Abstract
In poultry production, gut microbiota (GM) plays a pivotal role and influences different host functions related to the efficiency of production performances. Antimicrobial (AM) use is one of the main factors affecting GM composition and functions. Although several studies have focused their attention on the role of AMs as growth promoters in the modulation of GM in broilers, the consequences of higher AM concentrations administered during prophylactic treatments need to be better elucidated. For this purpose, 16S rRNA gene sequencing was performed to evaluate the impact of different prophylactic AM protocols on the composition and diversity of the broiler GM. Diversity analysis has shown that AM treatment significantly affects alpha diversity in ileum and beta diversity in both ileum and caecum. In ileal samples, the Enterobacteriaceae family has been shown to be particularly affected by AM treatments. AMs have been demonstrated to affect GM composition in broiler. These findings indicate that withdrawal periods were not enough for the restoral of the original GM. Further studies are needed for a better elucidation of the negative effects caused by an altered GM in broilers.
Keywords: prophylaxis, coccidiostat, broiler, 16S rRNA sequencing, ileum, caecum
1. Introduction
The intestinal microbiota is a complex microbial community that has established a symbiotic relationship with its animal host [1]. It is well known that gut microbiota (GM) plays an essential role as a first-line defense in the intestinal mucosal barrier [2]. This microbial community has evolved together with the animal host and through the colonization resistance ensures the prevention of animal health from the arousal of intestinal infectious disease [2,3]. In poultry production, GM plays a pivotal role and its several functions have been related to the efficiency of production performances [4,5,6]. Furthermore, GM influences different host functions, e.g., digestion of nutrients, immunity, and metabolism [1,7]. It has been demonstrated that GM perturbations could negatively affect intestinal health and lead to severe health consequences for animals, but also to economic losses for the farmer [7,8,9]. Antimicrobial (AM) use is one of the main factors affecting GM composition and functions. In recent years, there has been an increasing concern about antimicrobials resistance (AMR) and the role of AM use in the spreading of AMR, and the selection of multi-drug-resistant bacteria [10,11]. On the other hand, AM use leads to long-lasting disturbances of the commensal GM and may negatively affect broiler physiology [12]. Moreover, the broiler GM varies among the different intestinal tracts and its composition reflects their different functions [7]. Several studies focused their attention on the role of AMs as growth promoters in the modulation of GM in broilers [13,14,15], but the consequences of higher AM concentration on GM, e.g., during prophylactic treatments, need to be better elucidated. Therefore, the main aim of this study was to evaluate the impact of different prophylactic AM protocols on the composition and diversity of the broiler GM by 16S rRNA gene sequencing technology. In addition, we evaluated the different impact of AM treatment on the ileum and caecum microbiota.
2. Results
2.1. 16S rRNA Diversity Analysis
A total of 84 samples were analyzed both for ileal (n = 42) and caecal (n = 42) microbiota by 16S rRNA sequencing. A total of 8,036,623 raw reads (2 × 300 bp) were obtained after sequencing, with an average value of 95,674 reads/sample. After joint and quality filtering, a total of 4,963,263 reads passed the filters applied through the DADA2 plugin in the QIIME2 software package (https://qiime2.org; version: 2019.10), with an average value of 95,361 reads/sample. Following primer trimming and raw read quality filtering, ileal and caecal samples were analyzed separately. In order to avoid biases due to different sequencing depths, ileal samples and caecal samples were rarefied at 31,100 reads and 5600 reads, respectively; one sample from ileum was excluded from further analyses due to a low read count. After assigning taxonomies, a total of 3421 unique operational taxonomic units (OTUs) at 99% nucleotide sequence identity were identified in caecum samples, while 1153 unique OTUs were identified in ileum samples.
Alpha diversity analysis has shown that AM treatments have different consequences on ileum and caecum microbiota. Considering caecal samples, Pielou’s evenness (H = 5.38; p = 0.5) and Faith’s phylogenetic diversity (H = 7.69; p = 0.26) indicated no significant differences between samples. Instead, ileum samples were characterized by alpha diversity metrics statistically different with Pielou’s evenness (H = 13.53; p = 0.03) and Faith’s phylogenetic diversity (H = 13.12; p = 0.04).
