Abstract
Background
In recent years, emerging studies have demonstrated critical functions and potential clinical applications of long non-coding RNA (lncRNA) in osteosarcoma. To further validate the prognostic value of multiple lncRNAs, we have conducted this updated meta-analysis.
Methods
Literature retrieval was conducted by searching PubMed, Web of Science and the Cochrane Library (last update by October 2, 2019). A meta-analysis was performed to explore association between lncRNAs expression and overall survival (OS) of osteosarcoma patients. Relationships between lncRNAs expression and other clinicopathological features were also analyzed respectively.
Results
Overall, 4351 patients from 62 studies were included in this meta-analysis and 25 lncRNAs were identified. Pooled analyses showed that high expression of 14 lncRNAs connoted worse OS, while two lncRNAs were associated with positive outcome. Further, analysis toward osteosarcoma clinicopathologic features demonstrated that overexpression of TUG1 and XIST indicated poor clinical parameters of patients.
Conclusions
This meta-analysis has elucidated the prognostic potential of 16 lncRNAs in human osteosarcoma. Evidently, desperate expression and functional targets of these lncRNAs offer new approaches for prognosis and therapy of osteosarcoma.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12885-021-07882-w.
Keywords: LncRNAs, Osteosarcoma, Meta-analysis, Prognosis
Background
Osteosarcoma is the most common primary bone malignancy with an annual incidence of 3.1 per million [1]. Despite various treatments, such as chemotherapy, radiotherapy, surgery and targeted therapy, have been used for osteosarcoma, the prognosis remains poor [2, 3]. Of note, the 5-year survival rate for children and adults with non-metastatic osteosarcoma is 71.8%, while for patients with metastatic osteosarcoma dramatically decrease to 30.4% [4, 5]. Therefore, identification of new prognostic or therapeutic hallmarks are in urgent need to improve current situation. In fact, numerous studies have been conducted upon this issue in recent years, and some have shed light on the roles of multiple molecules, including RNAs, regulatory proteins, etc. [6–8]
With advancement of next-generation sequencing technologies, several kinds of non-coding RNAs (ncRNAs) have been discovered, such as the miRNA, siRNA, snoRNA, piRNA and lncRNA. LncRNAs, a cluster of non-coding RNA with more than 200 nucleotides, show no potential of protein coding but exert crucial functions in maintenance of the cellular homeostasis [9]. Mechanisms of lncRNAs in biological processes contain chromatin modifications, transcriptional modifications and post-transcriptional modifications that regulate the expression and features of other genes [10]. They have been elucidated to play critical roles in the development of various diseases, especially tumors [11]. Gouri et al. have reviewed the roles of lncRNAs in pancreatic ductal adenocarcinoma in which they demonstrated that lncRNAs closely associated with the tumorigenesis, partially through dysregulating the KRAS pathway. And it was noticed that the expression level of multiple lncRNAs were altered in tissue, plasma or serum specimens of pancreatic cancer patients, which support the idea that lncRNAs may serve as therapeutic biomarker for pancreatic ductal adenocarcinoma [12]. Moreover, researchers have demonstrated functional mechanisms of lncRNAs in regulating multiple physiological and pathophysiological processes by interacting with other intrinsic molecules [13]. Notably, roles of lncRNAs in progression, prognosis and metastasis of osteosarcoma have been broadly identified [14]. And circulating lncRNAs showed significant potential in osteosarcoma prognosis [15]. To further demonstrate the roles and prognostic potential of lncRNAs in osteosarcoma, we have conducted this meta-analysis.
Methods
Literature search strategy
Two independent researchers retrieved the published literature from database of Pubmed, Web of Science and Cochrane Library at the same time. Search terms used were the following: (osteosarcoma OR “osteogenic sarcoma”) AND (“long non-coding RNA” OR lncRNA OR “LINC RNA” OR “Long ncRNAs”). The last research time was October 2, 2019.
Selection criteria
A total of 550 articles were initially identified after removal of duplication. Two independent researchers (Wenchao Zhang and Xiaolei Ren) reviewed the title, abstract and full-text of all included articles. Articles that met the following criteria were included: (1) Research topic related to the lncRNAs expression and osteosarcoma prognosis; (2) the survival outcome was available in OS form and shown in table, Kaplan-Meier curve or HR value; (3) patients were divided into two groups based on the expression of lncRNAs (high versus low). The exclusive criteria were: (1) the review, case report, conference abstract, letters, sequencing data, bioinformatics analysis, retreated articles and meta-analysis; (2) survival data was obtained from online database such as the TCGA; (3) when more than one study reported on the same patient cohort, only the most recent one was included. Disagreements between the two reviewers were discussed to reach an consensus.
Data extraction and quality assessment
All articles were reviewed by two independent well-trained investigators to retrieve available data. The following information was listed for all articles: Name of first author, publication year, country where study conducted, detection method, sample number, lncRNA type, tumor stage, follow-up time, cut-off value, survival data (multivariate analysis was prioritized if both the univariate and multivariate analysis were provided), outcome measure, Hazard ratio (HR) of lncRNAs expression for OS and the corresponding 95% CI. If HR and 95% CI were not directly provided while a Kaplan-Meier curve was available, we retrieved the HR and 95% CI by using Engauge Digitizer version 4.1 and Tierney’s method as previously described [16]. Then, the quality of the included studies was assessed by two independent reviewers (Wenchao Zhang and Xiaolei Ren) by following the Newcastle-Ottawa Scale (NOS) [17].
Statistical analysis
Stata 12.0 (Stata Corporation, TX, USA) and Review Manager 5.3 (The Cochrane Collaboration, 2014.) were used in this meta-analysis to pool the HR and its 95% CI. Q-test was applied to evaluate the heterogeneity among the studies [18]. If the heterogeneity was substantial (I2 > 50%, P < 0.05), the random effect model would be adopted, otherwise the fixed effect model would be used. A pooled HR > 1 connoted a poor prognosis in patients with lncRNAs overexpression, while a pooled HR < 1 supported a better prognosis. P < 0.05 was considered statistically significant. LncRNAs that have been studied in more than two articles were included to further analyze the clinical parameters, aiming to figure out the association between clinical parameters and lncRNAs expression. Publication bias was measured by Begg’s and Egger’s tests [19].
Results
Characteristics of included studies
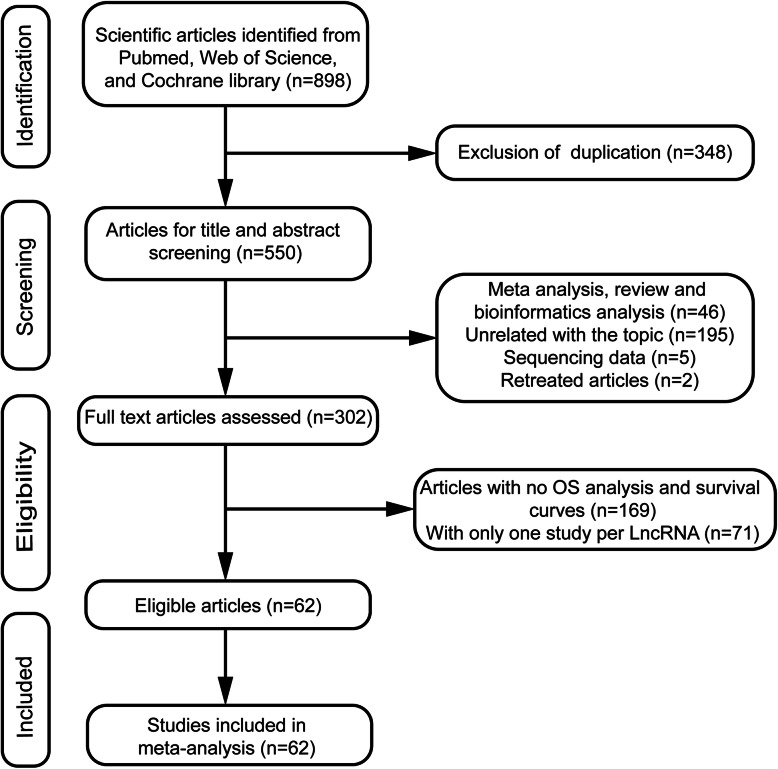
Overall, 62 articles comprising 4351 patients were recruited in our study after selection by following the criteria. The study filtrating process was shown in Fig. 1. Among the included 62 articles, 25 lncRNAs were identified and only four have been studied in more than three articles. The most extensively studied lncRNA was Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), which was shown in eight articles, followed by Taurine up-regulated gene 1 (TUG1), X-inactive specific transcript (XIST) and Nuclear enriched abundant transcript 1 (NEAT1). Almost all of the studies were conducted in China while only one study originated from Brazil. All the articles were published between 2015 and 2019, mostly in 2018 and 2019. The sample number in the selected studies ranged from 30 to 204. All studies used quantitative real-time polymerase chain reaction (qRT-PCR) to measure the expression of lncRNAs, and tissue was the most widely used sample. The cut-off values of lncRNAs expression varied among studies, mainly including median, optimal or mean value. The specific information was shown in Table 1. The quality evaluation showed that NOS scores of all the included studies were greater than 5 (Additional Figures S1 and S2).
Fig. 1.
Study flow chart
Table 1.
