Abstract
Insects in nature interact with a wide variety of microbial enemies including nematodes. These include entomopathogenic nematodes that contain mutualistic bacteria and together are able to infect a broad range of insects in order to complete their life cycle and multiply, filarial nematodes which are vectored by mosquitoes, and other parasitic nematodes. Entomopathogenic nematodes are commonly used in biological control practices and they form excellent research tools for understanding the genetic and functional bases of nematode pathogenicity and insect anti-nematode immunity. In addition, clarifying the mechanism of transmission of filarial nematodes by mosquitoes is critical for devising strategies to reduce disease transmission in humans. In all cases and in order to achieve these goals, it is vital to determine the number and type of insect host genes which are differentially regulated during infection and encode factors with anti-nematode properties. In this respect, the use of transcriptomic approaches has proven a key step for the identification of insect molecules with anti-nematode activity. Here, we review the progress in the field of transcriptomics that deals with the insect response to nematode infection. This information is important because it will expose conserved pathways of anti-nematode immunity in humans.
Keywords: insect, nematodes, infection, immunity, transcriptomics
1. Introduction
Insects occupy different natural habitats and face a constant battle for survival when confronting microbial challenges. They respond to microbial invaders by activating their innate immune system, which involves the detection of pathogens through pattern recognition receptors, the induction of intracellular immune signaling pathways, and the regulation of humoral and cellular immune mechanisms [1,2,3]. Insect host defenses include hemocyte-related functions and the production of antimicrobial peptides and other effector molecules, which are secreted into the hemolymph [4,5,6]. Understanding immune reactions in insects has been facilitated by the use of insect models that permit elegant genetic manipulations in the laboratory, but significant discoveries have also been made in insects of agricultural or medical importance [7,8,9,10].
Distinct attention has been given recently to the interaction between insect hosts and nematode parasites. Nematodes employ multiple strategies to disrupt the activation and normal function of insect physiological responses, many of which participate in the regulation of innate immune reactions [11]. For instance, the entomopathogenic nematodes belonging to the genera Heterorhabditis and Steinernema are inhabited by the Gram-negative bacteria Photorhabdus and Xenorhabdus, respectively, and they are capable of entering insects that mainly live in the soil in order to reproduce by feeding on various tissues before they search for other victims to infect [12]. In addition, filarial nematodes, which cause important diseases in humans, are transmitted primarily by mosquitos that mount antimicrobial peptide and melanotic encapsulation responses against the parasites [13]. In addition, other parasitic nematodes are able to infect diverse insects, like Howardula nematodes that infect mycophagus Drosophila species [14].
Elucidating the molecular interactions between insects and parasitic nematodes as well as the genetic basis of the insect response to nematode infection require methods that facilitate characterization of the global gene expression changes that occur at the organismal and tissue/cell-specific levels. Here, we review the recent progress on the implementation of various transcriptomic approaches into the identification and characterization of gene expression regulation and signaling pathway activation, which modulate anti-nematode processes in insects. Such information is essential for the discovery of molecules with anti-nematode activity.
2. Insect Transcriptomic Response to Entomopathogenic Nematodes
The majority of studies on the transcriptomic response of insect to entomopathogenic nematodes have been performed in Drosophila melanogaster because it is easy and inexpensive to maintain this insect model in the lab, the ability to obtain large numbers of individuals to conduct large genome-wide experiments, and the availability of a fully sequenced and annotated genome (Table 1).
Table 1.
Nematodes, their insect hosts, and transcriptomic approaches included in this review.
| Nematode Yype | Nematode Species | Insect Host | Transcriptomic Approach | Reference |
|---|---|---|---|---|
| Entomopathogenic | Heterorhabditis bacteriophora | Drosophila melanogaster | Affymetrix GeneChip | [15] |
| Heterorhabditis bacteriophora | Drosophila melanogaster | Illumina RNA-Seq | [16] | |
| Heterorhabditis bacteriophora | Heliothis virescens | Illumina RNA-Seq | [17] | |
| Steinernema carpocapsae | Drosophila melanogaster | Illumina RNA-Seq | [18] | |
| Filarial | Brugia malayi | Armigeres subalbatus | Expressed Sequence Tags | [19] |
| Dirofilaria immitis | Armigeres subalbatus | Expressed Sequence Tags | [19] | |
| Brugia malayi | Armigeres subalbatus | Microarray | [20,21] | |
| Brugia pahangi | Armigeres subalbatus | Microarray | [21] | |