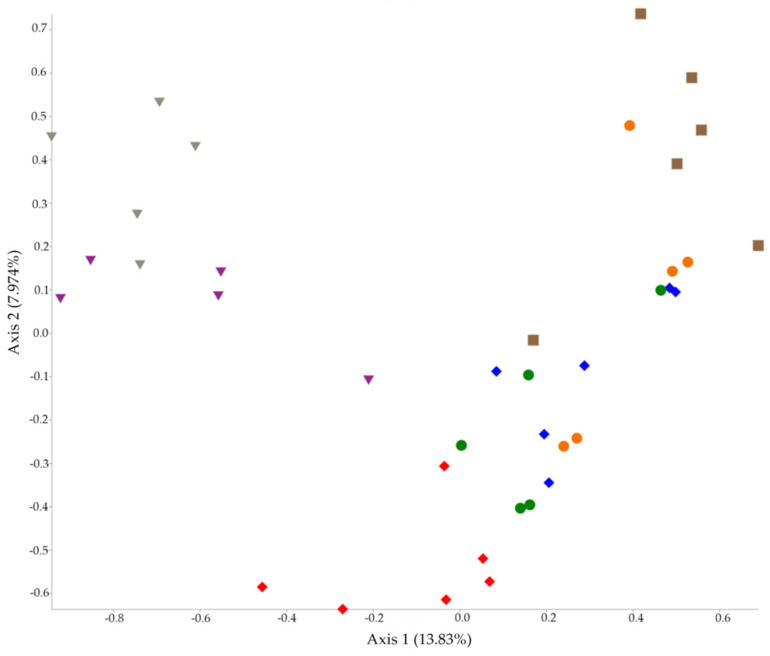
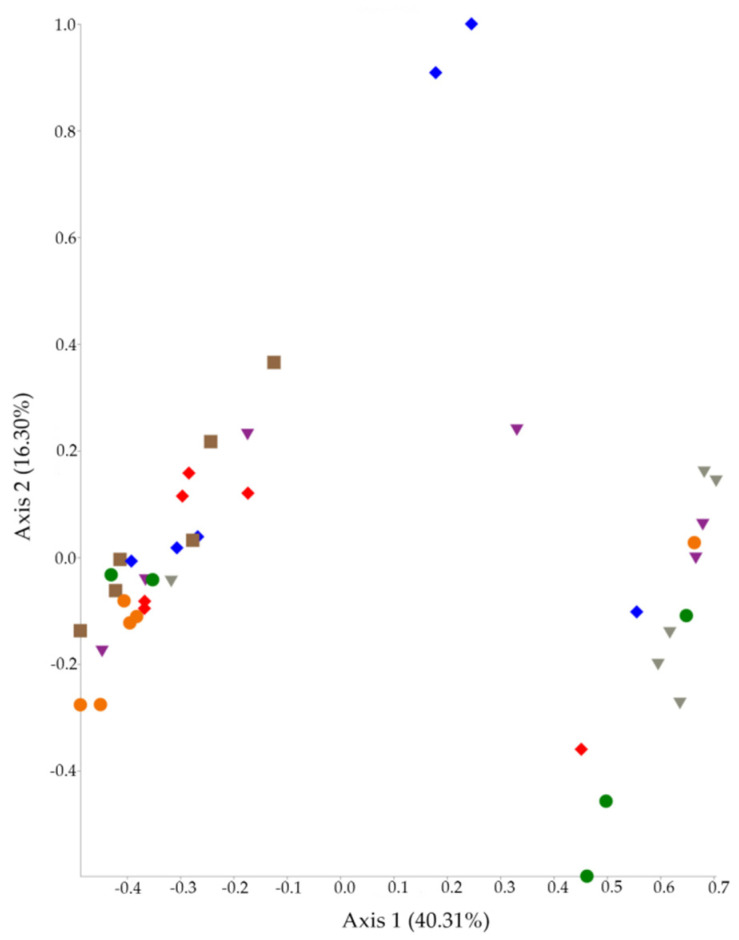
In addition, the Permutational Multivariate Analysis of Variance (PERMANOVA) test has shown that beta diversity metrics were always statistically different between groups (p = 0.001 for caecum and p = 0.002 for ileum). Principal Coordinate Analysis (PCoA) plots were generated both for caecum and ileum using beta diversity metrics and are presented in Figure 1 (caecum) and Figure 2 (ileum).
Figure 1.
PCoA plot of beta diversity analysis of caecal samples. Amoxicillin group (AMX) (red), amoxicillin + diclazuril group (AMX + DCZ) (blue), thiamphenicol group (THP) (purple), thiamphenicol + diclazuril group (THP + DCZ) (grey), sulfadiazine + trimethoprim group (TRIM) (brown), diclazuril group (DCZ) (orange) and control group (K) (green).
Figure 2.
PCoA plot of beta diversity analysis of ileal samples. Amoxicillin group (AMX) (red), amoxicillin + diclazuril group (AMX + DCZ) (blue), thiamphenicol group (THP) (purple), thiamphenicol + diclazuril group (THP + DCZ) (grey), sulfadiazine + trimethoprim group (TRIM) (brown), diclazuril group (DCZ) (orange) and control group (K) (green).
2.2. 16S rRNA Taxonomy Analysis
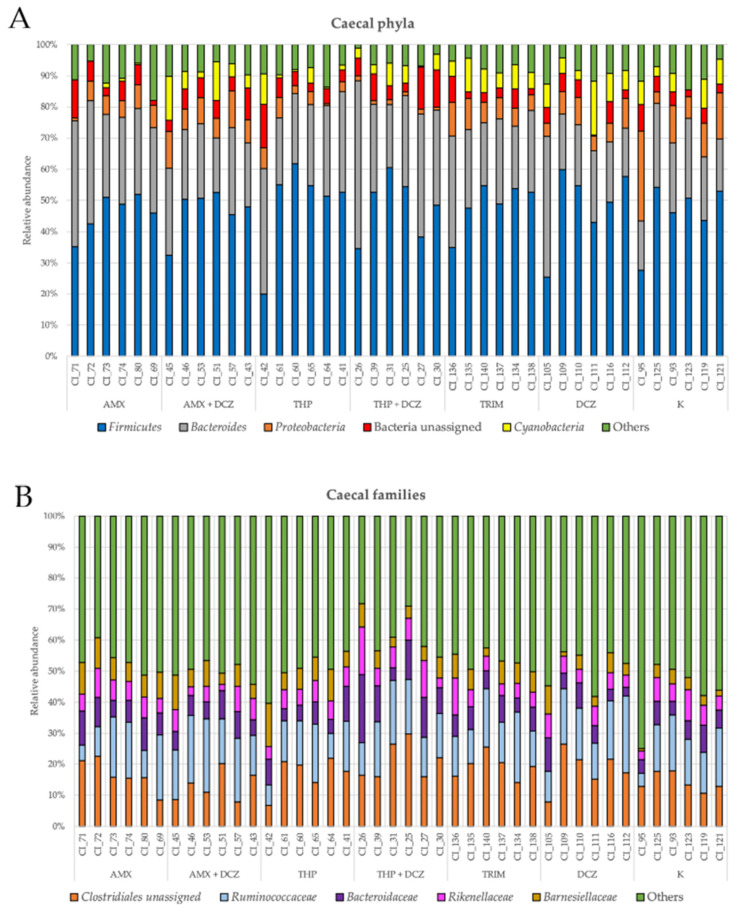
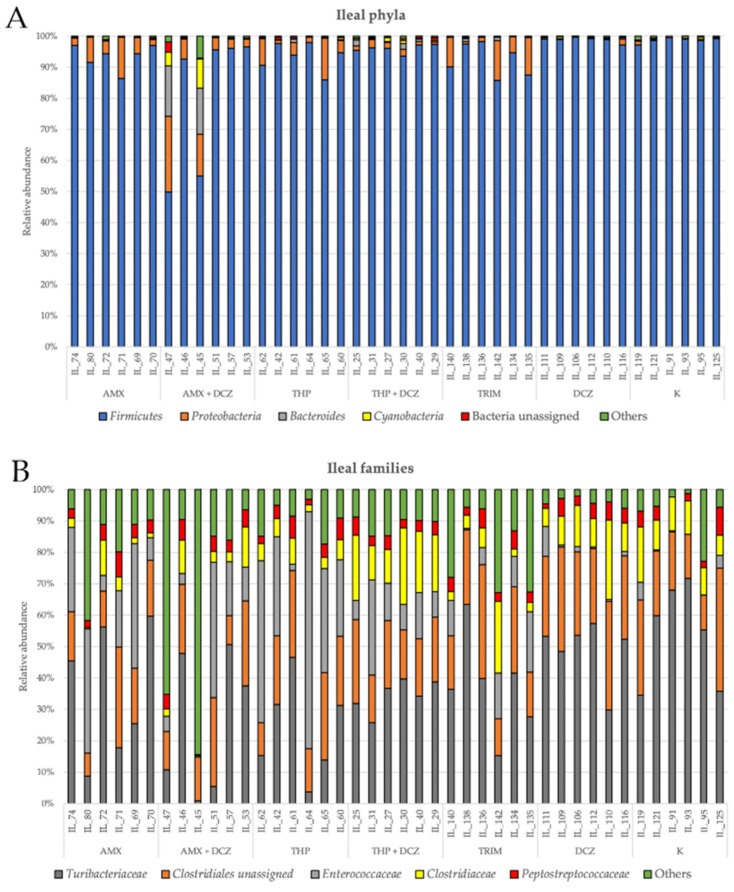
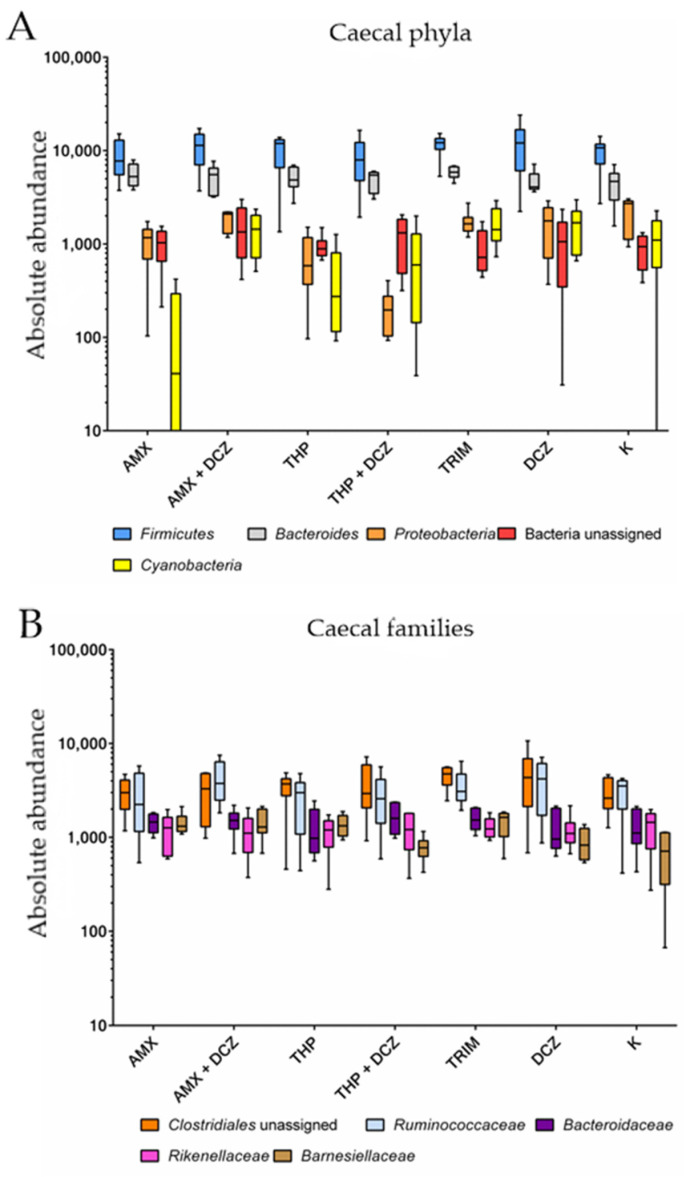
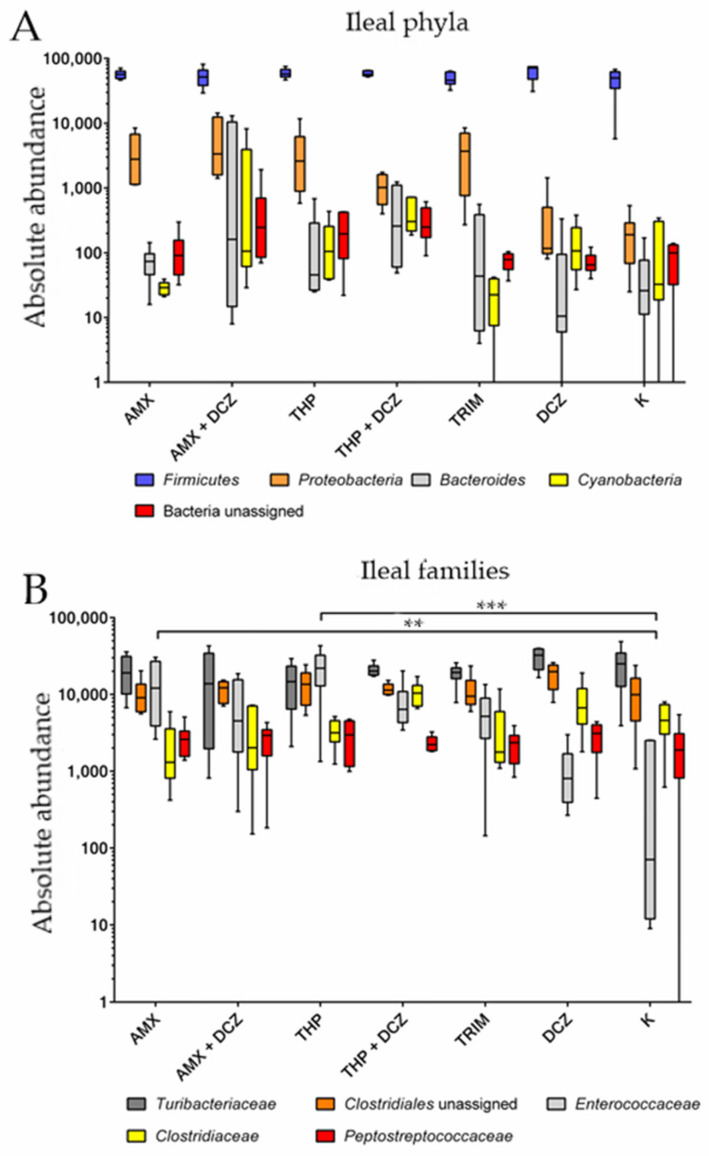
