Abstract
We investigated Staphylococcus aureus diversity, genetic factors, and humoral immune responses against antigens via genome analysis of S. aureus isolates from chronic rhinosinusitis (CRS) patients in a long-term follow-up. Of the 42 patients who provided S. aureus isolates and serum for a previous study, 34 could be included for follow-up after a decade. Clinical examinations were performed and bacterial samples were collected from the maxillary sinus and nares. S. aureus isolates were characterized by whole-genome sequencing, and specific anti-staphylococcal IgG in serum was determined using protein arrays. S. aureus was detected in the nares and/or maxillary sinus at both initial inclusion and follow-up in 15 of the 34 respondents (44%). Three of these (20%) had S. aureus isolates from the same genetic lineage as at inclusion. A low number of single-nucleotide polymorphisms (SNPs) were identified when comparing isolates from nares and maxillary sinus collected at the same time point. The overall change of antibody responses to staphylococcal antigens over time showed great variability, and no correlation was found between the presence of genes encoding antigens and the corresponding anti-staphylococcal IgG in serum; thus our findings did not support a role, in CRS, of the specific S. aureus antigens investigated.
Keywords: chronic rhinosinusitis, immunoglobulins, Staphylococcus aureus, whole-genome sequencing, enterotoxin, long-term, carriage, antigen
1. Introduction
Chronic rhinosinusitis (CRS) affects about 10% of the European population, and is a burdensome disease in which Staphylococcus aureus is suggested to have a potential role [1]. The presence of S. aureus in healthy maxillary sinuses is low [2], but it is commonly observed in patients with CRS [3,4,5]. S. aureus is generally considered to be a harmless commensal which colonizes the nares intermittently or persistently in 20–70% of healthy individuals [6,7,8]. However, it also has the potential to cause invasive diseases ranging from mild superficial skin disorders such as folliculitis to serious conditions including sepsis and infective endocarditis [9]. S. aureus superantigens has been shown to contribute to inflammatory diseases such as rheumatoid arthritis, asthma, and atopic dermatitis [10,11,12,13], and are suggested to have an impact on CRS [14,15,16]. The nares are regarded as the most important colonization site, since eradication from the nares can result in subsequent disappearance of S. aureus from other parts of the body [7].
Important factors related to pathogenicity include staphylococcal enterotoxins, enzymes, and cell-surface-associated virulence components [17,18,19]. Staphylococcal superantigens, such as enterotoxins, have been shown to enhance mucosal eosinophilia by Th-2 cytokine release and further T cell actions in CRS with nasal polyposis [20]. S. aureus within biofilm retains its capacity to release toxins, which can promote the inflammatory response in CRS [21,22]. Levels of IgG antibody against toxic shock syndrome toxin-1 (TSST-1) and LukF-PV, a subunit of the Panton–Valentine leukocidin (PVL), has shown to be significantly higher in CRS patients than in healthy controls, which could indicate that these specific S. aureus antigens affect the pathogenesis of CRS disease [23].
Further insight into microbial and host-specific factors could be of importance for the understanding of CRS, including the role of S. aureus in the pathogenesis [1]. Long-term studies of the anti-staphylococcal immune response in CRS patients and genetic characterization of S. aureus in the sinonasal cavities in cohorts of CRS patients are rare. We therefore aimed to study the persistence of S. aureus in the nares and maxillary sinus of CRS patients by performing genomic analyses of S. aureus and by determining serum antibody responses to specific staphylococcal antigens in a long-term follow-up of a Swedish cohort of patients with CRS.
2. Materials and Methods
2.1. Collection of Serum and Bacterial Samples from Patients with CRS
Primary inclusion took place as part of a previous study performed in 2004–2010 [4]. Subjects were of age >18 years and were diagnosed with CRS by an otorhinolaryngology (ORL) specialist. Two physicians specialized in ORL (authors UT and SH) performed all inclusion procedures. All patients had had surgery when the collection of specimens was performed. No patients were immunocompromised or had on-going antibiotic treatment at time for inclusion. Three patients had short course oral corticosteroid treatment at time of inclusion. S. aureus was isolated from the maxillary sinuses of 18/42 CRS patients (43%) and the nares of 24/42 CRS patients (57%), and serum samples from 29/42 CRS patients were collected and stored. The same cohort of patients, aside from two who were deceased, was contacted between 2017 and 2019 for a follow-up study. The first data collection is referred to as time point C1 and the second as C2. Bacterial samples were collected from the maxillary sinus and nares at C2 using the same protocol as at C1. Blood samples were collected from patients at C1 and sera were stored at −80 °C pending analyses. Additional blood samples were collected at C2. Sampling techniques and handling of specimens have been described previously [4]. Culturing and species verification of S. aureus were performed in accordance with routine diagnostic procedures at the Department of Laboratory Medicine, Clinical Microbiology, Örebro University Hospital. All isolates were stored at −80 °C in preservation medium (trypticase soy broth, BD Diagnostic Systems, Sparks, MD, USA) supplemented with 0.3% yeast extract (BD Diagnostic Systems) and 29% horse serum (SVA, Uppsala, Sweden). Thirty-four of the 40 patients from the cohort agreed to be included in the follow-up study.
2.2. Antibiotic Susceptibility Testing
S. aureus isolates obtained at C1 and C2 were suspended in sterile saline to 0.5 McFarland. Susceptibility testing was performed on Mueller-Hinton II agar 3.8% w/v (BD Diagnostic Systems, Sparks, MD, USA) using the standardized disk diffusion method in accordance with the European Committee on Antimicrobial Susceptibility Testing (www.eucast.org) guidelines for the following antibiotics: cefoxitin (30 µg), fusidic acid (10 µg), erythromycin (15 µg), clindamycin (2 µg), rifampicin (5 µg), gentamicin (10 µg), trimethoprim-sulfamethoxazole (25 µg), tetracycline (30 µg), and norfloxacin (10 µg). All disks were from Oxoid, Basingstoke, UK.
