Abstract
Background:
Mutations in the EGFR signaling pathway play an important role in the development of colorectal cancer (CRC). Mutations in these genes, like KRAS and BRAF, affect the treatment strategies and associated with poor prognosis and relative resistance to anti-EGFR therapies. Our aim was to conduct a systematic and meta-analysis on all studies that have been conducted on the prevalence of these gene mutations in Iranian CRC patients.
Methods:
Four science citation index databases (MEDLINE, EMBASE, Web of Science and Cochrane library) and local databases were searched up to March 2018 with related keywords. Two reviewers independently screened and extracted the data. Quality of all included studies was assessed using an adapted checklist from STROBE. A random-effect model was used to calculate the total prevalence of KRAS and BRAF mutations in CRC subjects by the event rate (ER). Meta-regression was utilized to explore heterogeneity causes.
Results:
In total, from 573 records, 23 eligible studies (2662 patients) were included for data extraction and analysis. In 18 of 23 included studies, the prevalence of KRAS mutations was 33.9% (95% CI=30.1-37.9) with I2=65.17 (p<0.001). The occurrence of KRAS mutations in codon 12 and 13 was 76.9% (95% CI = 70.4-82.3%) with I2=84.88 (p<0.001) and 23.5% (95% CI=17.9-30.3) with I2=85.85 (p<0.001), respectively. In 9 of 23 studies, the BRAF mutation rate was 3.2% (95% CI=0.003-13.6) with I2=88.61 (p<0.001).
Conclusion:
The prevalence of these mutations in CRC patients shows a significant difference in the different regions of Iran, which is probably due to environmental and racial factors.
Key Words: Colorectal cancer, CRC, Mutation, KRAS, BRAF, Iran
Colorectal cancer (CRC) is the third leading cause of cancer deaths around the world (1). It is the second most common cancer in the Iranian population (2,3). About half of the patients with CRC develop distant metastasis (4,5). Chemotherapy is one of the best strategies for metastatic colorectal cancer (mCRC) patients and several combinations of chemotherapeutic agents are utilized to extend survival for patients with mCRC (6). The development and progression of CRC is a multi-stage process that begins with polyps, and with the accumulation of extensive genetic alterations ultimately leads to malignant and metastatic tumors. The EGFR (Epidermal growth factor receptor) signaling pathway plays a significant role in regulating cell processes including proliferation, differentiation, cell motility and apoptosis. Mutations in oncogenic genes of this pathway such as KRAS and BRAF are commonly found in CRC patients and play a significant role in the development of metastatic colorectal cancer (7–9). In recent years, with the understanding of the molecular mechanisms involved in mCRC, therapeutic strategies have been rapidly improved with the development of molecular-targeted anti-cancer drugs. Regulating the EGFR expression has become a potential target for the prevention of mCRC progression (10).
For this purpose, the anti-EGFR antibodies, such as cetuximab and panitumumab, have been used in the treatment of patients with mCRC. To predict the patient's response to these therapies requires a series of prognostic biomarkers. The KRAS gene status is a prerequisite for the response to anti-EGFR therapy (11–13). It is known that KRAS gene mutations are associated with resistance to anti-EGFR therapy in CRC patients (14–16). Several studies have shown that KRAS mutations are present in over 30% of CRC patients and most of these occur in codon 12 (14,17–20). Moreover, other genetic alterations in the EGFR signaling pathway may be associated with poor prognosis and resistance to anti-EGFR therapy (14,21,22). The BRAF gene, like KRAS, is one of the downstream EGFR oncogenic genes, and its somatic mutations activate the EGF receptor signaling in tumor cells (23). Oncogenic BRAF mutations are associated with poor prognosis and survival in mCRC patients (24). Also, some studies have shown that mutations in this gene may be associated with resistance to anti-EGFR therapy (22,25). The prevalence of BRAF mutations is an average of 10% in CRC patients (20, 26). Several studies have already investigated the prevalence of these mutations in Iran. Most of these studies were limited by region and sample size. Therefore, systematic review of all studies can lead to a more accurate picture of the prevalence of KRAS and BRAF mutations in Iranian CRC patients. Also, one previous systematic review and meta-analysis conducted by Payandeh et.al. estimated the prevalence of KRAS mutations in Iranian CRC patients using 11 studies (27). Therefore, awareness of the prevalence of KRAS and BRAF mutations in different CRC patient groups may guide strategies for treatment and testing. Because of the small sample size or non-representative sample collection, current individual studies may not be suitable for estimating the overall rate of these mutations. The mutation rate reported in previous studies varies dramatically and the reported prevalence rates are thus unsuitable for applications to other populations. Our aim in this study is to systematically review and meta-analyze all studies that examined the frequency of KRAS and BRAF mutations in different regions of Iran to obtain a more accurate estimate of KRAS and BRAF mutation rate in Iranian CRC patients.
Methods
Search strategy of publication: This research is a systematic review study and was done according to PRISMA guidelines (28) and was based on a registered protocol in the PROSPERO database (CRD42016053577, available online at https://www.crd.york.ac.uk/prospero). In this study, all related papers until March 2018 indexed in science citation index databases (EMBASE, Medline, Web of Science and Cochrane library) and local databases (Irandoc and SID) were extracted and assessed. Searching strategy was performed in the English language and was done by the combination of two groups’ free-text words and Medical Subject Headings (MeSH)/EMTREE terms (group 1: colon cancer, colorectal cancer, colon tumor, colorectal tumor, rectum, metastatic colon cancer, metastatic colorectal cancer, CRC, mCRC, colon neoplasm, colorectal neoplasm, colorectal carcinoma and colon carcinoma; group 2: KRAS, K-RAS, c-KRAS, BRAF, B-RAF and c-BRAF) and "Iran". The details of our search strategy are provided in Supplementary table 1.
Table 1.