For caecal samples, the dominant phyla were in order: Firmicutes, Bacteroides, Proteobacteria, an unassigned bacterial phylum, and Cyanobacteria. At the family level, OTUs were assigned to an unassigned family of Clostridiales order, Ruminococcaceae, Bacteroidaceae, Rikinellaceae, and Barnesiellaceae. Among ileal samples, the main five phyla were in order: Firmicutes, Proteobacteria, Bacteroides, Cyanobacteria, and an unassigned bacterial phylum. At the family level, the main taxa identified were, respectively: Turibacteriaceae, an unassigned family of Clostridiales order, Enterococcaceae, Clostridiaceae, and Peptostreptococcaceae. Relative abundances of the aforementioned taxa are shown in bar plots of Figure 3 for caecum and Figure 4 for ileum.
Figure 3.
Relative abundance at the phylum (A) and family level (B) in caecum. Specific legends are reported for each bar plot.
Figure 4.
Relative abundance at the phylum (A) and family level (B) in ileum. Specific legends are reported for each bar plot.
Two-way ANOVA was performed on the abundance of OTU values belonging to the aforementioned phyla and families. Related p values are presented in Table 1. Detailed results of the two-way ANOVA are available in Table S1.
Table 1.
p-Values summary from two-way ANOVA analysis of the top five phyla and families in caecal and ileal samples.
| p-Values | ||||
|---|---|---|---|---|
| Source of Variation | Phyla | Families | ||
| Caecum | Ileum | Caecum | Ileum | |
| Interaction | 0.92 | 0.31 | 0.32 | <0.0001 |
| Groups | 0.64 | 0.32 | 0.81 | 0.17 |
| Taxa | <0.0001 | <0.0001 | <0.0001 | <0.0001 |