2.3. DNA Sequencing and Single-Nucleotide Polymorphism (SNP) Analysis
Whole-genome sequencing (WGS) of S. aureus was performed on genomic DNA extracted using the QIAGEN DNeasy Blood and Tissue Kit (QIAGEN, Hilden, Germany), with subsequent library construction using the Nextera XT Kit (Illumina, Little Chesterford, UK); specifically, a 300-cycle kit on the NextSeq platform (Illumina) according to the manufacturer’s instructions. The genomes were assembled using SPAdes v3.10.1 and MLST typed with the MLST command-line tool (https://github.com/tseemann/mlst).
A SNP-based phylogeny was created to assess the relationship between C1 and C2 isolates, using SNPs identified using NASP [24] with the BWA algorithm to align Illumina reads from individual isolates against the chromosome from the ST45 S. aureus isolate CA-347 (GenBank accession number: CP006044) [25] after removal of duplicated regions using NUCmer. Positions with less than 10-fold coverage and less than 90% unambiguous variant calls were excluded. A midpoint rooted maximum-likelihood phylogenetic tree was calculated in IQ-TREE [26].
For determining the presence of key genes in the assembled genomes, the QIAGEN CL Genomics Workbench v12.0 (QIAGEN, Aarhus, Denmark) was used to perform a BLASTN search with positive hits required to have >75% hit length and >85% sequence similarity.
2.4. Microarray-Based Immunoglobulin Analysis
The method regarding protein microarrays was previously described by Kloppot et al. [27] and Selle et al. [28] and from that source information about the antigens could be extracted. The antibody specificity patterns of the paired serum samples from C1 and C2 were analyzed using a protein microarray-based assay comprising 61 different S. aureus-specific antigens (Alere Technologies GmbH, Jena, Germany). All purified proteins were covalently immobilized to the array surface as duplicates using different concentrations ranging from 0.01 to 0.5 mg/mL. Purified IgG antibodies from different species (human, bovine, and murine) and one HRP-labeled protein served as positive controls.
Antibody detection using these protein microarrays was performed according to the following protocol. The microarrays were first incubated twice with washing buffer (1 × PBS/0.05%, Tween 20/0.25%, and TritonX100) at 37 °C and 400 rpm for 3 min. Next, they were incubated with blocking buffer (1 × PBS/0.05%, Tween 20/0.25%, TritonX100, and 2% milk powder) at 37 °C and 300 rpm for 5 min. The diluted serum samples (1:100) were incubated for 30 min at 37 °C and 300 rpm. After a washing step as described above (37 °C, 400 rpm, 5 min), the microarrays were incubated with a diluted (1:1000) HRP-labeled anti-human IgG-HPR antibody (Sigma-Aldrich, Taufkirchen, Germany) at 37 °C and 300 rpm for 30 min. The protein arrays were then washed twice with washing buffer (37 °C, 400 rpm, 3 min). Finally, the arrays were incubated with the substrate Seramun Green (Seramun, Heidesee, Germany) for 10 min without shaking at room temperature. The protein arrays were read out with the ArrayMate, and the data were analyzed using IconoClust software according to the manufacturer’s specifications (Alere Technologies GmbH, Jena, Germany).
Relative signal intensities of defined regions (at predefined spot coordinates) on the Staph-Toxin-Array were determined during readout. The signal intensities of the individual spots were normalized (NI). For this purpose, the average intensities were first calculated from all valid pixels of the local background (BG) and all valid pixels of the spot (M). The normalization of the spot intensity against the local background was then performed using the formula NI = 1 − (M/BG). Therefore, the NI values were between the value 0 (undetectable signal) and 1 (maximum signal).
2.5. Statistical Analysis
The Wilcoxon signed-rank test was used for matched paired data, the Mann–Whitney U-test was used for comparing two independent groups, and Fisher’s exact test was used for comparing percentages of independent groups. Holm–Bonferroni correction was applied to control for false discovery rate in multiple hypothesis tests [29]. A two-sided p-value < 0.05 was considered statistically significant. All statistical analyses were conducted in version 22 of IBM SPSS Statistics (IBM Corp., Armonk, NY, USA) and version 4.01 of the R statistical package (R Foundation for Statistical Computing, Vienna, Austria).
2.6. Ethical Approval
All procedures in this study were performed in accordance with the ethical standards of the national research committee and with the 1964 Helsinki declaration. The study was approved by the regional ethical review board in Uppsala, Sweden (refs: 2005:011/1, 2017:322).
3. Results
3.1. Presence of S. aureus in CRS Patients and Antibiotic Susceptibility
Thirty-four of 40 (85%) CRS patients could be included in the long-term follow-up (Figure 1). Of these, 14 (41%) had S. aureus in both the nares and maxillary sinus at initial inclusion (C1) and 13 (38%) had S. aureus in both locations at follow-up after approximately a decade (C2). In 15/34 (44%) patients, S. aureus was present in the nares and/or maxillary sinus at both initial inclusion and follow-up, and in 6/34 (18%) patients, S. aureus was present in both the nares and the maxillary sinus at both time points. Five of 34 (15%) patients had no S. aureus at C1 but had a positive culture at C2, and another 5/34 (15%) had a positive culture at C1 and a negative culture at C2 (Figure 1). Two patients had S. aureus only in the nares at follow-up.
Figure 1.