| Brugia malayi | Aedes aegypti | Microarray | [22] | |
| Brugia malayi | Aedes aegypti | RNA-Seq | [23,24] | |
| Wuchereria bancrofti | Culex quinquefasciatus | Microarray | [25] | |
| Parasitic | Shaerularia bombi | Bombus terrestris | RNA-Seq | [26] |
| Bursaphelenchus xylophilus | Monochamus alternatus | RNA-Seq | [27] | |
| Howardula aoronymphium | Drosophila neotestacea | RNA-Seq | [28] |
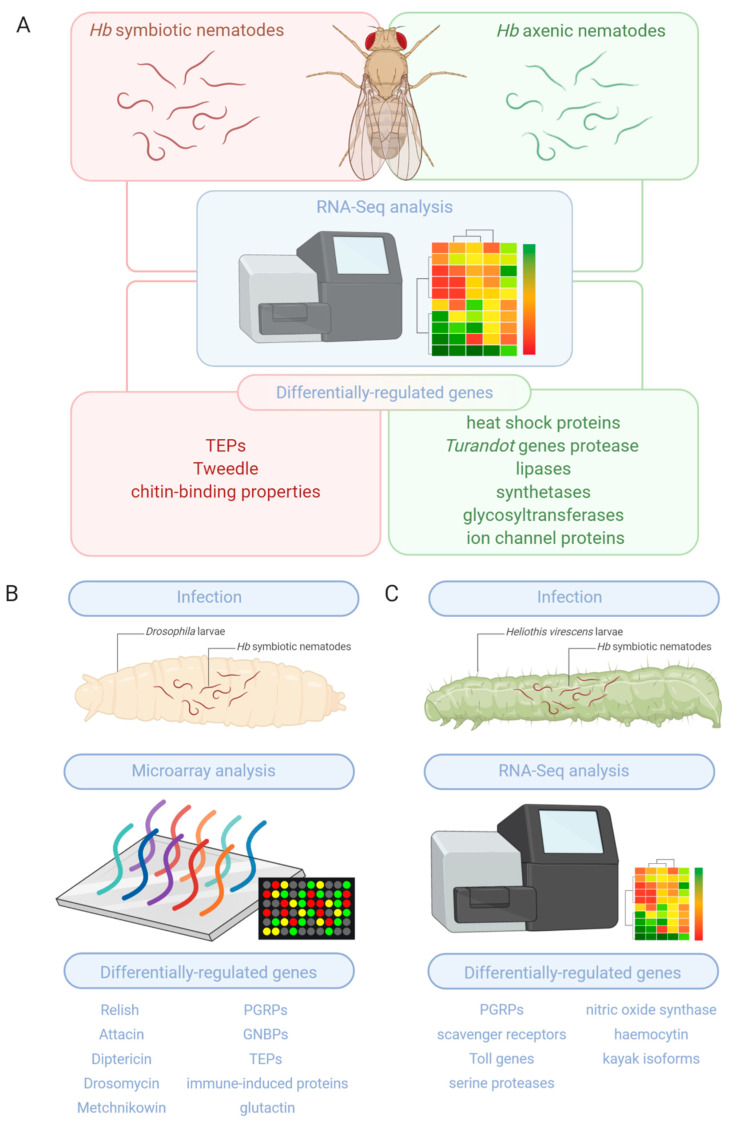
The first transcriptomic examination of the effect of entomopathogenic nematode infection on gene expression in a model insect host was carried out in D. melanogaster [15]. This study employed D. melanogaster larvae and the Affymetrix GeneChip microarray system to determine the number and identity of insect host genes, the expression of which is considerably affected following challenge with Heterorhabditis bacteriophora symbiotic (carrying their associated Photorhabdus luminescens bacteria) nematodes (Figure 1B). The experimental setup was prepared in a way to facilitate the comparison between gene expression changes in uninfected D. melanogaster larvae and those infected with symbiotic H. bacteriophora that had already released their P. luminescens bacteria for at least 6 h after infection. The results from the bioinformatic analyses revealed that 381 D. melanogaster larval transcripts were up-regulated and 104 transcripts were down-regulated exclusively upon H. bacteriophora infection when compared to transcriptomic data from previous studies involving infection with either bacteria or parasitoid wasps. Further inspection of the results produced a list of the most strongly regulated genes encoding D. melanogaster proteins with immune properties [e.g., members of the immune deficiency Imd pathway such as Relish, the antimicrobial peptides Attacin, Diptericin, Drosomycin, Metchnikowin, recognition proteins such as Peptidoglycan Recognition Proteins (PGRP) and Gram-Negative Binding Proteins (GNBP) as well as various immune-induced proteins and thioester containing proteins] [29]. Further, additional genes included those encoding factors that participate in developmental processes and particularly those controlled by the Wnt signaling pathway [30]. Importantly, larvae mutant for the recognition proteins PGRP-LF, GNBP-like 3, the thioester-containing protein 3, and the basement membrane component glutactin exhibited reduced survival ability against H. bacteriophora, indicating their participation in controlling entomopathogenic nematode infection in D. melanogaster.
Figure 1.
Differentially regulated genes of the insect immune response against entomopathogenic nematodes identified by (A) RNA-Seq analysis in D. melanogaster adult flies infected by H. bacteriophora symbiotic or axenic nematodes; (B) Microarray analysis in D. melanogaster larvae infected by H. bacteriophora symbiotic nematodes; (C) RNA-Seq analysis in H. virescens caterpillars infected by H. bacteriophora symbiotic nematodes.
Another investigation examined the transcriptomic response of D. melanogaster adult flies to H. bacteriophora entomopathogenic nematodes [16]. The novelty of this study was that it used for the first time Illumina RNA-Seq technology to interrogate the time-course interaction between symbiotic or axenic (lacking P. luminescens) nematodes on D. melanogaster physiological processes, including the innate immune response (Figure 1A). An important conclusion derived from this work was that H. bacteriohora nematodes, regardless of whether they contain or lack their related mutualistic bacteria, elicit a distinct transcriptomic response in the adult fly compared to infection with P. luminescens bacteria alone. Interestingly, the expression of a large number of fly genes was substantially downregulated upon infection with axenic H. bacteriophora, suggesting that nematodes deficient of their bacteria are capable of modifying several biological processes in the insect host. Infection of adult D. melanogaster with H. bacteriophora, and in particular axenic worms, also altered substantially the expression of Heat shock protein and Turandot (or Tot) genes [31], which indicates elevated levels of stress due to the infection. Similar upregulation was further observed for genes encoding proteins that participate in the regulation of metabolic activities (e.g., protease, lipase, and synthetase enzymes), tissue damage (e.g., glycosyltransferases), and genes involved in nociception (e.g., ion channel proteins). In agreement with the previous transcriptomic study in D. melanogaster larvae [15], H. bacteriophora symbiotic nematode infection altered the expression of Tep genes, which were recently shown to modulate the immune and metabolic response of D. melanogaster flies to Photorhabdus pathogens [32,33,34,35]; however, their role in the response of the fly to entomopathogenic nematodes is still unknown. Another unexplored question in insect anti-nematode immunity is the nature of proteins that participate in the detection of the parasites. This study reported the upregulation of Tweedle genes encoding proteins with chitin-binding properties, which might be involved in nematode recognition in the fly and possibly in other insects.
