Abstract
Phylogeny and taxonomy of the genus Haploporus were carried out based on a larger number of samples covering a wider geographic range including East Asia, South Asia, Europe, and America, and the species diversity of the genus is updated. Four species, Haploporus bicolor, H. longisporus, H. punctatus and H. srilankensis, are described as new species based on morphology and molecular phylogenetic analyses inferred from the internal transcribed spacer (ITS), the large subunit nuclear ribosomal RNA gene (nLSU), and the small subunit mitochondrial rRNA gene (mtSSU). Haploporus bicolor is characterized by the distinctly different colors between the pore surface and the tubes, small pores measuring 5–7 per mm, and narrow basidiospores measuring 10.5–11.9 × 4.5–5 µm; H. longisporus differs from other species in the genus by its large pores measuring 2–3 per mm, hyphae at dissepiment edge with simple septum, and the long basidiospores (up to 22 µm); H. punctatus is distinguished by its cushion-shaped basidiocarps, wide fusiform cystidioles with a simple septum at the tips, the absence of dendrohyphidia and the cylindrical to slightly allantoid basidiospores measuring 9–10.8 × 3.8–5 µm; H. srilankensis is characterized by its perennial habit, small pores measuring 4–5 per mm, dextrinoid skeletal hyphae, the presence of cystidioles and dendrohyphidia. An identification key to accepted species of Haploporus is provided.
Keywords: multi-marker analysis, phylogeny, polyporaceae, taxonomy, wood-rotting fungi
1. Introduction
Haploporus Bondartsev and Singer was established by Singer based on H. odorus (Sommerf.) Bondartsev and Singer [1]. The genus was characterized as having annual to perennial, resupinate to pileate basidiocarps, a dimitic to trimitic hyphal system with clamp connections on the generative hyphae, cyanophilous skeletal hyphae, cylindrical to subglobose, colorless, thick-walled, cyanophilous and ornamented basidiospores, and causing a white rot [1,2,3,4,5]. Kotlaba and Pouzar [2] described a genus Pachykytospora Kotl. and Pouzar based on Polyporus tuberculosus Fr., and it was treated as a synonym of Haploporus by Dai et al. [6]. This conclusion was confirmed by molecular analysis [5], and all species introduced in Pachykytospora have been transferred to Haploporus [2,5,6].
Recently, two species of Haploporus, H. brasiliensis Nogueira-Melo and Ryvarden and H. pileatus Ryvarden, were described from Brazil based on morphology only [7], and four species, H. angustisporus Meng Zhou and Y.C. Dai, H. crassus Meng Zhou and Y.C. Dai, H. gilbertsonii Meng Zhou, Vlasák and Y.C. Dai and H. microspores L.L. Shen, Y.C. Dai and B.K. Cui, were introduced as new based on morphological characteristics and molecular phylogenetic analyses [8,9]. So far, 19 species have been accepted in Haploporus [2,3,4,5,7,9,10,11].
The aim of the study is to explore the phylogeny and species diversity of Haploporus based on samples from Asia, Europe and America. The taxonomy and phylogeny are updated, and four new species, respectively, from China, Ecuador, and Sri Lanka, were confirmed to be members of the Haploporus. In this paper, we describe and illustrate those new species.
2. Materials and Methods
2.1. Morphological Studies
Specimens examined were deposited in the herbarium of the Institute of Microbiology, Beijing Forestry University (BJFC) and the private herbarium of Josef Vlasák (JV) which will forward specimens to the National Museum of Czech Republic in Prague (PRM). Macro-morphological descriptions were based on field notes. Sections of basidiocarps were studied microscopically according to Zhou and Cui [12] at magnifications 1000× using a Nikon Eclipse 80i microscope with phase contrast illumination. Drawings were made with the aid of a drawing tube. Microscopic features, measurements, and drawings were made from sections stained with Cotton Blue and Melzer’s reagent. Basidiospores were measured from sections cut from the tubes. To present basidiospores size variation, the 5% of measurements excluded from each end of the range are given in parentheses. Basidiospore spine lengths were not included in the measurements. Abbreviations include IKI = Melzer’s reagent, IKI+ = amyloid, KOH = 5% potassium hydroxide, CB = Cotton Blue, CB+ = cyanophilous, L = mean spore length (arithmetic average of all spores), W = mean basidiospores width (arithmetic average of all basidiospores), Q = the L/W ratio, and n = number of basidiospores measured from given number of specimens. Special color terms follow Petersen [13]. Herbarium abbreviations follow Thiers [14].
2.2. DNA Extraction and Sequencing
A cetyl trimethylammonium bromide (CTAB)rapid plant genome extraction kit (Aidlab Biotechnologies, Beijing, China) was used to obtain PCR products from dried specimens, according to the manufacturer’s instructions with some modifications [15,16]. The internal transcribed spacer (ITS) region was amplified with primer pair ITS5 (GGA AGT AAA AGT CGT AAC AAG G) and ITS4 (TCC TCC GCT TAT TGATAT GC) [17]. The large subunit nuclear ribosomal RNA gene (nLSU) region was amplified with primer pair LR0R (ACC CGC TGA ACT TAA GC) and LR7 (TAC TAC CAC CAA GAT CT) (http://www.biology.duke.edu/fungi/mycolab/primers.htm). The small subunit mitochondrial rRNA gene (mtSSU) region was amplified with primer pair MS1 (CAG CAG TCA AGA ATA TTA GTC AAT G) and MS2 (GCG GAT TAT CGA ATT AAA TAA C) [17]. The PCR procedure for ITS and mtSSU was as follows: initial denaturation at 95 °C for 3 min, followed by 34 cycles at 94 °C for 40 s, 54 °C for ITS and 55 °C for mtSSU for 45 s and 72 °C for 1 min, and a final extension of 72 °C for 10 min. The PCR procedure for nLSU was as follows: initial denaturation at 94 °C for 1 min, followed by 34 cycles at 94 °C for 30 s, 50 °C for 1 min and 72 °C for 1.5 min, and a final extension of 72 °C for 10 min [5]. The PCR products were purified with a Gel Extraction and PCR Purification Combo Kit (Spin-column) in Beijing Genomics Institute, Beijing, P.R. China. The purified products were then sequenced on an ABI-3730-XL DNA Analyzer (Applied Biosystems, Foster City, CA, USA) using the same primers as in the original PCR amplifications.
2.3. Phylogenetic Analysis
The sequences generated in this study were aligned with additional sequences downloaded from GenBank (Table 1) using ClustalX [18] and manually adjusted in BioEdit [19]. The sequence quality was checked following Nilsson et al. [20]. Perenniporia hainaniana B.K. Cui and C.L. Zhao and P. medulla-panis (Jacq.) Donk were used as outgroups, following Shen et al. [5]. Prior to phylogenetic analysis, ambiguous sequences at the start and the end were deleted and gaps were manually adjusted to optimize the alignment. Sequence alignment was deposited at TreeBase (http://purl.org/phylo/treebase; submission ID 27556).
Table 1.
Information on the sequences used in this study.