The main characteristics of the included studies in the meta-analysis
| ID | Study | country | LncRNA name | Sample number | Tumor stage | Distant metastasis | Follow-up (months) | Detected sample | Detection method | Cut-off value | Survival analysis | Outcome measure | HR (95% CI) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Ju L (2016) [20] | China | BCAR4 | 168 | IIA-IIB/III | 48 | 70 | tissue | qRT–PCR | median | Multivariate analysis | OS | 3.22 (0.89, 7.88) |
| 2 | Chen F (2016) [21] | China | BCAR4 | 60 | I-III | 13 | 60 | tissue | qRT–PCR | median | N/A | OS/RFS | 1.83 (0.68, 4.96) |
| 3 | Ruan R (2018) [22] | China | CCAT2 | 50 | IA-III | 16 | 70 | tissue | qRT–PCR | median | N/A | OS | 1.30 (0.60, 2.84) |
| 4 | Yan L (2018) [23] | China | CCAT2 | 40 | N/A | N/A | 60 | tissue | qRT–PCR | N/A | N/A | OS/RFS | 3.57 (1.25, 5.31) |
| 5 | Jiang N (2017) [24] | China | DANCR | 34 | N/A | 18 | 60 | tissue | qRT–PCR | N/A | Multivariate analysis | OS/DFS | 1.08 (1.23, 5.79) |
| 6 | Wang Y (2018) [25] | China | DANCR | 95 | I-III | 58 | 60 | tissue | qRT–PCR | Optimal | N/A | OS | 2.4 (0.85, 3.25) |
| 7 | Fei D (2018) [26] | China | FER1L4 | 48 | I-IV | 10 | 60 | tissue | qRT–PCR | median | N/A | OS/PFS | 0.45 (0.17, 0.97) |
| 8 | Chen Z (2018) [27] | China | FER1L4 | 73 | I-IV | 25 | 60 | tissue | qRT–PCR | N/A | N/A | OS/PFS | 0.43 (0.05, 0.86) |
| 9 | Ren Z (2019) [28] | China | FOXD2-AS1 | 35 | I-III | N/A | 60 | tissue | qRT–PCR | N/A | N/A | OS | 1.66 (0.47, 5.82) |
| 10 | Zhang H (2019) [29] | China | FOXD2-AS1 | 40 | N/A | N/A | 70 | tissue | qRT–PCR | N/A | N/A | OS | 1.55 (0.3, 7.86) |
| 11 | Cai L (2017) [30] | China | HNF1A-AS1 | 72 | IIA-III | 42 | <60 | tissue | qRT–PCR | median | Multivariate analysis | OS | 2.63 (1.55, 5.65) |
| 12 | Zhao H (2016) [31] | China | HNF1A-AS1 | 43 | I-III | 11 | 60 | tissue | qRT–PCR | median | Multivariate analysis | OS | 2.64 (1.39, 7.42) |
| 13 | Cao K (2019) [32] | China | HOXA11-AS | 61 | I-III | 17 | 60 | tissue | qRT–PCR | median | N/A | OS | 1.5 (0.43, 5.26) |
| 14 | Cui M (2017) [33] | China | HOXA11-AS | 51 | I-III | 13 | 60 | tissue | qRT–PCR | median | N/A | OS | 3.22 (0.31, 33.42) |
| 15 | Gu W (2018) [34] | China | HOXD-AS1 | 43 | I-III | N/A | 60 | tissue | qRT–PCR | N/A | N/A | OS | 1.39 (0.28, 7.02) |
| 16 | Qu Y (2018) [35] | China | HOXD-AS1 | 46 | I-IV | 11 | 60 | tissue | qRT–PCR | median | N/A | OS | 1.63 (1.19, 2.96) |
| 17 | Maciel Uzan (2016) [36] | Brazil | HULC | 33 | I-IV | 10 | 100 | tissue | qRT–PCR | optimal | Multivariate analysis | OS/EFS | 22.01 (2.26, 16.13) |
| 18 | Sun X (2015) [37] | China | HULC | 78 | IIA-III | 21 | 60 | tissue | qRT–PCR | median | Multivariate analysis | OS | 2.28 (1.48, 5.43) |
| 19 | He W (2019) [38] | China | LSINCT5 | 124 | I-III | 22 | 72 | tissue | qRT–PCR | median | Multivariate analysis | OS | 1.68 (1.02, 2.76) |
| 20 | Kong D (2018) [39] | China | LSINCT5 | 42 | I-IV | 18 | 60 | tissue | qRT–PCR | N/A | N/A | OS/DFS | 1.39 (1.13, 5.68) |
| 21 | Chen Y (2018) [40] | China | MALAT1 | 68 | N/A | N/A | 60 | tissue | qRT–PCR | N/A | Multivariate analysis | OS/DFS | 1.73 (1.10, 2.54) |
| 22 | Gao K (2016) [41] | China | MALAT1 | 162 | IIA-III | 44 | 70 | tissue | qRT–PCR | median | Multivariate analysis | OS | 2.8 (1.76, 7.84) |
| 23 | Huo Y (2017) [42] | China | MALAT1 | 68 | I-IV | 22 | 80 | serum | qRT–PCR | median | N/A | OS/PFS | 3.33 (1.43, 4.91) |
| 24 | Li Q (2017) [43] | China | MALAT1 | 64 | I-IV | 33 | 60 | tissue | qRT–PCR | optimal | Multivariate analysis | OS | 2.22 (0.3, 16.44) |
| 25 | Sun Y (2018) [44] | China | MALAT1 | 42 | I-III | 20 | 60 | tissue | qRT–PCR | N/A | N/A | OS | 2.2 (1.15, 4.21) |
| 26 | Sun Z (2019) [45] | China | MALAT1 | 76 | I-III | 46 | 120 | tissue | qRT–PCR | N/A | N/A | OS | 2.51 (0.79, 7.99) |
| 27 | Wang J (2017) [46] | China | MALAT1 | 70 | N/A | N/A | <60 | tissue | qRT–PCR | N/A | N/A | OS | 2.34 (0.55, 9.92) |
| 28 | Wang Y (2017b) [47] | China | MALAT1 | 55 | I-III | 31 | 60 | tissue | qRT–PCR | median | N/A | OS | 1.01 (0.25, 4.03) |
| 29 | Shen B (2019) [48] | China | MEG3 | 204 | I-III | 80 | 50 | tissue | qRT–PCR | median | Multivariate analysis | OS | 0.56 (0.36, 0.87) |
| 30 | Tian Z (2015) [49] | China | MEG3 | 64 | I-III | 17 | 60 | tissue | qRT–PCR | median | Multivariate analysis | OS | 0.45 (0.11, 1.81) |
| 31 | Ji S (2019) [50] | China | NEAT1 | 72 | N/A | N/A | 160 | tissue | qRT–PCR | median | N/A | OS | 2.09 (0.84, 5.22) |
| 32 | Li Y (2018) [51] | China | NEAT1 | 76 | I-III | 38 | 60 | tissue | qRT–PCR | median | N/A | OS | 1.88 (0.47, 7.49) |
| 33 | Tan H (2019) [52] | China | NEAT1 | 47 | I-III | 11 | 50 | tissue | qRT–PCR | median | N/A | OS | 1.84 (0.61, 5.57) |
| 34 | Zhu K (2019) [53] | China | OIP5-AS1 | 80 | N/A | N/A | 60 | tissue | qRT–PCR | N/A | N/A | OS | 1.48 (0.42, 5.15) |
| 35 | Dai J (2018) [54] | China | OIP5-AS1 | 48 | I-III | N/A | 60 | tissue | qRT–PCR | median | N/A | OS | 1.77 (1.17, 2.94) |
| 36 | Zhang C (2016) [55] | China | ODRUL | 60 | N/A | 12 | 80 | tissue | qRT–PCR | median | N/A | OS | 1.21 (0.36, 4.06) |
| 37 | Zhu K (2017) [56] | China | ODRUL | 80 | N/A | 48 | 100 | tissue | qRT–PCR | N/A | N/A | OS | 2.35 (1.08, 5.15) |
| 38 | Huang J (2018) [57] | China | PCAT1 | 62 | I-III | 35 | 60 | tissue | qRT–PCR | N/A | Multivariate analysis | OS/PFS | 1.53 (1.37, 2.92) |
| 39 | Zhang X (2018) [58] | China | PCAT1 | 30 | I-III | 17 | 60 | tissue | qRT–PCR | mean | Multivariate analysis | OS | 4.01 (1.56, 4.57) |
| 40 | Song J (2017) [59] | China | PVT1 | 46 | I-III | N/A | 70 | tissue | qRT–PCR | mean | N/A | OS | 1.63 (0.05, 5.39) |
| 41 | Zhou Q (2016) [60] | China | PVT1 | 53 | N/A | 13 | 60 | tissue | qRT–PCR | N/A | N/A | OS | 1.70 (0.66, 4.37) |
| 42 | Zhou B (2018) [61] | China | SNHG12 | 64 | N/A | N/A | 80 | tissue | qRT–PCR | N/A | N/A | OS | 2.10 (0.66, 6.64) |
| 43 | Zhou S (2018) [62] | China | SNHG12 | 31 | I-III | 21 | 60 | tissue | qRT–PCR | mean | N/A | OS | 1.52 (1.00, 18.48) |
| 44 | Liao S (2019) [63] | China | SNHG16 | 96 | I-III | 51 | 60 | tissue | qRT–PCR | mean | Multivariate analysis | OS | 1.58 (0.65, 3.86) |
| 45 | Wang X (2019) [64] | China | SNHG16 | 65 | I-III | 28 | 60 | tissue | qRT–PCR | median | N/A | OS | 2.62 (0.82, 8.36) |
