Abstract
Simple Summary
Intergenerational and transgenerational effects, in which exposure to stressors in a parental generation affects the phenotype of the offspring have been connected to anthropic impacts on biological systems. Therefore, environmental stress experienced inside a generation, particularly during gametogenesis, may lead to erroneous patterns in their offspring just emerging at early developmental stages. In this scenario, the sea urchin embryo represents a suitable model for integrating analyses of gene expression through embryogenesis with developmental alteration induced by environmental stressors. Herein we provide pieces of evidence for the alteration of the gene regulatory networks modulating embryo development after parental conditioning via non-lethal metal exposure. We show that the parentals’ conditioning does not affect viability but significantly impairs the developmental fate of the progeny and regulatory network across a generation. It is reasonable to suppose that changes in Paracentrotus lividus gonads may modify the expression of regulatory genes modulating synthesis and/or accumulation of maternal determinants, which, in turn, impaired the zygotic activation of GRNs responsible for proper embryo development.
Abstract
It is widely accepted that phenotypic traits can be modulated at the epigenetic level so that some conditions can affect the progeny of exposed individuals. To assess if the exposure of adult animals could result in effects on the offspring, the Mediterranean sea urchin and its well-characterized gene regulatory networks (GRNs) was chosen as a model. Adult animals were exposed to known concentrations of zinc and cadmium (both individually and in combination) for 10 days, and the resulting embryos were followed during the development. The oxidative stress occurring in parental gonads, embryo phenotypes and mortality, and the expression level of a set of selected genes, including members of the skeletogenic and endodermal GRNs, were evaluated. Increased oxidative stress at F0, high rates of developmental aberration with impaired gastrulation, in association to deregulation of genes involved in skeletogenesis (dri, hex, sm50, p16, p19, msp130), endodermal specification (foxa, hox11/13b, wnt8) and epigenetic regulation (kat2A, hdac1, ehmt2, phf8 and UBE2a) occurred either at 24 or 48 hpf. Results strongly indicate that exposure to environmental pollutants can affect not only directly challenged animals but also their progeny (at least F1), influencing optimal timing of genetic programme of embryo development, resulting in an overall impairment of developmental success.
Keywords: sea urchin, redox homeostasis, parental exposure, intergenerational effects, embryo development, gene expression profiles
1. Introduction
Intergenerational and transgenerational effects, in which exposure to stressors in a parental generation (F0) models the phenotype of the offspring (F1) [1,2], have been recently reported to be associated with the global climate change and anthropic impacts on biological systems [3,4,5].
Genome- or transcriptome-wide changes induced by environmental stressors on somatic cells can affect the physiology of exposed individuals; however, some alterations can be propagated to subsequent generations through the germline [6]. As such, modifications in the expression of maternal determinants may occur during the gametogenesis, making these alterations heritable [7,8,9]. Therefore, environmental stresses experienced by F0, in particular during gametogenesis, may lead to aberrant patterns in F1, which can become evident even since the early developmental stages.
It is known that a network of regulatory genes finely establishes the mechanisms of specification and differentiation through embryo development [10,11]. The eco-devo approach, integrating analyses of gene expression through embryogenesis with developmental alterations induced by environmental stressors, provides a tool for reframing the complexity of embryo development [12,13].
As unique invertebrate deuterostomes, echinoderms and, in particular, sea urchins, represent excellent research models in the study of the developmental biology of bilaterians [14,15], as well as fertilization and proliferation mechanisms [16,17,18] and genome evolution [19]. Moreover, they have been used as models in a lot of ecotoxicological studies for the assessment of the effects of anthropization, ocean warming and acidification [20,21,22,23,24].
The gene regulatory networks (GRNs) that control the sea urchin development represent one of the most complete models integrating embryogenesis and transcriptional regulatory switches [25,26]. Among the studied regulatory circuits, the skeletogenic and the endodermal regulatory networks are well conserved in sea urchin species. In the skeletogenic circuit the transcription factor Ets1, which is maternally inherited, activates the expression of genes encoding the transcription factors Alx1 and Hex. These activate dri and the three transcription factors cooperate and activate the expression of genes responsible for calcification and spicule formation, such as sm50, msp130, p16 and p19 [10,27]. Conversely, the endoderm specification starts with the early expression of wnt8 which turns on the expression of hox11/13b, foxa and bra [10,27].
In addition, developmental programs established by transcription factor networks are promoted, sustained, maintained and transmitted to daughter cells via epigenetic systems, also when the responsible transcription factors are not still active. These systems include post-transcriptional modifications of histone tails by chromatin writers (HAT, HDAC, HKMT, etc.), DNA methylation by DNMTs, long non-coding RNAs, and in turn recruit chromatin readers, such as chromatin remodelers [28,29,30,31]. Any alteration of these cascades may affect chromatin dynamics and deregulate a huge number of genes, causing aberrant developmental phenotypes.
In addition to Strongylocentrotus purpuratus, a few species contributed to define the GRNs. Among them, the Mediterranean sea urchin Paracentrotus lividus represents a widely studied species [32,33,34]. Interestingly, the high resolved expression profiles of the regulatory genes orchestrating developmental circuits are available also in P. lividus and the study of their perturbations due to pollutant exposure represents an important tool also to investigate mechanisms of toxicity [35,36].
The P. lividus, as an inhabitant of coastal waters, is exposed to a range of stressors, including pollutants from anthropogenic activities, throughout its life [37].
As a result of its lifestyle, the sea urchin often experiences different environmental conditions which may result in adaptive effects [38,39] and intergenerational effects affecting the phenotype of the offspring [40,41]. In this line, in early-stage Strongylocentrotus purpuratus embryos and larvae, high pCO2 has been shown to impact body size, metabolic rates, gene expression, internal acid-base balance and growth [42]. Recently, we showed that non-lethal metal conditioning heavily affected the sea urchin transcriptional profiles in reproductive tissues, thus suggesting effects on gametogenesis [43]. Accordingly, gametes from cadmium and manganese-exposed P. lividus females experienced difficulties in fertilisation and related embryos displayed abnormalities during the development [40]. Similarly, defects in hatching and larval morphology were also detected in Strongylocentrotus intermedius embryos from warm-conditioned F0 [44]; however, no data are currently available on the effects that metal exposure of either males or females could exert on the progeny.
Herein, we evaluated if the conditioning of parental P. lividus, via metal exposure could affect the developing offspring.
Toward this aim, adult sea urchins were challenged for 10 days with zinc (Zn) as a representative essential metal, and cadmium (Cd), as a representative xenobiotic metal with known toxic effects at environmentally relevant concentrations or similar to those used in other reports [43,45,46]. Since the simultaneous exposure of organisms to different metals may result in variable levels of toxicity due to neutralizing, additive or synergistic effects [22,47], two mixtures, consisting of Cd and Zn at different concentrations, were also used. The metal-conditioned gonads were analysed in order to assess reactive oxygen species (ROS) production and catalase activity, as well as protein levels of HSP70, HSP60 and HSP90.
In order to evaluate the influence of stress events experienced by parentals on the developing progeny, resulting embryos (F1) were let to develop until 48 h post-fertilization (hpf), and the morphology was analysed at various developmental stages. As a result that it is known that sea urchin exposure to metals affects the overall developmental regulatory networks, herein we wondered to investigate whether the parental exposure affected selected regulatory nodes. In particular, gene expression was examined in embryos from parents exposed to the low Zn and low metal mixture concentration treatments, because of high mortality rates observed in the other treatments. Therefore, to investigate molecular mechanisms perturbing the development, we analysed the transcriptional profiles of development-related genes, including selected members of the endoderm and skeletogenic GRNs (foxa, hox11/13b, wnt8, dri, hex, sm50, p16, p19, msp130). Moreover, we profiled also the transcriptional expression of genes associated with mechanisms of epigenetic regulation, including histone acetylase and deacetylase (kat2A and hdac1), histone lysine methylase and demethylase (ehmt2 and phf8) and histone ubiquitinase (UBE2a).
Overall, this study begins to unravel the potential for epigenetic mechanisms to play a role in the alterations in the timing or organisation of developmental processes that could also impair the rapid responses to environmental change in an ecologically important benthic marine invertebrate.
2. Materials and Methods
2.1. Sea Urchin Sampling and Exposure to Metals
Adult P. lividus specimens were collected nearby Capo Granitola, Trapani, Italy (Latitude 37°34′30.00″ N Longitude 12°40′47.26″ E) and rapidly transported to the laboratory. All the experimental workflow (including replicates) was performed in autumn/winter 2017. The animals were acclimated for 15 days in an aquarium with artificial seawater (ASW) (Instant Ocean Aquarium System), with 12 h:12 h light: dark photoperiod and continuous aeration. Salinity (35 ± 1‰), temperature (17.0 ± 1.0 °C) and pH (7.80 ± 0.10) were monitored daily. An amount of 3 g/animal of dehydrated macroalgae (Sera Marin Gourmet Nori) was provided every day during the acclimation, while feeding was interrupted 2 days before experimental sampling. After acclimation, P. lividus adults (3 males and 3 females per treatment) were exposed for 10 days to cadmium, zinc and combinations of the two pollutants in different tanks (10 L each) as reported in Table 1. Specimens in ASW were used as controls. Temperature and pH values were monitored daily. Each experiment was carried out three times. Each animal was treated just once, therefore the total number of males and females investigated in the experiments was 126 (3 males × 7 conditions (6 treatments + 1 control) × 3 replicates = 63 males treated plus 3 females × 7 conditions × 3 replicates = 63 females treated). Sulphate salts (Sigma-Aldrich, Milan, Italy) were used for the preparation of metal solutions. During exposure, animals were fed twice a week with 3 g/animal of dehydrated macroalgae.
