metabolome [56]. The first work including tumor patients reported the levels of 19 metabolites in addition to an analysis of the difference between CSF from normal individuals and brain tumor patients [57]. However, the lipid content of CSF was not included in these investigations until Srivastava et al. published phospholipids and total cholesterol resonance signals in CSF lipid extracts from brain tumor patients by 1H NMR; interestingly, those metabolites were not observable in the spectra acquired from healthy controls [48].
Leptomeningeal carcinomatosis is a disease with a median survival for diagnosed patients of approximately 2–4 months [58]; it encompasses the infiltration of the leptomeninges and CSF by malignant cells from a primary tumor such as breast cancer, lung cancer, and melanoma. Therefore, this disease provides a direct interaction between tumor cells and CSF that can be monitored through NMR analysis. CSF was collected at different time points after intradural cell implantation of F-98 cells into the parietal area of Fisher 344 rats. This biofluid was then analyzed by High Resolution Magic Angle Spinning (HR-MAS) to evaluate its ability as a reporter of disease, yielding sensitivity and specificity values of 89%. Higher levels of lactate, creatine and acetate were observed in the diseased animals while glucose levels were lower compared to those of the normal group [59]. The authors translated this approach into a human study involving 41 normal subjects and 26 patients. The goal was to improve the standard diagnostic evaluation which includes CSF cytology and contrast-enhanced MRI of the brain and spine. Multivariate analysis of the CSF-NMR profiles generated a classifier including myo-inositol, creatine, lactate, alanine and citrate which yielded an AUC (area under the curve) of 0.996 in a ROC (receiving operating characteristic) curve analysis [60]. Although this NMR-based diagnostic strategy offered better assessment than standardized procedures, the authors did not define these metabolic classifiers as pathognomonic, since similar findings can be attributed to other diseases.
2.4. Tissue
Tissue samples can be extracted and analyzed through standard water suppression sequences. Alternatively, the spectral acquisition can be carried out on the unextracted tissue using HR-MAS, thus preserving the sample for additional analysis such as histopathology. This method was introduced in 1996 [61], based on the work of Andrew and Lowe [62,63] and allows the acquisition of HR spectra but using intact tissue. The sample is usually inserted into a HR-MAS rotor and a small amount of deuterated solvent is added to provide a deuterium lock. This rotor is spun around an axis inclined at an angle of 54.7° (magic angle) to the static magnetic field (B0), which reduces the line-broadening that is typical of solid and semi-solid materials. As with solution NMR, the prominent water peak has to be suppressed, and as the sample contains both low molecular weight metabolites and macromolecules, broad macromolecular signals are usually reduced by the application of the CPMG sequence to allow visualization of overlapping signals from less concentrated small molecules.
Combination of the non-T2 filtered spectra, the CPMG sequence and a diffusion-edited acquisition that allows the visualization of macromolecules, can be utilized to retrieve all the spectral features from a single sample [64]. This approach was applied to discern glioma grading through the use of multivariate curve resolution [65] for metabolite quantification from the combined spectra [64]. This technique allows assignment of the spectral contributions of different metabolites when their resonances overlap, yielding a more accurate computation of metabolite levels. A similar question was addressed by Erb. et al. [66] in a HR-MAS metabolomics study of oligodendrogliomas, revealing higher levels of alanine and valine in high-grade, and elevated levels of proline, glutamate, glutamine, γ-aminobutyrate (GABA) and N-acetylaspartate (NAA) in low grade tumors. The authors assigned these differences to a switch from TCA to glycolysis metabolism attributable to a more hypoxic environment in high-grade oligodendrogliomas [66]. A recent investigation classified gliomas from different grades based on their 1H NMR profiles and found choline metabolism to be the most dysregulated pathway, and the levels of taurine to be the most powerful feature in classifying patients by O-PLS-DA when comparing low to high grades [67]. Likewise, a study [68] involving childhood brain and nervous system tumors found high levels of taurine in neuroblastomas as well as high phosphocholine (PC), along with its ratio to glycerophosphocholine (GPC). Additionally, phosphoethanolamine (PE) levels were significantly higher in medulloblastomas, in agreement with previous reports [69], and were also elevated in atypical meningiomas when compared to benign cases [70]. The metabolic profile of meningiomas has recently been explored by HR-MAS and the outcomes were correlated to the expression of the proliferation marker Ki-67, histological evaluation, and the expression of progesterone receptor [71]. These malignancies have been also investigated through solution 1H NMR, which reported higher levels of alanine and creatine in non-recurrent meningiomas compared to those experiencing rapid recurrence; furthermore, these two metabolites displayed significantly lower levels in grades II and III when compared to grade I [72]. Additional investigations [73] aiming at classifying tumors according to grade based on their HR-MAS profiles included studies of astrocytoma grades I, II and III, GBM and medulloblastoma specimens. Main features involved in the classification of each tumor type included NAA, lactate, creatine, myoinositol and glycine; however, some limitations arise from the low number of samples included for some tumor types, such as n = 2 for grade I astrocytoma and n = 3 for GBM [73]. Similarly, in a different study, HR-MAS was used to differentiate astrocytoma grade II, grade III gliomas, glioblastomas, metastases, meningiomas and lymphomas [74].
Vettukattil and coworkers [75] investigated the metabolic differences in Grade II astrocytoma and GBMs based on their 1H and 31P HR-MAS profiles. They found higher levels of GPC and myo-inositol in astrocytoma than in GBM and lower levels of PC, glycine, and lipids. Additionally, they subclassified GBMs according to recurrence, and reported higher levels of PC in non-recurrent tumors, in addition to higher levels of GPC, myo-inositol, and creatine, with lower levels of glycine.
Griffin and colleagues utilized a rat glioma model (BT4C cells on female BDIX rats) to quantify the 1H NMR-visible lipid resonances in vivo by MRS and ex vivo via HR-MAS during programmed cell death. They found that the concentration of polyunsaturated fatty acids increased three-fold during this process. The group identified the resonances attributable to the olefinic protons as the most significant in monitoring the dynamics of apoptosis [76]. Furthermore, their work has shown that both in vivo and in vitro glycine and creatine concentrations were strongly correlated with cell density, whereas choline-containing compounds were unaffected by cell loss [77]. Meanwhile, both saturated and unsaturated 1H NMR-visible lipids increased. However, choline-containing compounds were unaffected by a decrease in cell density. Taken together, the work of this group indicates the potential for 1H NMR to track the progress of cell death and drug efficacy in glioma cells through lipid analysis. The metabolism of these lipids was also suggested to be imbalanced in an investigation involving the NMR analysis of tissue from WHO grade I, II glioma (n = 7) WHO grade III, IV glioma (n = 12) and gliosis (n = 2) using a 900 MHz system [78]. Indeed, dysregulation of lipids associated with glioma grade was previously reported by Lehnhardt et al. [79], who showed an increase in PC and a decrease in sphingomyelin correlated with higher grades in a 31P analysis of extracted samples [79].
The combination of advanced multivariate analysis strategies together with the metabolic profiling of tumor tissue can provide classifiers to aid in the diagnosis of CNS malignancies. Constantin et al. performed multivariate analysis on HR-MAS spectra to distinguish between low grade gliomas that recurred at different grades. They applied several classification strategies such as linear discriminant analysis, logistic ridge regression, classification trees, support vector machines (SVM) and logit boost decision stumps. Features more frequently included in the classification models were myoinositol, 2HG, hypotaurine, choline compounds, glutathione (GSH), lipids and alanine [80]. SVM and neural networks were also implemented together with a novel 2D HR-MAS method, i.e. TOBSY (total through bond spectroscopy) [81] to classify tumor biopsies according to their type [82]. Jalbert et al. [83] developed a flow chart for classification of low-grade-gliomas using HR-MAS on brain tumor biopsies based on the levels of key metabolites. Similarly, a classification tree was built utilizing NMR spectra of 46 brain tumors biopsies for predicting the overall survival of patients in a retrospective study [84]. The authors found myo-inositol, GPC, glycine and alanine to be the key features for classification of patients according to short, intermediate and long survival.
Although the majority of investigations have been focused on the classification of the different types of brain malignancies, some researchers have also reported the variances between tumor and non-tumor tissues. Main differences were found in those metabolites usually identified with neural cells, such as NAA and GABA, that were found at lower levels in tumors [85]. NAA and aspartate were also found to be robust classifiers for clustering normal samples through SVM [82].
In contrast to DNA sequencing analysis, which only provides the underlying blueprint, ex vivo analysis of tissue by NMR provides a snapshot of the tumor biochemistry and through that snapshot, a direct measure of the functioning of the cell at that point in time. It is therefore ideal for measuring properties such as treatment response that change dynamically with time. However, it is limited by the sensitivity of the technique, which restricts the analysis to only the most abundant metabolites and tumor reprogramming which does not affect metabolism cannot be detected by this technique. Even for reprogramming which does involve metabolic changes, the alterations must result in a substantial change in the absolute concentration at steady state. A change induced by metabolic reprogramming which results in the increase of the relative concentration of a trace intermediate, for instance, or in increased flux through a little used pathway is difficult to detect reliably by ex vivo NMR.
Within this limited context, ex vivo NMR metabolomics has still proved useful in diverse areas of preclinical and clinical research. Common tasks in drug development such as identifying metabolic dysregulation in cancer, monitoring treatment response, flagging possible toxicity at the preclinical stage, among many others, have all benefited from the technique. The biomarkers first established by ex vivo NMR can also be used for the development of non-invasive MRS methods in clinical research. The main benefit of ex vivo NMR is its high reproducibility in comparison to other common analytical techniques which makes the quantification of even modest differences between groups possible [36]. Moreover, NMR is a non-destructive technique, an exceptional feature that is particularly relevant in the case of MAS, where the same tissue can be utilized for spectral acquisition and histopathology assessment. This can become a key component of the diagnosis process, mainly in IDH1 mutant tumors, where the presence of 2HG is a reporter of IDH1 mutational status in gliomas.
3. 13C-tracing for glioma metabolism
Isotope tracing may be employed to track the metabolic fate of a specific group within a metabolite, or the breakdown of a uniformly labeled substrate throughout different metabolic pathways. In view of the low natural abundance of 13C (ca. 1%), substrates have to be labeled in the positions of interest, so that 13C tracing can be applied to the study of glioma metabolism (Fig. 2A) to probe different routes, mainly based on its ability to provide information on positional isotopomers. A labeled tracer allows only the products of the labeled metabolite to be detected, eliminating contributions from other sources.
Fig. 2.
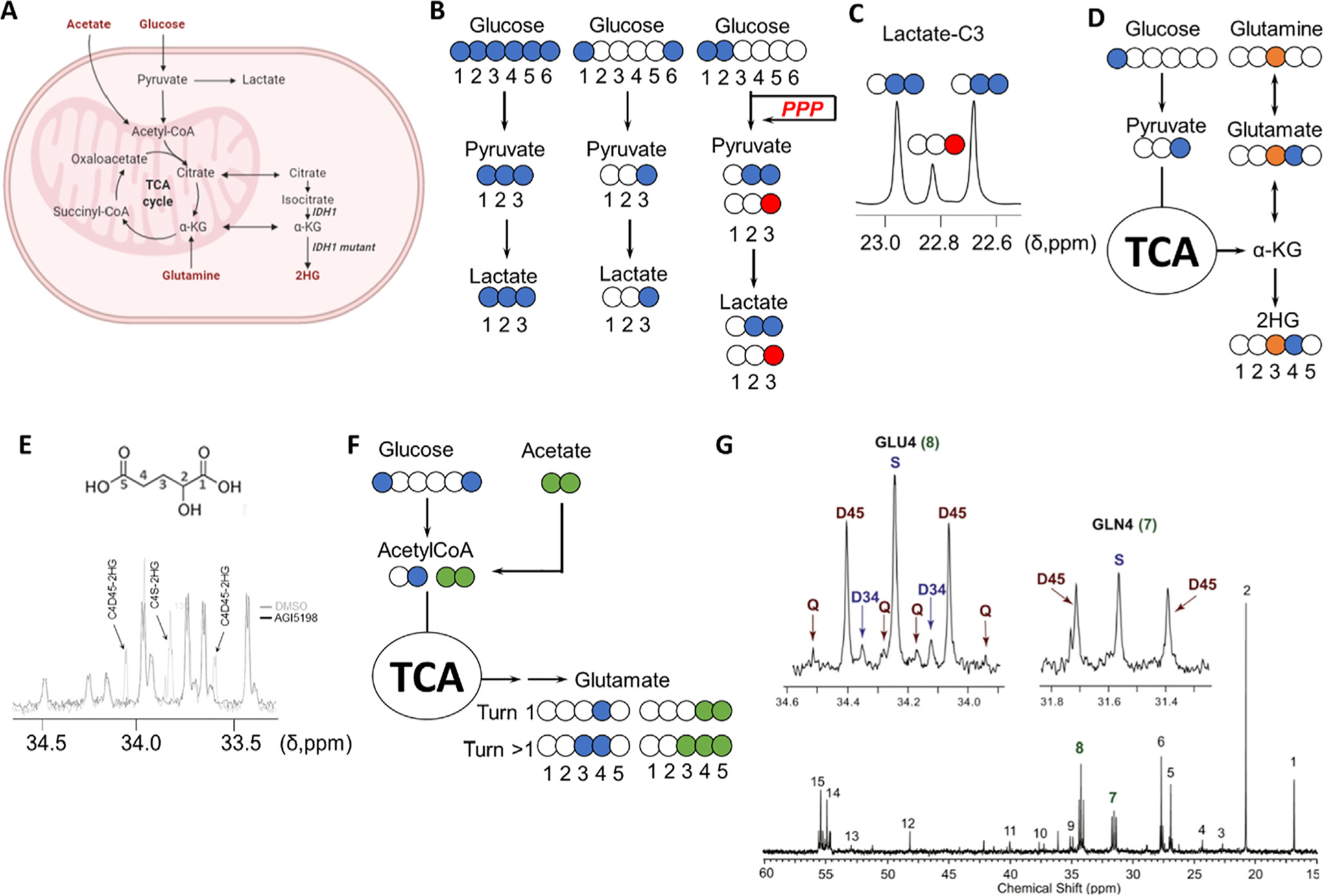
Investigation of metabolic rewiring in CNS malignancies. A, Key metabolic pathways involved in tumor metabolism. B, 13C tracing-based approaches to investigate the upregulation of glycolysis in cancer: blue colored circles denote the location of the 13C atoms in the substrates as well as in the subsequent metabolic products through glycolysis (blue) or pentose phosphate pathway (red). [1,2-13C]-glucose generates [2,3-13C]-lactate through glycolysis and [3-13C]-lactate if glucose-6P is diverted to the pentose phosphate pathway (PPP) by glucose-6P dehydrogenase, and once transformed into glyceraldehyde-3P, enters back into the glycolytic route. The contribution of each of those pathways to lactate formation can be quantified by comparing the areas under the resonance signals of lactate in a 13C spectrum, as the multiplet generated contains the information about the levels of both isotopologues. D, Analysis of the contribution of glucose and glutamine to 2HG synthesis in IDH1 mutant gliomas by examination of the resonances of C3 and C4 of 2HG from C3-13C-glutamine (orange circle) and C1-13C-glucose. E, Multiplet arising from 13C labeling of 2HG from TS603 cell line seeded in media containing [U-13C]-Glutamine and analyzed as described in [86]. 2HG structure with assignments displayed on top of the 13C spectrum [87]. F, Investigation of acetate contribution to the TCA through coadministration of [1,6-13C]-glucose (blue circles) and [1,2-13C]-acetate (green circles). (G) The multiplets attributable to incorporation of 13C into positions 3,4 and 5 (top) in both glutamine and glutamate C4 group serve to assign the specific contribution of each substrate (glucose and acetate) to the TCA. A 13C NMR GBM tumor spectrum after co-infusion is displayed. Chemical shift assignments can be found in the original article. Reproduced from [88] with permission.
3.1. Metabolic rewiring in glioma cell lines
13C tracers have been extensively used in cell culture to trace main metabolic pathways such as glycolysis and the TCA cycle. Tumors, including gliomas [89] have long been thought to primarily metabolize glucose as the principal nutrient, mainly to obtain energy through the so-called Warburg effect. In view of this enhanced glycolytic activity, initial research on glioma metabolism was focused on tracing glucose oxidation and consequent lactate production (Fig. 2B and 2C). One of the first investigations involving the 13C NMR analysis of brain tumor extracts to study the metabolic reprogramming associated with the malignancy was conducted by Brand et al. in 1992. The authors evaluated the labeling pattern of 13C labeled metabolites in C6 glioma and NIE-115 neuroblastoma cells by culturing these cells in [2-13C]- or [3-13C]-pyruvate [90]. The majority of research was subsequently performed through incubation of glioma cell lines with [3-13C]-lactate or [1-13C]-glucose and monitoring lactate production and its release into the media [91–93]. The relevance of assessing lactate in media is based on the assumption that the extracellular levels in vitro may mimic the hypoxic environments [94] usually encountered in brain tumors [95].
More sophisticated approaches appeared years later to explore the kinetics of metabolite exchange with the medium by incubating C6 glioma cells in 50% D2O and combinations of [1-13C]-glucose and [U-13C]-lactate [96]. The authors evaluated the formation of [2-13C]-pyruvate and the incorporation of 2H into lactate, since this isotope will be transferred from NADH(2H) to 13C pyruvate (derived from 13C lactate) in the LDH equilibrium. 13C Lactate resonances and the modified multiplet pattern arising from deuterium labeling of lactate were used to assess the rates of these reactions.
