Abstract
Tropical theileriosis is a tick-borne disease caused by hemoprotozoan parasites with considerable veterinary and economic impact worldwide. Ticks transmitting the disease belong to the Haemaphysalis, Rhipicephalus, and Hyalomma genera. The Hyalomma genus is very common in Sicily (Italy) and represents the main Theileria annulata vector in the island. Data concerning the molecular epidemiology of this pathogen are missing in the region. In 2018–2019, blood and serum samples were collected from 480 cows in seven Sicilian farms from four different provinces. Seroprevalence in the farms ranged from 22% to 71%. Three farms were selected for molecular analysis consisting of real-time PCR targeting the almost complete 18S ribosomal RNA (rRNA). Four amplicons per farm were sequenced and phylogenetic analyses were carried out. The four sequences were identical within each farm and showed 92–99% identity with the other farms and with sequences from Genbank. According to the phylogenetic analysis, these three sequences and an additional one from a laboratory-cultured Theileria annulata strain obtained in 1999 belonged to a single T. annulata clade with good bootstrap support with other sequences from Italy, India, and Iran, indicating limited geographical and temporal genetic variability of the parasite. This study represents the first phylogenetic analysis of T. annulata in Sicily, which will be useful to improve the strategies for theileriosis control and prevention.
Keywords: Piroplasmida, phylogenetic analysis, tick-borne pathogens, 18S rRNA, Sicily
1. Introduction
Parasites of the Theileria genus (order Piroplasmida, family Theileridae) infect a wide spectrum of domestic and wild animals and are transmitted by Ixodidae ticks belonging to the Haemaphysalis, Hyalomma, and Rhipicephalus genera [1]. In recent years, there has been a growing interest in the diseases caused by Theileria spp., also due to the increased accuracy of the diagnostic tests [2]. Tropical theileriosis in cattle is caused by Theileria annulata, and it is characterized by leukocyte proliferation following pathogen infection [3]. Other symptoms include fever, lymphadenopathy, and anemia. If not treated, animals die within 3–4 weeks of infection [4]. This pathology also causes a significant reduction in animal fertility. In Sicily, theileriosis is one of the most common tick-borne diseases with several outbreaks noticed each year [5]. The current control measures include the use of acaricides for vector control, therapy with buparvaquone, and vaccination [6]. Several pathogen molecules able to induce the host immune responses are currently under investigation as candidate vaccine antigens [7]. In Italy, vaccine prophylaxis against tropical theileriosis is not carried out since the currently available live-attenuated vaccine is not commercialized in Europe [8]. The success of the therapy depends on the timeliness of the execution of the pharmacological treatment [9]. For this reason, molecular biology represents a very efficient diagnostic approach to contrast the disease, since molecular methods, such as real-time PCR, allow rapidly detecting the pathogen and promptly initiating control actions [10].
Moreover, the advances in sequence data analysis allowed an improvement in pathogen identification and characterization. Ribosomal RNA (rRNA) is the most used genetic marker for phylogenetic analysis in eukaryotic organisms due to the conserved function and structure of the 18S rRNA molecule. This gene has been sequenced from several organisms, resulting in a large database for sequence comparisons.
The marker 18S rRNA gene is widely used for determining evolutionary patterns and similarity among the T. annulata species due to the presence of a conserved and hypervariable region (V4). Indeed, the molecule also possesses phylogenetically informative variable regions that are useful for determining relationships among strains [11].
T. annulata remains one of the most common tick-borne pathogens affecting domestic ruminants not only in Italy [5] but in several countries of Europe [12,13]. In Sicily, Hyalomma lusitanicum has been proposed as the main T. annulata vector in cattle, and this tick species is relatively prevalent in extensive cattle breeding [14,15]. Although the presence of T. annulata in Sicily is well known, phylogenetic studies concerning T. annulata have been carried out in other European countries but not in Italy [16,17,18].
This study was aimed at the phylogenetic characterization of T. annulata strains present in cattle in Sicily to better understand the epidemiology of the parasite on the island.
2. Results
Overall, serological prevalence in the Sicilian cattle was 26.0% (95% confidence intervals = 31.7–41.5). At the farm level, observed prevalence values ranged from 22% to 71% (Table 1).
Table 1.