The results show that an effect of the studied taxa was always significant on the total variance of the samples. Box plots are shown in Figure 5 (caecum) and Figure 6 (ileum). Most intriguingly, however, in Tukey’s post-test the ileal families Enterococcaceae have been shown to be significantly overrepresented in our samples in amoxicillin group (AMX) (p-value = 0.004) and in thiamphenicol group (THP) (p-value < 0.0001) groups in comparison to control group K.
Figure 5.
Box plots of two-way ANOVA analysis for caecal phyla (A) and families (B).
Figure 6.
Box plots of two-way ANOVA analysis for ileal phyla (A) and families (B). (** p-value < 0.01; *** p-value < 0.001).
3. Discussion
In the last two decades, AM use in veterinary medicine has undergone several changes and limitations mainly considering the threat of AMR. Prophylactic and therapeutic uses of AMs are still permitted, even if their way of use has been continuously revised in compliance with national and international laws. Poultry production is particularly concerned by these changes. It is well known that broiler GM is a favorable environment for the dissemination of AMR genes between commensal and pathogenic bacteria [16]. Moreover, the selective pressure of AM treatments can enforce the selection of resistant bacteria worsening the AMR threat in poultry productions [17]. On the other hand, any modifications of GM, also known as dysbiosis, could alter broiler physiology and increase susceptibility to infectious disease, or also metabolic and inflammatory dysfunctions [2].
