Abstract
CRISPR-based base editors (BEs) are widely used to induce nucleotide substitutions in living cells and organisms without causing the damaging DNA double-strand breaks and DNA donor templates. Cytosine BEs that induce C:G to T:A conversion and adenine BEs that induce A:T to G:C conversion have been developed. Various attempts have been made to increase the efficiency of both BEs; however, their activities need to be improved for further applications. Here, we describe a fluorescent reporter-based drug screening platform to identify novel chemicals with the goal of improving adenine base editing efficiency. The reporter system revealed that histone deacetylase inhibitors, particularly romidepsin, enhanced base editing efficiencies by up to 4.9-fold by increasing the expression levels of proteins and target accessibility. The results support the use of romidepsin as a viable option to improve base editing efficiency in biomedical research and therapeutic genome engineering.
INTRODUCTION
Clustered regularly interspaced short palindromic repeats (CRISPR)-based base editors (BEs) are widely used and revolutionary tools in biomedical research. BEs enable nucleotide substitution in living cells and organisms without DNA double-strand breaks and DNA donor templates (1). Two types of BEs have been developed; Cytosine BEs (CBEs) induce the C:G to T:A conversion and adenine BEs (ABEs) induce the A:T to G:C conversion. CBEs are formed by fusing a cytidine deaminase, such as APOBEC1, AID and CDA1, with a catalytically inactive Cas9 (2–5). Improved CBEs, such as BE3 and BE4, which inhibit uracil N-glycosylase via uracil glycosylase inhibitor and nick non-targeting DNA strands via Cas9 D10A nickase, increase cytosine base editing efficiencies (2,6). ABEs comprise Cas9 D10A nickase and an evolved Escherichia coli tRNA adenosine deaminase, which can target adenine in single strand DNAs (7). Engineered CBEs and ABEs that increase protein expression by codon optimization and nuclear targeting also improve base editing efficiencies (8,9).
Small molecules can improve the efficiency of Cas9-mediated genome editing by modulating DNA repair pathways. Inhibitors of the non-homologous end-joining pathway, such as Scr7 and NU7441, increase the efficiency of Cas9-mediated homology-directed repair (10–12). RS-1 is a stimulator of RAD51 that plays a major role in homologous recombination, and it also improves Cas9-mediated homology-directed repair (13,14). Inhibitions of the ATM and ATR signaling pathways using VE-822 and AZD-7762 enhance both knockout and knockin efficiencies of CRISPR–Cas12a in human pluripotent cells (15). These approaches using small molecules have great applications in biomedical research. However, no previous studies, to the best of our knowledge, have sought to improve the efficiency of CRISPR-based base editing.
We hypothesized that the inhibition of certain endogenous protein functions could affect and improve BE-mediated base editing efficiencies. Based on this hypothesis, we developed a fluorescent reporter-based drug screening platform to identify novel chemicals to improve adenine base editing efficiencies. Using the reporter system, we found that histone deacetylase (HDAC) inhibitors improved base editing efficiencies by increasing both the expression levels of proteins and target accessibility. Furthermore, HDAC inhibitors increased the genome editing efficiencies of various types of genome editing tools, including Cas9, CBE and recently improved BEs in human cells.
MATERIALS AND METHODS
Construction of plasmid DNA
The coding sequences of Cas9 in pCW-Cas9 (Addgene plasmid #50661) were exchanged by those of ABE7.10 in pCMV-ABE7.10 (Addgene plasmid #102919) to construct a plasmid DNA encoding reverse tetracycline-controlled transactivator (rt-TA)-dependent doxycycline-inducible ABE7.10 (i.e. pCW-ABE7.10). A plasmid DNA for the GFP reporter system, designated pLX- pLX-ΔEGFP-gRNA, was made by cloning the U6-gRNA and CMV-EGFP cassettes into a lentiviral vector and the artificial stop codon TAG was inserted into the third amino acid of EGFP. The gRNAs were cloned into a pRG2 (Addgene plasmid #104174) vector, and the target sequences are listed in Supplementary Table S1. p3s-Cas9HC (Addgene plasmid #43945), pCMV-BE3 (Addgene plasmid #73021), pCMV-ABE7.10 (Addgene plasmid #102919), pCMV-ABEmax (Addgene plasmid #112095), pCMV-BE4max (Addgene plasmid #112093), pCMV-ABEmax-P2A-GFP (Addgene plasmid #112101), and pCMV-BE4max-P2A-GFP (Addgene plasmid #112099) were used in the plasmid DNA transfection studies. To make GFP fusion proteins of Cas9, BE3 and ABE7.10, P2A-EGFP coding sequences were cloned before the DNA stop codons of the aforementioned plasmids using Gibson Assembly Master Mix (New England BioLabs). The sequence of two promoters, EFS (from Lenti-Cas9-Blast, Addgene plasmid #52962) and hPGK (from pCW-Cas9, Addgene plasmid #50661), were amplified and subcloned into an pCMV-ABE7.10-P2A-EGFP plasmid using Gibson Assembly Master Mix (New England BioLabs). Constructs made in this study have been deposited at Addgene. Their nucleotide sequences are summarized in Supplementary sequence 1.
Mammalian cell culture and generation of HAP1-ABEdox:GFP reporter cell lines
HEK293T/17 (ATCC CRL-11268) and HeLa (ATCC CCL-2) cells were maintained in Dulbecco's modified Eagle's medium and HAP1 cells were maintained in Iscove's modified Dulbecco's medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (Welgene). HAP1-ABEdox:GFP reporter cell lines were generated using two consecutive lentiviruses infections. First, to make the rt-TA dependent HAP1-ABEdox cell lines, 1 × 106 HEK293T/17 cells were seeded in the wells of six-well plates and transfected with a total of 5 μg of plasmid DNA (2.5 μg pCW-ABE7.10, 1.5 μg psPAX2, and 1 μg pMD2.G) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. The culture medium was changed 24 h after transfection, and lentiviruses were harvested and filtered using a 0.45 μM filter 48 h after transfection. Several different amounts of lentivirus supernatant were transduced into the HAP1 cells, and 2 μg/ml puromycin treatment was used 24 h after transduction to select the infected cells. The cell viability of each lentivirus condition was measured 48 h after puromycin selection and compared to that of cells without puromycin selection. Cells of lentivirus conditions with approximately 10% viability were selected to ensure that the ABEdox cassette was stably integrated into the cells with a low multiplicity of infection. Single clones of that HAP1-ABEdox cell lines were isolated as previously described (16). To make HAP1-ABEdox:GFP reporter cell lines, lentiviruses were generated using the pLX-ΔEGFP-gRNA vector as described above. Several different amounts of lentivirus supernatant were transduced in HAP1-ABEdox clone #2 (Supplementary Figure S1A), followed by treatment with 10 μg/ml blasticidin 24 h after transduction. Cell viability was measured 48 h after blasticidin selection, and cells with viabilities similar to cells without blasticidin selection were selected for further optimization (Supplementary Figure S1B) and chemical screening.
