Abstract
Biliary tract cancer (BTC) is comprised of intrahepatic cholangiocarcinoma (ICC), extrahepatic cholangiocarcinoma (EHC) and gallbladder cancer (GBC). These tumors arise in the biliary epithelium, share histological characteristics and are associated with grim prognosis even when diagnosed at early stages.Moreover, its relatively low incidence in developed countries has precluded the development of clinical trials addressing specific differences among BTC subgroups in terms of their biology, treatment response and clinical outcomes. In this scenario, the development of effective treatment strategies for patients has been rather modest. To date, the combination of cisplatin plus gemcitabine remains as the standard first line therapy in advanced disease and after progression to this regimen there are limited treatment options.Next generation sequencing (NGS) studies have assessed the distribution of driver genes and potentially actionable genomic alterations among ICC, EHC and GBC. Here, we outline genomic differences among these subsets and describe key milestones in order to develop novel targeted drugs against BTCs. Although the early results of several studies are promising, international collaboration is critical to conduct adequately-powered trials, enrolling patients from high-incidence countries.
Keywords: Gallbladder cancer (GBC), biliary tract cancer (BTC), genomics, next generation sequencing (NGS)
Introduction
Biliary tract cancer (BTC) is the second most common type of hepatobiliary cancer and is comprised of intrahepatic cholangiocarcinoma (ICC), extrahepatic cholangiocarcinoma (EHC) and gallbladder cancer (GBC). EHC can be further divided into perihilar, proximal or distal cholangiocarcinoma (1). In Western countries, ICC and EHC incidence are modest, ranging from 0.35 to 2 per 100,000/year. However, there are considerable regional differences associated with risk factors such as endemic liver fluke infection (Opisthorchis viverrini) in Asia (2). The highest incidence rates of GBC (up to 7.5 per 100,000 for men and 23 per 100,000 for women) are found among populations living in the Andean region (Chile, Bolivia), North-American Indians, Mexican Americans (3). As such, in Chile GBC is the second leading cause of death for women after breast cancer. Similarly, India accounts for a high burden of GBC worldwide with an age-standardized rate of 2.1 and 1.8 per 100,000 for incidence and mortality in women, respectively (4). GBC is closely associated with cholelithiasis and chronic gallbladder inflammation (cholecystitis). Other risk factors that might explain these regional differences in GBC incidence include chronic Salmonella or Helicobacter bilis infection, Native American ancestry and aflatoxin exposure (5-7).
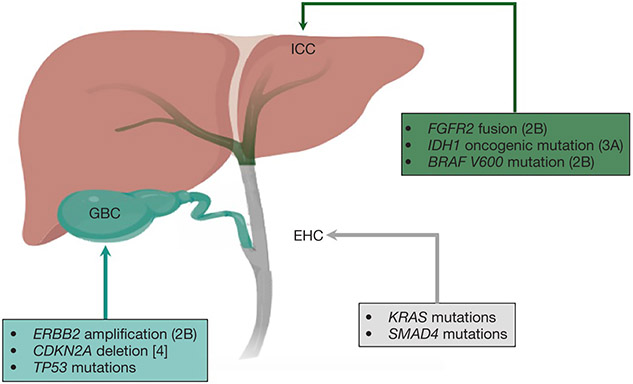
All BTCs arise in the biliary epithelium; they are associated with poor outcomes even when diagnosed at early stage (8,9). Although BTCs share some histological characteristics and features, there are important differences among them in terms of disease behavior, molecular profiles, and sensitivity to therapy (10). For example, GBC has been associated with worse prognosis compared to cholangiocarcinoma, however it has a greater response rate to chemotherapy (11). Historically, BTCs have been grouped together for the purpose of clinical trial accrual, however, genetic profiling has opened the opportunity to view them as molecularly distinct entities and enabled the targeting of specific genomic alterations. Herein we describe the genomic landscape of BTCs with a special comparative focus on relevant differences across BTC subgroups and biomarkers with potential clinical implications. Figure 1 summarizes the most prevalent genomic alterations observed across different BTC subtypes and their current actionability according to the OncoKB precision oncology database (12).
Figure 1.
Frequent genomic alterations in biliary tract cancer subtypes. Actionability of alterations is described according to the OncoKB precision oncology knowledge base.
Molecular landscape of BTC
Prevalent genomic alterations in BTC
Early BTC studies defined relevant oncogenes and tumor suppressor genes using polymerase chain reaction, Sanger sequencing, single-strand confirmation polymorphism and fluorescent in situ hybridization among other techniques. However, the actual frequency of these events and their relative distribution in different subgroups was difficult to assess. This can be explained by several reasons including sample heterogeneity, the application of a variety of different methodologies and a high stromal cell content in samples that may dilute out cancer-cell specific nucleic acids (10). Following the introduction of next generation sequencing (NGS), most of these issues have been surpassed (Table 1) (13-20). Indeed, NGS assays are highly sensitive, able to analyze a large panel of genes, and detect novel mutations, small insertions or deletions (indels), copy number alterations, and select gene fusions and rearrangements from small amounts of sample (21). A pioneer study used NGS to characterize 54 ICC Opisthorchis viverrini—related cholangiocarcinoma cases and found mutations in TP53 (44.4%), KRAS (16.7%), and SMAD4 (16.7%) (22). Subsequent studies have applied a variety of NGS platforms allowing a thorough comparison among BTC subgroups. A larger study analyzed 412 ICC, 57 EHC and 85 GBC cases that underwent genomic sequencing with the hybrid capture—based NGS platform FoundationOne (14). Interestingly, this study demonstrated significant differences among these 3 subtypes. Potentially actionable mutations were found in FGFR2 and IDH1/2, however, these were mostly limited to ICC with a prevalence of 11% and 20%, respectively. Also, ERBB2 amplification was more frequently observed in GBC (16%) and EHC (11%) versus ICC (3%). Similarly, a prospective study analyzed 195 cholangiocarcinomas using the targeted MSK-IMPACT platform (15) and found that FGFR2 fusions and IDH1, BAP1 and TP53 mutations occurred with greater frequency among ICCs, whereas KRAS, SMAD4, and STK11 alterations were more commonly seen in EHCs.