Characteristics of the KRAS screening studies included in the meta-analysis
| Author | Year | Location | Sex (%men) |
Mean age (%) | Sample size (N) |
Tumor Stage* 1 & 2 |
Tumor Stage* 3 & 4 |
Tumor Location* (colon) | Tumor Location* (rectum) | Tumor Grade* (poorly) | Tumor Grade* (Moderate) | Tumor Grade* (Well) | Method | Total KRAS Mutation (%) |
KRAS (codon 12) % | KRAS (codon 13) % | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aminifard [117] | 2016 | Kermanshah | 79.0 | 51.50 | 33 | 0.0 | 100 | 55.0 | 45.0 | 9.0 | 21.2 | 69.8 | Sequencing | 36.4 | 91.6 | 8.4 | |
| Bishehsari [31] | 2006 | Tehran | 56.6 | NR | 182 | NR | NR | 71.0 | 29.0 | NR | NR | NR | Sequencing | 37.4 | 66.0 | 32.5 | |
| Dolatkhah 1[48] | 2014 | Tabriz | 76.7 | 61.77 | 30 | 36.6 | 46.7 | NR | NR | 6.7 | 26.7 | 50.0 | Sequencing | 20.0 | NR | NR | |
| Dolatkhah 2[49] | 2016 | Tabriz | 65.0 | 61.90 | 100 | 37.0 | 29.0 | 72.0 | 28.0 | 3.0 | 22.0 | 49.0 | Sequencing | 26.0 | 61.5 | 34.6 | |
| Koochak [29] | 2016 | Tehran | 57.3 | 56.50 | 1000 | 0.0 | 100 | NR | NR | 16.4 | 38.4 | 43.9 | HRMA/ Pyrosequencing |
33.6 | 85.1 | 14.9 | |
| Lary [32] | 2011 | Tehran | 59.2 | 54.00 | 54 | 57.4 | 42.6 | 76.0 | 24.0 | 1.9 | 31.5 | 66.6 | RFLP | 35.2 | 57.9 | 52.6 | |
| Naghibalhossaini [50] | 2011 | Shiraz | 65.4 | NR | 110 | 64.5 | 35.5 | NR | NR | 43.6 | 40.0 | 4.5 | RFLP/SSCP/ Sequencing | 27.9 | NR | NR | |
| Naseri [33] | 2016 | Birjand | 70.0 | NR | 50 | NR | NR | 74.0 | 26.0 | 8.0 | 36.0 | 30.0 | Pyrosequencing | 28.0 | 71.4 | 28.6 | |
| Omidifar [34] | 2015 | Shiraz | 55.0 | 59.00 | 100 | NR | NR | NR | NR | NR | NR | NR | Sequencing | 32.0 | 75.0 | 28.2 | |
| Payandeh 1 [35] | 2016 | Kermanshah | 61.4 | 57.70 | 83 | 0.0 | 100 | 61.4 | 38.6 | 7.3 | 32.5 | 60.2 | HRMA/ AS-PCR/ Pyrosequencing | 44.6 | 81.1 | 18.9 | |
| Payandeh 2 [51] | 2015 | Kermanshah | 57.6 | 57.27 | 33 | 0.0 | 100 | 60.6 | 39.4 | NR | NR | NR | HRMA/ AS-PCR/ Pyrosequencing | 36.4 | 75.0 | 25.0 | |
| Roudbari [36] | 2016 | Tehran | 68.0 | 56.96 | 50 | NR | NR | NR | NR | NR | NR | NR | Reverse Dotbloting | 42.0 | 71.6 | 28.4 | |
| Saleh Jazi [37] | 2017 | Esfahan | 55.8 | 61.20 | 52 | 55.7 | 44.3 | 48.1 | 51.9 | 15.4 | 42.3 | 23.1 | Sequencing | 15.4 | 75.0 | 25.0 | |
| Shahriari Ahmadi [38] | 2018 | Tehran | 54.2 | 52.9 | 144 | NR | NR | 79.2 | 20.8 | 18.7 | 64.6 | 16.7 | RFLP/ HRMA/ Pyrosequencing | 52.1 | NR | NR | |
| Shemirani [39] | 2011 | Tehran | 76.8 | 49.00 | 48 | NR | NR | NR | NR | NR | NR | NR | Sequencing | 31.2 | 83.3 | 16.7 | |
| Sobhani [40] | 2011 | Tehran | 41.4 | 58.00 | 59 | NR | NR | 47.5 | 52.5 | NR | NR | NR | Sequencing | 20.3 | 83.3 | 16.7 | |
| Taghipour Zahir [41] | 2016 | Yazd | 61.2 | 56.00 | 90 | 47.7 | 52.3 | 76.7 | 23.3 | 2.2 | 38.9 | 35.5 | NR | 40.0 | NR | NR | |
| Vakil [42] | 2016 | Tehran | 57.8 | 57.00 | 99 | 38.4 | 61.6 | 100 | 0.0 | 9.1 | 35.3 | 55.5 | Pyrosequencing | 38.4 | 89.4 | 10.5 | |
N: Number, NR: Not reported, *: Percentage of all samples
N: Number, NR: Not reported, *: Percentage of all samples
To select the most relevant papers, searching through title, keywords and abstracts was done. The initial search included 601 articles which entered the review process for title and abstract (figure 1). Studies were selected if they met the following criteria: Cross-sectional, case series or cohort studies that evaluated the prevalence of KRAS or BRAF gene mutations in fresh frozen, FFPE or biopsy colorectal cancer samples; All studies in which the patient number with KRAS or BRAF mutations was more than one; All relevant data published by the same group or author; published data at valid international meetings; There was no limitation in methods used to detect mutations. The exclusion criteria were as follows: Studies that were unrelated to the prevalence of KRAS and BRAF mutations, such as those that focused solely on determining the clinical and pathological characteristics of the disease; Studies that examined only one of the 12 or 13 KRAS gene codons; reviews, case reports; Studies on colorectal cancer cell lines or animals, and studies that used KRAS-positive patients to monitor mutations in the BRAF gene (29).
Figure 1.
The flow chart for retrieving eligible studies used in the meta-analysis
Quality assessment and data extraction: Two reviewers (AY and MA) independently screened the titles and abstracts to avoid any kind of bias, and any discrepancies were resolved through discussion including a third reviewer (MJZ). The quality of studies was assessed using an adapted checklist from the STROBE (strengthening the reporting of observational studies in epidemiology) for the reporting of the quality of observational studies. Eventually, the studies that earned the minimum score entered the data extraction phase for meta-analysis. After removing unrelated, duplicated (using the Mendeley v.1.17.12 software) and low-quality articles by reviewing full papers, 23 studies (total of KRAS and BRAF) met the criteria and the following data were extracted from studies: author’s name, year of publication, location of study, sex (percentage of male), patient's mean age, sample size, clinicopathological information, mutation detection method, number of KRAS and BRAF mutations and number of KRAS mutant codons. One reviewer (AY) extracted data into Microsoft Excel 2016 sheet and second reviewer (MA) reviewed it.
Statistical analysis: Data analysis was performed using comprehensive meta-analysis 3.0 (CMA 3.0) software. Tests of homogeneity investigate whether the difference between studies in meta-analysis is only by chance. The degree of heterogeneity between the studies was performed using the Cochran Q heterogeneity test and the I2 statistics. A I2 value of >50% and or a Q-statistic value of p<0.05 suggests the presence of significant heterogeneity. It would be invalid to pool such data using the fixed-effects model. Meta-regression was conducted to investigate the effective factors on heterogeneity. These factors included publication year, location of study, patient's mean age, percentage of male, sample size, tumor grade, tumor stage and tumor location. Possibility of bias in reporting was checked using Begg test.
Results
Search results and study selection: In total, 601 records were retrieved by searching the databases. The flowchart of search records and screening process has been summarized in figure 1. After removing the unrelated and duplicated records (n=545), the 56 remaining records were screened using the title and abstract, and the 28 records that did not meet the inclusion criteria were excluded. Then, the full-text of the remaining 28 records was reviewed in detail, of which 5 records were excluded due to the reasons given in figure 1. Finally, 23 out of 28 studies met the inclusion criteria and were included in the meta-analysis. Of the 23 included studies, 14 studies were on KRAS mutations (29, 30, 39–42, 31–38), 5 on BRAF mutations (43–47), and 4 on mutations in both (48–51).
Study characteristics: Characteristics of KRAS and BRAF mutations studies are summarized in tables 1 and 2, respectively. The 23 eligible studies included 2662 patients with a mean age of 56.26, that KRAS and BRAF mutations were screened in 2317 and 590 patients, respectively. All studies were done in cross-sectional form. Bishesari et al. (31) conducted the earliest study in September 2006; while Shahriari-Ahmadi et al. (38) conducted the latest study in June 2018.
Table 2.