Transcriptomic responses to entomopathogenic nematodes in model insect hosts have also been expanded to infection by Steinernema carpocapsae, which shows increased pathogenicity towards D. melanogaster indicating potential differences in gene induction levels compared to H. bacteriophora infection [36]. Using an RNA-Seq analysis approach together with a method to generate S. carpocapsae axenic nematodes [37], the transcriptomic response of wild-type D. melanogaster larvae to this entomopathogenic nematode was tested at two time-points to estimate the time-course changes in gene expression pattern during infection [18]. Bioinformatic analysis showed that although the total number of induced genes at 6 and 24 h post infection was similar, symbiotic S. carpocapsae induced slightly higher number of genes than axenic nematodes. Preliminary analysis of these data unveiled that axenic and symbiotic S. carpocapsae are able to regulate the expression of multiple D. melanogaster genes which are shared between the two types of nematode infections or distinct genes which are specific to infection by one type of nematode. Gene Ontology analysis to determine the nature of the molecular pathways and biological activities they control showed that several immunity-related genes were mostly up-regulated in D. melanogaster larvae by symbiotic S. carpocapsae, whereas the expression of genes participating in chitin metabolism or neuroactive signaling interactions was mostly affected by axenic nematodes. With regard to immune genes, several genes in the Toll (e.g., GNBP3, Serpin-27A, and Drosomycin) and Imd (e.g., PGRP-LB, Kenny, Relish, and Attacin) pathways and fewer genes in the Janus kinase (Jak)-signal transducer and activator of transcription (Stat) (Jak/Stat) (e.g., Tot-A) and c-Jun N-terminal kinase (Jnk) (e.g., puckered) pathways as well as genes encoding factors that participate in cellular immune activities (e.g., Tep1, Tep2, and hemese) were differentially affected throughout the infection [5,38,39,40]. Interestingly, infection by axenic S. carpocapsae in particular also altered the expression of developmental genes in the imaginal disc growth factor family and genes regulated by the Notch and Wnt signaling pathways.
Understanding gene expression regulation in a natural insect host that is compromised by entomopathogenic nematode infection is important for designing innovative ways to eliminate noxious insect pests. For this, genome-wide changes in the gene expression profile of the tobacco budworm, Heliothis virescens, were determined during H. bacteriophora infection using Illumina RNA-Seq (Figure 1C) [17]. The approach involved transcriptomic analysis in H. virescens larvae at three stages of the infection process: nematode entry into the insect hemolymph, release of symbiotic P. temperata bacteria, and nematode encapsulation by hemocytes. Processing the sequencing information identified more than 200 transcripts annotated for immune or stress response. In general, the most striking changes in H. virescens gene transcript levels were noted at the nematode invasion and bacterial expulsion. More precisely, a large number of genes was up-regulated during the nematode invasion stage and this pattern was reversed upon release of P. temperata with most differentially regulated genes being down-regulated. When the infection was established, although the total number of genes with altered expression was significantly reduced compared to the previous stages of infection, it was observed that the number of up-regulated genes was 3-times more than those down-regulated. Up-regulated genes during the nematode invasion stage included PGRP and scavenger receptors (e.g., PGRP-2 and Scavenger receptor class b member 3), signaling pathway, and response molecules, as well as serine proteases and nitric oxide synthase, (e.g., Toll, Lebocin-4 precursor, and Serine protease easter-like). Down-regulated genes during the release of bacteria by H. bacteriophora included some recognition genes (e.g., Haemocytin), transcription factors (e.g., kayak isoforms), and serine proteases (e.g., Serine protease snake-like), whereas during a subsequent phase of infection, the expression of most genes remained unchanged (e.g., c-type lectin 11, Zinc finger protein 691-like, Serine protease Persephone-like isoform, Moricin-2 precursor, Chitinase 5 isoform). Of note, orthologs of most differentially regulated genes in H. virescens are also found in the genomes of other insects, including Bombyx mori, D. melanogaster, Tribolium castaneum, Aedes aegypti, Culex quinquefasciatus, Apis mellifera, and Diaphorina citri [17]. This demonstrates the conservation of genes regulated by entomopathogenic nematode infection the tobacco budworm.
3. Mosquito Transcriptomic Response to Filarial Nematodes
Filarial nematodes are important human pathogens because they cause human lymphatic filariasis. They are transmitted by mosquito species in which they develop from microfilariae to infective-stage larvae in approximately one week. Understanding their mode of transmission is paramount for blocking the spread of the parasites to human populations. To achieve this goal, transcriptomic analyses in mosquitoes has already generated important information on the regulation of genes during the development and migration of the nematodes in the infected vector (Table 1).