Table 1.
Main studies reporting molecular alterations associated with biliary tract cancers
| First author | Platform/genes analyzed | N of patients | Genomic alterations (%) |
|---|---|---|---|
| Nakamura et al. (13) | WES | 260 | ICC: KRAS 25%, FGFR2 8%, IDH1 6%; EHC: TP53 26%, KRAS 12%, SMAD4 10%; GBC: TP53 43%, ARID2 18%, EGFR 18% |
| Javle et al. (14) | HC NGS 182 | ICC 412; EHC 57; GBC 85 | ICC: TP53 27%, KRAS 22%, IDH1 16%; EHC: KRAS 42%, TP53 40%, SMAD4 21 %; GBC: TP53 59%, CDKN2A/B 19%, ERBB2 16% |
| Lowery et al. (15) | HC NGS 341/410 | ICC 158; EHC 37 | ICC: IDH1 30%, BAP1 20%, FGFR2 14%; EHC: KRAS 38%, TP53 49%, SMAD4 30% |
| Narayan et al. (16) | HC NGS 341/410 | GBC 81 | TP53 58%, SMAD4 31%, ARID1A 25% |
| Churi et al. (17) | AB NGS 46 | ICC 55; EHC 20 | ICC: TP53 29%, KRAS 24%, IDH 24%; EHC: KRAS 40%, TP53 45%, SMAD4 25% |
| Li et al. (18) | WES | GBC 57 | TP53 47%, ERBB3 12%, KRAS 8% |
| Ross et al. (19) | HC NGS 182 | ICC 28 | ARID1A 36%, IDH1/2 36%, TP53 36% |
| Zou et al. (20) | WES | ICC 102 | TP53 38%, KRAS 17%, ARID1A 7% |
WES, whole-exome sequencing; HC NGS, hybrid capture next generation sequencing; AB NGS, amplicon-based next generation sequencing; ICC, intrahepatic cholangiocarcinoma; EHC, extrahepatic cholangiocarcinoma; GBC, gallbladder cancer.
Molecular subtypes BTC
Additionally, NGS studies have contributed to refine etiologically distinct BTC subtypes. A study by the International Cancer Genome Consortium included a group of 489 ICC and EHC patients who underwent genomic, epigenomic, and transcriptomic analyses. Integrative clustering was primarily based on 71 patients who were assessed by whole-genome sequencing. This analysis found 4 clusters characterized by different clinical features including proportion of ICC and EHC and genomic alterations (23): Cluster 1 comprised mostly fluke-associated tumors that also displayed hypermethylation of CpG island-promoters, and enrichment of ARID1A and BRCA1/2 mutations. Cluster 2 was characterized by a mix of fluke-positive and fluke-negative tumors, and upregulated CTNNB1 and AKT1 expression. Clusters 1 and 2 were significantly enriched in TP53 mutations and ERBB2 amplifications. Clusters 3 and 4 comprised almost exclusively fluke-negative tumors. Cluster 3 displayed the highest level of copy number alterations, including enrichment of amplifications at 2p and 2q; these tumors also exhibited upregulation of immune checkpoint genes such as PD-1 and PD-L2. Cluster 4 was characterized by upregulation of FGFR family and PI3K pathways signatures along with frequent BAP1 and IDH1/2 mutations. Clusters 1 and 2 were enriched in EHC, whereas Clusters 3 and 4 were mostly ICC cases. Similarly, a Chinese study used a whole-exome sequencing approach and included 102 ICC patients. This study found that TP53-defective patients were more likely to be HBsAg-seropositive (P=0.021), also KRAS mutations were very infrequent in this group (20).
Germline alterations in BTC
Studies have determined that 10–20% of BTC patients display germline mutations on cancer-prone genes. Indeed, a recent study analyzed germline variants of selected genes in a cohort of 146 Japanese BTC patients. Germline variants were confirmed by the Sanger method and found a 11% of deleterious mutations in several genes including BRCA1, BRCA2, RAD51D, MLH1, or MSH2 (24). A similar study found a higher prevalence (20%) of germline pathogenic or likely pathogenic genomic alterations (25). In this study, a total of 78 patients (ICC =52, EHC =13 and GBC =13) were analyzed using the NGS MSK-IMPACT platform. This platform includes 76 genes associated with hereditary cancer predisposition. The most frequent germline mutations were found in BRCA2 (5.1%), APC (3.8%) and MUTYH (3.8%) genes. These findings support germline analysis in BTC patients in view of screening and therapeutic implications. Series of BTC patients with germline mutations in mismatch repair (MMR) genes such as MLH1 or MSH2 suggest that this phenomenon is infrequent and represent a small minority of patients with MMR-deficient tumors (26).
Potential prognostic and predictive genomic markers in BTC
FGFR
The FGFR family comprises four transmembrane tyrosine kinase receptors (namely FGFR1, −2, −3, and −4). Downstream signaling of activated FGFRs involves activation of the mitogen-activated protein kinase (MAPK) and/or phosphoinositide-3 kinase—Akt pathways. FGFR gene-fusions (particularly FGFR2) are considered a promising target for therapeutic inhibition. In contrast, the predictive role of FGFR1/2/3 mutations is more uncertain. As mentioned, FGFR2 fusions that involve its kinase domain are commonly seen in ICCs ranging from 8% to 14%. The most frequent fusion partners for FGFR2 is BICC1. However, a number of partners have been described in the literature, including KIAA1217, AHCYL1, TACC1, MGEA5, and PPHLN (15,27).