| Species | Sample No. | Location | GenBank Accession No. | Reference | ||
|---|---|---|---|---|---|---|
| ITS | nLSU | mt-SSU | ||||
| Haploporus alabamae | Dollinger 895 | USA | KY264038 | MK433606 | MW463004 | [9] |
| H. alabamae | JV 1704/75 | Costa Rica | MK429754 | MK433607 | MW463005 | [9] |
| H. angustisporus | Cui 9046 | China | KU941862 | KU941887 | [9] | |
| H. angustisporus | Dai 10951 | China | KX900634 | KX900681 | MW463006 | [9] |
| H. bicolor | Dai19951 | China | MW465684 | MW462995 | Present study | |
| H. crassus | Dai 13580 | China | MW465669 | KU941886 | [9] | |
| H. cylindrosporus | Dai 15643 | China | KU941853 | KU941877 | KU941902 | [5] |
| H. cylindrosporus | Dai 15664 | China | KU941854 | KU941878 | KU941903 | [5] |
| H. gilbertsonii | JV 1209/63-J | USA | MK429755 | MK433608 | [9] | |
| H. gilbertsonii | JV 1611/5-J | USA | MK429756 | MK433609 | MW463007 | [9] |
| H. latisporus | Dai 11873 | China | KU941847 | KU941871 | MW463008 | [5] |
| H. latisporus | Dai 10562 | China | KU941848 | KU941872 | KU941897 | [5] |
| H. longisporus | JV1906/C11-J | Ecuador | MW465685 | MW462996 | Present study | |
| H. microsporus | Dai 12147 | China | KU941861 | KU941885 | [5] | |
| H. nanosporus | LYAD 2044° | Gabon | KU941859 | KU941883 | KU941908 | [5] |
| H. nanosporus | LYAD 2044b | Gabon | KU941860 | KU941884 | [5] | |
| H. nepalensis | Dai 12937 | China | KU941855 | KU941879 | KU941904 | [5] |
| H. nepalensis | Cui 10729 | China | KU941856 | KU941880 | KU941905 | [5] |
| H. odorus | Dai 11296 | China | KU941845 | KU941869 | KU941894 | [5] |
| H. odorus | Yuan 2365 | China | KU941846 | KU941870 | KU941895 | [5] |
| H. odorus | KUC20121123-29 | Republic of Korea | KJ668537 | KJ668390 | ||
| H. papyraceus | Dai 10778 | China | KU941839 | KU941863 | KU941888 | [5] |
| H. papyraceus | Cui 8706 | China | KU941840 | KU941864 | KU941889 | [5] |
| H. pirongia | Dai 18659 | Australia | MH631017 | MH631021 | MW463009 | [9] |
| H. pirongia | Dai 18660 | Australia | MH631018 | MH631022 | MW463010 | [9] |
| H. punctatus | Dai19525 | Sri Lanka | MW465686 | MW462997 | Present study | |
| H. punctatus | Dai19628 | Sri Lanka | MW465687 | MW462998 | MW463011 | Present study |
| H. septatus | Dai 13581 | China | KU941843 | KU941867 | KU941892 | [5] |
| H. septatus | Cui 4100 | China | KU941844 | KU941868 | KU941893 | [5] |
| H. srilankensis | Dai19523 | Sri Lanka | MW465688 | MW462999 | MW463012 | Present study |
| H. srilankensis | Dai19524 | Sri Lanka | MW465689 | MW463000 | Present study | |
| H. srilankensis | Dai19526 | Sri Lanka | MW465690 | MW463001 | Present study | |
| H. srilankensis | Dai19530 | Sri Lanka | MW465691 | MW463002 | MW463013 | Present study |
| H. srilankensis | Dai19534 | Sri Lanka | MW465692 | MW463003 | MW463014 | Present study |
| H. subpapyraceus | Dai 9324 | China | KU941841 | KU941865 | KU941910 | [5] |
| H. subpapyraceus | Cui 2651 | China | KU941842 | KU941866 | KU941891 | [5] |
| H. subtrameteus | Dai 4222 | China | KU941849 | KU941873 | KU941898 | [5] |
| H. subtrameteus | Cui 10656 | China | KU941850 | KU941874 | KU941899 | [5] |
| H. subtrameteus | KUC20121102-36 | Republic of Korea | KJ668536 | KJ668389 | ||
| H. thindii | Cui 9373 | China | KU941851 | KU941875 | KU941900 | [5] |
| H. thindii | Cui 9682 | China | KU941852 | KU941876 | KU941901 | [5] |
| H. tuberculosus | 15559 | Sweden | KU941857 | KU941881 | KU941906 | [5] |
| H. tuberculosus | 15560 | Austria | KU941858 | KU941882 | KU941907 | [5] |
| H. tuberculosus | KA11 (GB) | Sweden | JX124705 | [21] | ||
| Perenniporia hainaniana | Cui 6364 | China | JQ861743 | JQ861759 | KF051044 | [16] |
| P. medulla-panis | Cui 3274 | China | JN112792 | JN112793 | KF051043 | [22] |
New sequences are shown in bold.
Maximum parsimony (MP), Maximum likelihood (ML) and Bayesian inference (BI) were employed to perform phylogenetic analysis of the three aligned datasets. The three phylogenetic analysis algorithms generated nearly identical topologies for each dataset, thus only the topology from the MP analysis is presented along with statistical values from the MP, ML and BI algorithms. Most parsimonious phylogenies were inferred from the ITS + nLSU + mtSSU, and their combinability was evaluated with the incongruence length difference (ILD) test [23] implemented in PAUP* 4.0b10 [24], under a heuristic search and 1000 homogeneity replicates giving a P value of 1.000, much greater than 0.01, which means there is no discrepancy among the two loci in reconstructing phylogenetic trees. Phylogenetic analysis approaches followed [22]. The tree construction procedure was performed in PAUP* version 4.0b10 [24]. All characters were equally weighted, and gaps were treated as missing data. Trees were inferred using the heuristic search option with TBR branch swapping and 1000 random sequence additions. Max-trees were set to 5000, branches of zero length were collapsed and all parsimonious trees were saved. Clade robustness was assessed using a bootstrap (BT) analysis with 1000 replicates [25]. Descriptive tree statistics tree length (TL), consistency index (CI), retention index (RI), rescaled consistency index (RC), and homoplasy index (HI) were calculated for each maximum parsimonious tree (MPT) generated.
jModeltest v.2.17 [26] was used to determine the best-fit evolution model of the combined dataset for Maximum likelihood (ML) and Bayesian inference (BI). Four unique partitions were established, GTR + I + G was the selected substitution model for each partition. RaxmlGUI 1.2 [27,28] was used for ML analysis. All parameters in the ML analysis used default settings. Statistical support values were obtained using non-parametric bootstrapping with 1000 replicates. The Bayesian inference (BI) was conducted with MrBayes 3.2.6 [29] in two independent runs, each of which had four chains for 10 million generations and started from random trees. Trees were sampled every 1000th generation. The first 25% of sampled trees were discarded as burn-in, whereas other trees were used to construct a 50% majority consensus tree and for calculating Bayesian posterior probabilities (BPPs).
Phylogenetic trees were visualized using Treeview [30]. Branches that received bootstrap support for Maximum likelihood (BS), Maximum parsimony (BP) and Bayesian posterior probabilities (BPP) greater than or equal to 75% (BS/BP) and 0.95 (BPP) were considered as significantly supported, respectively.
3. Results
3.1. Molecular Phylogeny
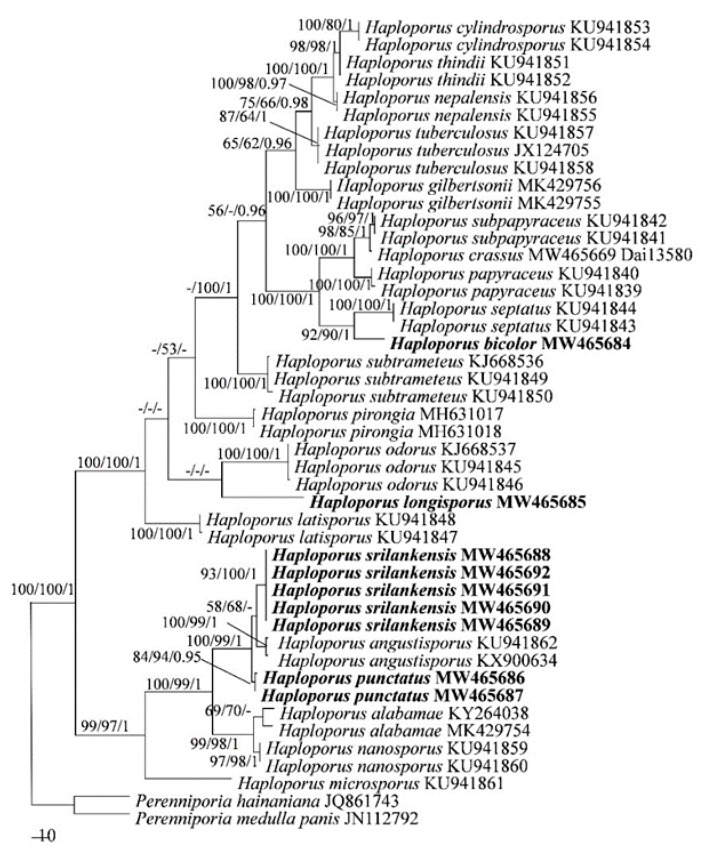
The ITS-based phylogeny included ITS sequences from 46 fungal collections representing 23 species. The dataset had an aligned length of 720 characters, of which 333 characters are constant, 63 are variable and parsimony-uninformative, and 324 are parsimony-informative. MP analysis yielded a tree (TL = 945, CI = 0. 624, RI = 0.872, RC = 0.544, HI = 0.376). The best model for the ITS sequences dataset estimated and applied in the BI was GTR+I+G. BI resulted in a similar topology with an average standard deviation of split frequencies = 0.006018 to MP analysis, and thus only the MP tree is provided. Both BT values (≥50%) and BPPs (≥0.90) are shown at the nodes (Figure 1).