Generation of HDAC1 and HDAC2 knockdown cell lines
We used pLKO_TRC001 (Addgene plasmid #10878) vector to generate plasmid DNA encoding HDAC1 (TRCN0000195467; target sequences: 5′-CGGTTAGGTTGCTTCAATCTA-3′) or HDAC2 (TRCN0000196590; target sequence: 5′-GACGGTATCATTCCATAAATA-3′)-targeting shRNA. To package each shRNA-encoding lentivirus, 1 × 106 HEK293T/17 cells were seeded in wells of a six-well plate 24 h before transfection, and a total of 5 μg (2.5 μg shRNA-expressing vector, 1.5 μg psPAX2 and 1 μg pMD2.G) plasmid DNA was transfected using Lipofectamine 2000 according to the manufacturer's protocol. The culture medium was changed 24 h after transfection, and lentiviruses were harvested and filtered 48 h after transfection. To generate HDAC1 or HDAC2 knockdown cell lines, 1 × 106 HEK293T/17 cells were infected with each lentivirus and 1 μg/ml puromycin was added to select shRNA-infected cells 24 h after transduction. To verify if HDAC1 or HDAC2 knockdown was successful, cells were harvested, and the protein expression levels were measured using western blotting.
Drug screening
In total, 414 compounds with anti-cancer properties (Selleckchem L3000) were screened. HAP1-ABEdox:GFP reporter cells were seeded at a density of 2 × 104 per well in a 96-well Bio-One 655090 microplate (Greiner) with 0.5 μg/ml doxycycline. After 1 h of incubation, 10 μl of the test compound was added to each well using a Janus liquid handler (PerkinElmer) to generate final concentrations of 100 and 500 nM. Since the potency of each compound varied, we chose two concentrations (100 and 500 nM) for the initial screening as per the manufacturer's instructions and other studies (17,18), and the concentrations of some candidate drugs was further optimized. The plates were incubated for 48 h and the cells were stained with Hoechst33342 dye. GFP expression levels were measured by capturing and analyzing the cell images with the Operetta High Contents Screening system (PerkinElmer). The hit compounds were selected according to the fold-change in GFP expression. Drug screens were performed with appropriate multiple controls (eight wells for dimethylsulfoxide [DMSO] as a negative control and eight wells for doxycycline as positive controls per plate). The mean of the Z-prime factor was 0.52 in the 100 nM screen and 0.09 in the 500 nM screen. The highest DMSO concentration in the screening was no more than 0.5%. All compounds were assessed using biological replicates.
Cas9 and ABE7.10 protein purification
Cas9 and ABE7.10 proteins were purified as described previously (19). Briefly, a plasmid encoding the His6-Cas9 or His6-ABE7.10 was used to transform BL21 Star (DE3)-competent Escherichia coli cells. A fresh colony was inoculated and cultured overnight in Luria–Bertani (LB) broth containing 50 μg/ml kanamycin with shaking at 37°C. Ten milliliter aliquots of overnight cultures of the cells were inoculated into 400 ml of LB broth containing 50 μg/ml kanamycin and cultured at 37°C until the optical density at 600 nm (OD600) reached approximately 0.65–0.70. Cells were cooled on ice, supplemented with 1 mM isopropyl β-d-1-thiogalactopyranoside, and cultured for 16 h with shaking at 18°C. For protein purification, cells were harvested using centrifugation at 6000 × g for 10 min at 4°C and resuspended in lysis buffer (50 mM NaH2PO4, 500 mM NaCl, 10 mM imidazole, 1% Triton X-100, 20% glycerol, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride, 1 mg/ml lysozyme, and 10 μM ZnCl2; pH 8.0). The cells were lysed using sonication for 9 min, and the soluble lysates were obtained using centrifugation at 15 000 × g for 20 min. The cleared lysates were applied to a nickel column (Ni-NTA agarose, Qiagen), washed, and eluted with 250 mM imidazole. For further purification, the eluted ABE7.10 protein solution was loaded onto a polypropylene column containing heparin agarose beads (Heparin Sepharose 6 Fast Flow, GE Healthcare) and washed. Bound ABE7.10 protein was eluted with elution buffer (50 mM NaH2PO4, 750 mM NaCl, 20% glycerol, 1 mM DTT, 10 μM ZnCl2; pH 8.0) and concentrated by Amicon ultra-centrifugal filter (Merck).
Transfection and drug treatments
To measure the base editing efficiency at endogenous sites, 1.6 × 105 HEK293T/17 cells or 8 × 104 HeLa cells were seeded in wells of 24-well plates one day before transfection. When confluency was 70%–80%, the cells were transfected with plasmids encoding BEs (ABE7.10, BE3, ABEmax and BE4max; 1.5 μg) or Cas9 (0.5 μg) and a plasmid encoding gRNA (500 ng) using Lipofectamine 2000 (3 μl for BEs and 2 μl for Cas9) as previously described (19). For the delivery of ribonucleoprotein (RNP), 10 μg of Cas9 or ABE7.10 proteins and 6 μg of synthetic gRNA (Integrated DNA Technologies) were incubated at 25°C to form the RNP complex. The RNP complex and HEK293T/17 cells (1.5 × 106) were resuspended in Neon electroporation R buffer and delivered using the Neon transfection system (Thermo Fisher Scientific) in two 20-ms pulses of 1300 V according to the manufacturer's protocol. Then, 5–10 nM of romidepsin (Selleckchem), abexinostat (Selleckchem), quisinostat (Selleckchem), 100 nM of trichostatin A (TSA, Sigma-Aldrich) and 0.5 μM of vorinostat (Sigma-Aldrich) were applied 6 h after plasmid DNA transfection or 1 h after RNP delivery at designated concentrations. gDNA was extracted 72 h after transfection using the DNeasy Blood and Tissue kit (Qiagen) according to the manufacturer's protocol. The base editing efficiency was determined as described in the next subsection.