In a recent phase II study, infigratinib, a selective FGFR1-3 kinase inhibitor, was studied in cholangiocarcinoma patients who harbored FGFR2 fusions or other FGFR alterations (28). The response rate among patients with FGFR2 fusions was 19%, however no responses were seen in patients with other genetic FGFR alterations. FGFR plays an important role in phosphate homeostasis. Consequently, hyperphosphatemia has been a relevant on target toxicity of this class of drugs. Indeed, this trial found a 16% of patients were affected by grade 3–4 hyperphosphatemia. A phase III trial evaluating first-line infigratinib versus chemotherapy in ICC patients with FGFR2 fusions has started accrual (NCT03773302). Erdafitinib is another oral pan-FGFR inhibitor that has also been evaluated in ICCs. The LUC2001 phase II trial (NCT02699606) evaluated 11 ICC patients with different genetic alterations in FGFR2 or FGFR3 (29). Given the abovementioned risk of hyperphosphatemia, investigators applied a dosing schedule based on serum phosphate levels. Within this group, 5 out of 11 patients showed partial responses. Interestingly, all responders had FGFR2 alterations. Even though investigators used an up-titration protocol, hyperphosphatemia was frequent (10/11 patients). Despite this, all patients continued their treatments. Interestingly, this drug has been recently approved by American Food and Drug Administration (FDA) for metastatic urothelial carcinoma, with susceptible FGFR3 or FGFR2 genetic alterations based on the results of the BLC2001 trial (30). A recent phase I trial evaluated the FGFR inhibitor DEBIO 1347 in biliary tract malignancies (31). Eight BTC patients were treated in the dose escalation cohort of this study and they reported a 12.5% response rate. Grade 3–4 hyperphosphatemia was seen in 21% of patients. The maximum tolerated dose was not reached; however, dermatologic toxicity became sometimes dose-limiting beyond the dose-limiting toxicity period. TAS-120 is highly selective, irreversible FGFR1-4 inhibitor that was also studied in a phase I trial including BTC patients harboring FGFR alterations. The outcomes of the subgroup of patients with FGFR2 gene fusions (n=28) were reported showing an objective response rate of 25% (32). This drug has also demonstrated preclinical activity against multiple FGFR2 mutations shown to confer resistance to other FGFR inhibitors such as FGFR2 V565F gatekeeper mutation. A recent study showed that TAS-120 was active in four patients with FGFR2-fusion-positive ICC who developed resistance to infigratinib or Debio 1347; two of these patients achieved a partial response and two achieved stable disease by RECIST v1.1 criteria (33). In 3 of these patients FGFR2 mutations associated with resistance to FGFR inhibitors were detected in tissue or circulating tumor DNA before starting TAS-120. Currently, the FOENIX-101 phase II trial is evaluating the efficacy of TAS-120 in patients with ICC harboring FGFR2 gene rearrangements (NCT02052778). The FGFR inhibitor pemigatinib has also shown promising results in a phase II trial that included a cohort with tumors with FGFR2 translocations (mostly ICC) (34). These results have provided foundation for a phase III trial assessing the efficacy of this drug versus gemcitabine plus cisplatin chemotherapy in first-line treatment of patients with cholangiocarcinoma with FGFR2 rearrangement (NCT03656536). Likewise, the FGFR inhibitor derazantinib had an encouraging 21% response rate in patients with advanced ICC with FGFR2 fusion in a phase I/II trial (35). A phase II trial assessing the efficacy of this drug in this population is ongoing (NCT03230318).
IDH1/2
Somatic mutations in IDH enzymes are observed in 5–36% of ICCs (27). These enzymes are part of the tricarboxylic acid cycle and catalyze the interconversion of isocitrate to alpha-ketoglutarate (36). Cancer-associated mutations in IDH1/2 result in high levels of (R)-2-hydroxyglutarate, an onco-metabolite that seems to be associated with alterations in several cell processes including redox state and DNA repair mechanisms, also modulating the activity of epigenetic tumor suppressor enzymes (37-39). In addition to its functional impact on cancer progression IDH1/2 mutations have been implicated in ICC prognosis. Some studies have found an association between IDH1/2 mutations and improved clinical outcomes (40,41). However, this is a controversial issue since other studies have not found significant differences (42). Both IDH1 and IDH2 inhibitors have shown robust activity in preclinical models for solid tumors and leukemia (43,44). Ivosidenib is a first-in-class mutant IDH1 inhibitor that has been evaluated in a phase I study in IDH1-mutant advanced cholangiocarcinoma (45). This trial included 73 BTC patients (dose escalation n=24 and expansion n=49) and reported an objective response rate of 5% and a 12-month progression-free survival of 22%. Also, 3% of patients displayed grade 3–4 toxicity. A randomized phase III trial (ClarIDHy) of ivosidenib in this population is currently ongoing (NCT02989857) and recently it has been reported in a press release that it has achieved its primary endpoint (46). ClarIDHy did not include GBC patients since IDH-1 mutations are very infrequent in this group. BAY 1436032 is another oral inhibitor against mutant IDH1, whose safety, tolerability and preliminary anti-tumor activity are under evaluation in a phase I basket trial (NCT02746081) including mutant IDH1 BTC patients (47). IDH2 mutations are found less frequently in ICC (3% to 5%) and they also have been described as an acquired resistance mutation to IDH1 inhibition in this disease (48). Enasidenib is a mutant IDH2 inhibitor that has been approved for IDH2-mutant acute myeloid leukaemia, however, there is not data supporting activity of this drug in IDH2-mutant BTC.
HER2
Amplification of ERBB2 (also known as HER2) occurs in 3% to 19% of BTCs and is more frequent in GBC and EHC versus ICC (14,49). The study of HER2 as an actionable target in BTC has been challenging. A phase II trial led by the American National Cancer Institute (NCT00478140) tried to assess the efficacy of trastuzumab in HER2-overexpressed BTC, however, this study was closed due to slow accrual. Subsequently, two phase II studies evaluating the activity lapatinib in BTC did not show any responses, however these studies included an unselected population (50,51). Currently, a multi-cohort phase II trial in China is set to evaluate the combination of gemcitabine and oxaliplatin with trastuzumab in ERBB2-amplified BTC (NCT02836847). A limited number of BTC patients with ERBB2 amplification have been recruited in basket trials demonstrating the efficacy of anti-HER2 therapies. For example, the MyPathway trial included seven HER2 overexpressed/amplified BTC patients treated with the combination of the anti-HER2 antibodies trastuzumab and pertuzumab. In these patients the objective response was 29% (52). A similar response rate (17%) was recently shown for trastuzumab emtansine in this population (1 in 6 patients) (53). The antibody drug conjugate trastuzumab deruxtecan and the bispecific HER2-targeted antibody ZW25 have shown interesting activity in non-breast non-gastric HER2-amplified tumors in phase I trials (54,55). HER2-amplified BTCs have not been adequately represented in these studies, however, it is an interesting population to be included in future trials.