Figure 1.
Maximum parsimony strict consensus tree illustrating the phylogeny of Haploporus based on internal transcribed spacer (ITS) sequences. Branches are labeled with parsimony bootstrap values ≥ 50% and Bayesian posterior probabilities ≥ 0.90.
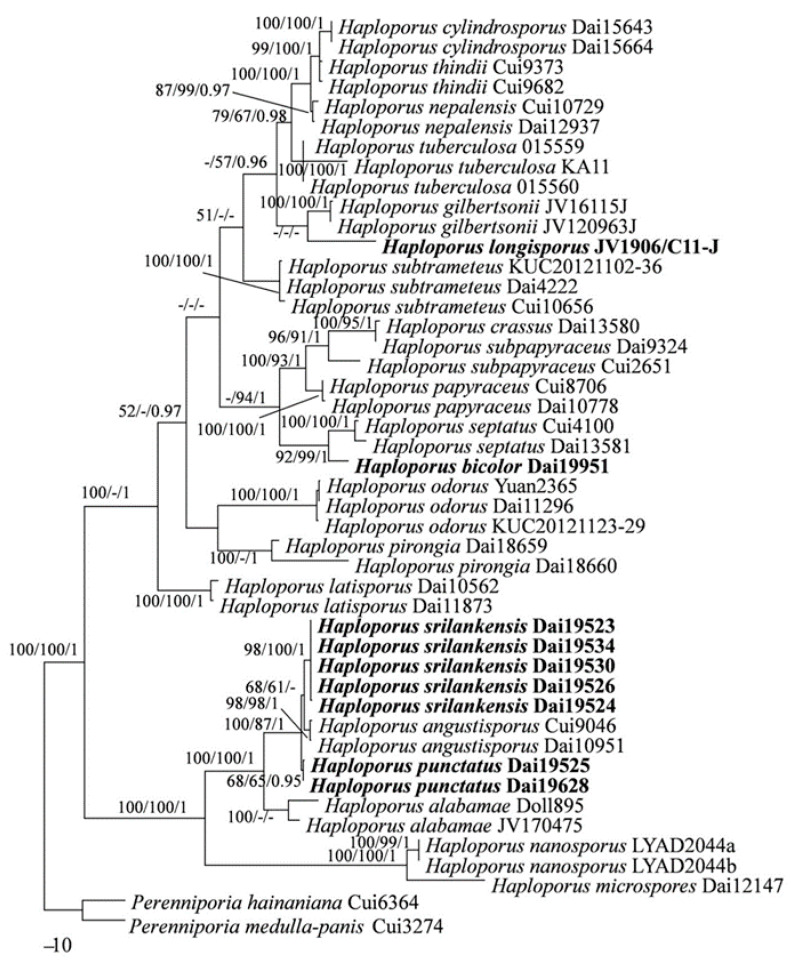
The combined three gene (ITS + nLSU + mtSSU) sequences dataset from 46 fungal specimens representing 23 taxa did not show any conflicts in tree topology for the reciprocal bootstrap trees, which allowed us to combine them (p > 0.01). The dataset had an aligned length of 2706 characters, of which 1792 characters are constant, 220 are variable and parsimony-uninformative, and 694 are parsimony-informative. Maximum parsimony analysis yielded a tree (TL = 1929, CI = 0.627, RI = 0.847, RC = 0.531, HI = 0.373). The best model for the combined ITS + nLSU + mtSSU dataset estimated and applied in the Bayesian analysis was GTR + I + G. Bayesian analysis resulted in a similar topology with an average standard deviation of split frequencies = 0.005181 to MP analysis, and thus only the MP tree is provided. Both BT values (≥50%) and BPPs (≥0.90) are shown at the nodes (Figure 2).
Figure 2.
Maximum parsimony strict consensus tree illustrating the phylogeny of Haploporus based on ITS + nLSU + mtSSU sequences. Branches are labeled with parsimony bootstrap values ≥ 50% and Bayesian posterior probabilities ≥ 0.90.
In both ITS + nLSU + mtSSU- and ITS-based phylogenies (Figure 1 and Figure 2), four new well-supported lineages were identified. Among them are two well-supported terminal clades and two isolated branches. Four specimens from Sri Lanka formed a well-supported clade (98% MP 100% ML and 1.00 BI), named as Haploporus srilankensis, sister to H. angustisporus. And two specimens of Haploporus punctatus nested in the same clade as H. srilankensis and H. angustisporus. In addition, samples of Haploporus bicolor and H. longisporus formed two distinct lineages, H. bicolor is sister to H. septatus L.L. Shen, Y.C. Dai and B.K. Cui; but H. longisporus is an independent lineage.
3.2. Taxonomy
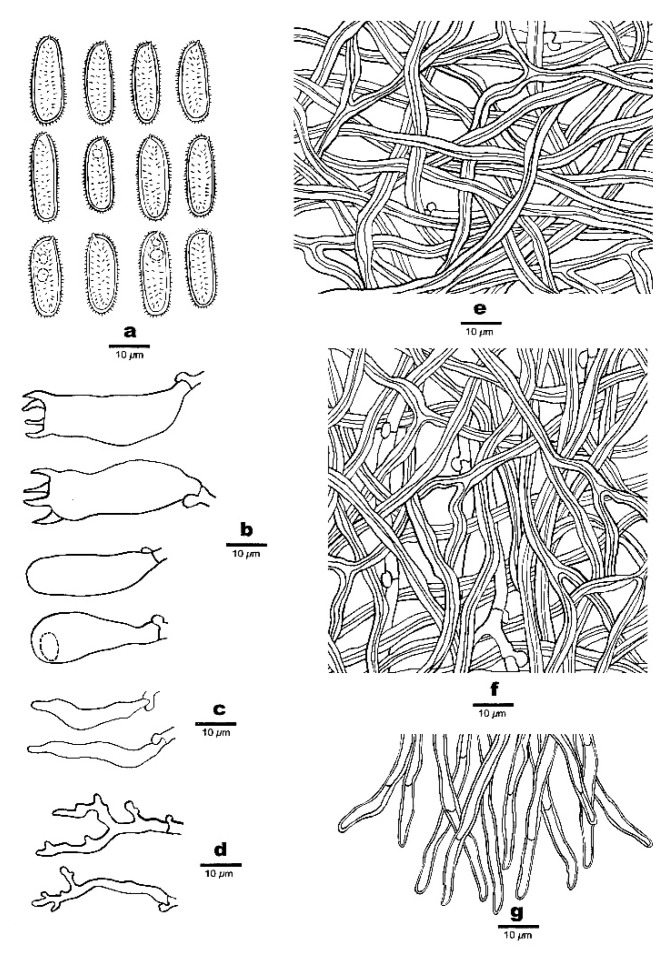
Haploporus bicolor Y.C. Dai, Meng Zhou and Yuan Yuan, sp. Nov. Figure 3 and Figure 4
Figure 3.
Basidiocarps of Haploporus bicolor (Holotype). Scale bar = 1.0 cm.
Figure 4.