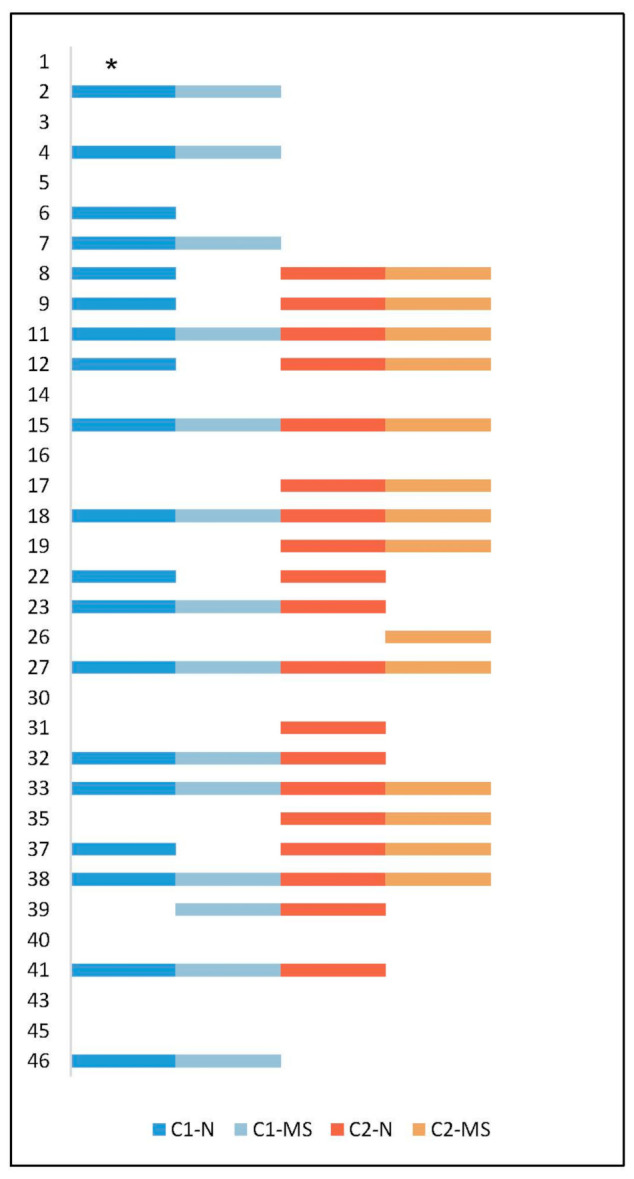
Presence of Staphylococcus aureus in the nares and maxillary sinus in 34 patients at initial collection (C1) and at follow-up (C2) after approximately 11 years. The Y-axis shows the study ID for all 34 patients. One step on the X-axis symbolizes S. aureus isolated at a specific time point and site. Sample sites: maxillary sinus (MS), nares (N). * One sample from nares at C1 was not available.
The investigated S. aureus isolates were almost fully susceptible to all tested antibiotics. Four isolates from four different patients displayed resistance. One nasal S. aureus isolate (clonal complex (CC)30) obtained at C1 was resistant to tetracycline, one isolate (CC8) from maxillary sinus at C1 was resistant to cefoxitin and thus methicillin-resistant but otherwise susceptible to all tested antibiotics, one isolate (CC12) obtained from the maxillary sinus at C1 was resistant to fusidic acid, and finally one nasal isolate (CC398) at C2 was resistant to both clindamycin and erythromycin.
3.2. SNP-Based Phylogenetic Analysis
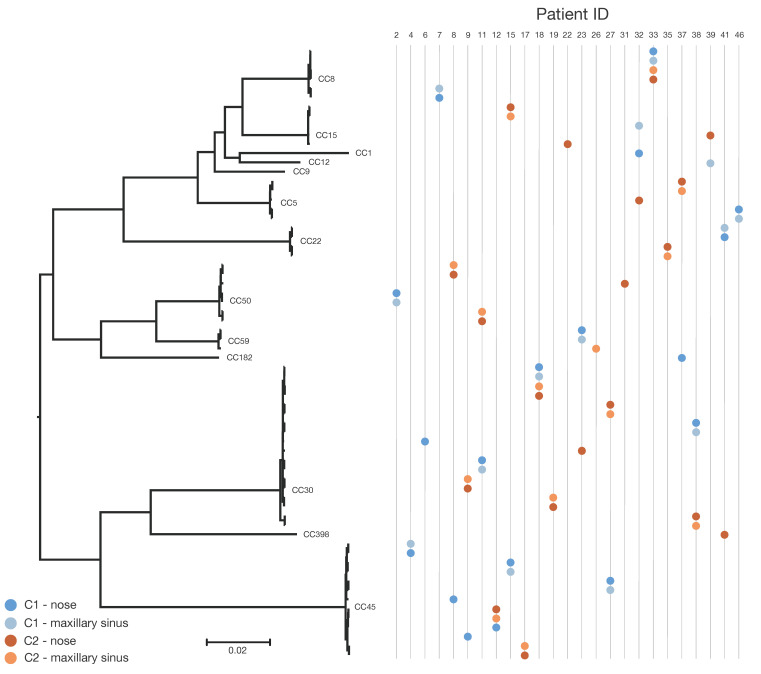
Among the 66 S. aureus isolates, 110,026 SNPs were identified across the conserved core genome of 77.5% (~2.21 Mbp), but generally a low number of SNPs was observed within hosts when comparing S. aureus isolates from the nares and maxillary sinus collected at the same time point. Figure 2 represent the relatedness of the S. aureus isolates based on SNP differences. All but one of the 27 paired S. aureus isolates collected from the nares and maxillary sinus collected at the same time point (C1 or C2) displayed the same clonal complex (CC). Three of the 15 patients with S. aureus in the maxillary sinus and/or nares at both time points presented identical S. aureus isolates (<67 SNPs over a decade). These patients (IDs 12, 18, and 33) were sampled 11, 11, and 13 years apart, respectively. Two of the three had S. aureus in all four samplings. The median number of SNPs for three strains with identical lineages at C1 and C2 was 48, with a range of 29–67. These persistent S. aureus isolates with same lineage showed no resistance to any antibiotics at either time point.
Figure 2.
Midpoint rooted phylogenetic tree based on core genome single-nucleotide polymorphisms (SNPs) at initial collection (C1) and follow-up after 8–15 years (C2; mean follow-up: 11 years) in S. aureus isolated from CRS patients. Clonal complexes are presented. Based on 110,026 SNPs in the ~2.21 Mbp conserved core genome of the reference chromosome. Scale bar indicates substitutions per site. Nasal sample collected at C1 for patient number 39 was missing.
Thirteen different CCs were found and the most prevalent CCs among the isolates were CC30 (27%, 18/66) and CC45 (20%, 13/66); details are given in Table 1. These findings are illustrated using a midpoint rooted phylogeny showing the clustering of isolates according to sequence types and clonal lineages (Figure 2). Genes encoding cell-wall-associated proteins (IsaA, Plc, Efb-C, and SCIN) were present in all isolates. One isolate was lacking the gene (scn) encoding SCIN. The tst-1 gene was only present in 24% of isolates, and lukF-PV was absent from all isolates.
Table 1.