Thereafter, several lines of research suggested the possibility that additional metabolic sources, rather than glucose, could account for the generation of energy and the supply of intermediates for tumor proliferation [97]. Therefore, investigations were conducted to resolve the contribution of these additional nutrients for cell growth. An NMR compatible perfusion system [98] was employed to compute the glycolytic and TCA activities using [1,6-13C]-glucose and [3-13C]-glutamine in SF188 cells in order to investigate the metabolism of proliferating GBM cells [99]. Thompson et al. found that lactate was produced from glutamine through the action of malic enzyme, which they linked to the concomitant production of NADPH [99]. Formation of [2-13C] glutamate from [1,6-13C]-glucose suggested complete TCA cycle activity and functional oxidative metabolism. Furthermore, the incorporation of [U-13C]-glucose and [3-13C]-glutamine into lipids was tracked through the analysis of the signals arising from the labeling of the acyl chain of the fatty acids in different positions. The triplets in ω-1 (second last position in the acyl chain) located at 23 ppm and ω-2 (32.3 ppm) in the 13C 1D NMR spectrum attributable to the labeling pattern R-13CH2-13CH2-13CH3 for ω-1 and R-13CH2-13CH2-13CH2-13CH3 for ω-2 reflect the contribution of two labeled units of acetyl-CoA to fatty acid synthesis. The labeling pattern analysis showed that 60% of the total signal was attributed to the triplet, revealing a preferential use of glucose for lipogenesis.
3.2. Tracking the effect of the IDH1 mutation on metabolic reprogramming by 13C NMR
The isocitrate dehydrogenase I gene (IDH1) has been the subject of particular attention in metabolic studies, both because of its high prevalence in gliomas as well as its value as a prognostic factor in diagnosis. IDH1 mutations occur in up to 80% of low-grade (WHO grade II) gliomas and about 5% of all glioblastomas (GBM) [100,101]. These mutations overwhelmingly occur at Arg132, which is most commonly mutated to histidine [102] and only a single copy of the gene seems to be affected in the vast majority of cases [102]. The resulting mutated enzyme gains the ability to produce the D enantiomer of 2-hydroxyglutarate (2HG) [103]. 2HG contributes to genomic instability by inhibiting the activity of ALKBH repair enzymes [104], which also sensitizes IDH1 tumors to alkylating agents [105]. 2HG is also an inhibitor of prolyl hydroxylases [106], which stabilizes the transcription factor HIF-1α that regulates the hypoxia response. 2HG also induces a multitude of epigenetic changes by impairing histone demethylation [107], for which the interested reader is referred to several excellent reviews [103,108].
By altering the flux through the isocitrate dehydrogenase pathway and by diverting NADPH to an alternative pathway, the IDH1 mutant reprograms the metabolism of these tumors, which may require a diversion of alternative pathways to generate TCA intermediates, since glutamine is now partially dedicated to the synthesis of 2HG and an alternate source of NADPH is needed to replace the lost flux from the decarboxylation reaction. One possible alternative to compensate for downregulated TCA intermediates is the upregulation of pyruvate carboxylase to generate oxaloacetate from pyruvate. This reaction can be tracked through [2-13C]-glucose by analyzing the levels of the resulting [3-13C]-glutamate resonance (27.6 ppm) against the [5-13C] one at 182 ppm. 13C NMR analysis of [2-13C]-glucose-derived 13C-labeled glutamate and glutamine in cell culture showed metabolic reprogramming by the IDH1 mutation which involves an increase in pyruvate carboxylase activity and downregulation of pyruvate dehydrogenase (PDH) [109]. This finding was further confirmed using hyperpolarized [2-13C]-pyruvate in the same biological systems [110]. Moreover, pyruvate carboxylase was also revealed as the main entry into the TCA cycle utilizing U-13C-glucose tracing in additional IDH1 mutant models [87]. Despite the reduction in PDH activity due to the IDH1 mutation, NHA (normal human astrocytes) harboring the IDH1 mutation and BT142 (an endogenous IDH1 mutant oligoastrocytoma cell line) still displayed a glucose to TCA flux higher than the GBM model investigated (U87). These observations reinforced the description of IDH1 mutant gliomas as being TCA-dependent, in contrast to the Warburg-like phenotype characteristic of GBMs [111]. 2HG synthesis has also been explored as it would involve a rewiring of the cellular metabolism to provide the substrate for the IDH1 mutant reaction. Accordingly, a combined analysis of [1-13C]-glucose and [3-13C]-glutamine was applied to the assignment of both glucose and glutamine as substrates of 2HG [110] (Fig. 2D). Recently, 13C tracing combined with LC-MS and NMR analyses has been applied to a GEMM (Genetically Engineered Mouse Model) of IDH1 mutant glioma [112]. 1D-HSQC spectra acquired from tumor tissue extracts infused with either [U-13C]-glucose or [U-13C]-glutamine revealed the latter as the main 2HG substrate in this model [113]. Additionally, its synthesis was also used as a reporter of response to treatment with XL765 [114], an inhibitor of the PI3K/mTOR axis [115]. Treatment with this agent in IDH1 mutant NHA and U87 models generated a distinct 1H NMR metabolic profile with 2HG as one of the key features dysregulated [114]. 13C NMR studies confirmed a decrease in 2HG synthesis in IDH1 mutant cell lines after inhibition of the PI3K/mTOR pathway as a result of a drop in the flux of glutamine and glucose towards its formation [114].
Phospholipid homeostasis has also been linked to IDH1 mutation; indeed, a dysregulation in phosphocholine/ethanolamine (PC and PE) and their adducts, GPC and glycerophosphorylethanolamine (GPE) was found in glioma xenografts harboring the IDH1 mutation by 31P MRS, and was further confirmed in IDH1-R132H-expressing U251 glioma cell lines by HR-MAS [116]. 31P NMR analysis of mouse xenografts revealed that NHA and U87 cell lines transduced with the mutant IDH1 R132H gene displayed lower levels of PC and PE in the polar extracts [117], and phosphatidylcholine (PtdC) and phosphatidylethanolamine (PtdE) were reduced in the lipid fraction [116,118]. Wild type and IDH1 mutant cell lines were cultured in media containing either [1,2-13C]-choline or [1,2-13C]-ethanolamine to track their incorporation into the correspondent phospholipids by 13C and 31P NMR over time [119] in order to estimate choline kinase (CK) and ethanolamine kinase (EK) activities [118]. The decreased synthesis of these metabolites was validated by treating endogenously mutant IDH1 and wild type cell lines with the selective IDH1 inhibitor AGI-5198 [120] (Fig. 2D). An analysis of orthotopic IDH1 mutant glioma xenografts after treatment showed decreased synthesis of PC, PtdC, PE and PtdEs, which the authors assigned to HIF-1α stabilization by 2HG [106]. Expression of the rate limiting enzymes of PtdC and PtdE biosynthesis, PC and PE cytidylyltransferases, was also downregulated as a function of 2HG concentration [118]. Since these enzymes reside in the endoplasmic reticulum (ER), the authors investigated the effect of 2HG in this organelle. Autophagy of the ER was also found to be triggered by the presence of 2HG [118], which inhibits collagen-4-prolyl hydroxylase activity, leading to accumulation of misfolded procollagen-IV.
3.3. Ex situ investigations of glioma metabolism by 13C NMR of extracted tissue
One of the first studies involving the quantification of 13C incorporation into metabolites within brain tumor tissue was conducted over 20 years ago in a C6 rat model of glioma. Animals were perfused for 1 h with a [1-13C]-glucose solution and the enrichment of glucose, lactate, alanine, GABA, glutamate, glutamine and aspartate were computed for tumor and non-tumor regions upon tissue retrieval [121]. In order to perform 13C tracing analysis on human patients, these subjects are usually infused through intravenous injection with a solution containing the 13C tracer prior to tumor resection [88,122,123]. Subsequently, collected tumor tissue is snap frozen and finally metabolites are extracted for NMR analysis. DeBerardinis et al. provided a protocol for 13C labeling in brain malignancies, including brain metastasis, which allowed the acquisition of very well resolved 13C resonance peaks. This approach serves to unambiguously resolve the multiplets of each signal, and thereby quantify the contribution of a specific pathway to the formation of a target metabolite. GBM human orthotopic tumors (HOTs) have also been analyzed by isolating cells from freshly resected tumors that are implanted directly into the NOD-SCID mouse brain, avoiding the cellular adaptation to in vitro culture [124]. Using [1,6-13C]- and [U-13C]-glucose and examining the multiplet appearance of labeled metabolites, Marin-Valencia et al. were able to show, based on the 2–3 doublet in glutamate-C2 from [U-13C]-glucose, that the TCA entry involving PC was active in tumors, although multiple turns of the TCA may also produce this doublet. To resolve this ambiguity, they used [3,4-13C]-glucose, since this probe only transfers the 13C into aspartate if PC is active. They observed labeling of C1 and C4 in aspartate and an excess of label in C1 relative to C5 in glutamate and glutamine. Together, the data suggested that PC contributes to anaplerosis in these tumors and that 13C-labeled glucose or lactate entered the tumor and was oxidized within the TCA cycle. When these authors investigated the fate of glutamine through [U-13C]-glutamine they observed that tumors display doublets at the glutamate C4 position arising from the labeling of C4 and C5 but not at C3, and thus the pattern cannot be produced from the direct conversion of [U-13C]-glutamine to [U-13C]-glutamate.
Based on the work of Henry et al. [125], demonstrating the utility of co-infusing [1,6-13C]-glucose and [1,2-13C]-acetate to explore the metabolism of both substrates simultaneously, and previous reports showing the metabolism of acetate in mouse brain [126], Mashimo et al. [88] reported the utilization of acetate in primary brain tumors and metastasis (Fig. 2F and 2G). Additionally, they confirmed that glutamine is not a preferential substrate for oxidation for the TCA in GBM. As a follow-up of this work, the same group investigated 13C patterns in glutamate and glutamine pools of tumor patients infused with [1,2-13C]-acetate to discriminate the intrinsic tumor acetate metabolism from that attributable to peripheral metabolism [127], which was reported by the presence of [2,3-13C]- and [1,2-13C]-lactate. The utilization of alternative substrates by brain tumors was also explored by De Feyter et al. [128] by evaluating the fate of [2,4-13C2]-β-hydroxybutyrate in RG2 and 9L glioma models through in vivo MRS and HR-NMR analysis ex vivo. These authors revealed how brain tumors have the ability to oxidize ketone bodies based on the labeling of the C4-glutamate signal.
4. Detection of biomarkers by in vivo Magnetic resonance spectroscopy
4.1. Methods for spatial localization of the signal by MRI
In contrast to solution NMR experiments, performing spectroscopy on a living sample requires a method of localizing the signal only to the region of interest. Fundamentally, spatial localization is accomplished by gradients applied across the sample that translate physical distances into field differences. A pulse acquire sequence without any gradient will give a signal reflective of the entire sample encased within the detection coil (in the case of a volume coil) or of the region in the immediate vicinity of the coil (in the case of a surface coil) and is usually only of interest when the entire sample can be encased within the coil (for example, mouse leg xenografts in a volume coil) or the tumor is at the surface (mouse subcutaneous tumors and surface coils). Acquiring a volume of interest (VOI) requires either the elimination of all magnetization outside the VOI or specific excitation of the VOI. Spoiling outside magnetization efficiently typically requires rf powers in excess of what can be generated safely clinically [129]. Volume-specific excitation is therefore the dominant method, although saturation pulses are often used to suppress signal bleed-through from outside the VOI.
For the acquisition of spatially localized MR spectra (i.e. spectra acquired from a single selected volume of interest), three major pulse sequence families (PRESS, STEAM and ISIS) are in widespread use and usually available on commercial MRI scanners. The PRESS sequence takes advantage of the fact that the Fourier transform of a sinc function is a 2D slab whose physical thickness is proportional to the ratio of the rf bandwidth to the magnitude of the field gradient in the direction perpendicular to the slice. Following a slab-selective pulse, two 180° refocusing pulses are then applied in the presence of gradients applied in orthogonal directions [130]. Only the VOI at the center of the three slices experiences all three pulses and is refocused for detection. Because of the dependence on the spin echo, the signal is weighted towards metabolites with less rapid T2 decay. The two echo times (subechoes) can be varied independently to alter the appearance of the spectrum to minimize or maximize specific metabolites based on the J-coupling.
Metabolites with short T2 values cannot be measured efficiently with the PRESS sequence. To alleviate this problem, the STEAM sequence substitutes the two spin echoes in the transverse plane with stimulated echoes in a 90° z-90° y-90° x sequence. The magnetization is stored along the z-axis after the second pulse, which eliminates T2 relaxation at that point. Hence, the STEAM sequence is preferred for fast relaxing species such as free lipids, and those with coupled-spin systems, including GABA and myoinositol, as shorter echoes can be employed, which limit spin evolution from T2 relaxation and J-coupling [131]. The use of 90° rather than 180° pulses also reduces power requirements, which need to be limited in some cases to prevent tissue heating. The drawback is the relative insensitivity of the technique. Half of the magnetization is not refocused in the final step, although this theoretical sensitivity advantage of STEAM over PRESS is often partially offset by greater signal loss with increasing echo time in the case of STEAM [132]. The signals obtained with both STEAM and PRESS are both acquired by refocusing the signal and are therefore T2 weighted. The ISIS sequence, by contrast, uses sequential acquisition of orthogonal slabs which are combined post-acquisition to form the VOI [133]. Since no echoes are used, this provides a possible alternative for fast relaxing species. Correct acquisition is dependent on the accurate subtraction of large signals and the method is prone to dynamic range problems, particularly for small VOIs [132]. Residual magnetization from imperfect pulses due to B1 inhomogeneity can also cause imperfect subtraction if the sample is not fully relaxed before the next step [129]. The result is often a selection profile that contains substantial contributions from outside the VOI.
An alternative approach, known as chemical shift imaging, produces an image where each volume element (or voxels) yields a spectrum. It is very slow in its standard form. However, a wide variety of techniques have been used to speed up acquisition including compressed sensing, low rank reconstruction, and multi-shot techniques. The interested reader is referred to other reviews on the subject for modern acquisition techniques [134].
4.2. Metabolic profiling by in vivo MRS
The first 1H MRS investigations of a brain tumor were carried out almost 30 years old ago [135]. Since then, numerous investigations have been conducted with a view to providing a characteristic phenotype associated with a specific brain tumor type (Fig. 3A) or to identifying metabolic markers of disease progression and response to treatment (see Table 2). Typically, the levels of a few metabolites such as lactate, lipids, NAA, Glx (glutamate and glutamine combined), total creatine (creatine + phosphocreatine), total choline (choline-containing compounds) and myo-inositol are evaluated as potential biomarkers for CNS malignancies. A major complication of characterizing the chemical composition of tumor tissue by MRS lies in the potential heterogeneity of the voxel selected. Tumor masses can include edema and necrotic cells which can contribute to the metabolic profile. Multivoxel MRSI partially resolves this complication by collecting spectra from different regions that can also include surrounding tissue.
Fig. 3.
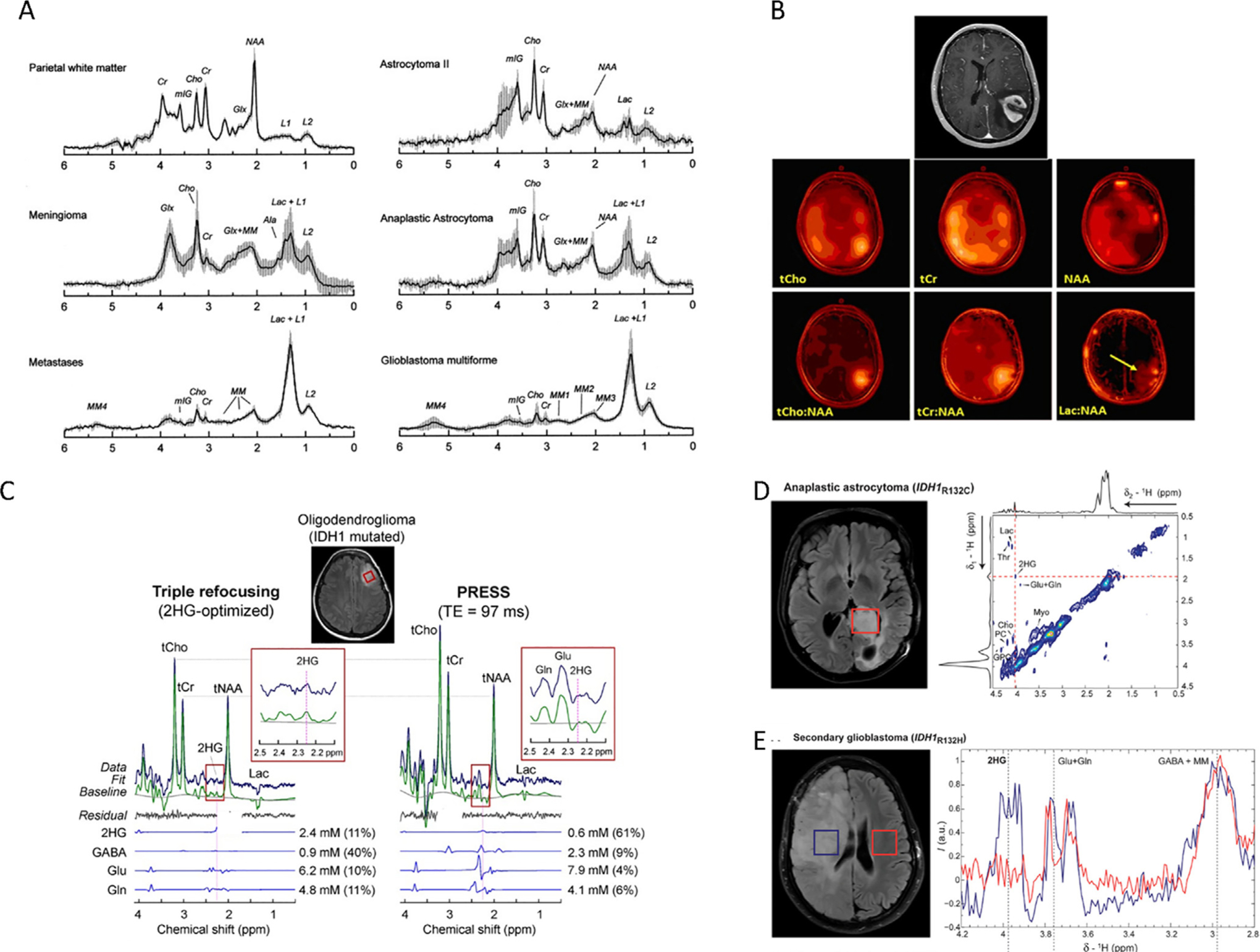
Metabolic characterization of brain tumors by in vivo MRS. A, Mean and SD (vertical lines) of normalized STEAM (TE = 30 ms) spectra for normal white matter (n = 6), meningioma (n = 8), metastases (n = 6), astrocytoma grade II (n = 5), anaplastic astrocytoma (n = 7) and glioblastoma (n = 13). Reproduced with permission from [204]. B, Contrast-enhanced axial MR image demonstrating an enhancing mass in the left parietal lobe (top) and corresponding multivoxel imaging data displayed as color maps displaying the spatial distributions of (left-to-right) total choline (tCho), total creatine (tCr), and N-acetylaspartate (NAA). Figure taken from [205]. C, In vivo spectra from an IDH1-mutated oligodendroglioma, obtained with triple refocusing (left) and PRESS TE = 97 ms (right). LCModel outputs and spectra of 2HG, GABA, Glu, and Gln are also shown. The voxel size and scan time were identical between the scans (2 × 2 × 2 cm3 and 5 min). Spectra were normalized to STEAM TE = 13 ms water. Insets show magnified spectra between 2.1 and 2.5 ppm to highlight 2HG resonance signal. Reproduced with permission from [206]. D, 2D LASER-COSY spectra in vivo of an anaplastic astrocytoma patient with IDH1-R132C at 3 T. The 2D LASER-COSY shows at 4.02/1.91 ppm the Hα-Hβ cross peak of 2HG. Projections along both spectral dimensions through 2HG cross peak indicate the SNR and spectral quality. The single voxel (3 × 3 × 3 cm3, red rectangle) was placed on the FLAIR images to include most of the tumor mass. Reproduced with permission from [207]. E, 1D MEGA-LASER spectra in vivo of a secondary GBM with IDH1-R132H at 3 T. Two voxels (3 × 3 × 3 cm3 each) were placed in both brain hemispheres, symmetrically from the middle line. Reproduced with permission from [207].