HER2 activating mutations have been described in between 1–8% of BTC and the potential therapeutic significance of these mutations is controversial. Interestingly, in a recent basket trial evaluating the efficacy of the pan-HER tyrosine kinase inhibitor neratinib in HER2-mutant tumors, nine BTC patients were enrolled. The reported response rate in this subgroup was 22% providing preliminary evidence of actionability (56).
BRAF
BRAF encodes a serine/threonine protein directly downstream from RAS in the MAPK cascade. The most frequent BRAF mutation is located in the V600 codon. In BTC patients BRAF mutations occur in 1% to 4% of ICC patients, while they are extremely infrequent in EHC or GBC (13,57). The phase II VE-BASKET trial included 8 BRAF-mutant BTC patients and reported 1 partial response (12%) (58). Subsequently, results of the ROAR Basket Trial demonstrated a 42% of response rate to the RAF inhibitor dabrafenib and MEK inhibitor trametinib in V600E BRAF-mutant BTC (59).
DDR
Alterations in DNA repair genes are relatively frequent in BTC including MSH6, BAP1, ATM, MLH1, MSH2, BRCA1 and BRCA2. Indeed, studies demonstrate that up to 16% of ICC and 45% of EHC patients display mutations in these genes (17). DNA-repair deficient cells (particularly BRCA1 or 2 mutants) can accumulate double-strand DNA breaks that result in genomic instability, leading to an increased sensitivity to DNA damaging therapies such as platinum compounds or poly ADP-ribose polymerase (PARP) inhibitors (60,61). Several studies have demonstrated the efficacy of PARP inhibitors in BRCA1/2 mutant breast and ovarian cancer (62-66). A recent series showed that 4 BRCA1/2-mutant cholangiocarcinoma patients who received PARP inhibitors had a favorable response. In fact, one of these patients had a progression free survival of 42 months (67). This specific patient had a known pathogenic germline BRCA2 mutation, however, the predictive features of germline versus somatic mutations remains to be elucidated. Anecdotal responses to PARP inhibitors have also been reported in metastatic BRCA1 mutant GBC (68). Interestingly, a recent study demonstrated that the accumulation of 2-Hydroxyglutarate associated with IDH mutations can suppress homologous recombination and thereby induces sensitivity to PARP inhibitors (69). These findings provide the foundations for a trial exploring the anti-tumoral activity of olaparib in solid tumors that harbor IDH1/2 mutations (NCT03212274).
EGFR
Epidermal growth factor receptor (EGFR) is a tyrosine kinase receptor that plays a key role in the cancer cell proliferation and is overexpressed in several solid tumors. Receptor-specific EGF ligands can bind onto EGFR (Erb1), Erb3, or Erb4 (70). Preclinical studies support the relevance of EGFR in the carcinogenesis of BTC, however results from early trials using anti-EGFR therapies have been disappointing (Table 2) (71-75), and to date no benefit has been demonstrated even in trials that have excluded RAS-mutant tumors.
Table 2.
Clinical trial efficacy data for targetable genomic alterations in biliary tract cancers
| Alteration | Therapy | N, patients | ORR (%) | Median PFS (months) | Reference |
|---|---|---|---|---|---|
| FGFR2 | Infigratinib | 61 | 15 | 5.8 | (28) |
| Erdafitinib | 11 | 46 | 5.6 | (29) | |
| Debio 1347 | 8 | 12.5 | NR | (31) | |
| TAS-120 | 28 | 25 | NR | (32) | |
| Derazantinib | 29 | 21 | 5.7 | (35) | |
| IDH1/2 | Ivosidenib | 73 | 5 | 3.8 | (45) |
| HER2 | Trast + P | 7 (amp) | 29 | NR | (52) |
| T-DM1 | 6 (amp) | 17 | NR | (53) | |
| Neratinib | 9 (mut) | 22 | NR | (56) | |
| BRAF | Vemurafenib | 8 | 12 | NR | (58) |
| D + T | 33 | 42 | 7.2 | (59) | |
| EGFR | Erl + Bev | 49 | 12 | 4.4 | (71) |
| GEMOX + Erl | 135 vs. 133 | 30 vs. 16‡ | 5.8 vs. 4.2† | (72) | |
| Gem + C | 44 | 20 | 5.8 | (73) | |
| GEMOX + C | 76 vs. 74 | 24 vs. 23† | 6.1 vs. 5.5† | (74) | |
| GEMOX + P | 46 | 33 | 8.3 | (75) |
not significant
statistically significant difference. Trast, trastuzumab; P, pertuzumab; T-DM1, trastuzumab emtansine; D, dabrafenib; T, trametinib; Erl, erlotinib; GEMOX, gemcitabine and oxaliplatin; C, cetuximab; P, panitumumab; NR, not reported; amp, amplified; mut, mutant.
Microsatellite instability (MSI)
MSI results from defects in the MMR system including MLH1, PMS2, MSH2, and MSH6 genes. The dysfunction of this repair system affects DNA proofreading after replication, leading to variable lengths of short tandem repeats, called microsatellites, within noncoding and coding regions (76). This phenomenon is known as MSI and can be attributed to germline mutations in one or several MMR genes that render a nonfunctional protein or to somatic hypermethylation of the MLH1 promoter that leads to epigenetic silencing. Studies show that 1–10% of BTCs can be categorized as MSI-high. These are more frequent in ICC compared to EHC or GBC (77,78). In localized disease, MSI-H GBC might be associated with better outcomes as it has been demonstrated in localized colorectal and gastric cancer, however, this needs further confirmation (79-81). MMR-deficiency has been shown to predict response of solid tumors to immunotherapy with PD-1 checkpoint inhibitors (82). Consequently, the FDA has provided a tissue/site-agnostic approval for pembrolizumab for this indication. Interestingly, in the pivotal KEYNOTE16 phase II study, a total of 4 MSI-high cholangiocarcinoma patients received pembrolizumab: 1 had a partial response and 3 had stable disease (83).