Characteristics of the BRAF screening studies included in the meta-analysis
| Author | Year | Location |
Sex
(%men) |
Mean age (%) | Sample size (N) |
Tumor Stage*
1 & 2 |
Tumor Stage*
3 & 4 |
Tumor Location* (colon) | Tumor Location* (rectum) | Tumor Grade* (poorly) | Tumor Grade* (Moderate) | Tumor Grade* (Well) | Method |
Total BRAF Mutation
(%) |
| Brim [43] | 2008 | Shiraz | 64.0 | 59.80 | 53 | 23.0 | 77.0 | NR | NR | 4.0 | 42.0 | 54.0 | Sequencing | 2.0 |
| Dolatkhah 1 [48] | 2014 | Tabriz | 76.7 | 61.77 | 30 | 36.6 | 46.7 | NR | NR | 6.7 | 26.7 | 50.0 | Sequencing | 0.0 |
| Dolatkhah 2 [49] | 2016 | Tabriz | 65.0 | 61.90 | 100 | 37.0 | 29.0 | 72.0 | 28.0 | 3.0 | 22.0 | 49.0 | Sequencing | 0.0 |
| Ghaffarpour [44] | 2011 | Tehran | 37.0 | 61.30 | 27 | 59.2 | 40.8 | NR | NR | 7.4 | 25.9 | 66.6 | Sequencing | 3.7 |
| Javadi [45] | 2014 | Shiraz | 55.0 | 59.80 | 100 | 66.0 | 34.0 | 100.0 | 0.0 | 0.0 | 17.0 | 83.0 | Sequencing | 0.0 |
| Naghibalhossaini [50] | 2011 | Shiraz | 65.4 | NR | 110 | 64.5 | 35.5 | NR | NR | 43.6 | 40.0 | 4.5 | PCR-RFLP/SSCP/ Sequencing | 0.0 |
| Mohammadi Asl [46] | 2014 | Ahwaz | 45.0 | 44.25 | 80 | NR | NR | NR | NR | NR | NR | NR | PCR-RFLP/sequencing | 46.3 |
| Molaie [47] | 2016 | Tehran | 56.4 | 51.00 | 85 | NR | NR | NR | NR | NR | NR | NR | Sequencing | 0.0 |
| Payandeh 2 [51] | 2015 | Kermanshah | 57.6 | 57.27 | 5 | 0.0 | 100 | 60.6 | 39.4 | NR | NR | NR | HRMA/AS-PCR/ pyrosequencing | 0.0 |
N: Number, NR: Not reported, *: Percentage of all samples
N: Number, NR: Not reported, *: Percentage of all samples
The sample size in the KRAS studies ranged from 33 to 1000 and in the BRAF studies from 5 to 110. Patients in ten studies were from Tehran (central) (29, 31, 32, 36, 38–40, 42, 44, 47), four studies from Shiraz (southwest) (34, 43, 45, 50), three from Kermanshah (west) (30, 35, 51), two from Tabriz (northwest) (48, 49), one study from Isfahan (central) (37), one from Yazd (central) (41), one from Ahwaz (southwest) (46) and one from Birjand (east) (33). In most studies (65.2%), FFPE samples were used and in one study, both FFPE and fresh frozen tissue were used (38). In 20 out of 23 (86.9%) studies, the incidence of CRC was higher in men than women. Different molecular methods were used for KRAS and BRAF mutations screening (tables 1 and 2); however, in most studies (60.8%), the screening method was based on polymerase chain reaction (PCR) and direct sequencing (30, 31, 47–50, 34, 37, 39, 40, 43–46). Also, the screening method was not reported in one study (41).
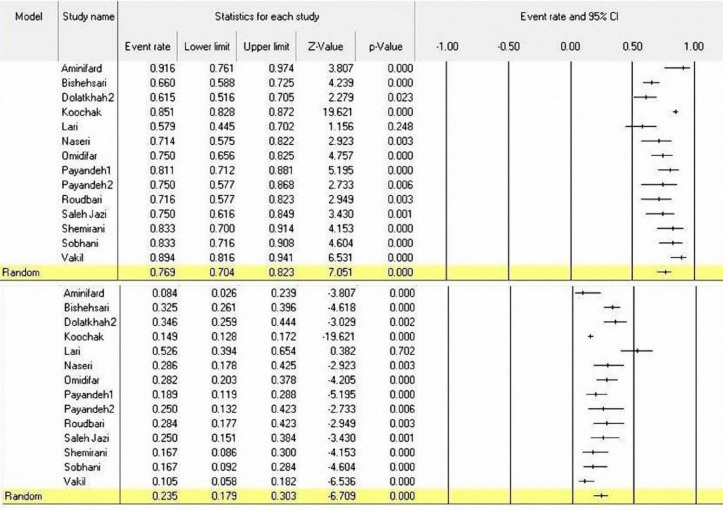
The prevalence of KRAS mutations in CRC patients: The prevalence of KRAS mutations has been reported in 18 of the 23 included studies (2317 patients). In these studies, the highest frequency of KRAS mutations reported by Shahriari-Ahmadi et al. was at a rate of 52.1% (95%CI: 44.0-60.1%) (38), and the lower frequency of KRAS mutations reported by Saleh Jazi et al. was at a rate of 15.4% (95%CI: 7.9-27.9%) (37). Using the random effect model, the overall prevalence of KRAS mutations was 33.9% (95%CI: 30.1-37.9%) with I2=65.17% and (p<0.001) (figure 2). Moreover, in 14 out of 18 studies, the frequency of KRAS mutations was reported in codons 12 and 13. Figure 3 shows the frequency of mutant codons among all KRAS mutations. The overall prevalence for codon 12 and 13 mutations was 76.9% (95% CI: 70.4-82.3%) with I2=84.88 (p<0.001) and 23.5% (95% CI=17.9-30.3) with I2=85.85 (p<0.001), respectively (figure 3).
Figure 2.
A Forest plot for the prevalence of KRAS mutation in Iranian CRC patients
Figure 3.
A Forest plot for the prevalence of KRAS mutant codons 12 & 13 in Iranian CRC patients
Moreover, the systematic review of included papers showed that the KRAS G12D and then KRAS G12V, were the most common amino acid change in KRAS sequence (data not shown). Given the heterogeneity of the findings of the studies and the differences in mutation rates in different regions of Iran, the possible factors causing heterogeneity were included in meta-regression analysis to find the main source of heterogeneity. The analysis showed that the year of publication, location and mean age variables were involved in heterogeneity. However, other variables such as gender, tumor grade, tumor stage, and sample size did not affect heterogeneity (table 3).
Table 3.
Effects of possible factors in the between-study heterogeneity in the prevalence of KRAF mutations (meta-regression model).
| Factors suspected of developing heterogeneity | Coefficient | Standard error | P-value | |
|---|---|---|---|---|
| Year | 2011 2014 2015 2016 2017 2018 |
-0.40 -0.87 -0.18 -0.08 -1.19 -0.60 |
0.25 0.51 0.28 0.21 0.45 0.29 |
0.11 0.09 0.53 0.70 0.008* 0.03* |
| Location | Birjand Kermanshah Shiraz Tabriz Tehran Yazd |
0.76 1.30 0.85 0.56 1.17 1.30 |
0.61 0.51 0.51 0.53 0.47 0.56 |
0.21 0.01* 0.09 0.30 0.01* 0.02* |
| Tumor grade | Poorly Moderately Well |
-1.29 0.52 0.26 |
1.74 0.46 0.64 |
0.46 0.25 0.68 |
| Tumor stage | 1 & 2 3 & 4 |
-0.62 0.68 |
0.43 0.36 |
0.14 0.06 |
| Tumor location | Colon Rectum |
1.67 -1.67 |
0.90 0.90 |
0.07 0.07 |
| Sex (male) | -0.29 | 1.11 | 0.79 | |
| Mean age | -0.07 | 0.03 | 0.01* | |
| Sample size | 0.0001 | 0.0004 | 0.81 | |
The prevalence of BRAF mutations in CRC patients: In 9 out of 23 studies (590 patients), the prevalence of BRAF mutations in CRC patients was analyzed using the random effect model. The prevalence rates of BRAF mutations were between 0% (42,47–51) and 46.3% (95% CI: 35.7-57.2%) (46). However, the frequency of mutation in most studies (66.6%) was 0%. In more than half of the studies (5 out of 9), the screening of BRAF mutations was based on the detection of BRAF-V600E mutation. The overall prevalence of BRAF mutations was 2.3% (95% CI: 0.003-13.6%) (Figure 4) with I2=88.61 and (p<0.001). The meta-regression analysis for finding the source of heterogeneity has shown that location and means age variables are likely to be effective in heterogeneity of studies (table 4).
Figure 4.
A Forest plot for the prevalence of BRAF mutation in Iranian CRC patients
Table 4.