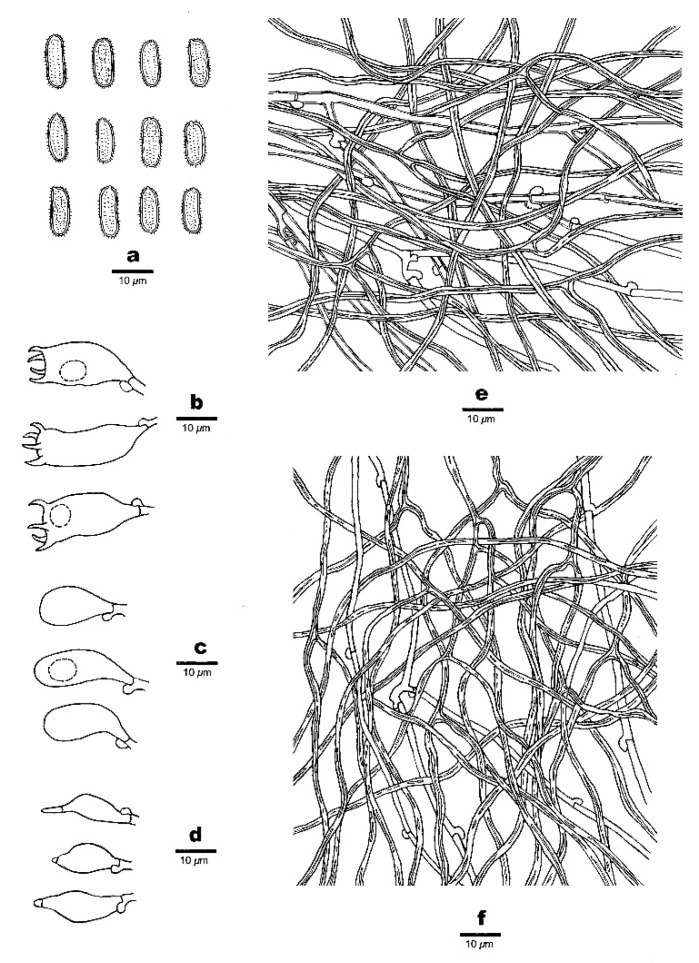
Microscopic structures of Haploporus bicolor (Holotype). (a). Basidiospores. (b). Basidia and basidioles. (c). Cystidioles. (d). Dendrohyphidia. (e). Hyphae from subiculum. (f). Hyphae from trama.
MycoBank: MB838450
Diagnosis—Differs from other Haploporus species by the distinct different colors between the pore surface and the tubes, no-dextrinoid skeletal hyphae, small pores measuring 5–7 per mm, the presence of fusiform cystidioles and abundant dendrohyphidia, and narrow basidiospores measuring 10.5–11.9 × 4.5–5 µm.
Type—China, Yunnan, Wenshan, Xichou County, Xiaoqiaogou Forest Farm, E 104°41′, N 23°21′, on a fallen angiosperm branch, 29 June 2019, Yu-Cheng Dai, Dai 19951 (Holotype BJFC 031625).
Etymology—Bicolor (Lat.): referring to the species having different colors between the pore surface and the tubes.
Fruiting body—Basidiocarp annual, resupinate, difficult to separate from the substrate, soft corky and white to cream when fresh, become buff when bruised, corky when dry, up to 5 cm long, 2 cm wide and 0.7 mm thick at the center. Pore surface cream to buff with peach tint when dry; sterile margin distinct, white, up to 1 mm; pores round to angular, 5–7 per mm; dissepiments thick, entire. Subiculum clay buff, corky, up to 0.4 mm thick. Tubes clay buff, corky, up to 0.3 mm long.
Hyphal structure—Hyphal system dimitic; generative hyphae bearing clamp connections, hyaline, thin-walled; skeletal hyphae dominant, thick-walled, frequently branched, IKI–, CB+; tissues unchanging in KOH.
Subiculum—Generative hyphae inconspicuous, hyaline, thin-walled, 1–2.5 µm in diameter (diam); skeletal hyphae dominant, distinctly thick-walled with a narrow lumen, frequently branched, flexuous, interwoven, 1–2.5 µm in diam.
Tubes—Generative hyphae infrequent, hyaline, thin-walled, 1–1.5 µm in diam; skeletal hyphae dominant, distinctly thick-walled with a narrow lumen, frequently branched, flexuous, interwoven, 1–2 µm in diam. Cystidia absent; cystidioles present, fusiform to ventricose, hyaline, thin-walled, 13.5–22 × 4–6.5 µm. Basidia barrel-shaped to pear-shaped with 4-sterigmata and a basal clamp connection, 14–25 × 6–11.5 µm; basidioles dominant, pear-shaped, slightly smaller than basidia. Dendrohyphidia abundant, hyaline, thin-walled. Some irregular-shaped crystals present among tube trama.
Spores—Basidiospores oblong ellipsoid to subcylindrical, hyaline, thick-walled with tuberculate ornamentation, IKI–, CB+, (10–)10.5–11.9(–12) × (4.1–)4.5–5(–5.1) µm, L = 11.10 µm, W = 4.86 µm, Q = 2.28 (n = 30/1).
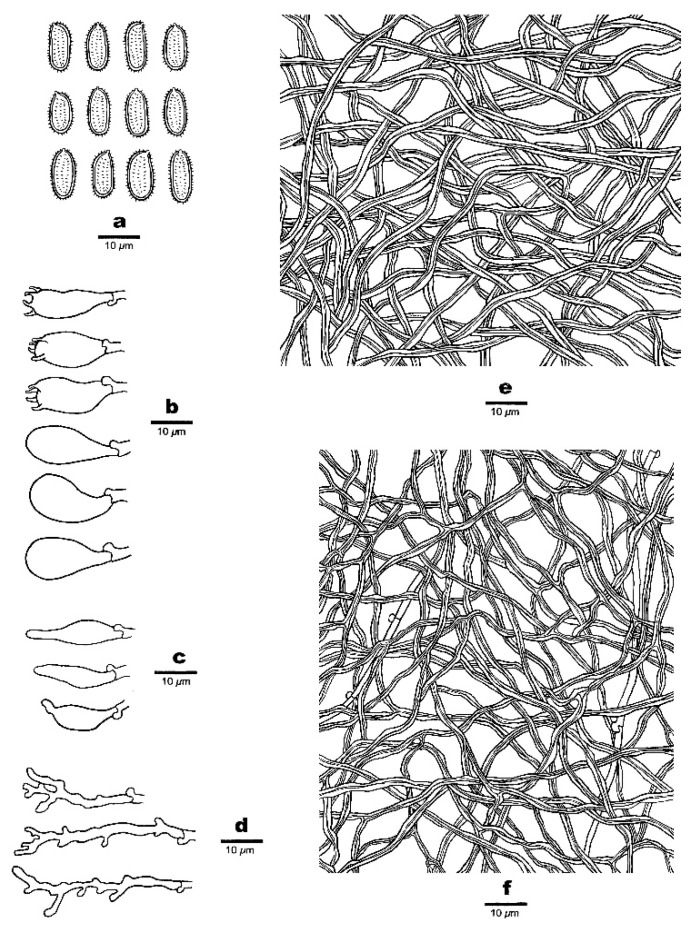
Haploporus longisporus Y.C. Dai, Meng Zhou and Vlasák, sp. Nov. Figure 5 and Figure 6
Figure 5.
Basidiocarp of Haploporus longisporus (Holotype). Scale bar = 1.0 cm.
Figure 6.
Microscopic structures of Haploporus longisporus (Holotype). (a). Basidiospores. (b). Basidia and basidioles. (c). Cystidioles. (d). Dendrohyphidia. (e). Hyphae from subiculum. (f). Hyphae from trama. (g). Hyphae at dissepiment edge.
MycoBank: MB838451
Diagnosis—Differs from other Haploporus species by the non-dextrinoid skeletal hyphae, large pores measuring 2–3 per mm, hyphae at dissepiment edge with simple septum, and the long cylindrical basidiospores measuring 18.2–22 × 7–9 µm.
Type—Ecuador, Pichincha, Vicodin svah volcan Pasochoa, W 78°29′, S 0°25′, on a dead angiosperm branch, June 2019, Josef Vlasák, JV1906/C11-J (Holotype PRM, isotypes BJFC 032989 and JV).
Etymology—Longisporus (Lat.): referring to the species having the distinct long basidiospores.
Fruiting body—Basidiocarp annual, resupinate, difficult to separate from the substrate, corky when dry, up to 10 cm long, 1.5 cm wide and 2 mm thick at the center. Pore surface cream to pale buff when dry; sterile margin indistinct; pores round to angular, 2–3 per mm; dissepiments thick, entire. Subiculum olivaceous buff, corky, up to 1 mm thick. Tubes olivaceous buff, corky, 1 mm long.