| 46 | Wang W (2018) [65] | China | SNHG20 | 32 | I-III | N/A | 60 | tissue | qRT–PCR | median | N/A | OS | 1.94 (1.19, 3.17) |
| 47 | Zhang J (2018) [66] | China | SNHG20 | 140 | I-III | 24 | 72 | tissue | qRT–PCR | N/A | Multivariate analysis | OS | 2.05 (0.53, 7.92) |
| 48 | Chen X (2019) [67] | China | TP73-AS1 | 132 | I-III | 22 | 72 | tissue | qRT–PCR | N/A | Multivariate analysis | OS | 1.89 (1.15, 3.13) |
| 49 | Yang G (2018) [68] | China | TP73-AS1 | 46 | I-III | 11 | 50 | tissue | qRT–PCR | mean | N/A | OS | 1.98 (0.61, 6.28) |
| 50 | Ma B (2016) [69] | China | TUG1 | 76 | I-III | 36 | 60 | tissue | qRT–PCR | optomal | Multivariate analysis | OS/PFS | 2.77 (1.29, 5.98) |
| 51 | Sheng K (2019) [70] | China | TUG1 | 40 | N/A | 26 | 60 | plasma | qRT–PCR | median | N/A | OS | 1.59 (1.36, 1.97) |
| 52 | Wang Q (2018) [71] | China | TUG1 | 94 | IIA -III | 26 | 70 | tissue | qRT–PCR | median | Multivariate3analysis | OS | 1.10 (0.96, 15.35) |
| 53 | Wang Y (2017a) [72] | China | TUG1 | 44 | I-III | 28 | 120 | tissue | qRT–PCR | N/A | N/A | OS | 2.12 (1.44, 3.67) |
| 54 | Yu X (2019) [73] | China | TUG1 | 40 | I-III | 11 | 60 | tissue | qRT–PCR | N/A | N/A | OS/RFS | 1.44 (1.15, 2.54) |
| 55 | Li W (2016) [74] | China | UCA1 | 135 | I-III | 34 | 60 | tissue | qRT–PCR | median | Multivariate analysis | OS | 2.19 (0.87, 5.55) |
| 56 | Wen J (2017) [75] | China | UCA1 | 151 | IIA -III | 79 | 60 | tissue | qRT–PCR | optimal | Multivariate analysis | OS | 2.52 (1.34, 4.83) |
| 57 | Li G (2017) [76] | China | XIST | 145 | I-IV | 44 | 60 | tissue | qRT–PCR | N/A | N/A | OS | 1.75 (1.00, 3.06) |
| 58 | Wang W (2019) [77] | China | XIST | 64 | IA -III | 15 | 70 | tissue | qRT–PCR | median | N/A | OS | 1.92 (1.25, 3.29) |
| 59 | Yang C (2018) [78] | China | XIST | 40 | I-III | N/A | 60 | tissue | qRT–PCR | N/A | N/A | OS | 3.07 (1.84, 11.15) |
| 60 | Zhang R (2017) [79] | China | XIST | 50 | N/A | 26 | 60 | tissue | qRT–PCR | N/A | N/A | OS | 1.59 (1.38, 2.88) |
| 61 | Li N (2017) [80] | China | ZFAS1 | 53 | I-III | 16 | 60 | tissue | qRT–PCR | median | Multivariate analysis | OS/RFS | 1.14 (0.23, 5.61) |
| 62 | Liu G (2017) [81] | China | ZFAS1 | 50 | N/A | N/A | 30 | tissue | qRT–PCR | median | N/A | OS | 1.59 (0.64, 3.95) |
DFS disease-free survival, PFS progression-free survival, RFS recurrence-free survival, OS overall survival, N/A not available
Overexpression of lncRNAs indicate different prognosis of osteosarcoma
In this meta-analysis, 25 lncRNAs were analyzed individually. The result showed that overexpression of 14 lncRNAs were associated with poor prognosis while two indicated a positive outcome. The overexpression of the rest nine lncRNAs were independent of osteosarcoma prognosis (Table 2).
Table 2.
meta-analysis results of 25 lncRNAs
| LncRNA name | No. of patients | No. of studies | HR(95%CI)-model | P value | heterogeneity | |
|---|---|---|---|---|---|---|
| I2 | P | |||||
| BCAR4 | 228 | 2 | 2.36 (1.13, 4.93)-fixed | 0.022 | 0.0% | 0.453 |
| CCAT2 | 90 | 2 | 2.18 (0.81, 5.86)-random | 0.123 | 71.2% | 0.062 |
| DANCR | 129 | 2 | 1.65 (0.76, 3.60)-random | 0.209 | 57.1% | 0.127 |
| FER1L4 | 121 | 2 | 0.44 (0.16, 0.72)-fixed | 0.032 | 0.0% | 0.945 |
| FOXD2-AS1 | 75 | 2 | 1.62 (0.60, 4.38)-fixed | 0.344 | 0.0% | 0.948 |
| HNF1A-AS1 | 115 | 2 | 2.63 (1.58, 4.39)-fixed | 0.000 | 0.0% | 0.994 |
| HOXA11-AS | 112 | 2 | 1.78 (0.59, 3.36)-fixed | 0.307 | 0.0% | 0.573 |
| HOXD-AS1 | 89 | 2 | 1.61 (1.04, 2.50)-fixed | 0.033 | 0.0% | 0.852 |
| HULC | 111 | 2 | 5.38 (0.62, 46.50)-random | 0.126 | 71.5% | 0.061 |
| LSINCT5 | 166 | 2 | 1.59 (1.04, 2.44)-fixed | 0.031 | 0.0% | 0.695 |
| MALAT1 | 605 | 8 | 2.15 (1.67, 2.76)-fixed | 0.000 | 0.0% | 0.676 |
| MEG3 | 268 | 2 | 0.55 (0.36, 0.84)-fixed | 0.005 | 0.0% | 0.770 |
| NEAT1 | 195 | 3 | 1.96 (1.05, 3.68)-fixed | 0.035 | 0.0% | 0.983 |
| ODRUL | 140 | 2 | 1.73 (1.12, 2.67)-fixed | 0.013 | 0.0% | 0.793 |
| OIP5-AS1 | 128 | 2 | 1.93 (1.00, 3.73)-fixed | 0.049 | 0.0% | 0.367 |
| PCAT1 | 92 | 2 | 2.43 (1.95, 6.24)-random | 0.065 | 87.9% | 0.004 |
| PVT1 | 99 | 2 | 1.69 (0.70, 4.06)-fixed | 0.241 | 0.0% | 0.974 |
| SNHG12 | 95 | 2 | 1.85 (0.75, 4.58)-fixed | 0.181 | 0.0% | 0.733 |
| SNHG16 | 161 | 2 | 1.91 (0.94, 3.86)-fixed | 0.074 | 0.0% | 0.498 |
| SNHG20 | 172 | 2 | 1.95 (1.23, 3.09)-fixed | 0.004 | 0.0% | 0.940 |
| TP73 | 178 | 2 | 1.90 (1.20, 3.02)-fixed | 0.006 | 0.0% | 0.943 |
| TUG1 | 294 | 5 | 1.64 (1.42, 1.92)-fixed | 0.000 | 0.0% | 0.435 |
| UCA1 | 286 | 2 | 2.41 (1.42, 4.07)-fixed | 0.001 | 0.0% | 0.809 |
| XIST | 299 | 4 | 1.79 (1.40, 2.30)-fixed | 0.000 | 0.0% | 0.601 |
| ZFAS1 | 103 | 2 | 1.47 (0.66, 3.23)-fixed | 0.343 | 0.0% | 0.723 |
CI confidence interval, HR hazard ratio
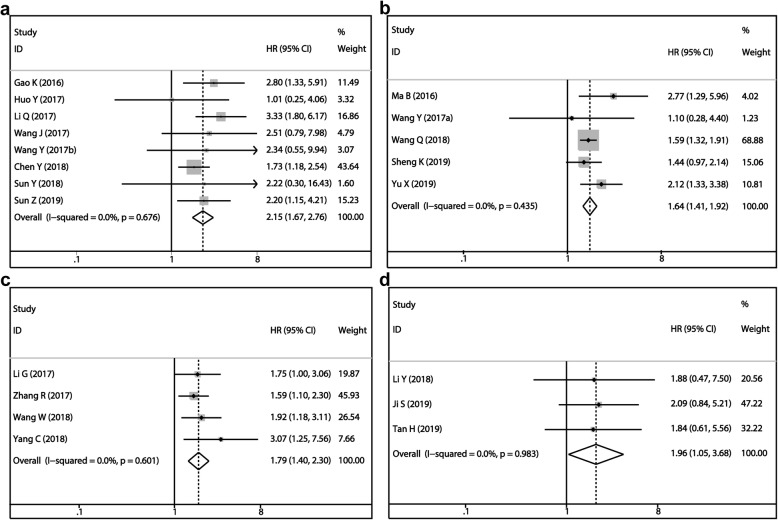
More attention was paid to the four most studied lncRNAs among all included studies, the MALAT1, TUG1, XIST and NEAT1. For MALAT1, eight articles included 605 patients were pooled. Overexpression of MALAT1 was a risk factor of osteosarcoma (HR = 2.15, 95%CI: 1.67–2.76, P < 0.001, Fig. 2a). We noticed that in one of these eight studies, the detected sample was serum rather than tissue. So, we analyzed the remaining seven studies after eliminating this one, and the predicted tendence of MALAT1 in osteosarcoma was not altered (HR = 2.20, 95%CI: 1.70–2.85, P < 0.001). Since no heterogeneity among studies was noted, we did not perform subgroup analysis (I2 = 0.0%, P = 0.676). Then, five studies focused on TUG1 containing 294 patients were analyzed, which showed that overexpression of TUG1 was associated with unfavorable clinical outcome of osteosarcoma patients (HR = 2.41, 95%CI: 1.42–4.07, P = 0.001, Fig. 2b). There was also a study that used plasma instead of tissue as the detected sample. Thus, we did another analysis without this study. The result showed the negative prediction role of TUG1 as well. (HR = 1.68, 95%CI: 1.43–1.99, P = 0.001). A negative association between XIST expression and OS of osteosarcoma patients was noticed (HR = 1.79, 95%CI: 1.40–2.30, P < 0.001, Fig. 2c) based on the analysis of four researches containing 299 patients. Finally, three studies incorporating 199 patients were analyzed to explore the association between NEAT1 expression and OS. It proved that the high expression of NEAT1 foreboded poor prognosis. (HR = 1.96, 95%CI: 1.05–3.68, P = 0.035, Fig. 2d).