Table 1.
Concentrations of considered metals.
| Treatment | Low (µg/L) L |
High (µg/L) H |
|---|---|---|
| Cd2+ | 10 | 100 |
| Zn2+ | 40 | 200 |
| Mix | Cd-Zn (10 + 40) | Cd-Zn (100 + 200) |
2.2. Protein Extraction, SDS-PAGE and Western Blot Analyses
Male gonads were pooled (0.3 g) and homogenized in 20 mM Tris–HCl buffer pH 7.5, 15% Glycerol, 1 mM EDTA, 0.1% Triton X-100. Similarly, 0.3 g of female gonads were also pooled and treated in the same manner. Therefore, a total of 42 samples (3 replicates/condition for males; 3 replicates/condition for females; 7 conditions) were analysed. The homogenates were centrifuged to remove any insoluble debris at 10,000 rpm for 10 min. The supernatants were collected and quantified using Qubit 2.0 Fluorometer (Thermo Fisher, San Jose, CA, USA) according to the manufacturer’s instructions and protein concentration was brought to 2.5 mg/mL in all samples. Proteins (50 µg) were separated on 10% SDS-PAGE under reducing conditions and transferred to nitrocellulose membranes (Thermo Fisher, CA, USA) at 50 V, in ice bath, for 2 h using transfer buffer (48 mM Tris, 39 mM glycine, pH 9.2, 20% methanol). After blocking in 5% BSA and 1× TBST (20 mM Tris-HCl pH 7.6, 137 mM NaCl, 0.1% Tween 20) for 1 h at room temperature, membranes were incubated overnight at 4 °C with different antibodies. In particular, monoclonal anti-Heat Shock Protein 70 (H5147, Sigma, Milan, Italy), monoclonal anti-Heat Shock Protein 60 (H-3524, Sigma, Milan, Italy), monoclonal anti-Heat Shock Protein 90 (SAB1305541, Sigma, Milan, Italy) and monoclonal anti-actin clone (A3853, Sigma, Milan, Italy) primary antibodies, were used and diluted 1:2000, 1:500, 1:800 and 1:500, respectively, in 1× TBST. The membranes were washed three times with TBST for 5 min and incubated for 1 h at room temperature with alkaline phosphatase-conjugated anti-mouse IgG (1:3000). The membranes were washed three times for 5 min with TBST and incubated with One step NBT/BCIP substrate solution (Thermo Fisher, CA, USA) and blots were acquired using the Molecular Imager VersaDoc system (Bio-Rad Laboratories, Hercules, CA, USA). The Image J software (Bio-Rad Laboratories) was used for densitometry analysis of the immunoblotted bands. The signals from each protein band were normalized against the β-actin content.
2.3. Reactive Oxygen Species (ROS) Detection and Catalase Activity
The catalase assay kit (CAT 100, Sigma, Milan, Italy) was used to measure the catalase activity in protein extracts. A standard curve was generated plotting the Red Quinoneimine dye absorbance at 520 nm versus different amounts of H2O2 (0.0125, 0.025, 0.05 and 0.075 µ/mol). Supernatants were added to the catalase colorimetric enzymatic reaction to determine the amount of H2O2 remaining in the mixture using the H2O2 standard curve. In the assay reaction, a volume amount corresponding to 20 µg of total protein content for each sample was added.
In particular, Equation (1) was used to calculate the difference in the amount of H2O2 (∆µ/moles) added to the colorimetric reaction between the blank and a given sample, while the catalase activity was calculated using the Equation (2).
| ∆μ/moles (H2O2) = µ/moles of H2O2 (blank) − µ/moles of H2O2 (sample) | (1) |
| Activity (µ(moles/min)/mL) = ∆μ/moles (H2O2) × d × 100)/(V × t) | (2) |
where d is the dilution of the original sample for catalase reaction; t is the catalase reaction duration (min); V is the sample volume in catalase reaction (mL); 100 is the dilution of an aliquot from catalase reaction in colorimetric reaction. To measure total ROS/RNS free radical activity present in P. lividus reproductive tissues the OxiSelect In Vitro ROS/RNS Assay Kit (Cell Biolabs, Inc. San Diego, CA, USA) was used according to the manufacturer’s instructions. Supernatants were incubated in a 96-well fluorescence plate with the DCFH probe to allow the oxidation reaction to proceed. Samples were then measured fluorometrically at λex 480 nm and λem 530 nm using the GloMax Discover Multimode Microplate Reader (Promega, Milan, Italy). The free radical content in samples was determined by comparison with a DCF standard curve (1 nM–10 µM).
2.4. Embryo Culture and Morphological Analysis
Exposed and control P. lividus animals were sacrificed and gonads were gently washed to allow gametes collection. In particular, for each experimental condition, eggs from treated females (3 females/treatment/experiment) were pooled and maintained in ASW until fertilisation with sperms (3 males/treatment/experiment) collected and diluted (1:500) in ASW. Similarly, pooled eggs from three untreated animals were fertilised in presence of pooled sperms from control males. All the performed fertilisation occurred in the condition of sperms/eggs ratio corresponding to 100:1. Eggs were rinsed after fertilization. At least 6000 embryos were allowed to develop in 250 mL tanks at 18 °C under gentle agitation in ASW and they were examined until the pluteus stage (48 hpf). The percentages of embryos showing normal or abnormal phenotype were determined by counting about 100 embryos/replicate in each treatment using the Olympus BX50 optical microscope.
2.5. RNA Purification and First-Strand cDNA Synthesis
Total RNA was purified from P. lividus embryos from parentals treated with the low concentration of Zn and the low concentration of the metal mixture (1000 embryos/treatment/experiment), at 24 and 48 hpf, using TRIzol Reagent (Invitrogen Corporation, Carlsbad, CA, USA) according to the manufacturer’s instructions. RNA concentrations and purity (Abs260, Abs260/280 and Abs260/230) were spectrophotometrically verified using Eppendorf 6131 BioPhotometer (Eppendorf AG, Hamburg, Germany). RNA integrity was evaluated on 1.5% agarose denaturing gel and RNA was stored at −80 °C for future use. An amount of 1 µg of total RNA was digested with Deoxyribonuclease I, Amplification Grade (Invitrogen Corporation, Carlsbad, CA, USA) to remove genomic DNA contamination, while DNase I was inactivated by adding 25 mM EDTA. First-strand cDNA was synthesised from 500 ng DNase I treated RNA using SuperScript VILO cDNA Synthesis Kit (Invitrogen Corporation, Carlsbad, CA, USA), according to the manufacturer’s instructions. The synthesised cDNAs were tested by PCR using 18S rRNA (Table 2) and then diluted 1:10 before use in real-time qPCR experiments.
Table 2.
Genes and oligonucleotide primers used in this study.
| Gene Name | Gene Symbol | Primer Sequence (5′–3′) |
|---|---|---|
| Forkhead box transcription factor a | foxa | CAGGTATGGGAAGCATGGGA GCGTATCTCATCGACATGGC |
| Homeodomain containing transcription factor Hox11/13b | hox11/13b | TTGCGACGTTCACAACAACT GTGAGATCGAGAGCCTGTGA |
| Wint8 | wnt8 | AAGTGTCATGGCGTCTCTGG ATGAGCTTGCCACTGACGAA |
| Deadringer-like | dri | TGGAAATAGACGAGAGGGGC GTGGTATCATGGTGGGTGGA |
| Hex homeodomain transcription factor | hex | TGAACCACCCTACTCCACTG GCCGCTCTATTTTGTCCGAG |
| Spicule matrix protein 50 | sm50 | GATGGCACACCAGCTTATCC CTGACGCTTCATGACTGGAG |
| Biomineralization protein p16 | p16 | AGCAGGAGCAGTCGGAGATAC CATCATCACTTCCCATATCGC |
| Biomineralization protein p19 | p19 | AGAGACCAGGCAGGAGACTAAG GTTGATGTCGAGCTTGTCTTTC |
| Mesenchyme-specific cell surface glycoprotein | msp130 | ATACATGGCAACCCAAGAAG CGATTCCAACGAAGATGAGT |
| Ubiquitin-conjugating enzyme E2 A | UBE2a | GATTTGAGGAGTGGAGGATTG AGCTGGCGGATCTTCTTGTA |
| Histone deacetylase | Hdac1 | TCACGCCAAGAAGTCAGAAG CCGTGGTGGATATCAATGTC |
| Histone acetyltransferase | kat2A | TGCAATGGATGGAAGAACC ATGTGCAGCAAGTTGATGG |
| Histone-lysine N-methyltransferase | EHMT2 | GTGCAGGAGCTCTTGGTAATG ACAGAGAGGGTCGGAAAGTG |
| Histone lysine demethylase | phf8 | AGCAGTTGCCATTCCTTTTC CGACCCATTCACATTCCAC |
| 18S ribosomal RNA | 18s | GAATGTCTGCCCTATCAACTTTCG TTGGATGTGGTAGCCGTTTCTC |
2.6. Gene Expression Profiling by Real-Time Quantitative Polymerase Chain Reaction (qPCR)
The qPCRs were carried out on the BIO-RAD CFX96 system using Power Sybr Green as the chemical detection method (Applied Biosystems, Forster City, CA, USA). The qPCRs were performed in 96-well plates in a 20 µL mixture containing 1 µL of a 1:10 dilution of the cDNAs, using the following PCR parameters: 95 °C for 10 min, followed by 40 cycles of 95 °C for 10 s and 60 °C for 35 s and melting curve from 65 to 95 °C. Amplicons were run on an agarose gel to confirm the specific gene amplification.