Single-nucleotide polymorphisms (SNPs) detected between the S. aureus isolated at both sample sites at initial collection (C1) and/or after 8–15 years (C2; mean follow-up: 11 years) based on a conserved core genome of ~2.23 Mbp across the collection. Sample site: maxillary sinus (MS), nares (N).
| Patient ID | Clonal Complex | Time Point | Sample Site | SNP Differences between C1 and C2 | SNP Differences Comparing C1 to C1 or C2 to C2 |
|---|---|---|---|---|---|
| 2 c | CC50 | C1 | N | Not applicable | 0 |
| 2 | CC50 | C1 | MS | ||
| 4 c | CC45 | C1 | N | Not applicable | 1 |
| 4 | CC45 | C1 | MS | ||
| 6 | CC30 | C1 | N | Not applicable | Not applicable |
| 7 c | CC8 | C1 | N | Not applicable | 2 |
| 7 | CC8 | C1 | MS | ||
| 8 | CC45 | C1 | N | 37,758–37,761 | 3 |
| 8 | CC50 | C2 | N | ||
| 8 | CC50 | C2 | MS | ||
| 9 | CC45 | C1 | N | 33,462–33,463 | 1 |
| 9 | CC30 | C2 | N | ||
| 9 | CC30 | C2 | MS | ||
| 11 | CC30 | C1 | N | 32,890–32,892 | 1 |
| 11 | CC30 | C1 | MS | ||
| 11 | CC50 | C2 | N | 3 | |
| 11 | CC50 | C2 | MS | ||
| 12 | CC45 | C1 | N | 29–30 a | 1 |
| 12 | CC45 | C2 | MS | ||
| 12 | CC45 | C2 | N | ||
| 15 | CC45 | C1 | N | 41,088–41,089 | 1 |
| 15 | CC45 | C1 | MS | ||
| 15 | CC15 | C2 | N | 2 | |
| 15 | CC15 | C2 | MS | ||
| 17 b | CC45 | C2 | N | Not applicable | 0 |
| 17 | CC45 | C2 | MS | ||
| 18 | CC30 | C1 | N | 65–67 a | 2 |
| 18 | CC30 | C1 | MS | ||
| 18 | CC30 | C2 | N | 0 | |
| 18 | CC30 | C2 | MS | ||
| 19 b | CC30 | C2 | N | Not applicable | 2 |
| 19 | CC30 | C2 | MS | ||
| 22 | CC9 | C1 | N | 18,896 | Not applicable |
| 22 | CC15 | C2 | N | ||
| 23 | CC59 | C1 | N | 32,884–32,885 | 3 |
| 23 | CC59 | C1 | MS | ||
| 23 | CC30 | C2 | N | ||
| 26 | CC59 | C2 | MS | Not applicable | Not applicable |
| 27 | CC45 | C1 | N | 33,500–33,505 | 1 |
| 27 | CC45 | C1 | MS | ||
| 27 | CC30 | C2 | N | 4 | |
| 27 | CC30 | C2 | MS | ||
| 31 | CC50 | C2 | N | Not applicable | Not applicable |
| 32 | CC1 | C1 | N | 13,939–17,065 | 15,884 |
| 32 | CC15 | C1 | MS | ||
| 32 | CC5 | C2 | N | ||
| 33 | CC8 | C1 | N | 48–51 a | 2 |
| 33 | CC8 | C1 | MS | ||
| 33 | CC8 | C2 | N | 5 | |
| 33 | CC8 | C2 | MS | ||
| 35 b | CC22 | C2 | N | Not applicable | 25 |
| 35 | CC22 | C2 | MS | ||
| 37 | CC182 | C1 | N | 29,807 | 0 |
| 37 | CC5 | C2 | N | ||
| 37 | CC5 | C2 | MS | ||
| 38 | CC30 | C1 | N | 817–818 | 2 |
| 38 | CC30 | C1 | MS | ||
| 38 | CC30 | C2 | N | 1 | |
| 38 | CC30 | C2 | MS | ||
| 39 | CC12 | C1 | MS | 12,182 | Not applicable |
| 39 | CC15 | C2 | N | ||
| 41 | CC22 | C1 | N | 33,882–33,883 | 1 |
| 41 | CC22 | C1 | MS | ||
| 41 | CC398 | C2 | N | ||
| 46 c | CC5 | C1 | N | Not applicable | 1 |
| 46 | CC5 | C1 | MS |
a Considered as persistent carriage. b No growth of S. aureus at time of initial inclusion. c No growth of S. aureus at follow up.
3.3. Anti-Staphylococcal Antibodies in CRS Patients
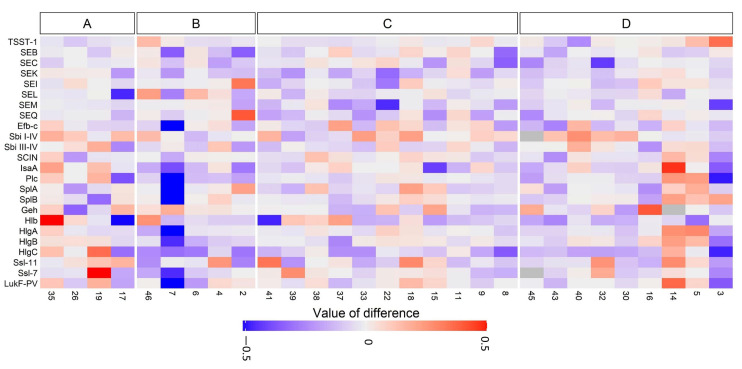
The humoral immune response towards 61 S. aureus-specific antigens was determined in sera from the 29/34 (85%) CRS patients whose serum samples were available from both time points. We focused on serum antibody responses against selected virulence factors that may be of importance for CRS, especially those directed at staphylococcal enterotoxins and microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) (Figure 3). The overall change in antibody levels showed great variability over time (Figure 3). Comparing specific antibody responses between sera from the five (5/29, 17%) patients who lost S. aureus colonization and the four (4/29, 14%) who became colonized with S. aureus showed no statistically significant change in antibody levels. Nine (31%) of the 29 patients showed no S. aureus at either C1 or C2, while 11 (38%) showed S. aureus at both time points. We found no statistically significant change in anti-staphylococcal IgG in serum when comparing these two groups.