Effects of possible factors in the between-study heterogeneity in the prevalence of BRAF mutations (meta-regression model)
| Factors suspected of developing heterogeneity | Coefficient | Standard error | P-value | |
|---|---|---|---|---|
| Year | 2011 2014 2015 2016 |
-0.36 1.01 1.49 -1.33 |
3.20 2.99 3.81 3.24 |
0.91 0.73 0.69 0.68 |
| Location | Kermanshah Shiraz Tabriz Tehran |
-2.25 -4.45 -4.56 -3.75 |
1.49 0.73 1.03 0.86 |
0.13 <0.001* <0.001* <0.001* |
| Tumor grade | Poorly Moderately Well |
-2.24 1.69 1.51 |
3.82 5.45 2.35 |
0.55 0.75 0.52 |
| Tumor stage | 1 & 2 3 & 4 |
-2.41 2.89 |
2.22 2.04 |
0.28 0.16 |
| Tumor location | Colon Rectum |
- - |
- - |
- - |
| Sex (male) | -8.43 | 6.82 | 0.21 | |
| Mean age | -0.22 | 0.06 | <0.001* | |
| Sample size | -0.02 | 0.03 | 0.50 | |
Publication bias: Begg funnel plot was performed to assess the reliability of the results. To investigate the presence of publication bias, a funnel plot of random effects calculated from individual studies examined the prevalence of KRAS (figure 5) and BRAF (figure 6) mutations in Iranian CRC patients. There was no strong indication of publication bias among the studies included in the meta-analysis for KRAS mutation, but there were some biases among BRAF mutation studies.
Figure 5.
Begg Funnel plot for publication bias of the prevalence of KRAS mutation in the studies
Figure 6.
Begg Funnel plot for publication bias of the prevalence of BRAF mutation in the studies
Discussion
The EGFR signaling pathway plays an important role in the development of colorectal cancer. The EGF receptor is responsible for the activation of genes involved in the RAS-RAF-MAPK and PI3K-AKT pathways (23,52). Nowadays, there is a significant improvement in the treatment of mCRC using specific monoclonal antibodies (mAbs) targeting the EGF receptor (53). However, it has been found that a significant number of patients with mCRC due to the activation of the EGFR signaling pathway by the mutation in the downstream oncogene genes of this pathway (KRAS, BRAF, NRAS and PIK3CA) was associated with a poor response to this treatment (7,54,55). Screening of the oncogenic mutations in the EGFR signaling pathway is an important part of determining the therapeutic strategy for the CRC patients and their mutation status may influence their response to anti- EGFR therapy (54).
Several meta-analyses have been performed to study KRAS and BRAF mutations in melanoma (56), non-small cell lung cancer (NSCLC) (57), colorectal cancer (58,59) and papillary thyroid cancer (60). In the present study, 23 studies, including 2662 CRC patients from different regions of Iran were analyzed for the prevalence of KRAS and BRAF somatic mutations. The rate of KRAS mutations was 33.9% (95% CI: 30.1-38.0%), which is close to published data from the United States (35.7% and 35%) (61,62) (31%) (63), China (32%) (64), Japan (33.5%) (65), Taiwan (33.5%) (66), Russia (35.9%) (67), France (33.8) (68), United Kingdom (36.9%) (69) and Brazil (36%) (70); although it differs from some published data from Germany (41%) (71), Italy (62.2%, 43%, 43% and 52.2%) (72–75), Turkey (44%) (76), India (20.5% and 23%) (77,78), Pakistan (13%) (79), Saudi Arabia (42.2% and 56%) (80,81), Morocco (24%) (82), Egypt (11% and 18.4%) (83,84), Thailand (23%) (85) and Korea (20.7%) (86). The prevalence of KRAS mutations in patients with CRC varies worldwide (11-66.1%) (59,61,81–83,85–91,63,92–101,64,102–104,66,68,75–77,80). The variability of the findings of studies may be related to ethnicity, geographical area and even lifestyle (10,71,94,105,106). Given the relatively high prevalence of KRAS mutations in the Iranian CRC patients, these findings alert physicians to patients that may be at elevated risk for carrying a tumor with KRAS mutation as the focus for screening.
Based on our results, most KRAS mutations occurred in codon 12 and then codon 13. These results are approximately similar to those of other studies (17,63,76,79,82). For example, in a study conducted in Belgium, the prevalence of KRAS mutations was 36.3%, with 91% of mutations in codons 12 and 13 (107). In the study of Dobre et al., the occurrence of KRAS mutations in codons 12 and 13 was 79.3% and 19.7%, respectively (108). An Indian study reported that KRAS mutations occur in 87% and 13% of cases in codons 12 and 13, respectively (109). This is also consistent with the reported data in a large series of 989 patients from Brazil where 87% of mutations were in codon 12 and 13% in codon 13 (70). However, a study in the Greek population reported a low frequency of KRAS mutations in codon 12 (29.3%) (110). Also, our findings showed that the KRAS G12D and then KRAS G12V were the most common amino acid change (data not shown),that were consistent with published data (86,100,103,104,108).
Iran is one of the largest world's country both by geographical area and by multi-ethnic populations and this can explain the diversity of the results of various studies. The meta-regression analysis of studies also identified the location as one of the factors contributing to the heterogeneity. Although, differences in the mutation status of KRAS in different regions of Iran due to the small sample size and the absence of comprehensive studies were observed, there is no definitive result of the geographical distribution effect on the prevalence of KRAS mutations. Therefore, more comprehensive researches are necessary to further describe the mechanisms involved. For example, a large-scale study from mCRC patients reported no significant difference in the mutant codons according to geographical areas (111). These findings suggest that the location may not have a great impact on the mutant codons. In addition, the heterogeneity of the data can be explained by the difference in the data collection method, the aim of data collection and the different time periods of sample collection.
BRAF is a part of the RAF gene family that plays a role like the KRAS in the EGFR pathway. BRAF mutations are less common compared to KRAS mutations. The BRAFV600E mutation is one of the most commonly genetic changes in colorectal cancer. The distribution of BRAF mutations significantly varies from 1. 1% to 25% across the globe (26, 59, 85–91, 93–95, 61, 96–101, 103, 104, 109, 112, 64, 66, 68, 71, 75, 76, 82). The most common BRAF mutation in this study was V600E, which occurs at c.1799T>A due to substitution. This mutation results in a constitutive activation of the MAPK pathway, which modulates the cell growth signals to transcriptional activity of regulatory genes in cell cycle (113). In the present study, the prevalence of BRAFV600E mutations in Iranian CRC subjects was 3.2% (95% CI: 0.003-13.6), which is consistent with the previous findings of Asian studies (1.1% to 4.9%) (66,86,89,104,114) but lower than several Western studies (9.5% to 15.8%) (61,68,87,103,112). Heterogeneity is a problem that may influence the interpretation of the findings of meta-analyses. This study showed a high degree of heterogeneity which is why a random effect model was utilized for analysis. The meta-regression analysis showed that study location and mean age are likely to contribute to the high levels of heterogeneity. Similarly to other studies, these differences in the frequency of mutations may be influenced by race, geographic distribution, life style, and other variables of study such as mean age (115,116). The geographic region can have a great impact on the tumor mutation patterns. In this meta-analysis, nine studies that had been done in different regions of Iran, were selected for geographical optimization.
This study offers several strengths. A key strength of our study is the large number and range of studies included for estimating the overall rate of KRAS and BRAF mutations in Iranian CRC patient groups. We carried out a systematic search strategy with suitable inclusion criteria, result in the large number of studies in such a meta-analysis conducted in Iranian CRC patients. These results can act as the reference point for future researches or treatment strategies. We utilized a suitable approach to choosing the random effect model for pooling studies by taking into account the presence or absence of heterogeneity. Furthermore, we utilized tests for publication bias assessing the reliability of the results.
There were several limitations in this study that should be considered in interpreting the findings. First, one of the most important limitations of this study was the form of data reporting incomplete data, and lack of response from authors. Although all 23 studies obtained the qualitative criteria for inclusion in the analysis, even in these articles, as shown in tables 1 and 2, some important characteristics such as patients mean age, distant metastasis, differentiation and tumor stage are not mentioned. Second, a search of the gray literature was not conducted, and therefore publication bias could not be removed entirely. Third, in many studies, several hotspot regions such as the exon 4 of the KRAS gene and exon 11 of the BRAF gene were not screened. Fourth, some studies reported the mutation prevalence only in mCRC patients. Fifth, the number of studies on BRAF mutations was low and in most studies, mutation screening conducted just for the BRAFV600E resulting in the possibility of bias. Also, the small sample size of published studies increased the risk of bias. Therefore, further studies on BRAF mutations seem necessary to prove our study results. Sixth, heterogeneity between studies like the study design and methods for diagnosis of KRAS and BRAF mutations may potentially affect the study results. Finally, we did not investigate the relationship between mutations and the clinical stage in many studies due to lack of data.