CDKN2A
Cyclin-dependent kinase inhibitor 2A (CDKN2A, also known as P16INK4A) is a tumor suppressor gene that encodes two proteins via an alternative splicing: p16INK4A and p14ARF. These proteins can act as inhibitors of cyclin-dependent kinase 4 (CDK4) and/or 6 (CDK6) thereby regulating the cell cycle. Altered CDKN2A is observed in many malignancies, however, its prognostic relevance varies by tumor type (84). In BTCs, CDKN2A is frequently silenced by hypermethylation, by homozygous deletion, or by inactivating mutations (10). Studies demonstrate that CDKN2A alterations are more frequently seen in GBC versus ICC or EHC (14) and have been associated with decreased survival (85). Although CDKN2A alterations are not considered actionable per se, homozygous CDKN2A loss is usually concomitant with methylthioadenosine phosphorylase (MTAP) gene deletions, which might be targetable. MTAP encodes a key enzyme in the methionine salvage pathway (86) and MTAP-deficient cells rely on protein arginine methyl-transferase 5 (PRMT5) for purine biosynthesis. Preclinical studies demonstrate that MTAP-deficient cancers depend on methionine adenosyltransferase II alpha (MAT2A) to produce S-adenosylmethionine, an essential PRMT5-substrate for survival of these cells (87). Currently, a phase I trial is set to evaluate the maximum tolerated dose, safety and efficacy of AG-270, a MAT2A inhibitor in solid tumors or lymphoma with homozygous loss of CDKN2A or MTAP (NCT03435250).
NTRK
The NTRK family includes three genes, NTRK1, NTRK2, and NTRK3, which encode three tropomyosin receptor kinase (TRK) proteins—TRKA, TRKB, and TRKC, respectively. Studies have identified NTRK fusions at low frequencies across various tumor types, including BTCs (88). Larotrectinib is now a FDA-approved selective small-molecule inhibitor of all three TRK proteins. Its use is approved for adult and pediatric patients with solid tumors that harbor NTRK gene fusions. Approval was based on the results of three studies: a phase I study in adults, a phase I/II study in children, and a phase II basket study in adolescents and adults. Altogether these studies included 55 NTRK fusion cases and reported a response rate of 80% (89). Two cholangiocarcinoma patients were included in these studies; one had a partial response and the other had progression of disease.
Conclusions
A deeper understanding of the molecular landscape and activated pathways involved in the carcinogenesis of BTCs has enabled to recognize BTCs subtypes as different entities. Furthermore, it has also guided the development of several novel drugs, which has derived into multiple ongoing phase II and III trials. Some of these trials have already shown preliminary efficacy on clinical outcomes and represent one step further towards becoming a standard treatment. Interestingly, some of the abovementioned targetable genomic alterations such as FGFR2 fusions, IDH1 and BRAF mutations, and ERBB2 amplifications have already been identified in circulating-tumor DNA (ctDNA) from ~20% of BTC patients (90). As has been discussed, BTCs are more prevalent in developing countries. One significant barrier to conduct adequately-powered trials to demonstrate definitive efficacy of these targeted therapies in BTC, is that the access to tissue and ctDNA NGS is limited in these countries. Consequently, international collaboration in coming years will play a key role in order to provide the expertise and resources to develop such studies. Hopefully, these initiatives, along with lowering costs of sequencing technologies, will increase the enrollment of BTC cases into molecularly-designed trials eventually leading to more treatment options for these patients in need.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- 1.Banales JM, Cardinale V, Carpino G, et al. Expert consensus document: Cholangiocarcinoma: current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA). Nat Rev Gastroenterol Hepatol 2016;13:261–80. [DOI] [PubMed] [Google Scholar]
- 2.Bridgewater JA, Goodman KA, Kalyan A, et al. Biliary Tract Cancer: Epidemiology, Radiotherapy, and Molecular Profiling. Am Soc Clin Oncol Educ Book 2016;35:e194–203. [DOI] [PubMed] [Google Scholar]
- 3.Wistuba II, Gazdar AF. Gallbladder cancer: lessons from a rare tumour. Nat Rev Cancer 2004;4:695–706. [DOI] [PubMed] [Google Scholar]
- 4.Are C, Ahmad H, Ravipati A, et al. Global epidemiological trends and variations in the burden of gallbladder cancer. J Surg Oncol 2017;115:580–90. [DOI] [PubMed] [Google Scholar]
- 5.Koshiol J, Wozniak A, Cook P, et al. Salmonella enterica serovar Typhi and gallbladder cancer: a case-control study and meta-analysis. Cancer Med 2016;5:3310–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Randi G, Franceschi S, La Vecchia C. Gallbladder cancer worldwide: geographical distribution and risk factors. Int J Cancer 2006;118:1591–602. [DOI] [PubMed] [Google Scholar]
- 7.Nogueira L, Foerster C, Groopman J, et al. Association of aflatoxin with gallbladder cancer in Chile. JAMA 2015;313:2075–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roa I, Ibacache G, Muñoz S, et al. Gallbladder cancer in Chile: Pathologic characteristics of survival and prognostic factors: analysis of 1,366 cases. Am J Clin Pathol 2014;141:675–82. [DOI] [PubMed] [Google Scholar]