Fig. 2.
Forest plot of overall survival for four most studied lncRNAs: a MALAT1, b TUG1, c XIST, d NEAT1
LncRNAs expression and osteosarcoma clinicopathologic features
Furthermore, the clinicopathologic features, including age, gender, clinical stage, tumor size and distant metastasis, were analyzed. We mainly focused on the MALAT1, TUG1 and XIST, lncRNAs that owned available data we needed in more than three articles. There were no significant differences in MALAT1 expression detected in different age (HR = 0.73, 95%CI: 0.43–1.24, P = 0.240), gender (HR = 0.73, 95%CI: 0.44–1.20, P = 0.210), clinical stage (HR = 1.48, 95%CI: 0.27–7.94, P = 0.650), tumor size (HR = 0.95, 95%CI: 0.50–1.81, P = 0.890) and distant metastasis (HR = 1.98, 95%CI: 0.32–12.05, P = 0.40). However, the distinction of TUG1 expression was observed in different clinical stage (HR = 4.66, 95%CI: 2.47–8.79, P < 0.001), tumor size (HR = 4.07, 95%CI: 2.33–7.12, P < 0.001) and distant metastasis (HR = 3.53, 95%CI: 1.20–10.41, P = 0.020). Osteosarcoma tissue derived from patients with higher clinical stage, larger tumor size and distant metastasis expressed high TUG1. Differences also have been found in XIST expression upon the clinical stage and metastasis. High clinical stage (HR = 3.92, 95%CI: 2.31–6.66, P < 0.001) and metastasis (HR = 3.15, 95%CI: 1.64–6.05, P < 0.001) were associated with high expression of XIST in tumor tissue. More detailed information was shown in Table 3.
Table 3.
Analysis of clinical features
| Outcome | No. of Studies | No. of Participants | OR (95% CI) | P value | Model | Heterogeneity Chi2, P-value, I2 |
|---|---|---|---|---|---|---|
| MALAT1 | ||||||
| Age | 3 | 259 | 0.73 (0.43, 1.24) | 0.24 | Fixed | 0.36, 0.84, 0% |
| Gender | 3 | 259 | 0.73 (0.44, 1.20) | 0.21 | Fixed | 0.16, 0.92, 0% |
| Clinical stage | 3 | 259 | 1.48 (0.27, 7.94) | 0.65 | Random | 15.91, 0.0004, 87% |
| Tumor size | 3 | 259 | 0.95 (0.50, 1.81) | 0.89 | Random | 10.77, 0.005, 81% |
| Distant metastasis | 3 | 259 | 1.98 (0.32, 12.05) | 0.46 | Random | 16.26, 0.0003, 88% |
| TUG1 | ||||||
| Age | 5 | 294 | 1.2 (0.48, 3.02) | 0.28 | Fixed | 1.97, 0.74, 0% |
| Gender | 5 | 294 | 1.02 (0.62, 1.65) | 0.95 | Fixed | 2.97, 0.56, 0% |
| Clinical stage | 4 | 254 | 4.66 (2.47, 8.79) | <0.00001 | Fixed | 0.45, 0.93, 0% |
| Tumor size | 4 | 254 | 4.07 (2.33, 7.12) | <0.00001 | Fixed | 2.96, 0.4, 0% |
| Distant metastasis | 5 | 294 | 3.53 (1.20, 10.41) | 0.02 | Random | 13.52, 0.009, 70% |
| XIST | ||||||
| Age | 2 | 209 | 1.18 (0.44, 3.15) | 0.74 | Random | 2.55, 0.11, 61% |
| Gender | 3 | 249 | 0.91 (0.56, 1.50) | 0.72 | Fixed | 1.56, 0.46, 0% |
| Clinical stage | 3 | 249 | 3.92 (2.31, 6.66) | <0.00001 | Fixed | 0.92, 0.63, 0% |
| Tumor size | 3 | 249 | 1.15 (0.41, 3.23) | 0.80 | Random | 6.74, 0.03, 70% |
| Distant metastasis | 2 | 209 | 3.10 (1.61, 5.95) | 0.0007 | Fixed | 0.58, 0.45, 0% |
CI confidence interval, OR odds ratio
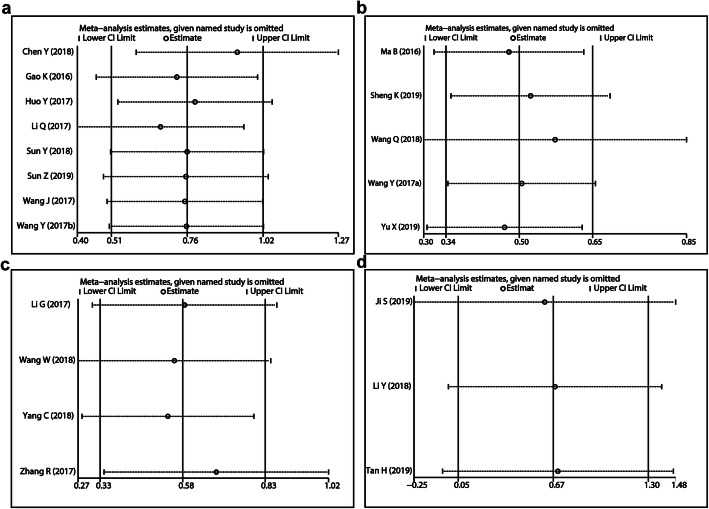
Sensitivity analysis
We did sensitivity analysis to the four lncRNAs which had studied in more than three articles respectively even though there was no heterogeneity detected (I2 = 0.0%, P > 0.05). The results showed that this meta-analysis was reliable (Fig. 3).
Fig. 3.
Sensitive analysis of lncRNAs for osteosarcoma: a MALAT1, b TUG1, c XIST, d NEAT1
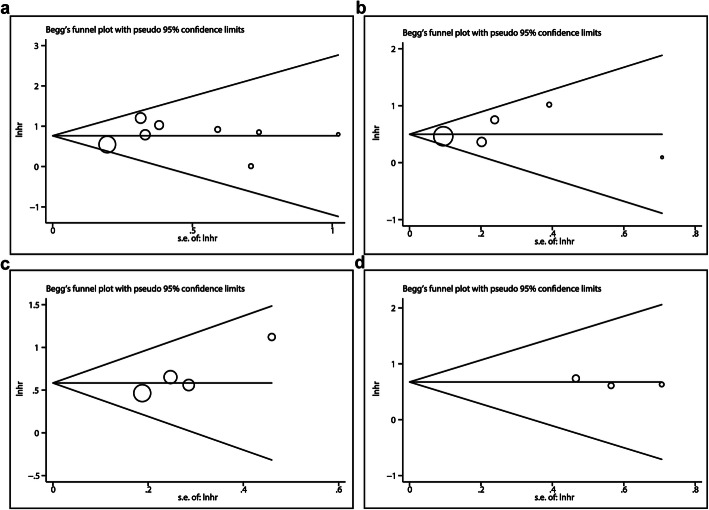
Publication Bias
Publication bias was measured by using the Begg’s and Egger’s test. We only analyzed the publication bias of lncRNAs that have been studied in more than three articles, including MALAT1, TUG1, XIST and NEAT1. No significant publication bias was found in any of the lncRNAs. Begg’s funnel plot was shown in Fig. 4. However, publication bias between different lncRNAs was subsistent since the number of published articles lacked consistence for them.
Fig. 4.
Funnel plots of the four most studied lncRNAs: a MALAT1, b TUG1, c XIST, d NEAT1
Discussion
Osteosarcoma remains intractable in clinical practice, and new approaches for prognostic evaluation and treatment of osteosarcoma are continuously requisite. Recently, targeted therapy and molecular biomarker diagnosis have emerged as the focus in cancers [82, 83]. LncRNAs, as indispensable regulators in a majority of biological processes [84], possess great potential for prognostic hallmarks. Further, advancement of technologies for structural and functional study enable us to unveil more evident features of lncRNAs serving as idea clinical biomarkers [85]. Considering the vast lncRNAs studied in osteosarcoma [14], we conducted the meta-analysis, with the aim to provide stronger evidences in this regard.