Figure 3.
Heat map presenting changes in IgG responses toward staphylococcal antigens in serum from 29 CRS patients at initial collection (C1) and after 8–15 years (C2; mean follow-up: 11 years). Serum samples were available from 29/34 (85%) CRS patients at both time points. (A) Patients with S. aureus only at C2, (B) patients with S. aureus only at C1, (C) patients with S. aureus at both C1 and C2, and (D) patients with no S. aureus at either C1 or C2. Patient ID is indicated on the X-axis.
When considering the differences for anti-staphylococcal IgG in all analyzed sera, patients who lost their S. aureus between C1 and C2 showed significantly lower median total IgG levels than patients who became colonized with S. aureus at C2 (p = 0.03). There was no significant difference in median total IgG levels in serum from patients without S. aureus at C1 and C2 and those with S. aureus at both time points (p = 1.0). Two of the three patients who were regarded as persistent carriers of identical S. aureus strains had provided sera for analysis (patient numbers 18 and 33). Neither of them differed from the rest of the patients in terms of antibody profile.
In an effort to determine a presumably direct connection between IgG responses and specific staphylococcal antigens, we used whole-genome sequencing to assess the presence of the corresponding genes in S. aureus isolates from the 29 patients for whom serum was available. There was no correlation regarding presence of a specific gene in the strain and existence of anti-staphylococcal antibody response towards the corresponding staphylococcal factor in the host. We found stable IgG responses against TSST-1 over time, except for one patient with a clearly increased level at C2. All 29 CRS patients showed antibodies toward TSST-1, but the tst-1 gene was only present in 24% of the S. aureus isolates. These isolates were collected from five patients, and belonged to CC30 (n = 6) and CC50 (n = 1). None of the collected isolates contained lukF-PV, which contributes to encoded PVL, but 89% of the patients (25/28) showed antibodies towards this antigen. Genes encoding cell-wall-associated proteins (IsaA, Plc, and Efb-C) were present in all isolates. The genes for enterotoxins (TSST-1, SEB, SEC, SEI, SEK, SEL, SEM, and SEQ), other secreted factors (Sbi, LukF-PV, HlgA, HlgB, HlgC, Hlb, SplA, SCIN, SplB, Ssl-11, Ssl-7), and cell-wall-associated proteins (IsaA, Plc, Sbi, Efb-C) showed no correlation with IgG levels in serum.
4. Discussion
S. aureus has been suggested to have an impact on CRS, and so this long-term follow-up of CRS patients aimed to investigate persistence of S. aureus in the nares and maxillary sinus and to determine the humoral immune response against S. aureus antigens. We also used whole-genome sequencing to examine the presence of the corresponding genes in sinonasal S. aureus. To the best of our knowledge, no previous studies have evaluated serum IgG responses to virulence factors in CRS or performed genetic analyses of persistent S. aureus over such a long time span. Studies of anti-staphylococcal immune response that use isolates from clinical settings rather than laboratory strains are rare. S. aureus was found in the nares and/or maxillary sinus of 44% of CRS patients at initial inclusion and follow-up.
The intra-host SNP variation was very low when comparing isolates from nares and maxillary sinus collected at the same time point, and these isolates were therefore considered to originate from the same strain given the mutation rate of 2–7 SNPs/year for S. aureus [30]. There was one patient at C1 with a nasal isolate belonging to CC1 and a maxillary sinus isolate belonging to CC15, but otherwise these highly consistent findings indicate colonization from the nares to the maxillary sinus as one joint sinonasal milieu. We could also demonstrate a persistence of specific S. aureus in the nares and maxillary sinus over a decade in three of 15 (20%) CRS patients. Other studies have shown a similar proportion of persistent S. aureus carriers in the anterior nares (12–30% of the population), but criteria for a classification as persistent carriage are not standardized. A minimum of two cultures, often within a few weeks of each other, has been suggested as a basis for classification [6,7,31,32]. In a follow-up study from 1999, 17 persistent nasal carriers were re-investigated after eight years and only three of them could still be regarded as persistent carriers [33]. This is in accordance with the S. aureus persistence rate in the present study. A strength of this study is that persistence of S. aureus is stated by identification of CC and not just by findings of S. aureus. In addition, 23/34 (29%) of patients were regarded as intermittent nasal carriers due to having S. aureus of different CCs.
All except four isolates were susceptible to all tested antibiotics. These four were from non-persisting S. aureus carriers and one of these was an MRSA. The low resistance rate of the isolates in our study likely also reflects the restricted use of antibiotics in Sweden. Basic treatment for CRS symptoms comprises topical application of corticosteroids together with daily nasal saline irrigations and antibiotics [34], but in Sweden antibiotics are seldom used; in selected cases, an isoxazolyl-penicillin is preferably used as first choice. Treatment of CRS with macrolides and doxycycline is common in other countries [35], and may result in emergence of resistance.
The two major clonal complexes identified in our collection were CC30 and CC45, which represented 47% of all S. aureus isolates in our study. None of them showed resistance to any of the antibiotics tested. These clonal complexes are also common among the general population [36,37], and so CRS does not seem to be associated with a particular S. aureus genotype. Genes for cell-wall-associated proteins were highly present in the isolates. Thus, genes important for colonization were present in both persisting strains and intermittent strains. The presence of the tst-1 gene in 24% of isolates and lukF-PV in none contradicts previous results indicating these two virulence factors to be of significance in CRS [23].
We found an overall large variability in antibody levels against staphylococcal antigens. Even if the genes for enterotoxins, other secreted factors, and cell-wall-associated proteins were detected, only a weak correlation to IgG levels in serum was observed. S. aureus colonization of other body sites, and previous infections in particular, may be a stronger stimulator of IgG responses possessing the gene encoding effector protein.
The most frequent carriage site of S. aureus in humans is the anterior nares, but S. aureus can also be found at other body sites such as the vagina, rectum, skin, and gastrointestinal tract [7,38,39]. The anterior nares seem to function as a reservoir for carriage and hence spread, and so elimination from the nares can result in subsequent loss of the bacteria from other parts of the body [7]. This indicates the importance of the nares for carriage. A study using a twin population demonstrated that host genetics does not strongly determine the microbiota, and suggested that S. aureus is a major part of the nasal community in some individuals and a distinct indicator of the ecological milieu in the nose of certain patients [8].