- 9.Jarnagin WR, Fong Y, DeMatteo RP, et al. Staging, resectability, and outcome in 225 patients with hilar cholangiocarcinoma. Ann Surg 2001;234:507–17; discussion 17-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hezel AF, Deshpande V, Zhu AX. Genetics of biliary tract cancers and emerging targeted therapies. J Clin Oncol 2010;28:3531–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckel F, Schmid RM. Chemotherapy in advanced biliary tract carcinoma: a pooled analysis of clinical trials. Br J Cancer 2007;96:896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakravarty D, Gao J, Phillips SM, et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol 2017;2017. doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakamura H, Arai Y, Totoki Y, et al. Genomic spectra of biliary tract cancer. Nat Genet 2015;47:1003–10. [DOI] [PubMed] [Google Scholar]
- 14.Javle M, Bekaii-Saab T, Jain A, et al. Biliary cancer: Utility of next-generation sequencing for clinical management. Cancer 2016;122:3838–47. [DOI] [PubMed] [Google Scholar]
- 15.Lowery MA, Ptashkin R, Jordan E, et al. Comprehensive Molecular Profiling of Intrahepatic and Extrahepatic Cholangiocarcinomas: Potential Targets for Intervention. Clin Cancer Res 2018;24:4154–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narayan RR, Creasy JM, Goldman DA, et al. Regional differences in gallbladder cancer pathogenesis: Insights from a multi-institutional comparison of tumor mutations. Cancer 2019;125:575–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Churi CR, Shroff R, Wang Y, et al. Mutation profiling in cholangiocarcinoma: prognostic and therapeutic implications. PLoS One 2014;9:e115383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li M, Zhang Z, Li X, et al. Whole-exome and targeted gene sequencing of gallbladder carcinoma identifies recurrent mutations in the ErbB pathway. Nat Genet 2014;46:872–6. [DOI] [PubMed] [Google Scholar]
- 19.Ross JS, Wang K, Gay L, et al. New routes to targeted therapy of intrahepatic cholangiocarcinomas revealed by next-generation sequencing. Oncologist 2014;19:235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zou S, Li J, Zhou H, et al. Mutational landscape of intrahepatic cholangiocarcinoma. Nat Commun 2014;5:5696. [DOI] [PubMed] [Google Scholar]
- 21.Atreya CE, Yaeger R, Chu E. Systemic Therapy for Metastatic Colorectal Cancer: From Current Standards to Future Molecular Targeted Approaches. Am Soc Clin Oncol Educ Book 2017;37:246–56. [DOI] [PubMed] [Google Scholar]
- 22.Ong CK, Subimerb C, Pairojkul C, et al. Exome sequencing of liver fluke-associated cholangiocarcinoma. Nat Genet 2012;44:690–3. [DOI] [PubMed] [Google Scholar]
- 23.Jusakul A, Cutcutache I, Yong CH, et al. Whole-Genome and Epigenomic Landscapes of Etiologically Distinct Subtypes of Cholangiocarcinoma. Cancer Discov 2017;7:1116–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wardell CP, Fujita M, Yamada T, et al. Genomic characterization of biliary tract cancers identifies driver genes and predisposing mutations. J Hepatol 2018;68:959–69. [DOI] [PubMed] [Google Scholar]
- 25.Lowery MA, Jordan E, Kemel Y, et al. A prospective analysis of germline alterations (GA) in biliary tract cancer (BTC). J Clin Oncol 2017;35:4085. [Google Scholar]
- 26.Cloyd JM, Chun YS, Ikoma N, et al. Clinical and Genetic Implications of DNA Mismatch Repair Deficiency in Biliary Tract Cancers Associated with Lynch Syndrome. J Gastrointest Cancer 2018;49:93–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valle JW, Lamarca A, Goyal L, et al. New Horizons for Precision Medicine in Biliary Tract Cancers. Cancer Discov 2017;7:943–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Javle M, Lowery M, Shroff RT, et al. Phase II Study of BGJ398 in Patients With FGFR-Altered Advanced Cholangiocarcinoma. J Clin Oncol 2018;36:276–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y, Park JO, Su W, et al. Preliminary results of a ph2a study to evaluate the clinical efficacy and safety of erdafitinib in Asian patients with biomarker-selected advanced cholangiocarcinoma (CCA). Ann Oncol 2018;29:viii205–70. [Google Scholar]
- 30.Loriot Y, Necchi A, Park SH, et al. Erdafitinib (ERDA; JNJ-42756493), a pan-fibroblast growth factor receptor (FGFR) inhibitor, in patients (pts) with metastatic or unresectable urothelial carcinoma (mUC) and FGFR alterations (FGFRa): Phase 2 continuous versus intermittent dosing. J Clin Oncol 2018;36:411. [Google Scholar]
- 31.Voss MH, Hierro C, Heist RS, et al. A Phase I, Open-Label, Multicenter, Dose-escalation Study of the Oral Selective FGFR Inhibitor Debio 1347 in Patients with Advanced Solid Tumors Harboring FGFR Gene Alterations. Clin Cancer Res 2019;25:2699–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meric-Bernstam F, Arkenau H, Tran B, et al. O-001Efficacy of TAS-120, an irreversible fibroblast growth factor receptor (FGFR) inhibitor, in cholangiocarcinoma patients with FGFR pathway alterations who were previously treated with chemotherapy and other FGFR inhibitors. Ann Oncol 2018;29. doi: 10.1093/annonc/mdy149. [DOI] [Google Scholar]
- 33.Goyal L, Shi L, Liu LY, et al. TAS-120 overcomes resistance to ATP-competitive FGFR inhibitors in patients with FGFR2 fusion-positive intrahepatic cholangiocarcinoma. Cancer Discov 2019. doi: 10.1158/2159-8290.CD-19-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hollebecque A, Borad M, Sahai V, et al. 756PInterim results of fight-202, a phase II, open-label, multicenter study of INCB054828 in patients (pts) with previously treated advanced/metastatic or surgically unresectable cholangiocarcinoma (CCA) with/without fibroblast growth factor (FGF)/FGF receptor (FGFR) genetic alterations. Ann Oncol 2018;29. doi: 10.1093/annonc/mdy282.139. [DOI] [Google Scholar]