Targeted deep sequencing and base editing efficiency analysis
The target sites were amplified using Phusion High-Fidelity DNA Polymerase (New England Biolabs) with appropriate primers for each target site. The amplicons were again amplified using TruSeq HT Dual index-containing primers to generate next-generation sequencing libraries. The libraries were subjected to paired-end Illumina Miniseq and iSeq 100 sequencing system with paired-end sequencing systems. The paired-end reads were joined using fastq-join tool (https://github.com/brwnj/fastq-join). The base editing efficiency was calculated as reads of base edited within the editing window divided by total reads using MAUND (https://github.com/ibs-cge/maund) as previously described (19). The results were also verified using BE-Analyzer (http://www.rgenome.net/be-analyzer/), a tool for the analysis of base editing efficiency (20). The primer sequences are listed in Supplementary Table S2.
Measurement of protein and gRNA expression levels
To determine the expression levels of GFP fusion protein, 2 μg of plasmids DNA encoding GFP fusion proteins were transfected in 1.6 × 105 HEK293T/17 cells and 10 nM romidepsin was applied 6 h after transfection. Cells were collected 72 h after transfection, and the GFP expression levels were analyzed using a BD FACSCanto II. Overall, 10 000 cells were analyzed and data were analyzed using Flowjo software. For western blot assay, whole cell lysate (WCL) was prepared using 1× RIPA buffer and assessed using the iWestern system (Thermo Fisher Scientific) according to the manufacturer's protocol. Briefly, 10 μg of WCL was separated in 4–12% Bis–Tris Plus Gels and transferred to a polyvinylidene difluoride membrane using the iBlot 2 Dry Blotting System. Immunoblotting was conducted with anti-Cas9 (Invitrogen, MA1–201), anti-HDAC1 (Cell Signaling Technology, #5356), anti-HDAC2 (Cell Signaling Technology, #5113), and anti-glyceraldehyde 3-phosphate dehydrogenase (anti-GAPDH; Santa Cruz Biotechnology, sc-47724) antibodies. Enhanced chemiluminescence was used to detect the immunoblotted proteins. The intensities of the detected bands were quantitated using the ImageJ Gel Analysis program. To determine the gRNA expression levels, total RNA was isolated using miRNeasy Mini Kit (Qiagen) and 1 μg of total RNA was reverse transcribed using miScript II RT Kit (Qiagen) according to the manufacturer's protocol. Quantitative PCR was preformed using iQ™ SYBR® Green Supermix (Bio-rad) on an CFX96 Touch Real-Time PCR Detection System (Bio-rad). The expression levels of gRNAs were normalized with those of housekeeping gene, GAPDH, and analyzed using median threshold cylce (Ct) value.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed using a Pierce Magnetic ChIP Kit (Thermo Fisher Scientific) according to the manufacturer's instructions. Briefly, HEK293T/17 cells were treated with 10 nM romidepsin for 72 h and fixed in 1% formaldehyde. After cell lysis, MNase digestion was performed according to the manufacturer's instructions and the lysates were sonicated to shear lengths of 200–1000 bp DNA fragments using three sets of 20-s pulses of sonication. Anti-normal rabbit IgG included in the kit or anti-acetyl-histone H3 Lys9 antibody (Upstate Biotechnology) were used to precipitate the DNA-protein complexes. The immunoprecipitated DNA was examined using qPCR. The specific primers are described in Supplementary Table S2.
RESULTS
Fluorescent reporter-based drug screen for enhancing ABE activity
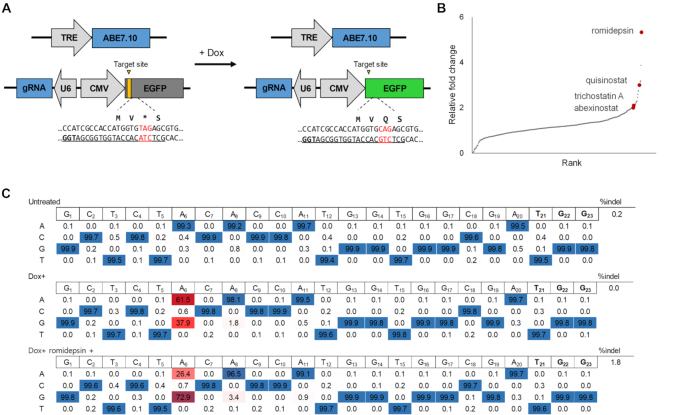
First, we generated doxycycline-inducible ABE7.10 cell lines (HAP1-ABEdox) for the drug screen. Eight single clones of HAP1-ABEdox were tested to determine whether doxycycline treatments could induce the expression of ABE7.10 to introduce A:T to G:C conversion. We demonstrated that rt-TA dependent expression of ABE7.10 induced A:T to G:C conversion at the endogenous CCR5 site (Supplementary Figure S1A). We selected HAP1-ABEdox clone #2 and developed GFP reporter cell lines (HAP1-ABEdox:GFP) to effectively measure the base editing efficiency in drug screens (Figure 1A). In the GFP reporter system, the premature stop codon (TAG) was inserted behind the start codon, which interfered with the expression of GFP. When the ABE7.10 proteins were expressed after doxycycline treatment, ABE7.10 and GFP targeting gRNAs converted the stop codon to glutamine (TAG to CAG) and the GFP proteins were expressed. We found that the GFP expression levels were dependent on the doxycycline concentration and the number of seeded cells (Supplementary Figure S1B).
Figure 1.
Drug screening with the HAP1-ABEdox:GFP reporter system. (A) Schematic overview of the doxycycline (Dox)-dependent HAP1-ABEdox:GFP reporter system. The EGFP-targeting gRNA was expressed by the U6 promoter, and EGFP with a premature stop codon (TAG) in the third amino acid position was expressed by the CMV promoter. The two cassettes, U6-gRNA and CMV-EGFP, were arranged in opposite orientations. The ABE7.10 expression was controlled by doxycycline and converted the premature stop codon into glutamine for EGFP expression. (B) The relative fold-change of GFP expression levels in 414 anti-cancer drug screens with a concentration of 100 nM. The fold-changes resulting from each drug were calculated relative to the GFP expression levels in cells treated with doxycycline alone. Four HDAC inhibitors are indicated (raw numbers are provided in Supplementary Table S3). All agents were assessed using biological duplicates. (C) The adenine base editing efficiencies at the EGFP target site were analyzed using targeted deep sequencing. The A:T to G:C substitution frequency at the A6 position was 72.9% in 10 nM romidepsin-treated cells as compared to 37.9% in cells treated with doxycycline alone.