There was a significant reduction of mean IgG toward staphylococcal antigens in sera from patients who lost S. aureus colonization between C1 and C2 compared to IgG levels in sera from patients displaying a positive S. aureus culture at follow-up. However, the lack of change of immune response in serum from patients without S. aureus at C1 and C2 compared to IgG levels in serum from patients with S. aureus at both time points provides conflicting results. Anti-staphylococcal antibody levels in serum reflect an ability to protect, to modify the course of infection, and to reduce the risk of complications of a staphylococcal infection [40,41], but our knowledge is limited regarding their role in chronic inflammatory disease. Antibody levels have previously been shown to be stable over years in healthy individuals [42]. Neutralizing serum antibodies are common but also widely variable in healthy populations [4,43,44]. However, higher IgG antibodies toward TSST-1 and SEA have been observed in persistent S aureus carriers than in non-carriers [40].
All of the 29 patients for whom serum and S. aureus isolates were available from C1 and C2 showed antibodies toward TSST-1. The tst-1 gene was only present in 24% of the S. aureus isolates. None of the collected isolates contained LukF-PV, encoding part of the PVL toxin, but 89% showed antibodies toward that antigen. Hence, the role of anti-staphylococcal antibodies in CRS is not obvious. A review by Holtfreter et al. concluded that antibodies against a wide range of staphylococcal enterotoxins are common in the healthy population, and increase during infection [45]. Also, Radke et al. identified a hierarchy of anti-staphylococcal proteins regarding which antigens the immune system was recognizing from S. aureus [46]. In addition, previous experience of staphylococcal infections such as skin and soft tissue infections with various S. aureus strains will probably also affect the immune response toward leucocidins and not only CRS. In addition, some individuals could be more prone to variation in antibody production. In a study by van Belkum et al., an increase in antibodies was only found in individuals with persistent nasal colonization by S. aureus [47]. Furthermore, it has been shown that raised antibody levels are present in S. aureus nasal carriers to a greater extent than in non-carriers [39]. Antibody responses in our study were not correlated to the presence of corresponding genes, indicating a previous infection or a stronger stimulator of immune response elsewhere. It would have been desirable to know the S. aureus carriage state at other body site as well as previous S. aureus infections between C1 and C2, since this could have affected the results, but this information is difficult to collect for such a long-term follow-up study. Information regarding gene expressions would also have been valuable.
5. Conclusions
In a Swedish cohort of CRS patients, we found decade-long persistence of S. aureus from the sinonasal site in 20% of our 34 patients, displaying a median of 37 SNP differences (range: 30–68). Carriage was intermittent in 35% of cases. A low number of SNPs across the core genome differed when comparing S. aureus from the nares and maxillary sinus collected at the same time point, indicating colonization of S. aureus from the nares to the maxillary sinus. The overall alterations of anti-staphylococcal antibodies over time showed great variability, and minor support for an impact of S. aureus on CRS. The vast majority of S. aureus isolates were susceptible to all tested antibiotics, including the S. aureus strains that had persisted for a decade.
Author Contributions
U.T., B.S., and S.H. participated in the conception and design of the study. U.T. and S.H. contacted the patients for recruitment, performed the inclusion procedures, and carried out the data acquisition. R.E., S.M., and E.M. performed the antibody array experiments. M.S. performed the whole-genome sequencing and SNP analyses. U.T., B.S., M.S., and R.E. interpreted the results. Y.C. and U.T. were involved in the computational statistical analysis of the data. U.T., M.S., and B.S. drafted the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by ALF founding Region Örebro County and the Research Committee of Örebro County Council OLL-929623, OLL-878861.
Institutional Review Board Statement
All procedures in this study were performed in accordance with the ethical standards of the national research committee and with the 1964 Helsinki declaration. The study was approved by the regional ethical review board in Uppsala, Sweden (refs: 2005:011/1, 2017:322).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Conflicts of Interest
The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Derycke L., Perez-Novo C., Van Crombruggen K., Corriveau M.N., Bachert C. Staphylococcus aureus and chronic airway disease. World Allergy Organ. J. 2010;3:223–228. doi: 10.1097/WOX.0b013e3181ecd8ae. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abou-Hamad W., Matar N., Elias M., Nasr M., Sarkis-Karam D., Hokayem N., Haddad A. Bacterial flora in normal adult maxillary sinuses. Am. J. Rhinol. Allergy. 2009;23:261–263. doi: 10.2500/ajra.2009.23.3317. [DOI] [PubMed] [Google Scholar]
- 3.Thanasumpun T., Batra P.S. Endoscopically-derived bacterial cultures in chronic rhinosinusitis: A systematic review. Am. J. Otolaryngol. 2015;36:686–691. doi: 10.1016/j.amjoto.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 4.Thunberg U., Soderquist B., Hugosson S. Bacterial findings in optimised sampling and characterisation of S. aureus in chronic rhinosinusitis. Eur. Arch. Otorhinolaryngol. 2017;274:311–319. doi: 10.1007/s00405-016-4239-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biel M.A., Brown C.A., Levinson R.M., Garvis G.E., Paisner H.M., Sigel M.E., Tedford T.M. Evaluation of the microbiology of chronic maxillary sinusitis. Pt 1Ann. Otol. Rhinol. Laryngol. 1998;107:942–945. doi: 10.1177/000348949810701107. [DOI] [PubMed] [Google Scholar]
