Abstract
MYB transcription factors are highly conserved from plants to vertebrates, indicating that their functions embrace fundamental mechanisms in the biology of cells and organisms. In humans, the MYB gene family is composed of three members: MYB, MYBL1 and MYBL2, encoding the transcription factors MYB, MYBL1, and MYBL2 (also known as c-MYB, A-MYB, and B-MYB), respectively. A truncated version of MYB, the prototype member of the MYB family, was originally identified as the product of the retroviral oncogene v-myb, which causes leukaemia in birds. This led to the hypothesis that aberrant activation of vertebrate MYB could also cause cancer. Despite more than three decades have elapsed since the isolation of v-myb, only recently investigators were able to detect MYB genes rearrangements and mutations, smoking gun evidence of the involvement of MYB family members in human cancer. In this review, we will highlight studies linking the activity of MYB family members to human malignancies and experimental therapeutic interventions tailored for MYB-expressing cancers.
Subject terms: Cancer, Oncogenes
Introduction
Vertebrate MYB genes encode transcription factors related to the v-myb oncogene, the transforming gene of avian retroviruses causing myelomas and lymphomas in birds1,2. AMV was originally identified as a virus that induces a disease in chickens similar to acute myelogenous leukaemia in humans3. The v-mybAMV oncogene product, a 45 kDa protein, was proved to be a truncated version of vertebrate MYB, the 75 kDa product of the proto-oncogene MYB, mainly expressed in haematopoietic tissues4,5. The v-myb oncogene was also found fused to a second oncogene, v-ets, in the E26 retrovirus that cause avian erythroblastosis6. Invertebrates carry only one MYB gene which, from a phylogenetical and functional point of view, is equivalent to vertebrate MYBL2, suggesting that this is the most ancient member of the family7,8. There is no homologue of the MYB gene in nematodes, although distantly related genes, such as Cdc5 and SNAPc, have been identified in Caenorhabditis elegans9,10.
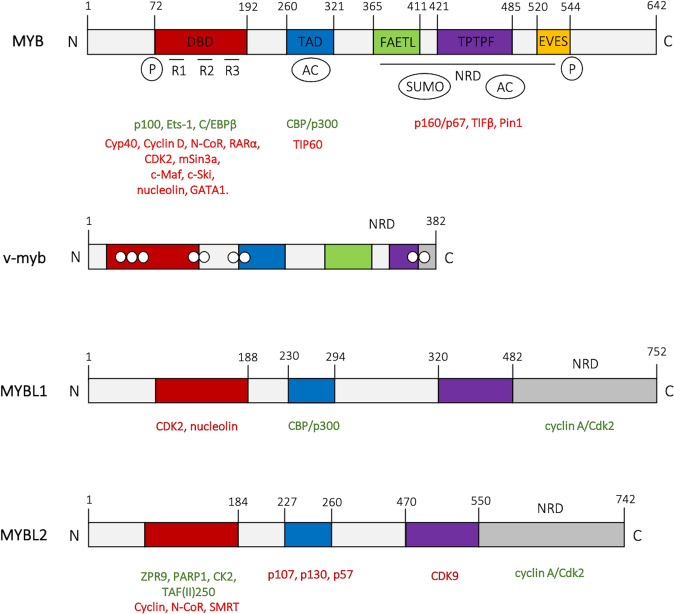
In humans and other mammals, the transcription factor MYB (encoded by MYB) is the prototype member of the family, which includes MYBL1 (encoded by MYBL1) and MYBL2 (encoded by MYBL2)11. Although similar in structure, the different MYB proteins interact with unique co-factors and their expression is often nonoverlapping, suggesting that they might have distinct biological roles (Fig. 1)12–15.
Fig. 1. MYB family members’ protein structures.
The v-myb DNA-binding domain is equivalent to amino acids 72–192 of MYB, except the introduction of four point mutations (I91N, L106H, V117D, and I181V) and the addition of six amino acids in N-terminal region derived from the retroviral Gag polyprotein187. The white dots on AMV v-myb structure indicate point mutations important for the ability of v-myb to transform cells188. MYB co-activators are listed in green and the co-repressors are listed in red. The DNA-binding domain (DBD) is comprised of three repeats (R1, R2, and R3). It is the binding site for a number of proteins including p100, PARP, c-Ski, N-CoR, RAR, Cyp40, C/EMPbeta, SMRT, and mSin3A, as depicted; the central transactivation domain (TAD) is the interaction site for CBP/p300; the negative regulatory domain (NRD) extends from the FAETL motif to the EVES peptide sequence (involved in intramolecular and intermolecular protein–protein interactions) and includes the binding sites for p160/p67, Pin1, and TIF1beta150,189–193. The post-translational modifications include phosphorylation (P), acetylation (AC), and sumoylation (SUMO)194–197.
MYB proteins structure and identification of target genes
MYB proteins contain a highly conserved helix-turn-helix (HTH) DNA-binding domain (DBD) at the N-terminus, encompassing three tandem repeated domains of ~50 amino acids containing tryptophan named R1, R2, and R316; a conserved C-terminal negative regulatory domain (NRD); a trans-activating domain (TAD) in the central portion of the protein. The latter includes an acidic region and a heptad leucine-zipper repeat only present in MYB and MYBL1 (Fig. 1)9.
All MYB family members recognise and bind the same DNA consensus sequence [PyAAC(G/T)G] to transactivate gene expression. This motif, firstly identified by the Klempnauer group using DNA footprinting assays, is known as the canonical MYB-binding site (MBS)17. The sequence was later confirmed to be present, and bound by v-myb, in the promoter region of the first MYB-target gene identified in vertebrates, mim-118. With the development of more advanced genomic technologies, different groups attempted the identification of MYB target genes at the global level. The Ness team found the c-MYB protein bound to over 10,000 promoters in the cancer breast cell line MCF-7, and validated known MYB target genes involved in the cell cycle, such as MYC and CCNB1, or identified new MYB target genes involved in stemness and transcription control such as JUN, KLF4, NANOG and SND119. Another study by the Gonda lab identified genes regulated by MYB in mouse myeloid progenitor cells. This study not only confirmed that MYB positively regulates promoters of key cell proliferation genes, such as Myc, but it can also work as a transcriptional repressor. Indeed, several key regulators of myeloid differentiation such as Runx1, Pu.1, Junb and Cebp were strongly suppressed by exogenous expression of MYB, suggesting a mechanism used by the transcription factor to suppress differentiation and promote self-renewal20. A selection of MYB target genes that have been shown to mediate physiological functions in normal or disease contexts is shown in Table 1.
Table 1.
Selected MYB target genes.
| Target gene | Protein | MYB member | References |
|---|---|---|---|
| ATR | Ataxia telangiectasia Rad3-related protein | MYB | 43 |
| BCL2 | B-cell lymphoma 2 | v-Myb, MYBL2,MYB | 137,198,199 |
| BIRC5 | Survivin | MYBL2, MYB | 136,200,201 |
| CCNA2 | Cyclin A2 | MYBL2 | 149,202 |
| CCNB1 | Cyclin B1 | MYB | 19,38,203 |
| CCND1 | Cyclin D1 | MYBL2 | 131,204 |
| CCNE1 | Cyclin E1 | MYB | 205,206 |
| CD34 | Haematopoietic progenitor cell antigen CD34 | MYB | 207–209 |
| CDK1 | Cyclin-dependent kinase 1 | MYBL2, MYB | 42,149,202 |
| CDK2 | Cyclin-dependent kinase 2 | MYBL2 | 210 |
| CDK6 | Cyclin-dependent kinase 6 | MYB | 211 |
| CLU | Apolipoprotein J/Clusterin | MYBL2 | 132,134,212 |
| CXCR4 | C-X-C chemokine receptor type 4 | v-Myb, MYB | 19,213 |
| IGF1R | Insulin-like growth factor 1 receptor | MYB | 106,107,109,113 |
| KIT | (c-)KIT/CD117 | MYB | 119,214,215 |
| MIM1 | Mitochondrial import protein 1 | v-Myb, MYB | 18,216,217 |
| MYC | (c-)MYC | MYBL2, MYB | 19,20,138,210,218 |
| NCAPH | Non-SMC Condensin I Complex Subunit H | MYBL2 | 169 |
| PLK1 | Polo-like kinase 1 | MYBL2 | 157,159,202 |
| TAL1 | T-cell acute lymphocytic leukaemia protein 1 | MYB | 27 |
| VEGF | Vascular endothelial growth factor | MYB | 118 |
The transcriptional activity of MYB proteins is regulated either positively or negatively by co-factors; cellular proteins physically interacting with the different MYB family members are indicated under their protein structures in Fig. 1. Structure–function relationships have been largely inferred by studying the prototype member of the family, MYB (c-MYB). For example, the TAD domain confers transactivating activity to MYB by recruiting CREB-binding domain protein (CBP) and p30021,22. The CAAT enhancer-binding protein (C/EBP) family member NF‐M cooperates with MYB in transcriptionally activating the mim‐1 promoter through an adjacent DNA-binding site and it is also co‐activated by CBP in a Ras‐dependent manner, suggesting that CBP might work by functionally linking MYB and NF-M22. Indeed, NF-M has been shown to affect the MYB-C/EBP interaction by disrupting the N-terminal region within the repeat domain R1 (amino acids 47–71), enhancing MYB oncogenic activity23.
MYB can cooperate, cross-regulate and compete with other transcription factors, such as members of the C/EBP family, the ETS family, and GATA124–26. Recently, it has been shown that in ALL patients aberrant recruitment of the histone acetyl transferase CBP/p300 by MYB in the enhancer region of the protooncogene TAL1 occurs via the formation of de novo MYB-binding elements27.
Alterations of MYB family genes in human cancer and experimental therapeutic approaches
MYB family members are often aberrantly expressed in human cancers, suggesting that they could be important for tumour initiation and/or maintenance. Since MYB proteins are essential for key cellular processes such as growth, differentiation and survival, it is likely that genomic mutations or alterations of gene expression might contribute to oncogenesis. Broadly expressed transcription factors are considered unsuitable therapeutic targets since their inactivation or downregulation could be detrimental to organism homoeostasis. Furthermore, it is inherently difficult to block the interaction of transcription factors with DNA using small molecules. Despite these caveats, therapeutic approaches aiming at inhibiting MYB oncoproteins, or their target genes, in cancer are under investigation in preclinical and clinical studies.
In the following paragraphs, we discuss studies in which MYB family members have been implicated in forms of human cancer. We also highlight laboratory experiments, or clinical trials, in which MYB, or MYB-regulated genes, have been targeted for therapeutic purposes.
MYB
Disruption of MYB causes embryonic lethality due to the failure of foetal hepatic haematopoiesis28. The key role of the MYB gene product in mammalian haematopoiesis is also indicated by its ability to regulate the expression of foetal haemoglobin and requirement for the maturation of T and B lymphocytes29–32. Although prevalently expressed in haematopoietic cells, MYB expression is detected also in neural tissues, as well as in colonic crypts and breast cells33–37.
MYB, similarly to the ubiquitous member of the family MYBL2, regulates cyclin-dependent kinases (Cdks) expression and activity, essential for cell duplication38,39. MYB autoregulates its own expression and is engaged in positive and negative regulatory loops with cyclins and Cdks, in both the G1 and G2 phases of the cell cycle38,40–42.
MYB alterations in cancer
Genetic mutations and augmented expression of MYB have been firstly noted in leukaemic cells, and only relatively recently in solid cancers. Overexpression of wild type MYB is insufficient for full transformation of human epithelial cells, supporting the hypothesis that it promotes tumourigenesis only in combination with additional genetic alterations43.
The first recurrent genomic rearrangements of the MYB locus were evidenced in acute T cell leukaemia, in which MYB overexpression is caused by gene duplication or translocation, juxtaposing strong enhancers from other genomic locations44. Summarising the information present in literature, it is possible to group MYB oncogenic alterations into three classes: overexpression, fusion with partner genes, and ectopic binding of the MYB oncoprotein to enhancer sequences caused by somatic mutations (i.e. TAL1 enhancer27). MYB gene amplification and overexpression have been observed in acute myeloid leukaemia (AML), non-Hodgkin lymphoma, colorectal cancer, and breast cancer5,45–48. Fusion with partner genes is mainly observed in solid tumours, as discussed in detail in the following sections.
MYB genomic alterations have been detected in multiple forms of human cancer, suggesting a causative role. Therefore, numerous studies have been conducted in which inhibition of MYB, or of its downstream genes, has been used as a potential therapeutic strategy. Preclinical studies and actionable MYB target genes are summarised in Table 2.
Table 2.