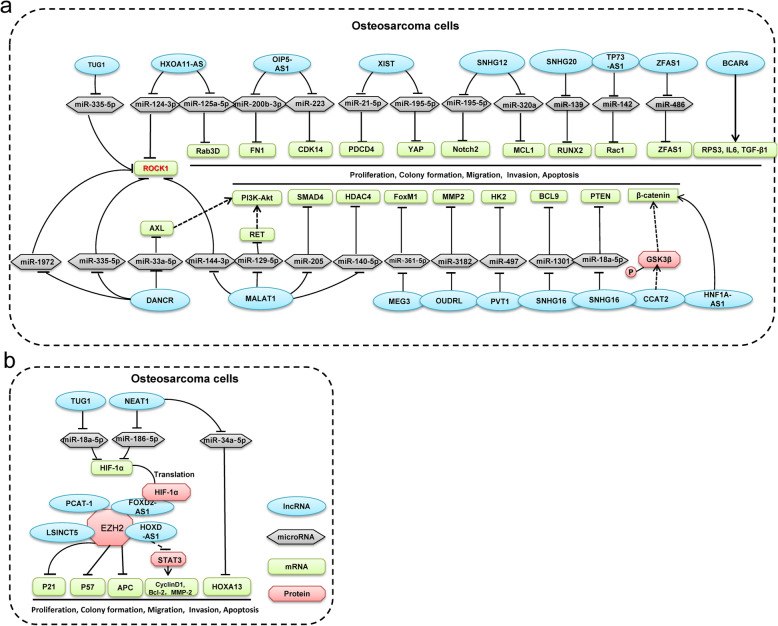
In this meta-analysis, a total of 4351 cases were included, and 25 lncRNAs were analyzed in which high expression of 14 lncRNAs connotes worse OS while two were associated with positive outcomes. Mechanisms involved in these lncRNAs are multifaced. BCAR4 promoted proliferation and migration by GLI2 target genes including RPS3, IL6, MUC5AC and TGF-β [20, 21]. HNF1A-AS1 targeted Wnt/β-catenin pathway to enhance proliferation and G1/S transition, migration and invasion by reducing the EMT [31]. Meanwhile, MALAT1 positively regulated RET to activate the PI3K-Akt signaling pathway by competitively binding with miR-129-5p, and thus enhancing stem cell-like properties [40]. Furthermore, MALAT1/miR-144-3p/ ROCK1 axis promoted the proliferation and metastasis of osteosarcoma [40]. Moreover, MALAT1 promoted proliferation and metastasis via miR-205/SMAD4 axis [43] and miR-140-5p/HDAC4 axis [44]. NEAT1 could up-regulate HOXA13 by decoying of miR-34a-5p, while NEAT1/ miR-186-5p/HIF-1α axis enhanced proliferation and reduced apoptosis [50–52]. Rho-associated protein kinase 1 (ROCK1), a serine/threonine kinase, is critical regulator of development and progression in various human malignant tumors. Importantly, TUG1 served as a ceRNA of miR-335-5p to affect ROCK1-mediated migration and invasion [72]. Besides, other important hallmarks of osteosarcoma demonstrate close association with TUG1. The effects of TUG1 overexpression on runt-related transcription factor 2 (RUNX2) expression were elucidated. It was noticed that overexpression of lncRNA TUG1 significantly down-regulated RUNX2 level [70]. Likewise, TUG1 could impede osteosarcoma cells proliferation, migration, and invasion by miR-140-5p/PFN2 axis [86]. XIST is another potential biomarker of osteosarcoma which has been reported to modulate osteosarcoma proliferation and invasion through miR-320b/RAP2B [87], miR-193a-3p/RSF1 [88], miR-21-5p/PDCD4 [79], and miR-195-5p/YAP axis [78]. In addition, SNHG16/miR-1301/BCL9 axis [64], MEG3/miR-361-5p/FoxM1 axis [48], SNHG20/miR-139/RUNX2 axis [65], TP73-AS1/miR-142/Rac1 axis [68] and SNHG12/miR-195-5p/Notch2 [62] axis worked as critical roles of enhancing proliferation, migration and invasion. Additionally, OIP5-AS1 and SNHG12 were involved in osteosarcoma doxorubicin resistance via miR-200b-3p/FN1 and miR-320a/MCL1 pathways, respectively [53, 61]. Further, enhancer of zeste homolog 2 (EZH2) was involved in DNA methylation and its mutations have been identified in various malignancies. HOXD-AS1 suppressed p57 expression by binding with EZH2 [34]. LSINCT5 binding with EZH2 inhibited APC transcription that could down-regulate the Wnt/ β-catenin pathway and activate the PI3K-Akt signaling pathway [39]. The detailed mechanisms are shown in Fig. 5.
Fig. 5.
Schematic diagram of lncRNAs mechanism in osteosarcoma cells
Previously, meta-analysis by Wang Y et al. in 2017 [47] and Chen D et al. in 2018 [89] have illustrated the relationship between osteosarcoma and lncRNAs. However, numbered lncRNAs (TUG1, UCA1, BCAR4, HULC, etc.) were analyzed, which led to significant limitation for their research. Among the 25 enrolled lncRNAs, four (MALAT1, TUG1, XIST and NEAT1) reported in more than three studies respectively have been the focus of our meta-analysis because efficacy confirmed in multiple datasets tend to be more convictive. Their high expression predicted poor prognosis of osteosarcoma (MALAT1 (HR = 2.15, 95%CI: 1.67–2.76, P < 0.001), TUG1 ((HR = 2.41, 95%CI: 1.42–4.07, P = 0.001), XIST (HR = 1.79, 95%CI: 1.40–2.30, P < 0.001), NEAT1 (HR = 1.96, 95%CI: 1.05–3.68, P = 0.035)). Specifically, we observed that, for lncRNA MALAT1 and TUG1, each contained one study that did not employ neoplastic tissue as the test item. Therefore, we did another analysis after eliminated them respectively in order to minimize the potential bias. Results showed no obvious difference compared to the previous analysis.
Besides, we have evaluated the relationship between lncRNAs expression and clinicopathological features of osteosarcoma. MALAT1 expression level was not associated with the age, gender, clinical stage, tumor size and metastasis. However, patients with elder age, larger tumor size and distant metastasis were accompanied by overexpression of TUG1 and XIST, which further demonstrated the negative role of lncRNA TUG1 and XIST in osteosarcoma progression. Furthermore, a series of lncRNAs have been elucidated to serve as important prognostic hallmarks in numerous tumors, for instance, MALAT1 in breast cancer and digestive system cancer [90, 91], XIST in various solid tumors [92], BCAR4 and SNHG16 in diverse human neoplasms [93–95].
To date, functional implications that support the prognostic roles of LncRNAs in human cancers have been expounded. Importantly, lncRNAs are capable of altering gene expression of cancer stem cells via interplaying with chromatin modification, transcriptional and post-transcriptional factors [96]. Cancer stem cells are critical initiators of tumors which are able to differentiate into heterogeneous lineages of cancer cells, thereby it is of great significance for neoplastic progress. Moreover, epithelial-mesenchymal transition, a prevalent process in tumors, is largely regulated by multiple lncRNAs transcriptionally or post-transcriptionally [84]. Besides, involvement of lncRNAs in regulating some key oncogenic factors such as p53 and MYC has provided evidence for their cancer-relevant functions [10]. And currently, the use of antisense oligonucleotides, small molecules for the targeting of lncRNAs, and tools based on CRISPR–Cas systems may provide new approaches for lncRNA-based targeted therapy [10]. However, adopting lncRNAs as the prognostic or therapeutic markers remains experimentally proposed since the lack of large sample trial to confirm their efficacy and safety. Our meta-analysis that pool and analyze the published dataset thus provide stronger evidence and somewhat promote the progress in this regard.
Ultimately, this meta-analysis yielded valuable results, but there were limitations: (1) Using different methods to extract data can lead to bias, and some HR values are obtained through the tool software indirectly, which makes the bias even greater. (2) Almost all of the included studies are from China, leading to bias caused by geographical differences. (3) Some enrolled studies have different follow-up time and cut-off value.
Conclusions
In conclusion, our study confirmed that lncRNAs are of significant potential in serving as molecular markers for prognosis of osteosarcoma. High expression of a set of lncRNAs predict positive prognosis while some indicate poor outcomes. This meta-analysis has laid a theoretical foundation for experimental exploration and clinical application of lncRNAs in the future.
Supplementary Information
Additional file 1 Study quality and bias in the retrospective cohort studies judged by the Newcastle-Ottawa Scale (NOS) checklist. Figure A.1 Quality assessment of all included studies. “Risk of bias summary” of all included studies. Figure A.2 Quality assessment of all included studies. “Risk of bias graph” of all included studies.
Acknowledgements
Not applicable.
Abbreviations
- LncRNA
Long non-coding RNA
- OS
overall survival
- HR
hazard ratio
- NOS
Newcastle-Ottawa Scale
- MALAT1
Metastasis-associated lung adenocarcinoma transcript 1
- TUG1
Taurine up-regulated gene 1
- XIST
X-inactive specific transcript
- NEAT1
nuclear enriched abundant transcript 1
- ROCK1
Rho-associated protein kinase 1
- EZH2
Enhancer of zeste homolog 2
Authors’ contributions
WCZ and XLR conceived and designed the work. Material preparation, data collection and analysis were performed by WCZ, XLR, LQ and CHZ. The first draft of the manuscript was written by WCZ and XLR and all authors commented on previous versions of the manuscript. CT revised the manuscript. ZHL contributed to article drafting, critical revision and final approval of the version to be published. All authors read and approved the final manuscript.
Funding
This study was supported by Natural Science Foundation of Hunan Province, China (2018JJ3716), National Natural Science Foundation of China (81902745), and the Hunan Provincial Science and Technology Association program (2017TJ-Q19).
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Wenchao Zhang and Xiaolei Ren contributed equally to this work.