The mosquito Armigeres subalbatus is a vector of the filarial nematode Brugia malayi and is capable of activating a distinct immune response against this parasite. In order to understand the details of the time-course molecular events that occur during filarial nematode infection and transmission, expressed sequence tags (ESTs) were generated from cDNA libraries, which were constructed from adult female mosquitoes that had previously been infected with either B. malayi through blood feeding or Dirofilaria immitis through injection [19]. Following annotation and Gene Ontology analysis, the results showed that several EST clusters encoded immune-related proteins that were subdivided into several categories based on their function. These sub-categories included Caspases, Lectins, Lysozymes, Prophenoloxidases, Thioester-containing proteins, Imd and Toll pathway members, and other unknown molecules, some of which contain homologous sequences in Anopheles gambiae, Ae. Aegypti, and C. quinquefasciatus mosquitoes as well as in the fruit fly D. melanogaster. This information is critical for future functional genomic studies to resolve vector competence of mosquitoes for filarial nematodes.
In a more detailed study to determine the specific molecular factors with anti-filarial immune properties in mosquito vectors, the time-course transcriptomic profile of A. subalbatus against B. malayi was assessed by adopting a natural exposure assay together with microarray analysis [20]. A curious observation was the limited overlap in the type of differentially regulated transcripts between time points. For example, at the early time points, PGRP encoding genes were mostly up-regulated but as the filarial infection progressed, a decrease in the abundance of C-type lectins was found. Because current information on filarial nematode detection in mosquitoes is limited, the observed variation in pattern recognition regulation warrants further attention. Similarly, several transcripts encoding serine proteases (e.g., Snake-like and Easter-like), which are also found in D. melanogaster and potentially take part in anti-filarial immune activities in mosquitoes, were either up- or down-regulated over the time-course of infection. Simultaneously, transcripts encoding antimicrobial peptides with sequence similarity to those found in A. gambiae (e.g., Cecropin, Defensin, Lysozyme, and Gambicin) were also differentially regulated upon B. malayi infection [13]. A novelty of this study was the detection of changes in several transcripts encoding factors with role in cytotoxic reactions (e.g., glutathione synthetase and transferase, cytochrome P450, and peroxidase), which indicates the activation of pathways responsible for metabolizing reactive intermediates upon B. malayi infection in A. subalbatus. In addition, more than 300 differentially regulated transcripts with no assigned function were described and many of those belong to conserved pathways with a putative immune role. Intriguingly, the type of differentially regulated transcripts in certain mosquito tissues and particularly in hemocytes varied substantially, suggesting a mosquito tissue-specific response against filarial infection.
A similar approach involving DNA microarrays hybridized with RNA probes that were constructed from female mosquitoes was followed to contrast the time-course transcriptome changes in A. subalbatus upon bloodmeal infection with either B. malayi or B. pahangi [21]. Major changes in gene transcript levels in both infection treatments were observed during the first 24 h of infection, which coincides with filarial nematode penetration into the mosquito midgut and invasion of muscle cells in the thorax. Most of the differentially expressed genes had unknown function and only 10% of those genes were related to known or putative immune processes. However, the development of Brugia parasites in A. subalbatus was accompanied by minor transcriptional changes in the mosquito vector and most differentially regulated transcripts encoded factors irrelevant to immune factors. A characteristic difference in transcriptional regulation between the two infection treatments was the increased number of A. subalbatus transcripts encoding reactive metabolites upon infection with B. malayi, but not with B. pahangi nematodes. This observation might imply a specific protective effect against oxidative damage that occurs by the mosquito melanization response to B. malayi parasites.
The transcriptomic response of the mosquito vector to the development of filarial nematodes has further been tested in Ae. aegypti during infection with B. malayi parasites. Using a genome microarray expression method, the global transcriptomic effects of B. malayi on Ae. aegypti were assessed at different stages of parasite development [22]. The results revealed a large variation in the number of induced mosquito genes and their level of induction at various developmental stages of the parasite. Similar to A. subalbatus, several genes with unknown function were downregulated during the early stages of infection, which correspond to penetration of the midgut and thoracic muscle cells. This illustrates the possibility that filarial nematodes have evolved strategies to counteract the mosquito recognition response upon invasion. When the parasites reach the L1 and subsequently the L2 and L3 stages, the direction of gene expression is reversed from down-regulation to up-regulation and the number of up-regulated genes increases as nematode development progresses. Most of the up-regulated genes encode antimicrobial peptides, mainly cecropins, recognition proteins, and signaling components, serine proteases, and melanization factors, as well as genes with putative immune role. This strongly indicates that development and migration of B. malayi in Ae. agypti induces a robust immune response.
To dissect the molecular interrelationship between Ae. aegypti and B. malayi, a dual Illumina RNA-Seq quantitative transcriptome profiling approach was established using tissues from filarial nematode-infected mosquitoes [23]. This study examined the genome-wide temporal gene expression changes that occur at various developmental stages in the parasite and until it reaches the infective stage in the mosquito vector (gene transcription dynamics in filarial nematodes will not be covered in this review). Analysis of the transcriptomic data in the mosquito uncovered two important aspects of the host response to filarial nematode infection. First, B. malayi infection over a 6–7-day period alters substantially the expression profile of genes coding for factors that participate in host metabolic activity (e.g., cAMP biosynthesis process, gluconeogenesis and oxidoreductase activity), suggesting a serious metabolic disturbance in Ae. aegypti due to parasite development. Second, changes in the expression of glutathione transferases and peroxidases inferred that during filarial nematode infection, the mosquito host is able to suspend toxic derivatives of oxygen metabolism, which forms a mechanism of protection against oxidative damage. Curiously, an Ae. aegypti strain with different genetic background was incapable of supporting B. malayi development, although the genetic basis of this mosquito-filarial nematode incompatibility was not further determined.