Preclinical and clinical therapeutic strategies based on inhibition of MYB or actionable MYB-target genes.
| Treatment | Target | Cancer type | References |
|---|---|---|---|
| AT7519, BE-09-LN53 (CDKi) | CDKs | ER + BC | 90 |
| ATRA | MYB | ACC | 111 |
| Celastrol | MYB-C/EBPβ-p300 | AML | 67 |
| Mebendazol | MYB | AML | 69 |
| miR-200b/c | EMT markers | ER + BC | 94 |
| Monensin A | MYB | AML, ACC | 110 |
| MYBMIM | MYB:CBP/p300 | AML | 71 |
| Naphthol AS-E phosphate | MYB-C/KIX(p300) | Leukaemia | 68 |
| Plumbagin | MYB/p300 | AML | 70 |
| TetMYB vaccine | MYB | CRC, ACC | 81 |
| VX-970 | ATR | ACC | 43 |
| Figitumumab | IGFR | ACC | 109 |
| Linsitinib | IGFR | ACC | 112 |
ATRA all trans retinoic acid, ACC adenoid cystic carcinoma, AML acute myeloid leukaemia, CDKs cyclin-dependent kinases, CRC colorectal cancer, EMT epithelial mesenchymal transition, ER + BC oestrogen receptor positive breast cancer.
MYB and leukaemia
In a cluster of acute lymphoblastic leukaemia (ALL) patients, mutations of the TAL1 enhancer create ex-novo MYB-binding sites. The leukaemias arising in these patients show MYB-dependency consequential to the aberrant activation of the TAL1 oncogene by MYB27. Through genomic screening of an independent set of 107 individuals with T cell ALL (T-ALL) and 12 T-ALL cell lines, Lahortiga et al. detected duplication of MYB in 9 of 107 (8.4%) cases and in five different cell lines49. The flanking genes HBS1L and AHI1 were duplicated in some patients, but the commonly duplicated region covered only the MYB gene. The duplication is associated with a threefold increase in MYB expression, and its knockdown initiates T cell differentiation. Thus, MYB duplications may be leukaemogenic in a subset of T-ALL patients49.
In acute basophilic leukaemia (ABL) the MYB locus is fused to another gene encoding the transcription factor GATA1. This rare subtype of acute myeloblastic leukaemia is characterised by the t(X;6)(p11;q23) translocation, leading to decrease or loss of GATA1 (located on chromosome X) expression50. Mice transgenically expressing the MYB–GATA1 fusion develop myelodysplasia and leukaemia when endogenous, wild-type GATA1 expression is concurrently downregulated51. Ducassou and co-workers showed that the fusion promotes not only haematopoietic progenitor cell self‐renewal, but also induces a bias toward granulocytic differentiation, consequently to sensitisation towards NGF- and IL-33-induced differentiation52. The skewing towards basophilic differentiation was confirmed in primary human CD34‐positive stem/progenitor cells, where the basophilic markers CD203c and FcϵRI were activated after MYB–GATA1 expression. In vivo experiments using NSG mice led to conclusive evidence that basophilic differentiation is a direct consequence of MYB-GATA1 expression, rather than loss of endogenous GATA152. The increased responsiveness to IL-33 could contribute to the leukaemic phenotype, as previously observed in other myeloproliferative malignancies53. Thus, MYB-GATA1 might promote cell growth, self-renewal and leukaemic transformation of basophilic progenitor cells52.
A case report described a Philadelphia-negative myeloproliferative neoplasm (Ph-MPN) with an uncommonly rapid leukaemic progression, linked to JAK2V617F mutation. This primary myelofibrosis (PMF)-patient developed a peculiar chromosomal rearrangement resulting in a fusion involving EWSR1 and MYB. There are only a few cases reporting fusion of EWSR1 in leukaemia, whereas it is common in soft tissue sarcoma54–56. EWSR1 is a FET (FUS, EWS, TAF15) family member whose function is to regulate transcription and mRNA splicing57. Therefore, it seems reasonable to speculate that the EWSR1-MYB fusion could lead to dysregulated MYB transcriptional activity. Indeed, expression of the MYB target gene BCL2 was deregulated in EWSR1-MYB positive PMF, suggesting that molecular alterations involving MYB could increase disease risk in PMF patients58.
AML is the most common form of acute leukaemia in adults59. Although recent advances in genomic characterisations have shed some light on the molecular patterns involved in this cancer, the 5-year survival rate is <70% in children and 35% in adults60,61.
AML is a heterogeneous disease, often characterised by the presence of gene fusions or recurrent mutations in a set of driver genes62. Genomic rearrangements involving the MLL gene, such as MLL–AF4 t(4;11)(q21;q23); MLL–AF9; t(9;11)(p22;q23); MLL–ENL; t(11;19)(q23;p13.3); MLL–AF10 t(10;11)(p12;q23) or MLL–AF6 t(6;11)(q27;q23) are associated with a very aggressive form of leukaemia63,64. MYB has been shown to be a key downstream effector of MLL fusion oncoproteins, suggesting that it could be a target for therapeutic interventions65. Since, as mentioned before, targeting transcription factors with small molecule inhibitors is difficult, the focus has been directed towards proteins that work as co-activators in the MYB network. p300 is a MYB transcriptional co-activator, required for leukaemogenesis66. The small molecule inhibitor Celastrol, a triterpenoid, was used to disrupt the MYB/p300 interaction, therefore interrupting MYB signalling in leukaemic cells. Celastrol did not change MYB expression but inhibited the interaction of the transactivation domain of MYB with the KIX domain of p300. Accordingly, Celastrol strongly inhibited MYB-dependent transcriptional activation of target genes. Celastrol enhanced survival of mice transplanted with patient-derived HoxA9/Meis1-driven AML, confirming that targeting MYB transcription function could be an effective strategy in this leukaemia67. Another compound used to disrupt the interaction between MYB and p300, Naphthol AS-E phosphate, inhibited the expression of the MYB gene itself, as well as that of several MYB-target genes, inducing myeloid differentiation and apoptosis68. The negative effect of Naphthol AS-E phosphate on MYB gene expression could be a consequence of the block of MYB gene autoregulation. Nicolaides et al. showed that human MYB maintains high levels of its expression through an autoregulatory mechanism involving MYB-binding sites in the 5′ flanking region of the MYB gene itself41.
The anti-helminth agent mebendazole exhibited anticancer activity in AML human cell lines by interfering with MYB activity. Short-term exposure to the drug induced changes in the expression level of MYB-regulated genes in cells expressing the MLL-AF9 fusion oncoprotein69. Expression of the MYB oncoprotein was drastically reduced in the presence of low concentrations of the drug in all cell lines analysed, whereas MYB mRNA levels were only reduced after exposure to very high mebendazole concentrations, and only in a few of the cell lines. This suggested that the drug acts mainly at the protein level. Indeed, inhibition of the proteasome reversed MYB protein loss, demonstrating that mebendazole causes proteasomal degradation of MYB by interfering with the heat shock protein 70 (HSP70) chaperone system. Importantly, mebendazole impaired AML cancer progression in vivo69.
5-hydroxy-2-methyl-1,4-naphthoquinone (also known as plumbagin) has been shown to target the transcriptional-activating domain (TAD) of MYB. By using the MYB TAD fused to the Gal4 DBD, the Klempnauer group observed that plumbagin inhibits transcription of a reporter gene containing GAL4-binding sites. Increasing the dosage of ectopically expressed p300, progressively antagonised the effect of plumbagin, demonstrating that the drug interfered with the p300–MYB interaction in AML cells70.
Recently, a peptidomimetic approach to block the activity of MYB was developed by designing an inhibitory peptide called MYBMIM. The MYBMIM inhibitory effect is caused by its ability to disrupt the MYB:CBP/p300 complex. MYBMIM directly binds to the KIX domain of CBP with an affinity similar to the naïve complex, causing its disassembly and reduced MYB-dependent expression of genes whose enhancers are occupied by it. NOD-scid mice engrafted with leukaemia cells treated with the peptide showed significant reduction of cancer burden, which was caused by mitochondrial apoptosis. Furthermore, ChIP analysis revealed a marked loss of the epigenetic mark H3K27ac on super-enhancers regulated by acetylation driven by p300:CBP, and consequent reduced expression of key MYB-regulated genes such as MYC and BCL271.
MYB and paediatric low-grade gliomas (PLGGs)
PLGGs typically present gene fusions, especially related to component of the MAPK pathway, such as BRAF72. MYB rearrangements have been recently discovered in the context of whole-genome sequencing (WGS) and/or RNA-sequencing (RNA-seq) of 249 samples of PLGGs, leading to the identification of recurrent MYB-QKI fusions in angiocentric gliomas73. MYB fused to the RNA-binding protein QKI confers oncogenic properties using three distinct mechanisms. Firstly, the alteration results in the translocation of a super enhancer located in the 3′ untranslated region of QKI upstream the MYB promoter, resulting in its activation. Secondly, the MYB-QKI fusion protein acts as transcription factor, binding and activating the MYB promoter through a positive feedback loop. Thirdly, hemizygous loss of QKI expression caused by the rearrangement of its locus contributes to oncogenesis since it functions as a tumour-suppressor gene74–76. Gene-set enrichment analysis (GSEA) revealed that the expression of MYB-QKI fusion was associated with MYB signature genes73. MYB protein structure and its modifications found in tumours are fundamental for its transforming ability. In fact, as already mentioned above, full-length MYB is not endowed with a strong oncogenic activity in vitro, whereas C-terminal truncations are required for its activation77. MYB-QKI breakpoints in MYB intron 9–15 result in C-terminal truncation and oncogenic activation of MYB73.
MYB and cancers of the gastrointestinal tract
80% of colorectal cancers are characterised by MYB overexpression, which is associated with tumour aggressiveness and poor prognosis78,79. MYB overexpression in colon cancer is a consequence of mutations in intron 1 regulatory sequence80. Given the broad presence of the oncoprotein in this cancer, investigators in the Australian Peter MacCallum Cancer Centre engineered a vaccine against the MYB antigen called TetMYB. It is composed of an inactivated MYB protein flanked by the tetanus toxin T cell epitopes cloned into the pVAX1 plasmid vector. The immunotherapeutic role of the pVAX1-Tet-human MYB DNA vaccine was investigated in colon and adenoid cystic carcinoma (ACC) patients, also in combination with the anti-PD-1 antibody BGB-A317 to assess safety and maximum tolerated dose (MTD) in a first-in-human clinical trial81. This approach should overcome limitations caused by epitope/MHC restriction when targeting an endogenous antigen, as its application will not depend upon a need to match the patient’s MHC subtype. The trial, if successful, could pave the way for vaccine treatment not only of colorectal cancers or ACC, but also other MYB-expressing cancers. This clinical trial is based upon preclinical studies of the same Australian group in mice transplanted with MC38 colon adenocarcinoma cells expressing high levels of MYB. Breaking peripheral tolerance with the vaccine strategy enhanced anti-tumour immunity mediated by both CD4+ and CD8+ T cells, without insurgence of autoimmunity, causing a significant suppression of MC38 cancer growth78. MYB alterations have been also observed in pancreatic cancer, where it has been shown to interact with genes required for proliferation, survival and metastasis82.
MYB and breast cancer
MYB has been found bound to more than 10,000 promoters in MCF-7 breast cancer cells and recognised as a key activator of downstream targets, including genes involved in cancer progression and metastasis, such as cyclooxygenase-2 (COX-2), BCL2, BCLXL, JUN, KLF4, NANOG, MYC, and CXCR419. Breast cancer is a heterogeneous disease with a clinical outcome strictly determined by molecular profiles83,84. Over 70% of human breast cancers are oestrogen receptor-positive (ER+) and express MYB85. Gonda and colleagues reported for the first time that inhibition of MYB expression severely impairs the proliferation of ER+, but not ER−, breast cancer cell lines37. The relationship between MYB and ER is also indicated by the expression of MYB in normal, ER+ murine mammary epithelial cells, suggesting a salient role of the MYB transcription factor in mammary cell proliferation and tumour development in the human and mouse systems37,86.
ER+ breast cancer benefits from endocrine therapy (ET), which can reduce local and distant cancer recurrence and mortality rate87,88. ET can be administrated as neoadjuvant, adjuvant or palliative treatment and includes aromatase inhibitors, selective ER modulators (SERMs) such as tamoxifen, and antagonists such as fulvestrant89.
In ER+ve breast cancer patients, MYB expression is oestrogen-dependent, since it was observed that MYB mRNA levels were 5-fold higher 24 h after stimulating breast cancer cells with beta-estradiol, suggesting a strong correlation between the proto-oncogene expression and ER status in cancer19. MYB expression in ER+ve breast cancer cells is regulated at the level of transcriptional elongation, leading to the hypothesis that CDK9 inhibitors could be used to indirectly target MYB in this cancer. Indeed, CDK9 inhibition resulted in apoptotic death of breast cancer cell lines, accompanied by dose-dependent inhibition of the MCL-1 gene and protein expression90. CDK9 inhibitors also impaired cell proliferation and cell cycle progression, inducing arrest at both the G1/S and G2/M phases of the cell cycle. Moreover, this led to the downregulation of MYB target genes involved in cell cycle progression such as CCNB1 and CCNE1, which was reversed by ectopic expression of MYB90.