The 18S ribosomal RNA was chosen as reference gene [43,48,49,50,51]. PCR efficiency of the target and reference genes was calculated using serial dilutions of pooled cDNAs from both control and treated samples. The measured amplification efficiency ranged from 1.8 to 2.1. Primer sequences used in this study are listed in Table 2.
Quantitative real-time PCRs were conducted according to the manufacturer’s recommended procedures, and every reaction was performed three times. Data analysis was carried out according to the 2−ΔΔCT method [52].
2.7. Statistical Analysis
Experiments were performed in triplicate. The results in the bar plots were expressed as a mean value ± SD. To compare treatment results in testis and ovary, ANOVA tests were performed using the R software, version 3.6.3 (Figure 1). Statistical analysis on phenotype evaluation (Figure 2) was done using t-test. For gene expression analysis, significant differences between the values of different treated groups and the reference control groups were also determined by t-test. The p-values less than 0.05 were considered statistically significant.
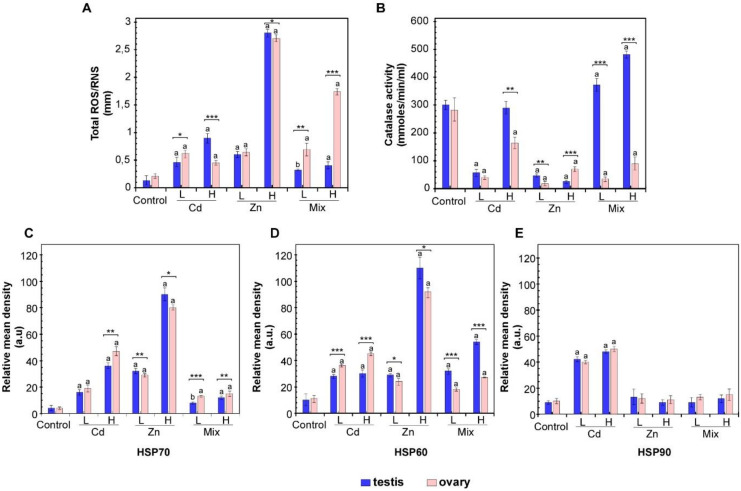
Figure 1.
Redox response system of P. lividus reproductive tissues (testis and ovary) after exposures to high (H), low (L) doses of Cd, Zn or a mix of both metals, as reported in Table 1. (A) Total ROS amount; (B) catalase activity; (C) relative HSP70 protein amount; (D) relative HSP60 protein amount; (E) relative HSP90 protein amount. Bars represent the mean values from the three different experiments ± SD. Statistical significance among testis and ovary results is shown as asterisks (*: p < 0.05; **: p < 0.01; ***: p < 0.001). Letters denote statistical significance of treated specimens vs. controls (b: p < 0.01; a: p < 0.001).
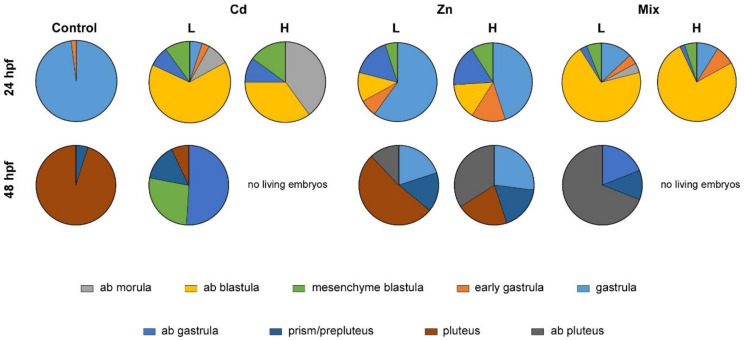
Figure 2.
Embryo morphological abnormalities at 24 and 48 hpf and percentages in response to the different treatments (Cd, Zn and Mix of them) experienced by parentals. Data corresponding to embryos from means of three experiments are shown; SD were lower than 5%. L: low concentration; H: high concentration; Ab: abnormal.
3. Results
3.1. Effects of Metal Exposures on the Stress Response of P. lividus Reproductive Tissues
P. lividus specimens were exposed to Cd, Zn and a mixture of both metals, each in two different concentrations, low and high for 10 days as described in the Material and Methods. In F0, no mortality or illness indicators, as dropped spines or reduced movements, were recorded among sea urchins during metal conditioning.
In order to evaluate the stress status of challenged P. lividus gonads, the production of ROS was evaluated in exposed and control animals. As shown in Figure 1A, a significant increase in ROS accumulation in male and female gonads was induced by metal exposure. Zn treatments resulted in a concentration-dependent increase in ROS contents in the testis and ovary. Evidence for gender-specific responses emerged for ROS production induced by metal mixes because ROS level increased more in ovaries than in testis.
The activity of catalases (CAT) was also profiled in order to evaluate the activation of the antioxidant system involved in ROS scavenging. As expected, catalase activity was inversely correlated with ROS levels. An overall reduction in CAT activity was found in female reproductive tissues in response to different Cd and Zn doses, as well as after combined exposures. A similar pattern was also observed in the testis, except for the unchanged CAT activity measured at the higher Cd dosage. Additionally, increased CAT activity in testis was measured in response to Mix L and Mix H (Figure 1B).
To evaluate the effects of metal-induced stress at the protein scale, we analysed the expression of HSP70, HSP60 and HSP90 proteins (Figure 1C–E and Supplementary Materials Figure S1). The densitometric analysis showed a concentration-dependent overexpression of HSP70 protein in male and female gonads exposed either to Cd and Zn (Figure 1C); while, evidence for gender-specific responses emerged for HSP60. In detail, although metal-induced HSP60 synthesis was found both in testis and ovarian tissue, exposure to increasing Cd and Zn levels resulted in dose-dependent HSP60 overexpression in the ovary; differently, in testis HSP60 upregulation was dose-dependent only in response to Zn (Figure 1D). For HSP90 increased levels were found in response to Cd, while Zn treatment did not induce significant changes (Figure 1E). To evaluate the effects of the combined exposures, the expression of these stress-related proteins was also measured in sea urchins exposed to Mix H and Mix L (Figure 1C–E). Densitometric analysis revealed that co-exposures induced HSP60 and HSP70, although to a lesser extent than observed with single metals. Moreover, albeit HSP60 increased especially in testis, HSP70 induction were higher in ovary. Conversely, HSP90 resulted uninduced.
3.2. Effects of Metal Exposures on the Offspring of Conditioned P. lividus
To evaluate the occurrence of intergenerational effects due to metal exposures of parentals (F0), eggs and sperms from exposed animals were allowed to fertilise and develop until 48 hpf (which corresponds to the pluteus stage in controls). No significant differences in fertilization success were observed across experiment conditions, and embryos were observed under a microscope at 24 and 48 hpf, corresponding to the gastrula and pluteus stages, respectively (Figure 2 and Supplementary Materials Figure S2).
Embryos from Cd (L) conditioned parentals showed several abnormalities. Retard in endoderm invagination occurred at 24 hpf and a high percentage of embryos (75%) were unable to proceed beyond the blastula stage. At 48 hpf abnormal blastulae, gastrulae and prepluteus stage embryos occurred (88%).
The high Cd concentration dramatically affected the development of the progeny. At 24 hpf, embryos showed abnormal development with blastomeres defects. Additionally, several embryos did not gastrulate and showed altered archenteron invagination. Moreover, apparently, there were no living embryos in the culture. Indeed, at 48 hpf, embryos completely degenerated.