- 6.Kluytmans J., van Belkum A., Verbrugh H. Nasal carriage of Staphylococcus aureus: Epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 1997;10:505–520. doi: 10.1128/CMR.10.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wertheim H.F., Melles D.C., Vos M.C., van Leeuwen W., van Belkum A., Verbrugh H.A., Nouwen J.L. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 2005;5:751–762. doi: 10.1016/S1473-3099(05)70295-4. [DOI] [PubMed] [Google Scholar]
- 8.Liu C.M., Price L.B., Hungate B.A., Abraham A.G., Larsen L.A., Christensen K., Stegger M., Skov R., Andersen P.S. Staphylococcus aureus and the ecology of the nasal microbiome. Sci. Adv. 2015;1:e1400216. doi: 10.1126/sciadv.1400216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lowy F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 10.Laudien M., Gadola S.D., Podschun R., Hedderich J., Paulsen J., Reinhold-Keller E., Csernok E., Ambrosch P., Hellmich B., Moosig F., et al. Nasal carriage of Staphylococcus aureus and endonasal activity in Wegener’s granulomatosis as compared to rheumatoid arthritis and chronic rhinosinusitis with nasal polyps. Clin. Exp. Rheumatol. 2010;28(Suppl. 57):51–55. [PubMed] [Google Scholar]
- 11.Seiti Yamada Yoshikawa F., Feitosa de Lima J., Notomi Sato M., Alefe Leuzzi Ramos Y., Aoki V., Leao Orfali R. Exploring the role of Staphylococcus aureus toxins in atopic dermatitis. Toxins. 2019;11:321. doi: 10.3390/toxins11060321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tabarya D., Hoffman W.L. Staphylococcus aureus nasal carriage in rheumatoid arthritis: Antibody response to toxic shock syndrome toxin-1. Ann. Rheum. Dis. 1996;55:823–828. doi: 10.1136/ard.55.11.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bachert C., Claeys S.E., Tomassen P., van Zele T., Zhang N. Rhinosinusitis and asthma: A link for asthma severity. Curr. Allergy Asthma Rep. 2010;10:194–201. doi: 10.1007/s11882-010-0096-0. [DOI] [PubMed] [Google Scholar]
- 14.Bachert C., Holtappels G. Pathophysiology of chronic rhinosinusitis, pharmaceutical therapy options. GMS Curr. Top. Otorhinolaryngol. Head Neck Surg. 2015;14:1–40. doi: 10.3205/cto000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernstein J.M., Kansal R. Superantigen hypothesis for the early development of chronic hyperplastic sinusitis with massive nasal polyposis. Curr. Opin. Otolaryngol. Head Neck Surg. 2005;13:39–44. doi: 10.1097/00020840-200502000-00010. [DOI] [PubMed] [Google Scholar]
- 16.Bachert C., Gevaert P., van Cauwenberge P. Staphylococcus aureus enterotoxins: A key in airway disease? Allergy. 2002;57:480–487. doi: 10.1034/j.1398-9995.2002.02156.x. [DOI] [PubMed] [Google Scholar]
- 17.Balaban N., Rasooly A. Staphylococcal enterotoxins. Int. J. Food Microbiol. 2000;61:1–10. doi: 10.1016/S0168-1605(00)00377-9. [DOI] [PubMed] [Google Scholar]
- 18.Foster T.J., McDevitt D. Surface-associated proteins of Staphylococcus aureus: Their possible roles in virulence. FEMS Microbiol. Lett. 1994;118:199–205. doi: 10.1111/j.1574-6968.1994.tb06828.x. [DOI] [PubMed] [Google Scholar]
- 19.Oliveira D., Borges A., Simoes M. Staphylococcus aureus toxins and their molecular activity in infectious diseases. Toxins. 2018;10:252. doi: 10.3390/toxins10060252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bachert C., Gevaert P., Holtappels G., Johansson S.G., van Cauwenberge P. Total and specific IgE in nasal polyps is related to local eosinophilic inflammation. J. Allergy Clin. Immunol. 2001;107:607–614. doi: 10.1067/mai.2001.112374. [DOI] [PubMed] [Google Scholar]
- 21.Foreman A., Holtappels G., Psaltis A.J., Jervis-Bardy J., Field J., Wormald P.J., Bachert C. Adaptive immune responses in Staphylococcus aureus biofilm-associated chronic rhinosinusitis. Allergy. 2011;66:1449–1456. doi: 10.1111/j.1398-9995.2011.02678.x. [DOI] [PubMed] [Google Scholar]
- 22.Foreman A., Jervis-Bardy J., Wormald P.J. Do biofilms contribute to the initiation and recalcitrance of chronic rhinosinusitis? Laryngoscope. 2011;121:1085–1091. doi: 10.1002/lary.21438. [DOI] [PubMed] [Google Scholar]
- 23.Thunberg U., Hugosson S., Fredlund H., Cao Y., Ehricht R., Monecke S., Muller E., Engelmann S., Söderquist B. Anti-staphylococcal humoral immune response in patients with chronic rhinosinusitis. Rhinol. Online. 2019;2:50–58. doi: 10.4193/RHINOL/19.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sahl J.W., Lemmer D., Travis J., Schupp J.M., Gillece J.D., Aziz M., Driebe E.M., Drees K.P., Hicks N.D., Williamson C.H.D., et al. NASP: An accurate, rapid method for the identification of SNPs in WGS datasets that supports flexible input and output formats. Microb. Genom. 2016;2:e000074. doi: 10.1099/mgen.0.000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stegger M., Driebe E.M., Roe C., Lemmer D., Bowers J.R., Engelthaler D.M., Keim P., Andersen P.S. Genome sequence of Staphylococcus aureus strain CA-347, a USA600 methicillin-resistant isolate. Genome Announc. 2013;1:e00517-13. doi: 10.1128/genomeA.00517-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kloppot P., Selle M., Kohler C., Stentzel S., Fuchs S., Liebscher V., Müller E., Kale D., Ohlsen K., Bröker B.M., et al. Microarray-based identification of human antibodies against Staphylococcus aureus antigens. Proteom. Clin. Appl. 2015;9:1003–1011. doi: 10.1002/prca.201400123. [DOI] [PubMed] [Google Scholar]