Breast cancer patients often develop resistance to treatment. Activation of epithelial–mesenchymal transition (EMT) is a mechanism by which breast cancer cells acquire resistance to targeted therapies91. Micro-RNAs have been implicated in the EMT process, particularly the miR-200 family92,93. Following ectopic over expression of miR-200b/c in drug-resistant cells, MYB expression levels decreased, indicating that it is a target of miR-200s. After silencing MYB in an ER+ve breast cancer cell line refractory to tamoxifen therapy, the authors of the study observed that the EMT markers vimentin, ZEB1, and ZEB2 were downregulated, further supporting the hypothesis that MYB is involved in EMT and drug resistance in breast cancer. Indeed, as expected, breast cell line sensitivity to tamoxifen therapy was increased after inhibiting MYB expression94.
MYB and ACC
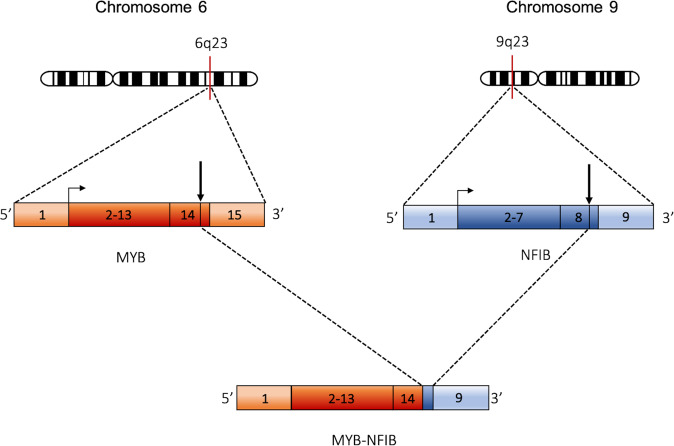
Stenman and colleagues discovered the translocation t(6;9)(q23;p23) as a genomic hallmark of ACC95. The translocation results in the fusion of the carboxyl-terminus of the MYB oncoprotein to five amino acids (SWYLG) encoded by the last exon of NFIB (Fig. 2)95,96. ACC is characterised by the presence of the MYB-NFIB fusion gene in 30–86% of cases, depending on the study97,98. An important consequence of chromosomal rearrangements in ACC is the translocation of strong enhancers near the MYB, or MYBL1, locus, which activates their transcription99. Rearrangements of the MYB locus have been observed in ACCs of the breast, lungs or glands in different body locations and in cylindromas, suggesting that MYB activation is frequent in exocrine gland tumours98,100–102. Another consequence of the chromosomal translocations detected in ACC and other gland tumours is, in some cases, loss of genetic material. In this regard, Mitani and colleagues theorised that two genetic events drive ACC pathogenesis: one involves the generation of fusion genes resulting from reciprocal translocation between chromosome 6q and 9p or other partners, and the other event constitutes a loss of genetic material, denoting the presence of one or more tumour suppressor genes103. Most ACCs do not acquire a large number of genetic changes, typical of other carcinomas104. Over half of ACC cases present chromosome 6 deletions, suggesting an important selection for these alterations in the molecular aetiology of these neoplasms. However, efforts to identify a tumour suppressor gene at these loci in ACC have been unsuccessful to date103,105.
Fig. 2. Schematic illustration of the MYB-NFIB fusion gene.
The t(6;9) translocation results in a MYB-NFIB gene fusion. Arrows indicate the breaking points.
MYB-NFIB is a putative oncoprotein, which has been shown to control ACC tumour cell proliferation and spherogenesis106. Intriguingly, the fusion gene is regulated by AKT-dependent signalling downstream of the IGF1 receptor and its expression can be downregulated by IGF1R-inhibition with linsitinib. Furthermore, EGFR and MET signalling also promote growth of ACC cells106. In line with these findings, evidence in patients or xenograft models indicate that monoclonal antibodies targeting IGF1 or EGF receptors could be effective drugs in ACCs expressing the fusion oncoprotein107–109. To investigate the implication of the MYB-NFIB fusion gene in ACC, Mitani and co-workers analysed a cohort of 123 salivary carcinomas, including primary ACCs of the salivary gland, metastatic ACCs, non-ACC salivary carcinomas, and normal salivary gland tissues103. Using RT-PCR, validated by fluorescence in situ hybridisation (FISH) analysis, they found that among 89 ACC cases (72 primary ACCs and 17 metastatic), 26 were positive for expression of the MYB-NFIB fusion transcript. Interestingly, none of the 34 non-ACC carcinomas were positive. In addition, 14 different fusion transcripts involving multiple exons of MYB and NFIB were identified. To provide further insights on the role of MYB in this cancer, expression of the wild type or fusion MYB transcripts was quantified. Unsurprisingly, MYB expression was elevated in MYB-NFIB fusion positive ACCs, probably caused by loss of the negative regulatory sequence at the 3′ untranslated region of MYB. Interestingly, the expression of wild type MYB was elevated >40-fold in fusion-negative ACCs compared to non-ACC carcinomas, and only 2-fold lower than fusion-positive ACCs. The authors concluded that whereas genomic rearrangement must be causative of MYB overexpression in fusion positive ACCs, alternative mechanisms may be responsible for MYB overexpression in fusion negative ACCs103. Thus, MYB overexpression is a frequent consequence of the MYB-NFIB fusion in glandular tumours, but can also occur via other mechanisms.
The polyether ionophore monensin was recently identified as a MYB inhibitor using a luciferase-based screen and tested on ACC cell lines derived from ACC patients. These cells were more sensitive to the anti-cancer agent than MYB-expression negative, control cell lines. Monensin suppressed both MYB-NFIB mRNA and protein levels. Moreover, the compound, and related polyether ionophores, also induced differentiation and promoted apoptosis of leukaemic cell lines, suggesting that MYB inhibitors can be effective against solid and liquid malignancies110. Using a chemical screen in Zebrafish, the group of Leonard Zon have demonstrated that retinoic acid is a suppressor of MYB in ACC. All trans retinoic acid (ATRA) treatment of mice bearing patient-derived ACC tumours showed reduced expression of MYB and binding of MYB at translocated enhancers. Importantly, ATRA inhibited the expression of cell cycle related, MYB-target genes. ATRA is used in the clinic for the treatment of promyelocytic leukaemia and has a known safety profile, suggesting that it will be soon used in the context of a clinical trial in ACC patients111.
Identification of actionable target genes downstream of MYB can be a reasonable alternative to avoid negative consequences caused by inactivation of the wild-type MYB transcription factor. Indeed, the potential haematologic toxicity of anti-MYB therapies could be further exacerbated in patients under regimens of chemotherapy and radiotherapy. An important gene axis regulated by MYB is the insulin growth factor and its receptor. Interestingly, insulin growth factor receptor (IGFR) signalling positively regulates MYB-NFIB in ACC, suggesting that MYB and IGFR are engaged in a feed forward loop in cancer112,113. Accordingly, it has been shown that the small molecule inhibitor Linsitinib or the therapeutic antibody Figitumumab reduce the growth of ACC tumours in mouse models and in patients, suggesting that targeting IGFR signalling could be an effective strategy in MYB overexpressing cancers109,112. In an effort to identify new MYB target genes in ACC, our group has generated retroviral vectors expressing wild-type MYB or two MYB-NFIB variants derived from ACC patients. The different MYB isoforms were ectopically expressed in immortalised breast MCF10A cells, and genes up or downregulated were identified by microarrays. GSEA revealed that ATR/BRCA was the top activated downstream pathway, with a significant upregulation of ATR gene expression43. ATR mRNA levels were increased in primary ACCs compared to normal salivary glands. Accordingly, the clinical ATR kinase inhibitor VX-970 caused apoptosis of primary ACC cells in vitro and significant shrinkage of ACC patient-derived xenografts. These results support the theory that acting on downstream target genes/proteins might be a worthy—and even safer—alternative to directly targeting the MYB gene itself43.
Surgery is the first line treatment for ACC, followed by cytotoxic chemotherapy and/or radiotherapy as adjuvant treatments to avoid recurrence. Unfortunately, standard treatments only provide limited benefit in advanced disease, which is usually lethal, with a high rate of recurrence and metastasis. Therefore, new and more effective treatments are urgently needed for these high-risk patients. Previous clinical trials have led to the approval of tyrosine kinase inhibitors (TKI) for the treatment of aggressive forms of solid malignancies, such as thyroid cancer refractory to radio therapy and unresectable hepatocellular carcinoma114,115. Most of the targeted tyrosine kinases are also MYB regulated, such as vascular endothelial growth factor receptors (VEGFRs), fibroblast growth factor receptors (FGFRs), the stem cell factor receptor KIT (c-KIT), FMS-like tyrosine kinase 3 (FLT3), platelet-derived growth factor receptors (PDGFRs), and the proto-oncogene RET95,115–119. Persson and co-workers have recently shown that VEGFA, FGF2, KIT and other genes encoding receptor tyrosine kinases are commonly overexpressed in ACC samples, leading to consider TKIs as credible candidates for the treatment of relapsed/metastatic ACC patients95. However, it has been observed an overall poor response in therapies against these targets in ACC, suggesting that other, more relevant MYB downstream genes should be clinically exploited in this tumour.
MYBL1
MYBL1 is predominantly expressed in the central nervous system (CNS), germinal B-lymphocytes, mammary gland ductal epithelium, and in the testis120,121. It has a key role in spermatogenesis, particularly in cell cycle progression of germ cells through pachynema121,122. MYBL1-null mice are viable, but exhibit growth abnormalities as well as defects in spermatogenesis and female breast development120.
MYBL1 alterations in cancer
MYBL1 rearrangements are a hallmark of low-grade gliomas (LGGs), the commonest paediatric CNS neoplasm, arising in children and adolescents114,123. Recent molecular characterisations through WGS have led to the identification of new genetic alterations in LGGs. These studies have identified activation of the MAPK/ERK pathway caused by the duplication of the tyrosine kinase domain (TKD) of the FGFR1 gene and frequent rearrangements of the MYB family members MYB and MYBL1 in diffuse cerebral LGGs124. 8q13.1 gain was observed as a significant recurrent event in diffuse astrocytoma grade IIs. This leads to a duplication of MYBL1 and truncation of its C-terminal NRD, resulting in anchorage-independent growth of NIH-3T3 cells and tumour formation in nude mice125. MYBL1 gene amplification is a distinct alteration of the subtype IDH-wt/H3-wt of diffuse gliomas, together with TERT and BRAF mutations, EGFR and FGFR1 alterations, and other chromosomal aberrations126. Although these alterations are rare, sequencing analysis of uncommon low-grade neuro-epithelial tumours revealed that these pathogenic mutations occur at a high frequency (78%) in this cohort114.
Patients with isomorphic diffuse glioma or astrocytoma can harbour copy number alterations of MYBL1 or MYB (13 out of 25 samples, 52%), as assessed with RNA sequencing. Gene fusions accounted for 50% of cases127.
ACC is characterised by the chromosomal translocation t(6;9), leading to the expression of the MYB-NFIB fusion gene95. Although MYB is the MYB family member most often involved in this cancer, it was recently demonstrated that a subset of ACCs contains the t(8;9) chromosomal translocation128. This results in the creation of a MYBL1-NFIB gene fusion, which probably functions in a manner similar to MYB-NFIB, given the structural analogies between MYBL1 and MYB. Indeed, tumours with MYB and MYBL1 translocations display overlapping gene expression profiles and clinical outcome, suggesting that the related MYB proteins are interchangeable oncogenic drivers in ACC. The research group that identified the translocation t(8;9), also highlighted a t(8;14) translocation, leading to the fusion of MYBL1 to the RAD51B gene128.
In MYB or MYB-NFIB negative subsets of breast ACC tumours, alternative genetic mechanisms of MYB activation have been demonstrated. RNA and WGS unveiled that these cancers could harbour MYBL1 rearrangements, including those between MYBL1-ACTN1 and MYBL1-NFIB102. In these rare triple negative breast cancers (TNBC), the histological pattern was identical to the MYB-NFIB-positive, salivary gland ACCs. The MYBL1 rearrangements were confirmed at genomic level by the FISH technique. The translocation results in an in-frame chimeric transcript containing the DNA-binding and transactivating domains, encoded by exons 1–14, of MYBL1 fused to the exon 9 of NFIB. In addition, another in-frame fusion between MYBL1-ACTN1 was also detected for the first time in ACC samples. The fusion leads to loss of the C-terminus region of MYBL1 due to the fusion of exons 1–8 of MYBL1 with exons 10–21 of ACTN1102.