The exposure of parentals to Zn also caused abnormality in the offspring. However, the percentages of abnormal embryos observed were lower than embryos from Cd conditioned gametes. At 24 hpf the 33% of embryos from Zn (L) conditioned parentals showed problems in gastrulation, such as abnormal endoderm invagination. Moreover, at 48 hpf (pluteus stage in the controls) developmental defects and skeletal malformations were more evident and variable morphologies were obtained. A significant number of embryos showed defects in spicule formation or elongation resembling gastrulae, prisms or plutei with arms shorter than controls. However, the mortality rate remained lower than Cd (L) exposed parentals. As expected, abnormalities were more severe in the offspring of parentals who experienced high Zn dosage either at 24 and 48 hpf.
Combined exposure to Cd and Zn (Mix L) caused significantly higher percentages of abnormalities (83% abnormal blastulae at 24 hpf) with respect to the treatment with Cd or Zn only. Living embryos displayed the worst phenotype, resulting unable to gastrulate and showing abnormal segmentation. Blastomeres were poorly connected and several isolated cells appeared in the background. At 48 hpf, significant increase in degenerated embryos occurred; additionally, shortened plutei with fractured ectoderm and incomplete skeletal rods were also present. Apparently, despite the aberrant phenotypes, the mortality rate was relatively low; conversely, no living embryos could be detected in the offspring of parentals who experienced Mix H.
Overall, parental exposure to metals can have profound consequences on gastrulation timing and embryo morphology.
3.3. Pathway-Centered Gene Expression Profiling
In the light of the recognized roles of precise genes in mechanisms of gene regulation and chromatin remodelling, we performed a pathway-centred gene expression analysis on developmental regulators of skeletogenic (dri, hex, sm50, p16, p19, msp130) and endodermal differentiation (foxa, hox11/13b, wnt8) [10,27], as well as on epigenetic regulators of chromatin accessibility (kat2A, hdac1, ehmt2, phf8 and UBE2a) (Figure 3 and Figure 4). The high deformity rate (higher than 50%) observed among the offspring of parentals who experienced Cd (L and H), the highest dosage of Zn and their combinations prompted us to analyse the transcriptional effects exclusively at the lowest Zn and Mix metal concentration whose low mortality rate do not expect to affect the RT-qPCR results.
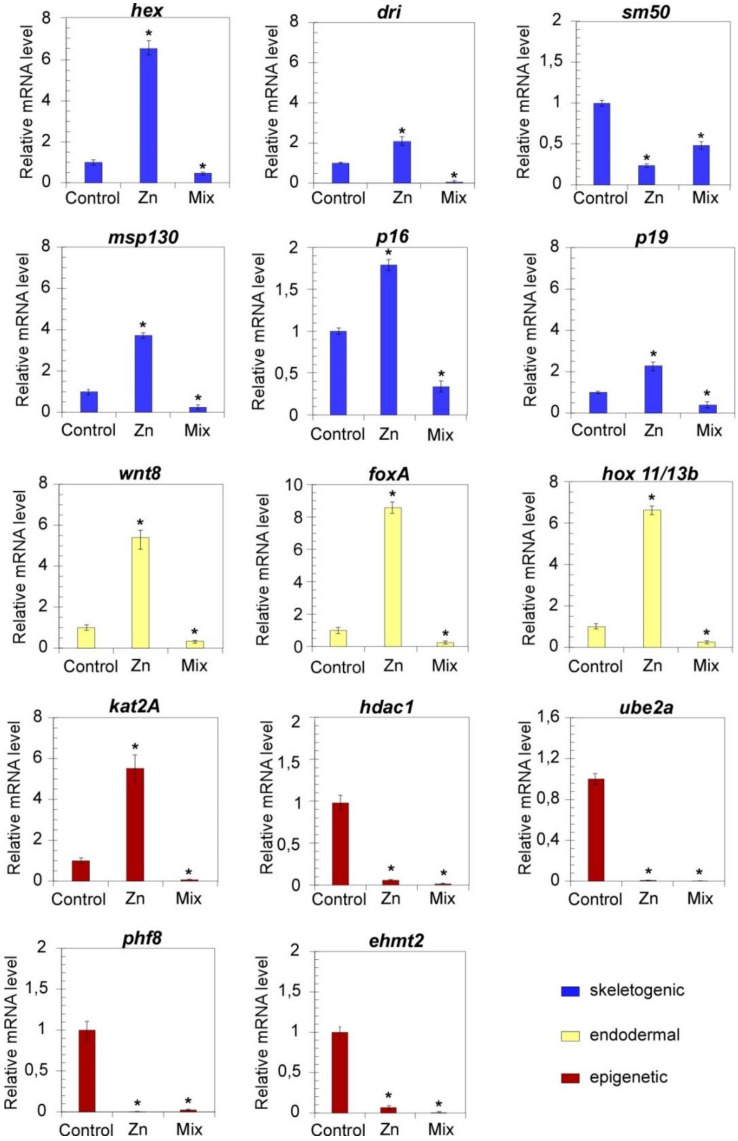
Figure 3.
Parental exposures to low doses of Zn and metal mixture (as defined in Table 1) induce alterations in the mRNA expression of development and epigenetic related genes at 24 hpf. RT-qPCR results showing the mRNA levels of indicated genes in P. lividus offspring, with respect to 18S at 24 hpf. Bars represent mean ± SD. Values were considered statistically significant at p less than 0.05 (*).
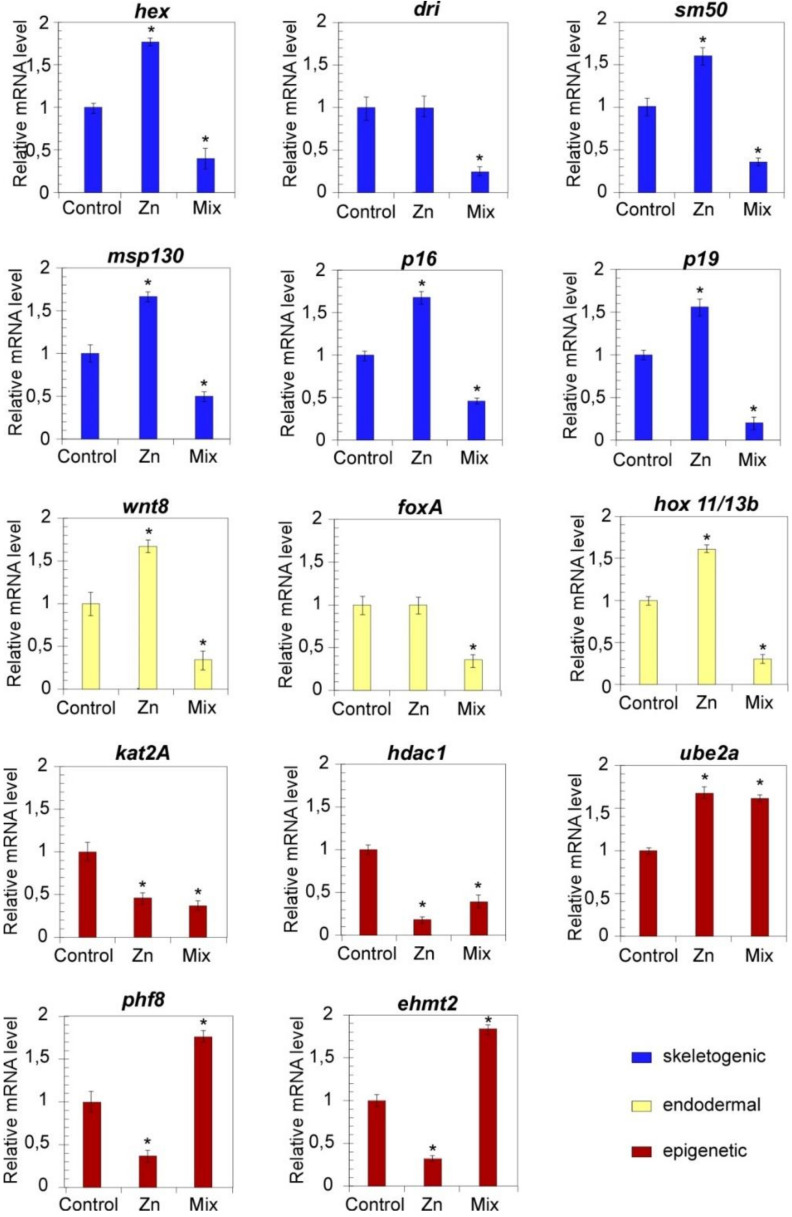
Figure 4.
Parental exposures to low doses of Zn and metal mixture (as defined in Table 1) induce alterations in the mRNA expression of development and epigenetic related genes at 48 hpf. RT-qPCR results showing the mRNA levels of indicated genes in P. lividus offspring, with respect to 18S at 48 hpf. Bars represent mean ± SD. Values were considered statistically significant at p less than 0.05 (*).
3.3.1. Expression Profiles of Development Regulators
In order to unveil possible variations in the transcriptional profile of members of the P. lividus GRNs, the mRNA levels of hex, dri, sm50, msp130, p16, p19, wnt8, foxa and hox11/13b were analysed in embryos (F1) from exposed parentals. The results are shown in Figure 3 (24 hpf) and Figure 4 (48 hpf).
