Abstract
The Nanfengmiju (Citrus reticulata cv. Nanfengmiju), a high-quality local variety of mandarin, is one of the major fruit crops in Jiangxi Province, China. Citrus melanose and stem-end rot, two common fungal diseases of Nanfengmiju, are both caused by Diaporthe spp. (syn. Phomopsis spp.). Identification of the Diaporthe species is essential for epidemiological studies, quarantine measures, and management of diseases caused by these fungi. Melanose disease was observed on Nanfengmiju fruit in Jiangxi Province of China in 2016. Based on morphological characterization and multi-locus phylogenetic analyses, three out of 39 isolates from diseased samples were identified as D. passifloricola. Since these three isolates did not cause melanose on citrus fruit in the pathogenicity tests, they were presumed to be endophytic fungi present in the diseased tissues. However, our results indicate that D. passifloricola may persist as a symptom-less endophyte in the peel of citrus fruit, yet it may cause stem-end if it invades the stem end during fruit storage. To the best of our knowledge, this is the first report of D. passifloricola as the causal agent of the stem-end rot disease in Citrus reticulata cv. Nanfengmiju.
Keywords: citrus, Diaporthe passifloricola, morphological characterization, multi-locus phylogenetic analyses
1. Introduction
As the earliest citrus producer in the world, China has over 4000 years of history of citrus cultivation. The citrus industry of China covers more than 20 provinces [1]. Recently, the cultivation area reached 2.5 million ha, and the production was about 38 million tons [2]. Melanose, one of the most common fungal diseases of citrus worldwide [3,4], generally occurs in many citrus-growing regions of China, such as Chongqing, Fujian, Guangdong, Guangxi, Hunan, Jiangxi, Shaanxi, Shanghai, Zhejiang, and so on [5,6,7]. All commercial citrus varieties are susceptible to melanose. Typical symptoms of melanose disease are small, discrete, sunken spots with a yellowish, reddish-brown to black color. Symptoms begin as tiny pustular lesions, then, pustular lesions disappear and become hardened gummed areas with a sandpaper-like surface [3,8,9]. Diaporthe spp. (syn. Phomopsis) are the causal agents of melanose and can also cause stem-end rots on fruit during the storage period. Since 95% of citrus is consumed as fresh fruit in China, melanose and stem-end rots diseases reduce the economic value of this crop seriously.
At present, Diaporthe citri is the only known causal agent of citrus melanose disease in the world. The species was first found as the causal agent of stem-end rot of citrus fruit in Florida, USA [10]. After that, D. citri was also associated with melanose of citrus fruit, leaves, and shoots and gummosis of perennial branches worldwide [11,12,13,14]. All Citrus species are susceptible to it [4]. In China, D. citri has been isolated in many citrus growing regions, including Guangxi [15], Guangdong [16], Fujian [17], Jiangxi [18], Sichuan [19], Taiwan, Guizhou, Yunnan, Hubei, Jiangsu [20], Zhejiang, and Shanghai [5]. In addition to D. citri, D. citriasiana, and D. citrichinensis have also been found to be pathogens of stem-end rot of citrus fruit in China. D. citriasiana distributes in Shaanxi and Jiangxi Provinces, China. D. citrichinensis is only found in Shaanxi Province, China [5].
The genus Diaporthe, belonging to the Diaporthaceae, Diaporthales, Ascomycota, shows high species diversity. Many species are harmful plant pathogens and exhibit broad host ranges [21,22,23,24,25]. A single species of Diaporthe is commonly associated with different hosts, while a single host may be infected by multiple species of Diaporthe [26,27]. Up to now, over 1020 names “Diaporthe” and around 950 names of the asexual morph “Phomopsis” are recorded in MycoBank lists (accessed July, 2020; http://www.mycobank.org), of which more than 100 Diaporthe or Phomopsis species have been reported in China [5,6,28,29,30,31,32,33]. In the past, morphological characteristics and host associations were the basis of the identification of Diaporthe species. The typical morphological characteristics of Diaporthe spp. are immersed ascomata and erumpent pseudostroma with elongated perithecial necks for the sexual morph [34] and black conidiomata with dimorphic conidia (alpha and beta conidia) for the asexual morph [35]. In some species, there are intermediates between alpha and beta conidia named gamma conidia [36]. However, morphological traits tend to vary in response to changes in environmental conditions, thus they may not be sufficiently reliable for the identification of Diaporthe at the species level [37]. With the development of molecular identification, multi-locus phylogenies combined with morphological characterization have been developed to identify Diaporthe species [21,24,30,37,38]. Nuclear ribosomal internal transcribed spacer regions (ITS), beta-tubulin gene (TUB), translation elongation factor 1-α gene (TEF), histone-3 gene (HIS), and calmodulin gene (CAL) are commonly employed markers to identify Diaporthe species [21,31,37,38].
The Nanfengmiju (Citrus reticulata cv. Nanfengmiju), a high-quality local variety of mandarin, is one of the major fruit crops in Jiangxi Province. The accumulation of dead citrus wood results in the increase of fungal inocula in orchards of Jiangxi. Currently, melanose has become the major fungal disease of Nanfengmiju, immensely reducing the commercial value of citrus production. The identification of Diaporthe spp. is essential for the epidemiology, quarantine measure, and management of citrus melanose and stem-end rot diseases. In this study, morphology, and sequences of five loci (ITS, TUB, TEF, HIS, and CAL) were employed to identify and characterize Diaporthe species on citrus fruit.
2. Results
2.1. Morphological Characterization of D. passifloricola
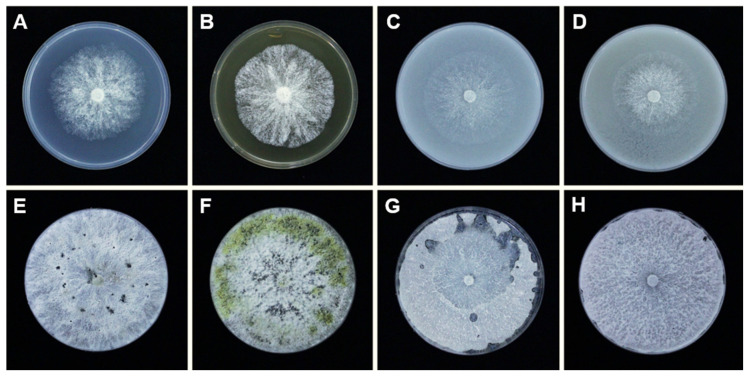
Thirty-nine isolates (Supplementary Figure S1), were obtained from 10 diseased citrus fruit with typical melanose symptoms. Of these, three isolates preliminarily identified as D. passifloricola with the ITS marker were designated as NFIF-3-11, NFIF-3-19, and NFIF-3-21, and sorted out for further study. All three isolates showed the same culture characteristics on four kinds of media. After three days of incubation, the diameter of colonies on potato dextrose agar (PDA), malt extract agar (MEA), corn meal agar (CMA), and oatmeal agar (OMA) media reached 53–69 mm ( = 60), 51–63 mm ( = 57), 43–56 mm ( = 51), and 44–51 mm ( = 49), respectively. The colonies were fluffy with smooth margins. After 30 days of incubation, the surface of colonies on PDA, CMA, and OMA media had a uniform whitish appearance, whereas the colony grown on MEA presented yellowish patches (Figure 1).
Figure 1.
The cultural characteristics of Diaporthe passifloricola (NFIF-3-21) on different media. The isolate was incubated at 25 °C in the dark. (A,E), PDA medium, (B,F), MEA medium, (C,G), CMA medium, (D,H), OMA medium. Note: A–D, Colonies after 3 days incubation, E–H, Colonies after 30 days incubation.
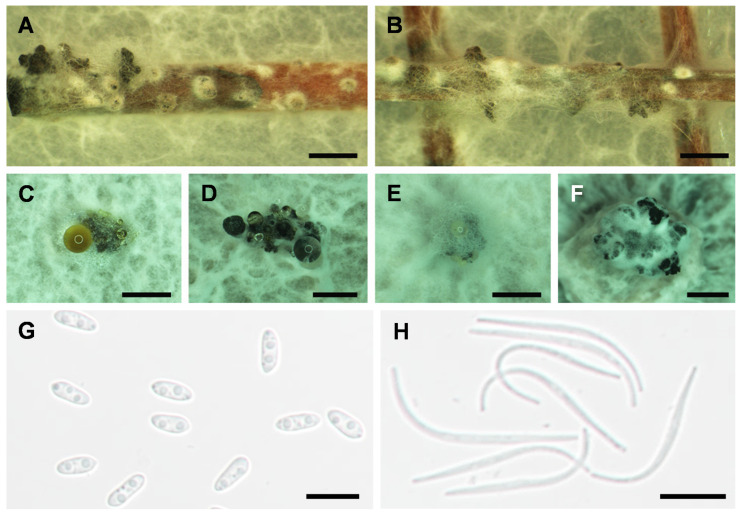
Sporulation was induced on PDA and 1/10 PDA medium supplemented with sterilized pine needles (PNA). Conidiomata (pycnidia) were solitary to aggregated, black, sub-globose to globose, up to 200 µm in diameter. Conidial masses were hyaline to creamy, yellowish. Conidial droplets were exuded from central ostioles. Pycnidial walls consisted of 3–6 layers, medium brown (Figure 2). All three isolates produced dimorphic conidia. Alpha (α) conidia were (6.9–) 7.2–8 (–8.2) µm × 3.1–4.1 µm (x = 7.6 × 3.6 µm², n = 30), aseptate, bi-guttulate, hyaline, fusoid, and ellipsoid, smooth, apex subrounded to rounded, base subtruncate to truncate. Beta (β) conidia were (22.3–) 23.7–26.6 (–27.9) µm × 1–2 µm (x = 25.1 × 1.5 µm², n = 30), aseptate, slightly curved to spindle-shaped, smooth, base truncate. Gamma (C) conidia were not observed.
Figure 2.
Asexual reproduction of Diaporthe passifloricola (NFIF-3-21). (A,B), conidiomata on PNA after 30 days incubation, (C–F), conidiomata on PDA after 30 days incubation, (G), alpha (α) conidia, (H), beta (β) conidia. Scale bars: A–B, 500 µm; C–F, 200 µm; G–H, 10 µm.