A peak at 3.03 ppm corresponding to creatine (Cr) and phosphocreatine (PCr) is normally found at both long and short echo times. High levels of Cr are found in the brain where it, in concert with PCr, serves to smooth transient changes in energy demand by buffering the ATP pool through the creatine-phosphate shuttle when the rate of ATP utilization temporarily outstrips the rate of production [136]. Total Cr level by itself is not believed to be a strong biomarker for tumor progression [137], although it emerged as a strong predictor of active tumor growth after surgery in a small study of pediatric brain tumors [138]. The prominent NAA peak at 2.01 ppm is commonly regarded as a neuronal marker, and NAA levels are decreased in disorders such as stroke and multiple sclerosis that are known to be associated with neuronal loss. However, some caution with its use as a neuronal marker is needed as in cultured cells NAA localizes not only to neurons; it has also been seen in oligodendrocyte-type-2 astrocyte progenitors and immature oligodendrocytes [139]. In addition, reversible decreases in NAA levels have been observed in response to cellular dysfunction short of death [140].
The broad lipid signal at 1.3 ppm, which is normally absent or only minimally present in healthy grey matter [141], often dominates the short echo time spectrum of CNS tumors. The lipid signal originates not from phospholipid membranes, where motion of lipids is predominantly constricted, and the signal is therefore broadened beyond detection, but from micrometer sized lipid droplets [142,143]. The formation of lipid droplets is upregulated as a protective mechanism in response to hypoxia [144] and other metabolic stressors [145]. Since such stressors often result in cell death, the mobile lipid signal is frequently found in regions of intense necrosis [146]. This necrosis may not be visibly evident on an anatomical MRI, as it can occur from small domains of necrotic tissue below the resolution of the image [146]. Necrosis is often, but not exclusively, associated with more aggressive phenotypes and an increase in mobile lipid signal correlates with higher grade tumors and poor survival chances [147–149]. Mobile lipids are a distinct characteristic of Grade IV gliomas [146], in particular of glioblastoma multiforme [146].
As described in section 3.1, lactate is the end product of anaerobic breakdown of glucose and a marker for metabolic reprogramming. The lactate signal appears in a crowded region of the spectrum and specific TE values are needed to discern it from that arising from the bulk CH2 moieties of fatty acids [150]. Since metabolic reprogramming is a relatively late feature in tumor development [151], there have been attempts to use lactate as a biomarker to distinguish high grade (II, III and IV) from low (I), with mixed results. Using a combination of short and long echo times to capture both the lactate and lipid signals at 1.5 T, Yamasaki et al. reported that the presence of a lactate peak in combination with the mobile lipid peak could stratify a diverse set of CNS tumors by WHO tumor grade by logistic discriminant with relatively high statistical confidence (p = 0.02 for distinguishing between grades II and III and p = 0.03 for distinguishing between grades III and IV) [152]. The method depended on qualitative assessment of the spectrum and the experimenters were not blinded. On the other hand, two early large multicenter studies found only infrequent occurrence of lactate in long TE single voxel studies with no obvious correlation with tumor grade [153,154] Other authors have reported statistically significant elevations at higher tumor grades [155].
Total choline is primarily detected from its singlet peak at 3.2 ppm. Choline is a major precursor of phospholipid synthesis and has been shown to correlate with tumor proliferation [156] (r2 = 0.81 measured against the Ki-67 labeling index) and cellularity [155]. CNS tumors typically display a metabolic profile characterized by increased choline and decreased NAA in tumor biopsy specimens, with the difference increasing with tumor grade [157–160]. When choline and NAA resonances were low, findings on surgical biopsy were more variable and the percentage of histologically confirmed tumors was lower. The ratio of these two metabolites (NAA and choline), known as CNI (total choline-to-NAA ratio index, normalized to the same ratio in the normal contralateral brain of the same patient) forms the basis for most diagnostic MRS methods [161,162] and it was utilized to detect high-grade gliomas [157–159], response to treatment [163,164] and disease progression [165]. Dowling et al. in an investigation of 29 patients who underwent 3D 1H MRS before undergoing surgery found that when choline and NAA resonances were low, findings on the surgical biopsy were more variable and the percentage of histologically confirmed tumors was lower [166]. Combined use of total choline (tCho) with other metabolites can increase accuracy. A classifier using a linear combination of tCho and lipids + lactate yielded an AUC = 0.97 in a ROC curve analysis of the separation of high and low grade pediatric brain tumors [160]. A comparable approach using levels of glutamine, lipids and scyllo-inositol and multivariate Cox regression was able to predict the survival in pediatric brain tumors yielding accuracy values similar to histological grading (p < 5×10−5) [167].
Other metabolites, although visible in short TE MRS, have found less wide application in clinical use. Resonances from target metabolites that cannot be unambiguously quantified through standard MRS pulse sequences have to be resolved through the application tailored difference editing sequences such as MEGA-PRESS or modulation of TE values. These metabolites can serve as markers of disease progression or response to treatment; for instance, the glycine resonance (3.51 ppm) normally overlaps with those of myo-inositol (3.5–3.6 ppm), that is present in much larger concentrations. However, in certain malignancies such as central neurocytoma, the glycine peak was easily observable [168], although its quantification and detection in other CNS malignancies was not straightforward; hence, optimization of the acquisition sequence had to be achieved. Utilization of TE1 = 60 ms and TE2 = 100 ms resolved the glycine peak in a 3 T system [169], and other approaches have also been reported [170,171] at different field strengths including at 1.5 T [172] and at 7 T [173,174]. In general, higher levels of glycine occur in high grade gliomas and a close association between glycine levels and rapid cell proliferation and post-contrast enhancement exists that is indicative of aggressive disease. By conducting simultaneous analyses of glycine and 2HG in IDH1 mutant tumors, it was found glycine/2HG was the best predictor of aggressiveness [175]; indeed, these discoveries are the basis of an ongoing clinical trial (NCT02388659). Taurine has also been proposed as a marker of primitive neuroectodermal tumors [176] after it was detected and quantified with a single voxel MRS approach and a short echo time (35 ms) to minimize its potential signal decay due to T2 relaxation. Myo-inositol is an osmolyte mainly located within glial cells [177], which has been detected at high levels in low-grade astrocytoma compared to controls and in lower levels in anaplastic astrocytomas and GBMs [178]. Lower intensity signals from additional metabolites have also proved useful in the differential diagnosis of tumor types. A combination of 1H MRS using STEAM (TE = 30 ms, TR = 2 s) and PRESS (TE = 288 ms, TR = 2 sec) was applied to the study of meningiomas, schwannomas and brain metastases to explore characteristic metabolic patterns in each tumor type [179]. Meningiomas displayed elevated glutamine/glutamate and low lipid levels in this study, while metastasis had long-T2 lipids and schwannomas show a more intense myo-inositol signal in comparison to normal tissue [179]. A tailored MEGA-PRESS sequence was utilized to quantify cystathionine since the edited pulses set for the selective detection of 2HG also resolved two signals at 2.7 and 3.9 ppm in IDH-mutated, 1p/19q codeleted gliomas [180]. Additional metabolites can also appear in MRS spectra due to treatment conditions, such as acetoacetate and acetone in brain tumors of patients undergoing a ketogenic diet [181].
The use of MRS in clinical diagnosis is not without controversy. Some of the studies showing positive predictive value for MRS tested its ability to distinguish between two specific selected conditions (i.e. high and low grade gliomas) [182] in the absence of other possible contradicting indications such as non-neoplastic lesions or inflammation [183]. This may not be a realistic simulation of clinical practice where differential diagnosis is rarely binary and multiple diagnoses are possible. In other studies, the predictive value of MRS was tested in isolation without quantifying the additional benefit MRS provides over anatomical imaging. In reality, MRS is dependent on the structural information from anatomical imaging. Hellstrom et al. investigated the combination of both MRI and MRS to classify tumors when both techniques were applied to the same patient [183]. Over 208 patients, MRS was revealed to benefit the assessment conducted by MRI in 31 cases (15%), although in most cases MRS did not add to the diagnostic value of MRI. Similarly, a study of 118 gliomas found that although NAA/Cr and Cho/Cr were statistically elevated in high grade gliomas, enhancement and necrosis evaluated by anatomical MRI were sufficient to distinguish between high- and low-grade gliomas with 96% sensitivity and 70% specificity.
Despite these issues, MRS remains a fairly popular technique in clinical practice. A survey of 220 imaging centers in Europe found 80% used MRS at least occasionally in clinical brain tumor imaging, although about a third expressed doubts about the clinical value and impact of MRS [184]. Lack of training and infrastructure were commonly cited reasons for lack of implementation [184], issues that may explain some of the variance in clinical studies as well. While some centers use clinical MRS as an adjuvant to MRI, the low specificity hinders the application of this technology as a diagnostic tool in the clinic.
4.3. 2-Hydroxyglutarate
As a consequence of the discovery of mutant IDH1 as the enzyme generating 2HG in tumors [185], efforts were made to address the detection of this metabolite non-invasively through MRS in view of the its potential prognostic value. Indeed, IDH1 status is a key feature for the modern classification of brain tumors. As a result, there are growing efforts to incorporate the measurement of 2HG non-invasively in the clinic (Fig. 3C). At least 5 clinical trials in the US (NCT03684109, NCT02388659, NCT02731521, NCT03677999, NCT02597335), in addition to our own center (NCT03952598) and another 4 in Europe (NCT02597335, NCT04233788, NCT02496741, NCT04233788) include the detection and quantification of 2HG via MRS in the clinic. While this technology is only available at few centers, potentially due to the lack of infrastructure and cost, the benefit to the patient in identifying the mutant IDH1 non-invasively early on is incomparable. Accurate quantification is important as 2HG has been reported to occur naturally [186,187] at very low concentrations, or as a consequence of 2-hydroxyglutaric aciduria [188,189]; indeed, first reports of 2HG detection in tissue were performed on patients suffering from this genetic disorder. In the context of brain malignancies, 2HG unambiguously defines the presence of the IDH1 mutation. Indeed, its detection through MRS is now part of the protocol for glioma classification in some clinical centers. The 2HG molecule has 5 nonexchangeable protons which give rise to a specific pattern in the MR spectrum, characterized by 3 multiplets located at approximately 4.02 ppm, 2.25 ppm, and 1.9 ppm. The first reports involved a classical approach on a 3 T scanner [190], utilizing PRESS with standard TR of 2 ms and TE of 30 ms. The spectra obtained did not resolve the 2HG signal completely but the increased intensity in the resonance at 2.25 ppm was utilized to measure 2HG/Cr levels that were found to be elevated in IDH1 mutant glioma patients and corroborated by LC-MS analysis ex vivo. A more tailored design was reported in parallel, involving prior calculation of optimum subecho times through quantum mechanical simulations for PRESS and difference editing sequences. An optimized PRESS sequence (both single voxel and multivoxel) was used for the detection and absolute quantification of 2HG in glioma patients, revealing much higher levels in IDH1 mutant patients [191]. This approach, which involves the acquisition of PRESS spectra with total TE = 97 ms (PRESS-97), was further validated in a cohort of 22 IDH1 mutant subjects in a 3 T scanner [192]; alternatively, higher-field systems such as 7 T would require the adjustment of key parameters to provide similar selectivity for 2HG detections at low concentrations [193]. The execution of this PRESS-97 strategy led to investigations aiming at generalizing this discovery and bringing it into the clinic. Indeed, a study including 89 patients reported a MR-based protocol for assessment of different tumor features (tumor infiltration, blood flow and blood vessel permeability) including 2HG determination. The authors found a correlation between 2HG levels and cellularity following the PRESS-97 protocol after including minor modifications in the two echo times TE1 and TE2 [6]. More recently, the PRESS-97 approach was compared to an edited MRS method using MEGA-PRESS (optimized to measure the 2HG signal at 4.02 ppm) applying both sequences to the same cohort of patients. Compared with PRESS-97, 2HG quantification through the edited method displayed a stronger correlation with the values obtained by ex vivo analysis, although it provided lower SNR. A different method involving 2D spectroscopy was applied by Andronesi et al. making use of their 2D LASER-COSY sequence [194] for identification of the coupling patterns arising from 2HG, as well as a 1D MEGA-LASER (localization by adiabatic selective refocusing) pulse sequence specifically adapted for 2HG for further quantification of this metabolite, although provided as the ratio to the sum of glutamine and glutamate. This approach overcame the lower SNR of the MEGA-PRESS sequence, providing 60% more signal and 90% of the signal of PRESS [195]. A similar method, semi-LASER [196], was applied at 3 T and 7 T including long TE (110 ms) to evaluate the performance of both systems for optimal combination of acquisition and quantification of 2HG [197]. All these advances in the detection and quantification of 2HG have made possible the utilization of this metabolite as a marker of IDH1 mutation either by itself [198–200] or in combination with other metabolites [201], as a marker of disease progression [202] and response to treatment in longitudinal studies [195,203]. While our center is currently employing the 2HG measurement via MRS in the clinic, this application is only available in a few centers in the US and Europe.
5. 13C Hyperpolarized magnetic resonance spectroscopy
Metabolic imaging of tumors is usually conducted through injection of 2-18F-fluoro-2-deoxy-d-glucose (18F-FDG-PET) and subsequent PET analysis. However, this technique does not inform about the enzymatic reactions following the uptake of glucose and its subsequent phosphorylation, and presents some limitations when tumors occur in tissues with an intrinsically active glucose uptake, such as the brain [208]. Additionally, 1H MRS only provides static information of brain metabolism, i.e. about concentrations of different endogenous metabolites, without tracking metabolic fluxes and pathways. Despite the many potential advantages of 13C tracer studies in this respect, the 13C technique was for many years largely restricted to in vitro cell culture studies, with relatively few, mostly preclinical, in vivo examples. 13C magnetic resonance is inherently insensitive because the low gyromagnetic ratio of 13C, that leads, even after isotopic enrichment, to a low population of excited spins in comparison to 1H. Combined with the limited turnover of a tracer within the imaging timeframe, single voxel acquisition is difficult and requires long infusions and acquisition times. 13C spectroscopic imaging at equilibrium polarization has, until recently [209,210], therefore been considered to be impossible [211].
These constraints led researchers to explore methods to enhance the sensitivity of detection. Some methods that have had considerable success in solution NMR, such as the use of micro and cryogenic coils, are difficult or impossible to adapt to the in vivo situation. Early efforts at signal enhancement took advantage of the Overhauser effect, where cross relaxation from the excited state of a paramagnetic contrast agent can be used to increase the population of the excited state of another nucleus at equilibrium [212,213]. Although the method has been successfully applied for oximetry [213,214] and redox imaging [215,216], the power requirements for the saturating pulse have limited the technique to ultralow fields where the SNR advantage is largely negated [212,213]. The efficiency of transfer is strongly temperature dependent and very high degrees of polarization can be reached at liquid helium temperatures. While liquid helium temperatures are obviously incompatible with in vivo samples, a breakthrough occurred in 2003 when it was demonstrated that the sample could be rapidly heated to room temperature by boiling water with a minimal loss of polarization [217]. Since a sterile closed path could be maintained during the procedure [218], the dissolution dynamic nuclear polarization technique (dDNP) was soon adapted to clinical use.