Hyphal structure—Hyphal system dimitic; generative hyphae bearing clamp connections, hyaline, thin-walled; skeletal hyphae dominant, thick-walled, frequently branched, IKI–, CB+; tissues unchanging in KOH.
Subiculum—Generative hyphae infrequent, hyaline, thin-walled, occasionally branched, 2–3 µm in diam; skeletal hyphae dominant, distinctly thick-walled with a narrow to wide lumen, frequently branched, flexuous, interwoven, 2.5–5 µm in diam.
Tubes—Generative hyphae hyaline, thin-walled, frequently branched, 1.5–3 µm in diam; skeletal hyphae dominant, distinctly thick-walled with a narrow to medium lumen, frequently branched, flexuous, interwoven, 2–5 µm in diam. Cystidia absent; cystidioles present, fusiform to slim clavate, hyaline, thin-walled, 20–33 × 3.5–5 µm. Basidia more or less barrel-shaped, sometimes constricted at middle, with 4-sterigmata and a basal clamp connection, 37–50 × 12–16 µm; basidioles dominant, clavate to pear-shaped, slightly smaller than basidia, sometimes with a large guttule. Hyphae at dissepiment edge usually thick-walled with a simple septum. Dendrohyphidia present, hyaline, thin-walled. Some irregular-shaped crystals present among tube trama.
Spores—Basidiospores cylindrical, hyaline, thick-walled with tuberculate ornamentation, sometimes with a few guttules, IKI–, CB+, (18–)18.2–22(–22.5) × (6.5–)7–9(–10) µm, L = 20.6 µm, W = 7.86 µm, Q = 2.62 (n = 30/1).
Haploporus punctatus Y.C. Dai, Meng Zhou and Yuan Yuan, sp. Nov. Figure 7 and Figure 8
Figure 7.
Basidiocarp of Haploporus punctatus (Holotype). Scale bar = 1.0 cm.
Figure 8.
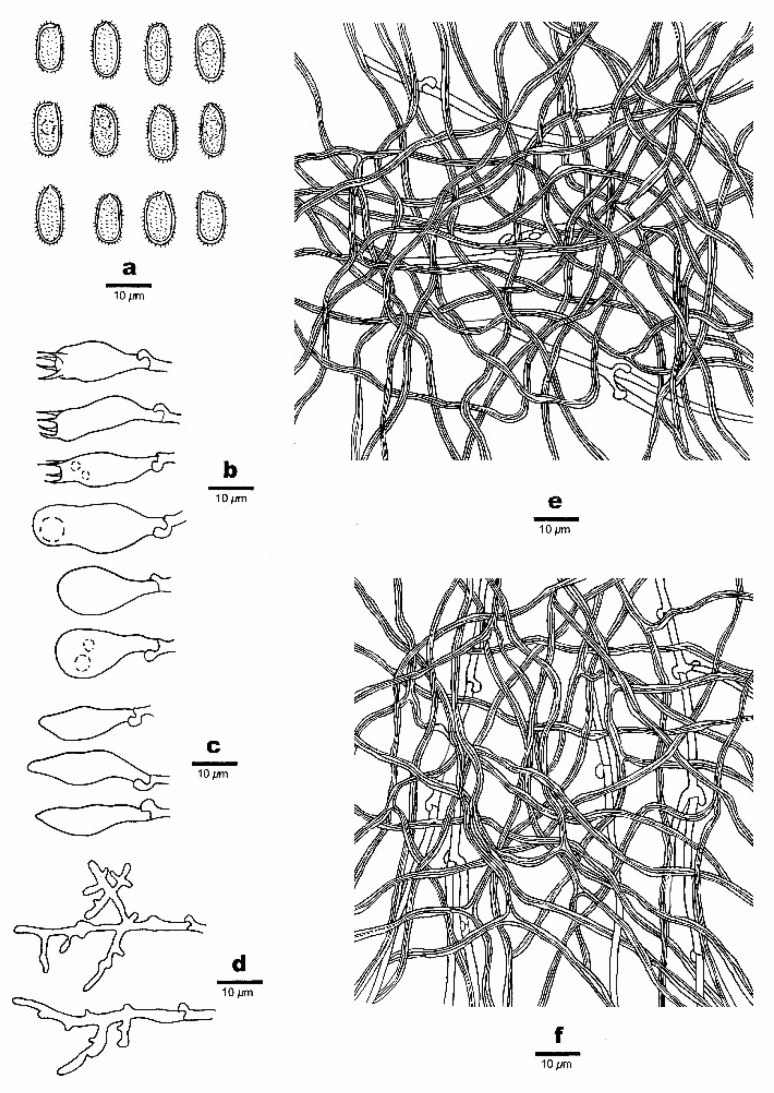
Microscopic structures of Haploporus punctatus (Holotype). (a). Basidiospores. (b). Basidia. (c). Basidioles. (d). Cystidioles. (e). Hyphae from subiculum. (f). Hyphae from trama.
MycoBank: MB 838452
Diagnosis—Differs from other Haploporus species by its cushion-shaped basidiocarps, dextrinoid skeletal hyphae and basidiospores, wide fusiform cystidioles with a simple septum at the tips, the absence of dendrohyphidia and the cylindrical to slightly allantoid basidiospores measuring 9–10.8 × 3.8–5 µm.
Type—Sri Lanka, Wadduwa, South Blogoda Lake, E 79°57′, N 6°41′, on a dead angiosperm branch, 28 Feburary 2019, Yu-Cheng Dai, Dai 19525 (Holotype BJFC 031204).
Etymology—Punctatus (Lat.): referring to the species having cushion-shaped basidiocarps.
Fruiting body—Basidiocarps annual, resupinate, difficult to separate from the substrate, corky when dry, cushion-shaped, distinctly thickened in center and receding at margin, up to 3 cm long, 3 cm wide and 4 mm thick at center. Pore surface pale buff when dry; sterile margin distinct, cream, up to 2 mm; pores round to angular, 3–5 per mm; dissepiments thick, entire. Subiculum olivaceous buff to clay buff, corky, up to 0.5 mm thick. Tubes buff, corky, up to 3.5 mm long.
Hyphal structure—Hyphal system dimitic; generative hyphae bearing clamp connections, hyaline, thin-to-slightly-thick-walled; skeletal hyphae dominant, thick-walled, frequently branched, dextrinoid, CB+; tissues unchanging in KOH.
Subiculum—Generative hyphae frequent, hyaline, thin-to-slightly-thick-walled, frequently branched, 1.5–2.5 µm in diam; skeletal hyphae dominant, hyaline, distinctly thick-walled with a narrow lumen to subsolid, frequently branched, flexuous, interwoven, 1.5–3 µm in diam.
Tubes—Generative hyphae frequent, hyaline, thin-walled, frequently branched, 1–2 µm in diam; skeletal hyphae dominant, distinctly thick-walled with a narrow lumen to subsolid, frequently branched, flexuous, interwoven, 1–2 µm in diam. Cystidia absent; cystidioles present, hyaline, thin-walled, wide fusiform, sometimes with a simple septum at the tips, 13–28 × 4–9 µm. Basidia barrel-shaped with 4-sterigmata and a basal clamp connection, occasionally with a large guttule, 16–28 × 6–10 µm; basidioles dominant, capitate to pear-shaped, distinctly smaller than basidia. Dendrohyphidia absent. Irregular-shaped crystals present abundantly among tube trama.
Spores—Basidiospores cylindrical to slightly allantoid, hyaline, thick-walled with tuberculate ornamentation, sometimes with a few guttules, dextrinoid, CB+, 9–10.8(–11) × 3.8–5(–5.3) µm, L = 10.36 µm, W = 4.36 µm, Q = 2.37–2.52 (n = 60/2).
Additional specimen examined (paratype)—Sri Lanka, Avissawella, Salgala Forest, E 80°16′, N 7°5′, on a fallen dead angiosperm branch, 28 February 2019, Dai 19628 (BJFC 031305).
Haploporus srilankensis Y.C. Dai, Meng Zhou and Yuan Yuan, sp. nov. Figure 9 and Figure 10
Figure 9.