2.2. Pathogenicity Test
In pathogenicity tests, non-wounded Nanfengmiju fruit were used to test the ability of three isolates to cause citrus melanose and stem-end rot diseases. At 15 days after inducing melanose symptom, three isolates of NFIF-3-11, NFIF-3-19, and NFIF-3-21 did not cause any symptoms, while the positive control D. citri strain caused typical reddish-brown to black lesion spots symptoms (Figure 3B). On the contrary, all the fruit inoculated with conidial suspension of isolates NFIF-3-11, NFIF-3-19, and NFIF-3-21, as well as positive control fruit inoculated with D. citri strain showed typical rot symptoms at 7 days after inoculation. No significant symptom was observed on negative control fruit inoculated with sterile water (Figure 3C). Re-isolation was performed following Koch’s postulation method. The strains were re-isolated from the experimentally inoculated fruit with stem-end rot symptoms. The identity of the re-isolated strains was confirmed by amplification and sequencing of ITS, TUB, TEF, HIS, and CAL molecular markers.
Figure 3.
(A) Nanfengmiju fruit from Jiangxi Province showing symptoms of melanose. (B) pathogenicity stimulating melanose symptoms on mandarin fruit. For each strain, 300 μL of conidial suspensions is dropped on a piece of cotton, and then placed on the bottom of the fruit. The inoculated fruit are placed in a plastic chamber maintain 95% relative humidity, incubated at 25 °C 12 h of light and 12 h of dark for 15 days. (C) pathogenicity stimulating stem-end rot symptoms on stem-end of mandarin fruit. The stems of citrus fruit are removed carefully, and 10 μL of conidial suspension of each strain is dropped there and incubated at 25 °C in the dark for 7 days. Note: B and C, from left to right are sterile water, conidial suspensions of D. citri (isolate NFHF-8-4) and conidia suspensions of D. passifloricola (isolate NFIF-3-21), respectively.
2.3. Phylogenetic Analyses
For preliminary identification, the MegaBlast search was performed for ITS region of three isolates in NCBI’s GenBank nucleotide database. All three isolates (NFIF-3-11, NFIF-3-19, and NFIF-3-21) showed 100% identity to Diaporthe ueckerae (KY565426) and Phomopsis sp. (KX510126, XP677503, KM229696, FJ233186, and GU595054), 99% identity to D. phaseolorum (LC360110), D. longicolla (KF577903), D. ueckerae (KY565424, KY565425), and D. passifloricola (NR_147595).
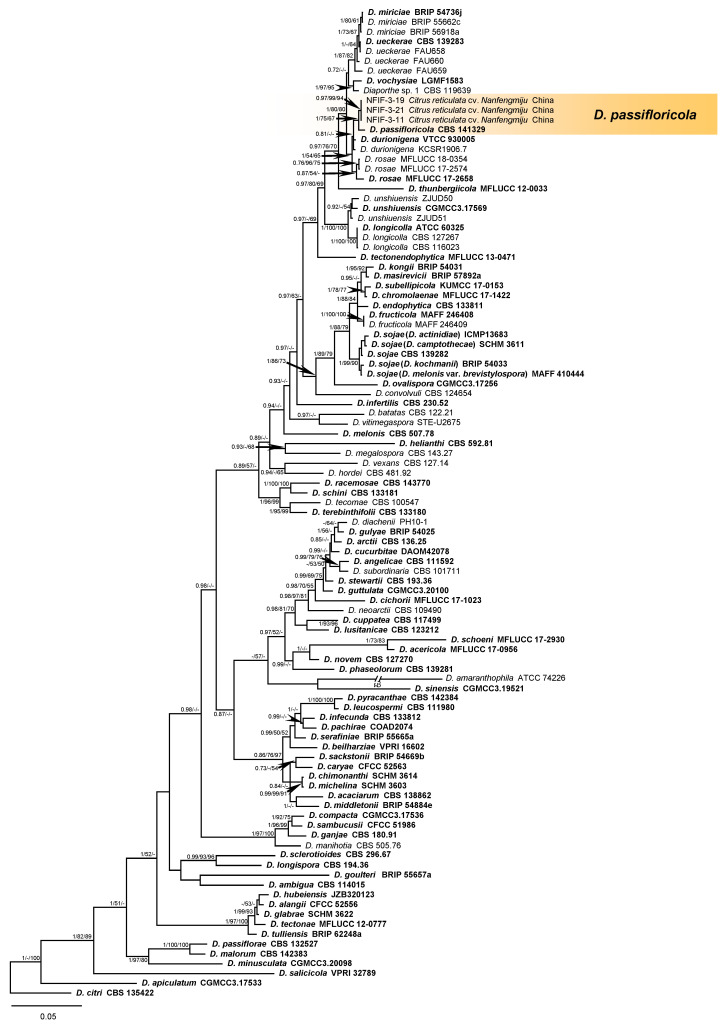
Multi-locus phylogenetic analyses were carried out based on the sequences of ITS, TUB, TEF, HIS, and CAL. To verify if these five loci were congruent and could be combined together, single locus analysis was also performed for each locus. The results indicated that the topology of single-locus trees was congruent (Supplementary Figures S2–S6). Fifteen new sequences were generated from three isolates in this study. Other published sequences of Diaporthe spp. were downloaded from GenBank database. In total, 2738 characters of 101 strains from 80 Diaporthe spp., including one outgroup species D. citri (CBS 135422), were employed for Bayesian Inference (BI), Maximum Likelihood (ML), and Maximum Parsimony (MP) analyses to construct phylogenetic tree. The dataset consisted of 611 characters of ITS (1–611), 868 characters of TUB (612–1479), 527 characters of TEF (1480–2006), 581 characters of HIS (2007–2587), and 578 characters of CAL (2588–3165), respectively. MP analyses of combined data generated a single most parsimonious tree (tree length (TL) = 5416, consistency index (CI) = 0.449, retention index (RI) = 0.739, rescaled consistency index (RC) = 0.332, and homoplasy index (HI) = 0.551). Of the 3165 analyzed characters, 1036 characters were parsimony-informative, 431 variable characters were parsimony uninformative, and 1698 characters were constant. Data of each region/loci were shown in Supplementary Table S1. Using the best scoring RA×ML analysis, a final optimization tree with a likelihood value of −30,716.492582 was generated. The matrix data had 1837 distinct alignment patterns in the ML analysis, with 39.30% of gaps and completely undetermined characters. Estimated base frequencies were as follows: A = 0.212443, C = 0.325722, G = 0.238041, T = 0.223795, with substitution rates AC = 1.252910, AG = 4.007552, AT = 1.250610, C = 1.175745, CT = 5.302300, GT = 1.000000. The gamma distribution shape parameter alpha = 0.938818 and the TL = 6.170537. The ML and MP tree of combined data had similar topology to BI tree. The posterior probabilities (PP) values calculated from BI, bootstrap support (BS) values calculated from ML and MP analyses were plotted in Figure 4 and Supplementary Figure S7. The combined loci analyses grouped three isolates (NFIF-3-11, NFIF-3-19, and NFIF-3-21) together with 0.97 of Bayesian posterior probabilities values (BIPP), 99% of Maximum likelihood bootstrap values (MLBS), and 94% of Maximum parsimony bootstrap values (MPBS), respectively. The isolates were classified as D. passifloricola with 1 of BIPP, 75% of MLBS, and 67% of MPBS, and distinct from D. durionigena, D. rosae, D. miriciae, and D. ueckerae. The analysis of polymorphic nucleotides in each locus of D. passifloricola, D. durionigene, and D. rosae also found 11, 4, 4, and 11 polymorphic nucleotides in ITS, TUB, TEF, and CAL, respectively (Supplementary Table S2). While there was no polymorphic nucleotide in HIS sequence of three species.
Figure 4.
Bayesian inference phylogenetic tree is generated from the analysis of the combined sequences of five loci (ITS, TUB, TEF, HIS, and CAL). Posterior probabilities support values ≥0.7 and Bootstrap support values ≥50%, Bayesian posterior probabilities values (BIPP)/ Maximum likelihood bootstrap values (MLBS)/ Maximum parsimony bootstrap values (MPBS) are displayed at the nodes. The tree is rooted with D. citri CBS 135422. Ex-type, ex-epitype, and holotype cultures are indicated in bold. The codes of isolates used for phylogenetic tree are given.
Materials examined: CHINA, Jiangxi Province, Fuzhou city, Nanfeng district, on fruit of Citrus reticulata cv. Nanfengmiju, August 2016, C. Chaisiri (living culture: CCTCC M 2020452 = NFIF-3-21).
3. Discussion
Diaporthe passifloricola was identified from leaf spots on Passiflora foetida in Malaysia [39]. The colonies of this species on MEA, OA, and PDA are dirty white. Alpha conidia are aseptate, hyaline, smooth, guttulate, fusoid-ellipsoid, tapering towards both ends, apex subobtuse, base subtruncate, (5–) 6–7 (–9) × 2.5 (–3) µm. Gamma conidia are not observed. Beta conidia are spindle shaped, aseptate, smooth, hyaline, apex acutely rounded, base truncate, tapering from lower third towards apex, curved, (20–) 22–25 (–27) × 1.5 (–2) µm. In this study, the colonies of the isolates on PDA were dirty white, which are similar to those of D. passifloricola [39], D. durionigena [40], D. rosae [41], and D. ueckerae [42], while that of D. miriciae is buff [23]. Morphological characteristics of alpha (bi-guttulate) and beta conidia of three isolates are consistent with those of D. passifloricola ex-type strain (CBS 141329) [39]. The sizes of alpha and beta conidia of three isolates are larger than those of D. durionigena [40] and D. rosae [41]. The alpha conidia of D. miriciae are not described of guttulate characterized [23], and the beta conidia of D. ueckerae are not observed in a previous study [42]. Thus, morphological characteristics of the three isolates are the most consistent with those of D. passifloricola. Taking into account that morphological characteristics sometimes vary with environmental conditions, they are not always reliable to identify the isolates to species level in genus of Diaporthe [37]. Thus, further molecular identification is necessary.
The sequence of the ITS region was once used alone to identify Diaporthe species. However, there are many intraspecific variations in ITS locus of certain Diaporthe species. Sometimes the intraspecific variation is even greater than interspecific variation, which makes it difficult to identify Diaporthe species with ITS sequence alone [43,44]. Currently, multi-locus phylogenetic analyses have been applied for the identification of Diaporthe species [37,45]. Thus, although ITS sequences of all three isolates showed 100% similarity with D. ueckerae (KY565426) in this study, it was unreliable, due to many intraspecific variations in ITS regions of Diaporthe species.
The combined use of the five loci (i.e., ITS, TUB, TEF, HIS, and CAL) is shown to be the best way to generate a phylogenetic tree to determine the boundaries of Diaporthe spp. [21,31,33,37,38,45]. After preliminary identification with ITS locus, four species of D. passifloricola, D. rosae, D. ueckerae, and D. miriciae were found to have high identity to the three isolates obtained in this study. Thus, five loci of ITS, TUB, TEF, HIS, and CAL were further employed to perform phylogenetic analysis.