The expression of all the genes was largely affected in embryos from Zn-conditioned parentals and a huge reduction of mRNA levels was measured in response to specific exposures. Except for the downregulation of sm50, the F1 of Zn-conditioned animals showed an overall transcriptional upregulation. Indeed, the RT-qPCR analysis revealed that the mRNA expression levels of the selected genes increased significantly in such embryos at 24 hpf with respect to untreated embryos. Even at 48 hpf, the general expression levels were still higher than controls albeit to a lesser extent, with a few exceptions, including dri and foxA, whose mRNA levels returned similar to controls. The mRNA levels of these genes were also evaluated in the offspring of animals exposed to Mix L. The qPCR analyses revealed that coexposure negatively affected the expression of examined transcripts since all the mRNA resulted downregulated both at 24 and 48 hpf.
3.3.2. Expression Profiles of Transcriptional Regulators
To evaluate the perturbation affecting the mRNA levels of genes involved in mechanisms of chromatin remodelling, the transcriptional profile of kat2A, hdac1, UBE2a, phf8 and ehmt2 was also analysed (Figure 3 and Figure 4).
At 24 hpf (Figure 3), the F1 from Zn conditioned sea urchins showed a significant general reduction in the transcriptional levels of the studied genes. However, upregulation events were also measured, since kat2A increased at 24 hpf (Figure 3), while significantly increased UBE2a mRNA levels were measured at 48 hpf (Figure 4).
To probe the effects of joint metal exposures, the transcriptional levels of this gene panel was also profiled in embryos from adults exposed to Mix L. Once again, a general transcriptional downregulation was observed at 24 hpf. Differently, ehmt2, phf8 and UBE2a resulted upregulated at 48 hpf.
4. Discussion
It has been reported that sea urchins can accumulate metals and they have been used as indicators of environmental pollution [53]. However, data reporting correlation between the environmental level of toxicants and accumulated metals do not comply with this intent. In particular, Cd and/or Zn content in P. lividus gonadal tissue from contaminated areas often does not reflect the environmental levels, as it does not result higher than that found in animals from low anthropic impact areas [54,55,56]. However, the possibility of effects on the offspring has been observed. In order to provide pieces of evidence on the transmission of parental stress to the progeny at the phenotypical and transcriptional level, we evaluated the effects of prolonged and non-lethal metal exposures on P. lividus adult individuals and the related F1. Recently, we have shown that sub-lethal metal exposures dramatically affect the canonical transcriptional profiles in reproductive tissue which, in turn, could alter spermatogenesis and oogenesis [43]. Herein we show that the exposure to Cd, Zn and their mixes induces an increase in ROS production similarly to those reported for different systems [57,58,59,60]; accordingly, catalase activity showed variations, presumably related to its role in the maintenance of the redox homeostasis, especially after Cd exposure. However, despite the increase in ROS levels, CAT activity was reduced in male and female gonads in response to Zn. These results are unusual [61,62,63] and suggest that other enzymes could be involved in the maintenance of the redox homeostasis. In response to mixes, CAT activity appears to reflect the amount of generated ROS so as to cope with the generated level of toxicity. In accordance with previous data on the specific gene expression pattern observed in testis and ovary [43], evidence for gender-specific responses to ROS emerged. Differences in stress susceptibility and tolerance to stress among sexes have been described elsewhere [64]; thus, it could be hypothesised that mechanisms of gonad maturation and gametogenesis may be likely responsible for the different responses.
Beyond this, a reduction in antioxidant activities was also reported in various organisms after metal exposure [65,66]. In this line, it appears that the redox response system of the sea urchin reproductive tissues may result impaired above certain ROS levels in response to experienced levels of toxicity. However, it is also conceivable that the prolonged treatments may result in possible acclimation and tolerance to stressors. In order to assess such an issue, we made efforts in the evaluation of the protein level of HSP60, HSP70 and HSP90. The HSPs exert a protective role as chaperones, by assisting protein folding and preventing their aggregation also in response to stressors including ROS, warming and metals [67,68,69,70,71]. Accordingly, their upregulation confirms the activation of a defence mechanism in the P. lividus gonads, which act to cope with the stress so as to tolerate, at least, the rate of ROS. Moreover, the pattern observed in response to mixes suggests that the coexistence of diverse toxicants simultaneously activating different signalling pathways may result in HSPs profiles different from those arising in response to a single contaminant.
It has been shown that the environmental stress experienced by parentals during gametogenesis may result in phenotypic changes of the offspring [72,73]. In this line, it has been reported that sea urchin maternal exposure to different conditions, including acidification and polycyclic aromatic hydrocarbons, affects the capacity of embryos to counteract oxidative stress conditions [74,75,76].
These findings prompted us to evaluate the possible occurrence of embryo defects originating from Cd and Zn conditioned parentals (both male and female sea urchin adults). Although Cd and Zn parental exposure diversely affected gonads, herein we show that the related offspring displayed significant difficulties in proceeding through the canonical developmental program. Additionally, combined metals exposure dramatically affected the fitness of the progeny, given the higher percentages of abnormalities and developmental failure rates recorded using the higher dosage. It appears that a common effect was a delay in cell differentiation and gastrulation events, probably caused by alterations of timing of specific pathways resulting in the abnormal relative order of events that could disrupt spatial relationships or signalling interactions and cause different aberrant phenotypes.
As a result of such phenotypic alterations, it was intriguing to profile the gene expression pattern of key factors that are responsible for the molecular mechanisms orchestrating the regulatory network of embryo development. It is well-known that accurate control of gene expression is mandatory for normal development. As a general rule, in the P. lividus skeletogenic lineage, the maternally inherited transcription factor Ets1 activates the expression of several genes including alx1, hex and dri which, in turn, cooperate to activate the expression of terminal differentiation genes, including the spicule matric gene sm50, msp130, p16 and p19 [10,27]. It has been reported that several stressors, including metals [77], X-rays [78], decadienal [79] and UVB [80] affect the expression of the skeletogenic genes.
Interestingly, we show that similar changes occurred also in the F1 of exposed animals (Figure 3 and Figure 4). In this line, it is reasonable to suppose that upregulation of dri and hex, as measured in the embryos from Zn exposed parentals, may rely on modifications in the pattern of maternal determinants that normally occur during gametogenesis. According to hex and dri aberrant expression, msp130, p16 and p19 resulted upregulated probably due to an excess of stimuli for gene activation, which may likely result in the observed skeleton formation in embryos. Beyond the amplitude of mRNA levels, it should be noted also the deregulation of sm50 expression timing. Typically, the architecture of a gene regulatory circuit activating sm50 is defined as a coherent feedforward structure based on Hex and Dri [27]. Although the upregulation of dri and hex occurred at 24 hpf a delay in the expression of sm50 was measured. Thus, it is reasonable to hypothesise that the exposure to metals at F0 affects the genetic program of F1 in terms of measured mRNA level and timing of developmental programs.
Similarly, the fine-tuning of endodermal signalling, based on the activation of the wnt8-hox11-13b-foxa node [81], were downregulated, which also explains the observed defects in the endoderm invagination.
It has been reported that several genes of the sea urchin embryo regulatory network are affected by Zn treatment both spatially and quantitatively. Among them, hex, p19 and wnt 8 were upregulated in S. purpuratus treated embryos, thus providing evidence that Zn perturbs the pattern of endomesoderm and skeletogenic signalling [82]. Therefore, it is reasonable to assume that the defects observed in the progeny of Zn-exposed parentals could be explained by the same mechanisms which could exert adverse effects on the regulatory networks of differentiation.
Interestingly, Zn and Cd co-exposure of parentals abrogated the transcriptional induction of all the analysed genes. These data confirm once again that the co-occurrence of different stressors, triggering multiple pathways, may provide gene expression profiles that were not overlapping to those occurring in response to a single contaminant also via an inherited manner.
Recently, several lines of evidence suggested that maternal exposure to different conditions affected the DNA methylation status of the offspring [83,84]. In particular, several genes involved in mechanisms of transcriptional regulation, including DNA modification and metabolism, as well in protein ubiquitination, were identified as differentially methylated in the offspring. Epigenetic regulation through histone modification may be altered by several environmental factors, including metals, that can affect histone and DNA writers and therefore influence the pattern of acetylation/methylation/ubiquitination of histones and DNA methylation [85,86,87,88]. Although these systems were not studied thoroughly in sea urchins, other models, including mouse embryonic stem cells, provide evidence for H3K27me1 reduction following exposure to different metals, including cadmium [89]. Therefore, it is not surprising that the genes involved in chromatin remodelling, as well as epigenetic regulators (kat2A, hdac1, ehmt2, phf8 and UBE2a), showed huge modifications in the expression profiles in the progeny from conditioned parentals, irrespective of the toxicity levels or experienced metals.
Additionally, it should be noted that, despite the transcriptional activation of GRN developmental regulators, yet at 24 hpf, the observed embryo phenotypes do not correlate with the molecular profiles. Thus, it appears that triggered specification and developmental processes are unable to be timely maintained and finalised probably due to the altered expression profiles of epigenetic regulators.