Results of serological analysis obtained in this study. ID, identifier.
| Farm ID | Province | Sampled Animals | Positive Animals | Prevalence |
|---|---|---|---|---|
| 1 | Ragusa | 216 | 48 | 22% |
| 2 | Ragusa | 53 | 52 | 23% |
| 3 | Siracusa * | 101 | 51 | 50% |
| 4 | Ragusa | 49 | 18 | 37% |
| 5 | Ragusa * | 21 | 15 | 71% |
| 6 | Enna * | 17 | 10 | 59% |
| 7 | Palermo | 24 | 8 | 33% |
* Farms selected for the molecular study.
According to the serological results, the three farms with the highest prevalence values and showing at least 10 animals serologically positive for T. annulata were selected for molecular studies (Farms 3, 5, and 6; Table 1).
For each of the three farms, four animals were selected for the sequencing of the almost complete 18S rRNA gene. Sequences obtained from the animals of the same farm were identical to each other and were considered as a single oligonucleotide sequence (Sequences 2–4; Table 2). These three sequences showed 98–100% identity.
Table 2.
Details of Theileria annulata sequences obtained in this study.
| Sequence ID | GenBank Accession Number (This Study) |
Provenience | Province | Number of Sequences from Positive Animals |
GenBank Accession Number of the Closest Sequence |
|---|---|---|---|---|---|
| Sequence 1 | MN944852 | Cultured Strain | Palermo (Sicily) |
1 | KF429799 |
| Sequence 2 | MT341815 | Field Strain | Ragusa (Sicily) |
4 | MK849885 |
| Sequence 3 | MT341857 | Field Strain | Syracuse (Sicily) | 4 | KT367871 |
| Sequence 4 | MT341858 | Field Strain | Enna (Sicily) |
4 | KT367868 |
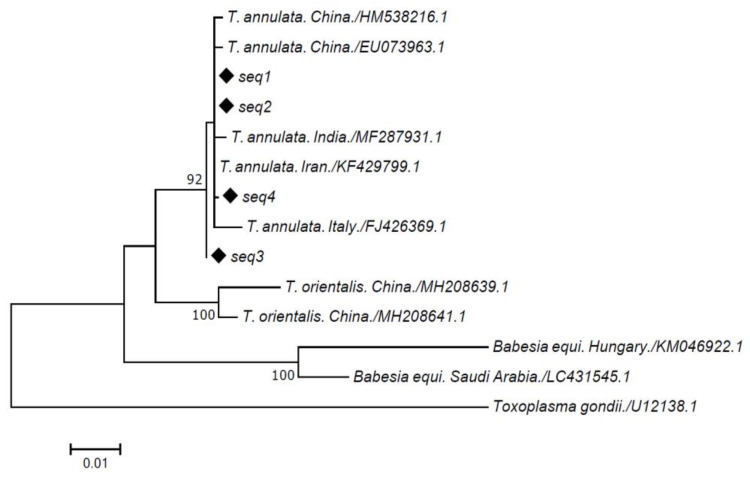
Moreover, a further 18S rRNA sequence, named sequence 1 (Seq1), was obtained from the T. annulata strain, isolated several years earlier at our institute and maintained in culture as described above. The four obtained sequences shared between 99.8% and 100% nucleotide sequence identity with sequences previously reported in GenBank (AY508463 and MN944852, respectively). Alignments of multiple 18S rRNA gene sequences revealed four haplotypes of T. annulata rRNA. The phylogenetic analysis (Figure 1) indicated that our sequences were classified in a single T. annulata clade with good bootstrap support with other sequences from Italy (FJ426369) and India and Iran (MF287931 and KF429799). Only sequence 3 rooted apart from the other sequences, indicating a likely different origin from the others. This finding calls for further investigation on the particularities of this farm.
Figure 1.
Phylogenetic tree obtained in this study using the 18S ribosomal RNA (rRNA) gene sequences.