The main molecular traits of D. passifloricola have been described in 2016 [39]. For ITS region, D. passifloricola (KX228292.1) shows 98% (556/567) similarity to D. miriciae (KJ197284.1) and 90% (466/519)–93% (402/430) similarity to five ‘Phomopsis tersa’ (e.g., KF516000.1 and JQ585648.1). For HIS sequence, D. passifloricola (KX228367.1) exhibits 100% identity (380/380) to D. absenteum (KP293559.1) and 99% identity (378/380) to ‘Diaporthe sp. 1 RG-2013’ (KC343687.1). Meanwhile, for TUB sequence, D. passifloricola (MB817057) is 99% similar to ‘Diaporthe sp. 1 RG-2013’ (KC344171.1 (513/517)) and D. miriciae (KJ197264.1 (589/595)). However, the difference among D. passifloricola and other two species D. durionigene and D.rosae, which have the closest genetic distance with D. passifloricola, has not been reported. In this study, polymorphic nucleotides in ITS, TUB, TEF, and CAL sequences of D. passifloricola, D. durionigene, and D. rosae are determined and can distinguish three species well.
The taxonomy of Diaporthe is complex. Many Diaporthe spp. were classified according to different criteria, i.e., host associations, morphological characteristics [26,28,46,47], or sequences of ITS region [22,26,48]. It is suggested that only those type strains, whose identification has been widely recognized, should be accepted as references for the taxonomy of this genus [37,49,50]. Moreover, several isolates included type strains from previous publications are selected for references with phylogenetic analysis in this study. While MegaBlast search was performed for each locus on NCBI, the Diaporthe species showing the highest similarity with the sequencing of each locus of the isolates were not the type strains. Thus, the species identified by us are different from those retrieved by a single locus MegaBlast search on NCBI.
Before this study, 22 Diaporthe spp. associated with citrus were known in the world [5,6,25,37,51,52]. They are either pathogens, endophytes, or saprobes on citrus [6,11,25,52,53,54]. This is the first time that D. passifloricola has been isolated from C. reticulata cv. Nanfengmiju.
In previous studies, 15 Diaporthe spp. have been reported to be associated with citrus in China [5,6]. Of them, three species are pathogens on citrus, i.e., D. citri, D. citriasiana, and D. citrichinensis. D. citri is identified as the causal agents of melanose disease as well as stem-end rot disease. In addition to being a pathogen, D. citri is also found as an endophyte in non-symptomatic twigs and as a saprobe on dead twigs. Two species, D. citriasiana, and D. citrichinensis, can only cause stem-end rot symptom on ponkan fruit (Citrus reticulata) [5]. The other 12 Diaporthe spp. were identified as endophytes or saprobes on citrus [6]. All of these indicate that the symbiotic relationship and ecological function of Diaporthe spp. with citrus plants is complex and variable.
Endophytes are defined as all organisms inhabiting plant organs which, at some time in their lives, can colonize internal plant tissues without causing significant damage to the host [55]. So defined, endophytes may also encompass asymptomatic latent pathogens. Sometimes asymptomatic fungi can cause diseases on their host plants under certain conditions. It’s reported that several Plectosphaerella spp. isolated from symptomless tomatoes and peppers can cause disease symptoms on tomato and pepper, and even basil and parsley when artificially inoculated [56,57]. Epichloë festucae is a well-known endophytic fungus of perennial ryegrass (Lolium perenne). However, a E. festucae noxA mutant is associated with severe stunting of the host as a result of hyphal hyper-branching and increased biomass [58]. Some fungal saprobes and pathogens can be isolated from rice (Oryza sativa) as endophytes [59]. In this study, since D. passifloricola isolates failed to cause melanose on citrus fruit, they are supposed to be the endophytic fungi colonizing diseased tissues with melanose symptoms. However, our results show that this species can induce stem-end rot symptoms on artificially inoculated citrus fruit. Thus, D. passifloricola could be a potential causal agent of stem-end rot disease during transportation and storage.
The disease spots of citrus melanose are formed by host hypersensitive response (HR). When the pathogens penetrate epidermal cells of the citrus, they are arrested and killed at the infection sites by hosts along with the development of melanose symptoms [60,61,62]. As a result, it is difficult to isolate pathogens in old disease spots. The disease spots were not newly formed, which might be the reason why we failed to isolate the pathogen causing melanose symptoms.
4. Materials and Methods
4.1. Fungal Isolation
In 2016, 10 citrus fruit of Nanfengmiju with typical symptoms of melanose were collected from a citrus orchard in Fuzhou City of Jiangxi Province (Figure 3A). The discrete and sunken black spots were observed on the fruit surface. Pieces of small sections about 5 mm2 from the margin of the lesion were cut off and soaked in 75% ethanol solution for 1 min. The sections were surface disinfested with 1% sodium hypochlorite solution (NaClO) for 1 min, rinsed three times with sterilized water, dried, and then incubated on PDA plates amended with 100 μg/mL streptomycin and 100 μg/mL ampicillin at 25 °C for 2 to 5 days. Hyphal tips growing from the pieces of the sample were transferred onto fresh PDA plates and incubated at 25 °C for 30 days as previous methods [7]. After sporulation, single-spore-isolation was performed as previously described [63]. All single-spore cultures were stored on half strength PDA slants in Eppendorf tubes at 4 °C, and on dried filter paper discs at −20 °C, respectively. A living culture of D. passifloricola in this study was deposited in China Center for Type Culture Collection (CCTCC), Wuhan, China.
4.2. Morphological Characterization
Sporulation was induced on PDA, MEA, CMA, OMA, and PNA. After inoculation, isolates were incubated at 25 °C with 12 h of light and 12 h of dark for 30 days. Conidia were harvested from the top of mature pycnidia. Pycnidia were picked up from pine needles with sterile toothpicks. The length and width of 30 conidia were measured with a stage micrometer under a Motic BA200 light microscope (Motic China Group Co., Ltd., Xiamen, China). The morphology of conidiomata was observed under OLYMPUS SZX16 stereo microscope (Olympus Corporation, Tokyo, Japan). Images of conidia were captured using a digital camera Nikon Eclipse 80i on a compound light microscope (Nikon Corporation, Tokyo, Japan) imaging system. Images of culture plates were captured using Cannon 600D digital camera (Cannon Inc., Tokyo, Japan). Colony and pycnidia color was investigated with a color chart according to the method of Rayner [64].
4.3. Pathogenicity Test
Pathogenicity tests were carried out on detached Nanfengmiju fruit (Citrus reticulata cv. Nanfengmiju). Non-wounded citrus fruit were washed with tap water, then surface disinfested with 75% of ethanol and rinsed with sterile water. Pycnidia with alpha conidia were induced as mentioned above and diluted to 106 conidia/mL with sterile water. To stimulate melanose symptoms, 300 μL of conidial suspensions was dropped on a piece of cotton, and then placed on the bottom of the fruit as previously described with a slight modification [65]. The inoculated fruit were placed in a plastic chamber with 95% relative humidity, incubated under the condition of 12 h of light and 12 h of dark at 25 °C for 15 days. Since Diaporthe spp. were the causal agents of both melanose and stem-end rot diseases on citrus fruit, their ability to cause stem-end rot symptom was also determined. The stems of citrus fruit were removed carefully, and 10 μl of conidial suspension (106 conidia/mL) of each strain was inoculated onto stem ends as previously described [5]. Then, the inoculated fruit were placed in a plastic chamber with wet towel tissues at the bottom. The chamber was wrapped with plastic film to maintain 95% relative humidity and incubated at 25 °C in the dark for 7 days. In all the pathogenicity tests, the conidial suspension (106 conidia/mL) of D. citri strain NFHF-8-4 [7] and sterile distilled water were used as positive and negative controls, respectively. Symptoms on fruit were observed. Four fruit were inoculated for each strain, and the experiments were repeated at least twice.
To authenticate the causal agent, tissue pieces from the margin of lesions on the experimentally inoculated and diseased fruit were placed on PDA to re-isolate the fungus. Molecular identification of the isolate was performed using the sequence of ITS, TUB, TEF, HIS, and CAL loci as mentioned below.
4.4. DNA Extraction, PCR Amplification, and Sequencing
DNA extraction was performed as previously described [66]. Fragments of ITS, TUB, TEF, HIS, and CAL were amplified by polymerase chain reaction (PCR) using primer pairs ITS1/ITS4 [67], Bt-2a/Bt-2b [68], EF1-728F/EF1-986R [69], CYLH3F/H3-1b [68,70], and CAL-228F/CAL-737R [69], respectively. Twenty-five microliters of PCR reaction included 1 μL genomic DNA (100–500 ng/μL), 1 μL (10 mM) of each primer, 9.5 μL double-distilled water, and 12.5 μL 2× Taq PCR Master Mix (Aidlab Biotechnologies Co., Ltd., Beijing, China). PCR amplification was carried out with an initial denaturation step at 95 °C for 3 min followed by 40 cycles, consisting of a denaturation step at 95 °C for 30 sec, an annealing step for 50 sec, an elongation step at 72 °C for 2 min, and a final step at 72 °C for 5 min. The annealing temperatures were 51 °C for the amplification of partial ITS, 55 °C for the amplification of partial TUB, TEF, and CAL, and 58 °C for the amplification of partial HIS, respectively, as mentioned previously [31]. The size of PCR products was verified by gel electrophoresis in Tris-borate-EDTA (TBE) buffer using 1% agarose gel. Sequencing was carried out at Wuhan Tianyi Huiyuan Biotechnology Co., Ltd., Wuhan, China.