In our study, we aim to investigate the impact of different treatments on broiler GM in ileum and caecum. Several studies have described the microbial communities that colonized the two different intestinal tracts [7,18]. Their microbial composition reflects the different functions of these organs, and their relationship has brought mutual benefits for each other. In particular, the ileum is the main site of nutrient absorption and it is generally colonized by bacteria that influence nutrients availability and host metabolism [6,7]. For example, Lactobacillus spp. is one of the main represented genera and its functions have been related to the production of essential vitamins, but also the recycling of intestinal bile acids [7]. On the other side, caecum is an important site of starch fermentation, water absorption, and urea recycling and these activities are strictly dependent on the metabolic capacity of caecum microbiota [6,19]. Caecal GM has always been reported as the one with the highest richness and abundance in bacterial composition and Bacteroides spp., Ruminococcus spp., and Faecalibacterium spp. are the main genera that have been found [7,18,20]. Our results highlighted that different AM treatments have a diverse impact on ileal and caecal communities.
Considering alpha diversity metrics, which are indicators of the abundance of a microbial community [21], our results show significantly different Pielou’s evenness (p = 0.03 for ileum; p = 0.5 for caecum) and Faith’s phylogenetic diversity (p = 0.04 for ileum; p = 0.26 for caecum) only in ileum. These results can suggest that the withdrawal periods could be not enough for the restoral of the abundance of microbial communities in the ileum. It is well known that broiler caecum harbors the richest and most diverse microbiota in comparison with other intestinal tracts [22,23,24]. Moreover, Mohd Shaufi et colleagues [25] have shown that caecum microbiota presented a significant difference in the expression of the bacterial colonization pathway (e.g., motility proteins, two-component system, and bacterial system). All these considerations can help to explain the alpha diversity metrics not affected in caecal samples.
On the other hand, beta diversity metrics are significantly affected by AM treatment (p = 0.001 in the caecum and p = 0.002 in the ileum). Beta indexes describe compositional changes between different microbial communities [21]. As expected, the six AM protocols caused different changes in GM of ileum and caecum. These significant results can also be visualized in PCoA plots (Figure 1 and Figure 2), where caecal control samples are clearly separated from the others. Otherwise, the ileal PCoA plot shows less separation between groups probably due to a lower diversity of ileal GM than the caecal one, as mentioned above. First, we can explain this result with the different broad-spectrum action of amoxicillin, thiamphenicol, and trimethoprim–sulfadiazine. In addition, the association with diclazuril can modulate the AM activity against gut bacteria. In our case, sulfadiazine–trimethoprim is not active against bacteria belonging to Enterococcus and Pseudomonas genera [26]. On the contrary, thiamphenicol and amoxicillin have a wider antibacterial activity than sulfadiazine–trimethoprim. Amoxicillin is often associated with clavulanic acid to improve antibacterial activity against β-lactamases producers [27]. In Italy, amoxicillin is only permitted as monotherapy in poultry production, and several pieces of evidence support that broiler GM is a reservoir of different β-lactamases producing bacteria, e.g., Extended-Spectrum-Beta-Lactamases producing Enterobacteriaceae [28,29].
In our study, taxonomy results were analyzed by two-way ANOVA to verify if a differentially abundant taxon related to the different AM treatment groups existed. The extremely abundant phylum in ileal samples was Firmicutes, and, due to this high prevalence, the phyla significantly affected the variance in our samples both in ileum and caecum. This result is in accordance with other published works, where Firmicutes is the main phyla in ileum accounting for more than 90% of the total OTUs and also the dominant one in caecum but less frequent than other phyla [4,7]. Moreover, in ileal samples, the Enterococcaceae family was shown to be significantly affected by AM treatments in groups AMX and THP. The increase in bacteria belonging to this family may be due to an increased resistance against amoxicillin and thiamphenicol. Enterococcaceae are known to present a naturally reduced susceptibility to penicillins, due to the expression of low-affinity penicillin-binding proteins [30,31]. Resistance to phenicols, instead, has been recently described in Enterococcus spp. harboring multi-drug resistance plasmids isolated from both animal and human samples [30,32].
4. Materials and Methods
4.1. Animals and Samples Collection
A total of 240 male broilers (Ross 308) were reared under the same conditions in the chicken broiler farm facility of the Department of Veterinary Sciences of the University of Turin as previously reported [33]. Briefly, chicks were randomly allocated into 7 groups. During production cycles, six groups received a different antimicrobial prophylactic program: thiamphenicol (Tirsan O.S. 200 mg/g) (THP), amoxicillin (Amoxid Polv 1430 g) (AMX), sulfadiazine + trimethoprim (Trimethosulfa orale) (TRIM), thiamphenicol (Tirsan O.S. 200 mg/g) + diclazuril (Coxiril) (THP + DCZ), amoxicillin (Amoxid Polv 1430 g) + diclazuril (Coxiril) (AMX + DCZ), and diclazuril (Coxiril) (DCZ). Untreated animals were used as a control group (K). Chickens received a starter diet from 0 to 25th day and a grower diet from 26th to 58th day of rearing and AMs were administered in both these periods. Water and feed were provided ad libitum during the whole cycle. The environmental conditions (lighting programs, temperature, relative humidity, and ventilation rates) were regulated accordingly to the Ross broiler management guidelines. AMs were administered via drinking water. Detailed prophylactic protocols are available in Table 2. The withdrawal periods were respected before slaughtering. At the end of the rearing cycle (58 days), 120 broilers were regularly slaughtered, in particular, 15 animals per each treated group and 30 animals of the untreated group. Intestinal contents from ileum and caecum were aseptically collected. Samples were immediately stored at −80 °C.
Table 2.