Using the HAP1-ABEdox:GFP system, we screened a commercially available drug library containing 414 chemicals to identify compounds that increased GFP expression levels. These compounds are used in cancer research and their targets have been validated (21). All compounds were tested at two concentrations (100 and 500 nM) and the GFP expression levels were measured 48 h after drug treatment using the high-throughput system. The fold-change in GFP expression levels in cells treated with each drug was calculated and compared with those in cells treated with doxycycline alone. Among the 414 agents, romidepsin, which is an HDAC inhibitor (22), had the greatest effect on the fold-change of the GFP expression levels (5.33- and 7.79-fold-change at 100 and 500 nM, respectively) (Figure 1B, Supplementary Figure S2, and Supplementary Table S3). In addition, most of the HDAC inhibitors (quisinostat, TSA, and abexinostat) in the chemical library ranked highly in the drug screen. Based on these results, we hypothesized that small-molecule inhibitors of HDAC improve adenine base editing efficiency.
To validate the change in GFP expression levels resulting from the ABE-mediated nucleotide substitutions, gDNAs were extracted from romidepsin-treated cells. The GFP target sequences were analyzed using targeted deep sequencing to verify the base editing efficiencies (Figure 1C). When ABE7.10 expression was induced using doxycycline in HAP1-ABEdox:GFP cells, the frequency of the A:T to G:C substitution at position 6 of protospacer sequences was 37.9%, which increased to 72.9% in romidepsin-treated cells. Overall, we concluded that romidepsin increased the A:T to G:C the base editing efficiency and GFP expression levels in HAP1-ABEdox:GFP cells.
HDAC inhibitor improves ABE activity at endogenous sites
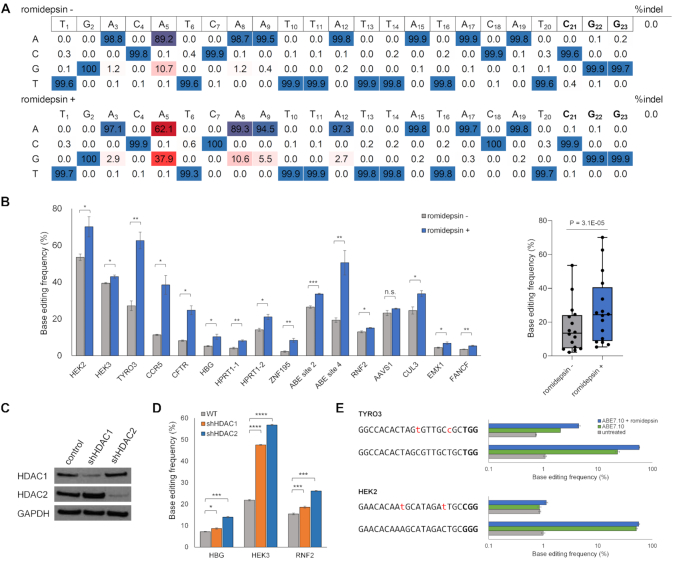
Next, we determined whether romidepsin could influence the base editing efficiency at endogenous target sites. Plasmid DNA encoding ABE7.10 and CCR5 targeting gRNA were transfected in HEK293T/17 cells and the cells were treated with 10 nM romidepsin (Figure 2A). In positions 4–7 of the target sites, which is the editing window of ABE7.10, the A:T to G:C substitution frequencies of A5 increased by >3.5-fold from 10.7% to 37.9%. In addition to A5, other positions showed increased A:T to G:C substitution frequencies, especially, the frequency at A8 increased from 1.2% to 10.6% (8.8-fold). We also tested various concentrations of romidepsin to determine the optimal concentration to enhance the base editing efficiency without cytotoxicity in HEK293T/17 cells (Supplementary Figure S3A and B). The findings showed that 5–10 nM of romidepsin was adequate to increase the base editing efficiency in HEK293T/17 cells. In addition, we evaluated both cell viability and the base editing efficiency for 2 weeks. The initial cell viability was slightly decreased in the presence of romidepsin. However, cell viability was similar to that of mock cells during further sub-culturing of the cells (Supplementary Figure S3C). Then, sixteen endogenous target sites were examined, and we found that romidepsin improved the A:T to G:C substitution frequencies at these sites by up to 3.8-fold (2.18–8.32% at the ZNF195 site) (Figure 2B). We also confirmed that romidepsin increased A:T to G:C substitution frequencies in HeLa cells by up to 4.6-fold (Supplementary Figure S4).
Figure 2.
Adenine base editing efficiencies at endogenous target sites are improved by romidepsin treatment. (A) Base editing efficiency at endogenous CCR5 target site. The A:T to G:C substitution frequency at the A5 position increased from 10.7% to 37.9% in 10 nM romidepsin-treated cells. (B) Base editing efficiencies of 16 endogenous target sites are shown in the left panel and the cumulative base editing efficiency across the target sites is depicted as the boxplot in the right panel. The P-value in the boxplot was calculated using the two-tailed Wilcoxon signed-rank test. Romidepsin improved the adenine base editing efficiency at almost all endogenous sites. The change was not statistically significant at the AAVS1 target site. (C) Western blot analysis showing HDAC1 and HDAC2 expression levels in HEK293T/17 cells stably expressing non-target (for negative control), HDAC1, and HDAC2 targeting shRNAs. GAPDH was used as a loading control. (D) Base editing efficiencies of three endogenous sites, HBG, HEK3 and RNF2 sites, in HDAC1 and HDAC2 knockdown HEK293T/17 cells. The ABE7.10-mediated base editing efficiency increased in both of the two knockdown cells in three endogenous target sites. (E) Base editing efficiencies of previously known off-target sites of TYRO3 and HEK2 targeting gRNAs. The two off-target sites had 2-bp mismatches with their gRNAs (shown in red lowercase). The adenine base editing efficiency increased in both on- and off-target sites. Error bars indicate SEM (n = 3). Ns, not significant; P ≥ 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 (using two-tailed Student's t-test).
In the drug screen, four HDAC inhibitors, including romidepsin, were ranked in the top thirty drugs that raised GFP expression levels. We examined whether the other three HDAC inhibitors—TSA, abexinostat, and quisinostat—could affect to the base editing activities, and found that these HDAC inhibitors also increased endogenous A:T to G:C substitution frequencies (Supplementary Figure S5A and B). Interestingly, vorinostat, another HDAC inhibitor in the library, was not ranked in the drug screening. Vorinostat operates at a concentration 10- to 100-fold higher than that used in this drug screening and we found that vorinostat also improves the base editing efficiency at 0.5 μM (Supplementary Figure S5B) (23). These small molecules, which increase ABE7.10-mediated base editing efficiency, are inhibitors of both HDAC1 and HDAC2 (20). To investigate the role of these HDACs in base editing efficiency, we generated HDAC1 or HDAC2 knockdown HEK293T/17 cells using shRNAs (Figure 2C) and measured the base editing efficiency at three endogenous target sites (Figure 2D). The results demonstrated that base editing efficiencies were significantly increased in HDAC1 and HDAC2 knockdown cells.