4.5. Phylogenetic Analyses
The preliminary identifications of the isolates obtained in this study were determined using newly generated ITS sequences with all available type-derived sequences listed in previous studies [6,24,25,37,51]. Based on the result of preliminary identification, Diaporthe species with the closest genetic distance to the isolates in this study were selected. Sequences (ITS, TUB, TEF, HIS, and CAL) of them were downloaded from NCBI’s GenBank nucleotide database (www.ncbi.nlm.nih.gov). All sequences used in this study are listed in Table 1, including 15 sequences of three new isolates. The reference isolates were selected from ex-type, ex-epitype, and holotype cultures. Five-locus phylogenetic analyses were conducted to identify isolates to species level according to previous studies [21,30,37]. Sequences of five loci (ITS, TUB, TEF, HIS, and CAL) were assembled. Alignments of assembled sequences were performed with L-INS-i iterative refinement method by MAFFT alignment, a version available online [71], and manual adjustment was conducted where it was necessary by BioEdit v.7.2.5 [72]. ML trees were generated with 1,000 replicates using RA×ML-HPC BlackBox v.8.2.10 [73], which was available on the CIPRES Science Gateway v.3.3 Web Portal [74]. The RAxML software selected general time reversible model of evolution including estimation of invariable sites (GTRGAMMA+I). MP analyses were carried out with 1,000 replicates using Phylogenetic Analyses Using Parsimony (PAUP*) v.4.0b10 [75], with tree bisection and reconnection (TBR) branch-swapping algorithm. All characters were weighted equally, and the alignment gaps were treated as missing characters. Descriptive tree statistics including TL, CI, RI, RC, and HI were calculated for parsimony analyses. MrModeltest v.2.3 [76] was used to perform statistical selection of the best-fit model of nucleotide substitution and the corrected Akaike information criterion (AIC) determined above was incorporated into evolutionary models in the analysis (Supplementary Table S1). BI analysis was performed by using MrBayes v.3.2.2, with Markov Chain Monte Carlo (MCMC) algorithm. Four simultaneous of MCMC chains were run for 20,000,000th generations, and trees were sampled frequency every 100th generations, resulting in a total of 20,000 trees, and started from a random tree topology. The calculation of BI analyses was stopped when the average standard deviation of split frequencies fell below 0.01. The first 10% of trees were discarded as burn-in phase of analysis, and the remaining 180,000 trees were summarized to calculate the PP in the majority rule consensus tree. Phylogenetic analyses and full alignment of datasets were submitted to TreeBASE (www.treebase.org) with the study ID: 27334.
Table 1.
GenBank accession numbers of isolates used in this study.
| Diaporthe Species | Culture No. | Host Species | Origin | GenBank No. | Reference(s) | ||||
|---|---|---|---|---|---|---|---|---|---|
| ITS | TUB | TEF | HIS | CAL | |||||
| D. acaciarum | CBS 138862 | Acacia tortilis | Tanzania | KP004460 | KP004509 | – | KP004504 | – | [77] |
| D. acericola | MFLUCC 17-0956 | Acer negundo | Italy | KY964224 | KY964074 | KY964180 | – | KY964137 | [78] |
| D. alangii | CFCC 52556 | Alangium kurzii | China | MH121491 | MH121573 | MH121533 | MH121451 | MH121415 | [31] |
| D. amaranthophila | ATCC 74226 | Amaranthus sp. | USA | AF079776 | – | – | – | – | [36] |
| D. ambigua | CBS 114015 | Pyrus communis | South Africa | KC343010 | KC343978 | KC343736 | KC343494 | KC343252 | [37] |
| D. angelicae | CBS 111592 | Heracleum sphondylium | Austria | KC343027 | KC343995 | KC343753 | KC343511 | KC343269 | [37] |
| D. apiculatum | CGMCC3.17533 | Camellia sinensis | China | KP267896 | KP293476 | KP267970 | – | – | [79] |
| D. arctii | CBS 136.25 | Arctium sp. | Unknown | KC343031 | KC343999 | KC343757 | KC343515 | KC343273 | [37] |
| D. batatas | CBS 122.21 | Ipomoea batatas | USA | KC343040 | KC344008 | KC343766 | KC343524 | KC343282 | [37] |
| D. beilharziae | VPRI 16602 | Indigofera australis | Australia | JX862529 | KF170921 | JX862535 | – | – | [80] |
| D. caryae | CFCC 52563 | Carya illinoensis | China | MH121498 | MH121580 | MH121540 | MH121458 | MH121422 | [31] |
| D. chimonanthi | SCHM 3614 | Chimonanthus praecox | China | AY622993 | – | – | – | – | [81] |
| D. chromolaenae | MFLUCC 17-1422 | Chromolaena odorata | Thailand | MT214362 | – | – | – | – | [82] |
| D. cichorii | MFLUCC 17-1023 | Cichorium intybus | Italy | KY964220 | KY964104 | KY964176 | – | KY964133 | [78] |
| D. citri | CBS 135422 | Citrus sp. | USA | KC843311 | KC843187 | KC843071 | MF418281 | KC843157 | [25,51] |
| D. compacta | CGMCC3.17536 | Camellia sinensis | China | KP267854 | KP293434 | KP267928 | KP293508 | – | [79] |
| D. convolvuli | CBS 124654 | Convolvulus arvensis | Turkey | KC343054 | KC344022 | KC343780 | KC343538 | KC343296 | [37] |
| D. cucurbitae | DAOM 42078 | Cucumis sativus | Canada | KM453210 | KP118848 | KM453211 | KM453212 | – | [42] |
| D. cuppatea | CBS 117499 | Aspalathus linearis | South Africa | KC343057 | KC344025 | KC343783 | KC343541 | KC343299 | [37] |
| D. diachenii | PH10-1 | Unknown | Lithuania | KR870866 | – | – | – | – | [83] |
| D. durionigena | VTCC 930005 | Durio zibethinus | Vietnam | MN453530 | MT276159 | MT276157 | – | – | [40] |
| D. durionigena | KCSR1906.7 | Durio zibethinus | Vietnam | MN453531 | MT276160 | MT276158 | – | – | [40] |
| D. endophytica | CBS 133811 | Schinus terebinthifolius | Brazil | KC343065 | KC344033 | KC343791 | KC343549 | KC343307 | [37] |
| D. fructicola | MAFF 246408 | Passiflora edulis x P. edulis f. flavicarpa | Japan | LC342734 | LC342736 | LC342735 | LC342737 | LC342738 | [84] |
| D. fructicola | MAFF 246409 | Passiflora edulis x P. edulis f. flavicarpa | Japan | LC342739 | LC342741 | LC342740 | LC342742 | LC342743 | [84] |
| D. ganjae | CBS 180.91 | Cannabis sativa | USA | KC343112 | KC344080 | KC343838 | KC343596 | KC343354 | [37] |
| D. glabrae | SCHM 3622 | Bougainvillea glabra | China | AY601918 | – | – | – | – | [85] |
| D. goulteri | BRIP 55657a | Helianthus annuus | Australia | KJ197289 | KJ197270 | KJ197252 | – | – | [23] |
| D. gulyae | BRIP 54025 | Helianthus annuus | Australia | JF431299 | KJ197271 | JN645803 | – | – | [23,86] |
| D. guttulata | CGMCC3.20100 | Unknown | China | MT385950 | MT424705 | MT424685 | MW022491 | MW022470 | [87] |
| D. helianthi | CBS 592.81 | Helianthus annuus | Serbia | KC343115 | KC344083 | KC343841 | KC343599 | KC343357 | [37] |
| D. hordei | CBS 481.92 | Hordeum vulgare | Norway | KC343120 | KC344088 | KC343846 | KC343604 | KC343362 | [37] |
| D. hubeiensis | JZB320123 | Vertis vinifera | China | MK335809 | MK500147 | MK523570 | – | MK500235 | [88] |
| D. infecunda | CBS 133812 | Schinus terebinthifolius | Brazil | KC343126 | KC344094 | KC343852 | KC343610 | KC343368 | [37] |
| D. infertilis | CBS 230.52 | Citrus sinensis | Suriname | KC343052 | KC344020 | KC343778 | KC343536 | KC343294 | [37] |
| D. kongii | BRIP 54031 | Helianthus annuus | Australia | JF431301 | KJ197272 | JN645797 | – | – | [23,86] |
| D. leucospermi | CBS 111980 | Leucospermum sp. | Australia | JN712460 | – | – | – | – | [89] |
| D. longicolla | ATCC 60325 | Glycine max | USA | KJ590728 | KJ610883 | KJ590767 | KJ659188 | KJ612124 | [42] |
| D. longicolla | CBS 127267 | Glycine max | Croatia | KC343199 | KC344167 | KC343925 | KC343683 | KC343441 | [42] |
| D. longicolla | CBS 116023 | Glycine max | USA | KC343198 | KC344166 | KC343924 | KC343682 | KC343440 | [42] |
| D. longispora | CBS 194.36 | Ribes sp. | Canada | KC343135 | KC344103 | KC343861 | KC343619 | KC343377 | [37] |
| D. lusitanicae | CBS 123212 | Foeniculum vulgare | Portugal | KC343136 | KC344104 | KC343862 | KC343620 | KC343378 | [37] |
| D. malorum | CBS 142383 | Malus domestica | Portugal | KY435638 | KY435668 | KY435627 | KY435648 | KY435658 | [90] |
| D. manihotia | CBS 505.76 | Manihot utilissima | Rwanda | KC343138 | KC344106 | KC343864 | KC343622 | KC343380 | [37] |
| D. masirevicii | BRIP 57892a | Helianthus annuus | Australia | KJ197277 | KJ197257 | KJ197239 | – | – | [23] |
| D. megalospora | CBS 143.27 | Sambucus canadensis | Unknown | KC343140 | KC344108 | KC343866 | KC343624 | KC343382 | [37] |
| D. melonis | CBS 507.78 | Cucumis melo | USA | KC343142 | KC344110 | KC343868 | KC343626 | KC343384 | [37] |
| D. michelina | SCHM 3603 | Michelia alba | China | AY620820 | – | – | – | – | [30] |
| D. middletonii | BRIP 54884e | Rapistrum rugostrum | Australia | KJ197286 | KJ197266 | KJ197248 | – | – | [23] |
| D. minusculata | CGMCC3.20098 | Unknown | China | MT385957 | MT424712 | MT424692 | MW022499 | MW022475 | [87] |
| D. miriciae | BRIP 54736j | Helianthus annuus | Australia | KJ197282 | KJ197262 | KJ197244 | – | – | [23] |
| D. miriciae | BRIP 55662c | Glycine max | Australia | KJ197283 | KJ197263 | KJ197245 | – | – | [23] |