3. Discussion
T. annulata remains one of the most common tick-borne pathogens affecting domestic ruminants in several European countries [5,12,13]. The observed seroprevalences are lower than those reported in other Mediterranean countries such as Tunisia or in other countries such as Sudan or Iraq [18,19,20,21,22]. Notwithstanding this, the parasite is actively circulating in the Sicilian cattle farms. In this study, the 18S rRNA gene partial sequences were used to evaluate the phylogenetic relationships of Sicilian T. annulata strains with other T. annulata strains, as well as with different Theileria species. Although the presence of T. annulata in Sicily is well known, phylogenetic studies concerning T. annulata have been carried out in other European countries but not in Italy [14,15,16,17,18].
In this study, Sicilian sequences formed a unique clade close to sequences from Italy, India, and Iran, confirming a molecular study conducted on a vaccine strain in Iran [23]. In that study, the 18S rRNA gene sequence of the T. annulata Iran vaccine strain showed from 97.9% to 99.9% identity with strains from China, Spain, and Italy. Moreover, a previous study concerning Theileria species in small ruminants in Italy also reported a closer association between the Italian Theileria ovis strain and Theileria spp. from Namibia and Iran [11]. These results may be the consequence of movements of animals and ticks between these countries, a hypothesis further strengthened by the usual practice of ancient peoples of migrating from these areas in the past, bringing with them some farm animals [24]. Furthermore, our data show that the sequences obtained from field samples are not substantially different from the sequence obtained from the strain isolated almost 20 years ago and cultured in our laboratory, suggesting a low rate of genetic variability of the pathogens.
4. Materials and Methods
4.1. Field Sample Collection
From September 2018 to March 2019, seven farms were investigated for T. annulata presence. Four farms were in the province of Ragusa, one in the province of Syracuse, and one each in the provinces of Enna and Palermo. All the selected farms were directed to the production of milk and meat. Animals of each farm, according to the regulations in force, are housed and cannot come into contact with the animals of other farms. Ethylenediaminetetraacetic acid (EDTA)-treated and untreated blood samples were obtained from each animal. The serum was separated by centrifugation and stored with EDTA-treated blood samples at −20° C.
4.2. In Vitro Cultivation of T. annulata Schizonts
Peripheral blood mononuclear cells (PBMCs) were isolated from heparinized blood samples as described elsewhere [25]. T. annulata schizonts from a positive blood sample obtained in the year 1999 were isolated and maintained in our laboratory in Roswell Park Memorial Institute (RPMI) 1640 medium added with 2 mM l-glutamine, 1% penicillin/streptomycin, and 10–20% fetal bovine serum (pH at 7.3) and incubated at 37° C in 5% CO2. Once the cell concentration of 5–9 × 105 cells/mL was reached, the culture was propagated in fresh soil.
4.3. Serological Analysis
Samples taken from animals housed on the seven farms were subjected to serological analysis. The presence of antibodies against T. annulata was assessed by immunofluorescence using a protocol modified from Burridge and Kimber [26].
4.4. Molecular Analysis and Phylogenetic Studies
For molecular analysis, samples belonging to the three farms with the highest prevalence values and showing at least 10 animals serologically positive for T. annulata were selected.
DNA was extracted using the PureLink Genomic DNA kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. DNA was extracted from the blood of field samples and PBMCs infected by T. annulata, and 200 ng of each purified genomic DNA was analyzed on 1% agarose gel (data not shown). Furthermore, the A206/A280 ratio was evaluated to verify the quality of the extracted DNA. Each sample was found suitable for further analysis with an A206/A280 ratio close to 2.0. T. annulata DNA was detected through real-time PCR [27]. For each positive farm, four samples, with a Ct value below 28, were selected to carry out the phylogenetic analysis.
4.5. Phylogenetic Study
4.5.1. 18S rRNA Gene Amplification and Sequencing
A portion of the 18S rRNA gene was obtained with specific primers 18S TBF and 18S TBR as described elsewhere [28]. The amplified product of approximately 1500 bp was used for sequencing analysis. DNA sequences were determined using the dideoxy chain termination method with a commercial DNA sequencing kit (BigDye™ Terminator v3.1 Cycle Sequencing Kit, Applied Biosystems™ (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. The obtained sequences were analyzed for nucleotide sequence identity by comparing them with reference strains in the GenBank database using the Basic Local Alignment Search Tool (BLAST). Multiple sequence alignments were performed using the ClustalW algorithm. Similarity matrices were then constructed from aligned sequence data by single distance using the Jukes and Cantor or the Kimura two-parameter model [29]. Phylogenetic trees were constructed using the neighbor-joining and the maximum parsimony methods as implemented in the Mega 2.1 package, deleting all the gap sites. Toxoplasma gondii was included as an outgroup [30]. The stability or accuracy of inferred topologies was assessed via bootstrap analysis of 1000 replicates.