Contributor Information
Chao Tu, Email: tuchao@csu.edu.cn.
Zhihong Li, Email: lizhihong@csu.edu.cn.
References
- 1.Gianferante DM, Mirabello L, Savage SA. Germline and somatic genetics of osteosarcoma - connecting aetiology, biology and therapy. Nat Rev Endocrinol. 2017;13(8):480–491. doi: 10.1038/nrendo.2017.16. [DOI] [PubMed] [Google Scholar]
- 2.Bishop MW, Janeway KA, Gorlick R. Future directions in the treatment of osteosarcoma. Curr Opin Pediatr. 2016;28(1):26–33. doi: 10.1097/MOP.0000000000000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whelan JS, Davis LE. Osteosarcoma, Chondrosarcoma, and Chordoma. J Clin Oncol. 2018;36(2):188–193. doi: 10.1200/JCO.2017.75.1743. [DOI] [PubMed] [Google Scholar]
- 4.Duchman KR, Gao Y, Miller BJ. Prognostic factors for survival in patients with high-grade osteosarcoma using the surveillance, epidemiology, and end results (SEER) program database. Cancer Epidemiol. 2015;39(4):593–599. doi: 10.1016/j.canep.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Anwar MA, El-Baba C, Elnaggar MH, Elkholy YO, Mottawea M, Johar D, Al Shehabi TS, Kobeissy F, Moussalem C, Massaad E, et al. Novel therapeutic strategies for spinal osteosarcomas. Semin Cancer Biol. 2019;64:83–92. doi: 10.1016/j.semcancer.2019.05.018. [DOI] [PubMed] [Google Scholar]
- 6.Bottani M, Banfi G, Lombardi G. Circulating miRNAs as diagnostic and prognostic biomarkers in common solid tumors: focus on lung, breast, prostate cancers, and osteosarcoma. J Clin Med. 2019;8(10):1661. doi: 10.3390/jcm8101661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu Y, Xie Z, Chen J, Chen J, Ni W, Ma Y, Huang K, Wang G, Wang J, Ma J, et al. Circular RNA circTADA2A promotes osteosarcoma progression and metastasis by sponging miR-203a-3p and regulating CREB3 expression. Mol Cancer. 2019;18(1):73. doi: 10.1186/s12943-019-1007-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mangelinck A, da Costa MEM, Stefanovska B, Bawa O, Polrot M, Gaspar N, Fromigue O. MT2A is an early predictive biomarker of response to chemotherapy and a potential therapeutic target in osteosarcoma. Sci Rep. 2019;9(1):12301. doi: 10.1038/s41598-019-48846-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mishra S, Verma SS, Rai V, Awasthee N, Chava S, Hui KM, Kumar AP, Challagundla KB, Sethi G, Gupta SC. Long non-coding RNAs are emerging targets of phytochemicals for cancer and other chronic diseases. Cell Mol Life Sci. 2019;76(10):1947–1966. doi: 10.1007/s00018-019-03053-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2020;22(2):96–118. doi: 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pandya G, Kirtonia A, Sethi G, Pandey AK, Garg M. The implication of long non-coding RNAs in the diagnosis, pathogenesis and drug resistance of pancreatic ductal adenocarcinoma and their possible therapeutic potential. Biochim Biophys Acta Rev Cancer. 1874;2020(2):188423. doi: 10.1016/j.bbcan.2020.188423. [DOI] [PubMed] [Google Scholar]
- 13.Ballantyne MD, McDonald RA, Baker AH. lncRNA/MicroRNA interactions in the vasculature. Clin Pharmacol Ther. 2016;99(5):494–501. doi: 10.1002/cpt.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Z, Li X, Yang Y, He Z, Qu X, Zhang Y. Long noncoding RNAs in the progression, metastasis, and prognosis of osteosarcoma. Cell Death Dis. 2016;7(9):e2389. doi: 10.1038/cddis.2016.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Botti G, Giordano A, Feroce F, De Chiara AR, Cantile M. Noncoding RNAs as circulating biomarkers in osteosarcoma patients. J Cell Physiol. 2019;234(11):19249–19255. doi: 10.1002/jcp.28744. [DOI] [PubMed] [Google Scholar]
- 16.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 18.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ju L, Zhou YM, Yang GS. Up-regulation of long non-coding RNA BCAR4 predicts a poor prognosis in patients with osteosarcoma, and promotes cell invasion and metastasis. Eur Rev Med Pharmacol Sci. 2016;20(21):4445–4451. [PubMed] [Google Scholar]
- 21.Chen F, Mo J, Zhang L. Long noncoding RNA BCAR4 promotes osteosarcoma progression through activating GLI2-dependent gene transcription. Tumor Biol. 2016;37(10):13403–13412. doi: 10.1007/s13277-016-5256-y. [DOI] [PubMed] [Google Scholar]
- 22.Ruan R, Zhao XL. LncRNA CCAT2 enhances cell proliferation via GSK3beta/beta-catenin signaling pathway in human osteosarcoma. Eur Rev Med Pharmacol Sci. 2018;22(10):2978–2984. doi: 10.26355/eurrev_201805_15053. [DOI] [PubMed] [Google Scholar]
- 23.Yan L, Wu X, Yin X, Du F, Liu Y, Ding X. LncRNA CCAT2 promoted osteosarcoma cell proliferation and invasion. J Cell Mol Med. 2018;22(5):2592–2599. doi: 10.1111/jcmm.13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang N, Wang X, Xie X, Liao Y, Liu N, Liu J, Miao N, Shen J, Peng T. lncRNA DANCR promotes tumor progression and cancer stemness features in osteosarcoma by upregulating AXL via miR-33a-5p inhibition. Cancer Lett. 2017;405:46–55. doi: 10.1016/j.canlet.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Zeng X, Wang N, Zhao W, Zhang X, Teng S, Zhang Y, Lu Z. Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Mol Cancer. 2018;17(1):89. doi: 10.1186/s12943-018-0837-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fei D, Zhang X, Liu J, Tan L, Xing J, Zhao D, Zhang Y. Long noncoding RNA FER1L4 suppresses tumorigenesis by regulating the expression of PTEN targeting miR-18a-5p in osteosarcoma. Cell Physiol Biochem. 2018;51(3):1364–1375. doi: 10.1159/000495554. [DOI] [PubMed] [Google Scholar]
- 27.Chen ZX, Chen CP, Zhang N, Wang TX. Low-expression of IncRNA FER1L4 might be a prognostic marker in osteosarcoma. Eur Rev Med Pharmacol Sci. 2018;22(8):2310–2314. doi: 10.26355/eurrev_201804_14820. [DOI] [PubMed] [Google Scholar]
- 28.Ren Z, Hu Y, Li G, Kang Y, Liu Y, Zhao H. HIF-1α induced long noncoding RNA FOXD2-AS1 promotes the osteosarcoma through repressing p21. Biomed Pharmacother. 2019;117:109104. doi: 10.1016/j.biopha.2019.109104. [DOI] [PubMed] [Google Scholar]
- 29.Zhang H, Lu Y, Wang J, Zhang T, Dong C, Li X, Wang X, Ma Q, Yang T, Zhou Y. Downregulation of the long non‑coding RNA FOXD2‑AS1 inhibits cell proliferation, migration and invasion in osteosarcoma. Mol Med Rep. 2019;20(1):292–302. doi: 10.3892/mmr.2019.10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai L, Lv J, Zhang Y, Li J, Wang Y, Yang H. The IncRNA HNF1A-AS1 is a negative prognostic factor and promotes tumorigenesis in osteosarcoma. J Cell Mol Med. 2017;21(11):2654–2662. doi: 10.1111/jcmm.12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao H, Hou W, Tao J, Zhao Y, Wan G, Ma C, Xu H. Upregulation of lncRNA HNF1A-AS1 promotes cell proliferation and metastasis in osteosarcoma through activation of the Wnt/beta-catenin signaling pathway. Am J Transl Res. 2016;8(8):3503–3512. [PMC free article] [PubMed] [Google Scholar]
- 32.Cao K, Fang Y, Wang H, Jiang Z, Guo L, Hu Y. The lncRNA HOXA11-AS regulates Rab3D expression by sponging miR-125a-5p promoting metastasis of osteosarcoma. Cancer Manag Res. 2019;11:4505–4518. doi: 10.2147/CMAR.S196025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui M, Wang J, Li Q, Zhang J, Jia J, Zhan X. Long non-coding RNA HOXA11-AS functions as a competing endogenous RNA to regulate ROCK1 expression by sponging miR-124-3p in osteosarcoma. Biomed Pharmacother. 2017;92:437–444. doi: 10.1016/j.biopha.2017.05.081. [DOI] [PubMed] [Google Scholar]
- 34.Gu W, Zhang E, Song L, Tu L, Wang Z, Tian F, Aikenmu K, Chu G, Zhao J. Long noncoding RNA HOXD-AS1 aggravates osteosarcoma carcinogenesis through epigenetically inhibiting p57 via EZH2. Biomed Pharmacother. 2018;106:890–895. doi: 10.1016/j.biopha.2018.06.173. [DOI] [PubMed] [Google Scholar]
- 35.Qu Y, Zheng S, Kang M, Dong R, Zhou H, Zhao D, Zhao J. Knockdown of long non-coding RNA HOXD-AS1 inhibits the progression of osteosarcoma. Biomed Pharmacother. 2018;98:899–906. doi: 10.1016/j.biopha.2018.01.024. [DOI] [PubMed] [Google Scholar]