Basidiocarp of Haploporus srilankensis (Holotype). Scale bar = 1.0 cm.
Figure 10.
Microscopic structures of Haploporus srilankensis (Holotype). (a). Basidiospores. (b). Basidia and basidioles. (c). Cystidioles. (d). Dendrohyphidia. (e). Hyphae from subiculum. (f). Hyphae from trama.
MycoBank: MB838453
Diagnosis—Differs from other Haploporus species by the combination of perennial habit, small pores measuring 4–5 per mm, dextrinoid skeletal hyphae, the presence of cystidioles and dendrohyphidia, and short basidiospores measuring 8.5–11.5 × 4–5.2 µm.
Type—Sri Lanka, Wadduwa, South Blogoda Lake, E 79°57′, N 6°41′, on a dead branch of a living angiosperm tree, 28 February 2019, Yu-Cheng Dai, Dai 19523 (Holotype BJFC 031202).
Etymology—Srilankensis (Lat.): referring to the species occurrence in Sri Lanka.
Fruiting body—Basidiocarps perennial, resupinate, difficult to separate from the substrate, corky when fresh and dry, up to 14 cm long, 4 cm wide and 2.5 mm thick at the center. Pore surface pale buff when fresh and pinkish buff to salmon when dry; sterile margin distinct, white to cream, up to 2 mm; pores round, 4–5 per mm; dissepiments thick, entire. Subiculum buff-yellow, corky, up to 0.5 mm thick. Tubes buff, corky, up to 2 mm long.
Hyphal structure—Hyphal system dimitic; generative hyphae bearing clamp connections, hyaline, thin-walled; skeletal hyphae dominant, thick-walled with a narrow lumen, frequently branched, dextrinoid, CB+; tissues unchanging in KOH.
Subiculum—Generative hyphae infrequent, hyaline, thin-walled, frequently branched, 1.5–2 µm in diam; skeletal hyphae dominant, hyaline, distinctly thick-walled with a narrow lumen, frequently branched, flexuous, interwoven, 1.5–2 µm in diam.
Tubes—Generative hyphae hyaline, thin-walled, frequently branched, 1–2 µm in diam; skeletal hyphae dominant, distinctly thick-walled with a narrow lumen, frequently branched, flexuous, interwoven, 1–2.5 µm in diam. Cystidia absent; cystidioles present, distinctly fusiform, hyaline, thin-walled, 17–24 × 4–6 µm. Basidia clavate to ampullaceous with 4-sterigmata and a basal clamp connection, occasionally with a few small guttules, 18–28 × 5–9 µm; basidioles dominant, capitate to pear-shaped, distinctly wider than basidia. Dendrohyphidia present, hyaline, thin-walled. Some irregular-shaped crystals present among tube trama.
Spores—Basidiospores oblong ellipsoid, hyaline, thick-walled, with tuberculate ornamentation, occasionally with a large guttule, dextrinoid, CB+, (8.3–)8.5–11(–12) × (3.9–)4–5.2(–5.5) µm, L = 9.80 µm, W = 4.56 µm, Q = 2.07–2.21 (n = 90/3).
Additional specimens examined (paratypes)—Sri Lanka, Wadduwa, South Blogoda Lake, E 79°57′, N 6°41′, on a dead angiosperm branch, 28 Feb. 2019, Dai 19524 (BJFC 031203), Dai 19526 (BJFC 031205), Dai 19530 (BJFC 031209), Dai 19534 (BJFC 031213).
4. Discussion
In both ITS and ITS + nLSU + mtSSU-based phylogenies (Figure 1 and Figure 2), Haploporus bicolor is sister to H. septatus. Morphologically, Haploporus bicolor resembles H. septatus by sharing resupinate basidiocarps with approximately the same-sized small pores (5–7 per mm), but H. septatus differs from H. bicolor by wider basidiospores (8.5–11 × 5–6 µm vs. 10.5–11.9 × 4.5–5 µm), dextrinoid skeletal hyphae and absence of dendrohyphidia [5]. In addition, Haploporus bicolor is also related to H. crassus, H. papyraceus (Cooke) Y.C. Dai and Niemelä, H. subpapyraceus L.L. Shen, Y.C. Dai and B.K. Cui in our phylogenies (Figure 1 and Figure 2). However, these three species have larger pores (3–5 per mm) and wider basidiospores (width > 5 µm), in which they obviously differ from Haploporus bicolor [5,9,31].
Phylogenetically, Haploporus longisporus is an independent lineage. Morphologically, Haploporus longisporus, H. crassus, H. pirongia (G. Cunn.) Meng Zhou, Y.C. Dai and T.W. May and H. septatus share similar hyphae at dissepiment edge (thick-walled with a simple septum). However, Haploporus crassus is distinguished from H. longisporus by its thick-walled basidia and shorter basidiospores (13.5–16.5 µm vs. 18.2–22 µm) [9]. Haploporus pirongia differs from H. longisporus in having smaller basidiospores (11–14 × 5.2–7 µm vs. 18.2–22 × 7–9 µm) [9]. Haploporus septatus differs from H. longisporus by the dextrinoid skeletal hyphae and smaller pores and basidiospores (5–6 per mm, 8.5–11 × 5–6 μm) [5].
In both ITS- and ITS + nLSU + mtSSU-based phylogenies (Figure 1 and Figure 2), the studied Sri Lankan samples formed two independent lineages, and they are represented by two new species: Haploporus punctatus and H. srilankensis. Both new species are closely related to Haploporus angustisporus and these three species are nested in a clade (Figure 1 and Figure 2). Morphologically, Haploporus angustisporus differs from H. punctatus by having longer basidiospores (10–13.5 × 4–5 µm vs. 9–10.8 × 3.8–5 µm) and obviously narrow fusiform cystidioles without simple septum at the tips [9]. Haploporus angustisporus differs from H. srilankensis by its annual habit, the absence of dendrohyphidia, relatively longer basidiospores (10–13.5 × 4–5 µm vs. 8.5–11.5 × 4–5 µm) [9]. Although both Haploporus punctatus and H. srilankensis occur in Sri Lanka, there are 17 base pairs differences between H. punctatus and H. srilankensis, which amounts to > 2% nucleotide differences in the ITS regions. In addition, Haploporus srilankensis differs from H. punctatus by its perennial habit, the presence of dendrohyphidia and cystidioles without septa.
Haploporus punctatus may be confused with H. subpapyraceus and H. crassus in having approximately the same pores size (3–5 per mm) and cystidioles with a simple septum at the tips. However, Haploporus subpapyraceus has wider basidiospores (9–12 × 5.5–8 μm vs. 9–10.8 × 3.8–5 µm) [5]. Haploporus crassus differs from H. punctatus by its longer basidiospores (13.5–16.5 × 4–5 µm vs. 9–10.8 × 3.8–5 µm) and the unique thick-walled basidia [9].
All the species of Haploporus with molecular evidence and close to our new species are discussed above. Furthermore, two species of Haploporus, H. brasiliensis and H. pileatus, were described from Brazil based on morphology only [7]. Haploporus brasiliensis distinctly differs from our four new species by its obviously smaller basidiospores and the absence of cystidioles and dendrohyphidia [7]. Haploporus pileatus is distinguished from all the species in this genus by its distinct pileate basidiocarps [7].
Currently, 23 species are accepted in Haploporus, and their main morphological characteristics are listed in Table 2. A key to accepted species of Haploporus is provided as follows.
Table 2.