| D. miriciae | BRIP 56918a | Vigna radiata | Australia | KJ197284 | KJ197264 | KJ197246 | – | – | [23] |
| D. neoarctii | CBS 109490 | Ambrosia trifida | USA | KC343145 | KC344113 | KC343871 | KC343629 | KC343387 | [37] |
| D. novem | CBS 127270 | Glycine max | Croatia | KC343156 | KC344124 | KC343882 | KC343640 | KC343398 | [37] |
| D. ovalispora | CGMCC3.17256 | Citrus limon | China | KJ490628 | KJ490449 | KJ490507 | KJ490570 | – | [6] |
| D. pachirae | COAD2074 | Pachira glabra | Brazil | MG559537 | MG559541 | MG559539 | – | MG559535 | [91] |
| D. passiflorae | CBS 132527 | Passiflora edulis | South America | JX069860 | KY435674 | KY435633 | KY435654 | KY435664 | [92] |
| D. passifloricola | CBS 141329 | Passiflora foetida | Malaysia | KX228292 | KX228387 | – | KX228367 | – | [39] |
| D. passifloricola | NFIF-3-11 | Citrus reticulata cv. Nanfengmiju | China | MG786598 | MG925398 | MG925401 | MK238998 | MK238995 | This study |
| D. passifloricola | NFIF-3-19 | Citrus reticulata cv. Nanfengmiju | China | MG786599 | MG925399 | MG925402 | MK238999 | MK238996 | This study |
| D. passifloricola | NFIF-3-21 | Citrus reticulata cv. Nanfengmiju | China | MG786600 | MG925400 | MG925403 | MK239000 | MK238997 | This study |
| D. phaseolorum | CBS 139281 | Phaseolus vulgaris | USA | KJ590738 | KJ610893 | KJ590739 | KJ659220 | KJ612135 | [42] |
| D. pyracanthae | CBS 142384 | Pyracantha coccinea | Portugal | KY435635 | KY435666 | KY435625 | KY435645 | KY435656 | [90] |
| D. racemosae | CBS 143770 | Euclea racemosa | South Africa | MG600223 | MG600227 | MG600225 | MG600221 | MG600219 | [93] |
| D. rosae | MFLUCC 17-2658 | Rosa sp. | Thailand | MG828894 | MG843878 | – | – | MG829273 | [41] |
| D. rosae | MFLUCC 18-0354 | Magnolia champaca | Thailand | MG906792 | MG968951 | MG968953 | – | – | [94] |
| D. rosae | MFLUCC 17-2574 | Senna siamea | Thailand | MG906793 | MG968952 | MG968954 | – | – | [94] |
| D. sackstonii | BRIP 54669b | Helianthus annuus | Australia | KJ197287 | KJ197267 | KJ197249 | – | – | [23] |
| D. salicicola | VPRI 32789 | Salix purpurea | Australia | JX862531 | KF170923 | JX862537 | – | – | [80] |
| D. sambucusii | CFCC 51986 | Sambucus williamsii | China | KY852495 | KY852511 | KY852507 | KY852503 | KY852499 | [95] |
| D. schini | CBS 133181 | Schinus terebinthifolius | Brazil | KC343191 | KC344159 | KC343917 | KC343675 | KC343433 | [37] |
| D. schoeni | MFLUCC 17-2930 | Schoenus nigricans | Italy | KY964226 | KY964109 | KY964182 | – | KY964139 | [78] |
| D. sclerotioides | CBS 296.67 | Cucumis sativus | Netherlands | KC343193 | KC344161 | KC343919 | KC343677 | KC343435 | [37] |
| D. serafiniae | BRIP 55665a | Helianthus annuus | Australia | KJ197274 | KJ197254 | KJ197236 | – | – | [23] |
| D. sinensis | CGMCC3.19521 | Amaranthus sp. | China | MK637451 | MK660447 | MK660449 | MK660451 | – | [96] |
| D. sojae | CBS 139282 | Glycine max | USA | KJ590719 | KJ610875 | KJ590762 | KJ659208 | KJ612116 | [42] |
| D. sojae (D. actinidiae) | ICMP13683 | Actinidia deliciosa | New Zealand | KC145886 | – | KC145941 | – | – | [97] |
| D. sojae (D. camptothecae) | SCHM 3611 | Camptotheca acuminate | China | AY622996 | – | – | – | – | [81] |
| D. sojae (D. kochmanii) | BRIP 54033 | Helianthus annuus | Australia | JF431295 | – | JN645809 | – | – | [42,86] |
| D. sojae (D. melonis var. brevistylospora) | MAFF 410444 | Cucumis melo | Japan | KJ590714 | KJ610870 | KJ590757 | KJ659203 | KJ612111 | [42] |
| D. stewartii | CBS 193.36 | Cosmos bipinnatus | Unknown | FJ889448 | JX275421 | GQ250324 | – | JX197415 | [44,45] |
| D. subellipicola | KUMCC 17-0153 | Unknown | China | MG746632 | MG746634 | MG746633 | – | – | [98] |
| D. subordinaria | CBS 101711 | Plantago lanceolata | New Zealand | KC343213 | KC344181 | KC343939 | KC343697 | KC343455 | [37] |
| D. tecomae | CBS 100547 | Tabebuia sp. | Brazil | KC343215 | KC344183 | KC343941 | KC343699 | KC343457 | [37] |
| D. tectonae | MFLUCC 12-0777 | Tectona grandis | Thailand | KU712430 | KU743977 | KU749359 | – | KU749345 | [99] |
| D. tectonendophytica | MFLUCC 13-0471 | Tectona grandis | Thailand | KU712439 | KU743986 | KU749367 | – | KU749354 | [99] |
| D. terebinthifolii | CBS 133180 | Schinus terebinthifolius | Brazil | KC343216 | KC344184 | KC343942 | KC343700 | KC343458 | [37] |
| D. thunbergiicola | MFLUCC 12-0033 | Thunbergia laurifolia | Thailand | KP715097 | – | KP715098 | – | – | [100] |
| D. tulliensis | BRIP 62248a | Theobroma cacao | Australia | KR936130 | KR936132 | KR936133 | – | – | [101] |
| D. ueckerae | CBS 139283 | Cucumis melo | USA | KJ590726 | KJ610881 | KJ590747 | KJ659215 | KJ612122 | [42] |
| D. ueckerae | FAU659 | Cucumis melo | USA | KJ590724 | KJ610879 | KJ590745 | KJ659213 | KJ612120 | [42] |
| D. ueckerae | FAU658 | Cucumis melo | USA | KJ590725 | KJ610880 | KJ590746 | KJ659214 | KJ612119 | [42] |
| D. ueckerae | FAU660 | Cucumis melo | USA | KJ590723 | KJ610878 | KJ590744 | KJ659212 | KJ612121 | [42] |
| D. unshiuensis | CGMCC3.17569 | Citrus unshiu | China | KJ490587 | KJ490408 | KJ490466 | KJ490529 | – | [6] |
| D. unshiuensis | ZJUD51 | Fortunella margarita (Lour.) Swingle | China | KJ490586 | KJ490407 | KJ490465 | KJ490528 | – | [6] |
| D. unshiuensis | ZJUD50 | Fortunella margarita (Lour.) Swingle | China | KJ490585 | KJ490406 | KJ490464 | KJ490527 | – | [6] |
| D. vexans | CBS 127.14 | Solanum melongena | USA | KC343229 | KC344197 | KC343955 | KC343713 | KC343471 | [37] |
| D. vitimegaspora | STE-U2675 | Vitis vinifera | Taiwan | AF230749 | – | – | – | – | [26] |
| D. vochysiae | LGMF1583 | Vochysia divergens | Brazil | MG976391 | MK007527 | MK007526 | MK033323 | MK007528 | [102] |
| Diaporthe sp. 1 | CBS 119639 | Man, abscess | Germany | KC343202 | KC344170 | KC343928 | KC343686 | KC343444 | [37] |
| Diaporthella corylina | CBS 121124 | Corylus sp. | China | KC343004 | KC343972 | KC343730 | KC343488 | KC343246 | [37] |
a ATCC: American Type Culture Collection, Manassas, Virginia, USA; BRIP: Plant Pathology Herbarium, Department of Employment, Economic, Development and Innovation, Queensland, Australia; CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; CFCC: China Forestry Culture Collection Center, Beijing, China; CGMCC: China General Microbiological Culture Collection, Beijing, China; COAD: Coleção Octávio Almeida Drummond, Universidade Ferderal de Viçosa, Viçosa, Brazil; DAOM: Plant Research Institute, Department of Agriculture (Mycology), Ottawa, Canada; FAU: Isolates in culture collection of Systematic Mycology and Microbiology Laboratory, USDA-ARS, Beltsville, Maryland, USA; ICMP: International Collection of Micro-organisms from Plants, Landcare Research, Auckland, New Zealand; JZB: Culture collection of Institute of Plant and Environment Protection, Beijing Academy of Agriculture and Forestry Sciences, Beijing, China; KCSR, VTCC: Vietnam Type Culture Collection, Institute of Microbiology and Biotechnology (IMBT), Vietnam National University, Hanoi, Vietnam; HUMCC: Kunming Institute of Botany Culture Collection, Yunnan, China; LGMF: Culture collection of Laboratory of Genetics of Microorganisms, Federal University of Parana, Curitiba, Brazil; MAFF: Ministry of Agriculture, Forestry and Fisheries, Tsukuba, Ibaraki, Japan; MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand; SCHM: Mycological Herbarium of South China Agricultural University, Guangzhou, China; STE-U: Culture collection of the Department of Plant Pathology, University of Stellenbosch, South Africa; VPRI: Victorian Plant Pathogen Herbarium, Bundoora, Australia; ZJUD: Diaporthe species culture collection at the Institute of Biotechnology, Zhejiang University, Hangzhou, China; Ex-type, ex-epitype, and holotype cultures are indicated in bold. Isolates obtained in this study are indicated in italics. b ITS: Nuclear ribosomal internal transcribed spacer regions; TUB: Beta-tubulin gene; TEF: Translation elongation factor 1-α gene; HIS: Histone-3 gene; and CAL: Calmodulin gene. Sequences generated in this study are indicated in italics.
5. Conclusions
Our results indicate that D. passifloricola, may occur as an asymptomatic endophyte in the peel of citrus fruit. If is manages to invade the fruit stalk, however, it may induce typical stem-end rot symptoms during transportation and storage. To the best of our knowledge, this is the first time D. passifloricola has been isolated from Citrus reticulata cv. Nanfengmiju in China and identified as a causal agent of stem-end rot disease in this crop.
Acknowledgments
We thank Kevin D. Hyde (Center of Excellence in Fungal Research, Mae Fah Luang University, Thailand) and Jian-Kui Liu (Fungal Research Laboratory, University of Electronic Science and Technology of China, China) for technical assistance and invaluable advice. The authors sincerely thank the reviewers and editor for their contributions to improve the manuscript during the revision process.