Another organ in which ACC neoplasms can originate is the lung. Primary tracheobronchial ACC is one of the rarest types of lung cancer, accounting for <1% of cases. Pei et al. analysed 7 lung ACCs, documenting that 7 out of 7 cases presented MYB or MYBL1 genes fused with NFIB or, less frequently, with RAD51B101. Primary cutaneous ACCs display a genetic landscape similar to those of salivary glands, showing fusions of either MYB or MYBL1 with the common partner NFIB129.
MYBL2
MYBL2, encoding the transcription factor MYBL2, is ubiquitous and often co-expressed with other MYB members. It has been shown to regulate cell cycle progression, cell survival and differentiation being an essential component of the DREAM complex130–133. It is also a promoter of cell survival by activating antiapoptotic genes such as BIRC5 (survivin), CLU (ApoJ/clusterin) and BCL2134–137. MYBL2 has been shown to aid repair of DNA double-strand breaks, supporting genome stability in haematopoietic and pluripotent stem cells138,139. Expression of MYBL2 is important for both normal and transformed cell homoeostasis. This concept is supported by the early embryonic lethal phenotype of MYBL2 knockout mice, due to impaired inner cell mass formation, or suppression of cell cycle progression and cell survival in oesophageal, hepatic, colorectal, and sympathetic nervous system cancer cells in which the expression of MYBL2 has been downregulated140–145. The activity of MYBL2 is highly regulated at transcriptional and post-transcriptional levels. Cyclins and their catalytic partners, the cyclin-dependent kinases (Cdks), function as key regulators of the cell cycle146. Cyclin D1 with Cdk4 or Cdk6 has been shown to play an important role at the ‘restriction point’ in the G1 phase of the cell cycle before cells enter into the mitotic cycle, whereas, for the transition from G1 to S phase, cyclin E–Cdk2 complexes are the most critical, and cyclin A–Cdk2 complexes are required during S phase146. MYBL2 is regulated by the transcription factor E2F and required for the expression of cyclin B and cdc2 in G2/M147–149. When overexpressed, the tumour suppressor protein p53 induces Waf1/Cip1/p21 protein-dependent cell-cycle arrest and activation of MYBL2 allows cells to escape this block, suggesting that MYBL2 acts at a later stage than Waf1/Cip1/p21 during cell-cycle progression150,151. MYBL2 is a substrate for cyclin A/E–Cdk2 kinase activity and its transcriptional activity is regulated by phosphorylation148,150.
Genes involved in the G2/M phase of the cell cycle are activated by MYBL2 switching from the repressive DREAM to the MuvB (MMB) complex136,152,153. MYBL2 is transcriptionally repressed in G1, activated by cyclin A/Cdk2-mediated phosphorylation during S-phase, and subsequently degraded in late G2 in a ubiquitin-dependent manner147,148,154,155. Phosphorylation of MYBL2 occurs at Serine or Threonine residues followed by Proline156. Pin1 isomerase recognises the pSer/pThr-Pro residues altering functions of the MYBL2 protein by inducing conformation changes. Cdk-dependent phosphorylation and Pin1 isomerization induce Plk1 kinase binding to MYBL2. Plk1 phosphorylates the region of MYBL2 containing the transcriptional activation domain (TAD), suggesting that PLK1-induced modification of MYBL2 is crucially required for transcriptional activation of pro-mitotic genes157. Consistent with an important role in cell cycle progression, down-regulation of MYBL2 leads to spindle and centrosome defects, arrest in the G2/M phase of the cell cycle, failure in cytokinesis, polyploidy and apoptosis132,158.
MYBL2 alterations in cancer
The DREAM complex [DP, RB-like, E2F4 and MuvB (synMuv genes, class B)] is a master coordinator of cell cycle-dependent gene expression and the balance between repressive DREAM and activating MYB-MuvB (MMB) complexes is frequently perturbed in cancer159–162. Increased expression of several components of the MMB complex, including MYBL2 and FOXM1, correlates with aggressive tumour features and poor prognosis144,163. To investigate the clinical relevance of the MYBL2/FOXM1/CDK/PLK1 axis, Werwein et al. used a pan-cancer resource of expression signatures that correlate cancer gene expression and clinical prognosis data, called PRECOG157. Interestingly, among the 50 genes (and their products) analysed, 44 (including MYBL2, FOXM1, CCNA2, and PLK1) were found to be targets of DREAM-mediated repression, while 29 of them were also targets of MMB activation157. Overexpression of MYBL2 disturbs myeloid differentiation and promotes the progression of solid cancers where it is also an indicator of poor prognosis133,142,164–166. MYBL2 is frequently overexpressed in malignancies including breast cancer, non-small-cell lung cancer (NSCLC), AML, colorectal cancer, pancreatic ductal adenocarcinoma, and neuroblastoma167–175.
The molecular mechanisms causing increased expression of MYBL2 in multiple human cancers are still not fully elucidated.
MYBL2 and breast cancer
Alterations of gene expression might be caused by amplification of the MYBL2 locus located in chromosome 20q13176–178. 20q13 amplification or copy gains are common in breast cancer and are usually associated with poor prognosis176. Notably, a MYBL2 germline polymorphism causing the Serine-to-Glycine amino acid change S427G is correlated to a high risk of basal-like breast cancer133. Moreover, MYBL2 overexpression was noted in HER2+/ER− and luminal B breast cancer samples, but not in luminal A or normal breast tissue, strongly suggesting a correlation between MYBL2 expression and aggressiveness of breast cancer177. A recent review explains the molecular mechanisms of MYBL2 amplification and new therapeutic opportunities in breast cancers179.
MYBL2, clear cell renal cell carcinoma (ccRCC) and NSCLC
ccRCC is the most frequent renal malignancy180. MYBL2 expression could be used as a biomarker to predict patients’ prognosis in this cancer. MYBL2 was found upregulated in a cohort of 530 ccRCC patients compared to healthy tissues. Of note, upregulated MYBL2 was significantly associated with age and sex of cancer patients, advanced T stage, lymph node and distant metastases, clinical stage and histological grade181. Moreover, a significant correlation between high MYBL2 expression and worse prognosis was established by Kaplan–Meier analysis, indicating that MYBL2 expression is an independent biomarker of progression in ccRCC181.
Another neoplasia characterised by deregulation of MYBL2 is non-small-cell lung cancer (NSCLC). Analysis of MCODE clusters highlighted genes involved in “driver networks” for NSCLC, which include the transcription factors FOXM1, TFDP1, E2F4, SIN3, and MYBL2182. A further study confirmed the potential oncogenic role of MYBL2 in NSCLC. Through chromatin immunoprecipitation (ChIP) assay, researchers identified a direct binding between MYBL2 and the gene Non-SMC CondensinIComplex Subunit H (NCAPH), well-known to have oncogenic properties in lung cancer. A significant correlation between high NCAPH expression and poor prognosis was confirmed, suggesting that targeting the MYBL2-regulated gene could be of potential therapeutic value in this setting169.
MYBL2 and leukaemia
MYBL2 overexpression is a prognostic factor in AML, defining a subset of patients with poor prognosis170,183. This could be linked to the reduced expression of miR-30a, miR-30b and miR-30c, involved in the regulation of haematopoiesis and cell differentiation, which were shown to be expressed at lower levels in MYBL2high AML samples170,184,185. The strong correlation between overexpression of MYBL2 and downregulation of the miR-30 cluster suggests that the micro-RNAs antagonise the expression of MYBL2, or that the latter suppresses miRNAs expression in AML170.
A recent study published in Cell revealed a link between MYBL2 and the protein phosphatase 2A (PP2A) in leukaemia. Morita and co-workers identified a class of small molecule that they called iHAPs—improved heterocyclic activators of PP2A—able to activate a PP2A complex, which suppresses tumour progression. PP2A is an enzyme formed by different subunits; among them, PPP2R1A, PPP2CA, and PPP2R5E are strictly required for antitumor activity186. Using isotope-labelled amino acids (SILAC) and mass spectrometry analysis, substrates dephosphorylated by PP2A in the presence or absence of iHAPs were identified, among which MYBL2. The researchers were able to activate the PP2A complex, usually present in an inactive form in cancers due to the overexpression of inhibitory proteins, and observe dephosphorylation of MYBL2 on Ser241, required for transactivation of cell cycle-related genes, resulting in an irreversible growth arrest of multiple cancer cells. Thus, MYBL2 is centrally involved in cancer cell proliferation and can be indirectly targeted by small molecule-mediated reactivation of the PP2A tumour suppressor protein186.
Conclusions
The MYB transcription factors are a point of convergence of numerous signalling pathways essential for multiple cellular functions, and their deregulation has been associated with aggressive behaviour of cancer cells. Reflecting the high similarity of protein structures, MYBL1, MYBL2, and MYB are all involved in the control of cell survival, proliferation and differentiation. One could hypothesise that spatio-temporal differences in gene expression during organogenesis and in pathological conditions may determine specific MYB requirements in cells. The ever-expanding number of studies reporting deregulation of MYB family members in the pathogenesis of human cancers is instigating researchers to find new and more efficient methods to target these transcription factors. Direct pharmacological inhibition of MYB or its product MYB, is emerging as a potential therapeutic strategy for both liquid and solid malignancies. Nevertheless, inhibiting MYB could potentially lead to haematopoietic toxicity, indicating that targeting downstream target genes and coactivator molecules might make more clinical sense. Further studies will be required to develop effective therapeutic interventions aiming at suppressing MYB signalling in tumours while minimising risks to patients.