4.5.2. Sequence Accession Numbers
The GenBank accession numbers for the gene sequences obtained in this study are MN944852, MT341815, MT341857, and MT341858.
5. Conclusions
As for other tick-borne pathogens, the distribution of Theileria spp. infections is related to several factors, including the presence of the tick vector species, the host, and the reservoir. Our data showed the presence of T. annulata in sampled animals, confirming that this parasite is prevalent in intensive cattle breeding.
Furthermore, our data show that the sequences obtained from field samples are not substantially different from the sequence obtained from the strain isolated almost 20 years ago and cultured in our laboratory, suggesting a low rate of genetic variability of the pathogen.
Serological investigations and characterization of T. annulata strains circulating in an area are useful to identify possible routes of entry of new strains into a territory, as well as identify the most appropriate therapeutic approaches, including vaccination strategies, for the prevention and control of the pathogen.
Acknowledgments
The authors would like to thank Rosalia D’Agostino for her support in the cell culture laboratory.
Author Contributions
Conceptualization, D.V. and V.G.; methodology, V.G.; software, S.D.C.; validation, V.G.; investigation, V.G., V.B., A.G., D.G., F.L.R., and G.S.; resources, D.V.; data curation, V.G., V.B., and S.D.C.; writing—original draft preparation, V.G. and V.B.; writing—review and editing, V.B. and J.M.; visualization, D.V.; supervision, A.T. and J.M.; project administration, D.V.; funding acquisition, D.V. All authors read and agreed to the published version of the manuscript.
Funding
This research was funded by the Italian Ministry of Health RC IZSSI 20/16 and 08/19.
Institutional Review Board Statement
The study did not involve any animal experiments. Blood samples were taken from cattle that were naturally infected or in which tick-borne disease was suspected. Blood sampling was necessary in order to perform laboratory analysis and did not involve any suffering of the animals sampled.
Informed Consent Statement
Not applicable.
Data Availability Statement
10–20% GenBank accession numbers of the sequences obtained in this study are MN944852, MT341815, MT341857, and MT341858, available at https://www.ncbi.nlm.nih.gov/genbank/.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lempereur L., Beck R., Antunes S., Baneth G., Silaghi C., Holman P., Zintl A., Fonseca I., Marques C., Duarte A., et al. Guidelines for the Detection of Babesiaand Theileria Parasites. Vector-Borne Zoonotic Dis. 2017;17:51–65. doi: 10.1089/vbz.2016.1955. [DOI] [PubMed] [Google Scholar]
- 2.Galuppi R., Aureli S., Bonoli C., Caffara M., Tampieri M. Detection and Molecular Characterization of Theileria sp. in Fallow Deer (Dama dama) and Ticks from an Italian Natural Preserve. Res. Vet. Sci. 2011;91:110–115. doi: 10.1016/j.rvsc.2010.07.029. [DOI] [PubMed] [Google Scholar]
- 3.Torina A., Villari S., Blanda V., Vullo S., La Manna M.P., Azgomi M.S.S., Di Liberto D., De La Fuente J., Sireci G. Innate Immune Response to Tick-Borne Pathogens: Cellular and Molecular Mechanisms Induced in the Hosts. Int. J. Mol. Sci. 2020;21:5437. doi: 10.3390/ijms21155437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nene V., Morrison I. Approaches to Vaccination Against Theileria Parva and Theileria Annulata. Parasite Immunol. 2016;38:724–734. doi: 10.1111/pim.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loria G.R., Riili S., Vitale F., Greco A., Sparagano O. Clinical and Laboratory Studies on Theileriosis Outbreaks in Sicily, Italy. Parassitol. 1999;41:63–67. [PubMed] [Google Scholar]