Supplementary Materials
The following are available online at https://www.mdpi.com/2223-7747/10/2/218/s1, Table S1 nucleotide substitution models, MP and ML alignment properties, Table S2 Polymorphic nucleotides in ITS, TUB, TEF, and CAL sequences of D. passifloricola, D. durionigene, and D. rosae, Figure S1. The prevalence of Diaporthe species on citrus in Jiangxi Province, China based on phylogenetic identification. Numbers (%) indicate the number of obtained isolates of certain species and the percentage among the total 140 isolates [1]. Yellow color indicate 39 isolates of Diaporthe sp. were found in this study, Figure S2. The phylogenetic tree is generated from the analysis of sequences of ITS locus. A, Maximum likelihood and B, Maximum parsimony. Bootstrap support values ≥50%, (MLBS/MPBS) are displayed at the nodes. The tree is rooted with Diaporthella corylina CBS 121124. Ex-type, ex-epitype and ex-isotype cultures are indicated in bold. The codes of isolates used for phylogenetic tree are given, Figure S3. The phylogenetic tree is generated from the analysis of sequences of TUB locus. A, Maximum likelihood and B, Maximum parsimony. Bootstrap support values ≥50%, (MLBS/MPBS) are displayed at the nodes. The tree is rooted with Diaporthella corylina CBS 121124. Ex-type, ex-epitype and ex-isotype cultures are indicated in bold. The codes of isolates used for phylogenetic tree are given, Figure S4. The phylogenetic tree is generated from the analysis of sequences of TEF locus. A, Maximum likelihood and B, Maximum parsimony. Bootstrap support values ≥50%, (MLBS/MPBS) are displayed at the nodes. The tree is rooted with Diaporthella corylina CBS 121124. Ex-type, ex-epitype and ex-isotype cultures are indicated in bold. The codes of isolates used for phylogenetic tree are given, Figure S5. The phylogenetic tree is generated from the analysis of sequences of HIS locus. A, Maximum likelihood and B, Maximum parsimony. Bootstrap support values ≥50%, (MLBS/MPBS) are displayed at the nodes. The tree is rooted with Diaporthella corylina CBS 121124. Ex-type, ex-epitype and ex-isotype cultures are indicated in bold. The codes of isolates used for phylogenetic tree are given, Figure S6. The phylogenetic tree is generated from the analysis of sequences of CAL locus. A, Maximum likelihood and B, Maximum parsimony. Bootstrap support values ≥50%, (MLBS/MPBS) are displayed at the nodes. The tree is rooted with Diaporthella corylina CBS 121124. Ex-type, ex-epitype and ex-isotype cultures are indicated in bold. The codes of isolates used for phylogenetic tree are given, Figure S7. The phylogenetic tree is generated from the analysis of the combined sequences of five loci (ITS, TUB, TEF, HIS, and CAL). A, Maximum likelihood and B, Maximum parsimony, bootstrap support values ≥50%, (MLBS/MPBS) are displayed at the nodes. The tree is rooted with D. citri CBS 135422. Ex-type, ex-epitype and holotype cultures are indicated in bold. The codes of isolates used for phylogenetic tree are given.
Author Contributions
Conceptualization, C.C., Y.L. and C.-X.L.; validation, C.C., X.-Y.L., Y.L. and C.-X.L.; formal analysis, C.C. and X.-Y.L.; investigation and resources, C.C., X.-Y.L., W.-X.Y. and Y.L.; data curation, C.C., X.-Y.L., Y.L., W.-X.Y. and C.-X.L.; writing, C.C., C.-X.L. and Y.L.; funding acquisition, Y.L. and C.-X.L. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Key Research and Development Program of China (number 2017YFD020200103).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Alignment data generated in the current study are available in TreeBASE (accession http://purl.org/phylo/treebase/phylows/study/TB2:S27334). All sequence data are available in NCBI GenBank following the accession numbers in the manuscript.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Deng X.X., Peng C.J., Chen Z.S., Deng Z.N., Xu J.G., Li J. Citrus Varieties in China. China Agriculture Press; Beijing, China: 2008. [Google Scholar]
- 2.FAO . Citrus Fruit—Fresh and Processed Statistical Bulletin 2016. Food and Agriculture Organization of the United Nations; Rome, Italy: 2017. [Google Scholar]
- 3.Kucharek T., Whiteside J., Brown E. Plant Pathology Fact Sheet. Florida Cooperative Extension Service, Institute of Food and Agricultural Sciences, University of Florida; Florida, FL, USA: 2000. Melanose and phomopsis stem-end rot of citrus; pp. 26–30. [Google Scholar]
- 4.Timmer L.W., Garnsey S.M., Graham J.H. Scab Diseases, revised edition: 31–32 ed. American Phytopathological Society Press; St. Paul, MN, USA: 2000. p. 92. [Google Scholar]
- 5.Huang F., Hou X., Dewdney M.M., Fu Y.S., Chen G.Q., Hyde K.D., Li H.Y. Diaporthe species occurring on citrus in China. Fungal Divers. 2013;61:237–250. doi: 10.1007/s13225-013-0245-6. [DOI] [Google Scholar]
- 6.Huang F., Udayanga D., Wang X.H., Hou X., Mei X.F., Fu Y.S., Hyde K.D., Li H.Y. Endophytic Diaporthe associated with Citrus: A phylogenetic reassessment with seven new species from China. Fungal Biol. 2015;119:331–347. doi: 10.1016/j.funbio.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Chaisiri C., Liu X.Y., Lin Y., Li J.B., Xiong B., Luo C.X. Phylogenetic analysis and development of molecular tool for detection of Diaporthe citri causing melanose disease of citrus. Plants. 2020;9:329. doi: 10.3390/plants9030329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bach W.J., Wolf F.A. The isolation of the fungus that causes citrus melanose and the pathological anatomy of the host. J. Agric. Res. 1928;37:243–252. [Google Scholar]
- 9.Gopal K., Lakshmi L.M., Sarada G., Nagalakshmi T., Sankar T.G., Gopi V., Ramana K.T.V. Citrus melanose (Diaporthe citri Wolf): A review. Int. J. Curr. Microbiol. App. Sci. 2014;3:113–124. [Google Scholar]
- 10.Fawcett H.S. The cause of stem-end rot of citrus fruits (Phomopsis citri n. sp.) Phytopathology. 1912;2:109–113. [Google Scholar]
- 11.Mondal S.N., Vicent A., Reis R.F., Timmer L.W. Saprophytic colonization of citrus twigs by Diaporthe citri and factors affecting pycnidial production and conidial survival. Plant. Dis. 2007;91:387–392. doi: 10.1094/PDIS-91-4-0387. [DOI] [PubMed] [Google Scholar]
- 12.Chen G.Q., Jiang L.Y., Xu F.S., Li H.Y. In vitro and in vivo screening of fungicides for controlling citrus melanose caused by Diaporthe citri. J. Zhejiang Univ. (Agric. Life Sci.) 2010;36:440–444. (In Chinese) [Google Scholar]
- 13.Jiang L.Y., Xu F.S., Huang Z.D., Huang F., Chen G.Q., Li H.Y. Occurrence and control of citrus melanose caused by Diaporthe citri. Acta Agric. Zhejiangensis. 2012;24:647–653. (In Chinese) [Google Scholar]
- 14.Udayanga D., Manamgoda D.S., Liu X.Z., Chukeatirote E., Hyde K.D. What are the common anthracnose pathogens of tropical fruits? Fungal Divers. 2013;61:165–179. doi: 10.1007/s13225-013-0257-2. [DOI] [Google Scholar]
- 15.Huang L. The diseases of Citrus spp. of Guangxi. Guangxi Nong Xun. 1943;4:27–61. (In Chinese) [Google Scholar]
- 16.Guangdong Station of Plant Quarantine The preliminary investigation results of Citrus spp. insects and diseases in East Guangdong province. 1955. pp. 1–3. (In Chinese)
- 17.Qiu W.F. The records of diseases and insects of economic plants in Fujian (1, 2, 3) Xin Nong Ji Kan. 1941;1:70–75, 161–166, 209–229. (In Chinese) [Google Scholar]
- 18.Plant quarantine Station. Department of Agriculture. Jiangxi Academy of Agriculture . The Records of Plant Insects and Diseases in Jiangxi, the Part of Plant Diseases. Jiangxi People’s Publishing House; Nanchang, China: 1960. pp. 1–247. (in Chinese) [Google Scholar]
- 19.Li L. The index of the parasitic fungi of Szechwan, China. Pl. Dis. Rep. Suppl. 1948;173:1–38. [Google Scholar]
- 20.Chinese Research Institute of Pomelogy and Citrus . The Records of Chinese Fruit Trees’ Diseases and Pests. China Agriculture Press; Beijing, China: 1994. (In Chinese) [Google Scholar]
- 21.Guarnaccia V., Groenewald J.Z., Woodhall J., Armengol J., Cinelli T., Eichmeier A., Ezra D., Fontaine F., Gramaje D., Gutierrez-Aguirregabiria A., et al. Diaporthe diversity and pathogenicity revealed from a broad survey of grapevine diseases in Europe. Persoonia. 2018;40:135–153. doi: 10.3767/persoonia.2018.40.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santos J.M., Phillips A.J.L. Resolving the complex of Diaporthe (Phomopsis) species occurring on Foeniculum vulgare in Portugal. Fungal Divers. 2009;34:111–125. [Google Scholar]
- 23.Thompson S.M., Tan Y.P., Shivas R.G., Neate S.M., Morin L., Bissett A., Aitken E.A.B. Green and brown bridges between weeds and crops reveal novel Diaporthe species in Australia. Persoonia. 2015;35:39–49. doi: 10.3767/003158515X687506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Udayanga D., Castlebury L.A., Rossman A.Y., Chukeatirote E., Hyde K.D. Insights into the genus Diaporthe: Phylogenetic species delimitation in the D. eres species complex. Fungal Divers. 2014;67:203–229. doi: 10.1007/s13225-014-0297-2. [DOI] [Google Scholar]
- 25.Guarnaccia V., Crous P.W. Emerging citrus diseases in Europe caused by species of Diaporthe. Ima Fungus. 2017;8:317–334. doi: 10.5598/imafungus.2017.08.02.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mostert L., Crous P.W., Kang J.C., Phillips A.J.L. Species of Phomopsis and a Libertella sp. occurring on grapevines with specific reference to South Africa: Morphological, cultural, molecular and pathological characterization. Mycologia. 2001;93:146–167. doi: 10.1080/00275514.2001.12061286. [DOI] [Google Scholar]