Main morphological characters of Haploporus species.
| Species | Type Locality | Basidiomata | Pore Surface | Pores/mm | Hyphae | Cystidioles | Basidiospores | Dendrohy-Phidia | References |
|---|---|---|---|---|---|---|---|---|---|
| H. alabamae | USA | annual, resupinate | cream when fresh, pale brown when dry | 3–5 | trimitic, skeletal hyphae IKI– | fusiform | ovoid to ellipsoid, 10–12 × 4–6 μm | absent | [2,8] |
| H. angustisporus | China | annual, resupinate | cream to pale yellowish brown when fresh, brownish when bruised, olivaceous buff to pale brown when dry | 3–5 | dimitic, skeletal hyphae dextrinoid | fusiform | oblong ellipsoid, 10–13.5 × 4–5 µm | absent | [9] |
| H. bicolor | China | annual, resupinate | cream to buff with peach tint when dry | 5-7 | dimitic, skeletal hyphae IKI– | fusiform to ventricose | oblong ellipsoid to subcylindrical, 10.5–11.9 × 4.5–5 µm | present | Present study |
| H. brasiliensis | Brazil | annual, resupinate | ochraceous buff | 1-3 | dimitic, skeletal hyphae dextrinoid | absent | oblong ellipsoid, 6–8 × 4–5 µm | absent | [7] |
| H. crassus | China | annual, resupinate | white to cream when fresh, buff-yellow when dry | 3–5 | dimitic, skeletal hyphae IKI– | ventricose with a simple septum | oblong ellpsoid, 13.5–16.5 × 7.5–9.5 µm | absent | [9] |
| H. cylindrosporus | China | annual, resupinate | white to cream when fresh, pinkish buff to clay-buff when dry | 4–5 | dimitic, skeletal hyphae dextrinoid | absent | cylindrical, 10–11.5 × 4.5–5 μm | absent | [5] |
| H. gilbertsonii | USA | annual, resupinate | pale buff to buff when dry | 2–3 | dimitic, skeletal hyphae IKI– | fusiform | oblong ellipsoid, 12–15 × 6–8 µm | absent | [9] |
| H. latisporus | China | annual, resupinate | white to cream when fresh, clay-buff when dry | 1–3 | dimitic, skeletal hyphae IKI– | fusiform | oblong ellipsoid to ellipsoid, 13–18.5 × 9–10 μm | absent | [3,8] |
| H. longisporus | Ecuador | annual, resupinate | cream to pale buff when dry | 2–3 | dimitic, skeletal hyphae IKI– | fusiform to clavate | cylindrical, 18.2–22 × 7–9 µm | present | Present study |
| H. microsporus | China | annual, resupinate | pinkish buff to clay-buff when dry | 7–9 | dimitic, skeletal hyphae dextrinoid | fusiform | ellipsoid, 5.3–6.7 × 3–4.1 µm | present | [8] |
| H. nanosporus | Gabon | annual to perennial, resupinate | cream when fresh, buff to clay-buff when dry | 7–8 | trimitic, skeletal hyphae dextrinoid | absent | ellipsoid, 5–6 × 3–4 μm | absent | [4] |
| H. nepalensis | Nepal | annual, resupinate | white to cream when fresh, clay-pink to clay-buff when dry | 2–3 | dimitic, skeletal hyphae IKI– | fusiform | ellipsoid to short cylindrical, 8.5–11.5 × 4.5–6.5 μm | absent | [8,32] |
| H. odorus | China | perennial, effused-reflexed to pileate | white to cream when fresh, pale buff to light brown when dry | 3–5 | trimitic, skeletal hyphae IKI– | absent | ovoid, 5.5–6 × 4–5 μm | present | [5,8] |
| H. papyraceus | USA | annual, resupinate | white to cream when fresh, cream to buff when dry | 3–4 | dimitic, skeletal hyphae dextrinoid | fusiform | cylindrical, 13–15 × 5–6 μm | present | [31] |
| H. pileatus | Brazil | annual, pileate | ochraceous buff | 3–4 | dimitic, skeletal hyphae dextrinoid | absent | cylindrical, 9–10 × 4–5 µm | absent | [7] |
| H. pirongia | New Zealand | annual, resupinate | white to cream when fresh, pale brownish when bruised, pinkish buff to clay-buff when dry | 3–4 | trimitic, skeletal hyphae IKI– | fusiform | oblong ellipsoid to cylindrical, 11–14 × 5.2–7 µm | absent | [9] |
| H. punctatus | Sri Lanka | annual, resupinate | pale buff when dry | 3–5 | dimitic, skeletal hyphae dextrinoid | fusiform occasionally with a simple septum | cylindrical to slightly allantoid, 9–10.8 × 3.8–5 µm | absent | Present study |
| H. srilankensis | Sri Lanka | perennial, resupinate | pale buff when fresh, pinkish buff to salmon when dry | 4–5 | dimitic, skeletal hyphae dextrinoid | fusiform | oblong ellipsoid, 8.5–11 × 4–5.2 µm | present | Present study |
| H. septatus | China | annual, resupinate | white to cream when fresh, light buff when dry | 5–6 | dimitic, skeletal hyphae dextrinoid | fusiform | oblong to ellipsoid, 8.5–11 × 5–6 μm | absent | [5] |
| H. subpapyraceus | China | annual, resupinate | white to cream when fresh, cream to buff- yellow when dry | 3–5 | dimitic, skeletal hyphae dextrinoid | clavate with a simple septum | widely ellipsoid, 9–12 × 5.5–8 μm | absent | [5] |
| H. subtrameteus | Russia | perennial, resupinate | incarnadine when fresh, light reddish brown when dry | 3–4 | dimitic to trimitic, skeletal hyphae IKI– | fusiform | oblong ellipsoid to ellipsoid, 8.5–11 × 4.5–6.5 μm | absent | [5] |
| H. thindii | India | annual to perennial, resupinate | cream when fresh, clay-buff when dry | 3–4 | dimitic, skeletal hyphae IKI– | fusiform | cylindrical, 10.5–14.5 × 5.2–6.5 μm | absent | [5,8] |
| H. tuberculosus | Belarus | annual to perennial resupinate | cream when fresh, olivaceous buff when dry | 2–3 | trimitic, skeletal hyphae dextrinoid | fusiform | oblong ellipsoid, 13–15 × 6–8.5 μm | absent | [32] |
Key to Accepted Species of Haploporus
| 1. Basidiospores < 8 μm in length | 2 |
| 1. Basidiospores > 8 μm in length | 5 |
| 2. Pores 7–9 per mm | 3 |
| 2. Pores < 6 per mm | 4 |
| 3. Cystidioles absent | H. nanosporus |
| 3. Cystidioles present | H. microsporus |
| 4. Pores1–3 per mm; skeletal hyphae dextrinoid | H. brasiliensis |
| 4. Pores 4–5 per mm; skeletal hyphae non-dextrinoid | H. odorus |
| 5. Pores 5–7 per mm | 6 |
| 5. Pores < 5 per mm | 7 |
| 6. Skeletal hyphae dextrinoid; dendrohyphidia absent | H. septatus |
| 6. Skeletal hyphae non-dextrinoid; dendrohyphidia present | H. bicolor |
| 7. Basidiocarps pileate | H. pileatus |
| 7. Basidiocarps resupinate | 8 |
| 8. Pores 2–3 per mm | 9 |
| 8. Pores > 3 per mm | 13 |
| 9. Hyphal system trimitic; skeletal hyphae dextrinoid | H. tuberculosus |
| 9. Hyphal system dimitic; skeletal hyphae non-dextrinoid | 10 |
| 10. Basidiospores < 16 μm in length | 11 |
| 10. Basidiospores >16 μm in length | 12 |
| 11. Basidiospores ellipsoid to short cylindrical, <12 μm in length | H. nepalensis |
| 11. Basidiospores oblong ellipsoid, >12 μm in length | H. gilbertsonii |
| 12. Dendrohyphidia absent | H. latisporus |
| 12. Dendrohyphidia present | H. longisporus |
| 13. Basidiocarps perennial | 14 |
| 13. Basidiocarps annual | 16 |
| 14. Skeletal hyphae dextrinoid | H. srilankensis |
| 14. Skeletal hyphae non-dextrinoid | 15 |
| 15. Pore surface light reddish brown when dry; basidiospores ellipsoid | H. subtrameteus |
| 15. Pore surface clay-buff when dry; basidiospores cylindrical | H. thindii |
| 16. Cystidioles absent | H. cylindrosporus |
| 16. Cystidioles present | 17 |
| 17. Skeletal hyphae dextrinoid | 18 |
| 17. Skeletal hyphae non-dextrinoid | 21 |
| 18. Basidiospores broadly ellipsoid, >5 μm in width | 19 |
| 18. Basidiospores oblong ellipsoid or cylindrical to slightly allantoid, <5 μm in width | 20 |
| 19. Cystidioles fusiform without apically simple septa | H. papyraceus |
| 19. Cystidioles slim clavate with apically simple septa | H. subpapyraceus |
| 20. Basidiospores oblong ellipsoid | H. angustisporus |
| 20. Basidiospores cylindrical to slightly allantoid | H. punctatus |
| 21. Hyphae at dissepiment edge non-septate | H. alabamae |
| 21. Hyphae at dissepiment edge with a simple septum | 22 |
| 22. Basidia thick-walled; basidiospores > 7 μm in width | H. crassus |
| 22. Basidia thin-walled; basidiospores < 7 μm in width | H. pirongia |
Author Contributions
Conceptualization, Y.-C.D. and M.Z.; methodology, M.Z.; performing the experiment, M.Z.; formal analysis, M.Z.; validation, M.Z., Y.-C.D., J.V. and Y.Y.; resources, Y.-C.D. and J.V.; writing—original draft preparation, M.Z.; writing—review and editing, Y.-C.D., J.V. and Y.Y.; visualization, M.Z.; supervision, Y.-C.D. and Y.Y.; project administration, Y.Y.; funding acquisition, Y.-C.D. All authors have read and agreed to the published version of the manuscript.