Dissolution DNP can give polarizations up to ~ 70% [219] and signal enhancements of many thousands depending on the substrate (Fig. 4A). The theory and applications of DNP [220] and dDNP [221,222] have been extensively covered elsewhere. To be an effective 13C probe for dDNP, the probe solution should preferably: (a) be non-toxic at high bolus concentrations for in vivo administration; (b) have a carbon atom in the molecule with T1 longer than 20 s; (c) have high aqueous solubility (>5 M); (d) form a glass when mixed with the paramagnetic trityl radicals and frozen to enable homogenous mixing of the radical. While the glassing and concentration requirements can be alleviated, the rapid and irreversible relaxation to equilibrium forms a more formidable barrier to widespread clinical adoption. The rapid decay leads to a requirement for the polarization to happen on-site and in the immediate vicinity of the scanner, which greatly increases the expense and complicates the logistics of the procedure [221].
Fig. 4.
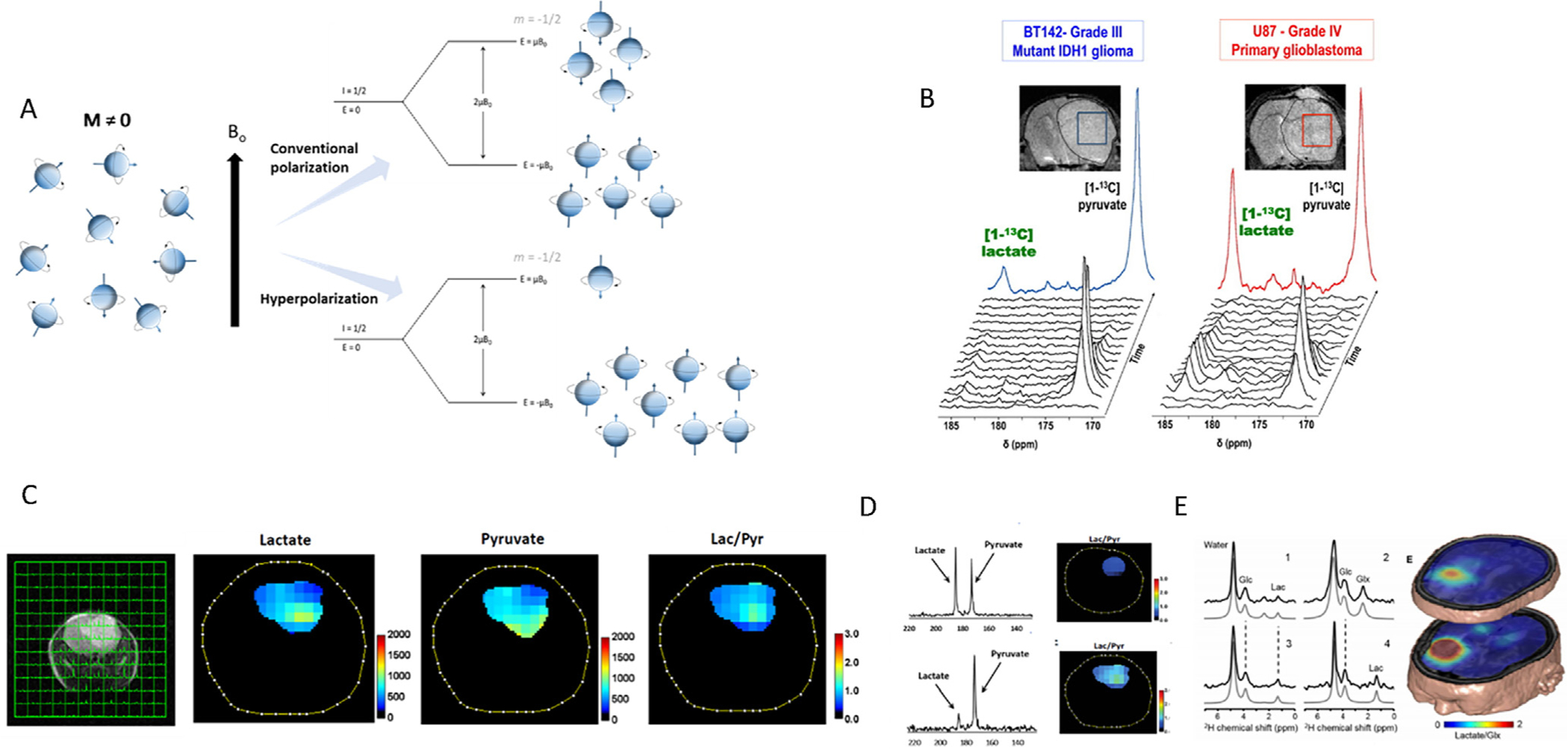
Assessment of metabolic fluxes in brain tumors through hyperpolarized and deuterium MRSI. A, Comparison of the magnetization resulting from conventional experimentation and when samples are subjected to hyperpolarization. The increasing difference of nuclei between the energy states allows for a remarkable increment in the intensity of the resulting NMR signal. B, T2-weighted MRIs of BT142 (left) and U87 (right) tumor-bearing mice and positioning of tumor voxels (U87 red; BT142 blue). The tumor is circled in dashed lines. Corresponding stack plot of hyperpolarized 13C MR spectra obtained at 14.1 T following intravenous injection of hyperpolarized [1-13C]-pyruvate (δ = 172.9 ppm). Production of hyperpolarized [1-13C]-lactate could be detected in both tumor types at δ = 185 ppm, although the level of hyperpolarized [1-13C]-lactate was lower in BT142 tumors as compared to U87. The colored spectra represent the sum of the spectra over time (U87 red; BT142 blue). Reproduced with permission from [223]. C, Multivoxel MRS of a mouse model of glioma after infusion with hyperpolarized [1-13C]-pyruvate and the heatmaps displaying the levels of the metabolites of interest within the tumor area. D, Averaged 13C spectra of lactate and pyruvate in an aggressive model of glioma (top) and an indolent one (bottom) revealing how the most aggressive model presents a more active glycolytic pathway. Reproduced with permission from [224]. E, 2H-MRS investigations of tumor metabolism in a CNS tumor to examine the glycolytic activity in tissue (1 and 3) within the lesion as determined through MRI, (2) from normal-appearing occipital lobe, and (4) containing cerebrospinal fluid from the left lateral ventricle. Figure reproduced with permission from [225].
5.1. [1/2-13C]- Pyruvate
The first report of utilization of HP [1-13C]-pyruvate showing its conversion to lactate in human brain cancers was published by Miloushev et al. [226]. They found higher levels of HP lactate in a recurrent glioblastoma patient compared to those found in the normal brain, in contrast to a treated anaplastic oligodendroglioma patient who displayed low levels of HP lactate within the malignant tissue. In another study, the correlation of IDH1 mutation status and glycolytic activity was explored utilizing [1-13C]-hyperpolarized pyruvate. While first investigations described IDH1 mutant gliomas as non-glycolytic [223] (Fig. 4B), recent reports have revealed that these tumors can actually have active glycolysis as they become more aggressive [224] (Fig. 4C and D). In addition to lactate, PDH generates acetyl-CoA and HP 13C CO2, which is subsequently converted to HP 13C bicarbonate serving as a marker of mitochondrial metabolism; indeed, in a small cohort of 10 brain tumor patients, the signal attributable to HP 13C bicarbonate was not detected in the tumors as opposed to normal tissue [227], indicating a defect in mitochondrial oxidative metabolism. The development of this methodology has made it possible to conduct clinical trials involving [1-13C]-pyruvate for assessing brain tumors (NCT03067467 and NCT03565367) and monitor response to treatment in brain metastasis (NCT03324360).
The utilization of hyperpolarized [2-13C]-pyruvate serves to assess the entry of this metabolite into the TCA cycle, since glutamate gets labelled either in position 3 or 5 depending of the activities of pyruvate carboxylase or pyruvate dehydrogenase respectively. Although investigations involving this probe demonstrated its applicability in mouse models of glioma [228], revealing that TCA cycle metabolites levels were reduced due to downregulation of PDH associated to IDH1 mutation [110], it was not until recently that it was successfully applied to the human brain [229].
5.2. [1-13C]-α-Ketoglutarate
[1-13C]-α-Ketoglutarate (α-KG) has been utilized to detect IDH1 mutant activity in engineered IDH1 mutant orthotopic tumors. However, the natural abundance of [5-13C]-α-KG resulted in a signal overlapping to that of [1-13C]-2HG at about 184 ppm in vivo. Therefore, Chaumeil et al. [230] employed a multi-band variable flip angle excitation scheme [231] optimized to monitor 2HG production by preserving the α-KG hyperpolarization and selecting the time frame at which the build-up of 2HG generated the strongest signal (20 s post injection), thus making it discernable from that of [5-13C]-α-KG, which decays earlier, after reaching its maximum intensity (10 s post injection). Furthermore, α-KG is one of the main substrates involved in transamination, since it acts as the nitrogen acceptor yielding glutamate. Consequently, HP [1-13C]-α-KG can be employed to monitor the activity of transaminases in vivo, such as BCAT (Branched Chain Amino Acids Transaminase), which has been reported to be specifically inhibited by 2HG in brain tumors [232]. Moreover, the presence of IDH1 mutation in a U87-IDH1 mutant mouse model was found to be correlated with decreased levels of several transaminases; consequently, the hyperpolarized [1-13C]-glutamate resonance located at 177.5 ppm was not observed in this model [233].
5.3. [1,3-13C2]- Acetoacetate
Following the work by Miller et al. [234] establishing the feasibility of utilizing hyperpolarized acetoacetate to monitor ketone body metabolism in vivo, Najac and coworkers [235] described a method for application in brain tumors. [1,3-13C]-Acetoacetate was useful to determine the activity of β-hydroxybutyrate dehydrogenase (BHD) since it is involved in the conversion of acetoacetate into β-hydroxybutyrate (β-HB). The products of this probe yield distinct resonance signals including those of [1,3-13C]-acetoacetate (C3 = 210.9 ppm and C1 = 175.5 ppm) and [1-13C]-HB (181.1 ppm), in accordance with previous reports [236]. Additionally, this reaction utilizes NADH, and could serve as a reporter of the NAD+/NADH ratio. In the work by Najac et al. [235], the authors observed reduced β-HB levels and through a spectrophotometric assay detected higher NAD+/NADH in the tumor models (U87), revealing a lower BHD activity and no correlation with the IDH1 status.
5.4. [U-2H, U-13C]-Glucose
Hyperpolarized glucose is an attractive alternative to pyruvate for imaging tumor metabolism as it allows the possibility of measuring the flux through the entire glycolytic pathway, potentially including diversions into the pentose phosphate pathway. After early measurements in vitro in yeast cells [237], [U-2H, U-13C]-glucose was successfully used to measure glycolysis in murine lymphoma and lung tumors [211]. Low but measurable levels of the glycolytic intermediates DHAP and 6PG were observed, along with HCO3−, which was used to establish flux to the pentose phosphate pathway on the assumption that decarboxylation of pyruvate in the TCA cycle was minimal in tumors. A decrease in lactate production was observed after treatment with the chemotherapeutic drug etopside. The method was later extended to cerebral metabolism in healthy mice by Mishkovsky et al. [238].
5.5. γ-Glutamyl-[1- 13C]-Glycine
The oxidative stress in tumors is mitigated by upregulated production of glutathione. Central to this process is γ-glutamyl-transferase (GGT), which plays a critical role in the catalysis of glutathione to glutamate and cysteinylglycine, an essential step for the import of glutathione into the cell. The activity of GGT can be monitored by γ-glutamyl-[1-13C]-glycine by examining the formation of [1-13C]-glycine at 177.5 ppm [239]. Measurements in GBM tumors confirmed higher GGT flux within the malignant tissue, in line with higher expression levels of this enzyme [240].
5.6. Assessment of glioma metabolism through 2H magnetic resonance
Given the expensive infrastructure and complexity of 13C hyperpolarized experiments, an alternative approach is to utilize 2H for MRSI, as shown in recent studies of human brain tumors [225], following earlier studies in rats [241]. The main advantage of 2H MRS over 13C MRS for metabolic labeling studies is the rapid T1 relaxation. While longer T1 values are usually preferred for hyperpolarization experiments to limit the decay of hyperpolarization before the transfer of the 13C-labeled group of interest to further metabolites, in non-hyperpolarized (thermal equilibrium) experiments short T1 times allow the acquisition of many scans without saturating the signal. In this first report [225] of 2H imaging in brain tumors, GBM patients subjected to [6,6′−2H]-glucose infusion displayed lower levels of labeled glutamate/glutamine and higher levels of labeled lactate in the tumor (Fig. 4E) in addition to higher acetate levels in the malignant tissue after [2,2,2′−2H]-acetate infusion. However, that approach only allowed investigations in the steady state. This limitation was overcome by a different approach involving infusion with 2H tracers and the spectral acquisition of 1H MRS data following the computation of the loss of 1H signal (Fig. 4F) due to 2H incorporation [242]. The authors utilized an intracranial animal model involving rats harboring F98 cells and reported the same observations as previous studies, in which the conversion of glucose into lactate was enhanced in tumor tissue.
6. Conclusions
NMR-based techniques can provide metabolic information using biofluids as reporters of systemic consequences of disease processes or as metabolic signatures of disease. Additionally, NMR analysis of tumor tissue, both in vivo and ex vivo can also report on the metabolism involved in the disease process. Infusion with 13C tracers and subsequent in situ MRS or ex vivo analysis through HR-NMR are key methodologies for measuring metabolic fluxes and dissecting the metabolic reprogramming associated with malignancies. 13C MRS studies have also played an important role in delineating how alternative substrates such as acetate and ketone bodies can support brain tumor growth. Wider application of NMR-based 13C tracing to study human disease faces considerable obstacles, particularly cost, the need for 13C substrate infusions and monitoring, and most importantly, the low sensitivity of 13C MRS, which can be overcome through newly developed post-processing algorithms or by applying hyperpolarized MRSI, although increasing the costs considerably. Despite these difficulties, 13C MRS has been successfully applied to answer questions in a variety of brain tumors and can even surpass standard methods such as 18FDG-PET in some situations. The increasing availability of higher field human MRI systems and advances in NMR techniques, including 2H-MRS methods, have the potential to make isotope labeling studies accessible to a much wider range of clinical research applications. In addition, temporal resolution from hyperpolarization could be combined with pathway information from steady state analysis to obtain reliable metabolic maps related to CNS malignancies.
Acknowledgements
This was funded in part by the Intramural Research Program, Center for Cancer Research, National Cancer Institute, and National Institutes of Health, USA. We would like to thank Fernando Gonzalez-Rodado for his assistance with the graphical abstract.
Glossary
- 2HG
2-Hydroxyglutarate
- 6PG
Glucose 6-Phosphate
- αKG
α-Ketoglutarate
- βHB
β-Hydroxybutyrate
- AUC
Area Under the Curve
- BCAT
Branched Chain Amino Acids Transaminase
- BDH
β-Hydroxybutyrate Dehydrogenase
- CNS
Central Nervous System
- COSY
Correlation Spectroscopy
- CPMG
Carr-Purcell-Meiboom-Gill
- CSF
Cerebrospinal Fluid
- DHA
Dihydroxyacetone Phosphate
- DNP
Dynamic Nuclear Polarization
- GABA
Gamma Aminobutyric Acid
- GBM
Glioblastoma
- GEMM
Genetically Engineered Mouse Model
- GGT
γ-Glutamyl Transferase
- GPC
Glycerophosphocholine
- GSH
Glutathione
- HIF-1α
Hypoxia Inducible Factor
- HOT
Human Orthotopic Tumors
- HP
Hyperpolarized
- HR
High Resolution
- IDH
Isocitrate Dehydrogenase
- LASER
Localization by Adiabatic Selective Refocusing
- MAS
Magic Angle Spinning
- MRI
Magnetic Resonance Imaging
- MRS
Magnetic Resonance Spectroscopy
- MRSI
Magnetic Resonance Spectroscopic Imaging
- NAA
N-acetyl Aspartate
- NHA
Normal Human Astrocytes
- NMR
Nuclear Magnetic Resonance
- NOD-SCID
Nonobese Diabetic-Severe Combined Immunodeficiency
- NOESY
Nuclear Overhauser Effect Spectroscopy
- O-PLS-DA
Orthogonal-Partial Least Squares-Discriminant Analysis
- PC
Phosphocholine
- PDH
Pyruvate Dehydrogenase
- PE
Phosphoethanolamine
- ROC
Receiving Operation Characteristic
- SE
Short Echo
- SNR
Signal-to-Noise Ratio
- SVM
Support Vector Machine
- TCA cycle
Tricarboxylic Acids cycle
- TE
Echo Time
- TR
Repetition Time
- TOBSY
Total Through-Bond Correlation Spectroscopy
- TOCSY
Total Correlation Spectroscopy
- VOI
Volume of Interest
- WHO
World Health Organization
Footnotes
Declaration of Competing Interest
The authors declare that there is no conflict of interest.