Acknowledgements
Y.C. and A.S. are supported by a grant from the Oracle Cancer Trust.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lipsick JS, Wang DM. Transformation by v-Myb. Oncogene. 1999;18:3047–3055. doi: 10.1038/sj.onc.1202745. [DOI] [PubMed] [Google Scholar]
- 2.Roussel M, et al. Three new types of viral oncogene of cellular origin specific for haematopoietic cell transformation. Nature. 1979;281:452–455. doi: 10.1038/281452a0. [DOI] [PubMed] [Google Scholar]
- 3.Hall WJ, Bean CW, Pollard M. Transmission of fowl leukosis through chick embryos and young chicks. Am. J. Vet. Res. 1941;2:272–279. [Google Scholar]
- 4.Boyle WJ, Lipsick JS, Reddy EP, Baluda MA. Identification of the leukemogenic protein of avian myeloblastosis virus and of its normal cellular homologue. Proc. Natl Acad. Sci. USA. 1983;80:2834–2838. doi: 10.1073/pnas.80.10.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Westin EH, et al. Differential expression of the amv gene in human hematopoietic cells. Proc. Natl Acad. Sci. USA. 1982;79:2194–2198. doi: 10.1073/pnas.79.7.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nunn MF, Seeburg PH, Moscovici C, Duesberg PH. Tripartite structure of the avian erythroblastosis virus E26 transforming gene. Nature. 1983;306:391–395. doi: 10.1038/306391a0. [DOI] [PubMed] [Google Scholar]
- 7.Davidson CJ, Tirouvanziam R, Herzenberg LA, Lipsick JS. Functional evolution of the vertebrate Myb gene family: B-Myb, but neither A-Myb nor c-Myb, complements Drosophila Myb in hemocytes. Genetics. 2005;169:215–229. doi: 10.1534/genetics.104.034132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lipsick JS. The C-MYB story-is it definitive? Proc. Natl Acad. Sci. USA. 2010;107:17067–17068. doi: 10.1073/pnas.1012402107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lipsick JS, et al. Functional evolution of the Myb oncogene family. Blood Cells Mol. Dis. 2001;27:456–458. doi: 10.1006/bcmd.2001.0404. [DOI] [PubMed] [Google Scholar]
- 10.Ohi R, et al. Myb-related Schizosaccharomyces pombe cdc5p is structurally and functionally conserved in eukaryotes. Mol. Cell. Biol. 1998;18:4097–4108. doi: 10.1128/MCB.18.7.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davidson CJ, Guthrie EE, Lipsick JS. Duplication and maintenance of the Myb genes of vertebrate animals. Biol. Open. 2013;2:101–110. doi: 10.1242/bio.20123152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nomura N, et al. Isolation of human cDNA clones of myb-related genes, A-myb and B-myb. Nucleic Acids Res. 1988;16:11075–11089. doi: 10.1093/nar/16.23.11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gewirtz AM, Calabretta B. A c-myb antisense oligodeoxynucleotide inhibits normal human hematopoiesis in vitro. Science. 1988;242:1303–1306. doi: 10.1126/science.2461588. [DOI] [PubMed] [Google Scholar]
- 14.O’Rourke JP, Ness SA. Alternative RNA splicing produces multiple forms of c-Myb with unique transcriptional activities. Mol. Cell. Biol. 2008;28:2091–2101. doi: 10.1128/MCB.01870-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nat. Rev. Cancer. 2008;8:523–534. doi: 10.1038/nrc2439. [DOI] [PubMed] [Google Scholar]
- 16.Ogata K, et al. Solution structure of a DNA-binding unit of Myb: a helix-turn-helix-related motif with conserved tryptophans forming a hydrophobic core. Proc. Natl Acad. Sci. USA. 1992;89:6428–6432. doi: 10.1073/pnas.89.14.6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biedenkapp H, Borgmeyer U, Sippel AE, Klempnauer KH. Viral myb oncogene encodes a sequence-specific DNA-binding activity. Nature. 1988;335:835–837. doi: 10.1038/335835a0. [DOI] [PubMed] [Google Scholar]
- 18.Ness SA, Marknell A, Graf T. The v-myb oncogene product binds to and activates the promyelocyte-specific mim-1 gene. Cell. 1989;59:1115–1125. doi: 10.1016/0092-8674(89)90767-8. [DOI] [PubMed] [Google Scholar]
- 19.Quintana AM, Liu F, O’Rourke JP, Ness SA. Identification and regulation of c-Myb target genes in MCF-7 cells. BMC Cancer. 2011;11:30. doi: 10.1186/1471-2407-11-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao L, et al. Integrated genome-wide chromatin occupancy and expression analyses identify key myeloid pro-differentiation transcription factors repressed by Myb. Nucleic Acids Res. 2011;39:4664–4679. doi: 10.1093/nar/gkr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dai P, et al. CBP as a transcriptional coactivator of c-Myb. Genes Dev. 1996;10:528–540. doi: 10.1101/gad.10.5.528. [DOI] [PubMed] [Google Scholar]
- 22.Oelgeschlager M, Janknecht R, Krieg J, Schreek S, Luscher B. Interaction of the co-activator CBP with Myb proteins: effects on Myb-specific transactivation and on the cooperativity with NF-M. EMBO J. 1996;15:2771–2780. doi: 10.1002/j.1460-2075.1996.tb00637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oelgeschlager M, Kowenz-Leutz E, Schreek S, Leutz A, Luscher B. Tumorigenic N-terminal deletions of c-Myb modulate DNA binding, transactivation, and cooperativity with C/EBP. Oncogene. 2001;20:7420–7424. doi: 10.1038/sj.onc.1204922. [DOI] [PubMed] [Google Scholar]
- 24.Mink S, Kerber U, Klempnauer KH. Interaction of C/EBPbeta and v-Myb is required for synergistic activation of the mim-1 gene. Mol. Cell. Biol. 1996;16:1316–1325. doi: 10.1128/MCB.16.4.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shapiro LH. Myb and Ets proteins cooperate to transactivate an early myeloid gene. J. Biol. Chem. 1995;270:8763–8771. doi: 10.1074/jbc.270.15.8763. [DOI] [PubMed] [Google Scholar]
- 26.Bartunek P, Kralova J, Blendinger G, Dvorak M, Zenke M. GATA-1 and c-myb crosstalk during red blood cell differentiation through GATA-1 binding sites in the c-myb promoter. Oncogene. 2003;22:1927–1935. doi: 10.1038/sj.onc.1206281. [DOI] [PubMed] [Google Scholar]
- 27.Mansour MR, et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346:1373–1377. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mucenski ML, et al. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65:677–689. doi: 10.1016/0092-8674(91)90099-K. [DOI] [PubMed] [Google Scholar]
- 29.Stadhouders R, et al. HBS1L-MYB intergenic variants modulate fetal hemoglobin via long-range MYB enhancers. J. Clin. Investig. 2014;124:1699–1710. doi: 10.1172/JCI71520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang J, et al. cMYB is involved in the regulation of fetal hemoglobin production in adults. Blood. 2006;108:1077–1083. doi: 10.1182/blood-2006-01-008912. [DOI] [PubMed] [Google Scholar]
- 31.Gautam S, et al. The transcription factor c-Myb regulates CD8(+) T cell stemness and antitumor immunity. Nat. Immunol. 2019;20:337–349. doi: 10.1038/s41590-018-0311-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas MD, Kremer CS, Ravichandran KS, Rajewsky K, Bender TP. c-Myb is critical for B cell development and maintenance of follicular B cells. Immunity. 2005;23:275–286. doi: 10.1016/j.immuni.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 33.Kastan MB, Slamon DJ, Civin CI. Expression of protooncogene c-myb in normal human hematopoietic cells. Blood. 1989;73:1444–1451. doi: 10.1182/blood.V73.6.1444.1444. [DOI] [PubMed] [Google Scholar]
- 34.Ess KC, Witte DP, Bascomb CP, Aronow BJ. Diverse developing mouse lineages exhibit high-level c-Myb expression in immature cells and loss of expression upon differentiation. Oncogene. 1999;18:1103–1111. doi: 10.1038/sj.onc.1202387. [DOI] [PubMed] [Google Scholar]
- 35.Malaterre J, et al. c-Myb is required for neural progenitor cell proliferation and maintenance of the neural stem cell niche in adult brain. Stem Cells. 2008;26:173–181. doi: 10.1634/stemcells.2007-0293. [DOI] [PubMed] [Google Scholar]
- 36.Malaterre J, et al. c-Myb is required for progenitor cell homeostasis in colonic crypts. Proc. Natl Acad. Sci. USA. 2007;104:3829–3834. doi: 10.1073/pnas.0610055104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drabsch Y, et al. Mechanism of and requirement for estrogen-regulated MYB expression in estrogen-receptor-positive breast cancer cells. Proc. Natl Acad. Sci. USA. 2007;104:13762–13767. doi: 10.1073/pnas.0700104104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakata Y, et al. c-Myb contributes to G2/M cell cycle transition in human hematopoietic cells by direct regulation of cyclin B1 expression. Mol. Cell. Biol. 2007;27:2048–2058. doi: 10.1128/MCB.01100-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pilkinton M, Sandoval R, Song J, Ness SA, Colamonici OR. Mip/LIN-9 regulates the expression of B-Myb and the induction of cyclin A, cyclin B, and CDK1. J. Biol. Chem. 2007;282:168–175. doi: 10.1074/jbc.M609924200. [DOI] [PubMed] [Google Scholar]
- 40.Lei W, Liu F, Ness SA. Positive and negative regulation of c-Myb by cyclin D1, cyclin-dependent kinases, and p27 Kip1. Blood. 2005;105:3855–3861. doi: 10.1182/blood-2004-08-3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicolaides NC, Gualdi R, Casadevall C, Manzella L, Calabretta B. Positive autoregulation of c-myb expression via Myb binding sites in the 5’ flanking region of the human c-myb gene. Mol. Cell. Biol. 1991;11:6166–6176. doi: 10.1128/MCB.11.12.6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ku DH, et al. c-myb transactivates cdc2 expression via Myb binding sites in the 5’-flanking region of the human cdc2 gene. J. Biol. Chem. 1993;268:2255–2259. doi: 10.1016/S0021-9258(18)53990-9. [DOI] [PubMed] [Google Scholar]
- 43.Andersson MK, et al. ATR is a MYB regulated gene and potential therapeutic target in adenoid cystic carcinoma. Oncogenesis. 2020;9:5. doi: 10.1038/s41389-020-0194-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clappier E, et al. The C-MYB locus is involved in chromosomal translocation and genomic duplications in human T-cell acute leukemia (T-ALL), the translocation defining a new T-ALL subtype in very young children. Blood. 2007;110:1251–1261. doi: 10.1182/blood-2006-12-064683. [DOI] [PubMed] [Google Scholar]
- 45.Okada M, et al. c-myb gene analysis in T-cell malignancies with del(6q) Cancer Genet. Cytogenet. 1990;48:229–236. doi: 10.1016/0165-4608(90)90125-T. [DOI] [PubMed] [Google Scholar]
- 46.Torelli G, et al. Expression of c-myb protooncogene and other cell cycle-related genes in normal and neoplastic human colonic mucosa. Cancer Res. 1987;47:5266–5269. [PubMed] [Google Scholar]
- 47.Thompson MA, Flegg R, Westin EH, Ramsay RG. Microsatellite deletions in the c-myb transcriptional attenuator region associated with over-expression in colon tumour cell lines. Oncogene. 1997;14:1715–1723. doi: 10.1038/sj.onc.1201007. [DOI] [PubMed] [Google Scholar]
- 48.Li Y, et al. c-Myb enhances breast cancer invasion and metastasis through the Wnt/beta-Catenin/Axin2 pathway. Cancer Res. 2016;76:3364–3375. doi: 10.1158/0008-5472.CAN-15-2302. [DOI] [PubMed] [Google Scholar]
- 49.Lahortiga I, et al. Duplication of the MYB oncogene in T cell acute lymphoblastic leukemia. Nat. Genet. 2007;39:593–595. doi: 10.1038/ng2025. [DOI] [PubMed] [Google Scholar]
- 50.Quelen C, et al. Identification of a transforming MYB-GATA1 fusion gene in acute basophilic leukemia: a new entity in male infants. Blood. 2011;117:5719–5722. doi: 10.1182/blood-2011-01-333013. [DOI] [PubMed] [Google Scholar]
- 51.Belloni E, et al. In vivo expression of an aberrant MYB-GATA1 fusion induces leukemia in the presence of GATA1 reduced levels. Leukemia. 2011;25:733–736. doi: 10.1038/leu.2010.317. [DOI] [PubMed] [Google Scholar]
- 52.Ducassou S, et al. MYB-GATA1 fusion promotes basophilic leukaemia: involvement of interleukin-33 and nerve growth factor receptors. J. Pathol. 2017;242:347–357. doi: 10.1002/path.4908. [DOI] [PubMed] [Google Scholar]
- 53.Mager LF, et al. IL-33 signaling contributes to the pathogenesis of myeloproliferative neoplasms. J. Clin. Investig. 2015;125:2579–2591. doi: 10.1172/JCI77347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bailly RA, et al. DNA-binding and transcriptional activation properties of the EWS-FLI-1 fusion protein resulting from the t(11;22) translocation in Ewing sarcoma. Mol. Cell. Biol. 1994;14:3230–3241. doi: 10.1128/MCB.14.5.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eliazer S, Spencer J, Ye D, Olson E, Ilaria RL., Jr. Alteration of mesodermal cell differentiation by EWS/FLI-1, the oncogene implicated in Ewing’s sarcoma. Mol. Cell. Biol. 2003;23:482–492. doi: 10.1128/MCB.23.2.482-492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schoch C, et al. Twenty-three cases of acute lymphoblastic leukemia with translocation t(4;11)(q21;q23): the implication of additional chromosomal aberrations. Ann. Hematol. 1995;70:195–201. doi: 10.1007/BF01700375. [DOI] [PubMed] [Google Scholar]