- 36.Uzan VR, Lengert A, Boldrini E, Penna V, Scapulatempo-Neto C, Scrideli CA, Filho AP, Cavalcante CE, de Oliveira CZ, Lopes LF, et al. High expression of HULC is associated with poor prognosis in osteosarcoma patients. PLoS One. 2016;11(6):e0156774. doi: 10.1371/journal.pone.0156774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun XH, Yang LB, Geng XL, Wang R, Zhang ZC. Increased expression of lncRNA HULC indicates a poor prognosis and promotes cell metastasis in osteosarcoma. Int J Clin Exp Pathol. 2015;8(3):2994–3000. [PMC free article] [PubMed] [Google Scholar]
- 38.He W, Lu M, Xiao D. LSINCT5 predicts unfavorable prognosis and exerts oncogenic function in osteosarcoma. Biosci Rep. 2019;39(5):BSR20190612. doi: 10.1042/BSR20190612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kong D, Li C, Yang Q, Wei B, Wang L, Peng C. Long noncoding RNA LSINCT5 acts as an oncogene via increasing EZH2-induced inhibition of APC expression in osteosarcoma. Biochem Biophys Res Commun. 2018;507(1–4):193–197. doi: 10.1016/j.bbrc.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 40.Chen Y, Huang W, Sun W, Zheng B, Wang C, Luo Z, Wang J, Yan W. LncRNA MALAT1 promotes cancer metastasis in osteosarcoma via activation of the PI3K-Akt signaling pathway. Cell Physiol Biochem. 2018;51(3):1313–1326. doi: 10.1159/000495550. [DOI] [PubMed] [Google Scholar]
- 41.Gao KT, Lian D. Long non-coding RNA MALAT1 is an independent prognostic factor of osteosarcoma. Eur Rev Med Pharmacol Sci. 2016;20(17):3561–3565. [PubMed] [Google Scholar]
- 42.Huo Y, Li Q, Wang X, Jiao X, Zheng J, Li Z, Pan X. MALAT1 predicts poor survival in osteosarcoma patients and promotes cell metastasis through associating with EZH2. Oncotarget. 2017;8(29):46993–47006. doi: 10.18632/oncotarget.16551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Q, Pan X, Wang X, Jiao X, Zheng J, Li Z, Huo Y. Long noncoding RNA MALAT1 promotes cell proliferation through suppressing miR-205 and promoting SMAD4 expression in osteosarcoma. Oncotarget. 2017;8(63):106648–106660. doi: 10.18632/oncotarget.20678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun Y, Qin B. Long noncoding RNA MALAT1 regulates HDAC4-mediated proliferation and apoptosis via decoying of miR-140-5p in osteosarcoma cells. Cancer Med. 2018;7(9):4584–4597. doi: 10.1002/cam4.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun Z, Zhang T, Chen B. Long non-coding RNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) promotes proliferation and metastasis of osteosarcoma cells by targeting c-met and SOX4 via miR-34a/c-5p and miR-449a/b. Med Sci Monit. 2019;25:1410–1422. doi: 10.12659/MSM.912703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, Sun G. FOXO1-MALAT1-miR-26a-5p feedback loop mediates proliferation and migration in osteosarcoma cells. Oncol Res. 2017;25(9):1517–1527. doi: 10.3727/096504017X14859934460780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Huang Y, Xiang P, Tian W. LncRNA expression and implication in osteosarcoma: a systematic review and meta-analysis. Onco Targets Ther. 2017;10:5355–5361. doi: 10.2147/OTT.S149889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen B, Zhou N, Hu T, Zhao W, Wu D, Wang S. LncRNA MEG3 negatively modified osteosarcoma development through regulation of miR-361-5p and FoxM1. J Cell Physiol. 2019;234(8):13464–13480. doi: 10.1002/jcp.28026. [DOI] [PubMed] [Google Scholar]
- 49.Tian ZZ, Guo XJ, Zhao YM, Fang Y. Decreased expression of long non-coding RNA MEG3 acts as a potential predictor biomarker in progression and poor prognosis of osteosarcoma. Int J Clin Exp Pathol. 2015;8(11):15138–15142. [PMC free article] [PubMed] [Google Scholar]
- 50.Ji S, Wang S, Zhao X, Lv L. Long noncoding RNA NEAT1 regulates the development of osteosarcoma through sponging miR-34a-5p to mediate HOXA13 expression as a competitive endogenous RNA. Mol Genet Genomic Med. 2019;7(6):e673.. [DOI] [PMC free article] [PubMed]
- 51.Li Y, Cheng C. Long noncoding RNA NEAT1 promotes the metastasis of osteosarcoma via interaction with the G9a-DNMT1-snail complex. Am J Cancer Res. 2018;8(1):81–90. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Tan H, Zhao L. lncRNA nuclear-enriched abundant transcript 1 promotes cell proliferation and invasion by targeting miR-186-5p/HIF-1 alpha in osteosarcoma. J Cell Biochem. 2019;120(4):6502–6514. doi: 10.1002/jcb.27941. [DOI] [PubMed] [Google Scholar]
- 53.Zhu K-P, Zhang C-L, Ma X-L, Zhang L. Fibronectin-1 modulated by the long noncoding RNA OIP5-AS1/miR-200b-3p axis contributes to doxorubicin resistance of osteosarcoma cells. J Cell Physiol. 2019;234(5):6927–6939. doi: 10.1002/jcp.27518. [DOI] [PubMed] [Google Scholar]
- 54.Dai J, Xu L, Hu X, Han G, Jiang H, Sun H, Zhu G, Tang X. Long noncoding RNA OIP5-AS1 accelerates CDK14 expression to promote osteosarcoma tumorigenesis via targeting miR-223. Biomed Pharmacother. 2018;106:1441–1447. doi: 10.1016/j.biopha.2018.07.109. [DOI] [PubMed] [Google Scholar]
- 55.Zhang CL, Zhu KP, Shen GQ, Zhu ZS. A long non-coding RNA contributes to doxorubicin resistance of osteosarcoma. Tumour Biol. 2016;37(2):2737–2748. doi: 10.1007/s13277-015-4130-7. [DOI] [PubMed] [Google Scholar]
- 56.Zhu KP, Ma XL, Zhang CL. LncRNA ODRUL contributes to osteosarcoma progression through the miR-3182/MMP2 Axis. Mol Ther. 2017;25(10):2383–2393. doi: 10.1016/j.ymthe.2017.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57.Huang J, Deng G, Liu T, Chen W, Zhou Y. Long noncoding RNA PCAT-1 acts as an oncogene in osteosarcoma by reducing p21 levels. Biochem Biophys Res Commun. 2018;495(4):2622–2629. doi: 10.1016/j.bbrc.2017.12.157. [DOI] [PubMed] [Google Scholar]
- 58.Zhang X, Zhang Y, Mao Y, Ma X. The lncRNA PCAT1 is correlated with poor prognosis and promotes cell proliferation, invasion, migration and EMT in osteosarcoma. OncoTargets Ther. 2018;11:629–638. doi: 10.2147/OTT.S152063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Song J, Wu X, Liu F, Li M, Sun Y, Wang Y, Wang C, Zhu K, Jia X, Wang B, et al. Long non-coding RNA PVT1 promotes glycolysis and tumor progression by regulating miR-497/HK2 axis in osteosarcoma. Biochem Biophys Res Commun. 2017;490(2):217–224. doi: 10.1016/j.bbrc.2017.06.024. [DOI] [PubMed] [Google Scholar]
- 60.Zhou Q, Chen F, Zhao J, Li B, Liang Y, Pan W, Zhang S, Wang X, Zheng D. Long non-coding RNA PVT1 promotes osteosarcoma development by acting as a molecular sponge to regulate miR-195. Oncotarget. 2016;7(50):82620–82633. doi: 10.18632/oncotarget.13012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou B, Li L, Li Y, Sun H, Zeng C. Long noncoding RNA SNHG12 mediates doxorubicin resistance of osteosarcoma via miR-320a/MCL1 axis. Biomed Pharmacother. 2018;106:850–857. doi: 10.1016/j.biopha.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 62.Zhou S, Yu L, Xiong M, Dai G. LncRNA SNHG12 promotes tumorigenesis and metastasis in osteosarcoma by upregulating Notch2 by sponging miR-195-5p. Biochem Biophys Res Commun. 2018;495(2):1822–1832. doi: 10.1016/j.bbrc.2017.12.047. [DOI] [PubMed] [Google Scholar]
- 63.Liao S, Xing S, Ma Y. LncRNA SNHG16 sponges miR-98-5p to regulate cellular processes in osteosarcoma. Cancer Chemother Pharmacol. 2019;83(6):1065–1074. doi: 10.1007/s00280-019-03822-5. [DOI] [PubMed] [Google Scholar]
- 64.Wang X, Hu K, Chao Y, Wang L. LncRNA SNHG16 promotes proliferation, migration and invasion of osteosarcoma cells by targeting miR-1301/BCL9 axis. Biomed Pharmacother. 2019;114:108798. doi: 10.1016/j.biopha.2019.108798. [DOI] [PubMed] [Google Scholar]
- 65.Wang W, Luo P, Guo W, Shi Y, Xu D, Zheng H, Jia L. LncRNA SNHG20 knockdown suppresses the osteosarcoma tumorigenesis through the mitochondrial apoptosis pathway by miR-139/RUNX2 axis. Biochem Biophys Res Commun. 2018;503(3):1927–1933. doi: 10.1016/j.bbrc.2018.07.137. [DOI] [PubMed] [Google Scholar]
- 66.Zhang J, Ju C, Zhang W, Xie L. LncRNA SNHG20 is associated with clinical progression and enhances cell migration and invasion in osteosarcoma. IUBMB Life. 2018;70(11):1115–1121. doi: 10.1002/iub.1922. [DOI] [PubMed] [Google Scholar]