5. Conclusions
In the last years, a great deal of interest has been generated in the characterization of the phenotypic alterations in the offspring from parentals that had experienced environmental stresses, especially during gametogenesis. Herein, we show that F0 conditioning that did not affect the viability of the exposed individuals, significantly impaired the developmental fate of the progeny and altered various regulatory gene networks across a generation. It is reasonable to suppose that changes in P. lividus gonads may modify the expression of specific gene sets required for synthesis and accumulation of maternal determinants which, in turn, impaired the zygotic activation of GRNs responsible for proper embryo development. Further research should address whether maternal or paternal exposures could differentially affect germ cells and thus embryo development. Similarly, comparison of epigenetic landscape among generations and studies on the localisation of maternal determinants should provide novel insights in the fine mechanisms integrating embryo physiology and environment.
Supplementary Materials
The following are available online at https://www.mdpi.com/2079-7737/10/2/103/s1, Figure S1: original Western blot results for HSP70, HSP60, HSP90 expression in testis and ovary. Figure S2: phenotypes of P. lividus embryos from conditioned parentals.
Author Contributions
Conceptualization, A.C., A.N. and M.A.R.; funding acquisition, A.C.; investigation, T.M., G.B. and M.D.N.; methodology, T.M., G.B., M.D.N., C.D.B. and M.M.; supervision, A.N.; validation, M.T., M.A.R., S.C. and A.N.; writing the original draft, A.C., A.N., M.A.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Comitato interministeriale per la programmazione economica (CIPE), Centro Internazionale di Studi avanzati su Ambiente e Salute-CISAS to Angela Cuttitta.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data herein presented are available in this article and Supplementary Material.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Marshall D.J. Transgenerational plasticity in the sea: Context-dependent maternal effects across the life history. Ecology. 2008;89:418–427. doi: 10.1890/07-0449.1. [DOI] [PubMed] [Google Scholar]
- 2.Bonduriansky R., Day T. Nongenetic Inheritance and Its Evolutionary Implications. Annu. Rev. Ecol. Evol. Syst. 2009;40:103–125. doi: 10.1146/annurev.ecolsys.39.110707.173441. [DOI] [Google Scholar]
- 3.Shama L.N.S., Mark F.C., Strobel A., Lokmer A., John U., Mathias Wegner K. Transgenerational effects persist down the maternal line in marine sticklebacks: Gene expression matches physiology in a warming ocean. Evol. Appl. 2016;9:1096–1111. doi: 10.1111/eva.12370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donelson J.M., Munday P.L. Transgenerational plasticity mitigates the impact of global warming to offspring sex ratios. Glob. Chang. Biol. 2015;21:2954–2962. doi: 10.1111/gcb.12912. [DOI] [PubMed] [Google Scholar]
- 5.Goncalves P., Anderson K., Thompson E.L., Melwani A., Parker L.M., Ross P.M., Raftos D.A. Rapid transcriptional acclimation following transgenerational exposure of oysters to ocean acidification. Mol. Ecol. 2016;25:4836–4849. doi: 10.1111/mec.13808. [DOI] [PubMed] [Google Scholar]
- 6.Klosin A., Lehner B. Mechanisms, timescales and principles of trans-generational epigenetic inheritance in animals. Curr. Opin. Genet. Dev. 2016;36:41–49. doi: 10.1016/j.gde.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Munday P.L. Transgenerational acclimation of fishes to climate change and ocean acidification. F1000Prime Rep. 2014;6:99. doi: 10.12703/P6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonduriansky R. Rethinking heredity, again. Trends Ecol. Evol. 2012;27:330–336. doi: 10.1016/j.tree.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Bertoldo M.J., Locatelli Y., O’Neill C., Mermillod P. Impacts of and interactions between environmental stress and epigenetic programming during early embryo development. Reprod. Fertil. Dev. 2015;27:1125–1136. doi: 10.1071/RD14049. [DOI] [PubMed] [Google Scholar]
- 10.Ben-Tabou De-Leon S., Davidson E.H. Gene regulation: Gene control network in development. Annu. Rev. Biophys. Biomol. Struct. 2007;36:191–212. doi: 10.1146/annurev.biophys.35.040405.102002. [DOI] [PubMed] [Google Scholar]
- 11.Davidson E.H. Emerging properties of animal gene regulatory networks. Nature. 2010;468:911–920. doi: 10.1038/nature09645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sultan S.E. Development in context: The timely emergence of eco-devo. Trends Ecol. Evol. 2007;22:575–582. doi: 10.1016/j.tree.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert S.F. Ecological developmental biology: Environmental signals for normal animal development. Evol. Dev. 2012;14:20–28. doi: 10.1111/j.1525-142X.2011.00519.x. [DOI] [PubMed] [Google Scholar]
- 14.Costa S., Nicosia A., Cuttitta A., Gianguzza F., Ragusa M.A. An intronic cis-regulatory element is crucial for the alpha tubulin Pl-Tuba1a gene activation in the ciliary band and animal pole neurogenic domains during sea urchin development. PLoS ONE. 2017;12:e0170969. doi: 10.1371/journal.pone.0170969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howard-Ashby M., Materna S.C., Brown C.T., Chen L., Cameron R.A., Davidson E.H. Gene families encoding transcription factors expressed in early development of Strongylocentrotus purpuratus. Dev. Biol. 2006;300:90–107. doi: 10.1016/j.ydbio.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 16.Briggs E., Wessel G.M. In the beginning... Animal fertilization and sea urchin development. Dev. Biol. 2006;300:15–26. doi: 10.1016/j.ydbio.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 17.Knudson A.G. Of sea urchins and worms: Development and cancer. Cell Death Differ. 2004;11:11–12. doi: 10.1038/sj.cdd.4401295. [DOI] [PubMed] [Google Scholar]
- 18.Ragusa M.A., Nicosia A., Costa S., Casano C., Gianguzza F. A Survey on Tubulin and Arginine Methyltransferase Families Sheds Light on P. lividus Embryo as Model System for Antiproliferative Drug Development. Int. J. Mol. Sci. 2019;20:2136. doi: 10.3390/ijms20092136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davidson E. The Regulatory Genome. Elsevier Inc.; Amsterdam, The Netherlands: 2006. [Google Scholar]
- 20.Hofmann G.E., Todgham A.E. Living in the Now: Physiological Mechanisms to Tolerate a Rapidly Changing Environment. Annu. Rev. Physiol. 2010;72:127–145. doi: 10.1146/annurev-physiol-021909-135900. [DOI] [PubMed] [Google Scholar]
- 21.Dupont S., Ortega-Martínez O., Thorndyke M. Impact of near-future ocean acidification on echinoderms. Ecotoxicology. 2010;19:449–462. doi: 10.1007/s10646-010-0463-6. [DOI] [PubMed] [Google Scholar]
- 22.Ragusa M.A., Costa S., Cuttitta A., Gianguzza F., Nicosia A. Coexposure to sulfamethoxazole and cadmium impairs development and attenuates transcriptional response in sea urchin embryo. Chemosphere. 2017;180:275–284. doi: 10.1016/j.chemosphere.2017.04.030. [DOI] [PubMed] [Google Scholar]
- 23.Ragusa M., Nicosia A., Costa S., Cuttitta A., Gianguzza F. Metallothionein Gene Family in the Sea Urchin Paracentrotus lividus: Gene Structure, Differential Expression and Phylogenetic Analysis. Int. J. Mol. Sci. 2017;18:812. doi: 10.3390/ijms18040812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matranga V., Pinsino A., Randazzo D., Giallongo A., Dubois P. Long-term environmental exposure to metals (Cu, Cd, Pb, Zn) activates the immune cell stress response in the common European sea star (Asterias rubens) Mar. Environ. Res. 2012;76:122–127. doi: 10.1016/j.marenvres.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 25.Peter I.S., Davidson E.H. Assessing regulatory information in developmental gene regulatory networks. Proc. Natl. Acad. Sci. USA. 2017;114:5862–5869. doi: 10.1073/pnas.1610616114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garfield D.A., Runcie D.E., Babbitt C.C., Haygood R., Nielsen W.J., Wray G.A. The Impact of Gene Expression Variation on the Robustness and Evolvability of a Developmental Gene Regulatory Network. PLoS Biol. 2013;11:e1001696. doi: 10.1371/journal.pbio.1001696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gildor T., De-Leon S.B.-T. Comparative Study of Regulatory Circuits in Two Sea Urchin Species Reveals Tight Control of Timing and High Conservation of Expression Dynamics. PLoS Genet. 2015;11:e1005435. doi: 10.1371/journal.pgen.1005435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henikoff S., Greally J.M. Epigenetics, cellular memory and gene regulation. Curr. Biol. 2016;26:R644–R648. doi: 10.1016/j.cub.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 29.John R.M., Rougeulle C. Developmental epigenetics: Phenotype and the flexible epigenome. Front. Cell Dev. Biol. 2018;6:130. doi: 10.3389/fcell.2018.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J., Wang Y., Wang C., Hu J.F., Li W. LncRNA Functions as a New Emerging Epigenetic Factor in Determining the Fate of Stem Cells. Front. Genet. 2020;11:277. doi: 10.3389/fgene.2020.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lyko F. The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 2018;19:81–92. doi: 10.1038/nrg.2017.80. [DOI] [PubMed] [Google Scholar]