In conclusion despite some limitations, we achieved remarkable results in this meta-analysis. This meta-analysis showed that the total prevalence of KRAS and BRAF mutations varies in different regions of Iran. Moreover, comparing the results of this study with other studies showed that the prevalence of these mutations in Iranian patients with CRC is also diverse in comparison with other populations, which may be related to ethnicity, geographical distribution and lifestyle.
To targeted treatment process of CRC patients, reduces undesirable effects of treatment and prevents waste of resources, estimating an accurate overall rate of KRAS and BRAF mutations in Iranian patients is important. Finally, this study recommends that due to high rate of KRAS mutations in Iranian patients with CRC, it should be considered in determining treatment strategies.
Authors’ contributions
AY contributed to the concept and design of the study. AY and MA designed and carried out search strategies. AA and MN conducted the meta-analysis and interpretation of data. AY, AA and MA drafted the manuscript. MJZ supervised the findings of this work and reviewed the final manuscript. All authors read, revised and confirmed the manuscript.
Acknowledgments
The authors thank Prof. AliAkbar Haghdoost for systematic review guidance, and Dr. Nasrollah Saleh-gohari for reviewing the manuscript.
Funding:
The authors have not declared any specific grant for this work from any funding organization in the public, commercial or not-for-profit sectors.
Conflict of interest:
The authors declare that they have no conflict of interest.
References
- 1.Zeuner A, Todaro M, Stassi G, De Maria R. Colorectal cancer stem cells: from the crypt to the clinic. Cell Stem Cell. 2014;15:692–705. doi: 10.1016/j.stem.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 2.Mohebbi E, Nahvijou A, Hadji M, et al. Iran cancer statistics in 2012 and projection of cancer incidence by 2035. Basic Clin Cancer Res. 2018;9:3–22. [Google Scholar]
- 3.Ansari R, Mahdavinia M, Sadjadi A, et al. Incidence and age distribution of colorectal cancer in Iran: Results of a population-based cancer registry. Cancer Lett. 2006;240:143–7. doi: 10.1016/j.canlet.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 5.Parkin DM, Bray F, Ferlay J, Pisani P. Global Cancer Statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 6.Meyerhardt JA, Mayer RJ. Systemic therapy for colorectal cancer. N Engl J Med. 2005;352:476–87. doi: 10.1056/NEJMra040958. [DOI] [PubMed] [Google Scholar]
- 7.McCubrey JA, Steelman LS, Abrams SL, et al. Roles of the RAF/MEK/ERK and PI3K/PTEN/AKT pathways in malignant transformation and drug resistance. Adv Enzyme Regul. 2006;46:249–79. doi: 10.1016/j.advenzreg.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–62. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 9.Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer. 2001;37:9–15. doi: 10.1016/s0959-8049(01)00231-3. [DOI] [PubMed] [Google Scholar]
- 10.Van Cutsem E, Köhne CH, Láng I, et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2011;29:2011–9. doi: 10.1200/JCO.2010.33.5091. [DOI] [PubMed] [Google Scholar]
- 11.Sorbye H, Dragomir A, Sundström M, et al. High BRAF mutation frequency and marked survival differences in subgroups according to KRAS/BRAF mutation status and tumor tissue availability in a prospective population-based metastatic colorectal cancer cohort. PLoS One . 2015;10:e0131046. doi: 10.1371/journal.pone.0131046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bardelli A, Siena S. Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer. J Clin Oncol. 2010;28:1254–61. doi: 10.1200/JCO.2009.24.6116. [DOI] [PubMed] [Google Scholar]
- 13.Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, et al. Oncogenic activation of the RAS/ RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res. 2007;67:2643–8. doi: 10.1158/0008-5472.CAN-06-4158. [DOI] [PubMed] [Google Scholar]
- 14.Wilson PM, LaBonte MJ, Lenz HJ. Molecular markers in the treatment of metastatic colorectal cancer. Cancer J. 2010;16:262–72. doi: 10.1097/PPO.0b013e3181e07738. [DOI] [PubMed] [Google Scholar]
- 15.Misale S, Yaeger R, Hobor S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–6. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allegra CJ, Jessup JM, Somerfield MR, et al. American society of clinical oncology provisional clinical opinion: Testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091–6. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- 17.Bos JL, Fearon ER, Hamilton SR, et al. Prevalence of ras gene mutations in human colorectal cancers. Nature. 1987;327:293–7. doi: 10.1038/327293a0. [DOI] [PubMed] [Google Scholar]
- 18.Bos JL. Ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–9. [PubMed] [Google Scholar]
- 19.Andreyev HJN, Norman AR, Cunningham D, et al. Kirsten ras mutations in patients with colorectal cancer: the ‘RASCAL II’ study. Br J Cancer. 2001;85:692–6. doi: 10.1054/bjoc.2001.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang JY, Richardson BC. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 2005;6:322–7. doi: 10.1016/S1470-2045(05)70168-6. [DOI] [PubMed] [Google Scholar]
- 21.Laurent-Puig P, Cayre A, Manceau G, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol. 2009;27:5924–30. doi: 10.1200/JCO.2008.21.6796. [DOI] [PubMed] [Google Scholar]
- 22.Smith CG, Fisher D, Claes B, et al. Somatic profiling of the epidermal growth factor receptor pathway in tumors from patients with advanced colorectal cancer treated with chemotherapy ± cetuximab. Clin Cancer Res . 2013;19:4104–13. doi: 10.1158/1078-0432.CCR-12-2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peyssonnaux C, Eychène A. The Raf/MEK/ERK pathway: new concepts of activation. Biol Cell. 2001;93:53–62. doi: 10.1016/s0248-4900(01)01125-x. [DOI] [PubMed] [Google Scholar]
- 24.Yokota T, Ura T, Shibata N, et al. BRAF mutation is a powerful prognostic factor in advanced and recurrent colorectal cancer. Br J Cancer. 2011;104:856–62. doi: 10.1038/bjc.2011.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen Y, Wang J, Han X, et al. Effectors of Epidermal growth factor receptor pathway: the genetic profiling of kras, braf, pik3ca, nras mutations in colorectal cancer characteristics and personalized medicine. PLoS One. 2013;8:e81628. doi: 10.1371/journal.pone.0081628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 27.Payandeh M, Amirifard N, Sadeghi M, et al. The prevalence of KRAS mutation in colorectal cancer patients in Iranian population: A systematic review and meta-analysis study. Biomed Res Ther. 2017;4:1693–704. [Google Scholar]
- 28.Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA Statement Checklist of items to include when reporting. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koochak A, Rakhshani N, Karbalaie Niya MH, et al. Mutation analysis of kras and braf genes in metastatic colorectal cancer: a first large scale study from Iran. Asian Pac J Cancer Prev . 2016;17:603–8. doi: 10.7314/apjcp.2016.17.2.603. [DOI] [PubMed] [Google Scholar]
- 30.Amirifard N, Sadeghi E, Farshchian N, et al. Evaluation of KRAS gene mutations in metastatic colorectal cancer patients in Kermanshah province. Asian Pacific J Cancer Prev. 2016;17:3085–8. [PubMed] [Google Scholar]