Transcriptomic studies in mosquito vectors have proved useful for the identification of genomic regions that confer resistance to filarial nematodes. An excellent example is described in a study that examines the genetic basis of Ae. aegypti immune response to B. malayi. [24]. For this, both genomics to identify the Ae. aegypti locus responsible for conferring anti-nematode resistance and Illumina RNA-Seq to analyze gene expression changes in resistant and susceptible mosquitos were used. Initial whole-genome sequencing of an Ae. aegypti population from Kenya located a single dominant locus and a subset of candidate genes that affect susceptibility to B. malayi. Subsequent comparison between the transcriptomic profile of resistant and susceptible Ae. aegypti to filarial nematodes during the first 12 h of infection showed strong similarities in the number of differentially regulated genes in the two mosquito populations. Those differentially regulated genes included genes encoding antimicrobial peptides, recognition proteins such as lectins, signaling components in the Imd, Toll, and Jak/Stat signaling pathways, and serine proteases. A small number of genes involved in metabolism, digestion, nutrient transport, egg production, and translation, as well as genes induced through infection by Wolbachia were also differentially regulated. Crucially, due to suppressive activity by the parasite or physiological impairment in the host at 48 h post B. malayi infection, susceptible mosquitoes were found to down-regulate a group of immune genes encoding antimicrobial peptides and prophenoloxidase. Overall, these results point to the conclusion that resistant Ae. aegypti genotypes exhibit enhanced immune response to B. malayi nematodes.
The notion that filarial nematodes are able to interfere with immune activities in their mosquito vectors has further been supported by transcriptome microarrays in C. quinquefasciatus infected by Wuchereria bancrofti [25]. Meta-analysis of the data showed that W. bancrofti parasites transmitted by C. quinquefasciatus are able to regulate a large number of genes with unknown function and a small number of genes related to immune processes. The latter mainly included genes coding for serine proteases, cuticular proteins, and heat shock proteins. Thus, W. bancrofti infection possibly induces melanization, tissue repair, and stress responses in C. quinquefasciatus; however, this filarial nematode, like Brugia parasites, can develop efficiently in the vector mosquito through evading its immune response.
4. Insect Transcriptomic Response to Infection by Parasitic Nematodes
The transcriptomic response of insects to nematode parasites other than entomopathogenic or filarial nematodes is mostly unexplored. The limited number of investigations have concentrated mostly on Drosophila neotestacea flies infected by Howardula nematodes as well as B. terrestris bumblebees and M. alternatus beetles infected by Sphaerularia and Bursaphelenchus nematodes, respectively (Table 1).
Bumblebees B. terrestris are naturally infected by the widely distributed nematode parasite S. bombi, which confers phenotypic changes to the host, it affects host population dynamics and may enhance the transmission of the parasite. To define the genome-wide transcriptional profile in B. terrestris queens upon parasitism by S. bombi during overwintering diapause and at later lifecycle stages, a quantitative Illumina RNA-Seq approach was followed [26]. The analysis confirmed that S. bombi infection impacts gene expression during B. terrestris diapause, but it has a stronger effect on the transcriptional regulation of its host after diapause. Interestingly, distinct sets of genes were differentially expressed in S. bombi-infected B. terrestris during and after diapause. Differentially expressed genes during diapause were associated with transcriptional and diapause regulation as well as energy metabolism. Those differentially expressed after diapause included genes involved in circadian rhythm, mitochondrial function, and some encoded components of the Toll signaling pathway. Focusing on the transcriptionally affected genes with immune function in B. terrestris, those were mostly induced by S. bombi after the diapause stage. These included genes encoding immune recognition proteins (e.g., scavenger receptor), signaling regulation components (e.g., domeless), effector molecules (e.g., thioester-containing proteins), and genes with role in phagocytosis (e.g., draper). The outcome from this research suggests that immune activity is modified in B. terrestris upon infection with the nematode S. bombi and a future challenge will be to determine whether the observed gene expression variation in the infected bumblebees is caused by the parasite per se or it is the result of side effects due to gene dysregulation in the host.
Certain invasive parasitic nematodes are vectored by insects in order to gain access to plants and trees. For example, pine wilt is a disease of pine caused by the pinewood nematode, B. xylophilus. The pinewood nematode, which is vectored by the pine sawyer beetle M. alternatus, is native to North America and is not considered a primary pathogen of native pines but is the cause of pine wilt in some non-native species [41]. Using global transcriptome information from previous studies that analyzed the response of M. alternatus to heat stress and pesticides [27,42], a recent comparative study expanded these findings by probing immune-related gene transcript changes in M. alternatus infected by B. xylophilus parasites [43]. Using high-throughput Illumina RNA-Seq, it was thoroughly demonstrated that the presence of B. xylophilus in M. alternatus adults alters the expression profile of a large number of genes with known or putative immune function. Most of these genes were highly conserved in the red flour beetle T. castaneum and also showed lower conservation in the mosquitoes Ae. aegypti and A. gambiae. The conserved genes encoded pathogen recognition proteins (e.g., PGRP, β-glucan recognition proteins, thioester, containing proteins), extracellular signal modulation molecules (e.g., serine proteases and serine protease inhibitors), intracellular signal pathway components (e.g., Toll-like receptors, Spaetzle, MyD88, Relish, Domeless), and various effectors (antimicrobial peptides and prophenoloxidases). This information is pivotal for tackling ecological issues on the association between insects and their natural nematode parasites.