References
- [1].Ostrom QT, Gittleman H, Liao P, Vecchione-Koval T, Wolinsky Y, Kruchko C, Barnholtz-Sloan JS, CBTRUS Statistical Report: Primary brain and other central nervous system tumors diagnosed in the United States in 2010–2014, Neuro Oncol 19 (suppl_5) (2017), v1–v88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW, The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary, Acta Neuropathol 131 (6) (2016) 803–820. [DOI] [PubMed] [Google Scholar]
- [3].Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, WHO Classification of Tumours of the Central Nervous System (revised 4th edition), World Health Organization, Lyon, 2016. [DOI] [PubMed] [Google Scholar]
- [4].Barnholtz-Sloan JS, Sloan AE, Davis FG, Vigneau FD, Lai P, Sawaya RE, Incidence proportions of brain metastases in patients diagnosed (1973 to 2001) in the Metropolitan Detroit Cancer Surveillance System, J. Clin. Oncol 22 (14) (2004) 2865–2872. [DOI] [PubMed] [Google Scholar]
- [5].National Comprehensive Cancer Network. Central nervous system cancers (version 1.2018) [https://www.nccn.org/professionals/physician_gls/pdf/cns.pdf]. [DOI] [PubMed]
- [6].de la Fuente MI, Young RJ, Rubel J, Rosenblum M, Tisnado J, Briggs S, Arevalo-Perez J, Cross JR, Campos C, Straley K, et al. , Integration of 2-hydroxyglutarate-proton magnetic resonance spectroscopy into clinical practice for disease monitoring in isocitrate dehydrogenase-mutant glioma, Neuro Oncol 18 (2) (2016) 283–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lapointe S, Perry A, Butowski NA, Primary brain tumours in adults, Lancet 392 (10145) (2018) 432–446. [DOI] [PubMed] [Google Scholar]
- [8].Pavlova NN, Thompson CB, The Emerging Hallmarks of Cancer Metabolism, Cell Metab 23 (1) (2016) 27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ward PS, Thompson CB, Metabolic Reprogramming: A Cancer Hallmark Even Warburg Did Not Anticipate, Cancer Cell 21 (3) (2012) 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ye F, Zhang Y, Liu Y, Yamada K, Tso JL, Menjivar JC, Tian JY, Yong WH, Schaue D, Mischel PS, et al. , Protective properties of radio-chemoresistant glioblastoma stem cell clones are associated with metabolic adaptation to reduced glucose dependence, PLoS ONE 8 (11) (2013) e80397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Heintz A, Constans JM: Role of Magnetic Resonance Spectroscopy in Clinical Management of Brain Tumors. In: Atlas of Clinical Cases on Brain Tumor Imaging Edited by Özsunar Y, Şenol U: Springer, Cham; 2020. [Google Scholar]
- [12].Bhatt AN, Chauhan A, Khanna S, Rai Y, Singh S, Soni R, Kalra N, Dwarakanath BS, Transient elevation of glycolysis confers radio-resistance by facilitating DNA repair in cells, Bmc Cancer 15 (2015) 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Floberg JM, Schwarz JK, Manipulation of Glucose and Hydroperoxide Metabolism to Improve Radiation Response, Semin Radiat Oncol 29 (1) (2019) 33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Barton RH, Nicholson JK, Elliott P, Holmes E, High-throughput 1H NMR-based metabolic analysis of human serum and urine for large-scale epidemiological studies: validation study, Int. J. Epidemiol 37 (Suppl 1) (2008) i31–i40. [DOI] [PubMed] [Google Scholar]
- [15].Dumas ME, Maibaum EC, Teague C, Ueshima H, Zhou B, Lindon JC, Nicholson JK, Stamler J, Elliott P, Chan Q, et al. , Assessment of analytical reproducibility of 1H NMR spectroscopy based metabonomics for large-scale epidemiological research: the INTERMAP Study, Anal. Chem 78 (7) (2006) 2199–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Keun HC, Ebbels TM, Antti H, Bollard ME, Beckonert O, Schlotterbeck G, Senn H, Niederhauser U, Holmes E, Lindon JC, et al. , Analytical reproducibility in (1)H NMR-based metabonomic urinalysis, Chem. Res. Toxicol 15 (11) (2002) 1380–1386. [DOI] [PubMed] [Google Scholar]
- [17].Schreier C, Kremer W, Huber F, Neumann S, Pagel P, Lienemann K, Pestel S, Reproducibility of NMR analysis of urine samples: impact of sample preparation, storage conditions, and animal health status, Biomed Res. Int 2013 (2013) 878374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kruk J, Doskocz M, Jodlowska E, Zacharzewska A, Lakomiec J, Czaja K, Kujawski J, NMR Techniques in Metabolomic Studies: A Quick Overview on Examples of Utilization, Appl. Magn. Reson 48 (1) (2017) 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zheng G, Price WS, Solvent signal suppression in NMR, Prog. Nucl. Magn. Reson. Spectrosc 56 (3) (2010) 267–288. [DOI] [PubMed] [Google Scholar]
- [20].Aranibar N, Ott KH, Roongta V, Mueller L, Metabolomic analysis using optimized NMR and statistical methods, Anal. Biochem 355 (1) (2006) 62–70. [DOI] [PubMed] [Google Scholar]
- [21].Mckay RT, How the 1D-NOESY suppresses solvent signal in metabonomics NMR spectroscopy: An examination of the pulse sequence components and evolution, Concepts in Magnetic Resonance Part A 38A (5) (2011) 197–220. [Google Scholar]
- [22].Ogg RJ, Kingsley PB, Taylor JS, WET, a T1- and B1-insensitive water-suppression method for in vivo localized 1H NMR spectroscopy, J. Magn. Reson B 104 (1) (1994) 1–10. [DOI] [PubMed] [Google Scholar]
- [23].Hwang TL, Shaka AJ, Water Suppression That Works. Excitation Sculpting Using Arbitrary Wave-Forms and Pulsed-Field Gradients, J. Magn. Reson., Ser A 112 (2) (1995) 275–279. [Google Scholar]
- [24].Piotto M, Saudek V, Sklenar V, Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions, J. Biomol. NMR 2 (6) (1992) 661–665. [DOI] [PubMed] [Google Scholar]
- [25].Carr HY, Purcell EM, Effects of Diffusion on Free Precession in Nuclear Magnetic Resonance Experiments, Phys. Rev 94 (3) (1954) 630–638. [Google Scholar]
- [26].Meiboom S, Gill D, Modified Spin-Echo Method for Measuring Nuclear Relaxation Times, Rev. Sci. Instrum 29 (8) (1958) 688–691. [Google Scholar]
- [27].Nagana Gowda GA, Raftery D, Quantitating metabolites in protein precipitated serum using NMR spectroscopy, Anal. Chem 86 (11) (2014) 5433–5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Beckonert O, Keun HC, Ebbels TMD, Bundy J, Holmes E, Lindon JC, Nicholson JK, Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts, Nat. Protoc 2 (11) (2007) 2692–2703. [DOI] [PubMed] [Google Scholar]
- [29].Lacy P, McKay RT, Finkel M, Karnovsky A, Woehler S, Lewis MJ, Chang D, Stringer KA, Signal intensities derived from different NMR probes and parameters contribute to variations in quantification of metabolites, PLoS ONE 9 (1) (2014) e85732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z et al. : BioMagResBank. Nucleic Acids Res 2008, 36(Database issue):D402–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vazquez-Fresno R, Sajed T, Johnson D, Li C, Karu N et al. : HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res 2018, 46(D1):D608–D617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S et al. : HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res 2009, 37(Database issue):D603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Gipson GT, Tatsuoka KS, Sweatman BC, Connor SC, Weighted least-squares deconvolution method for discovery of group differences between complex biofluid 1H NMR spectra, J. Magn. Reson 183 (2) (2006) 269–277. [DOI] [PubMed] [Google Scholar]
- [34].Hao J, Liebeke M, Astle W, De Iorio M, Bundy JG, Ebbels TM, Bayesian deconvolution and quantification of metabolites in complex 1D NMR spectra using BATMAN, Nat. Protoc 9 (6) (2014) 1416–1427. [DOI] [PubMed] [Google Scholar]
- [35].Vu TN, Laukens K, Getting your peaks in line: a review of alignment methods for NMR spectral data, Metabolites 3 (2) (2013) 259–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Nagana Gowda GA, Raftery D, Can NMR solve some significant challenges in metabolomics?, J Magn. Reson 260 (2015) 144–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Asiago VM, Nagana Gowda GA, Zhang S, Shanaiah N, Clark J, Raftery D, Use of EDTA to minimize ionic strength dependent frequency shifts in the 1H NMR spectra of urine, Metabolomics 4 (4) (2008) 328. [Google Scholar]
- [38].Rist MJ, Muhle-Goll C, Gorling B, Bub A, Heissler S, Watzl B, Luy B, Influence of Freezing and Storage Procedure on Human Urine Samples in NMR-Based Metabolomics, Metabolites 3 (2) (2013) 243–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Schleif FM, Riemer T, Borner U, Schnapka-Hille L, Cross M, Genetic algorithm for shift-uncertainty correction in 1-D NMR-based metabolite identifications and quantifications, Bioinformatics 27 (4) (2011) 524–533. [DOI] [PubMed] [Google Scholar]
- [40].Forshed J, Schuppe-Koistinen I, Jacobsson SP, Peak alignment of NMR signals by means of a genetic algorithm, Anal. Chim. Acta 487 (2) (2003) 189–199. [Google Scholar]
- [41].Ravanbakhsh S, Liu P, Bjorndahl TC, Mandal R, Grant JR, Wilson M, Eisner R, Sinelnikov I, Hu X, Luchinat C, et al. , Accurate, fully-automated NMR spectral profiling for metabolomics, PLoS ONE 10 (5) (2015) e0124219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Worley B, Powers R, Multivariate Analysis in Metabolomics, Curr Metabolomics 1 (1) (2013) 92–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ruiz-Rodado V, Luque-Baena RM, te Vruchte D, Probert F, Lachmann RH, Hendriksz CJ, Wraith JE, Imrie J, Elizondo D, Sillence DJ, et al. , H-1 NMR-Linked Urinary Metabolic Profiling of Niemann-Pick Class C1 (NPC1) Disease: Identification of Potential New Biomarkers using Correlated Component Regression (CCR) and Genetic Algorithm (GA) Analysis Strategies, Curr Metabol 2 (2) (2014) 88–121. [Google Scholar]
- [44].Peeling J, Sutherland G, Marat K, Tomchuk E, Bock E, 1H and 13C nuclear magnetic resonance studies of plasma from patients with primary intracranial neoplasms, J. Neurosurg 68 (6) (1988) 931–937. [DOI] [PubMed] [Google Scholar]
- [45].Aru V, Lam C, Khakimov B, Hoefsloot HCJ, Zwanenburg G, Lind MV, Schafer H, van Duynhoven J, Jacobs DM, Smilde AK et al. : Quantification of lipoprotein profiles by nuclear magnetic resonance spectroscopy and multivariate data analysis (vol 94, pg 210, 2017). Trac-Trend Anal Chem 2019, 119. [Google Scholar]
- [46].Kelimu A, Xie R, Zhang K, Zhuang Z, Mamtimin B, Sheyhidin I, Metabonomic signature analysis in plasma samples of glioma patients based on (1)H-nuclear magnetic resonance spectroscopy, Neurol India 64 (2) (2016) 246–251. [DOI] [PubMed] [Google Scholar]
- [47].Baranovicova E, Galanda T, Galanda M, Hatok J, Kolarovszki B, Richterova R, Racay P, Metabolomic profiling of blood plasma in patients with primary brain tumours: Basal plasma metabolites correlated with tumour grade and plasma biomarker analysis predicts feasibility of the successful statistical discrimination from healthy subjects - a preliminary study, IUBMB Life 71 (12) (2019) 1994–2002. [DOI] [PubMed] [Google Scholar]
- [48].Srivastava NK, Pradhan S, Gowda GA, Kumar R, In vitro, high-resolution 1H and 31P NMR based analysis of the lipid components in the tissue, serum, and CSF of the patients with primary brain tumors: one possible diagnostic view, NMR Biomed 23 (2) (2010) 113–122. [DOI] [PubMed] [Google Scholar]
- [49].Klos KJ, O’Neill BP, Brain metastases, Neurologist 10 (1) (2004) 31–46. [DOI] [PubMed] [Google Scholar]
- [50].Schellinger PD, Meinck HM, Thron A, Diagnostic accuracy of MRI compared to CCT in patients with brain metastases, J. Neurooncol 44 (3) (1999) 275–281. [DOI] [PubMed] [Google Scholar]
- [51].Larkin JR, Dickens AM, Claridge TDW, Bristow C, Andreou K, Anthony DC, Sibson NR, Early Diagnosis of Brain Metastases Using a Biofluids-Metabolomics Approach in Mice, Theranostics 6 (12) (2016) 2161–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Nicholson JK, Wilson ID, High resolution proton magnetic resonance spectroscopy of biological fluids, Prog Nucl Mag Res Sp 21 (4) (1989) 449–501. [Google Scholar]
- [53].Bell JD, Brown JC, Sadler PJ, Macleod AF, Sonksen PH, Hughes RD, Williams R, High resolution proton nuclear magnetic resonance studies of human cerebrospinal fluid, Clin Sci (Lond) 72 (5) (1987) 563–570. [DOI] [PubMed] [Google Scholar]
- [54].Petroff OAC, Yu RK, Ogino T, High-Resolution Proton Magnetic Resonance Analysis of Human Cerebrospinal Fluid, J. Neurochem 47 (4) (1986) 1270–1276. [DOI] [PubMed] [Google Scholar]
- [55].Sweatman BC, Farrant RD, Holmes E, Ghauri FY, Nicholson JK, Lindon JC, 600 MHz 1H-NMR spectroscopy of human cerebrospinal fluid: Effects of sample manipulation and assignment of resonances, J. Pharm. Biomed. Anal 11 (8) (1993) 651–664. [DOI] [PubMed] [Google Scholar]
- [56].Wishart DS, Lewis MJ, Morrissey JA, Flegel MD, Jeroncic K, Xiong Y, Cheng D, Eisner R, Gautam B, Tzur D, et al. , The human cerebrospinal fluid metabolome, J. Chromatogr. B Analyt. Technol. Biomed Life Sci 871 (2) (2008) 164–173. [DOI] [PubMed] [Google Scholar]
- [57].Koschorek F, Offermann W, Stelten J, Braunsdorf WE, Steller U, Gremmel H, Leibfritz D, High-resolution 1H NMR spectroscopy of cerebrospinal fluid in spinal diseases, Neurosurg. Rev 16 (4) (1993) 307–315. [DOI] [PubMed] [Google Scholar]
- [58].Clarke JL, Perez HR, Jacks LM, Panageas KS, DeAngelis LM, Leptomeningeal metastases in the MRI era, Neurology 74 (18) (2010) 1449–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Cho HR, Wen H, Ryu YJ, An YJ, Kim HC, Moon WK, Han MH, Park S, Choi SH, An NMR metabolomics approach for the diagnosis of leptomeningeal carcinomatosis, Cancer Res 72 (20) (2012) 5179–5187. [DOI] [PubMed] [Google Scholar]
- [60].An YJ, Cho HR, Kim TM, Keam B, Kim JW, Wen H, Park CK, Lee SH, Im SA, Kim JE, et al. , An NMR metabolomics approach for the diagnosis of leptomeningeal carcinomatosis in lung adenocarcinoma cancer patients, Int. J. Cancer 136 (1) (2015) 162–171. [DOI] [PubMed] [Google Scholar]
- [61].Maas WE, Laukien FH, Cory DG, Gradient, high resolution, magic angle sample spinning NMR, J. Am. Chem. Soc 118 (51) (1996) 13085–13086. [Google Scholar]
- [62].Andrew ER, Bradbury A, Eades RG, Removal of Dipolar Broadening of Nuclear Magnetic Resonance Spectra of Solids by Specimen Rotation, Nature 183 (4678) (1959) 1802–1803. [Google Scholar]
- [63].Lowe IJ, Free Induction Decays of Rotating Solids, Phys. Rev. Lett 2 (7) (1959) 285–287. [Google Scholar]
- [64].Righi V, Cavallini N, Valentini A, Pinna G, Pavesi G, Rossi MC, Puzzolante A, Mucci A, Cocchi M, A metabolomic data fusion approach to support gliomas grading, NMR Biomed 33 (3) (2020). [DOI] [PubMed] [Google Scholar]
- [65].de Juan A, Tauler R, Multivariate Curve Resolution-Alternating Least Squares for Spectroscopic Data, Data Handl Sci. Techn 30 (2016) 5–51. [Google Scholar]
- [66].Erb G, Elbayed K, Piotto M, Raya J, Neuville A, Mohr M, Maitrot D, Kehrli P, Namer IJ, Toward improved grading of malignancy in oligodendrogliomas using metabolomics, Magnet Reson. Med 59 (5) (2008) 959–965. [DOI] [PubMed] [Google Scholar]
- [67].Jothi J, Janardhanam VA, Krishnaswamy R, Metabolic Variations between Low-Grade and High-Grade Gliomas-Profiling by (1)H NMR Spectroscopy, J. Proteome Res 19 (6) (2020) 2483–2490. [DOI] [PubMed] [Google Scholar]
- [68].Wilson M, Davies NP, Brundler MA, McConville C, Grundy RG, Peet AC, High resolution magic angle spinning 1H NMR of childhood brain and nervous system tumours, Molecular Cancer 8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Tugnoli V, Schenetti L, Mucci A, Nocetti L, Toraci C, Mavilla L, Basso G, Rovati R, Tavani F, Zunarelli E, et al. , A comparison between in vivo and ex vivo HR-MAS H-1 MR spectra of a pediatric posterior fossa lesion, Int. J. Mol. Med 16 (2) (2005) 301–307. [DOI] [PubMed] [Google Scholar]