- 31.Bishehsari F, Mahdavinia M, Malekzadeh R, et al. Patterns of K-ras mutation in colorectal carcinomas from Iran and Italy (a Gruppo Oncologico dell’Italia Meridionale study): Influence of microsatellite instability status and country of origin. Ann Oncol. 2006;17:91–6. doi: 10.1093/annonc/mdl959. [DOI] [PubMed] [Google Scholar]
- 32.Lary S, Ghaffarzadegan K, Afsharnezhad S, Hoeinkhani S, Mohammadi G. Evaluation of K-ras mutations in colorectal carcinoma (CRC) patients of East-North of Iran. Clin. Biochem . 2011;44:S200–1. [Google Scholar]
- 33.Mohsen N, Ahmadreza S, Fatemeh H, Fatemeh H, Fariba ER. Frequency of K-RAS and N-RAS gene mutations in colorectal cancers in Southeastern Iran. Asian Pac. J. Cancer Prev. 2016;17:4511–15. [PubMed] [Google Scholar]
- 34.Omidifar N, Geramizadeh B, Mirzai M. K-ras mutation in colorectal cancer, a report from Southern Iran. Iran J Med Sci. 2015;40:454–60. [PMC free article] [PubMed] [Google Scholar]
- 35.Payandeh M, Shazad B, Sadeghi M, Shahbazi M. Correlation between RAS test results and prognosis of metastatic colorectal cancer patients: A report from western Iran. Asian Pac J Cancer Prev. 2016;17:1729–32. doi: 10.7314/apjcp.2016.17.4.1729. [DOI] [PubMed] [Google Scholar]
- 36.Roudbari F, Poopak B, Sheikhsofla F, et al. Evaluation of frequency of kirsten rat sarcoma gene mutations in Iranian colorectal cancer. Tehran Univ Med J. 2016;74:392–9. [Google Scholar]
- 37.Jazi MS, Zahri S, Latif-Navid S, Talebi A. Relationship between common KRAS gene mutations and clinicopathological features of patients with colorectal cancer in Isfahan, Iran. Govaresh. 2017;22:39–46. [Google Scholar]
- 38.Shahriari-Ahmadi A, Ansarinejad N, Fardad F, Abbaszadeh M, Sadeghi M. KRAS and NRAS testing in metastatic colorectal cancer in Central Iran (Tehran): A review on literature of the middle east. Indian J Med Paediatr Oncol. 2018;39:210–14. [Google Scholar]
- 39.Shemirani AI, Haghighi MM, Milanizadeh S, et al. The role of kras mutations and MSI status in diagnosis of colorectal cancer. Gastroenterol Hepatol Bed Bench. 2011;4:70–5. [PMC free article] [PubMed] [Google Scholar]
- 40.Sobhani S, Ghaffarpour M, Mostakhdemin HZ, et al. The Prevalence Of Common Mutation Frequency In K-Ras Codons 12, 13 In Iranian Colorectal Cancer Patients. Genetics In The 3rd Millennium. 2010;8:2011–18. [Google Scholar]
- 41.Zahir ST, Nazemian-Yazdi M, Arasteh P, et al. Clinicopathological features and survival rate of colorectal adenocarcinoma patients with and without a KRAS mutation: A five-year study in Yazd, Iran. Asian Pacific J. Cancer Prev. 2016;17:3417–22. [PubMed] [Google Scholar]
- 42.Vakil L, Najafipour R, Rakhshani N, et al. Investigation of FIH-1 and SOCS3 expression in KRAS mutant and wild-type patients with colorectal cancer. Tumor Biol. 2016;37:8841–8. doi: 10.1007/s13277-015-4723-1. [DOI] [PubMed] [Google Scholar]
- 43.Brim H, Mokarram P, Naghibalhossaini F, et al. Impact of BRAF, MLH1 on the incidence of microsatellite instability high colorectal cancer in populations based study. Mol Cancer. 2008;7:68. doi: 10.1186/1476-4598-7-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghaffarpour M, Pargoo EM, Sobhani S, et al. Abstract 3799: PIK3CA and BRAF mutational status and association with clinicopathological features in Iranian colonrectal cancer patients. Cancer Res. 2011;71:3799. [Google Scholar]
- 45.Javadi F, Geramizadeh B, Mirzai M. BRAF gene mutation analysis in colorectal cancer in South of Iran. Ann Color Res. 2014;2:e19917. [Google Scholar]
- 46.Mohammadi Asl J, Almasi S, Tabatabaiefar MA. High frequency of BRAF proto-oncogene hot spot mutation V600E in cohort of colorectal cancer patients from Ahvaz City, southwest Iran. Pakistan J Biol Sci. 2014;17:565–9. doi: 10.3923/pjbs.2014.565.569. [DOI] [PubMed] [Google Scholar]
- 47.Molaei M, Farahani RK, Maftouh M, et al. Lack of BRAFV600E mutation in stage I and II of colorectal cancer. Gastroenterol Hepatol Bed Bench. 2016;9:94–9. [PMC free article] [PubMed] [Google Scholar]
- 48.Dolatkhah R, Somi MH, Bonyadi MJ, et al. Colorectal cancer in Iran: molecular epidemiology and screening strategies. J Cancer Epidemiol. 2015;2015:643020. doi: 10.1155/2015/643020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dolatkhah R, Somi MH, Kermani IA, et al. Association between proto-oncogene mutations and clinicopathologic characteristics and overall survival in colorectal cancer in East Azerbaijan, Iran. Onco Targets Ther. 2016;9:7385–95. doi: 10.2147/OTT.S116373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Naghibalhossaini F, Hosseini HM, Mokarram P, Zamani M. High frequency of genes’ promoter methylation, but lack of BRAF V600E Mutation among Iranian colorectal cancer patients. Pathol Oncol Res. 2011;17:819–25. doi: 10.1007/s12253-011-9388-5. [DOI] [PubMed] [Google Scholar]
- 51.Payandeh M, Sadeghi M, Sadeghi E, Gholami F. Analysis of KRAS , BRAF and NRAS in patients with colorectal cancer: the First Report of Western Iran. Am J Cancer Prev. 2015;3:19–22. [Google Scholar]
- 52.Harari PM, Allen GW, Bonner JA. Biology of interactions: Antiepidermal growth factor receptor agents. J Clin Oncol. 2007;25:4057–65. doi: 10.1200/JCO.2007.11.8984. [DOI] [PubMed] [Google Scholar]
- 53.Scaltriti M, Baselga J. The epidermal growth factor receptor pathway: a model for targeted therapy. Clin. Cancer Res. 2006;12:5268–72. doi: 10.1158/1078-0432.CCR-05-1554. [DOI] [PubMed] [Google Scholar]
- 54.De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–62. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 55.De Roock W, Piessevaux H, De Schutter J, et al. KRAS wild-type state predicts survival and is associated to early radiological response in metastatic colorectal cancer treated with cetuximab. Ann Oncol. 2008;19:508–15. doi: 10.1093/annonc/mdm496. [DOI] [PubMed] [Google Scholar]
- 56.Safaee Ardekani G, Jafarnejad SM, Tan L, Saeedi A, Li G. The prognostic value of BRAF mutation in colorectal cancer and melanoma: A systematic review and meta-analysis. PLoS One. 2012;7:e47054. doi: 10.1371/journal.pone.0047054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen D, Zhang LQ, Huang JF, et al. BRAF mutations in patients with non-small cell lung cancer: a systematic review and meta-analysis. PLoS One. 2014;9:e101354. doi: 10.1371/journal.pone.0101354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang ZY, Wu XY, Huang YF, et al. Promising biomarkers for predicting the outcomes of patients with KRAS wild-type metastatic colorectal cancer treated with anti-epidermal growth factor receptor monoclonal antibodies: A systematic review with meta-analysis. Int J Cancer. 2013;133:1914–25. doi: 10.1002/ijc.28153. [DOI] [PubMed] [Google Scholar]
- 59.Therkildsen C, Bergmann TK, Henrichsen-Schnack T, et al. The predictive value of KRAS, NRAS, BRAF, PIK3CA and PTEN for anti-EGFR treatment in metastatic colorectal cancer: A systematic review and meta-analysis. Acta Oncol. 2014;53:852–64. doi: 10.3109/0284186X.2014.895036. [DOI] [PubMed] [Google Scholar]