An area of particular interest in the field of insect immunity involves the protective role of endosymbionts against pathogenic infection [44]. With regard to insect parasitic nematodes, Spiroplasma endosymbionts have been shown to provide protection to the mushroom-feeding fruit fly Drosophila neotestacea upon infection with the natural nematode parasite H. aoronymphium [45]. In order to determine the molecular basis of the Spiroplasma-modulated priming effect, whole transcriptome Illumina RNA-Seq was performed in D. neotestacea flies to identify enhanced immune gene activity that would possibly account for elevated immune response against H. aoronymphium infection [28]. Nearly 700 differentially regulated transcripts were detected with more than 300 being up-regulated in response to nematode exposure rather than to Spiroplasma infection. Gene Ontology analysis of the immunity-related genes demonstrated that the up-regulated transcripts included some clotting factors (e.g., fondue), lectins and proteases, fibrinogen-like domain-containing proteins and molecules regulating chitin metabolism, and a single isoform of the antimicrobial peptide-encoding gene defensin. The up-regulated transcripts with Gene Ontology terms for immune response, innate immunity or response to stress were highly conserved in D. melanogaster [46]. A few detected down-regulated genes were not related to immune function and mostly encoded proteins participating in translation, cell proliferation, and egg development. Finally, D. neotestacea flies contain Spiroplasma, a small number of immune genes encoding proteases, and a few prophenoloxidases. Improving defensive symbiosis in insects is an attractive alternative for combating parasitic nematode infection and the information gained from this investigation contributes towards this direction.
5. Conclusions and Future Directions
Interactions between nematodes and insects are very common in nature. Nematodes infect a variety of insect species in order to gain access to necessary resources that will allow them to produce their progeny before they move to another suitable host. In addition, nematodes exploit insects for their dispersal and transmission to mammalian hosts where they cause serious infectious diseases. In the case of entomopathogenic nematodes which carry their own species-specific mutualistic bacteria, the interactions implicating the two pathogens and the insect host physiological response are complex and dynamic. To define the kind of interactions that take place at different stages of the infection process and determine whether these interactions impact the insect immune response, the use of molecular and functional techniques has generated exciting information that help us understand the mechanisms involved. An important strategy involves the application of transcriptomic approaches, which have started to decipher the identity of genes in the insect host that participate in the host defense against nematode parasites. Although transcriptomics may have certain limitations such as low resolution, sensitivity and specificity, consideration of partial transcript structure for gene expression, and identification of already recognized genes [47], the use of transcriptomics has revolutionized the field of insect-nematode immunity, because it has started to reveal candidate molecules, the functional role of which can be further confirmed in mechanistic assays. Additionally, a combination of proteomic approaches and systems biology strategies will undoubtedly circumvent the main deficit of transcriptomics, that mRNA transcript levels do not directly correspond to protein levels or function [48]. Future approaches are expected to deploy a combination of transcriptomics and genetic engineering techniques, such as CRISPR gene editing, to unravel the precise role of candidate genes and their products in the interaction of insects with parasitic nematodes.
Acknowledgments
We thank members of the Department of Biological Sciences at The George Washington University for critical reading of the manuscript.
Author Contributions
I.E.; Writing—Original draft preparation, C.H.; Writing—Review, editing and preparation of the figure. All authors have read and agreed to the published version of the manuscript.
Funding
Research on Drosophila transcriptomics was supported by a start-up fund from the Columbian College of Arts and Sciences at The George Washington University to I.E.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Castillo J.C., Reynolds S.E., Eleftherianos I. Insect immune responses to nematode parasites. Trends Parasitol. 2011;27:537–547. doi: 10.1016/j.pt.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Wang C., Ren M., Liu X., Xia H., Chen K. Peptidoglycan recognition proteins in insect immunity. Mol. Immunol. 2019;106:69–76. doi: 10.1016/j.molimm.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 3.Hillyer J.F. Insect immunology and hematopoiesis. Dev. Comp. Immunol. 2016;58:102–118. doi: 10.1016/j.dci.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark K.D. Insect Hemolymph Immune Complexes. Subcell. Biochem. 2020;94:123–161. doi: 10.1007/978-3-030-41769-7_5. [DOI] [PubMed] [Google Scholar]