- 6.Saruhan B., Paşa S. [Therapeutic efficacy of buparvaquone (buparvon) in cattle with theileriosis] Turk. J. Parasitol. 2008;32:317–321. [PubMed] [Google Scholar]
- 7.Torina A., Blanda V., Villari S., Piazza A., La Russa F., Grippi F., La Manna M.P., Di Liberto D., De La Fuente J., Sireci G. Immune Response to Tick-Borne Hemoparasites: Host Adaptive Immune Response Mechanisms as Potential Targets for Therapies and Vaccines. Int. J. Mol. Sci. 2020;21:8813. doi: 10.3390/ijms21228813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.International Office of Epizootics . Biological Standards Commission Manual of Diagnostic Tests and Vaccines for Terrestrial Animals: (Mammals, Birds and Bees)—Chapter 3.4.14. Theileriosis. 8th ed. World Organisation for Animal Health; Paris, France: 2018. [Google Scholar]
- 9.Hasheminasab S.S., Moradi P., Wright I. A Four Year Epidemiological and Chemotherapy Survey of Babesiosis and Theileriosis, and Tick Vectors in Sheep, Cattle and Goats in Dehgolan, Iran. Ann. Parasitol. 2018;64:43–48. doi: 10.17420/ap6401.131. [DOI] [PubMed] [Google Scholar]
- 10.Dandasena D., Bhandari V., Sreenivasamurthy G.S., Murthy S., Roy S., Bhanot V., Arora J.S., Singh S., Sharma P. A Real-Time PCR Based Assay for Determining Parasite to Host Ratio and Parasitaemia in the Clinical Samples of Bovine Theileriosis. Sci. Rep. 2018;8:1–7. doi: 10.1038/s41598-018-33721-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sparagano O., Špitalská E., Namavari M., Torina A., Cannella V., Caracappa S. Phylogenetics of Theileria Species in Small Ruminants. Ann. N. Y. Acad. Sci. 2006;1081:505–508. doi: 10.1196/annals.1373.075. [DOI] [PubMed] [Google Scholar]
- 12.Antunes S., Ferrolho J., Domingues N., Santos A.S., Santos-Silva M.M., Domingos A. Anaplasma Marginale and Theileria Annulata in Questing Ticks from Portugal. Exp. Appl. Acarol. 2016;70:79–88. doi: 10.1007/s10493-016-0057-y. [DOI] [PubMed] [Google Scholar]
- 13.Calleja-Bueno L., Sainz A., García-Sancho M., Rodríguez-Franco F., González-Martín J.V., Villaescusa A. Molecular, Epidemiological, Haematological and Biochemical Evaluation in Asymptomatic Theileria Annulata Infected Cattle from an Endemic Region in Spain. Ticks Tick Borne Dis. 2017;8:936–941. doi: 10.1016/j.ttbdis.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 14.Torina A., Alongi A., Scimeca S., Vicente J., Caracappa S., De La Fuente J. Prevalence of Tick-Borne Pathogens in Ticks in Sicily. Transbound. Emerg. Dis. 2010;57:46–48. doi: 10.1111/j.1865-1682.2010.01101.x. [DOI] [PubMed] [Google Scholar]
- 15.Torina A., Blanda V., Caracappa S., Blanda M., Auteri M., La Russa F., Scimeca S., D’Agostino R., DiSclafani R., Villari S., et al. A Geographical Information System Based Approach for Integrated Strategies of Tick Surveillance and Control in the Peri-Urban Natural Reserve of Monte Pellegrino (Palermo, Southern Italy) Int. J. Environ. Res. Public Health. 2018;15:404. doi: 10.3390/ijerph15030404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Criado-Fornelio A., Martinez-Marcos A., Buling-Saraña A., Barba-Carretero J. Molecular studies on Babesia, Theileria and Hepatozoon in Southern Europe. Vet. Parasitol. 2003;113:189–201. doi: 10.1016/S0304-4017(03)00078-5. [DOI] [PubMed] [Google Scholar]
- 17.Gomes J., Salgueiro P., Inácio J., Amaro A., Pinto J., Tait A., Shiels B., Da Fonseca I.P., Santos-Gomes G., Weir W. Population Diversity of Theileria Annulata in Portugal. Infect. Genet. Evol. 2016;42:14–19. doi: 10.1016/j.meegid.2016.04.023. [DOI] [PubMed] [Google Scholar]