- 60.Li C, Lee KC, Schneider EB, et al. BRAF V600E mutation and its association with clinicopathological features of papillary thyroid cancer: a meta-analysis. J Clin Endocrinol Metab. 2012;97:4559–70. doi: 10.1210/jc.2012-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ogino S, Nosho K, Kirkner GJ, et al. PIK3CA mutation is associated with poor prognosis among patients with curatively resected colon cancer. J Clin Oncol. 2009;27:1477–84. doi: 10.1200/JCO.2008.18.6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liao X, Lochhead P, Nishihara R, et al. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med. 2012;367:1596–1606. doi: 10.1056/NEJMoa1207756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Phipps AI, Buchanan DD, Makar KW, et al. KRAS-mutation status in relation to colorectal cancer survival: the joint impact of correlated tumour markers. Br J Cancer. 2013;108:1757–64. doi: 10.1038/bjc.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berg M, Danielsen SA, Ahlquist T, et al. DNA sequence profiles of the colorectal cancer critical gene set KRAS-BRAF-PIK3CA-PTEN-TP53 related to age at disease onset. PLoS One . 2010;5:e13978. doi: 10.1371/journal.pone.0013978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakanishi R, Harada J, Tuul M, et al. Prognostic relevance of KRAS and BRAF mutations in Japanese patients with colorectal cancer. Int J Clin Oncol. 2013;18:1042–8. doi: 10.1007/s10147-012-0501-x. [DOI] [PubMed] [Google Scholar]
- 66.Hsieh LL, Er TK, Chen CC, et al. Characteristics and prevalence of KRAS, BRAF, and PIK3CA mutations in colorectal cancer by high-resolution melting analysis in Taiwanese population. Clin Chim Acta. 2012;413:1605–11. doi: 10.1016/j.cca.2012.04.029. [DOI] [PubMed] [Google Scholar]
- 67.Yanus GA, Belyaeva AV, Ivantsov AO, et al. Pattern of clinically relevant mutations in consecutive series of Russian colorectal cancer patients. Med Oncol. 2013;30:686. doi: 10.1007/s12032-013-0686-5. [DOI] [PubMed] [Google Scholar]
- 68.Barault L, Veyrie N, Jooste V, et al. Mutations in the RAS-MAPK, PI(3)K (phosphatidylinositol-3-OH kinase) signaling network correlate with poor survival in a population-based series of colon cancers. Int J Cancer. 2008;122:2255–9. doi: 10.1002/ijc.23388. [DOI] [PubMed] [Google Scholar]
- 69.Souglakos J, Philips J, Wang R, et al. Prognostic and predictive value of common mutations for treatment response and survival in patients with metastatic colorectal cancer. Br J Cancer. 2009;101:465–72. doi: 10.1038/sj.bjc.6605164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zalis M, Vieira F, Zalcberg Renaunt J, et al. KRAS mutation profile in colorectal cancer patients in Brazil: A cohort of 989 individuals. J Clin Oncol. 2009;27:e15017. [Google Scholar]
- 71.Baldus SE, Schaefer KL, Engers R, et al. Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clin Cancer Res. 2010;16:790–9. doi: 10.1158/1078-0432.CCR-09-2446. [DOI] [PubMed] [Google Scholar]
- 72.Miglio U, Mezzapelle R, Paganotti A, et al. Mutation analysis of KRAS in primary colorectal cancer and matched metastases by means of highly sensitivity molecular assay. Pathol Res Pract. 2013;209:233–6. doi: 10.1016/j.prp.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 73.Simi L, Pratesi N, Vignoli M, et al. High-resolution melting analysis for rapid detection of KRAS, BRAF, and PIK3CA gene mutations in colorectal cancer. Am J Clin Pathol. 2008;130:247–53. doi: 10.1309/LWDY1AXHXUULNVHQ. [DOI] [PubMed] [Google Scholar]
- 74.Bozzao C, Varvara D, Piglionica M, et al. Survey of KRAS, BRAF and PIK3CA mutational status in 209 consecutive Italian colorectal cancer patients. Int J Biol Markers. 2012;27:366–74. doi: 10.5301/JBM.2012.9765. [DOI] [PubMed] [Google Scholar]
- 75.Cappuzzo F, Varella-Garcia M, Finocchiaro G, et al. Primary resistance to cetuximab therapy in EGFR FISH-positive colorectal cancer patients. Br J Cancer. 2008;99:83–89. doi: 10.1038/sj.bjc.6604439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Selcukbiricik F, Erdamar S, Ozkurt CU, et al. The role of K-RAS and B-RAF mutations as biomarkers in metastatic colorectal cancer. J BUON. 2013;18:116–23. [PubMed] [Google Scholar]
- 77.Patil H, Korde R, Kapat A. KRAS gene mutations in correlation with clinicopathological features of colorectal carcinomas in Indian patient cohort. Med Oncol. 2013;30:617. doi: 10.1007/s12032-013-0617-5. [DOI] [PubMed] [Google Scholar]
- 78.Bisht S, Ahmad F, Sawaimoon S, Bhatia S, Das BR. Molecular spectrum of KRAS, BRAF, and PIK3CA gene mutation: determination of frequency, distribution pattern in Indian colorectal carcinoma. Med Oncol. 2014;31:124. doi: 10.1007/s12032-014-0124-3. [DOI] [PubMed] [Google Scholar]
- 79.Murtaza BN, Bibi A, Rashid MU, et al. Spectrum of K ras mutations in Pakistani colorectal cancer patients. Brazilian J Med Biol Res. 2013;47:35–41. doi: 10.1590/1414-431X20133046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bader T, Ismail A. Higher prevalence of KRAS mutations in colorectal cancer in Saudi Arabia: Propensity for lung metastasis. Alexandria J Med. 2014;50:203–9. [Google Scholar]
- 81.Zahrani A, Kandil M, Badar T, et al. Clinico-pathological study of K-ras mutations in colorectal tumors in Saudi Arabia. Tumori. 2014;100:75–9. doi: 10.1700/1430.15819. [DOI] [PubMed] [Google Scholar]
- 82.Marchoudi N, Amrani Hassani Joutei H, Jouali F, Fekkak J, Rhaissi H. Distribution of KRAS and BRAF mutations in Moroccan patients with advanced colorectal cancer. Pathol Biol. 2013;61:273–6. doi: 10.1016/j.patbio.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 83.Soliman AS, Bondy ML, El-Badawy SA, et al. Contrasting molecular pathology of colorectal carcinoma in Egyptian and Western patients. Br J Cancer. 2001;85:1037–46. doi: 10.1054/bjoc.2001.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chan AO, Soliman AS, Zhang Q, et al. Differing DNA methylation patterns and gene mutation frequencies in colorectal carcinomas from Middle Eastern countries. Clin Cancer Res . 2005;11:8281–7. doi: 10.1158/1078-0432.CCR-05-1000. [DOI] [PubMed] [Google Scholar]
- 85.Chaiyapan W, Duangpakdee P, Boonpipattanapong T, Kanngern S, Sangkhathat S. Somatic mutations of K-Ras and BRAF in Thai Colorectal cancer and their prognostic value. Asian Pacific J Cancer Prev. 2013;14:329–32. doi: 10.7314/apjcp.2013.14.1.329. [DOI] [PubMed] [Google Scholar]
- 86.Kwon MJ, Lee SE, Kang SY, Choi YL. Frequency of KRAS, BRAF, and PIK3CA mutations in advanced colorectal cancers: Comparison of peptide nucleic acid-mediated PCR clamping and direct sequencing in formalin-fixed, paraffin-embedded tissue. Pathol Res Pract. 2011;207:762–8. doi: 10.1016/j.prp.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 87.Simi L, Pratesi N, Vignoli M, et al. High-resolution melting analysis for rapid Detection of KRAS, BRAF, and PIK3CA gene mutations in colorectal cancer. Am J Clin Pathol. 2008;130:247–53. doi: 10.1309/LWDY1AXHXUULNVHQ. [DOI] [PubMed] [Google Scholar]
- 88.Mao C, Zhou J, Yang Z, et al. KRAS, BRAF and PIK3CA mutations and the loss of PTEN expression in Chinese patients with colorectal cancer. PLoS One. 2012;7:e36653. doi: 10.1371/journal.pone.0036653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bando H, Yoshino T, Shinozaki E, et al. Simultaneous identification of 36 mutations in KRAS codons 61and 146, BRAF, NRAS, and PIK3CAin a single reaction by multiplex assay kit. BMC Cancer. 2013;13:405. doi: 10.1186/1471-2407-13-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Soeda H, Shimodaira H, Watanabe M, et al. Clinical usefulness of KRAS, BRAF, and PIK3CA mutations as predictive markers of cetuximab efficacy in irinotecan- and oxaliplatin-refractory Japanese patients with metastatic colorectal cancer. Int J Clin Oncol . 2013;18:670–7. doi: 10.1007/s10147-012-0422-8. [DOI] [PubMed] [Google Scholar]