- [70].Monleon D, Morales JM, Gonzalez-Darder J, Talamantes F, Cortes O, Gil-Benso R, Loezin-Gines C, Cerda-Nicolas M, Celda B, Benign and atypical meningioma metabolic signatures by high-resolution magic-angle spinning molecular profiling, J. Proteome Res 7 (7) (2008) 2882–2888. [DOI] [PubMed] [Google Scholar]
- [71].Bender L, Somme F, Ruhland E, Cicek AE, Bund C, Namer IJ, Metabolomic Profile of Aggressive Meningiomas by Using High-Resolution Magic Angle Spinning Nuclear Magnetic Resonance, J. Proteome Res 19 (1) (2020) 292–299. [DOI] [PubMed] [Google Scholar]
- [72].Pfisterer WK, Nieman RA, Scheck AC, Coons SW, Spetzler RF, Preul MC, Using ex vivo proton magnetic resonance spectroscopy to reveal associations between biochemical and biological features of meningiomas, Neurosurg. Focus 28 (1) (2010). [DOI] [PubMed] [Google Scholar]
- [73].Chen WX, Lou HY, Zhang HP, Nie X, Lan WX, Yang YX, Xiang Y, Qi JP, Lei H, Tang HR, et al. , Grade classification of neuroepithelial tumors using high-resolution magic-angle spinning proton nuclear magnetic resonance spectroscopy and pattern recognition, Sci China Life Sci 54 (7) (2011) 606–616. [DOI] [PubMed] [Google Scholar]
- [74].Wright AJ, Fellows GA, Griffiths JR, Wilson M, Bell BA, Howe FA, Ex-vivo HRMAS of adult brain tumours: metabolite quantification and assignment of tumour biomarkers, Molecular Cancer 9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Vettukattil R, Gulati M, Sjobakk TE, Jakola AS, Kvernmo NA, Torp SH, Bathen TF, Gulati S, Gribbestad IS, Differentiating diffuse World Health Organization grade II and IV astrocytomas with ex vivo magnetic resonance spectroscopy, Neurosurgery 72 (2) (2013) 186–195, discussion 195. [DOI] [PubMed] [Google Scholar]
- [76].Griffin JL, Lehtimaki KK, Valonen PK, Grohn OHJ, Kettunen MI, Yla-Herttuala S, Pitkanen A, Nicholson JK, Kauppinen RA, Assignment of H-1 nuclear magnetic resonance visible polyunsaturated fatty acids in BT4C gliomas undergoing ganciclovir-thymidine kinase gene therapy-induced programmed cell death, Cancer Res 63 (12) (2003) 3195–3201. [PubMed] [Google Scholar]
- [77].Lehtimaki KK, Valonen PK, Griffin JL, Vaisanen TH, Grohn OHJ, Kettunen MI, Vepsalainen J, Yla-Herttuala S, Nicholson J, Kauppinen RA, Metabolite changes in BT4C rat gliomas undergoing ganciclovir-thymidine kinase gene therapy-induced programmed cell death as studied by H-1 NMR spectroscopy in vivo, ex vivo, and in vitro, J. Biol. Chem 278 (46) (2003) 45915–45923. [DOI] [PubMed] [Google Scholar]
- [78].Lee JE, Jeun SS, Kim SH, Yoo CY, Baek HM, Yang SH, Metabolic profiling of human gliomas assessed with NMR, J. Clin. Neurosci 68 (2019) 275–280. [DOI] [PubMed] [Google Scholar]
- [79].Lehnhardt FG, Rohn G, Ernestus RI, Grune M, Hoehn M, 1H- and (31)P-MR spectroscopy of primary and recurrent human brain tumors in vitro: malignancy-characteristic profiles of water soluble and lipophilic spectral components, NMR Biomed 14 (5) (2001) 307–317. [DOI] [PubMed] [Google Scholar]
- [80].Constantin A, Elkhaled A, Jalbert L, Srinivasan R, Cha S, Chang SM, Bajcsy R, Nelson SJ, Identifying malignant transformations in recurrent low grade gliomas using high resolution magic angle spinning spectroscopy, Artif. Intell. Med 55 (1) (2012) 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Andronesi OC, Mintzopoulos D, Struppe J, Black PM, Tzika AA, Solid-state NMR adiabatic TOBSY sequences provide enhanced sensitivity for multidimensional high-resolution magic-angle-spinning H-1 MR spectroscopy, J. Magn. Reson 193 (2) (2008) 251–258. [DOI] [PubMed] [Google Scholar]
- [82].Andronesi OC, Blekas KD, Mintzopoulos D, Astrakas L, Black PM, Tzika AA, Molecular classification of brain tumor biopsies using solid-state magic angle spinning proton magnetic resonance spectroscopy and robust classifiers, Int. J. Oncol 33 (5) (2008) 1017–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Jalbert LE, Elkhaled A, Phillips JJ, Neill E, Williams A, Crane JC, Olson MP, Molinaro AM, Berger MS, Kurhanewicz J, et al. , Metabolic Profiling of IDH Mutation and Malignant Progression in Infiltrating Glioma, Sci. Rep 7 (2017) 44792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Gandia-Gonzalez ML, Cerdan S, Barrios L, Lopez-Larrubia P, Feijoo PG, Palpan A, Roda JM, Solivera J, Assessment of Overall Survival in Glioma Patients as Predicted by Metabolomic Criteria, Frontiers Oncology 9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Righi V, Roda JM, Paz J, Mucci A, Tugnoli V, Rodriguez-Tarduchy G, Barrios L, Schenetti L, Cerdan S, Garcia-Martin ML, H-1 HR-MAS and genomic analysis of human tumor biopsies discriminate between high and low grade astrocytomas, NMR Biomed 22 (6) (2009) 629–637. [DOI] [PubMed] [Google Scholar]
- [86].Pichumani K, Mashimo T, Baek HM, Ratnakar J, Mickey B, DeBerardinis RJ, Maher EA, Bachoo RM, Malloy CR, Kovacs Z, Conditions for C-13 NMR detection of 2-hydroxyglutarate in tissue extracts from isocitrate dehydrogenase-mutated gliomas, Anal. Biochem 481 (2015) 4–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Ruiz-Rodado V, Lita A, Dowdy T, Celiku O, Saldana AC, Wang H, Yang CZ, Chari R, Li A, Zhang W, et al. , Metabolic plasticity of IDH1-mutant glioma cell lines is responsible for low sensitivity to glutaminase inhibition, Cancer & metabolism 8 (2020) 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Mashimo T, Pichumani K, Vemireddy V, Hatanpaa KJ, Singh DK, Sirasanagandla S, Nannepaga S, Piccirillo SG, Kovacs Z, Foong C, et al. , Acetate Is a Bioenergetic Substrate for Human Glioblastoma and Brain Metastases, Cell 159 (7) (2014) 1603–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Oudard S, Arvelo F, Miccoli L, Apiou F, Dutrillaux AM, Poisson M, Dutrillaux B, Poupon MF, High glycolysis in gliomas despite low hexokinase transcription and activity correlated to chromosome 10 loss, Br. J. Cancer 74 (6) (1996) 839–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Brand A, Engelmann J, Leibfritz D, A 13C NMR study on fluxes into the TCA cycle of neuronal and glial tumor cell lines and primary cells, Biochimie 74 (9–10) (1992) 941–948. [DOI] [PubMed] [Google Scholar]
- [91].Portais JC, Schuster R, Merle M, Canioni P, Metabolic flux determination in C6 glioma cells using carbon-13 distribution upon [1–13C]glucose incubation, Eur. J. Biochem 217 (1) (1993) 457–468. [DOI] [PubMed] [Google Scholar]
- [92].Bouzier AK, Goodwin R, de Gannes FM, Valeins H, Voisin P, Canioni P, Merle M: Compartmentation of lactate and glucose metabolism in C6 glioma cells. A 13c and 1H NMR study. J Biol Chem 1998, 273(42):27162–27169. [DOI] [PubMed] [Google Scholar]
- [93].Bouzier AK, Voisin P, Goodwin R, Canioni P, Merle M, Glucose and lactate metabolism in C6 glioma cells: evidence for the preferential utilization of lactate for cell oxidative metabolism, Dev. Neurosci 20 (4–5) (1998) 331–338. [DOI] [PubMed] [Google Scholar]
- [94].Yasui H, Kawai T, Matsumoto S, Saito K, Devasahayam N, Mitchell JB, Camphausen K, Inanami O, Krishna MC, Quantitative imaging of pO2 in orthotopic murine gliomas: hypoxia correlates with resistance to radiation, Free Radic Res 51 (9–10) (2017) 861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Colwell N, Larion M, Giles AJ, Seldomridge AN, Sizdahkhani S, Gilbert MR, Park DM, Hypoxia in the glioblastoma microenvironment: shaping the phenotype of cancer stem-like cells, Neuro Oncol 19 (7) (2017) 887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Rodrigues TB, Gray HL, Benito M, Garrido S, Sierra A, Geraldes CF, Ballesteros P, Cerdan S, Futile cycling of lactate through the plasma membrane of C6 glioma cells as detected by (13C, 2H) NMR, J. Neurosci. Res 79 (1–2) (2005) 119–127. [DOI] [PubMed] [Google Scholar]
- [97].Vander Heiden MG, Cantley LC, Thompson CB, Understanding the Warburg effect: the metabolic requirements of cell proliferation, Science 324 (5930) (2009) 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Mancuso A, Beardsley NJ, Wehrli S, Pickup S, Matschinsky FM, Glickson JD, Real-time detection of 13C NMR labeling kinetics in perfused EMT6 mouse mammary tumor cells and betaHC9 mouse insulinomas, Biotechnol. Bioeng 87 (7) (2004) 835–848. [DOI] [PubMed] [Google Scholar]
- [99].DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB, Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis, Proc Natl Acad Sci U S A 104 (49) (2007) 19345–19350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. , IDH1 and IDH2 mutations in gliomas, N. Engl. J. Med 360 (8) (2009) 765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A, Analysis of the IDH1 codon 132 mutation in brain tumors, Acta Neuropathol 116 (6) (2008) 597–602. [DOI] [PubMed] [Google Scholar]
- [102].Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. , An integrated genomic analysis of human glioblastoma multiforme, Science 321 (5897) (2008) 1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Ye D, Guan KL, Xiong Y, Metabolism, Activity, and Targeting of D- and L-2-Hydroxyglutarates, Trends Cancer 4 (2) (2018) 151–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Chen F, Bian K, Tang Q, Fedeles BI, Singh V, Humulock ZT, Essigmann JM, Li D, Oncometabolites d- and l-2-Hydroxyglutarate Inhibit the AlkB Family DNA Repair Enzymes under Physiological Conditions, Chem. Res. Toxicol 30 (4) (2017) 1102–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Cairncross JG, Wang M, Jenkins RB, Shaw EG, Giannini C, Brachman DG, Buckner JC, Fink KL, Souhami L, Laperriere NJ, et al. , Benefit from procarbazine, lomustine, and vincristine in oligodendroglial tumors is associated with mutation of IDH, J. Clin. Oncol 32 (8) (2014) 783–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, Yu W, Li Z, Gong L, Peng Y, et al. , Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha, Science 324 (5924) (2009) 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. , IDH mutation impairs histone demethylation and results in a block to cell differentiation, Nature 483 (7390) (2012) 474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Dang L, Su SM, Isocitrate Dehydrogenase Mutation and (R)-2-Hydroxyglutarate: From Basic Discovery to Therapeutics Development, Annu. Rev. Biochem 86 (2017) 305–331. [DOI] [PubMed] [Google Scholar]
- [109].Izquierdo-Garcia JL, Cai LM, Chaumeil MM, Eriksson P, Robinson AE, Pieper RO, Phillips JJ, Ronen SM, Glioma cells with the IDH1 mutation modulate metabolic fractional flux through pyruvate carboxylase, PLoS ONE 9 (9) (2014) e108289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Izquierdo-Garcia JL, Viswanath P, Eriksson P, Cai L, Radoul M, Chaumeil MM, Blough M, Luchman HA, Weiss S, Cairncross JG, et al. , IDH1 Mutation Induces Reprogramming of Pyruvate Metabolism, Cancer Res 75 (15) (2015) 2999–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Chesnelong C, Chaumeil MM, Blough MD, Al-Najjar M, Stechishin OD, Chan JA, Pieper RO, Ronen SM, Weiss S, Luchman HA, et al. , Lactate dehydrogenase A silencing in IDH mutant gliomas, Neuro Oncol 16 (5) (2014) 686–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Amankulor NM, Kim Y, Arora S, Kargl J, Szulzewsky F, Hanke M, Margineantu DH, Rao A, Bolouri H, Delrow J, et al. , Mutant IDH1 regulates the tumor-associated immune system in gliomas, Genes Dev 31 (8) (2017) 774–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Ruiz-Rodado V, Seki T, Dowdy T, Lita A, Zhang M, Han S, Yang C, Cherukuri MK, Gilbert MR, Larion M, Metabolic Landscape of a Genetically Engineered Mouse Model of IDH1 Mutant Glioma, Cancers (Basel) 12 (6) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Batsios G, Viswanath P, Subramani E, Najac C, Gillespie AM, Santos RD, Molloy AR, Pieper RO, Ronen SM, PI3K/mTOR inhibition of IDH1 mutant glioma leads to reduced 2HG production that is associated with increased survival, Sci. Rep 9 (1) (2019) 10521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Yu P, Laird AD, Du X, Wu J, Won KA, Yamaguchi K, Hsu PP, Qian F, Jaeger CT, Zhang W, et al. , Characterization of the activity of the PI3K/mTOR inhibitor XL765 (SAR245409) in tumor models with diverse genetic alterations affecting the PI3K pathway, Mol. Cancer Ther 13 (5) (2014) 1078–1091. [DOI] [PubMed] [Google Scholar]
- [116].Esmaeili M, Hamans BC, Navis AC, van Horssen R, Bathen TF, Gribbestad IS, Leenders WP, Heerschap A, IDH1 R132H mutation generates a distinct phospholipid metabolite profile in glioma, Cancer Res 74 (17) (2014) 4898–4907. [DOI] [PubMed] [Google Scholar]
- [117].Viswanath P, Radoul M, Izquierdo-Garcia JL, Luchman HA, Gregory Cairncross J, Pieper RO, Phillips JJ, Ronen SM, Mutant IDH1 gliomas downregulate phosphocholine and phosphoethanolamine synthesis in a 2-hydroxyglutarate-dependent manner, Cancer & metabolism 6 (2018) 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Viswanath P, Radoul M, Izquierdo-Garcia JL, Ong WQ, Luchman HA, Cairncross JG, Huang B, Pieper RO, Phillips JJ, Ronen SM, 2-Hydroxyglutarate-Mediated Autophagy of the Endoplasmic Reticulum Leads to an Unusual Downregulation of Phospholipid Biosynthesis in Mutant IDH1 Gliomas, Cancer Res 78 (9) (2018) 2290–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Gabellieri C, Beloueche-Babari M, Jamin Y, Payne GS, Leach MO, Eykyn TR, Modulation of choline kinase activity in human cancer cells observed by dynamic 31P NMR, NMR Biomed 22 (4) (2009) 456–461. [DOI] [PubMed] [Google Scholar]
- [120].Popovici-Muller J, Saunders JO, Salituro FG, Travins JM, Yan S, Zhao F, Gross S, Dang L, Yen KE, Yang H, et al. , Discovery of the First Potent Inhibitors of Mutant IDH1 That Lower Tumor 2-HG in Vivo, ACS Med. Chem. Lett 3 (10) (2012) 850–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Bouzier AK, Quesson B, Valeins H, Canioni P, Merle M, [1-(13)C]glucose metabolism in the tumoral and nontumoral cerebral tissue of a glioma-bearing rat, J. Neurochem 72 (6) (1999) 2445–2455. [DOI] [PubMed] [Google Scholar]
- [122].Beckmann N, Turkalj I, Seelig J, Keller U, 13C NMR for the assessment of human brain glucose metabolism in vivo, Biochemistry 30 (26) (1991) 6362–6366. [DOI] [PubMed] [Google Scholar]
- [123].Maher EA, Marin-Valencia I, Bachoo RM, Mashimo T, Raisanen J, Hatanpaa KJ, Jindal A, Jeffrey FM, Choi C, Madden C, et al. , Metabolism of [U-13 C]glucose in human brain tumors in vivo, NMR Biomed 25 (11) (2012) 1234–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Marin-Valencia I, Yang C, Mashimo T, Cho S, Baek H, Yang XL, Rajagopalan KN, Maddie M, Vemireddy V, Zhao Z, et al. , Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo, Cell Metab 15 (6) (2012) 827–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Deelchand DK, Nelson C, Shestov AA, Ugurbil K, Henry PG, Simultaneous measurement of neuronal and glial metabolism in rat brain in vivo using co-infusion of [1,6-C-13(2)]glucose and [1,2-C-13(2)]acetate, J. Magn. Reson 196 (2) (2009) 157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Marin-Valencia I, Good LB, Ma Q, Malloy CR, Patel MS, Pascual JM, Cortical metabolism in pyruvate dehydrogenase deficiency revealed by ex vivo multiplet C-13 NMR of the adult mouse brain, Neurochem. Int 61 (7) (2012) 1036–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Pichumani K, Mashimo T, Vemireddy V, Ijare OB, Mickey BE, Malloy CR, Marin-Valencia I, Baskin DS, Bachoo RM, Maher EA, Measurement of C-13 turnover into glutamate and glutamine pools in brain tumor patients, FEBS Lett 591 (21) (2017) 3548–3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].De Feyter HM, Behar KL, Rao JU, Madden-Hennessey K, Ip KL, Hyder F, Drewes LR, Geschwind JF, de Graaf RA, Rothman DL, A ketogenic diet increases transport and oxidation of ketone bodies in RG2 and 9L gliomas without affecting tumor growth, Neuro Oncol 18 (8) (2016) 1079–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Keevil SF, Spatial localization in nuclear magnetic resonance spectroscopy, Phys. Med. Biol 51 (16) (2006) R579–R636. [DOI] [PubMed] [Google Scholar]