- 57.Schwartz JC, Cech TR, Parker RR. Biochemical properties and biological functions of FET proteins. Annu. Rev. Biochem. 2015;84:355–379. doi: 10.1146/annurev-biochem-060614-034325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pierini T, et al. MYB deregulation from a EWSR1-MYB fusion at leukemic evolution of a JAK2 (V617F) positive primary myelofibrosis. Mol. Cytogenet. 2016;9:68. doi: 10.1186/s13039-016-0277-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamamoto JF, Goodman MT. Patterns of leukemia incidence in the United States by subtype and demographic characteristics, 1997–2002. Cancer Causes Control. 2008;19:379–390. doi: 10.1007/s10552-007-9097-2. [DOI] [PubMed] [Google Scholar]
- 60.Rowe JM. Will new agents impact survival in AML? Best. Pract. Res. Clin. Haematol. 2019;32:101094. doi: 10.1016/j.beha.2019.101094. [DOI] [PubMed] [Google Scholar]
- 61.Taga T, Tomizawa D, Takahashi H, Adachi S. Acute myeloid leukemia in children: current status and future directions. Pediatr. Int. 2016;58:71–80. doi: 10.1111/ped.12865. [DOI] [PubMed] [Google Scholar]
- 62.Estey EH. Acute myeloid leukemia: 2019 update on risk-stratification and management. Am. J. Hematol. 2018;93:1267–1291. doi: 10.1002/ajh.25214. [DOI] [PubMed] [Google Scholar]
- 63.Huret JL, Dessen P, Bernheim A. An atlas of chromosomes in hematological malignancies. Example: 11q23 and MLL partners. Leukemia. 2001;15:987–989. doi: 10.1038/sj.leu.2402135. [DOI] [PubMed] [Google Scholar]
- 64.Meyer C, et al. The MLL recombinome of acute leukemias. Leukemia. 2006;20:777–784. doi: 10.1038/sj.leu.2404150. [DOI] [PubMed] [Google Scholar]
- 65.Hess JL, et al. c-Myb is an essential downstream target for homeobox-mediated transformation of hematopoietic cells. Blood. 2006;108:297–304. doi: 10.1182/blood-2005-12-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pattabiraman DR, et al. Interaction of c-Myb with p300 is required for the induction of acute myeloid leukemia (AML) by human AML oncogenes. Blood. 2014;123:2682–2690. doi: 10.1182/blood-2012-02-413187. [DOI] [PubMed] [Google Scholar]
- 67.Uttarkar S, et al. Targeting acute myeloid leukemia with a small molecule inhibitor of the Myb/p300 interaction. Blood. 2016;127:1173–1182. doi: 10.1182/blood-2015-09-668632. [DOI] [PubMed] [Google Scholar]
- 68.Uttarkar S, et al. Naphthol AS-E phosphate inhibits the activity of the transcription factor Myb by blocking the interaction with the KIX domain of the coactivator p300. Mol. Cancer Ther. 2015;14:1276–p1285. doi: 10.1158/1535-7163.MCT-14-0662. [DOI] [PubMed] [Google Scholar]
- 69.Walf-Vorderwulbecke V, et al. Targeting acute myeloid leukemia by drug-induced c-MYB degradation. Leukemia. 2018;32:882–889. doi: 10.1038/leu.2017.317. [DOI] [PubMed] [Google Scholar]
- 70.Uttarkar S, et al. Small-molecule disruption of the Myb/p300 cooperation targets acute myeloid leukemia cells. Mol. Cancer Ther. 2016;15:2905–2915. doi: 10.1158/1535-7163.MCT-16-0185. [DOI] [PubMed] [Google Scholar]
- 71.Ramaswamy K, et al. Peptidomimetic blockade of MYB in acute myeloid leukemia. Nat. Commun. 2018;9:110. doi: 10.1038/s41467-017-02618-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pfister S, et al. BRAF gene duplication constitutes a mechanism of MAPK pathway activation in low-grade astrocytomas. J. Clin. Investig. 2008;118:1739–1749. doi: 10.1172/JCI33656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bandopadhayay P, et al. MYB-QKI rearrangements in angiocentric glioma drive tumorigenicity through a tripartite mechanism. Nat. Genet. 2016;48:273–282. doi: 10.1038/ng.3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang G, et al. RNA-binding protein quaking, a critical regulator of colon epithelial differentiation and a suppressor of colon cancer. Gastroenterology. 2010;138:231–240 e231-235. doi: 10.1053/j.gastro.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mulholland PJ, et al. Genomic profiling identifies discrete deletions associated with translocations in glioblastoma multiforme. Cell Cycle. 2006;5:783–791. doi: 10.4161/cc.5.7.2631. [DOI] [PubMed] [Google Scholar]
- 76.Ichimura K, et al. Small regions of overlapping deletions on 6q26 in human astrocytic tumours identified using chromosome 6 tile path array-CGH. Oncogene. 2006;25:1261–1271. doi: 10.1038/sj.onc.1209156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gonda TJ, Buckmaster C, Ramsay RG. Activation of c-myb by carboxy-terminal truncation: relationship to transformation of murine haemopoietic cells in vitro. EMBO J. 1989;8:1777–1783. doi: 10.1002/j.1460-2075.1989.tb03571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Williams BB, et al. Induction of T cell-mediated immunity using a c-Myb DNA vaccine in a mouse model of colon cancer. Cancer Immunol. Immunother. 2008;57:1635–1645. doi: 10.1007/s00262-008-0497-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ramsay RG, et al. Myb expression is higher in malignant human colonic carcinoma and premalignant adenomatous polyps than in normal mucosa. Cell Growth Differ. 1992;3:723–730. [PubMed] [Google Scholar]
- 80.Hugo H, et al. Mutations in the MYB intron I regulatory sequence increase transcription in colon cancers. Genes Chromosomes Cancer. 2006;45:1143–1154. doi: 10.1002/gcc.20378. [DOI] [PubMed] [Google Scholar]
- 81.Pham T, et al. First-in-human phase I clinical trial of a combined immune modulatory approach using TetMYB vaccine and Anti-PD-1 antibody in patients with advanced solid cancer including colorectal or adenoid cystic carcinoma: The MYPHISMO study protocol ( NCT03287427) Contemp. Clin. Trials Commun. 2019;16:100409. doi: 10.1016/j.conctc.2019.100409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Srivastava SK, et al. MYB is a novel regulator of pancreatic tumour growth and metastasis. Br. J. Cancer. 2015;113:1694–1703. doi: 10.1038/bjc.2015.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vuong D, Simpson PT, Green B, Cummings MC, Lakhani SR. Molecular classification of breast cancer. Virchows Arch. 2014;465:1–14. doi: 10.1007/s00428-014-1593-7. [DOI] [PubMed] [Google Scholar]
- 84.Goldhirsch A, et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann. Oncol. 2013;24:2206–2223. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang RM, et al. MYB regulates the DNA damage response and components of the homology-directed repair pathway in human estrogen receptor-positive breast cancer cells. Oncogene. 2019;38:5239–5249. doi: 10.1038/s41388-019-0789-3. [DOI] [PubMed] [Google Scholar]
- 86.Miao RY, et al. MYB is essential for mammary tumorigenesis. Cancer Res. 2011;71:7029–7037. doi: 10.1158/0008-5472.CAN-11-1015. [DOI] [PubMed] [Google Scholar]
- 87.Early Breast Cancer Trialists’ Collaborative G. et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet. 2011;378:771–784. doi: 10.1016/S0140-6736(11)60993-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Early Breast Cancer Trialists’ Collaborative G. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 89.Early Breast Cancer Trialists’ Collaborative G. Aromatase inhibitors versus tamoxifen in early breast cancer: patient-level meta-analysis of the randomised trials. Lancet. 2015;386:1341–1352. doi: 10.1016/S0140-6736(15)61074-1. [DOI] [PubMed] [Google Scholar]
- 90.Mitra P, Yang RM, Sutton J, Ramsay RG, Gonda TJ. CDK9 inhibitors selectively target estrogen receptor-positive breast cancer cells through combined inhibition of MYB and MCL-1 expression. Oncotarget. 2016;7:9069–9083. doi: 10.18632/oncotarget.6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ward A, et al. Re-expression of microRNA-375 reverses both tamoxifen resistance and accompanying EMT-like properties in breast cancer. Oncogene. 2013;32:1173–1182. doi: 10.1038/onc.2012.128. [DOI] [PubMed] [Google Scholar]
- 92.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial–mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gao Y, Zhang W, Liu C, Li G. miR-200 affects tamoxifen resistance in breast cancer cells through regulation of MYB. Sci. Rep. 2019;9:18844. doi: 10.1038/s41598-019-54289-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Persson M, et al. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc. Natl Acad. Sci. USA. 2009;106:18740–18744. doi: 10.1073/pnas.0909114106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brill LB, II, et al. Analysis of MYB expression and MYB-NFIB gene fusions in adenoid cystic carcinoma and other salivary neoplasms. Mod. Pathol. 2011;24:1169–1176. doi: 10.1038/modpathol.2011.86. [DOI] [PubMed] [Google Scholar]
- 97.Togashi Y, et al. MYB and MYBL1 in adenoid cystic carcinoma: diversity in the mode of genomic rearrangement and transcripts. Mod. Pathol. 2018;31:934–946. doi: 10.1038/s41379-018-0008-8. [DOI] [PubMed] [Google Scholar]
- 98.Magers MJ, et al. MYB-NFIB gene fusion in prostatic basal cell carcinoma: clinicopathologic correlates and comparison with basal cell adenoma and florid basal cell hyperplasia. Mod. Pathol. 2019;32:1666–1674. doi: 10.1038/s41379-019-0297-6. [DOI] [PubMed] [Google Scholar]
- 99.Drier Y, et al. An oncogenic MYB feedback loop drives alternate cell fates in adenoid cystic carcinoma. Nat. Genet. 2016;48:265–272. doi: 10.1038/ng.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fehr A, et al. The MYB-NFIB gene fusion-a novel genetic link between adenoid cystic carcinoma and dermal cylindroma. J. Pathol. 2011;224:322–327. doi: 10.1002/path.2909. [DOI] [PubMed] [Google Scholar]
- 101.Pei J, et al. Detecting MYB and MYBL1 fusion genes in tracheobronchial adenoid cystic carcinoma by targeted RNA-sequencing. Mod. Pathol. 2019;32:1416–1420. doi: 10.1038/s41379-019-0277-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kim J, et al. MYBL1 rearrangements and MYB amplification in breast adenoid cystic carcinomas lacking the MYB-NFIB fusion gene. J. Pathol. 2018;244:143–150. doi: 10.1002/path.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mitani Y, et al. Comprehensive analysis of the MYB-NFIB gene fusion in salivary adenoid cystic carcinoma: Incidence, variability, and clinicopathologic significance. Clin. Cancer Res. 2010;16:4722–4731. doi: 10.1158/1078-0432.CCR-10-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ho AS, et al. The mutational landscape of adenoid cystic carcinoma. Nat. Genet. 2013;45:791–798. doi: 10.1038/ng.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rutherford S, Yu Y, Rumpel CA, Frierson HF, Jr., Moskaluk CA. Chromosome 6 deletion and candidate tumor suppressor genes in adenoid cystic carcinoma. Cancer Lett. 2006;236:309–317. doi: 10.1016/j.canlet.2005.05.049. [DOI] [PubMed] [Google Scholar]
- 106.Andersson M. K., Afshari M. K., Andren Y., Wick M. J., & Stenman G. Targeting the oncogenic transcriptional regulator MYB in adenoid cystic carcinoma by inhibition of IGF1R/AKT signaling. J. Natl Cancer Inst.109, (2017). [DOI] [PubMed]
- 107.Morelli MP, et al. Prioritizing phase I treatment options through preclinical testing on personalized tumorgraft. J. Clin. Oncol. 2012;30:e45–e48. doi: 10.1200/JCO.2011.36.9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mahadevan D, et al. Phase 1b study of safety, tolerability and efficacy of R1507, a monoclonal antibody to IGF-1R in combination with multiple standard oncology regimens in patients with advanced solid malignancies. Cancer Chemother. Pharm. 2014;73:467–473. doi: 10.1007/s00280-013-2372-x. [DOI] [PubMed] [Google Scholar]
- 109.Calvo E, et al. A Phase I clinical trial and independent patient-derived xenograft study of combined targeted treatment with dacomitinib and figitumumab in advanced solid tumors. Clin. Cancer Res. 2017;23:1177–1185. doi: 10.1158/1078-0432.CCR-15-2301. [DOI] [PubMed] [Google Scholar]
- 110.Yusenko MV, et al. Monensin, a novel potent MYB inhibitor, suppresses proliferation of acute myeloid leukemia and adenoid cystic carcinoma cells. Cancer Lett. 2020;479:61–70. doi: 10.1016/j.canlet.2020.01.039. [DOI] [PubMed] [Google Scholar]
- 111.Mandelbaum J, et al. Zebrafish blastomere screen identifies retinoic acid suppression of <ovid:i>MYB</ovid:i> in adenoid cystic carcinoma. J. Exp. Med. 2018;215:2673–2685. doi: 10.1084/jem.20180939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Andersson KM, Afshari MK, Andrén Y, Wick MJ, Stenman G. Targeting the oncogenic transcriptional regulator MYB in adenoid cystic carcinoma by inhibition of IGF1R/AKT signaling. J. Natl Cancer Inst. 2017;109:djx017. doi: 10.1093/jnci/djx017. [DOI] [PubMed] [Google Scholar]