- 27.Rehner S.A., Uecker F.A. Nuclear ribosomal internal transcribed spacer phylogeny and host diversity in the coelomycete Phomopsis. Can. J. Bot. 1994;72:1666–1674. doi: 10.1139/b94-204. [DOI] [Google Scholar]
- 28.Chi P.K., Jiang Z.D., Xiang M.M. Flora Fungorum Sinicorum. Volume 34. Science Press; Beijing, China: 2007. (In Chinese) [Google Scholar]
- 29.Dissanayake A.J., Zhang W., Liu M., Hyde K.D., Zhao W.S., Li X.H., Yan J.Y. Diaporthe species associated with peach tree dieback in Hubei, China. Mycosphere. 2017;8:533–549. doi: 10.5943/mycosphere/8/5/2. [DOI] [Google Scholar]
- 30.Gao Y.H., Liu F., Duan W.J., Crous P.W., Cai L. Diaporthe is paraphyletic. Ima Fungus. 2017;8:153–187. doi: 10.5598/imafungus.2017.08.01.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Q., Fan X.L., Guarnaccia V., Tian C.M. High diversity of Diaporthe species associated with dieback diseases in China, with twelve new species described. MycoKeys. 2018;39:97–149. doi: 10.3897/mycokeys.39.26914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan X.L., Yang Q., Bezerra J.D.P., Alvarez L.V., Tian C.M. Diaporthe from walnut tree (Juglans regia) in China, with insight of the Diaporthe eres complex. Mycol. Prog. 2018;17:841–853. doi: 10.1007/s11557-018-1395-4. [DOI] [Google Scholar]
- 33.Guo Y.S., Crous P.W., Bai Q., Fu M., Yang M.M., Wang X.H., Du Y.M., Hong N., Xu W.X., Wang G.P. High diversity of Diaporthe species associated with pear shoot canker in China. Persoonia. 2020;45:132–162. doi: 10.3767/persoonia.2020.45.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wehmeyer L.E. The genus Diaporthe Nitschke and its segregates. Univ. Mich. Stud. Sci. Ser. 1933;9:1–349. [Google Scholar]
- 35.Sutton B.C. The Coelomycetes. Fungi Imperfecti with Pycnidia, Acervuli and Stromata. Commonwealth Mycological Institute; London, UK: 1980. [Google Scholar]
- 36.Rosskopf E.N., Charudattan R., Shabana Y.M., Benny G.L. Phomopsis amaranthicola, a new species from amaranthus sp. Mycologia. 2000;92:114–122. doi: 10.1080/00275514.2000.12061135. [DOI] [Google Scholar]
- 37.Gomes R.R., Glienke C., Videira S.I.R., Lombard L., Groenewald J.Z., Crous P.W. Diaporthe: A genus of endophytic, saprobic and plant pathogenic fungi. Persoonia. 2013;31:1–41. doi: 10.3767/003158513X666844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santos L., Alves A., Alves R. Evaluating multi-locus phylogenies for species boundaries determination in the genus Diaporthe. PeerJ. 2017;5:1–26. doi: 10.7717/peerj.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crous P.W., Wingfield M.J., Richardson D.M., Le Roux J.J., Strasberg D., Edwards J., Roets F., Hubka V., Taylor P.W.J., Heykoop M., et al. Fungal planet description sheets: 400–468. Persoonia. 2016;36:316–458. doi: 10.3767/003158516X692185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crous P.W., Wingfield M.J., Chooi Y.H., Gilchrist C.L.M., Lacey E., Pitt J.I., Roets F., Swart W.J., Cano-Lira J.F., Valenzuela-Lopez N., et al. Fungal planet description sheets: 1042–1111. Persoonia. 2020;44:301–459. doi: 10.3767/persoonia.2020.44.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wanasinghe D.N., Phukhamsakda C., Hyde K.D., Jeewon R., Lee H.B., Jones E.B.G., Tibpromma S., Tennakoon D.S., Dissanayake A.J., Jayasiri S.C., et al. Fungal diversity notes 709–839: Taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers. 2018;89:1–60. doi: 10.1007/s13225-018-0395-7. [DOI] [Google Scholar]
- 42.Udayanga D., Castlebury L.A., Rossman A.Y., Chukeatirote E., Hyde K.D. The Diaporthe sojae species complex: Phylogenetic re-assessment of pathogens associated with soybean, cucurbits and other field crops. Fungal Biol. 2015;119:383–407. doi: 10.1016/j.funbio.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 43.Farr D.F., Castlebury L.A., Rossman A.Y., Putnam M.L. A new species of Phomopsis causing twig dieback of Vaccinium vitis-idaea (lingonberry) Mycol. Res. 2002;106:745–752. doi: 10.1017/S095375620200583X. [DOI] [Google Scholar]
- 44.Santos J.M., Correia V.G., Phillips A.J.L., Spatafora J.W. Primers for mating-type diagnosis in Diaporthe and Phomopsis: Their use in teleomorph induction in vitro and biological species definition. Fungal Biol. 2010;114:255–270. doi: 10.1016/j.funbio.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 45.Udayanga D., Liu X.Z., Crous P.W., McKenzie E.H.C., Chukeatirote E., Hyde K.D. A multi-locus phylogenetic evaluation of Diaporthe (Phomopsis) Fungal Divers. 2012;56:157–171. doi: 10.1007/s13225-012-0190-9. [DOI] [Google Scholar]
- 46.Uecker F.A. A world list of Phomopsis names with notes on nomenclature, morphology and biology. Mycol. Mem. 1988;13:1–231. [Google Scholar]
- 47.Brayford D. Variation in Phomopsis isolates from Ulmus species in the British Isles and Italy. Mycol. Res. 1990;94:691–697. doi: 10.1016/S0953-7562(09)80670-9. [DOI] [Google Scholar]
- 48.van Niekerk J.M., Groenewald J.Z., Farr D.F., Fourie P.H., Halleen F., Crous P.W. Reassessment of Phomopsis species on grapevine. Australas. Plant. Pathol. 2005;34:27–39. doi: 10.1071/AP04072. [DOI] [Google Scholar]
- 49.Rossman A.Y., Adams G.C., Cannon P.F., Castlebury L.A., Crous P.W., Gryzenhout M., Jaklitsch W.M., Mejia L.C., Stoykov D., Udayanga D., et al. Recommendations of generic names in Diaporthales competing for protection or use. Ima Fungus. 2015;6:145–154. doi: 10.5598/imafungus.2015.06.01.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dissanayake A.J., Phillips A.J.L., Hyde K.D., Yan J.Y., Li X.H. The current status of species in Diaporthe. Mycosphere. 2017;8:1106–1156. doi: 10.5943/mycosphere/8/5/5. [DOI] [Google Scholar]
- 51.Udayanga D., Castlebury L.A., Rossman A.Y., Hyde K.D. Species limits in Diaporthe: Molecular re-assessment of D. citri, D. cytosporella, D. foeniculina and D. rudis. Persoonia. 2014;32:83–101. doi: 10.3767/003158514X679984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guarnaccia V., Crous P.W. Species of Diaporthe on Camellia and Citrus in the Azores Islands. Phytopathol. Mediterr. 2018;57:307–319. [Google Scholar]
- 53.Douanla-Meli C., Langer E., Mouafo F.T. Fungal endophyte diversity and community patterns in healthy and yellowing leaves of Citrus limon. Fungal Ecol. 2013;6:212–222. doi: 10.1016/j.funeco.2013.01.004. [DOI] [Google Scholar]
- 54.Murali T.S., Suryanarayanan T.S., Geeta R. Endophytic Phomopsis species: Host range and implications for diversity estimates. Can. J. Microbiol. 2006;52:673–680. doi: 10.1139/w06-020. [DOI] [PubMed] [Google Scholar]
- 55.Petrini O. Fungal endophytes of tree leaves. In: Andrews J.H., Hirano S.S., editors. Microbial Ecology of Leaves. Springer; New York, NY, USA: 1991. pp. 179–197. (Brock/Springer Series in Contemporary Bioscience). [Google Scholar]
- 56.Raimondo M.L., Carlucci A. Characterization and pathogenicity of Plectosphaerella spp. collected from basil and parsley in Italy. Phytopathol. Mediterr. 2018;57:284–295. [Google Scholar]
- 57.Raimondo M.L., Carlucci A. Characterisation and pathogenicity assessment of Plectosphaerella species associated with stunting disease on tomato and pepper crops in Italy. Plant. Pathol. 2018;67:626–641. doi: 10.1111/ppa.12766. [DOI] [Google Scholar]
- 58.Takemoto D., Tanaka A., Kayano Y., Saikia S., Wrenn R., Scott B. Reactive oxygen as a signal in grass-Epichloë symbiosis. In: Young C.A., Aiken G.E., McCullry R.L., Strickland J.R., Schardl C.L., editors. Epichloae, Endophytes of Cool Season Grasses: Implications, Utilization and Biology. Proceedings of the 7th International Symposium on Fungal Endophytes of Grasses. Samuel Roberts Noble Foundation; Lexington, KY, USA: 2012. pp. 109–112. [Google Scholar]
- 59.Fisher P.J., Petrini O. Fungal saprobes and pathogens as endophytes of rice (Oryza sativa L.) New Phytol. 1992;120:137–143. doi: 10.1111/j.1469-8137.1992.tb01066.x. [DOI] [Google Scholar]
- 60.Akai S. Histology of defense in plants. In: Horsfall J.G., Dimond A.E., editors. Plant Pathology. Volume 1. Academic Press; New York, NY, USA: 1959. pp. 435–467. [Google Scholar]
- 61.Arimoto Y., Homma Y., Misato T. Studies on citrus melanose and citrus stem-end rot by Diaporthe citri (Faw.) Wolf. Part 2. Infection mode of D. citri to citrus leaf. Ann. Phytopath. Soc. Jpn. 1980;46:575–581. doi: 10.3186/jjphytopath.46.575. [DOI] [Google Scholar]
- 62.Arimoto Y., Homma Y., Misato T. Studies on citrus melanose and citrus stem-end rot by Diaporthe citri (Faw.) Wolf. Part 3. Leaf against infection of D. citri. Ann. Phytopath. Soc. Jpn. 1982;48:559–569. doi: 10.3186/jjphytopath.48.559. [DOI] [Google Scholar]
- 63.Yin L.F., Chen S.N., Chen G.K., Schnabel G., Du S.F., Chen C., Li G.Q., Luo C.X. Identification and characterization of three Monilinia species from plum in China. Plant. Dis. 2015;99:1775–1783. doi: 10.1094/PDIS-12-14-1308-RE. [DOI] [PubMed] [Google Scholar]
- 64.Rayner R.W. A Mycological Colour Chart. Commonwealth Mycological Institute and British Mycological Society; Kew, Surrey, UK: 1970. [Google Scholar]