To examine whether romidepsin affected A:T to G:C substitution frequencies at the potential off-target sites, we examined two endogenous target sites, TYRO3 and HEK2, which have previously identified to have their off-target sites. We transfected two gRNAs targeting these genes with ABE7.10 and analyzed base editing efficiencies using targeted deep sequencing (7,19). We found that romidepsin improved ABE7.10 activity in both on- and off-target sites (Figure 2E). Taken together, these findings verified that HDAC inhibitors, including romidepsin, improve the base editing efficiency at endogenous target sites by inhibiting HDAC1 and HDAC2 expression.
HDAC inhibitor increases the expression levels of proteins and gRNAs
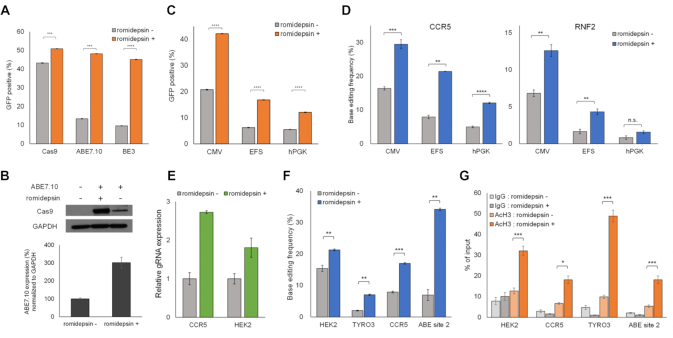
We then explored how romidepsin increased base editing efficiencies and speculated that the expression levels of ABE7.10 might affect them. Nucleosomes formed on non-integrated plasmid DNA transfected into mammalian cells and HDAC inhibitors enhance CMV promoter-driven gene expression (22,23). The expression levels of ABE protein are critical for adenine base editing (9). Thus, we examined whether romidepsin influences the expression levels of ABE7.10 and increases its base editing efficiencies. We constructed plasmid DNA encoding ABE7.10-2A-EGFP proteins to determine the expression levels using fluorescence measurements. HEK293T/17 cells were transfected with the plasmids and were subsequently analyzed using flow cytometry (Figure 3A). GFP-positive cells comprised 48.2% of the romidepsin-treated cells and 13.4% of the untreated cells, which means that romidepsin increased the expression levels of ABE7.10 by 3.6-fold. The increase in ABE7.10 protein expression levels was also verified using western blotting (Figure 3B). We also cloned spCas9–2A-EGFP and BE3-2A-EGFP constructs and analyzed their protein expression levels. GFP-positive cells were increased up to 1.2- and 4.7-fold in cells transfected with spCas9–2A-EGFP and BE3-2A-EGFP, respectively (Figure 3A). Since romidepsin increased the protein expression levels of both spCas9 and BE3, we investigated whether it also improved spCas9- and BE3-mediated mutation efficiencies at endogenous target sites. Targeted deep sequencing revealed that romidepsin increased spCas9-mediated indel frequencies at 12 different endogenous loci by up to 1.8-fold (27.2–49.4% at the HBG site) and BE3-mediated C:G to T:A conversion efficiencies by up to 4.4-fold (3.16–13.8% at the ZNF195 site) (Supplementary Figure S6A and B). To investigate whether romidepsin increased alternative promoter-driven ABE7.10 expression levels and the base editing efficiency, we further constructed plasmids with ABE7.10-2A-EGFP expression using EF-1α short (EFS) and human phosphoglycerate kinase (hPGK) promoters, and found that romidepsin increased both EFS and hPGK promoter-driven ABE7.10 expression levels (6.2–16.8% and 5.5–12.1%, respectively) and base editing efficiency (Figure 3C, D). We also verified that romidepsin also increased U6 promoter-driven gRNA expression as previously described, and HDAC inhibitors enhanced RNA polymerase III promoter-driven small RNA expression (Figure 3E) (24). Collectively, these findings demonstrated that romidepsin increased the base editing efficiency by affecting both protein and gRNA expression levels.
Figure 3.
Romidepsin improves base editing efficiency by affecting the protein and gRNA expression levels and the chromatin state. (A) Flow cytometry analysis of the expression levels of ABE7.10–2A-EGFP, spCas9–2A-EGFP and BE3–2A-EGFP proteins in the presence and absence of romidepsin. The proportion of GFP-positive cells increased following romidepsin treatment. (B) Western blot assay to demonstrate the ABE7.10 protein expression levels in HEK293T/17 cells. The expression levels of ABE7.10 proteins increased in the presence of romidepsin. GAPDH is shown as a loading control. (C) Flow cytometry analysis of the expression levels of CMV, EFS, and hPGK promoter-driven ABE7.10-2A-EGFP proteins in the presence and absence of romidepsin. Romidepsin improved both EFS and hPGK promoter-driven protein expression. (D) Targeted deep sequencing analysis showed that romidepsin improved the base editing efficiency at both CCR5 and RNF2 sites in HEK293T/17 cells transfected with EFS- and hPGK promoter-driven ABE7.10 expressing plasmids. (E) Quantitative real-time PCR analysis for detecting gRNA expression levels. Romidepsin increased U6 promoter-driven gRNA expression levels. (F) Base editing efficiencies at four endogenous target sites in RNP delivered HEK293T/17 cells. (G) ChIP assay results at four endogenous target sites. The acetylation percentage at the four sites increased with romidepsin treatment. Error bars indicate SEM (n = 3); ns, not significant; P ≥ 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 (using two-tailed Student's t-test).
HDAC inhibitor improves ABE activity by inhibiting histone deacetylation at the target locus
We performed RNP delivery of ABE7.10 to verify that romidepsin increases base editing efficiency by affecting protein expression levels. RNP delivery uses already quantified proteins and gRNAs. Thus, this method is unaffected by protein expression levels in mammalian cells. We electroporated RNPs that targeted four endogenous sites into HEK293T/17 cells (Figure 3F). Remarkably, romidepsin treatments increased the base editing efficiencies of all four endogenous target sites by up to 4.9-fold (6.9–34.2% at ABE site 2). Based on the previous demonstration that mutagenesis by Cas9 was affected by chromatin accessibility (25–28), we evaluated whether increased base editing efficiency because of romidepsin was also affected by similar factors. Romidepsin is a HDAC inhibitor that can induce an open chromatin state through histone hyperacetylation, which may affect base editing efficiency. ChIP assays using an antibody against histone H3 acetylation revealed that romidepsin treatment increased H3 acetylation at the four endogenous target sites (Figure 3G). These results showed that romidepsin inhibits histone deacetylation, resulting in the formation of a euchromatin state, which, in turn, increased base editing efficiencies at the target locus.