- 113.Reiss K, et al. The protooncogene c-myb increases the expression of insulin-like growth factor 1 and insulin-like growth factor 1 receptor messenger RNAs by a transcriptional mechanism. Cancer Res. 1991;51:5997–6000. [PubMed] [Google Scholar]
- 114.Qaddoumi I, et al. Genetic alterations in uncommon low-grade neuroepithelial tumors: BRAF, FGFR1, and MYB mutations occur at high frequency and align with morphology. Acta Neuropathol. 2016;131:833–845. doi: 10.1007/s00401-016-1539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Locati LD, et al. A phase II study of sorafenib in recurrent and/or metastatic salivary gland carcinomas: translational analyses and clinical impact. Eur. J. Cancer. 2016;69:158–165. doi: 10.1016/j.ejca.2016.09.022. [DOI] [PubMed] [Google Scholar]
- 116.Tchekmedyian V, et al. Phase II study of lenvatinib in patients with progressive, recurrent or metastatic adenoid cystic carcinoma. J. Clin. Oncol. 2019;37:1529–1537. doi: 10.1200/JCO.18.01859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Okamoto K, et al. Antitumor activities of the targeted multi-tyrosine kinase inhibitor lenvatinib (E7080) against RET gene fusion-driven tumor models. Cancer Lett. 2013;340:97–103. doi: 10.1016/j.canlet.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 118.Lutwyche JK, Keough RA, Hunter J, Coles LS, Gonda TJ. DNA binding-independent transcriptional activation of the vascular endothelial growth factor gene (VEGF) by the Myb oncoprotein. Biochem. Biophys. Res. Commun. 2006;344:1300–1307. doi: 10.1016/j.bbrc.2006.04.045. [DOI] [PubMed] [Google Scholar]
- 119.Ferrao P, Gonda TJ, Ashman LK. Expression of constitutively activated human c-Kit in Myb transformed early myeloid cells leads to factor independence, histiocytic differentiation, and tumorigenicity. Blood. 1997;90:4539–4552. doi: 10.1182/blood.V90.11.4539. [DOI] [PubMed] [Google Scholar]
- 120.Toscani A, et al. Arrest of spermatogenesis and defective breast development in mice lacking A-myb. Nature. 1997;386:713–717. doi: 10.1038/386713a0. [DOI] [PubMed] [Google Scholar]
- 121.Mettus RV, et al. Murine A-myb: evidence for differential splicing and tissue-specific expression. Oncogene. 1994;9:3077–3086. [PubMed] [Google Scholar]
- 122.Bolcun-Filas E, et al. A-MYB (MYBL1) transcription factor is a master regulator of male meiosis. Development. 2011;138:3319–3330. doi: 10.1242/dev.067645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Armstrong GT, et al. Survival and long-term health and cognitive outcomes after low-grade glioma. Neuro Oncol. 2011;13:223–234. doi: 10.1093/neuonc/noq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang J, et al. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat. Genet. 2013;45:602–612. doi: 10.1038/ng.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ramkissoon LA, et al. Genomic analysis of diffuse pediatric low-grade gliomas identifies recurrent oncogenic truncating rearrangements in the transcription factor MYBL1. Proc. Natl Acad. Sci. USA. 2013;110:8188–8193. doi: 10.1073/pnas.1300252110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ellison DW, et al. cIMPACT-NOW update 4: diffuse gliomas characterized by MYB, MYBL1, or FGFR1 alterations or BRAF(V600E) mutation. Acta Neuropathol. 2019;137:683–687. doi: 10.1007/s00401-019-01987-0. [DOI] [PubMed] [Google Scholar]
- 127.Wefers AK, et al. Isomorphic diffuse glioma is a morphologically and molecularly distinct tumour entity with recurrent gene fusions of MYBL1 or MYB and a benign disease course. Acta Neuropathol. 2020;139:193–209. doi: 10.1007/s00401-019-02078-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Brayer KJ, Frerich CA, Kang H, Ness SA. Recurrent fusions in MYB and MYBL1 define a common, transcription factor-driven oncogenic pathway in salivary gland adenoid cystic carcinoma. Cancer Discov. 2016;6:176–187. doi: 10.1158/2159-8290.CD-15-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kyrpychova L, et al. Small subset of adenoid cystic carcinoma of the skin is associated with alterations of the MYBL1 gene similar to their extracutaneous counterparts. Am. J. Dermatopathol. 2018;40:721–726. doi: 10.1097/DAD.0000000000001091. [DOI] [PubMed] [Google Scholar]
- 130.Iness AN, et al. The cell cycle regulatory DREAM complex is disrupted by high expression of oncogenic B-Myb. Oncogene. 2019;38:1080–1092. doi: 10.1038/s41388-018-0490-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sala A, Calabretta B. Regulation of BALB/c 3T3 fibroblast proliferation by B-myb is accompanied by selective activation of cdc2 and cyclin D1 expression. Proc. Natl Acad. Sci. USA. 1992;89:10415–10419. doi: 10.1073/pnas.89.21.10415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Santilli G, et al. Temperature-dependent modification and activation of B-MYB: implications for cell survival. J. Biol. Chem. 2005;280:15628–15634. doi: 10.1074/jbc.M411747200. [DOI] [PubMed] [Google Scholar]
- 133.Thorner AR, et al. In vitro and in vivo analysis of B-Myb in basal-like breast cancer. Oncogene. 2009;28:742–751. doi: 10.1038/onc.2008.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cervellera M, et al. Direct transactivation of the anti-apoptotic gene apolipoprotein J (clusterin) by B-MYB. J. Biol. Chem. 2000;275:21055–21060. doi: 10.1074/jbc.M002055200. [DOI] [PubMed] [Google Scholar]
- 135.Lang G, Gombert WM, Gould HJ. A transcriptional regulatory element in the coding sequence of the human Bcl-2 gene. Immunology. 2005;114:25–36. doi: 10.1111/j.1365-2567.2004.02073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Knight AS, Notaridou M, Watson RJ. A Lin-9 complex is recruited by B-Myb to activate transcription of G2/M genes in undifferentiated embryonal carcinoma cells. Oncogene. 2009;28:1737–1747. doi: 10.1038/onc.2009.22. [DOI] [PubMed] [Google Scholar]
- 137.Grassilli E, Salomoni P, Perrotti D, Franceschi C, Calabretta B. Resistance to apoptosis in CTLL-2 cells overexpressing B-Myb is associated with B-Myb-dependent bcl-2 induction. Cancer Res. 1999;59:2451–2456. [PubMed] [Google Scholar]
- 138.Lorvellec M, et al. B-Myb is critical for proper DNA duplication during an unperturbed S phase in mouse embryonic stem cells. Stem Cells. 2010;28:1751–1759. doi: 10.1002/stem.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Garcia P, Frampton J. The transcription factor B-Myb is essential for S-phase progression and genomic stability in diploid and polyploid megakaryocytes. J. Cell Sci. 2006;119:1483–1493. doi: 10.1242/jcs.02870. [DOI] [PubMed] [Google Scholar]
- 140.Tanaka Y, Patestos NP, Maekawa T, Ishii S. B-myb is required for inner cell mass formation at an early stage of development. J. Biol. Chem. 1999;274:28067–28070. doi: 10.1074/jbc.274.40.28067. [DOI] [PubMed] [Google Scholar]
- 141.Gualdrini F, et al. Addiction of MYCN amplified tumours to B-MYB underscores a reciprocal regulatory loop. Oncotarget. 2010;1:278–288. doi: 10.18632/oncotarget.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Qin H, et al. Prognostic implications and oncogenic roles of MYBL2 protein expression in esophageal squamous-cell carcinoma. Onco Targets Ther. 2019;12:1917–1927. doi: 10.2147/OTT.S190145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Calvisi DF, et al. Activation of v-Myb avian myeloblastosis viral oncogene homolog-like2 (MYBL2)-LIN9 complex contributes to human hepatocarcinogenesis and identifies a subset of hepatocellular carcinoma with mutant p53. Hepatology. 2011;53:1226–1236. doi: 10.1002/hep.24174. [DOI] [PubMed] [Google Scholar]
- 144.Wei T, et al. YAP-dependent induction of UHMK1 supports nuclear enrichment of the oncogene MYBL2 and proliferation in liver cancer cells. Oncogene. 2019;38:5541–5550. doi: 10.1038/s41388-019-0801-y. [DOI] [PubMed] [Google Scholar]
- 145.Lee YJ, et al. Ginkgetin induces G2-phase arrest in HCT116 colon cancer cells through the modulation of bMyb and miRNA34a expression. Int. J. Oncol. 2017;51:1331–1342. doi: 10.3892/ijo.2017.4116. [DOI] [PubMed] [Google Scholar]
- 146.MacLachlan TK, Sang N, Giordano A. Cyclins, cyclin-dependent kinases and cdk inhibitors: implications in cell cycle control and cancer. Crit. Rev. Eukaryot. Gene Expr. 1995;5:127–156. doi: 10.1615/CritRevEukarGeneExpr.v5.i2.20. [DOI] [PubMed] [Google Scholar]
- 147.Lam EW, Watson RJ. An E2F-binding site mediates cell-cycle regulated repression of mouse B-myb transcription. EMBO J. 1993;12:2705–2713. doi: 10.1002/j.1460-2075.1993.tb05932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sala A, et al. Activation of human B-MYB by cyclins. Proc. Natl Acad. Sci. USA. 1997;94:532–536. doi: 10.1073/pnas.94.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhu W, Giangrande PH, Nevins JR. E2Fs link the control of G1/S and G2/M transcription. EMBO J. 2004;23:4615–4626. doi: 10.1038/sj.emboj.7600459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ziebold U, Bartsch O, Marais R, Ferrari S, Klempnauer KH. Phosphorylation and activation of B-Myb by cyclin A-Cdk2. Curr. Biol. 1997;7:253–260. doi: 10.1016/S0960-9822(06)00121-7. [DOI] [PubMed] [Google Scholar]
- 151.Lin D, et al. Constitutive expression of B-myb can bypass p53-induced Waf1/Cip1-mediated G1 arrest. Proc. Natl Acad. Sci. USA. 1994;91:10079–10083. doi: 10.1073/pnas.91.21.10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Litovchick L, et al. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol. Cell. 2007;26:539–551. doi: 10.1016/j.molcel.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 153.Schmit F, et al. LINC, a human complex that is related to pRB-containing complexes in invertebrates regulates the expression of G2/M genes. Cell Cycle. 2007;6:1903–1913. doi: 10.4161/cc.6.15.4512. [DOI] [PubMed] [Google Scholar]
- 154.Zwicker J, Liu N, Engeland K, Lucibello FC, Muller R. Cell cycle regulation of E2F site occupation in vivo. Science. 1996;271:1595–1597. doi: 10.1126/science.271.5255.1595. [DOI] [PubMed] [Google Scholar]
- 155.Robinson C, et al. Cell-cycle regulation of B-Myb protein expression: specific phosphorylation during the S phase of the cell cycle. Oncogene. 1996;12:1855–1864. [PubMed] [Google Scholar]
- 156.Bartsch O, Horstmann S, Toprak K, Klempnauer KH, Ferrari S. Identification of cyclin A/Cdk2 phosphorylation sites in B-Myb. Eur. J. Biochem. 1999;260:384–391. doi: 10.1046/j.1432-1327.1999.00191.x. [DOI] [PubMed] [Google Scholar]
- 157.Werwein E, Cibis H, Hess D, Klempnauer KH. Activation of the oncogenic transcription factor B-Myb via multisite phosphorylation and prolyl cis/trans isomerization. Nucleic Acids Res. 2019;47:103–121. doi: 10.1093/nar/gky935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Tarasov KV, et al. B-MYB is essential for normal cell cycle progression and chromosomal stability of embryonic stem cells. PLoS ONE. 2008;3:e2478. doi: 10.1371/journal.pone.0002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sadasivam S, Duan S, DeCaprio JA. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev. 2012;26:474–489. doi: 10.1101/gad.181933.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wolter P, Hanselmann S, Pattschull G, Schruf E, Gaubatz S. Central spindle proteins and mitotic kinesins are direct transcriptional targets of MuvB, B-MYB and FOXM1 in breast cancer cell lines and are potential targets for therapy. Oncotarget. 2017;8:11160–11172. doi: 10.18632/oncotarget.14466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Fischer M, Müller GA. Cell cycle transcription control: DREAM/MuvB and RB-E2F complexes. Crit. Rev. Biochem. Mol. Biol. 2017;52:638–662. doi: 10.1080/10409238.2017.1360836. [DOI] [PubMed] [Google Scholar]
- 162.Iness AN, Litovchick L. MuvB: a key to cell cycle control in ovarian cancer. Front. Oncol. 2018;8:223. doi: 10.3389/fonc.2018.00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Liao GB, et al. Regulation of the master regulator FOXM1 in cancer. Cell Commun. Signal. 2018;16:57. doi: 10.1186/s12964-018-0266-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Engelhard A, Campbell K, Calabretta B. B-myb alters the response of myeloid precursor cells to G-CSF. Exp. Cell Res. 2000;254:153–162. doi: 10.1006/excr.1999.4742. [DOI] [PubMed] [Google Scholar]