- [130].Bottomley PA, Spatial localization in NMR spectroscopy in vivo, Ann. N. Y. Acad. Sci 508 (1987) 333–348. [DOI] [PubMed] [Google Scholar]
- [131].Moonen CT, von Kienlin M, van Zijl PC, Cohen J, Gillen J, Daly P, Wolf G, Comparison of single-shot localization methods (STEAM and PRESS) for in vivo proton NMR spectroscopy, NMR Biomed 2 (5–6) (1989) 201–208. [DOI] [PubMed] [Google Scholar]
- [132].Yongbi NM, Payne GS, Collins DJ, Leach MO, Quantification of signal selection efficiency, extra volume suppression and contamination for ISIS, STEAM and PRESS localized 1H NMR spectroscopy using an EEC localization test object, Phys. Med. Biol 40 (7) (1995) 1293–1303. [DOI] [PubMed] [Google Scholar]
- [133].Ordidge RJ, Connelly A, Lohman JAB: Image-selected in Vivo spectroscopy (ISIS). A new technique for spatially selective nmr spectroscopy. Journal of Magnetic Resonance (1969) 1986, 66(2):283–294. [Google Scholar]
- [134].Bogner W, Otazo R, Henning A, Accelerated MR spectroscopic imaging-a review of current and emerging techniques, NMR Biomed (2020), e4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Bruhn H, Frahm J, Gyngell ML, Merboldt KD, Hanicke W, Sauter R, Hamburger C, Noninvasive Differentiation of Tumors with Use of Localized H-1 Spectroscopy Invivo - Initial Experience in Patients with Cerebral-Tumors - Response, Invest. Radiol 25 (9) (1990) 1049–1050. [DOI] [PubMed] [Google Scholar]
- [136].Greenhaff PL, The creatine-phosphocreatine system: there’s more than one song in its repertoire, J. Physiol 537 (Pt 3) (2001) 657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Bulik M, Jancalek R, Vanicek J, Skoch A, Mechl M, Potential of MR spectroscopy for assessment of glioma grading, Clin. Neurol. Neurosurg 115 (2) (2013) 146–153. [DOI] [PubMed] [Google Scholar]
- [138].Tzika AA, Zurakowski D, Poussaint TY, Goumnerova L, Astrakas LG, Barnes PD, Anthony DC, Billett AL, Tarbell NJ, Scott RM, et al. , Proton magnetic spectroscopic imaging of the child’s brain: the response of tumors to treatment, Neuroradiology 43 (2) (2001) 169–177. [DOI] [PubMed] [Google Scholar]
- [139].Urenjak J, Williams SR, Gadian DG, Noble M, Specific expression of N-acetylaspartate in neurons, oligodendrocyte-type-2 astrocyte progenitors, and immature oligodendrocytes in vitro, J. Neurochem 59 (1) (1992) 55–61. [DOI] [PubMed] [Google Scholar]
- [140].Demougeot C, Garnier P, Mossiat C, Bertrand N, Giroud M, Beley A, Marie C, N-Acetylaspartate, a marker of both cellular dysfunction and neuronal loss: its relevance to studies of acute brain injury, J. Neurochem 77 (2) (2001) 408–415. [DOI] [PubMed] [Google Scholar]
- [141].van der Graaf M, In vivo magnetic resonance spectroscopy: basic methodology and clinical applications, Eur. Biophys. J 39 (4) (2010) 527–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Barba I, Cabanas ME, Arus C, The relationship between nuclear magnetic resonance-visible lipids, lipid droplets, and cell proliferation in cultured C6 cells, Cancer Res 59 (8) (1999) 1861–1868. [PubMed] [Google Scholar]
- [143].Remy C, Fouilhe N, Barba I, Sam-Lai E, Lahrech H, Cucurella MG, Izquierdo M, Moreno A, Ziegler A, Massarelli R, et al. , Evidence that mobile lipids detected in rat brain glioma by 1H nuclear magnetic resonance correspond to lipid droplets, Cancer Res 57 (3) (1997) 407–414. [PubMed] [Google Scholar]
- [144].Bensaad K, Favaro E, Lewis CA, Peck B, Lord S, Collins JM, Pinnick KE, Wigfield S, Buffa FM, Li JL, et al. , Fatty acid uptake and lipid storage induced by HIF-1alpha contribute to cell growth and survival after hypoxiareoxygenation, Cell Rep 9 (1) (2014) 349–365. [DOI] [PubMed] [Google Scholar]
- [145].Petan T, Jarc E, Jusovic M, Lipid Droplets in Cancer: Guardians of Fat in a Stressful World, Molecules 23 (8) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Li X, Lu Y, Pirzkall A, McKnight T, Nelson SJ, Analysis of the spatial characteristics of metabolic abnormalities in newly diagnosed glioma patients, J. Magn. Reson. Imaging 16 (3) (2002) 229–237. [DOI] [PubMed] [Google Scholar]
- [147].Opstad KS, Ladroue C, Bell BA, Griffiths JR, Howe FA, Linear discriminant analysis of brain tumour H-1 MR spectra: a comparison of classification using whole spectra versus metabolite quantification, NMR Biomed 20 (8) (2007) 763–770. [DOI] [PubMed] [Google Scholar]
- [148].Crawford FW, Khayal IS, Mcgue C, Saraswathy S, Pirzkall A, Cha S, Lamborn KR, Chang SM, Berger MS, Nelson SJ, Relationship of pre-surgery metabolic and physiological MR imaging parameters to survival for patients with untreated GBM, J Neuro-Oncol 91 (3) (2009) 337–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Murphy PS, Rowland JI, Viviers L, Brada M, Leach MO, Dzik-Jurasz ASK, Could assessment of glioma methylene lipid resonance by in vivo H-1-MRS be of clinical value?, Brit J. Radiol 76 (907) (2003) 459–463. [DOI] [PubMed] [Google Scholar]
- [150].Yablonskiy DA, Neil JJ, Raichle ME, Ackerman JJH, Homonuclear J coupling effects in volume localized NMR spectroscopy: Pitfalls and solutions, Magnet Reson. Med 39 (2) (1998) 169–178. [DOI] [PubMed] [Google Scholar]
- [151].Faubert B, Solmonson A, DeBerardinis RJ, Metabolic reprogramming and cancer progression, Science 368 (2020) 6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Yamasaki F, Takaba J, Ohtaki M, Abe N, Kajiwara Y, Saito T, Yoshioka H, Hama S, Akimitsu T, Sugiyama K, et al. , Detection and differentiation of lactate and lipids by single-voxel proton MR spectroscopy, Neurosurg. Rev 28 (4) (2005) 267–277. [DOI] [PubMed] [Google Scholar]
- [153].Negendank WG, Sauter R, Brown TR, Evelhoch JL, Falini A, Gotsis ED, Heerschap A, Kamada K, Lee BC, Mengeot MM, et al. , Proton magnetic resonance spectroscopy in patients with glial tumors: a multicenter study, J. Neurosurg 84 (3) (1996) 449–458. [DOI] [PubMed] [Google Scholar]
- [154].Ott D, Hennig J, Ernst T, Human brain tumors: assessment with in vivo proton MR spectroscopy, Radiology 186 (3) (1993) 745–752. [DOI] [PubMed] [Google Scholar]
- [155].Li X, Vigneron DB, Cha S, Graves EE, Crawford F, Chang SM, Nelson SJ, Relationship of MR-derived lactate, mobile lipids, and relative blood volume for gliomas in vivo, AJNR Am. J. Neuroradiol 26 (4) (2005) 760–769. [PMC free article] [PubMed] [Google Scholar]
- [156].Herminghaus S, Pilatus U, Moller-Hartmann W, Raab P, Lanfermann H, Schlote W, Zanella FE, Increased choline levels coincide with enhanced proliferative activity of human neuroepithelial brain tumors, NMR Biomed 15 (6) (2002) 385–392. [DOI] [PubMed] [Google Scholar]
- [157].Guo J, Yao CJ, Chen H, Zhuang DX, Tang WJ, Ren G, Wang Y, Wu JS, Huang FP, Zhou LF, The relationship between Cho/NAA and glioma metabolism: implementation for margin delineation of cerebral gliomas, Acta Neurochir 154 (8) (2012) 1361–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].McKnight TR, Noworolski SM, Vigneron DB, Nelson SJ, An automated technique for the quantitative assessment of 3D-MRSI data from patients with glioma, J. Magn. Reson. Imaging 13 (2) (2001) 167–177. [DOI] [PubMed] [Google Scholar]
- [159].Weybright P, Sundgren PC, Maly P, Hassan DG, Nan B, Rohrer S, Junck L, Differentiation between brain tumor recurrence and radiation injury using MR spectroscopy, Am. J. Roentgenol 185 (6) (2005) 1471–1476. [DOI] [PubMed] [Google Scholar]
- [160].Astrakas LG, Zurakowski D, Tzika AA, Zarifi MK, Anthony DC, De Girolami U, Tarbell NJ, Black PM, Noninvasive magnetic resonance spectroscopic imaging biomarkers to predict the clinical grade of pediatric brain tumors, Clin. Cancer Res 10 (24) (2004) 8220–8228. [DOI] [PubMed] [Google Scholar]
- [161].Moller-Hartmann W, Herminghaus S, Krings T, Marquardt G, Lanfermann H, Pilatus U, Zanella FE, Clinical application of proton magnetic resonance spectroscopy in the diagnosis of intracranial mass lesions, Neuroradiology 44 (5) (2002) 371–381. [DOI] [PubMed] [Google Scholar]
- [162].Wang Q, Zhang H, Zhang J, Wu C, Zhu W, Li F, Chen X, Xu B, The diagnostic performance of magnetic resonance spectroscopy in differentiating high-from low-grade gliomas: A systematic review and meta-analysis, Eur. Radiol 26 (8) (2016) 2670–2684. [DOI] [PubMed] [Google Scholar]
- [163].Nelson SJ, Kadambi AK, Park I, Li Y, Crane J, Olson M, Molinaro A, Roy R, Butowski N, Cha S, et al. , Association of early changes in H-1 MRSI parameters with survival for patients with newly diagnosed glioblastoma receiving a multimodality treatment regimen, Neuro-Oncology 19 (3) (2017) 430–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Nelson SJ, Li Y, Lupo JM, Olson M, Crane JC, Molinaro A, Roy R, Clarke J, Butowski N, Prados M, et al. , Serial analysis of 3D H-1 MRSI for patients with newly diagnosed GBM treated with combination therapy that includes bevacizumab, J. Neuro-Oncol 130 (1) (2016) 171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Anwar M, Molinaro AM, Morin O, Chang SM, Haas-Kogan DA, Nelson SJ, Lupo JM, Identifying Voxels at Risk for Progression in Glioblastoma Based on Dosimetry, Physiologic and Metabolic MRI, Radiat Res 188 (3) (2017) 303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Dowling C, Bollen AW, Noworolski SM, McDermott MW, Barbaro NM, Day MR, Henry RG, Chang SM, Dillon WP, Nelson SJ, et al. , Preoperative proton MR spectroscopic imaging of brain tumors: Correlation with histopathologic analysis of resection specimens, Am. J. Neuroradiol 22 (4) (2001) 604–612. [PMC free article] [PubMed] [Google Scholar]
- [167].Wilson M, Cummins CL, MacPherson L, Sun Y, Natarajan K, Grundy RG, Arvanitis TN, Kauppinen RA, Peet AC, Magnetic resonance spectroscopy metabolite profiles predict survival in paediatric brain tumours, Eur. J. Cancer 49 (2) (2013) 457–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Yeh IB, Xu MS, Ng WH, Ye J, Yang D, Lim CCT, Central neurocytoma: typical magnetic resonance spectroscopy findings and atypical ventricular dissemination, Magn. Reson. Imaging 26 (1) (2008) 59–64. [DOI] [PubMed] [Google Scholar]
- [169].Choi C, Ganji SK, DeBerardinis RJ, Dimitrov IE, Pascual JM, Bachoo R, Mickey BE, Malloy CR, Maher EA, Measurement of Glycine in the Human Brain in Vivo by H-1-MRS at 3 T: Application in Brain Tumors, Magnet Reson Med 66 (3) (2011) 609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Tiwari V, An ZX, Ganji SK, Baxter J, Patel TR, Pan E, Mickey BE, Maher EA, Pinho MC, Choi CH, Measurement of glycine in healthy and tumorous brain by triple-refocusing MRS at 3T in vivo, NMR Biomed 30 (9) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Hattingen E, Lanfermann H, Quick J, Franz K, Zanella FE, Pilatus U, H-1 MR spectroscopic imaging with short and long echo time to discriminate glycine in glial tumours, Magn. Reson. Mater. Phy 22 (1) (2009) 33–41. [DOI] [PubMed] [Google Scholar]
- [172].Davies NP, Wilson M, Natarajan K, Sun Y, MacPherson L, Brundler MA, Arvanitis TN, Grundy RG, Peet AC, Non-invasive detection of glycine as a biomarker of malignancy in childhood brain tumours using in-vivo H-1 MRS at 1.5 Tesla confirmed by ex-vivo, high-resolution magic-angle spinning NMR, NMR Biomed 23 (1) (2010) 80–87. [DOI] [PubMed] [Google Scholar]
- [173].Gambarota G, Mekle R, Xin LJ, Hergt M, van der Zwaag W, Krueger G, Gruetter R, In vivo measurement of glycine with short echo-time H-1 MRS in human brain at 7 T, Magn Reson Mater Phy 22 (1) (2009) 1–4. [DOI] [PubMed] [Google Scholar]
- [174].Choi C, Douglas D, Hawesa H, Jindal A, Storey C, Dimitrov I, Measurement of Glycine in Human Prefrontal Brain by Point-Resolved Spectroscopy at 7.0 Tesla In Vivo, Magnet Reson Med 62 (5) (2009) 1305–1310. [DOI] [PubMed] [Google Scholar]
- [175].Tiwari V, Daoud EV, Hatanpaa KJ, Gao A, Zhang S, An Z, Ganji SK, Raisanen JM, Lewis CM, Askari P, et al. , Glycine by MR spectroscopy is an imaging biomarker of glioma aggressiveness, Neuro Oncol 22 (7) (2020) 1018–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Kovanlikaya A, Panigrahy A, Krieger MD, Gonzalez-Gomez I, Ghugre N, McComb JG, Gilles FH, Nelson MD, Bluml S, Untreated pediatric primitive neuroectodermal tumor in vivo: Quantitation of taurine with MR spectroscopy, Radiology 236 (3) (2005) 1020–1025. [DOI] [PubMed] [Google Scholar]
- [177].Brand A, Richterlandsberg C, Leibfritz D, Multinuclear Nmr-Studies on the Energy-Metabolism of Glial and Neuronal Cells, Dev Neurosci-Basel 15 (3–5) (1993) 289–298. [DOI] [PubMed] [Google Scholar]
- [178].Castillo M, Smith JK, Kwock L, Correlation of myo-inositol levels and grading of cerebral astrocytomas, Am J Neuroradiol 21 (9) (2000) 1645–1649. [PMC free article] [PubMed] [Google Scholar]
- [179].Cho YD, Choi GH, Lee SP, Kim JK, H-1-MRS metabolic patterns for distinguishing between meningiomas and other brain tumors, Magn. Reson. Imaging 21 (6) (2003) 663–672. [DOI] [PubMed] [Google Scholar]
- [180].Branzoli F, Pontoizeau C, Tchara L, Di Stefano AL, Kamoun A, Deelchand DK, Valabregue R, Lehericy S, Sanson M, Ottolenghi C, et al. , Cystathionine as a marker for 1p/19q codeleted gliomas by in vivo magnetic resonance spectroscopy, Neuro-Oncology 21 (6) (2019) 765–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Artzi M, Liberman G, Vaisman N, Bokstein F, Vitinshtein F, Aizenstein O, Ben Bashat D, Changes in cerebral metabolism during ketogenic diet in patients with primary brain tumors: H-1-MRS study, J Neuro-Oncol 132 (2) (2017) 267–275. [DOI] [PubMed] [Google Scholar]
- [182].Law M, Yang S, Wang H, Babb JS, Johnson G, Cha S, Knopp EA, Zagzag D, Glioma grading: sensitivity, specificity, and predictive values of perfusion MR imaging and proton MR spectroscopic imaging compared with conventional MR imaging, AJNR Am. J. Neuroradiol 24 (10) (2003) 1989–1998. [PMC free article] [PubMed] [Google Scholar]
- [183].Hellstrom J, Romanos Zapata R, Libard S, Wikstrom J, Ortiz-Nieto F, Alafuzoff I, Raininko R, The value of magnetic resonance spectroscopy as a supplement to MRI of the brain in a clinical setting, PLoS ONE 13 (11) (2018) e0207336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Thust SC, Heiland S, Falini A, Jager HR, Waldman AD, Sundgren PC, Godi C, Katsaros VK, Ramos A, Bargallo N, et al. , Glioma imaging in Europe: A survey of 220 centres and recommendations for best clinical practice, Eur. Radiol 28 (8) (2018) 3306–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. , Cancer-associated IDH1 mutations produce 2-hydroxyglutarate, Nature 462 (7274) (2009) 739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Intlekofer AM, Dematteo RG, Venneti S, Finley LW, Lu C, Judkins AR, Rustenburg AS, Grinaway PB, Chodera JD, Cross JR, et al. , Hypoxia Induces Production of L-2-Hydroxyglutarate, Cell Metab 22 (2) (2015) 304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Fan J, Teng X, Liu L, Mattaini KR, Looper RE, Vander Heiden MG, Rabinowitz JD, Human phosphoglycerate dehydrogenase produces the oncometabolite D-2-hydroxyglutarate, ACS Chem. Biol 10 (2) (2015) 510–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Sener RN, L-2 hydroxyglutaric aciduria: Proton magnetic resonance spectroscopy and diffusion magnetic resonance imaging findings, J Comput Assist Tomo 27 (1) (2003) 38–43. [DOI] [PubMed] [Google Scholar]