- 65.Kanematsu S., Kobayashi T., Kudo A., Ohtsu Y. Conidial morphology, pathogenicity and culture characteristics of Phomopsis isolates from peach, Japanese pear and apple in Japan. Jpn J. Phytopathol. 1999;65:264–273. doi: 10.3186/jjphytopath.65.264. [DOI] [Google Scholar]
- 66.Hu M.J., Cox K.D., Schnabel G., Luo C.X. Monilinia species causing brown rot of peach in China. PLoS ONE. 2011;6:e24990. doi: 10.1371/journal.pone.0024990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.White T.J., Bruns T., Lee S., Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. PCR Protocols: A Guide to Methods and Applications. Academic Press; San Diego, CA, USA: 1990. pp. 315–322. [Google Scholar]
- 68.Glass N.L., Donaldson G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microb. 1995;61:1323–1330. doi: 10.1128/AEM.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carbone I., Kohn L.M. A method for desianing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91:553–556. doi: 10.1080/00275514.1999.12061051. [DOI] [Google Scholar]
- 70.Crous P.W., Groenewald J.Z., Risède J.M., Simoneau P., Hywel-Jones N.L. Calonectria species and their Cylindrocladium anamorphs: Species with clavate vesicles. Stud. Mycol. 2004;50:415–430. doi: 10.3114/sim.55.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2017;bbx108:1–7. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hall A.T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/nt. Nucleic Acids Res. 1999;41:95–98. [Google Scholar]
- 73.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miller M.A., Pfeiffer W., Schwartz T. TeraGrid Conference: Extreme Digital Discovery. Volume 41. San Diego Supercomputer Center; San Diego, CA, USA: 2011. The CIPRES science gateway: A community resource for phylogenetic analyses; pp. 1–8. [Google Scholar]
- 75.Swofford D.L. PAUP* Phylogenetic Analysis Using Parsimony, (*and Other Methods). Version 4.0 b10. Sinauer Associates; Sunderland, MA, USA: 2003. [Google Scholar]
- 76.Nylander J.A.A. MrModeltest v.2. Program Distributed by the Author. Evolitionary Biology Centre, Uppsala Univeristy; Uppsala, Sweden: 2004. [Google Scholar]
- 77.Crous P.W., Wingfield M.J., Schumacher R.K., Summerell B.A., Giraldo A., Gené J., Guarro J., Wanasinghe D.N., Hyde K.D., Camporesi E., et al. Fungal planet description sheets: 281–319. Persoonia. 2014;33:212–289. doi: 10.3767/003158514X685680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dissanayake A.J., Camporesi E., Hyde K.D., Zhang W., Yan J.Y., Li X.H. Molecular phylogenetic analysis reveals seven new Diaporthe species from Italy. Mycosphere. 2017;8:853–877. doi: 10.5943/mycosphere/8/5/4. [DOI] [Google Scholar]
- 79.Gao Y.H., Liu F., Cai L. Unravelling Diaporthe species associated with Camellia. Syst. Biodivers. 2016;14:102–117. doi: 10.1080/14772000.2015.1101027. [DOI] [Google Scholar]
- 80.Tan Y.P., Edwards J., Grice K.R.E., Shivas R.G. Molecular phylogenetic analysis reveals six new species of Diaporthe from Australia. Fungal Divers. 2013;61:251–260. doi: 10.1007/s13225-013-0242-9. [DOI] [Google Scholar]
- 81.Chang C.Q., Xi P.G., Xiang M.M., Jiang Z.D., Chi P.K. New species of Phomopsis on woody plants in Hunan province. Mycosystema. 2005;24:145–154. [Google Scholar]
- 82.Mapook A., Hyde K.D., McKenzie E.H.C., Jones E.B.G., Bhat D.J., Jeewon R., Stadler M., Samarakoon M.C., Malaithing M., Tanunchai B., et al. Taxonomic and phylogenetic contributions to fungi associated with the invasive weed Chromolaena odorata (siam weed) Fungal Divers. 2020;101:1–175. doi: 10.1007/s13225-020-00444-8. [DOI] [Google Scholar]
- 83.Souza A.R.C., Baldoni D.B., Lima J., Porto V., Marcuz C., Machado C., Ferraz R.C., Kuhn R.C., Jacques R.J.S., Guedes J.V.C., et al. Selection, isolation, and identification of fungi for bioherbicide production. Braz. J. Microbiol. 2017;48:101–108. doi: 10.1016/j.bjm.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Crous P.W., Carnegie A.J., Wingfield M.J., Sharma R., Mughini G., Noordeloos M.E., Santini A., Shouche Y.S., Bezerra J.D.P., Dima B., et al. Fungal planet description sheets: 868–950. Persoonia. 2019;42:291–473. doi: 10.3767/persoonia.2019.42.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chang C.Q., Cheng Y.H., Xiang M.M., Jiang Z.D., Chi P.K. New species of Phomopsis in woody plants in Fujian provice. Mycosystema. 2005;24:6–11. [Google Scholar]
- 86.Thompson S.M., Tan Y.P., Young A.J., Neate S.M., Aitken E.A.B., Shivas R.G. Stem cankers on sunflower (Helianthus annuus) in Australia reveal a complex of pathogenic Diaporthe (Phomopsis) species. Persoonia. 2011;27:80–89. doi: 10.3767/003158511X617110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dissanayake A.J., Chen Y.Y., Liu J.K. Unravelling Diaporthe species associated with woody hosts from Karst Formations (Guizhou) in China. J. Fungi. 2020;6:251. doi: 10.3390/jof6040251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Manawasighe I.S., Dissanayake A.J., Li X.H., Liu M., Wanasinghe D.N., Xu J.P., Zhao W.S., Zhang W., Zhou Y.Y., Hyde K.D., et al. High genetic diversity and species complexity of Diaporthe associated with grapevine dieback in China. Front. Microbiol. 2019;10:1936. doi: 10.3389/fmicb.2019.01936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Crous P.W., Summerell B.A., Swart L., Denman S., Taylor J.E., Bezuidenhout C.M., Palm M.E., Marincowitz S., Groenewald J.Z. Fungal pathogens of Proteaceae. Persoonia. 2011;27:20–45. doi: 10.3767/003158511X606239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Santos L., Phillips A.J.L., Crous P.W., Alves A. Diaporthe species on Rosaceae with descriptions of D. pyracanthae sp. nov. and D. malorum sp. nov. Mycosphere. 2017;8:485–511. doi: 10.5943/mycosphere/8/5/1. [DOI] [Google Scholar]
- 91.Milagres C.A., Belisário R., Silva M.A., Lisboa D.O., Pinho D.B., Furtado G.Q. A novel species of Diaporthe causing leaf spot in Pachira glabra. Trop. Plant. Pathol. 2018;43:460–467. doi: 10.1007/s40858-018-0242-0. [DOI] [Google Scholar]
- 92.Crous P.W., Summerell B.A., Shivas R.G., Burgess T.I., Decock C.A., Dreyer L.L., Granke L.L., Guest D.I., Hardy G.E.S.J., Hausbeck M.K., et al. Fungal planet description sheets: 107–127. Persoonia. 2012;28:138–182. doi: 10.3767/003158512X652633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Marin-Felix Y., Hernández-Restrepo M., Wingfield M.J., Akulov A., Carnegie A.J., Cheewangkoon R., Gramaje D., Groenewald J.Z., Guarnaccia V., Halleen F., et al. Genera of phytopathogenic fungi: GOPHY 2. Stud. Mycol. 2019;92:47–133. doi: 10.1016/j.simyco.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gao Y.H., Su Y.Y., Sun W., Cai L. Diaporthe species occurring on Lithocarpus glabra in China, with descriptions of five new species. Fungal Biol. 2015;119:295–309. doi: 10.1016/j.funbio.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 95.Yang Q., Du Z., Tian C.M. Phylogeny and morphology reveal two new species of Diaporthe from traditional chinese medicine in Northeast China. Phytotaxa. 2018;336:159–170. doi: 10.11646/phytotaxa.336.2.3. [DOI] [Google Scholar]
- 96.Feng X.X., Chen J.J., Wang G.R., Cao T.T., Zheng Y.L., Zhang C.L. Diaporthe sinensis, a new fungus from Amaranthus sp. in China. Phytotaxa. 2019;425:259–268. doi: 10.11646/phytotaxa.425.5.1. [DOI] [Google Scholar]
- 97.Sommer N.F., Beraha L. Diaporthe actinidiae, a new species causing stem-end rot of chinese gooseberry fruits. Mycologia. 1975;67:650–653. doi: 10.1080/00275514.1975.12019787. [DOI] [Google Scholar]
- 98.Hyde K.D., Chaiwan N., Norphanphoun C., Boonmee S., Camporesi E., Chethana K.W.T., Dayarathne M.C., de Silva N.I., Dissanayake A.J., Ekanayaka A.H., et al. Mycosphere notes 169–224. Mycosphere. 2018;9:271–430. doi: 10.5943/mycosphere/9/2/8. [DOI] [Google Scholar]
- 99.Doilom M., Dissanayake A.J., Wanasinghe D.N., Boonmee S., Liu J.K., Bhat D.J., Taylor J.E., Bahkali A.H., McKenzie E.H.C., Hyde K.D. Microfungi on Tectona grandis (teak) in Northern Thailand. Fungal Divers. 2016;82:107–182. doi: 10.1007/s13225-016-0368-7. [DOI] [Google Scholar]
- 100.Liu J.K., Hyde K.D., Jones E.B.G., Ariyawansa H.A., Bhat D.J., Boonmee S., Maharachchikumbura S.S.N., McKenzie E.H.C., Phookamsak R., Phukhamsakda C., et al. Fungal diversity notes 1–110: Taxonomic and phylogenetic contributions to fungal species. Fungal Divers. 2015;72:1–197. doi: 10.1007/s13225-015-0324-y. [DOI] [Google Scholar]
- 101.Crous P.W., Wingfield M.J., Roux J.J.L., Richardson D.M., Strasberg D., Shivas R.G., Alvarado P., Edwards J., Moreno G., Sharma R., et al. Fungal planet description sheets: 371–399. Persoonia. 2015;35:264–327. doi: 10.3767/003158515X690269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Noriler S.A., Savi D.C., Ponomareva L.V., Rodrigues R., Rohr J., Thorson J.S., Glienke C., Shaaban K.A. Vochysiamides a and b: Two new bioactive carboxamides produced by the new species Diaporthe vochysiae. Fitoterapia. 2019;138:104273. doi: 10.1016/j.fitote.2019.104273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Alignment data generated in the current study are available in TreeBASE (accession http://purl.org/phylo/treebase/phylows/study/TB2:S27334). All sequence data are available in NCBI GenBank following the accession numbers in the manuscript.