Antimicrobial (AM) prophylactic treatment protocols applied during broiler rearing. The animals were divided into seven groups and slaughtered after a withdrawal period.
| Group | AMs | Dosing Protocols per os | Time-Lapse of Treatments (Rearing Days) | Withdrawal (Days) |
|---|---|---|---|---|
| AMX | Amoxicillin | 30 mg·kg−1 b.w. twice/day | 20–22/53–56 | 1 |
| AMX + DCZ | Amoxicillin Diclazuril |
30 mg·kg−1 b.w. twice/day 1 mg/kg |
20–22/53–56 0–52 |
1 5 |
| THP | Thiamphenicol | 65 mg·kg−1 b.w./day | 23–25/47–51 | 6 |
| THP + DCZ | Thiamphenicol Diclazuril |
65 mg·kg−1 b.w./day 1 mg/kg |
23–25/47–51 0–52 |
6 5 |
| TRIM | Sulfadiazine Trimethoprim |
20 mg·kg−1 b.w./day 4 mg b.w./day |
21–25/50–54 | 3 |
| DCZ | Diclazuril | 1 mg/kg | 0–52 | 5 |
| K | - | - | - | - |
b.w.: body weight.
4.2. DNA Extraction and 16S rRNA Sequencing
Genomic DNA was isolated from 6 samples, randomly selected in each group, both for ileal and caecal content. DNAzol (Thermo Fisher Scientific, Waltham, MA, USA) and DNeasy PowerClean Pro Cleanup Kit (QIAGEN, Hilden, Germany) were, respectively, used for DNA extraction and purification according to the manufacturer’s procedures. DNA was quantified using a Nanodrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Library preparation and 16S rRNA gene sequencing were performed by an external laboratory (BMR Genomics, Padova, Italy). The variable V3 and V4 regions were amplified with universal prokaryote primers Pro341F and Pro805R [34]. A Nextera XT Index kit (Illumina, San Diego, CA, USA) was used for libraries’ preparation. Finally, amplicons were sequenced with the Illumina MiSeq platform using a 2 × 300 bp paired-end protocol.
4.3. Bioinformatic Analyses
Raw reads were divided according to their intestinal origin (ileum and caecum) and analyzed separately with the same bioinformatic pipeline. The quality of raw reads was checked with FastQC v. 0.11.9 software. Then, raw reads were processed following Qiime2 v. 2019.10 pipeline [35]. Primers were trimmed using Cutadapt [36]. Low-quality reads with a q-score < 20 were removed. After trimming and filtering steps, clean reads were processed with DADA2 [16]. The amplicon sequence variant (ASV) table was used for generating the rooted and unrooted phylogenetic trees. Afterwards, alpha (Pielou’s evenness and Faith’s phylogenetic Diversity) and beta diversity indexes (unweighted UniFrac, weighted UniFrac, Jaccard, and Bray-Curtis distances) were determined. PCoA plots were generated using Emperor tool in Qiime2. Statistical analyses were conducted using Qiime2 plugins. A pairwise Kruskall–Wallis test and nonparametric Permutational Multivariate Analysis of Variance (PERMANOVA) tests were conducted, respectively, for alpha and beta metrics. The alpha-rarefaction curve was generated to understand if the sequencing depth was enough to describe all microbial communities. Finally, taxonomy was assigned to the obtained ASV table using the Greengenes database clustered at 99% identity. The Greengenes database was previously trained to the region targeted by sequencing primers. Box plots were generated for taxonomy results visualization.
4.4. Taxonomy Analyses
Data from the taxonomic assignment of OTUs were tested for differentially abundant taxa for both phyla and families. Statistical analyses were performed by two-way ANOVA using GraphPad Prism v.6 (GraphPad Software, San Diego, CA, USA). The two effects evaluated were the AM treatment group (AMX, AMX + DCZ, THP, THP + DCZ, TS, DCZ, and K) and the top five phyla and families represented in ileal and caecal samples. We tested multiple comparison corrections with Tukey’s test when two-way ANOVA showed a p-value < 0.05 or a significant interaction was revealed.
5. Conclusions
In conclusion, our study demonstrates that AM prophylactic protocols, as they are normally adopted in poultry production, are responsible for GM compositional alterations. Moreover, the respected withdrawal periods were not enough for the restoral of a GM similar to the K group’s one. This conclusion could seem to be in accordance with Elokil et al. [37], who recently published a paper where adult broilers had negative consequences on GM after enrofloxacin administration. However, this study is not comparable with ours because the age of broilers and the applied AM protocols are different. These changes to GM can lead to negative consequences for broilers, and, in particular, future studies should focus on how GM alterations affect host intestinal physiology. Moreover, further investigations focusing on the selective pressure applied by AM treatments on the expression of AMR genes in GM need to be carried out.
Acknowledgments
The authors are grateful to Elena Pagani (Monge & C, Monasterolo di Savigliano, Italy) for providing technical support.
Supplementary Materials
The following are available online at https://www.mdpi.com/2079-6382/10/2/146/s1, Table S1: two-way ANOVA results.
Author Contributions
Conceptualization, F.T.C.; Data curation, M.C., P.P. and S.D.; Formal analysis, M.C., S.R., D.G., E.G. and F.T.C.; Funding acquisition, F.T.C.; Investigation, M.C. and S.D.; Methodology, S.R., E.G., P.P. and F.T.C.; Project administration, F.T.C.; Supervision, F.T.C.; Validation, D.G.; Writing—original draft, M.C., D.G. and S.D.; Writing—review and editing, M.C., S.D. and F.T.C. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by Ministero dell’Istruzione, dell’Università e della Ricerca (MIUR) under the program “Dipartimenti di Eccellenza ex L.232/2016” to the Department of Veterinary Science, University of Turin, Italy. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Institutional Review Board Statement
All applicable international, national and/or institutional guidelines for care and use of animals were followed. The authors further specify that intestinal contents were collected at the end of the rearing cycle during a regular slaughter.