- 91.Ling Y, Ying JM, Qiu T, Shan L, Guo L, Lü N. Detection of KRAS, BRAF, PIK3CA and EGFR gene mutations in colorectal carcinoma. Zhonghua Bing Li Xue Za Zhi. 2012;41:590–4. doi: 10.3760/cma.j.issn.0529-5807.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 92.White Y, Bagchi A, Van Ziffle J, et al. KRAS insertion mutations are oncogenic and exhibit distinct functional properties. Nat Commun. 2016;7:10647. doi: 10.1038/ncomms10647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lièvre A, Merlin JL, Sabourin JC, et al. RAS mutation testing in patients with metastatic colorectal cancer in French clinical practice: A status report in 2014. Dig Liver Dis. 2018;50:507–12. doi: 10.1016/j.dld.2017.12.029. [DOI] [PubMed] [Google Scholar]
- 94.Guedes JG, Veiga I, Rocha P, et al. High resolution melting analysis of KRAS, BRAF and PIK3CA in KRASexon 2 wild-type metastatic colorectal cancer. BMC Cancer. 2013;13:169. doi: 10.1186/1471-2407-13-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lurkin I, Stoehr R, Hurst CD, et al. Two Multiplex Assays That Simultaneously Identify 22 Possible Mutation Sites in the KRAS, BRAF, NRAS and PIK3CA Genes. Jones C, editor. PLoS One. 2010;5:e8802. doi: 10.1371/journal.pone.0008802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bai B, Shan L, Xie B, et al. Mutations in KRAS codon 12 predict poor survival in Chinese patients with metastatic colorectal cancer. Oncol Lett. 2017;15:3161–6. doi: 10.3892/ol.2017.7709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fumagalli D, Gavin PG, Taniyama Y, et al. A rapid, sensitive, reproducible and cost-effective method for mutation profiling of colon cancer and metastatic lymph nodes. BMC Cancer . 2010;10:101. doi: 10.1186/1471-2407-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Khan SA, Zeng Z, Shia J, Paty PB. EGFR Gene Amplification and KRAS mutation predict response to combination targeted therapy in metastatic colorectal cancer. Pathol Oncol Res . 2017;23:673–7. doi: 10.1007/s12253-016-0166-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Balschun K, Haag J, Wenke A-K, et al. KRAS, NRAS, PIK3CA Exon 20, and BRAF genotypes in synchronous and metachronous primary colorectal cancers: diagnostic and therapeutic implications. J Mol Diagn. 2011;13:436–45. doi: 10.1016/j.jmoldx.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vaughn CP, Zobell SD, Furtado LV, Baker CL, Samowitz WS. Frequency of KRAS, BRAF, and NRAS mutations in colorectal cancer. Genes Chromosom Cancer. 2011;50:307–12. doi: 10.1002/gcc.20854. [DOI] [PubMed] [Google Scholar]
- 101.Fransén K, Klintenäs M, Österström A, et al. Mutation analysis of the BRAF, ARAF and RAF-1 genes in human colorectal adenocarcinomas. Carcinogenesis. 2004;25:527–33. doi: 10.1093/carcin/bgh049. [DOI] [PubMed] [Google Scholar]
- 102.Sinha R, Hussain S, Mehrotra R, et al. Kras gene mutation and RASSF1A, FHIT and MGMT gene promoter hypermethylation: indicators of tumor staging and metastasis in adenocarcinomatous sporadic colorectal cancer in indian population. PLoS One. 2013;8:e60142. doi: 10.1371/journal.pone.0060142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Neumann J, Zeindl-Eberhart E, Kirchner T, Jung A. Frequency and type of KRAS mutations in routine diagnostic analysis of metastatic colorectal cancer. Pathol Res Pract. 2009;205:858–62. doi: 10.1016/j.prp.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 104.Zhang J, Zheng J, Yang Y, et al. Molecular spectrum of KRAS, NRAS, BRAF and PIK3CA mutations in Chinese colorectal cancer patients: analysis of 1,110 cases. Sci Rep . 2015;5:18678. doi: 10.1038/srep18678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jayasekara H, English DR, Haydon A, et al. Associations of alcohol intake, smoking, physical activity and obesity with survival following colorectal cancer diagnosis by stage, anatomic site and tumor molecular subtype. Int J Cancer. 2018;142:238–50. doi: 10.1002/ijc.31049. [DOI] [PubMed] [Google Scholar]
- 106.Ogino S, Stampfer M. Lifestyle factors and microsatellite instability in colorectal cancer: The evolving field of molecular pathological epidemiology. J Natl Cancer Inst. 2010;102:365–7. doi: 10.1093/jnci/djq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–62. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 108.Dobre M, Dinu DE, Panaitescu E, et al. KRAS gene mutations-prognostic factor in colorectal cancer? Rom J Morphol Embryol. 2015;56:671–8. [PubMed] [Google Scholar]
- 109.Bagadi SB, Sanghvi M, Nair SB, Das BR, et al. Combined mutational analysis of KRAS, NRAS and BRAF genes in Indian patients with colorectal carcinoma. Int J Biol Markers. 2012;27:27–33. doi: 10.5301/JBM.2012.9108. [DOI] [PubMed] [Google Scholar]
- 110.Symvoulakis EK, Zaravinos A, Panutsopulos D, et al. Highly conserved sequence of exon 15 braf gene and kras codon 12 mutation among greek patients with colorectal cancer. Int J Biol Markers. 2007;22:12–18. doi: 10.1177/172460080702200102. [DOI] [PubMed] [Google Scholar]
- 111.Gil Ferreira C, Aran V, Zalcberg-Renault I, et al. KRAS mutations: variable incidences in a Brazilian cohort of 8,234 metastatic colorectal cancer patients. BMC Gastroenterol. 2014;14:73. doi: 10.1186/1471-230X-14-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Velho S, Oliveira C, Ferreira A, et al. The prevalence of PIK3CA mutations in gastric and colon cancer. Eur J Cancer. 2005;41:1649–54. doi: 10.1016/j.ejca.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 113.Sharma A, Trivedi NR, Zimmerman MA, et al. Mutant V599E B-Raf Regulates growth and vascular development of malignant melanoma tumors mutant V599E B-Raf Regulates growth and vascular development of malignant melanoma tumors. Cancer Res. 2005;65:2412–21. doi: 10.1158/0008-5472.CAN-04-2423. [DOI] [PubMed] [Google Scholar]
- 114.Yip WK, Choo CW, Shian Leong VC, et al. Molecular alterations of Ras-Raf-mitogen-activated protein kinase and phosphatidylinositol 3-kinase-Akt signaling pathways in colorectal cancers from a tertiary hospital at Kuala Lumpur, Malaysia. APMIS . 2013;121:954–66. doi: 10.1111/apm.12152. [DOI] [PubMed] [Google Scholar]
- 115.Rozek LS, Herron CM, Greenson JK, et al. Smoking, gender, and ethnicity predict somatic BRAF mutations in colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:838–43. doi: 10.1158/1055-9965.EPI-09-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Slattery ML, Curtin K, Sweeney C, et al. Diet and lifestyle factor associations with CpG island methylator phenotype and BRAF mutations in colon cancer. Int J Cancer. 2007;120:656–63. doi: 10.1002/ijc.22342. [DOI] [PubMed] [Google Scholar]
- 117.Amirifard N, Sadeghi E, Farshchian N, et al. Evaluation of KRAS Gene Mutations in Metastatic Colorectal Cancer Patients in Kermanshah Province. Asian Pac J Cancer Prev. 2016;17:3085–8. [PubMed] [Google Scholar]