- 5.Sheehan G., Garvey A., Croke M., Kavanagh K. Innate humoral immune defences in mammals and insects: The same, with differences? Virulence. 2018;9:1625–1639. doi: 10.1080/21505594.2018.1526531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vlisidou I., Wood W. Drosophila blood cells and their role in immune responses. FEBS J. 2015;282:1368–1382. doi: 10.1111/febs.13235. [DOI] [PubMed] [Google Scholar]
- 7.Wojda I., Staniec B., Sułek M., Kordaczuk J. The greater wax moth Galleria mellonella: Biology and use in immune studies. Pathog. Dis. 2020;78:ftaa057. doi: 10.1093/femspd/ftaa057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar A., Srivastava P., Sirisena P., Dubey S.K., Kumar R., Shrinet J., Sunil S. Mosquito Innate Immunity. Insects. 2018;9:95. doi: 10.3390/insects9030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchon N., Silverman N., Cherry S. Immunity in Drosophila melanogaster—From microbial recognition to whole-organism physiology. Nat. Rev. Immunol. 2014;14:796–810. doi: 10.1038/nri3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Getanjaly V.L.R., Sharma P., Kushwaha R. Beneficial insects and their value to agriculture. Res. J. Agric. Forest. Sci. 2015;3:25–30. [Google Scholar]
- 11.Cooper D., Eleftherianos I. Parasitic Nematode Immunomodulatory Strategies: Recent Advances and Perspectives. Pathogens. 2016;5:58. doi: 10.3390/pathogens5030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Labaude S., Griffith C.T. Transmission Success of Entomopathogenic Nematodes Used in Pest Control. Insects. 2018;9:72. doi: 10.3390/insects9020072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Famakinde D.O. Mosquitoes and the Lymphatic Filarial Parasites: Research Trends and Budding Roadmaps to Future Disease Eradication. Trop. Med. Infect. Dis. 2018;3:4. doi: 10.3390/tropicalmed3010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perlman S.J., Spicer G.S., Shoemaker D.D., Jaenike J. Associations between mycophagous Drosophila and their Howardula nematode parasites: A worldwide phylogenetic shuffle. Mol. Ecol. 2003;12:237–249. doi: 10.1046/j.1365-294X.2003.01721.x. [DOI] [PubMed] [Google Scholar]
- 15.Arefin B., Kucerova L., Dobes P., Markus R., Strnad H., Wang Z., Hyrsl P., Zurovec M., Theopold U. Genome-wide transcriptional analysis of Drosophila larvae infected by entomopathogenic nematodes shows involvement of complement, recognition and extracellular matrix proteins. J. Innate Immun. 2014;6:192–204. doi: 10.1159/000353734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castillo J.C., Creasy T., Kumari P., Shetty A., Shokal U., Tallon L.J., Eleftherianos I. Drosophila anti-nematode and antibacterial regulators revealed by RNA-Seq. BMC Genom. 2015;16:519. doi: 10.1186/s12864-015-1690-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.An R., Suri K.S., Jurat-Fuentes J.L., Grewal P.S. Dynamics of transcriptomic response to infection by the nematode Heterorhabditis bacteriophora and its bacterial symbiont Photorhabdus temperata in Heliothis virescens larvae. Insect Mol. Biol. 2017;26:584–600. doi: 10.1111/imb.12321. [DOI] [PubMed] [Google Scholar]
- 18.Yadav S., Daugherty S., Shetty A.C., Eleftherianos I. RNAseq Analysis of the Drosophila Response to the Entomopathogenic Nematode Steinernema. G3. 2017;7:1955–1967. doi: 10.1534/g3.117.041004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayhew G.F., Bartholomay L.C., Kou H.-Y., Rocheleau T.A., Fuchs J.F., Aliota M.T., Tsao I.-Y., Huang C.-Y., Liu T.-T., Hsiao K.-J., et al. Construction of an expressed sequences tag library for the mosquito vector Armigeres subalbatus. BMC Genom. 2007;8:462. doi: 10.1186/1471-2164-8-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aliota M.T., Fuchs J.F., Mayhew G.F., Che C.-C., Christensen B.M. Mosquito transcriptome changes and filarial worm resistance in Armigeres subalbatus. BMC Genom. 2007;8:463. doi: 10.1186/1471-2164-8-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aliota M.T., Fuchs J.F., Rocheleau T.A., Clark A.K., Hillyer J.F., Chen C.-C., Christensen B.M. Mosquito transcriptome profiles and filarial worm susceptibility in Armigeres subalbatus. PLoS Negl. Trop. Dis. 2010;4:e666. doi: 10.1371/journal.pntd.0000666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erickson S.M., Xi Z., Mayhew G.F., Ramirez J.L., Aliota M.T., Christensen B.M., Dimopoulos G. Mosquito infection responses to developing filarial worms. PLoS Negl. Trop. Dis. 2009;3:e529. doi: 10.1371/journal.pntd.0000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi Y.-J., Aliota M.T., Mayhew G.F., Erickson S.M., Christensen B.M. Dual RNA-seq of parasite and host reveals gene expression dynamics during filarial worm-mosquito interactions. PLoS Negl. Trop. Dis. 2014;8:e2905. doi: 10.1371/journal.pntd.0002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juneja P., Ariani C.V., Ho Y.S., Akorli J., Palmer W.J., Pain A., Jiggins F.M. Exome and transcriptome sequencing of Aedes aegypti identifies a locus that confers resistance to Brugia malayi and alters the immune response. PLoS Pathog. 2015;11:e1004765. doi: 10.1371/journal.ppat.1004765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartholomay L.C., Waterhouse R.M., Mayhew G.F., Campbell C.L., Michel K., Zou Z., Ramirez J.L., Das S., Alvarez K., Arensburger P., et al. Pathogenomics of Culex quinquefasciatus and meta-analysis of infection responses to diverse pathogens. Science. 2010;330:88–90. doi: 10.1126/science.1193162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colgan T.J., Carolan J.C., Sumner S., Blaxter M.L., Brown M.J.F. Infection by the castrating parasitic nematode Sphaerularia bombi changes gene expression in Bombus terrestris bumblebee queens. Insect Mol. Biol. 2019;29:170–182. doi: 10.1111/imb.12618. [DOI] [PubMed] [Google Scholar]