- 18.Ferrolho J., Antunes S., Santos A.S., Velez R., Padre L., Cabezas-Cruz A., Santos A.S., Domingos A. Detection and Phylogenetic Characterization of Theileria spp. and Anaplasma Marginale in Rhipicephalus Bursa in Portugal. Ticks Tick Borne Dis. 2016;7:443–448. doi: 10.1016/j.ttbdis.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 19.Rjeibi M.R., Darghouth M.A., Gharbi M. Prevalence of Theileria and Babesia Species in Tunisian Sheep. Onderstepoort J. Vet. Res. 2016;83:6. doi: 10.4102/ojvr.v83i1.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Darghouth M.A., Boulter N.R., Gharbi M., Sassi L., Tait A., Hall R. Vaccination of Calves with an Attenuated Cell Line of Theileria Annulata and the Sporozoite Antigen SPAG-1 Produces a Synergistic Effect. Vet. Parasitol. 2006;142:54–62. doi: 10.1016/j.vetpar.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 21.Mohammed-Ahmed G., Hassan S., El Hussein A., Salih D. Molecular, Serological and Parasitological Survey of Theileria Annulata in North Kordofan State, Sudan. Vet. Parasitol. Reg. Stud. Rep. 2018;13:24–29. doi: 10.1016/j.vprsr.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 22.Al-Saeed A.T.M., Omer L.T., Abdo J., Habibi G., Salih D.A., Seitzer U., Ahmed J.S. Epidemiological Studies on Tropical Theileriosis (Theileria Annulata Infection of Cattle) in Kurdistan Region, Iraq. Parasitol. Res. 2009;106:403–407. doi: 10.1007/s00436-009-1675-7. [DOI] [PubMed] [Google Scholar]
- 23.Habibi G. Phylogenetic Analysis of Theileria annulata Infected Cell Line S15 Iran Vaccine Strain. Iran. J. Parasitol. 2012;7:73–81. [PMC free article] [PubMed] [Google Scholar]
- 24.Sardina M.T., Ballester M., Marmi J., Finocchiaro R., Van Kaam J.B.C.H.M., Portolano B., Folch J.M. Phylogenetic Analysis of Sicilian Goats Reveals a New mtDNA Lineage. Anim. Genet. 2006;37:376–378. doi: 10.1111/j.1365-2052.2006.01451.x. [DOI] [PubMed] [Google Scholar]
- 25.Agnone A., La Manna M.P., Vesco G., Gargano V., Macaluso G., Dieli F., Sireci G., Villari S. Analysis of Interferon-Gamma Producing Cells During Infections by Yersinia Enterocolitica O:9 and Brucella Abortus in cattle. Vet. Ital. 2019;55:149–155. doi: 10.12834/VetIt.1374.7538.2. [DOI] [PubMed] [Google Scholar]
- 26.Torina A., Cordaro A., Blanda V., D’Agostino R., Scimeca S., Scariano E.M., Sireci G., Lelli R. A Promising New ELISA Diagnostic Test for Cattle Babesiosis Based on Babesia Bigemina Apical Membrane Antigen? Vet. Ital. 2016;52:63–69. doi: 10.12834/VetIt.74.237.2. [DOI] [PubMed] [Google Scholar]
- 27.Ros-García A., Nicolás A., Garcia-Perez A.L., Juste R.A., Hurtado A. Development and evaluation of a real-time PCR assay for the quantitative detection of Theileria annulata in cattle. Parasites Vectors. 2012;5:171. doi: 10.1186/1756-3305-5-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagore D., García-Sanmartín J., García-Pérez A.L., Juste R.A., Hurtado A. Identification, Genetic Diversity and Prevalence of Theileria and Babesia Species in a Sheep Population from Northern Spain. Int. J. Parasitol. 2004;34:1059–1067. doi: 10.1016/j.ijpara.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 29.Jukes T.H., Cantor C.R. Mammalian Protein Metabolism. Elsevier BV; Amsterdam, The Netherlands: 1969. Evolution of Protein Molecules; pp. 21–132. [Google Scholar]
- 30.Tamura K., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
10–20% GenBank accession numbers of the sequences obtained in this study are MN944852, MT341815, MT341857, and MT341858, available at https://www.ncbi.nlm.nih.gov/genbank/.