- 67.Chen X, Zhou Y, Liu S, Zhang D, Yang X, Zhou Q, Song Y, Liu Y. LncRNA TP73-AS1 predicts poor prognosis and functions as oncogenic lncRNA in osteosarcoma. J Cell Biochem. 2018;120(2):2569–2575. doi: 10.1002/jcb.27556. [DOI] [PubMed] [Google Scholar]
- 68.Yang G, Song R, Wang L, Wu X. Knockdown of long non-coding RNA TP73-AS1 inhibits osteosarcoma cell proliferation and invasion through sponging miR-142. Biomed Pharmacother. 2018;103:1238–1245. doi: 10.1016/j.biopha.2018.04.146. [DOI] [PubMed] [Google Scholar]
- 69.Ma B, Li M, Zhang L, Huang M, Lei JB, Fu GH, Liu CX, Lai QW, Chen QQ, Wang YL. Upregulation of long non-coding RNA TUG1 correlates with poor prognosis and disease status in osteosarcoma. Tumour Biol. 2016;37(4):4445–4455. doi: 10.1007/s13277-015-4301-6. [DOI] [PubMed] [Google Scholar]
- 70.Sheng K, Li Y. LncRNA TUG1 promotes the development of osteosarcoma through RUNX2. Exp Ther Med. 2019;18(4):3002–3008. doi: 10.3892/etm.2019.7880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Q, Chen Q. Role of taurine upregulated gene 1 as a predictor of poor outcome in osteosarcoma. J Cancer Res Ther. 2018;14(Supplement):S405–s409. doi: 10.4103/0973-1482.172585. [DOI] [PubMed] [Google Scholar]
- 72.Wang Y, Yang T, Zhang Z, Lu M, Zhao W, Zeng X, Zhang W. Long non-coding RNA TUG1 promotes migration and invasion by acting as a ceRNA of miR-335-5p in osteosarcoma cells. Cancer Sci. 2017;108(5):859–867. doi: 10.1111/cas.13201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu X, Hu L, Li S, Shen J, Wang D, Xu R, Yang H. Long non-coding RNA Taurine upregulated gene 1 promotes osteosarcoma cell metastasis by mediating HIF-1alpha via miR-143-5p. Cell Death Dis. 2019;10(4):280. doi: 10.1038/s41419-019-1509-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li W, Xie P, Ruan WH. Overexpression of lncRNA UCA1 promotes osteosarcoma progression and correlates with poor prognosis. J Bone Oncol. 2016;5(2):80–85. doi: 10.1016/j.jbo.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wen JJ, Ma YD, Yang GS, Wang GM. Analysis of circulating long non-coding RNA UCA1 as potential biomarkers for diagnosis and prognosis of osteosarcoma. Eur Rev Med Pharmacol Sci. 2017;21(3):498–503. [PubMed] [Google Scholar]
- 76.Li GL, Wu YX, Li YM, Li J. High expression of long non-coding RNA XIST in osteosarcoma is associated with cell proliferation and poor prognosis. Eur Rev Med Pharmacol Sci. 2017;21(12):2829–2834. [PubMed] [Google Scholar]
- 77.Wang W, Shen H, Cao G, Huang J. Long non-coding RNA XIST predicts poor prognosis and promotes malignant phenotypes in osteosarcoma. Oncol Lett. 2019;17(1):256–262. doi: 10.3892/ol.2018.9596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang C, Wu K, Wang S, Wei G. Long non-coding RNA XIST promotes osteosarcoma progression by targeting YAP via miR-195-5p. J Cell Biochem. 2018;119(7):5646–5656. doi: 10.1002/jcb.26674. [DOI] [PubMed] [Google Scholar]
- 79.Zhang R, Xia T. Long non-coding RNA XIST regulates PDCD4 expression by interacting with miR-21-5p and inhibits osteosarcoma cell growth and metastasis. Int J Oncol. 2017;51(5):1460–1470. doi: 10.3892/ijo.2017.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li N, Sun ZH, Fang M, Xin JY, Wan CY. Long non-coding RNA ZFAS1 sponges miR-486 to promote osteosarcoma cells progression and metastasis in vitro and vivo. Oncotarget. 2017;8(61):104160–104170. doi: 10.18632/oncotarget.22032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu G, Wang L, Han H, Li Y, Lu S, Li T, Cheng C. LncRNA ZFAS1 promotes growth and metastasis by regulating BMI1 and ZEB2 in osteosarcoma. Am J Cancer Res. 2017;7(7):1450–1462. [PMC free article] [PubMed] [Google Scholar]
- 82.Sayles LC, Breese MR, Koehne AL, Leung SG, Lee AG, Liu HY, Spillinger A, Shah AT, Tanasa B, Straessler K, et al. Genome-informed targeted therapy for osteosarcoma. Cancer Discov. 2019;9(1):46–63. doi: 10.1158/2159-8290.CD-17-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shaikh AB, Li F, Li M, He B, He X, Chen G, Guo B, Li D, Jiang F, Dang L, et al. Present advances and future perspectives of molecular targeted therapy for osteosarcoma. Int J Mol Sci. 2016;17(4):506. doi: 10.3390/ijms17040506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen H, Shan G. The physiological function of long-noncoding RNAs. Noncoding RNA Res. 2020;5(4):178–184. doi: 10.1016/j.ncrna.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chillón I, Marcia M. The molecular structure of long non-coding RNAs: emerging patterns and functional implications. Crit Rev Biochem Mol Biol. 2020;55(6):662–690. doi: 10.1080/10409238.2020.1828259. [DOI] [PubMed] [Google Scholar]
- 86.Zhao ZY, Zhao YC, Liu W. Long non-coding RNA TUG1 regulates the progression and metastasis of osteosarcoma cells via miR-140-5p/PFN2 axis. Eur Rev Med Pharmacol Sci. 2019;23(22):9781–9792. doi: 10.26355/eurrev_201911_19541. [DOI] [PubMed] [Google Scholar]
- 87.Lv GY, Miao J, Zhang XL. Long noncoding RNA XIST promotes osteosarcoma progression by targeting Ras-related protein RAP2B via miR-320b. Oncol Res. 2018;26(6):837–846. doi: 10.3727/096504017X14920318811721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu D, Nie X, Ma C, Liu X, Liang X, An Y, Zhao B, Wu X. RSF1 functions as an oncogene in osteosarcoma and is regulated by XIST/miR-193a-3p axis. Biomed Pharmacother. 2017;95:207–214. doi: 10.1016/j.biopha.2017.08.068. [DOI] [PubMed] [Google Scholar]
- 89.Chen D, Wang H, Zhang M, Jiang S, Zhou C, Fang B, Chen P. Abnormally expressed long non-coding RNAs in prognosis of osteosarcoma: a systematic review and meta-analysis. J Bone Oncol. 2018;13:76–90. doi: 10.1016/j.jbo.2018.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang Z, Katsaros D, Biglia N, Shen Y, Fu Y, Loo LWM, Jia W, Obata Y, Yu H. High expression of long non-coding RNA MALAT1 in breast cancer is associated with poor relapse-free survival. Breast Cancer Res Treat. 2018;171(2):261–271. doi: 10.1007/s10549-018-4839-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Song W, Zhang RJ, Zou SB. Long noncoding RNA MALAT1 as a potential novel biomarker in digestive system cancers: a meta-analysis. Minerva Med. 2016;107(4):245–250. [PubMed] [Google Scholar]
- 92.Mao H, Wang K, Feng Y, Zhang J, Pan L, Zhan Y, Sheng H, Luo G. Prognostic role of long non-coding RNA XIST expression in patients with solid tumors: a meta-analysis. Cancer Cell Int. 2018;18:34. doi: 10.1186/s12935-018-0535-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhao W, Wang Z, Fang X, Li N, Fang J. Long noncoding RNA breast cancer antiestrogen resistance 4 is associated with cancer progression and its significant prognostic value. J Cell Physiol. 2019;234(8):12956–12963. doi: 10.1002/jcp.27962. [DOI] [PubMed] [Google Scholar]
- 94.Zhang C, Ren X, He J, Wang W, Tu C, Li Z. The prognostic value of long noncoding RNA SNHG16 on clinical outcomes in human cancers: a systematic review and meta-analysis. Cancer Cell Int. 2019;19:261. doi: 10.1186/s12935-019-0971-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tu C, Ren X, He J, Zhang C, Chen R, Wang W, Li Z. The value of LncRNA BCAR4 as a prognostic biomarker on clinical outcomes in human cancers. J Cancer. 2019;10(24):5992–6002. doi: 10.7150/jca.35113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ma Z, Wang YY, Xin HW, Wang L, Arfuso F, Dharmarajan A, Kumar AP, Wang H, Tang FR, Warrier S, et al. The expanding roles of long non-coding RNAs in the regulation of cancer stem cells. Int J Biochem Cell Biol. 2019;108:17–20. doi: 10.1016/j.biocel.2019.01.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 Study quality and bias in the retrospective cohort studies judged by the Newcastle-Ottawa Scale (NOS) checklist. Figure A.1 Quality assessment of all included studies. “Risk of bias summary” of all included studies. Figure A.2 Quality assessment of all included studies. “Risk of bias graph” of all included studies.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.