- 35.Mazzaferro V, El-Rayes BF, Droz Dit Busset M, et al. Derazantinib (ARQ 087) in advanced or inoperable FGFR2 gene fusion-positive intrahepatic cholangiocarcinoma. Br J Cancer 2019;120:165–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Losman JA, Kaelin WG Jr. What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev 2013;27:836–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reitman ZJ, Yan H. Isocitrate dehydrogenase 1 and 2 mutations in cancer: alterations at a crossroads of cellular metabolism. J Natl Cancer Inst 2010;102:932–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yen KE, Bittinger MA, Su SM, et al. Cancer-associated IDH mutations: biomarker and therapeutic opportunities. Oncogene 2010;29:6409–17. [DOI] [PubMed] [Google Scholar]
- 39.Molenaar RJ, Maciejewski JP, Wilmink JW, et al. Wild-type and mutated IDH1/2 enzymes and therapy responses. Oncogene 2018;37:1949–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang P, Dong Q, Zhang C, et al. Mutations in isocitrate dehydrogenase 1 and 2 occur frequently in intrahepatic cholangiocarcinomas and share hypermethylation targets with glioblastomas. Oncogene 2013;32:3091–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cercek A, Kemeny NE, Boerner T, et al. A bi-institutional phase II study of hepatic arterial infusion (HAI) with floxuridine (FUDR) and dexamethasone (Dex) combined with systemic gemcitabine and oxaliplatin (GemOx) for unresectable intrahepatic cholangiocarcinoma (ICC). J Clin Oncol 2018;36:4092. [Google Scholar]
- 42.Goyal L, Govindan A, Sheth RA, et al. Prognosis and Clinicopathologic Features of Patients With Advanced Stage Isocitrate Dehydrogenase (IDH) Mutant and IDH Wild-Type Intrahepatic Cholangiocarcinoma. Oncologist 2015;20:1019–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rohle D, Popovici-Muller J, Palaskas N, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science 2013;340:626–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang F, Travins J, DeLaBarre B, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science 2013;340:622–6. [DOI] [PubMed] [Google Scholar]
- 45.Lowery MA, Abou-Alfa GK, Burris HA, et al. Phase I study of AG-120, an IDH1 mutant enzyme inhibitor: Results from the cholangiocarcinoma dose escalation and expansion cohorts. J Clin Oncol 2017;35:4015. [Google Scholar]
- 46.Agios Announces the Randomized Phase 3 ClarIDHy Trial of TIBSOVO® (ivosidenib) Achieved its Primary Endpoint in Previously Treated IDH1 Mutant Cholangiocarcinoma Patients. Available online: https://agiospharmaceuticalsinc.gcs-web.com/news-releases/news-release-details/agios-announces-randomized-phase-3-claridhy-trial-tibsovor. Accessed May 25, 2019.
- 47.Pellino A, Loupakis F, Cadamuro M, et al. Precision medicine in cholangiocarcinoma. Transl Gastroenterol Hepatol 2018;3:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harding JJ, Lowery MA, Shih AH, et al. Isoform Switching as a Mechanism of Acquired Resistance to Mutant Isocitrate Dehydrogenase Inhibition. Cancer Discov 2018;8:1540–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Galdy S, Lamarca A, McNamara MG, et al. HER2/HER3 pathway in biliary tract malignancies; systematic review and meta-analysis: a potential therapeutic target? Cancer Metastasis Rev 2017;36:141–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peck J, Wei L, Zalupski M, et al. HER2/neu may not be an interesting target in biliary cancers: results of an early phase II study with lapatinib. Oncology 2012;82:175–9. [DOI] [PubMed] [Google Scholar]
- 51.Ramanathan RK, Belani CP, Singh DA, et al. A phase II study of lapatinib in patients with advanced biliary tree and hepatocellular cancer. Cancer Chemother Pharmacol 2009;64:777–83. [DOI] [PubMed] [Google Scholar]
- 52.Hainsworth JD, Meric-Bernstam F, Swanton C, et al. Targeted Therapy for Advanced Solid Tumors on the Basis of Molecular Profiles: Results From MyPathway, an Open-Label, Phase IIa Multiple Basket Study. J Clin Oncol 2018;36:536–42. [DOI] [PubMed] [Google Scholar]
- 53.Li BT, Makker V, Buonocore DJ, et al. A multi-histology basket trial of ado-trastuzumab emtansine in patients with HER2 amplified cancers. J Clin Oncol 2018;36:2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iwata H, Tamura K, Doi T, et al. Trastuzumab deruxtecan (DS-8201a) in subjects with HER2-expressing solid tumors: Long-term results of a large phase 1 study with multiple expansion cohorts. J Clin Oncol 2018;36:2501. [Google Scholar]
- 55.Meric-Bernstam F, Beeram M, Mayordomo JI, et al. Single agent activity of ZW25, a HER2-targeted bispecific antibody, in heavily pretreated HER2-expressing cancers. J Clin Oncol 2018;36:2500. [Google Scholar]
- 56.Hyman DM, Piha-Paul SA, Won H, et al. HER kinase inhibition in patients with HER2- and HER3-mutant cancers. Nature 2018;554:189–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goeppert B, Frauenschuh L, Renner M, et al. BRAF V600E-specific immunohistochemistry reveals low mutation rates in biliary tract cancer and restriction to intrahepatic cholangiocarcinoma. Mod Pathol 2014;27:1028–34. [DOI] [PubMed] [Google Scholar]
- 58.Hyman DM, Puzanov I, Subbiah V, et al. Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 Mutations. N Engl J Med 2015;373:726–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wainberg ZA, Lassen UN, Elez E, et al. Efficacy and safety of dabrafenib (D) and trametinib (T) in patients (pts) with BRAF V600E-mutated biliary tract cancer (BTC): A cohort of the ROAR basket trial. J Clin Oncol 2019;37:187. [Google Scholar]
- 60.Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005;434:917–21. [DOI] [PubMed] [Google Scholar]
- 61.Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005;434:913–7. [DOI] [PubMed] [Google Scholar]