Funding
The research is supported by the National Natural Science Foundation of China (Project No. U1802231).
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: [https://www.ncbi.nlm.nih.gov/; https://www.mycobank.org/page/Simple%20names%20search; http://purl.org/phylo/treebase, submission ID 27556].
Conflicts of Interest
The authors declare that there are no conflicts of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Singer R. Notes on taxonomy and nomenclature of the polypores. Mycologia. 1944;36:65–69. doi: 10.1080/00275514.1944.12017529. [DOI] [Google Scholar]
- 2.Dai Y.C., Niemelä T., Kinnunen J. The polypore genera Abundisporus and Perenniporia (Basidiomycota) in China, with notes on Haploporus. Ann. Bot. Fenn. 2002;39:169–182. doi: 10.3852/14-216. [DOI] [Google Scholar]
- 3.Li J., Dai Y.C., Yuan H.S. A new species of Haploporus (Basidiomycotina) from China. Mycotaxon. 2007;99:181–187. [Google Scholar]
- 4.Piątek M. Taxonomic position and world distribution of Pachykytospora nanospora (Polyporaceae) Ann. Bot. Fenn. 2005;42:23–25. [Google Scholar]
- 5.Shen L.L., Chen J.J., Wang M., Cui B.K. Taxonomy and multi-gene phylogeny of Haploporus (Polyporales, Basidiomycota) Mycol. Prog. 2016;15:731–742. doi: 10.1007/s11557-016-1203-y. [DOI] [Google Scholar]
- 6.Kotlaba F., Pouzar Z. A new genus of the polypores—Pachykytospora gen. nov. Česká Mykol. 1963;17:27–34. [Google Scholar]
- 7.Lira C.R.S., Nogueira-Melo G.S., Ryvarden L., Gibertoni T.B. Two new species of Haploporus from Brazil. Synop. Fungorum. 2018;38:62–65. [Google Scholar]
- 8.Cui B.K., Li H.J., Ji X., Zhou J.L., Song J., Si J., Yang Z.L., Dai Y.C. Species diversity, taxonomy and phylogeny of Polyporaceae (Basidiomycota) in China. Fungal Divers. 2019;97:137–392. doi: 10.1007/s13225-019-00427-4. [DOI] [Google Scholar]
- 9.Zhou M., Wang L., May T.W., Vlasák J., Chen J.J., Dai Y.C. Phylogeny and diversity of Haploporus (Polyporaceae, Basidiomycota) MycoKeys. 2019;54:77–98. doi: 10.3897/mycokeys.54.34362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai Y.C., Kashiwadani H. Haploporus subtrameteus (Polyporaceae, Basidiomycota) found in Japan. Mycoscience. 2009;50:452–454. doi: 10.1007/S10267-009-0498-9. [DOI] [Google Scholar]
- 11.Hattori T., Adhikari M.K., Suda T., Doi Y. A list of polypores (Basidiomycotina, Aphyllophorales) collected in Jumla. Nepal. Bull. Natl. Sci. Mus. 2002;28:27–38. doi: 10.1007/s13225-012-0177-6. [DOI] [Google Scholar]
- 12.Zhou J.L., Cui B.K. Phylogeny and taxonomy of Favolus (Basidiomycota) Mycologia. 2017;109:766–779. doi: 10.1080/00275514.2017.1409023. [DOI] [PubMed] [Google Scholar]
- 13.Petersen J.H. Farvekort. The Danish Mycological Society´s Colour-Chart. Foreningen til Svampekundskabens Fremme; Greve, Italy: 1996. pp. 1–6. [Google Scholar]
- 14.Thiers B. Index Herbariorum: A Global Directory of Public Herbaria and Associated Staff. New York Botanical Garden’s Virtual Herbarium; New York, NY, USA: 2018. [(accessed on 15 January 2021)]. Available online: http://sweetgum.nybg.org/science/ih/ [Google Scholar]
- 15.Cao Y., Wu S.H., Dai Y.C. Species clarification of the prize medicinal Ganoderma mushroom “Lingzhi”. Fungal Divers. 2012;56:49–62. doi: 10.1007/s13225-012-0178-5. [DOI] [Google Scholar]
- 16.Zhao C.L., Cui B.K. Morphological and molecular identification of four new resupinate species of Perenniporia (Polyporales) from southern China. Mycologia. 2013;105:945–958. doi: 10.3852/12-201. [DOI] [PubMed] [Google Scholar]
- 17.White T.J., Bruns T., Lee S., Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gefand D.H., Sninsky J.J., White M.J.T., editors. PCR Protocols: A Guide to Methods and Applications. Academic Press; San Diego, FL, USA: 1990. pp. 315–322. [DOI] [Google Scholar]
- 18.Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The CLUSTAL X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall T.A. Bioedit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 20.Nilsson R.H., Tedersoo L., Abarenkov K., Ryberg M., Kristiansson E., Hartmann M., Schoch C.L., Nylander J.A.A., Bergsten J., Porter T.M., et al. Five simple guidelines for establishing basic authenticity and reliability of newly generated fungal ITS sequences. MycoKeys. 2012;4:37–63. doi: 10.3897/mycokeys.4.3606. [DOI] [Google Scholar]
- 21.Binder M., Justo A., Riley R., Salamov A., Lopez-Giraldez F., Sjökvist E., Copeland A., Foster B., Sun H., Larsson E., et al. Phylogenetic and phylogenomic overview of the Polyporales. Mycologia. 2013;105:1350–1373. doi: 10.3852/13-003. [DOI] [PubMed] [Google Scholar]
- 22.Zhao C.L., Chen H., Song J., Cui B.K. Phylogeny and taxonomy of the genus Abundisporus (Polyporales, Basidiomycota) Mycol. Prog. 2015;14:38. doi: 10.1007/s11557-015-1062-y. [DOI] [Google Scholar]
- 23.Farris J.S., Källersjö M., Kluge A.G., Bult C. Testing significance of incongruence. Cladistics. 1994;10:315–319. doi: 10.1111/j.1096-0031.1994.tb00181.x. [DOI] [Google Scholar]
- 24.Swofford D.L. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Cmethods) Sinauer Associates; Sunderland, MA, USA: 2002. [Google Scholar]
- 25.Felsenstein J. Confidence intervals on phylogenetics: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 26.Darriba D., Taboada G.L., Doallo R., Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analysis with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 28.Silvestro D., Michalak I. raxmlGUI: A graphical front-end for RAxML. Org. Divers. Evol. 2012;12:335–337. doi: 10.1007/s13127-011-0056-0. [DOI] [Google Scholar]
- 29.Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Hőhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Page R.D.M. TreeView: Application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. Cabios. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 31.Gerber A.L., Loguercio-Leite C. New records of polypores (Aphyllophorales) from Southern Brazil. Mycotaxon. 1997;62:305. [Google Scholar]
- 32.Piątek M. Haploporus tuberculosus, a new polypore genus and species in Belarus, with a new combination in Haploporus. Pol. Bot. J. 2003;48:81–83. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: [https://www.ncbi.nlm.nih.gov/; https://www.mycobank.org/page/Simple%20names%20search; http://purl.org/phylo/treebase, submission ID 27556].