- [189].Goffette SM, Duprez TP, Nassogne MCL, Vincent MFA, Jakobs C, Sindic CJ, L-2-hydroxyglutaric aciduria: clinical, genetic, and brain MRI characteristics in two adult sisters, Eur. J. Neurol 13 (5) (2006) 499–504. [DOI] [PubMed] [Google Scholar]
- [190].Pope WB, Prins RM, Thomas MA, Nagarajan R, Yen KE, Bittinger MA, Salamon N, Chou AP, Yong WH, Soto H, et al. , Non-invasive detection of 2-hydroxyglutarate and other metabolites in IDH1 mutant glioma patients using magnetic resonance spectroscopy, J Neuro-Oncol 107 (1) (2012) 197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Choi C, Ganji SK, DeBerardinis RJ, Hatanpaa KJ, Rakheja D, Kovacs Z, Yang XL, Mashimo T, Raisanen JM, Marin-Valencia I, et al. , 2-hydroxyglutarate detection by magnetic resonance spectroscopy in subjects with IDH-mutated gliomas, Nat. Med 18 (4) (2012) 624–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Choi CH, Ganji S, Hulsey K, Madan A, Kovacs Z, Dimitrov I, Zhang S, Pichumani K, Mendelsohn D, Mickey B, et al. , A comparative study of short-and long-TE H-1 MRS at 3 T for in vivo detection of 2-hydroxyglutarate in brain tumors, NMR Biomed 26 (10) (2013) 1242–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Ganji SK, An ZX, Tiwari V, McNeil S, Pinho MC, Pan E, Mickey BE, Maher EA, Choi CH, In Vivo Detection of 2-Hydroxyglutarate in Brain Tumors by Optimized Point-Resolved Spectroscopy (PRESS) at 7T, Magnet Reson Med 77 (3) (2017) 936–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Andronesi OC, Ramadan S, Mountford CE, Sorensen AG, Low-Power Adiabatic Sequences for In Vivo Localized Two-Dimensional Chemical Shift Correlated MR Spectroscopy, Magnet Reson Med 64 (6) (2010) 1542–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Andronesi OC, Loebel F, Bogner W, Marjanska M, Vander Heiden MG, Iafrate AJ, Dietrich J, Batchelor TT, Gerstner ER, Kaelin WG, et al. , Treatment Response Assessment in IDH-Mutant Glioma Patients by Noninvasive 3D Functional Spectroscopic Mapping of 2-Hydroxyglutarate, Clin. Cancer Res 22 (7) (2016) 1632–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Scheenen TWJ, Klomp DWJ, Wijnen JP, Heerschap A, Short echo time H-1-MRSI of the human brain at 3T with minimal chemical shift displacement errors using adiabatic refocusing pulses, Magnet Reson Med 59 (1) (2008) 1–6. [DOI] [PubMed] [Google Scholar]
- [197].Berrington A, Voets NL, Larkin SJ, de Pennington N, Mccullagh J, Stacey R, Schofield CJ, Jezzard P, Clare S, Cadoux-Hudson T et al. : A comparison of 2-hydroxyglutarate detection at 3 and 7T with long-TE semi-LASER. Nmr Biomed 2018, 31(3). [DOI] [PubMed] [Google Scholar]
- [198].Emir UE, Larkin SJ, de Pennington N, Voets N, Plaha P, Stacey R, Al-Qahtani K, Mccullagh J, Schofield CJ, Clare S, et al. , Noninvasive Quantification of 2-Hydroxyglutarate in Human Gliomas with IDH1 and IDH2 Mutations, Cancer Res 76 (1) (2016) 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Leather T, Jenkinson MD, Das K, Poptani H, Magnetic Resonance Spectroscopy for Detection of 2-Hydroxyglutarate as a Biomarker for IDH Mutation in Gliomas, Metabolites 7 (2) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Natsumeda M, Motohashi K, Igarashi H, Nozawa T, Abe H, Tsukamoto Y, Ogura R, Okada M, Kobayashi T, Aoki H, et al. , Reliable diagnosis of IDH-mutant glioblastoma by 2-hydroxyglutarate detection: a study by 3-T magnetic resonance spectroscopy, Neurosurg. Rev 41 (2) (2018) 641–647. [DOI] [PubMed] [Google Scholar]
- [201].Nagashima H, Tanaka K, Sasayama T, Irino Y, Sato N, Takeuchi Y, Kyotani K, Mukasa A, Mizukawa K, Sakata J, et al. , Diagnostic value of glutamate with 2-hydroxyglutarate in magnetic resonance spectroscopy for IDH1 mutant glioma, Neuro-Oncology 18 (11) (2016) 1559–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Natsumeda M, Igarashi H, Nomura T, Ogura R, Tsukamoto Y, Kobayashi T, Aoki H, Okamoto K, Kakita A, Takahashi H et al. : Accumulation of 2-hydroxyglutarate in gliomas correlates with survival: a study by 3.0-tesla magnetic resonance spectroscopy. Acta Neuropathol Commun 2014, 2:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Choi CH, Raisanen JM, Ganji SK, Zhang S, McNeil SS, An ZX, Madan A, Hatanpaa KJ, Vemireddy V, Sheppard CA, et al. , Prospective Longitudinal Analysis of 2-Hydroxyglutarate Magnetic Resonance Spectroscopy Identifies Broad Clinical Utility for the Management of Patients With IDH-Mutant Glioma, J. Clin. Oncol 34 (33) (2016) 4030–U4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Howe FA, Barton SJ, Cudlip SA, Stubbs M, Saunders DE, Murphy M, Wilkins P, Opstad KS, Doyle VL, McLean MA, et al. , Metabolic profiles of human brain tumors using quantitative in vivo H-1 magnetic resonance spectroscopy, Magnet Reson Med 49 (2) (2003) 223–232. [DOI] [PubMed] [Google Scholar]
- [205].Chiang GC, Kovanlikaya I, Choi C, Ramakrishna R, Magge R, Shungu DC, Magnetic Resonance Spectroscopy, Positron Emission Tomography and Radiogenomics-Relevance to Glioma, Front. Neurol 9 (2018) 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].An Z, Ganji SK, Tiwari V, Pinho MC, Patel T, Barnett S, Pan E, Mickey BE, Maher EA, Choi C, Detection of 2-hydroxyglutarate in brain tumors by triple-refocusing MR spectroscopy at 3T in vivo, Magnet Reson Med 78 (1) (2017) 40–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Andronesi OC, Kim GS, Gerstner E, Batchelor T, Tzika AA, Fantin VR, Vander Heiden MG, Sorensen AG, Detection of 2-Hydroxyglutarate in IDH-Mutated Glioma Patients by In Vivo Spectral-Editing and 2D Correlation Magnetic Resonance Spectroscopy, Sci. Transl. Med 4 (116):116ra114–116ra114 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Kim MM, Parolia A, Dunphy MP, Venneti S, Non-invasive metabolic imaging of brain tumours in the era of precision medicine, Nat. Rev. Clin. Oncol 13 (12) (2016) 725–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Brender JR, Kishimoto S, Merkle H, Reed G, Hurd RE, Chen AP, Ardenkjaer-Larsen JH, Munasinghe J, Saito K, Seki T, et al. , Dynamic Imaging of Glucose and Lactate Metabolism by 13C-MRS without Hyperpolarization, Sci. Rep 9 (2019) 3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Kishimoto S, Brender JR, Crooks DR, Matsumoto S, Seki T, Oshima N, Merkle H, Lin P, Reed G, Chen AP et al. : Imaging of glucose metabolism by 13C-MRI distinguishes pancreatic cancer subtypes in mice. Elife 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Rodrigues TB, Serrao EM, Kennedy BWC, Hu DE, Kettunen MI, Brindle KM, Magnetic resonance imaging of tumor glycolysis using hyperpolarized C-13-labeled glucose, Nat. Med 20 (1) (2014) 93–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Golman K, Leunbach I, Ardenkjaer-Larsen JH, Ehnholm GJ, Wistrand LG, Petersson JS, Jarvi A, Vahasalo S, Overhauser-enhanced MR imaging (OMRI), Acta Radiol 39 (1) (1998) 10–17. [DOI] [PubMed] [Google Scholar]
- [213].Krishna MC, English S, Yamada K, Yoo J, Murugesan R, Devasahayam N, Cook JA, Golman K, Ardenkjaer-Larsen JH, Subramanian S, et al. , Overhauser enhanced magnetic resonance imaging for tumor oximetry: coregistration of tumor anatomy and tissue oxygen concentration, Proc Natl. Acad Sci. U.S.A 99 (4) (2002) 2216–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Grucker D, Guiberteau T, Eclancher B, Oximetry by dynamic nuclear polarization, Magn. Reson. Med 34 (2) (1995) 219–226. [DOI] [PubMed] [Google Scholar]
- [215].Yamato M, Shiba T, Naganuma T, Ichikawa K, Utsumi H, Yamada K, Overhauser-enhanced magnetic resonance imaging characterization of mitochondria functional changes in the 6-hydroxydopamine rat model, Neurochem. Int 59 (6) (2011) 804–811. [DOI] [PubMed] [Google Scholar]
- [216].Yamato M, Shiba T, Yamada K, Watanabe T, Utsumi H, Noninvasive assessment of the brain redox status after transient middle cerebral artery occlusion using Overhauser-enhanced magnetic resonance imaging, J. Cereb. Blood Flow Metab 29 (10) (2009) 1655–1664. [DOI] [PubMed] [Google Scholar]
- [217].Ardenkjaer-Larsen JH, Fridlund B, Gram A, Hansson G, Hansson L, Lerche MH, Servin R, Thaning M, Golman K, Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR, P. Natl. Acad Sci. USA 100 (18) (2003) 10158–10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Ardenkjaer-Larsen JH, Leach AM, Clarke N, Urbahn J, Anderson D, Skloss TW, Dynamic nuclear polarization polarizer for sterile use intent, NMR Biomed 24 (8) (2011) 927–932. [DOI] [PubMed] [Google Scholar]
- [219].Capozzi A, Patel S, Wenckebach WT, Karlsson M, Lerche MH, Ardenkjaer-Larsen JH, Gadolinium Effect at High-Magnetic-Field DNP: 70% (13)C Polarization of [U-(13)C] Glucose Using Trityl, J. Phys. Chem. Lett 10 (12) (2019) 3420–3425. [DOI] [PubMed] [Google Scholar]
- [220].Lilly Thankamony AS, Wittmann JJ, Kaushik M, Corzilius B, Dynamic nuclear polarization for sensitivity enhancement in modern solid-state NMR, Prog. Nucl. Magn. Reson. Spectrosc 102–103 (2017) 120–195. [DOI] [PubMed] [Google Scholar]
- [221].Kurhanewicz J, Vigneron DB, Ardenkjaer-Larsen JH, Bankson JA, Brindle K, Cunningham CH, Gallagher FA, Keshari KR, Kjaer A, Laustsen C, et al. , Hyperpolarized (13)C MRI: Path to Clinical Translation in Oncology, Neoplasia 21 (1) (2019) 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Keshari KR, Wilson DM, Chemistry and biochemistry of 13C hyperpolarized magnetic resonance using dynamic nuclear polarization, Chem. Soc. Rev 43 (5) (2014) 1627–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Chaumeil MM, Radoul M, Najac C, Eriksson P, Viswanath P, Blough MD, Chesnelong C, Luchman HA, Cairncross JG, Ronen SM, Hyperpolarized 13C MR imaging detects no lactate production in mutant IDH1 gliomas: Implications for diagnosis and response monitoring, NeuroImage: Clinical 12 (2016) 180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Ruiz-Rodado V, Malta TM, Seki T, Lita A, Dowdy T, Celiku O, Cavazos-Saldana A, Li A, Liu Y, Han S, et al. , Metabolic Reprogramming Associated with Aggressiveness Occurs in the G-CIMP-High Molecular Subtypes of IDH1mut Lower Grade Gliomas, Neuro Oncol 22 (4) (2019) 480–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].De Feyter HM, Behar KL, Corbin ZA, Fulbright RK, Brown PB, McIntyre S, Nixon TW, Rothman DL, de Graaf RA, Deuterium metabolic imaging (DMI) for MRI-based 3D mapping of metabolism in vivo, Sci. Adv 4 (8) (2018), eaat7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Miloushev VZ, Granlund KL, Boltyanskiy R, Lyashchenko SK, DeAngelis LM, Mellinghoff IK, Brennan CW, Tabar V, Yang TJ, Holodny AI, et al. , Metabolic Imaging of the Human Brain with Hyperpolarized (13)C Pyruvate Demonstrates (13)C Lactate Production in Brain Tumor Patients, Cancer Res 78 (14) (2018) 3755–3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Park I, Larson PEZ, Gordon JW, Carvajal L, Chen HY, Bok R, Van Criekinge M, Ferrone M, Slater JB, Xu D, et al. , Development of methods and feasibility of using hyperpolarized carbon-13 imaging data for evaluating brain metabolism in patient studies, Magn. Reson. Med 80 (3) (2018) 864–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Park JM, Josan S, Jang T, Merchant M, Watkins R, Hurd RE, Recht LD, Mayer D, Spielman DM, Volumetric spiral chemical shift imaging of hyperpolarized [2-(13) c]pyruvate in a rat c6 glioma model, Magn. Reson. Med 75 (3) (2016) 973–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Chung BT, Chen HY, Gordon J, Mammoli D, Sriram R, Autry AW, Le Page LM, Chaumeil MM, Shin P, Slater J, et al. , First hyperpolarized [2-(13)C] pyruvate MR studies of human brain metabolism, J. Magn. Reson 309 (2019) 106617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Chaumeil MM, Larson PE, Yoshihara HA, Danforth OM, Vigneron DB, Nelson SJ, Pieper RO, Phillips JJ, Ronen SM, Non-invasive in vivo assessment of IDH1 mutational status in glioma, Nat. Commun 4 (2013) 2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Larson PE, Kerr AB, Chen AP, Lustig MS, Zierhut ML, Hu S, Cunningham CH, Pauly JM, Kurhanewicz J, Vigneron DB, Multiband excitation pulses for hyperpolarized 13C dynamic chemical-shift imaging, J. Magn. Reson 194 (1) (2008) 121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].McBrayer SK, Mayers JR, DiNatale GJ, Shi DD, Khanal J, Chakraborty AA, Sarosiek KA, Briggs KJ, Robbins AK, Sewastianik T, et al. , Transaminase Inhibition by 2-Hydroxyglutarate Impairs Glutamate Biosynthesis and Redox Homeostasis in Glioma, Cell 175 (1) (2018) 101–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Chaumeil M, Larson P, Woods S, Cai L, Eriksson P, Robinson A, Lupo J, Vigneron D, Nelson S, Pieper R, et al. , Hyperpolarized [1-C-13] Glutamate: A Metabolic Imaging Biomarker of Idh1 Mutational Status in Glioma, Neuro-Oncology 16 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Miller JJ, Ball DR, Lau AZ, Tyler DJ, Hyperpolarized ketone body metabolism in the rat heart, NMR Biomed 31 (6) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Najac C, Radoul M, Le Page LM, Batsios G, Subramani E, Viswanath P, Gillespie AM, Ronen SM, In vivo investigation of hyperpolarized [1,3-C-13(2)] acetoacetate as a metabolic probe in normal brain and in glioma, Sci Rep-Uk 9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].von Morze C, Ohliger MA, Marco-Rius I, Wilson DM, Flavell RR, Pearce D, Vigneron DB, Kurhanewicz J, Wang ZJ, Direct assessment of renal mitochondrial redox state using hyperpolarized C-13-acetoacetate, Magnet Reson Med 79 (4) (2018) 1862–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Timm KN, Hartl J, Keller MA, Hu DE, Kettunen MI, Rodrigues TB, Ralser M, Brindle KM, Hyperpolarized [U-H-2, U-C-13] Glucose Reports on Glycolytic and Pentose Phosphate Pathway Activity in EL4 Tumors and Glycolytic Activity in Yeast Cells, Magn. Reson. Med 74 (6) (2015) 1543–1547. [DOI] [PubMed] [Google Scholar]
- [238].Mishkovsky M, Anderson B, Karlsson M, Lerche MH, Sherry AD, Gruetter R, Kovacs Z, Comment A, Measuring glucose cerebral metabolism in the healthy mouse using hyperpolarized (13)C magnetic resonance, Sci. Rep 7 (1) (2017) 11719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Nishihara T, Yoshihara HA, Nonaka H, Takakusagi Y, Hyodo F, Ichikawa K, Can E, Bastiaansen JA, Takado Y, Comment A, et al. , Direct Monitoring of gamma-Glutamyl Transpeptidase Activity In Vivo Using a Hyperpolarized (13) C-Labeled Molecular Probe, Angew. Chem. Int. Ed. Engl 55 (36) (2016) 10626–10629. [DOI] [PubMed] [Google Scholar]
- [240].Batsios G, Najac C, Cao P, Viswanath P, Subramani E, Saito Y, Gillespie AM, Yoshihara HAI, Larson P, Sando S, et al. , In vivo detection of gamma-glutamyl-transferase up-regulation in glioma using hyperpolarized gamma-glutamyl-[1-(13)C]glycine, Sci. Rep 10 (1) (2020) 6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Lu M, Zhu XH, Zhang Y, Mateescu G, Chen W, Quantitative assessment of brain glucose metabolic rates using in vivo deuterium magnetic resonance spectroscopy, J Cerebr Blood F Met 37 (11) (2017) 3518–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Rich LJ, Bagga P, Wilson NE, Schnall MD, Detre JA, Haris M, Reddy R, (1) H magnetic resonance spectroscopy of (2)H-to-(1)H exchange quantifies the dynamics of cellular metabolism in vivo, Nat. Biomed. Eng 4 (3) (2020) 335–342. [DOI] [PMC free article] [PubMed] [Google Scholar]