HDAC inhibitor increases base editing activity of both ABEmax and BE4max
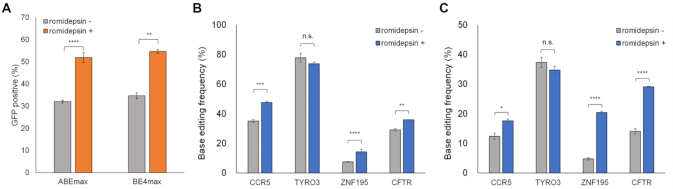
Improved ABE and CBE (ABEmax and BE4max, respectively) were recently developed by optimizing protein expression and their nuclear localization sequences (9). Presently, we investigated whether romidepsin could also improve the base editing efficiencies of ABEmax and BE4max. We transfected plasmid DNA encoding ABEmax-2A-GFP and BE4max-2A-GFP into HEK293T/17 cells to verify the protein expression levels of those two BEs in the presence romidepsin. Flow cytometry analysis showed that romidepsin increased the expression levels of both ABEmax (32.1–51.9%) and BE4max (34.7–54.7%) (Figure 4A). We then analyzed the base editing efficiencies at four endogenous target sites, and found that romidepsin also increased the base editing efficiencies of ABEmax and BE4max by up to 4.3-fold (4.8–20.4% at the ZNF195 site) (Figure 4B, C). These results indicated that romidepsin improved the base editing efficiencies of enhanced BEs.
Figure 4.
Romidepsin improves base editing efficiencies of both ABEmax and BE4max. (A) Flow cytometry analysis of GFP-positive cells in ABEmax-2A-GFP and BE4max-2A-GFP transfected HEK293T/17 cells. The expression levels of ABEmax-2A-GFP and BE4max-2A-GFP proteins were improved by romidepsin treatment. (B, C) The effects of romidepsin treatment on ABEmax- (B) and BE4max- (C) mediated base editing efficiency in HEK293T/17 cells were analyzed using targeted deep sequencing. Error bars indicate SEM (n = 3), ns (not significant), P ≥ 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 (using two-tailed Student's t-test).
DISCUSSION
Here, we have developed a doxycycline-inducible ABE-mediated fluorescent reporter system that can be used to easily determine the adenine base editing efficiencies using fluorescent signaling. The fluorescent reporter system enables the detection of chemical-dependent adenine base editing efficiency. Using this system, we performed chemical screens to identify substances that enhanced adenine base editing efficiencies and found that HDAC inhibitors, including romidepsin, achieved this effect by increasing the expression levels of protein and gRNA and by causing euchromatin states of the target sites.
Although previous studies show that the chromatin state influences Cas9 activity (25–28), there have been no reports, to the best of our knowledge, that the chromatin state influences the activity of BEs. Recently, Liu et al. showed that the inhibition of HDAC1 and HDAC2 expression enhances Cas9-based genome editing (29). They found that attenuation of HDAC1 and HDAC2 activity leads to an open chromatin state and facilitates Cas9 binding to the target DNA, resulting in an increase in genome editing efficiencies. Our finding that romidepsin improves Cas9 activity are consistent with the results of prior studies. We have further demonstrated that HDAC inhibitors, including romidepsin, improve adenine base editing efficiency by inhibiting HDAC1 and HDAC2 expression. These HDACs are regulators of DNA damage response (30). Thus, HDAC inhibitors might affect both recruitment of the BE protein to target sites and DNA repair machinery after targeted deamination at the target sites.
To determine whether the product purity affected by the DNA repair mechanism was changed by romidepsin, we analyzed the ABE- and CBE-mediated base editing product purity. As found in a previous study, ABE induced adenine base editing with very high product purity (7), and romidepsin did not affect their product purity. However, romidepsin significantly improved the base editing purity of BE3 at the HEK2 site, which is CBE-mediated base editing with low product purity, and at the CCR5 site, which was detected with low product purity (Supplementary Figure S7) (6). These findings indicated that romidepsin might also be widely used for CBE-mediated genome editing to improve base editing efficiency as well as product purity.
Koblan et al. have demonstrated that the poor expression of ABE7.10 and BE3 is a bottleneck to the efficiency of BEs, and they also developed improved BEs that were designated ABEmax and BE4max (9). Presently, We found that romidepsin increased the protein expression level of ABE7.10 and BE3 by up to 4.0-fold, and ABE7.10- and BE3-mediated base editing efficiency at endogenous sites by up to 4.9-fold. These results are consistent with those of previous studies. Although romidepsin significantly improved ABEmax and BE4max expression levels and increased base editing efficiency at low activity sites, it did not increase base editing efficiency at highly active sites. For example, at the TYRO3 site, although romidepsin strongly influenced the chromatin state at the target sites and improved base editing efficiency of both ABE7.10 and BE3 by plasmid delivery, the base editing efficiency of ABEmax and BE4max did not increase. These findings indicate that the effect of romidepsin on the protein expression levels and chromatin status is complex and depends on the target sites or delivery materials.
RNP delivery allows the delivery of defined amounts of protein–gRNA complexes to cells. Romidepsin significantly improved ABE7.10 activities in RNP delivery. Since the quantified protein–gRNA complexes cleave target DNA almost immediately after delivery and are degraded rapidly in cells, the chromatin state may highly affect the base editing efficiency in RNP delivery. The direct delivery of RNPs has many advantages. These include the rapid action of the RNP complex to target DNA, high gene editing activity, and reduced off-target effects, toxicity, and immune response. We believe that HDAC inhibitors can be used for RNP-mediated therapeutic applications in vivo.