- 28.Selle M., Hertlein T., Oesterreich B., Klemm T., Kloppot P., Müller E., Ehricht R., Stentzel S., Bröker B.M., Engelmann S., et al. Global antibody response to Staphylococcus aureus live-cell vaccination. Sci. Rep. 2016;6:24754. doi: 10.1038/srep24754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holm S. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 1979;6:65–70. [Google Scholar]
- 30.Duchêne S., Holt K.E., Weill F.X., Le Hello S., Hawkey J., Edwards D.J., Fourment M., Holmes E.C. Genome-scale rates of evolutionary change in bacteria. Microb. Genom. 2016;2:e000094. doi: 10.1099/mgen.0.000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wertheim H.F., Verveer J., Boelens H.A., van Belkum A., Verbrugh H.A., Vos M.C. Effect of mupirocin treatment on nasal, pharyngeal, and perineal carriage of Staphylococcus aureus in healthy adults. Antimicrob. Agents Chemother. 2005;49:1465–1467. doi: 10.1128/AAC.49.4.1465-1467.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eriksen N.H., Espersen F., Rosdahl V.T., Jensen K. Carriage of Staphylococcus aureus among 104 healthy persons during a 19-month period. Epidemiol. Infect. 1995;115:51–60. doi: 10.1017/S0950268800058118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.VandenBergh M.F., Yzerman E.P., van Belkum A., Boelens H.A., Sijmons M., Verbrugh H.A. Follow-up of Staphylococcus aureus nasal carriage after 8 years: Redefining the persistent carrier state. J. Clin. Microbiol. 1999;37:3133–3140. doi: 10.1128/JCM.37.10.3133-3140.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harvey R.J., Snidvongs K., Kalish L.H., Oakley G.M., Sacks R. Corticosteroid nasal irrigations are more effective than simple sprays in a randomized double-blinded placebo-controlled trial for chronic rhinosinusitis after sinus surgery. Int. Forum. Allergy Rhinol. 2018;8:461–470. doi: 10.1002/alr.22093. [DOI] [PubMed] [Google Scholar]
- 35.Lees K.A., Orlandi R.R., Oakley G., Alt J.A. The role of macrolides and doxycycline in chronic rhinosinusitis. Immunol. Allergy Clin. N. Am. 2020;40:303–315. doi: 10.1016/j.iac.2019.12.005. [DOI] [PubMed] [Google Scholar]
- 36.Holtfreter S., Grumann D., Schmudde M., Nguyen H.T., Eichler P., Strommenger B., Kopron K., Kolata J., Giedrys-Kalemba S., Steinmetzet I., et al. Clonal distribution of superantigen genes in clinical Staphylococcus aureus isolates. J. Clin. Microbiol. 2007;45:2669–2680. doi: 10.1128/JCM.00204-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rasmussen G., Monecke S., Ehricht R., Soderquist B. Prevalence of clonal complexes and virulence genes among commensal and invasive Staphylococcus aureus isolates in Sweden. PLoS ONE. 2013;8:e77477. doi: 10.1371/journal.pone.0077477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Acton D.S., Plat-Sinnige M.J., van Wamel W., de Groot N., van Belkum A. Intestinal carriage of Staphylococcus aureus: How does its frequency compare with that of nasal carriage and what is its clinical impact? Eur. J. Clin. Microbiol. Infect. Dis. 2009;28:115–127. doi: 10.1007/s10096-008-0602-7. [DOI] [PubMed] [Google Scholar]
- 39.Dancer S.J., Noble W.C. Nasal, axillary, and perineal carriage of Staphylococcus aureus among women: Identification of strains producing epidermolytic toxin. J. Clin. Pathol. 1991;44:681–684. doi: 10.1136/jcp.44.8.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verkaik N.J., de Vogel C.P., Boelens H.A., Grumann D., Hoogenboezem T., Vink C., Hooijkaas H., Foster T., Verbrugh H., Van Belkum A., et al. Anti-staphylococcal humoral immune response in persistent nasal carriers and noncarriers of Staphylococcus aureus. J. Infect. Dis. 2009;199:625–632. doi: 10.1086/596743. [DOI] [PubMed] [Google Scholar]
- 41.Soderquist B., Danielsson D., Holmberg H., Vikerfors T. Granulocyte colony-stimulating factor (G-CSF) and interleukin (IL)-8 in sera from patients with Staphylococcus aureus septicemia. Clin. Microbiol. Infect. 1995;1:101–109. doi: 10.1111/j.1469-0691.1995.tb00452.x. [DOI] [PubMed] [Google Scholar]
- 42.Dryla A., Prustomersky S., Gelbmann D., Hanner M., Bettinger E., Kocsis B., Kustos T., Henics T., Meinke A., Nagy E. Comparison of antibody repertoires against Staphylococcus aureus in healthy individuals and in acutely infected patients. Clin. Diagn. Lab. Immunol. 2005;12:387–398. doi: 10.1128/CDLI.12.3.387-398.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Banks M.C., Kamel N.S., Zabriskie J.B., Larone D.H., Ursea D., Posnett D.N. Staphylococcus aureus express unique superantigens depending on the tissue source. J. Infect. Dis. 2003;187:77–86. doi: 10.1086/345874. [DOI] [PubMed] [Google Scholar]
- 44.Holtfreter S., Bauer K., Thomas D., Feig C., Lorenz V., Roschack K., Friebe E., Selleng K., Lövenich S., Greve T., et al. egc-encoded superantigens from Staphylococcus aureus are neutralized by human sera much less efficiently than are classical staphylococcal enterotoxins or toxic shock syndrome toxin. Infect. Immun. 2004;72:4061–4071. doi: 10.1128/IAI.72.7.4061-4071.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holtfreter S., Kolata J., Broker B.M. Towards the immune proteome of Staphylococcus aureus—The anti-S. aureus antibody response. Int. J. Med. Microbiol. 2010;300:176–192. doi: 10.1016/j.ijmm.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 46.Radke; E. E.; Brown, S.M.; Pelzek, A.J.; Fulmer, Y.; Hernandez, D.N.; Torres, V.J.; Thomsen, I.P.; Chiang, W.K.; Miller, A.O.; Shopsin, B.; et al. Hierarchy of human IgG recognition within the Staphylococcus aureus immunome. Sci. Rep. 2018;8:13296. doi: 10.1038/s41598-018-31424-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Belkum A., Verkaik N., De Vogel C.P., Boelens H.A., Verveer J., Nouwen J.L., Verbrugh H.A., Wertheim H.F.L. Reclassification of Staphylococcus aureus nasal carriage types. J. Infect. Dis. 2009;199:1820–1826. doi: 10.1086/599119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.