- 32.Duboc V., Röttinger E., Lapraz F., Besnardeau L., Lepage T. Left-right asymmetry in the sea urchin embryo is regulated by nodal signaling on the right side. Dev. Cell. 2005;9:147–158. doi: 10.1016/j.devcel.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 33.Bessodes N., Haillot E., Duboc V., Röttinger E., Lahaye F., Lepage T. Reciprocal Signaling between the Ectoderm and a Mesendodermal Left-Right Organizer Directs Left-Right Determination in the Sea Urchin Embryo. PLoS Genet. 2012;8:e1003121. doi: 10.1371/journal.pgen.1003121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lapraz F., Haillot E., Lepage T. A deuterostome origin of the Spemann organiser suggested by Nodal and ADMPs functions in Echinoderms. Nat. Commun. 2015;6:1–20. doi: 10.1038/ncomms9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anello L., Cavalieri V., Di Bernardo M. Developmental effects of the protein kinase inhibitor kenpaullone on the sea urchin embryo. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2018;204:36–44. doi: 10.1016/j.cbpc.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 36.Turturici G., La Fiora V., Terenzi A., Barone G., Cavalieri V. Perturbation of Developmental Regulatory Gene Expression by a G-Quadruplex DNA Inducer in the Sea Urchin Embryo. Biochemistry. 2018;57:4391–4394. doi: 10.1021/acs.biochem.8b00551. [DOI] [PubMed] [Google Scholar]
- 37.Boudouresque F.C., Verlaque M. Chapter 13 Ecology of Paracentrotus lividus. Dev. Aquac. Fish. Sci. 2007;37:243–285. doi: 10.1016/S0167-9309(07)80077-9. [DOI] [Google Scholar]
- 38.Evans T.G., Chan F., Menge B.A., Hofmann G.E. Transcriptomic responses to ocean acidification in larval sea urchins from a naturally variable pH environment. Mol. Ecol. 2013;22:1609–1625. doi: 10.1111/mec.12188. [DOI] [PubMed] [Google Scholar]
- 39.Evans T.G., Pespeni M.H., Hofmann G.E., Palumbi S.R., Sanford E. Transcriptomic responses to seawater acidification among sea urchin populations inhabiting a natural pH mosaic. Mol. Ecol. 2017;26:2257–2275. doi: 10.1111/mec.14038. [DOI] [PubMed] [Google Scholar]
- 40.Migliaccio O., Castellano I., Cirino P., Romano G., Palumbo A. Maternal exposure to cadmium and manganese impairs reproduction and progeny fitness in the sea urchin Paracentrotus lividus. PLoS ONE. 2015;10:e0131815. doi: 10.1371/journal.pone.0131815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong J.M., Johnson K.M., Kelly M.W., Hofmann G.E. Transcriptomics reveal transgenerational effects in purple sea urchin embryos: Adult acclimation to upwelling conditions alters the response of their progeny to differential pCO2 levels. Mol. Ecol. 2018;27:1120–1137. doi: 10.1111/mec.14503. [DOI] [PubMed] [Google Scholar]
- 42.Strader M.E., Wong J.M., Hofmann G.E. Ocean acidification promotes broad transcriptomic responses in marine metazoans: A literature survey. Front. Zool. 2020;17:1–23. doi: 10.1186/s12983-020-0350-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di Natale M., Bennici C., Biondo G., Masullo T., Monastero C., Tagliavia M., Torri M., Costa S., Ragusa M.A., Cuttitta A., et al. Aberrant gene expression profiles in Mediterranean sea urchin reproductive tissues after metal exposures. Chemosphere. 2019;216:48–58. doi: 10.1016/j.chemosphere.2018.10.137. [DOI] [PubMed] [Google Scholar]
- 44.Zhao C., Zhang L., Shi D., Ding J., Yin D., Sun J., Zhang B., Zhang L., Chang Y. Transgenerational effects of ocean warming on the sea urchin Strongylocentrotus intermedius. Ecotoxicol. Environ. Saf. 2018;151:212–219. doi: 10.1016/j.ecoenv.2018.01.014. [DOI] [PubMed] [Google Scholar]
- 45.Varotto L., Domeneghetti S., Rosani U., Manfrin C., Cajaraville M.P., Raccanelli S., Pallavicini A., Venier P. DNA Damage and Transcriptional Changes in the Gills of Mytilus galloprovincialis Exposed to Nanomolar Doses of Combined Metal Salts (Cd, Cu, Hg) PLoS ONE. 2013;8:e54602. doi: 10.1371/journal.pone.0054602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martínez-Soto M.C., Tovar-Sánchez A., Sánchez-Quiles D., Rodellas V., Garcia-Orellana J., Basterretxea G. Seasonal variation and sources of dissolved trace metals in Maó Harbour, Minorca Island. Sci. Total Environ. 2016;565:191–199. doi: 10.1016/j.scitotenv.2016.03.244. [DOI] [PubMed] [Google Scholar]
- 47.Wah Chu K., Chow K.L. Synergistic toxicity of multiple heavy metals is revealed by a biological assay using a nematode and its transgenic derivative. Aquat. Toxicol. 2002;61:53–64. doi: 10.1016/S0166-445X(02)00017-6. [DOI] [PubMed] [Google Scholar]
- 48.Stamateris R.E., Rafiq K., Ettensohn C.A. The expression and distribution of Wnt and Wnt receptor mRNAs during early sea urchin development. Gene Expr. Patterns. 2010;10:60–64. doi: 10.1016/j.gep.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 49.Ragusa M.A., Costa S., Gianguzza M., Roccheri M.C., Gianguzza F. Effects of cadmium exposure on sea urchin development assessed by SSH and RT-qPCR: Metallothionein genes and their differential induction. Mol. Biol. Rep. 2013;40:2157–2167. doi: 10.1007/s11033-012-2275-7. [DOI] [PubMed] [Google Scholar]
- 50.Vergara-Amado J., Silva A.X., Manzi C., Nespolo R.F., Cárdenas L. Differential expression of stress candidate genes for thermal tolerance in the sea urchin Loxechinus albus. J. Therm. Biol. 2017;68:104–109. doi: 10.1016/j.jtherbio.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 51.Nicosia A., Bennici C., Biondo G., Costa S., Di Natale M., Masullo T., Monastero C., Ragusa M.A., Tagliavia M., Cuttitta A. Characterization of Translationally Controlled Tumour Protein from the Sea Anemone Anemonia viridis and Transcriptome Wide Identification of Cnidarian Homologues. Genes. 2018;9:30. doi: 10.3390/genes9010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 53.Warnau M., Biondo R., Temara A., Bouquegneau J.M., Jangoux M., Dubois P. Distribution of heavy metals in the echinoid Paracentrotus lividus from the Mediterranean Posidonia oceanica ecosystem: Seasonal and geographical variations. J. Sea Res. 1998;39:267–280. doi: 10.1016/S1385-1101(97)00064-6. [DOI] [Google Scholar]
- 54.Mostafa M.H., Collins K.J. Heavy Metal Concentrations in Sea Urchin Tissues From Egypt, Ireland and United Kingdom. Chem. Ecol. 1995;10:181–190. doi: 10.1080/02757549508035340. [DOI] [Google Scholar]
- 55.Soualili D., Dubois P., Gosselin P., Pernet P., Guillou M. Assessment of seawater pollution by heavy metals in the neighbourhood of Algiers: Use of the sea urchin, Paracentrotus lividus, as a bioindicator. ICES J. Mar. Sci. 2008;65:132–139. doi: 10.1093/icesjms/fsm183. [DOI] [Google Scholar]
- 56.La Torre G.L., Cicero N., Bartolomeo G., Rando R., Vadalà R., Santini A., Durazzo A., Lucarini M., Dugo G., Salvo A. Assessment and Monitoring of Fish Quality from a Coastal Ecosystem under High Anthropic Pressure: A Case Study in Southern Italy. Int. J. Environ. Res. Public Health. 2020;17:3285. doi: 10.3390/ijerph17093285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Du C., Anderson A., Lortie M., Parsons R., Bodnarn A. Oxidative damage and cellular defense mechanisms in sea urchin models of aging. Free Radic. Biol. Med. 2013;63:254–263. doi: 10.1016/j.freeradbiomed.2013.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng J.L., Zeng L., Xu M.Y., Shen B., Wu C.W. Different effects of low- and high-dose waterborne zinc on Zn accumulation, ROS levels, oxidative damage and antioxidant responses in the liver of large yellow croaker Pseudosciaena crocea. Fish. Physiol. Biochem. 2017;43:153–163. doi: 10.1007/s10695-016-0275-6. [DOI] [PubMed] [Google Scholar]