Informed Consent Statement
Not Applicable.
Data Availability Statement
The 16S rRNA sequencing raw reads presented in this study can be found online at https://www.ncbi.nlm.nih.gov/sra under the accession number PRJNA698460.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Le Roy C.I., Woodward M.J., Ellis R.J., La Ragione R.M., Claus S.P. Antibiotic treatment triggers gut dysbiosis and modulates metabolism in a chicken model of gastro-intestinal infection. BMC Vet. Res. 2019;15:1–13. doi: 10.1186/s12917-018-1761-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iacob S., Iacob D.G. Infectious Threats, the Intestinal Barrier, and Its Trojan Horse: Dysbiosis. Front. Microbiol. 2019;10:1–17. doi: 10.3389/fmicb.2019.01676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kers J.G., Velkers F.C., Fischer E.A.J., Hermes G.D.A., Stegeman J.A., Smidt H. Host and environmental factors affecting the intestinal microbiota in chickens. Front. Microbiol. 2018;9:235. doi: 10.3389/fmicb.2018.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yeoman C.J., Chia N., Jeraldo P., Sipos M., Goldenfeld N.D., White B.A. The microbiome of the chicken gastrointestinal tract. Anim. Health Res. Rev. 2012;13:89–99. doi: 10.1017/S1466252312000138. [DOI] [PubMed] [Google Scholar]
- 5.Wei S., Morrison M., Yu Z. Bacterial census of poultry intestinal microbiome. Poult. Sci. 2013;92:671–683. doi: 10.3382/ps.2012-02822. [DOI] [PubMed] [Google Scholar]
- 6.Stanley D., Hughes R.J., Moore R.J. Microbiota of the chicken gastrointestinal tract: Influence on health, productivity and disease. Appl. Microbiol. Biotechnol. 2014;98:4301–4310. doi: 10.1007/s00253-014-5646-2. [DOI] [PubMed] [Google Scholar]
- 7.Xiao Y., Xiang Y., Zhou W., Chen J., Li K., Yang H. Microbial community mapping in intestinal tract of broiler chicken. Poult. Sci. 2017;96:1387–1393. doi: 10.3382/ps/pew372. [DOI] [PubMed] [Google Scholar]
- 8.Fletcher O.J., Mansell R., Martin M.P., Borst L.B., Barnes H.J., Gonzalez L.M. Gross morphometry, histomorphometry, and immunohistochemistry confirm early and persistent jejunal crypt hyperplasia in poults with enteritis and depressed growth. Avian Dis. 2018;62:163–170. doi: 10.1637/11759-101717-Reg.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Awad W.A., Hess C., Hess M. Enteric pathogens and their toxin-induced disruption of the intestinal barrier through alteration of tight junctions in chickens. Toxins. 2017;9:60. doi: 10.3390/toxins9020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiong W., Wang Y., Sun Y., Ma L., Zeng Q., Jiang X., Li A., Zeng Z., Zhang T. Antibiotic-mediated changes in the fecal microbiome of broiler chickens define the incidence of antibiotic resistance genes. Microbiome. 2018;6:1–11. doi: 10.1186/s40168-018-0419-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mehdi Y., Létourneau-Montminy M.P., Gaucher M.L., Chorfi Y., Suresh G., Rouissi T., Brar S.K., Côté C., Ramirez A.A., Godbout S. Use of antibiotics in broiler production: Global impacts and alternatives. Anim. Nutr. 2018;4:170–178. doi: 10.1016/j.aninu.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgun A., Dzutsev A., Dong X., Greer R.L., Sexton D.J., Ravel J., Schuster M., Hsiao W., Matzinger P., Shulzhenko N. Uncovering effects of antibiotics on the host and microbiota using transkingdom gene networks. Gut. 2015;64:1732–1743. doi: 10.1136/gutjnl-2014-308820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi J.H., Lee K., Kim D.W., Kil D.Y., Kim G.B., Cha C.J. Influence of dietary avilamycin on ileal and cecal microbiota in broiler chickens. Poult. Sci. 2018;97:970–979. doi: 10.3382/ps/pex360. [DOI] [PubMed] [Google Scholar]
- 14.Gadde U.D., Oh S., Lillehoj H.S., Lillehoj E.P. Antibiotic growth promoters virginiamycin and bacitracin methylene disalicylate alter the chicken intestinal metabolome. Sci. Rep. 2018;8:3592. doi: 10.1038/s41598-018-22004-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Costa M.C., Bessegatto J.A., Alfieri A.A., Weese J.S., Filho J.A.B., Oba A. Different antibiotic growth promoters induce specific changes in the cecal microbiota membership of broiler chicken. PLoS ONE. 2017;12:e171642. doi: 10.1371/journal.pone.0171642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y., Hu Y., Liu F., Cao J., Lv N., Zhu B., Zhang G., Fu Gao G. Integrated metagenomic and metatranscriptomic profiling reveals differentially expressed resistomes in human, chicken, and pig gut microbiomes. Environ. Int. 2020;138:105649. doi: 10.1016/j.envint.2020.105649. [DOI] [PubMed] [Google Scholar]
- 17.Guardabassi L., Apley M., Olsen J.E., Toutain P.-L., Weese S. Optimization of Antimicrobial Treatment to Minimize Resistance Selection. Microbiol. Spectr. 2018;6:637–673. doi: 10.1128/microbiolspec.arba-0018-2017. [DOI] [PubMed] [Google Scholar]