The off-target effect is a significant issue for CRISPR-based BEs. BEs induce unwanted mutations in non-target loci in the genome and transcriptome (19,31–33). We found that ABE used with romidepsin treatment increased base editing in both on- and off-target sites. These results may reflect the ability of HDAC inhibitors to enhance ABE activity by increasing the expression of BEs and modulating the chromatin state in a genome-wide manner. Further studies regarding the off-target effects with the treatments of HDAC inhibitors will be illuminating. To reduce unwanted off-target effects, recently developed BEs, which reduce unwanted off-target effects, in combination with HDAC inhibitor treatment may be a good option (34–36). As shown in Figure 2A, romidepsin treatments increased the base editing efficiency at the A8 and A9 positions, which were outside the base editing window. To determine whether romidepsin affects the range of the base editing window, we compared the fold-change of base editing efficiencies using romidepsin for all adenines in the protospacer sequence used in this study. There was no tendency for altered range of the base editing window (Supplementary Figure S8A). We also transfected different amounts of ABE7.10 in the presence and absence of romidepsin and analyzed the base editing window (Supplementary Figure S8B). Comparing the two samples with similar base editing efficiency (e.g. 300 ng and 75 ng ABE7.10 in the presence or absence of romidepsin, respectively) did not reveal a significant difference between the range of the base editing windows.
Overall, HDAC inhibitors offer increased base editing efficiency in endogenous target loci. This improvement may provide insights into improved efficiency of the base editing toolkit for precision genome editing. Food and Drug Administration approved HDAC inhibitors, such as romidepsin and vorinostat, can be valuable for in vivo targeted base editing in biomedical and therapeutic research.
DATA AVAILABILITY
The sequencing data for this study are available from the Sequencing Read Archive (https://www.ncbi.nlm.nih.gov/sra) under accession numbers PRJNA635270 and PRJNA675418. All flow cytometry data has been uploaded to Flow Repository (Repository ID: FR-FCM-Z3DV).
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Center for Genome Engineering, Institute for Basic Science for their assistance and resources during the high-throughput sequencing. We would like to thank the Scientific Publications Team at Asan Medical Center for editorial assistance in preparing the paper. Also, we thank the High-Throughput Screening core facility at the ConveRgence mEDIcine research cenTer (CREDIT), Asan Medical Center, for screening support and instrumentation.
Notes
Present address: Heon Seok Kim, Department of Medicine, Stanford University School of Medicine, Stanford, CA, USA.
Contributor Information
Ha Rim Shin, Department of Biomedical Sciences, Asan Medical Institute of Convergence Science and Technology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Republic of Korea; Stem Cell Immunomodulation Research Center, University of Ulsan College of Medicine, Seoul, Republic of Korea.
Ji-Eun See, Department of Biomedical Sciences, Asan Medical Institute of Convergence Science and Technology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Republic of Korea; Stem Cell Immunomodulation Research Center, University of Ulsan College of Medicine, Seoul, Republic of Korea.
Jiyeon Kweon, Department of Biomedical Sciences, Asan Medical Institute of Convergence Science and Technology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Republic of Korea; Stem Cell Immunomodulation Research Center, University of Ulsan College of Medicine, Seoul, Republic of Korea.
Heon Seok Kim, Center for Genome Engineering, Institute for Basic Science, Daejeon, Republic of Korea; Department of Chemistry, Seoul National University, Seoul, Republic of Korea.
Gi-Jun Sung, Department of Biomedical Sciences, Asan Medical Institute of Convergence Science and Technology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Republic of Korea; Stem Cell Immunomodulation Research Center, University of Ulsan College of Medicine, Seoul, Republic of Korea.
Sojung Park, Convergence Medicine Research Center (CREDIT)/Biomedical Research Center, Asan Institute for Life Sciences, Seoul, Republic of Korea; Department of Convergence Medicine, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Republic of Korea.
An-Hee Jang, Department of Biomedical Sciences, Asan Medical Institute of Convergence Science and Technology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Republic of Korea; Stem Cell Immunomodulation Research Center, University of Ulsan College of Medicine, Seoul, Republic of Korea.
Gayoung Jang, Department of Biomedical Sciences, Asan Medical Institute of Convergence Science and Technology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Republic of Korea; Stem Cell Immunomodulation Research Center, University of Ulsan College of Medicine, Seoul, Republic of Korea.
Kyung‐Chul Choi, Department of Biomedical Sciences, Asan Medical Institute of Convergence Science and Technology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Republic of Korea; Stem Cell Immunomodulation Research Center, University of Ulsan College of Medicine, Seoul, Republic of Korea; Department of Pharmacology, Asan Medical Institute of Convergence Science and Technology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Republic of Korea.
Inki Kim, Convergence Medicine Research Center (CREDIT)/Biomedical Research Center, Asan Institute for Life Sciences, Seoul, Republic of Korea; Department of Convergence Medicine, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Republic of Korea.
Jin-Soo Kim, Center for Genome Engineering, Institute for Basic Science, Daejeon, Republic of Korea; Department of Chemistry, Seoul National University, Seoul, Republic of Korea.
Yongsub Kim, Department of Biomedical Sciences, Asan Medical Institute of Convergence Science and Technology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Republic of Korea; Stem Cell Immunomodulation Research Center, University of Ulsan College of Medicine, Seoul, Republic of Korea.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation of Korea [2017M3A9B4062419, 2020R1F1A1075508, 2018R1A5A2020732]. Funding for open access charge: National Research Foundation of Korea [2017M3A9B4062419].
Conflict of interest statement. J-E.S. and Y.K. have filed a patent application based on this work.