- 59.Viarengo A., Canesi L., Pertica M., Poli G., Moore M.N., Orunesu M. Heavy metal effects on lipid peroxidation in the tissues of mytilus gallopro vincialis lam. Comp. Biochem. Physiol. Part C Comp. 1990;97:37–42. doi: 10.1016/0742-8413(90)90168-9. [DOI] [Google Scholar]
- 60.Bishop G.M., Dringen R., Robinson S.R. Zinc stimulates the production of toxic reactive oxygen species (ROS) and inhibits glutathione reductase in astrocytes. Free Radic. Biol. Med. 2007;42:1222–1230. doi: 10.1016/j.freeradbiomed.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 61.Livingstone D.R. Contaminant-stimulated reactive oxygen species production and oxidative damage in aquatic organisms. Mar. Pollut. Bull. 2001;42:656–666. doi: 10.1016/S0025-326X(01)00060-1. [DOI] [PubMed] [Google Scholar]
- 62.Winston G.W., Di Giulio R.T. Prooxidant and antioxidant mechanisms in aquatic organisms. Aquat. Toxicol. 1991;19:137–161. doi: 10.1016/0166-445X(91)90033-6. [DOI] [Google Scholar]
- 63.Nicosia A., Salamone M., Mazzola S., Cuttitta A. Transcriptional and biochemical effects of cadmium and manganese on the defense system of octopus vulgaris paralarvae. Biomed. Res. Int. 2015;2015 doi: 10.1155/2015/437328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schäfer S., Abele D., Weihe E., Köhler A. Sex-specific biochemical and histological differences in gonads of sea urchins (Psammechinus miliaris) and their response to phenanthrene exposure. Mar. Environ. Res. 2011;71:70–78. doi: 10.1016/j.marenvres.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 65.Peters L.D., Livingstone D.R. Antioxidant enzyme activities in embryologic and early larval stages of turbot. J. Fish. Biol. 1996;49:986–997. doi: 10.1111/j.1095-8649.1996.tb00095.x. [DOI] [Google Scholar]
- 66.Cao L., Huang W., Liu J., Yin X., Dou S. Accumulation and oxidative stress biomarkers in Japanese flounder larvae and juveniles under chronic cadmium exposure. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2010;151:386–392. doi: 10.1016/j.cbpc.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 67.Roccheri M.C., Agnello M., Bonaventura R., Matranga V. Cadmium induces the expression of specific stress proteins in sea urchin embryos. Biochem. Biophys. Res. Commun. 2004;321:80–87. doi: 10.1016/j.bbrc.2004.06.108. [DOI] [PubMed] [Google Scholar]
- 68.Clayton M.E., Steinmann R., Fent K. Different expression patterns of heat shock proteins hsp 60 and hsp 70 in zebra mussels (Dreissena polymorpha) exposed to copper and tributyltin. Aquat. Toxicol. 2000;47:213–226. doi: 10.1016/S0166-445X(99)00022-3. [DOI] [Google Scholar]
- 69.Gupta S.C., Sharma A., Mishra M., Mishra R.K., Chowdhuri D.K. Heat shock proteins in toxicology: How close and how far? Life Sci. 2010;86:377–384. doi: 10.1016/j.lfs.2009.12.015. [DOI] [PubMed] [Google Scholar]
- 70.Navarro A., Faria M., Barata C., Pina B. Transcriptional response of stress genes to metal exposure in zebra mussel larvae and adults. Environ. Pollut. 2011;159:100–107. doi: 10.1016/j.envpol.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 71.Harianto J., Nguyen H.D., Holmes S.P., Byrne M. The effect of warming on mortality, metabolic rate, heat-shock protein response and gonad growth in thermally acclimated sea urchins (Heliocidaris erythrogramma) Mar. Biol. 2018;165:96. doi: 10.1007/s00227-018-3353-8. [DOI] [Google Scholar]
- 72.Hamdoun A., Epel D. Embryo stability and vulnerability in an always changing world. Proc. Natl. Acad. Sci. USA. 2007;104:1745–1750. doi: 10.1073/pnas.0610108104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Byrne M. Climate Alert: Climate Change Monitoring and Strategy. University of Sydney Press; Sydney, Australia: 2010. Impact of Climate Change Stressors on Marine Invertebrate Life Histories with a Focus on the Mollusca and Echinodermata; pp. 142–185. [Google Scholar]
- 74.Lister K.N., Lamare M.D., Burritt D.J. Dietary pollutants induce oxidative stress, altering maternal antioxidant provisioning and reproductive output in the temperate sea urchin Evechinus chloroticus. Aquat. Toxicol. 2016;177:106–115. doi: 10.1016/j.aquatox.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 75.Lister K.N., Lamare M.D., Burritt D.J. Maternal antioxidant provisioning mitigates pollutant-induced oxidative damage in embryos of the temperate sea urchin Evechinus chloroticus. Sci. Rep. 2017;7:1–10. doi: 10.1038/s41598-017-02077-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Clark M.S., Suckling C.C., Cavallo A., Mackenzie C.L., Thorne M.A.S., Davies A.J., Peck L.S. Molecular mechanisms underpinning transgenerational plasticity in the green sea urchin Psammechinus miliaris. Sci. Rep. 2019;9:1–12. doi: 10.1038/s41598-018-37255-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Morroni L., Sartori D., Costantini M., Genovesi L., Magliocco T., Ruocco N., Buttino I. First molecular evidence of the toxicogenetic effects of copper on sea urchin Paracentrotus lividus embryo development. Water Res. 2019;160:415–423. doi: 10.1016/j.watres.2019.05.062. [DOI] [PubMed] [Google Scholar]
- 78.Matranga V., Zito F., Costa C., Bonaventura R., Giarrusso S., Celi F. Embryonic development and skeletogenic gene expression affected by X-rays in the Mediterranean sea urchin Paracentrotus lividus. Ecotoxicology. 2010;19:530–537. doi: 10.1007/s10646-009-0444-9. [DOI] [PubMed] [Google Scholar]
- 79.Varrella S., Romano G., Costantini S., Ruocco N., Ianora A., Bentley M.G., Costantini M. Toxic Diatom Aldehydes Affect Defence Gene Networks in Sea Urchins. PLoS ONE. 2016;11:e0149734. doi: 10.1371/journal.pone.0149734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bonaventura R., Poma V., Costa C., Matranga V. UVB radiation prevents skeleton growth and stimulates the expression of stress markers in sea urchin embryos. Biochem. Biophys. Res. Commun. 2005;328:150–157. doi: 10.1016/j.bbrc.2004.12.161. [DOI] [PubMed] [Google Scholar]
- 81.De-Leon S.B.T., Davidson E.H. Information processing at the foxa node of the sea urchin endomesoderm specification network. Proc. Natl. Acad. Sci. USA. 2010;107:10103–10108. doi: 10.1073/pnas.1004824107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Poustka A.J., Kühn A., Groth D., Weise V., Yaguchi S., Burke R.D., Herwig R., Lehrach H., Panopoulou G. A global view of gene expression in lithium and zinc treated sea urchin embryos: New components of gene regulatory networks. Genome Biol. 2007;8:R85. doi: 10.1186/gb-2007-8-5-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Strader M.E., Kozal L.C., Leach T.S., Wong J.M., Chamorro J.D., Housh M.J., Hofmann G.E. Examining the Role of DNA Methylation in Transcriptomic Plasticity of Early Stage Sea Urchins: Developmental and Maternal Effects in a Kelp Forest Herbivore. Front. Mar. Sci. 2020;7:205. doi: 10.3389/fmars.2020.00205. [DOI] [Google Scholar]
- 84.Strader M.E., Wong J., Kozal L., Leach T., Hofmann G. Parental environments alter DNA methylation in offspring of the purple sea urchin, Strongylocentrotus purpuratus. J. Exp. Mar. Biol. Ecol. 2019;517:54–64. doi: 10.1016/j.jembe.2019.03.002. [DOI] [Google Scholar]
- 85.Dai H., Wang Z. Histone Modification Patterns and Their Responses to Environment. Curr. Environ. Health Rep. 2014;1:11–21. doi: 10.1007/s40572-013-0008-2. [DOI] [Google Scholar]
- 86.Martinez-Zamudio R., Ha H.C. Environmental epigenetics in metal exposure. Epigenetics. 2011;6:820–827. doi: 10.4161/epi.6.7.16250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Genchi G., Sinicropi M.S., Lauria G., Carocci A., Catalano A. The Effects of Cadmium Toxicity. Int. J. Environ. Res. Public Health. 2020;17:3782. doi: 10.3390/ijerph17113782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cavalieri V., Spinelli G. Environmental epigenetics in zebrafish. Epigenet. Chromatin. 2017;10:1–11. doi: 10.1186/s13072-017-0154-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gadhia S.R., Calabro A.R., Barile F.A. Trace metals alter DNA repair and histone modification pathways concurrently in mouse embryonic stem cells. Toxicol. Lett. 2012;212:169–179. doi: 10.1016/j.toxlet.2012.05.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data herein presented are available in this article and Supplementary Material.