- 62.Robson M, Im SA, Senkus E, et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N Engl J Med 2017;377:523–33. [DOI] [PubMed] [Google Scholar]
- 63.Litton JK, Rugo HS, Ettl J, et al. Talazoparib in Patients with Advanced Breast Cancer and a Germline BRCA Mutation. N Engl J Med 2018;379:753–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moore K, Colombo N, Scambia G, et al. Maintenance Olaparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N Engl J Med 2018;379:2495–505. [DOI] [PubMed] [Google Scholar]
- 65.Coleman RL, Oza AM, Lorusso D, et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017;390:1949–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mirza MR, Monk BJ, Herrstedt J, et al. Niraparib Maintenance Therapy in Platinum-Sensitive, Recurrent Ovarian Cancer. N Engl J Med 2016;375:2154–64. [DOI] [PubMed] [Google Scholar]
- 67.Golan T, Raitses-Gurevich M, Kelley RK, et al. Overall Survival and Clinical Characteristics of BRCA-Associated Cholangiocarcinoma: A Multicenter Retrospective Study. Oncologist 2017;22:804–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xie Y, Jiang Y, Yang XB, et al. Response of BRCA1-mutated gallbladder cancer to olaparib: A case report. World J Gastroenterol 2016;22:10254–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sulkowski PL, Corso CD, Robinson ND, et al. 2-Hydroxyglutarate produced by neomorphic IDH mutations suppresses homologous recombination and induces PARP inhibitor sensitivity. Sci Transl Med 2017;9. doi: 10.1126/scitranslmed.aal2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oyasiji T, Zhang J, Kuvshinoff B, et al. Molecular Targets in Biliary Carcinogenesis and Implications for Therapy. Oncologist 2015;20:742–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lubner SJ, Mahoney MR, Kolesar JL, et al. Report of a multicenter phase II trial testing a combination of biweekly bevacizumab and daily erlotinib in patients with unresectable biliary cancer: a phase II Consortium study. J Clin Oncol 2010;28:3491–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee J, Park SH, Chang HM, et al. Gemcitabine and oxaliplatin with or without erlotinib in advanced biliary-tract cancer: a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2012;13:181–8. [DOI] [PubMed] [Google Scholar]
- 73.Borbath I, Ceratti A, Verslype C, et al. Combination of gemcitabine and cetuximab in patients with advanced cholangiocarcinoma: a phase II study of the Belgian Group of Digestive Oncology. Ann Oncol 2013;24:2824–9. [DOI] [PubMed] [Google Scholar]
- 74.Malka D, Cervera P, Foulon S, et al. Gemcitabine and oxaliplatin with or without cetuximab in advanced biliary-tract cancer (BINGO): a randomised, open-label, non-comparative phase 2 trial. Lancet Oncol 2014;15:819–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jensen LH, Lindebjerg J, Ploen J, et al. Phase II marker-driven trial of panitumumab and chemotherapy in KRAS wild-type biliary tract cancer. Ann Oncol 2012;23:2341–6. [DOI] [PubMed] [Google Scholar]
- 76.Vilar E, Tabernero J. Molecular dissection of microsatellite instable colorectal cancer. Cancer Discov 2013;3:502–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Winkelmann R, Schneider M, Hartmann S, et al. Microsatellite Instability Occurs Rarely in Patients with Cholangiocarcinoma: A Retrospective Study from a German Tertiary Care Hospital. Int J Mol Sci 2018;19. doi: 10.3390/ijms19051421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Silva VW, Askan G, Daniel TD, et al. Biliary carcinomas: pathology and the role of DNA mismatch repair deficiency. Chin Clin Oncol 2016;5:62. [DOI] [PubMed] [Google Scholar]
- 79.Roa JC, Roa I, Correa P, et al. Microsatellite instability in preneoplastic and neoplastic lesions of the gallbladder. J Gastroenterol 2005;40:79–86. [DOI] [PubMed] [Google Scholar]
- 80.Sargent DJ, Marsoni S, Monges G, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol 2010;28:3219–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Choi YY, Kim H, Shin SJ, et al. Microsatellite Instability and Programmed Cell Death-Ligand 1 Expression in Stage II/III Gastric Cancer: Post Hoc Analysis of the CLASSIC Randomized Controlled study. Ann Surg 2019;270:309–16. [DOI] [PubMed] [Google Scholar]
- 82.Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao R, Choi BY, Lee MH, et al. Implications of Genetic and Epigenetic Alterations of CDKN2A (p16(INK4a)) in Cancer. EBioMedicine 2016;8:30–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ueki T, Hsing AW, Gao YT, et al. Alterations of p16 and prognosis in biliary tract cancers from a population-based study in China. Clin Cancer Res 2004;10:1717–25. [DOI] [PubMed] [Google Scholar]
- 86.Zhang H, Chen ZH, Savarese TM. Codeletion of the genes for p16INK4, methylthioadenosine phosphorylase, interferon-alpha1, interferon-beta1, and other 9p21 markers in human malignant cell lines. Cancer Genet Cytogenet 1996;86:22–8. [DOI] [PubMed] [Google Scholar]
- 87.Marjon K, Cameron MJ, Quang P, et al. MTAP Deletions in Cancer Create Vulnerability to Targeting of the MAT2A/PRMT5/RIOK1 Axis. Cell Rep 2016;15:574–87. [DOI] [PubMed] [Google Scholar]
- 88.Stransky N, Cerami E, Schalm S, et al. The landscape of kinase fusions in cancer. Nat Commun 2014;5:4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Drilon A, Laetsch TW, Kummar S, et al. Efficacy of Larotrectinib in TRK Fusion-Positive Cancers in Adults and Children. N Engl J Med 2018;378:731–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mody K, Kasi PM, Yang J, et al. Circulating Tumor DNA Profiling of Advanced Biliary Tract Cancers. JCO Precis Oncol 2019. doi: 10.1200/PO.18.00324. [DOI] [PubMed] [Google Scholar]