- 27.Li H., Zhao X., Qiao H., He X., Tan J., Hao D. Comparative Transcriptome Analysis of the Heat Stress Response in Monochamus alternatus Hope (Coleoptera: Cerambycidae) Front. Physiol. 2020;10:1568. doi: 10.3389/fphys.2019.01568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamilton P.T., Leong J.S., Koop B.F., Perlman S.J. Transcriptional responses in a Drosophila defensive symbiosis. Mol. Ecol. 2014;23:1558–1570. doi: 10.1111/mec.12603. [DOI] [PubMed] [Google Scholar]
- 29.Lemaitre B., Hoffmann J. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 30.Bejsovec A. Wingless/Wnt signaling in Drosophila: The pattern and the pathway. Mol. Reprod. Dev. 2013;80:882–894. doi: 10.1002/mrd.22228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ekengren S., Hultmark D. A family of Turandot-related genes in the humoral stress response of Drosophila. Biochem. Biophys. Res. Commun. 2001;284:998–1003. doi: 10.1006/bbrc.2001.5067. [DOI] [PubMed] [Google Scholar]
- 32.Shokal U., Kopydlowski H., Harsh S., Eleftherianos I. Thioester-Containing Proteins 2 and 4 Affect the Metabolic Activity and Inflammation Response in Drosophila. Infect. Immuni. 2018;86:e00810-17. doi: 10.1128/IAI.00810-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shokal U., Eleftherianos I. The Drosophila Thioester containing Protein-4 participates in the induction of the cellular immune response to the pathogen Photorhabdus. Dev. Comp. Immunol. 2017;76:200–208. doi: 10.1016/j.dci.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 34.Shokal U., Kopydlowski H., Eleftherianos I. The distinct function of Tep2 and Tep6 in the immune defense of Drosophila melanogaster against the pathogen Photorhabdus. Virulence. 2017;12:1–15. doi: 10.1080/21505594.2017.1330240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shokal U., Eleftherianos I. Thioester-Containing Protein-4 Regulates the Drosophila Immune Signaling and Function against the Pathogen Photorhabdus. J. Innate Immun. 2017;9:83–93. doi: 10.1159/000450610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peña J.M., Carillo M.A., Hallem E.A. Variation in the susceptibility of Drosophila to different entomopathogenic nematodes. Infect. Immun. 2015;83:1130–1138. doi: 10.1128/IAI.02740-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yadav S., Shokal U., Forst S., Eleftherianos I. An improved method for generating axenic entomopathogenic nematodes. BMC Res. Notes. 2015;8:461. doi: 10.1186/s13104-015-1443-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myllymäki H., Rämet M. JAK/STAT pathway in Drosophila immunity. Scand. J. Immunol. 2014;79:377–385. doi: 10.1111/sji.12170. [DOI] [PubMed] [Google Scholar]
- 39.Tafesh-Edwards G., Eleftherianos I. JNK signaling in Drosophila immunity and homeostasis. Immunol. Lett. 2020;226:7–11. doi: 10.1016/j.imlet.2020.06.017. [DOI] [PubMed] [Google Scholar]
- 40.Gold K.S., Brückner K. Macrophages and cellular immunity in Drosophila melanogaster. Semin. Immunol. 2015;27:357–368. doi: 10.1016/j.smim.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao L., Mota M., Vieira P., Butcher R.A., Sun J. Interspecific communication between pinewood nematode, its insect vector, and associated microbes. Trends Parasitol. 2014;30:299–308. doi: 10.1016/j.pt.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 42.Wu S., Zhu X., Liu Z., Shao E., Carballar-Lejarazú R., Guo Y., Xiong Y., Mou Y., Xu R., Hu X. Identification of Genes Relevant to Pesticides and Biology from Global Transcriptome Data of Monochamus alternatus Hope (Coleptera: Cerambycidae) Larvae. PLoS ONE. 2016;11:e0147855. doi: 10.1371/journal.pone.0147855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou J., Yu H.-Y., Zhang W., Ahmad F., Hu S.-N., Zhao L.-L., Zou Z., Sun J.-H. Comparative analysis of the Monochamus alternatus immune system. Insect Sci. 2018;25:581–603. doi: 10.1111/1744-7917.12453. [DOI] [PubMed] [Google Scholar]
- 44.Eleftherianos I., Atri J., Accetta J., Castillo J.C. Endosymbiotic bacteria in insects: Guardians of the immune system? Front. Physiol. 2013;4:46. doi: 10.3389/fphys.2013.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jaenike J., Unckless R., Cockburn S.N., Boelio L.M., Perlman S.J. Adaptation via symbiosis: Recent spread of a Drosophila defensive symbiont. Science. 2010;329:212–215. doi: 10.1126/science.1188235. [DOI] [PubMed] [Google Scholar]
- 46.Hanson M.A., Hamilton P.T., Perlman S.J. Immune genes and divergent antimicrobial peptides in flies of the subgenus Drosophila. BMC Evol. Biol. 2016;16:228. doi: 10.1186/s12862-016-0805-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z., Gerstein M., Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feder M.E., Walser J.-C. The biological limitations of transcriptomics in elucidating stress and stress responses. J. Evol. Biol. 2005;18:901–910. doi: 10.1111/j.1420-9101.2005.00921.x. [DOI] [PubMed] [Google Scholar]