- 18.Kers J.G., Fischer E.A.J., Stegeman J.A., Smidt H., Velkers F.C. Comparison of different invasive and non-invasive methods to characterize intestinal microbiota throughout a production cycle of broiler chickens. Microorganisms. 2019;7:431. doi: 10.3390/microorganisms7100431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clavijo V., Flórez M.J.V. The gastrointestinal microbiome and its association with the control of pathogens in broiler chicken production: A review. Poult. Sci. 2018;97:1006–1021. doi: 10.3382/ps/pex359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemos M.P.L., Saraiva M.M.S., Leite E.L., Silva N.M.V., Vasconcelos P.C., Giachetto P.F., Freitas Neto O.C., Givisiez P.E.N., Gebreyes W.A., Oliveira C.J.B. The posthatch prophylactic use of ceftiofur affects the cecal microbiota similar to the dietary sanguinarine supplementation in broilers. Poult. Sci. 2020;99:6013–6021. doi: 10.1016/j.psj.2020.06.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagner B.D., Grunwald G.K., Zerbe G.O., Mikulich-Gilbertson S.K., Robertson C.E., Zemanick E.T., Harris J.K. On the use of diversity measures in longitudinal sequencing studies of microbial communities. Front. Microbiol. 2018;9:1–11. doi: 10.3389/fmicb.2018.01037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shang Y., Kumar S., Oakley B., Kim W.K. Chicken gut microbiota: Importance and detection technology. Front. Vet. Sci. 2018;5:254. doi: 10.3389/fvets.2018.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ngunjiri J., Taylor K., Abundo M., Jang H., Elaish M., Mahesh K., Ghorbani A., Wijeratne S., Weber B., Johnson T., et al. Farm Stage, Bird Age, and Body Site Dominantly Affect the Quantity, Taxonomic Composition, and Dynamics of Respiratory and Gut Microbiota of Commercial Layer Chickens. Appl. Environ. Microbiol. 2019;85:1–17. doi: 10.1128/AEM.03137-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glendinning L., Watson K.A., Watson M. Development of the duodenal, ileal, jejunal and caecal microbiota in chickens. Anim. Microbiome. 2019;1:1–11. doi: 10.1186/s42523-019-0017-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohd Shaufi M.A., Sieo C.C., Chong C.W., Gan H.M., Ho Y.W. Deciphering chicken gut microbial dynamics based on high-throughput 16S rRNA metagenomics analyses. Gut Pathog. 2015;7:4–12. doi: 10.1186/s13099-015-0051-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalkut G. Sulfonamides and trimethoprim. Cancer Investig. 1998;16:612–615. doi: 10.3109/07357909809032892. [DOI] [PubMed] [Google Scholar]
- 27.Bush K., Bradford P.A. Beta-Lactams and beta-Lactamase Inhibitors: An Overview. Cold Spring Harb. Perspect. Med. 2016;6:a025247. doi: 10.1101/cshperspect.a025247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saliu E.M., Vahjen W., Zentek J. Types and prevalence of extended-spectrum beta-lactamase producing Enterobacteriaceae in poultry. Anim. Health Res. Rev. 2017;18:46–57. doi: 10.1017/S1466252317000020. [DOI] [PubMed] [Google Scholar]
- 29.Apostolakos I., Mughini-Gras L., Fasolato L., Piccirillo A. Assessing the occurrence and transfer dynamics of ESBL/pAmpC-producing Escherichia coli across the broiler production pyramid. PLoS ONE. 2019;14:e217174. doi: 10.1371/journal.pone.0217174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torres C., Alonso C.A., Ruiz-Ripa L., León-Sampedro R., del Campo R., Coque T.M. Antimicrobial Resistance in Enterococcus spp. of animal origin. Antimicrob. Resist. Bact. Livest. Companion Anim. 2018;6:185–227. doi: 10.1128/microbiolspec.arba-0032-2018. [DOI] [PubMed] [Google Scholar]
- 31.Gagetti P., Bonofiglio L., García Gabarrot G., Kaufman S., Mollerach M., Vigliarolo L., von Specht M., Toresani I., Lopardo H.A. Resistance to β-lactams in enterococci. Rev. Argent. Microbiol. 2019;51:179–183. doi: 10.1016/j.ram.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 32.Kohler V., Vaishampayan A., Grohmann E. Broad-host-range Inc18 plasmids: Occurrence, spread and transfer mechanisms. Plasmid. 2018;99:11–21. doi: 10.1016/j.plasmid.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 33.Giannuzzi D., Biolatti B., Longato E., Divari S., Starvaggi Cucuzza L., Pregel P., Scaglione F.E., Rinaldi A., Chiesa L.M., Cannizzo F.T. Application of RNA-sequencing to identify biomarkers in broiler chickens prophylactic administered with antimicrobial agents. Animal. 2020 doi: 10.1016/j.animal.2020.100113. in press. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi S., Tomita J., Nishioka K., Hisada T., Nishijima M. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS ONE. 2014;9:e105592. doi: 10.1371/journal.pone.0105592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Gonzalez Peña A., Goodrich J.K., Gordon J.I., et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 37.Elokil A.A., Abouelezz K.F.M., Ahmad H.I., Pan Y., Li S. Investigation of the Impacts of Antibiotic Exposure on the Diversity of the Gut Microbiota in Chicks. Animals. 2020;10:896. doi: 10.3390/ani10050896. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S rRNA sequencing raw reads presented in this study can be found online at https://www.ncbi.nlm.nih.gov/sra under the accession number PRJNA698460.