- 165.Guan Z, Cheng W, Huang D, Wei A. High MYBL2 expression and transcription regulatory activity is associated with poor overall survival in patients with hepatocellular carcinoma. Curr. Res. Transl. Med. 2018;66:27–32. doi: 10.1016/j.retram.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 166.Pattschull G, et al. The Myb-MuvB complex is required for YAP-dependent transcription of mitotic genes. Cell Rep. 2019;27:3533–3546 e3537. doi: 10.1016/j.celrep.2019.05.071. [DOI] [PubMed] [Google Scholar]
- 167.Chen J, Chen X. MYBL2 is targeted by miR-143-3p and regulates breast cancer cell proliferation and apoptosis. Oncol. Res. 2018;26:913–922. doi: 10.3727/096504017X15135941182107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Jin Y, et al. B-Myb is up-regulated and promotes cell growth and motility in non-small cell lung cancer. Int. J. Mol. Sci. 2017;18:860. doi: 10.3390/ijms18060860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Xiong YC, Wang J, Cheng Y, Zhang XY, Ye XQ. Overexpression of MYBL2 promotes proliferation and migration of non-small-cell lung cancer via upregulating NCAPH. Mol. Cell. Biochem. 2020;468:185–193. doi: 10.1007/s11010-020-03721-x. [DOI] [PubMed] [Google Scholar]
- 170.Fuster O, et al. Adverse prognostic value of MYBL2 overexpression and association with microRNA-30 family in acute myeloid leukemia patients. Leuk. Res. 2013;37:1690–1696. doi: 10.1016/j.leukres.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 171.Heinrichs S, et al. MYBL2 is a sub-haploinsufficient tumor suppressor gene in myeloid malignancy. Elife. 2013;2:e00825. doi: 10.7554/eLife.00825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ren F, et al. MYBL2 is an independent prognostic marker that has tumor-promoting functions in colorectal cancer. Am. J. Cancer Res. 2015;5:1542–1552. [PMC free article] [PubMed] [Google Scholar]
- 173.Yu R, et al. Clinicopathologic features and prognostic implications of MYBL2 protein expression in pancreatic ductal adenocarcinoma. Pathol. Res. Pract. 2017;213:964–968. doi: 10.1016/j.prp.2017.04.024. [DOI] [PubMed] [Google Scholar]
- 174.Raschella G, et al. Expression of B-myb in neuroblastoma tumors is a poor prognostic factor independent from MYCN amplification. Cancer Res. 1999;59:3365–3368. [PubMed] [Google Scholar]
- 175.Raschella G, et al. Requirement of b-myb function for survival and differentiative potential of human neuroblastoma cells. J. Biol. Chem. 1995;270:8540–8545. doi: 10.1074/jbc.270.15.8540. [DOI] [PubMed] [Google Scholar]
- 176.Shi H, et al. Single nucleotide polymorphisms in the 20q13 amplicon genes in relation to breast cancer risk and clinical outcome. Breast Cancer Res. Treat. 2011;130:905–916. doi: 10.1007/s10549-011-1600-5. [DOI] [PubMed] [Google Scholar]
- 177.Tanner MM, et al. Frequent amplification of chromosomal region 20q12-q13 in ovarian cancer. Clin. Cancer Res. 2000;6:1833–1839. [PubMed] [Google Scholar]
- 178.Forozan F, et al. Comparative genomic hybridization analysis of 38 breast cancer cell lines: a basis for interpreting complementary DNA microarray data. Cancer Res. 2000;60:4519–4525. [PubMed] [Google Scholar]
- 179.Bayley R, Ward C, Garcia P. MYBL2 amplification in breast cancer: Molecular mechanisms and therapeutic potential. Biochim. Biophys. Acta Rev. Cancer. 2020;1874:188407. doi: 10.1016/j.bbcan.2020.188407. [DOI] [PubMed] [Google Scholar]
- 180.Ljungberg B, et al. European association of urology guidelines on renal cell carcinoma: the 2019 update. Eur. Urol. 2019;75:799–810. doi: 10.1016/j.eururo.2019.02.011. [DOI] [PubMed] [Google Scholar]
- 181.Sun SS, Fu Y, Lin JY. Upregulation of MYBL2 independently predicts a poorer prognosis in patients with clear cell renal cell carcinoma. Oncol. Lett. 2020;19:2765–2772. doi: 10.3892/ol.2020.11408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ahmed F. Integrated network analysis reveals FOXM1 and MYBL2 as key regulators of cell proliferation in non-small cell lung cancer. Front. Oncol. 2019;9:1011. doi: 10.3389/fonc.2019.01011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Clarke M, et al. MYBL2 haploinsufficiency increases susceptibility to age-related haematopoietic neoplasia. Leukemia. 2013;27:661–670. doi: 10.1038/leu.2012.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 185.Cammarata G, et al. Differential expression of specific microRNA and their targets in acute myeloid leukemia. Am. J. Hematol. 2010;85:331–339. doi: 10.1002/ajh.21667. [DOI] [PubMed] [Google Scholar]
- 186.Morita K, et al. Allosteric activators of protein phosphatase 2A display broad antitumor activity mediated by dephosphorylation of MYBL2. Cell. 2020;181:702–715.e720. doi: 10.1016/j.cell.2020.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 187.Leverson JD, Ness SA. Point mutations in v-Myb disrupt a cyclophilin-catalyzed negative regulatory mechanism. Mol. Cell. 1998;1:203–211. doi: 10.1016/S1097-2765(00)80021-0. [DOI] [PubMed] [Google Scholar]
- 188.Introna M, et al. Mutations in v-myb alter the differentiation of myelomonocytic cells transformed by the oncogene. Cell. 1990;63:1289–1297. doi: 10.1016/0092-8674(90)90424-D. [DOI] [PubMed] [Google Scholar]
- 189.Takahashi T, et al. Inhibitory interaction of c-Myb and GATA-1 via transcriptional co-activator CBP. Oncogene. 2000;19:134–140. doi: 10.1038/sj.onc.1203228. [DOI] [PubMed] [Google Scholar]
- 190.Rushton JJ, Ness SA. The conserved DNA binding domain mediates similar regulatory interactions for A-Myb, B-Myb, and c-Myb transcription factors. Blood Cells Mol. Dis. 2001;27:459–463. doi: 10.1006/bcmd.2001.0405. [DOI] [PubMed] [Google Scholar]
- 191.Bergholtz S, et al. The highly conserved DNA-binding domains of A-, B- and c-Myb differ with respect to DNA-binding, phosphorylation and redox properties. Nucleic Acids Res. 2001;29:3546–3556. doi: 10.1093/nar/29.17.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Luscher B, Christenson E, Litchfield DW, Krebs EG, Eisenman RN. Myb DNA binding inhibited by phosphorylation at a site deleted during oncogenic activation. Nature. 1990;344:517–522. doi: 10.1038/344517a0. [DOI] [PubMed] [Google Scholar]
- 193.Cervellera MN, Sala A. Poly(ADP-ribose) polymerase is a B-MYB coactivator. J. Biol. Chem. 2000;275:10692–10696. doi: 10.1074/jbc.275.14.10692. [DOI] [PubMed] [Google Scholar]
- 194.Bies J, Markus J, Wolff L. Covalent attachment of the SUMO-1 protein to the negative regulatory domain of the c-Myb transcription factor modifies its stability and transactivation capacity. J. Biol. Chem. 2002;277:8999–9009. doi: 10.1074/jbc.M110453200. [DOI] [PubMed] [Google Scholar]
- 195.Sano Y, Ishii S. Increased affinity of c-Myb for CREB-binding protein (CBP) after CBP-induced acetylation. J. Biol. Chem. 2001;276:3674–3682. doi: 10.1074/jbc.M006896200. [DOI] [PubMed] [Google Scholar]
- 196.Aziz N, et al. Modulation of c-Myb-induced transcription activation by a phosphorylation site near the negative regulatory domain. Proc. Natl Acad. Sci. USA. 1995;92:6429–6433. doi: 10.1073/pnas.92.14.6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Tomita A, et al. c-Myb acetylation at the carboxyl-terminal conserved domain by transcriptional co-activator p300. Oncogene. 2000;19:444–p451. doi: 10.1038/sj.onc.1203329. [DOI] [PubMed] [Google Scholar]
- 198.Frampton J, Ramqvist T, Graf T. v-Myb of E26 leukemia virus up-regulates bcl-2 and suppresses apoptosis in myeloid cells. Genes Dev. 1996;10:2720–2731. doi: 10.1101/gad.10.21.2720. [DOI] [PubMed] [Google Scholar]
- 199.Salomoni P, Perrotti D, Martinez R, Franceschi C, Calabretta B. Resistance to apoptosis in CTLL-2 cells constitutively expressing c-Myb is associated with induction of BCL-2 expression and Myb-dependent regulation of bcl-2 promoter activity. Proc. Natl Acad. Sci. USA. 1997;94:3296–3301. doi: 10.1073/pnas.94.7.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zhou F, Zhang L, van Laar T, van Dam H, Ten Dijke P. GSK3beta inactivation induces apoptosis of leukemia cells by repressing the function of c-Myb. Mol. Biol. Cell. 2011;22:3533–3540. doi: 10.1091/mbc.e11-06-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Down CF, Millour J, Lam EW, Watson RJ. Binding of FoxM1 to G2/M gene promoters is dependent upon B-Myb. Biochim. Biophys. Acta. 2012;1819:855–862. doi: 10.1016/j.bbagrm.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 202.Osterloh L, et al. The human synMuv-like protein LIN-9 is required for transcription of G2/M genes and for entry into mitosis. EMBO J. 2007;26:144–157. doi: 10.1038/sj.emboj.7601478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Okada M, Akimaru H, Hou DX, Takahashi T, Ishii S. Myb controls G(2)/M progression by inducing cyclin B expression in the Drosophila eye imaginal disc. EMBO J. 2002;21:675–684. doi: 10.1093/emboj/21.4.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Bartusel T, Schubert S, Klempnauer KH. Regulation of the cyclin D1 and cyclin A1 promoters by B-Myb is mediated by Sp1 binding sites. Gene. 2005;351:171–180. doi: 10.1016/j.gene.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 205.Srivastava SK, et al. Myb overexpression overrides androgen depletion-induced cell cycle arrest and apoptosis in prostate cancer cells, and confers aggressive malignant traits: potential role in castration resistance. Carcinogenesis. 2012;33:1149–1157. doi: 10.1093/carcin/bgs134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Cheasley D, et al. Defective Myb function ablates cyclin E1 expression and perturbs intestinal carcinogenesis. Mol. Cancer Res. 2015;13:1185–1196. doi: 10.1158/1541-7786.MCR-15-0014. [DOI] [PubMed] [Google Scholar]
- 207.Siu G, Wurster AL, Lipsick JS, Hedrick SM. Expression of the CD4 gene requires a Myb transcription factor. Mol. Cell. Biol. 1992;12:1592–1604. doi: 10.1128/MCB.12.4.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Melotti P, Ku DH, Calabretta B. Regulation of the expression of the hematopoietic stem cell antigen CD34: role of c-myb. J. Exp. Med. 1994;179:1023–1028. doi: 10.1084/jem.179.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ma XP, Calabretta B. DNA binding and transactivation activity of A-myb, a c-myb-related gene. Cancer Res. 1994;54:6512–6516. [PubMed] [Google Scholar]
- 210.Papetti M, Augenlicht LH. MYBL2, a link between proliferation and differentiation in maturing colon epithelial cells. J. Cell. Physiol. 2011;226:785–791. doi: 10.1002/jcp.22399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.De Dominici M, et al. Targeting CDK6 and BCL2 Exploits the “MYB Addiction” of Ph(+) acute lymphoblastic leukemia. Cancer Res. 2018;78:1097–1109. doi: 10.1158/0008-5472.CAN-17-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Frau M, et al. Mybl2 expression is under genetic control and contributes to determine a hepatocellular carcinoma susceptible phenotype. J. Hepatol. 2011;55:111–119. doi: 10.1016/j.jhep.2010.10.031. [DOI] [PubMed] [Google Scholar]
- 213.Liu F, Lei W, O’Rourke JP, Ness SA. Oncogenic mutations cause dramatic, qualitative changes in the transcriptional activity of c-Myb. Oncogene. 2006;25:795–805. doi: 10.1038/sj.onc.1209105. [DOI] [PubMed] [Google Scholar]
- 214.Hogg A, et al. Inactivation of a c-Myb/estrogen receptor fusion protein in transformed primary cells leads to granulocyte/macrophage differentiation and down regulation of c-kit but not c-myc or cdc2. Oncogene. 1997;15:2885–2898. doi: 10.1038/sj.onc.1201472. [DOI] [PubMed] [Google Scholar]
- 215.Ratajczak MZ, et al. Myb and ets proteins are candidate regulators of c-kit expression in human hematopoietic cells. Blood. 1998;91:1934–1946. doi: 10.1182/blood.V91.6.1934. [DOI] [PubMed] [Google Scholar]
- 216.Dudek H, Tantravahi RV, Rao VN, Reddy ES, Reddy EP. Myb and Ets proteins cooperate in transcriptional activation of the mim-1 promoter. Proc. Natl Acad. Sci. USA. 1992;89:1291–1295. doi: 10.1073/pnas.89.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Wilczek C, Chayka O, Plachetka A, Klempnauer KH. Myb-induced chromatin remodeling at a dual enhancer/promoter element involves non-coding rna transcription and is disrupted by oncogenic mutations of v-myb. J. Biol. Chem. 2009;284:35314–35324. doi: 10.1074/jbc.M109.066175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Nakagoshi H, Kanei-Ishii C, Sawazaki T, Mizuguchi G, Ishii S. Transcriptional activation of the c-myc gene by the c-myb and B-myb gene products. Oncogene. 1992;7:1233–1240. [PubMed] [Google Scholar]