REFERENCES
- 1. Rees H.A., Liu D.R.. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018; 19:770–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R.. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016; 533:420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hess G.T., Fresard L., Han K., Lee C.H., Li A., Cimprich K.A., Montgomery S.B., Bassik M.C.. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat. Methods. 2016; 13:1036–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ma Y., Zhang J., Yin W., Zhang Z., Song Y., Chang X.. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat. Methods. 2016; 13:1029–1035. [DOI] [PubMed] [Google Scholar]
- 5. Nishida K., Arazoe T., Yachie N., Banno S., Kakimoto M., Tabata M., Mochizuki M., Miyabe A., Araki M., Hara K.Y.et al.. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016; 353:aaf8729. [DOI] [PubMed] [Google Scholar]
- 6. Komor A.C., Zhao K.T., Packer M.S., Gaudelli N.M., Waterbury A.L., Koblan L.W., Kim Y.B., Badran A.H., Liu D.R.. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci Adv. 2017; 3:eaao4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I., Liu D.R.. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature. 2017; 551:464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zafra M.P., Schatoff E.M., Katti A., Foronda M., Breinig M., Schweitzer A.Y., Simon A., Han T., Goswami S., Montgomery E.et al.. Optimized base editors enable efficient editing in cells, organoids and mice. Nat. Biotechnol. 2018; 36:888–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Koblan L.W., Doman J.L., Wilson C., Levy J.M., Tay T., Newby G.A., Maianti J.P., Raguram A., Liu D.R.. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 2018; 36:843–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chu V.T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K., Kuhn R.. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015; 33:543–548. [DOI] [PubMed] [Google Scholar]
- 11. Maruyama T., Dougan S.K., Truttmann M.C., Bilate A.M., Ingram J.R., Ploegh H.L.. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 2015; 33:538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Robert F., Barbeau M., Ethier S., Dostie J., Pelletier J.. Pharmacological inhibition of DNA-PK stimulates Cas9-mediated genome editing. Genome Med. 2015; 7:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Song J., Yang D., Xu J., Zhu T., Chen Y.E., Zhang J.. RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat. Commun. 2016; 7:10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pinder J., Salsman J., Dellaire G.. Nuclear domain ‘knock-in’ screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Res. 2015; 43:9379–9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ma X., Chen X., Jin Y., Ge W., Wang W., Kong L., Ji J., Guo X., Huang J., Feng X.H.et al.. Small molecules promote CRISPR-Cpf1-mediated genome editing in human pluripotent stem cells. Nat. Commun. 2018; 9:1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kweon J., Jang A.H., Shin H.R., See J.E., Lee W., Lee J.W., Chang S., Kim K., Kim Y.. 2020) A CRISPR-based base-editing screen for the functional assessment of BRCA1 variants. Oncogene. 39:30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Inglese J., Auld D.S., Jadhav A., Johnson R.L., Simeonov A., Yasgar A., Zheng W., Austin C.P.. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:11473–11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Macarron R., Hertzberg R.P.. Design and implementation of high throughput screening assays. Mol. Biotechnol. 2011; 47:270–285. [DOI] [PubMed] [Google Scholar]
- 19. Kim D., Kim D.E., Lee G., Cho S.I., Kim J.S.. Genome-wide target specificity of CRISPR RNA-guided adenine base editors. Nat. Biotechnol. 2019; 37:430–435. [DOI] [PubMed] [Google Scholar]
- 20. Hwang G.H., Park J., Lim K., Kim S., Yu J., Yu E., Kim S.T., Eils R., Kim J.S., Bae S.. Web-based design and analysis tools for CRISPR base editing. BMC Bioinformatics. 2018; 19:542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pattabiraman D.R., Bierie B., Kober K.I., Thiru P., Krall J.A., Zill C., Reinhardt F., Tam W.L., Weinberg R.A.. Activation of PKA leads to mesenchymal-to-epithelial transition and loss of tumor-initiating ability. Science. 2016; 351:aad3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. West A.C., Johnstone R.W.. New and emerging HDAC inhibitors for cancer treatment. J. Clin. Invest. 2014; 124:30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tiffon C., Adams J., van der Fits L., Wen S., Townsend P., Ganesan A., Hodges E., Vermeer M., Packham G.. The histone deacetylase inhibitors vorinostat and romidepsin downmodulate IL-10 expression in cutaneous T-cell lymphoma cells. Br. J. Pharmacol. 2011; 162:1590–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yen M.C., Weng T.Y., Chen Y.L., Lin C.C., Chen C.Y., Wang C.Y., Chao H.L., Chen C.S., Lai M.D.. An HDAC inhibitor enhances cancer therapeutic efficiency of RNA polymerase III promoter-driven IDO shRNA. Cancer Gene Ther. 2013; 20:351–357. [DOI] [PubMed] [Google Scholar]
- 25. Chen X., Rinsma M., Janssen J.M., Liu J., Maggio I., Goncalves M.A.. Probing the impact of chromatin conformation on genome editing tools. Nucleic Acids Res. 2016; 44:6482–6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen X., Liu J., Janssen J.M., Goncalves M.. The chromatin structure differentially impacts high-specificity CRISPR-Cas9 nuclease strategies. Mol Ther Nucleic Acids. 2017; 8:558–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yarrington R.M., Verma S., Schwartz S., Trautman J.K., Carroll D.. Nucleosomes inhibit target cleavage by CRISPR-Cas9 in vivo. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:9351–9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kallimasioti-Pazi E.M., Thelakkad Chathoth K., Taylor G.C., Meynert A., Ballinger T., Kelder M.J.E., Lalevee S., Sanli I., Feil R., Wood A.J.. Heterochromatin delays CRISPR-Cas9 mutagenesis but does not influence the outcome of mutagenic DNA repair. PLoS Biol. 2018; 16:e2005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu B., Chen S., Rose A., Chen D., Cao F., Zwinderman M., Kiemel D., Aissi M., Dekker F.J., Haisma H.J.. Inhibition of histone deacetylase 1 (HDAC1) and HDAC2 enhances CRISPR/Cas9 genome editing. Nucleic Acids Res. 2020; 48:517–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Roos W.P., Krumm A.. The multifaceted influence of histone deacetylases on DNA damage signalling and DNA repair. Nucleic Acids Res. 2016; 44:10017–10030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim D., Lim K., Kim S.T., Yoon S.H., Kim K., Ryu S.M., Kim J.S.. Genome-wide target specificities of CRISPR RNA-guided programmable deaminases. Nat. Biotechnol. 2017; 35:475–480. [DOI] [PubMed] [Google Scholar]
- 32. Jin S., Zong Y., Gao Q., Zhu Z., Wang Y., Qin P., Liang C., Wang D., Qiu J.L., Zhang F.et al.. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science. 2019; 364:292–295. [DOI] [PubMed] [Google Scholar]
- 33. Zuo E., Sun Y., Wei W., Yuan T., Ying W., Sun H., Yuan L., Steinmetz L.M., Li Y., Yang H.. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science. 2019; 364:289–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grunewald J., Zhou R., Iyer S., Lareau C.A., Garcia S.P., Aryee M.J., Joung J.K.. CRISPR DNA base editors with reduced RNA off-target and self-editing activities. Nat. Biotechnol. 2019; 37:1041–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rees H.A., Wilson C., Doman J.L., Liu D.R.. Analysis and minimization of cellular RNA editing by DNA adenine base editors. Sci Adv. 2019; 5:eaax5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhou C., Sun Y., Yan R., Liu Y., Zuo E., Gu C., Han L., Wei Y., Hu X., Zeng R.et al.. Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature. 2019; 571:275–278. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data for this study are available from the Sequencing Read Archive (https://www.ncbi.nlm.nih.gov/sra) under accession numbers PRJNA635270 and PRJNA675418. All flow cytometry data has been uploaded to Flow Repository (Repository ID: FR-FCM-Z3DV).