Abstract
Sensitive detection of microsatellite instability (MSI) in tissue or liquid biopsies using next generation sequencing (NGS) has growing prognostic and predictive applications in cancer. However, the complexities of NGS make it cumbersome as compared to established multiplex-PCR detection of MSI. We present a new approach to detect MSI using inter-Alu-PCR followed by targeted NGS, that combines the practical advantages of multiplexed-PCR with the breadth of information provided by NGS. Inter-Alu-PCR employs poly-adenine repeats of variable length present in every Alu element and provides a massively-parallel, rapid approach to capture poly-A-rich genomic fractions within short 80–150bp amplicons generated from adjacent Alu-sequences. A custom-made software analysis tool, MSI-tracer, enables Alu-associated MSI detection from tissue biopsies or MSI-tracing at low-levels in circulating-DNA. MSI-associated indels at somatic-indel frequencies of 0.05–1.5% can be detected depending on the availability of matching normal tissue and the extent of instability. Due to the high Alu copy-number in human genomes, a single inter-Alu-PCR retrieves enough information for identification of MSI-associated-indels from ∼100 pg circulating-DNA, reducing current limits by ∼2-orders of magnitude and equivalent to circulating-DNA obtained from finger-sticks. The combined practical and informational advantages of inter-Alu-PCR make it a powerful tool for identifying tissue-MSI-status or tracing MSI-associated-indels in liquid biopsies.
INTRODUCTION
Tumors with microsatellite instability (MSI) accumulate high numbers of somatic microsatellite (MS) insertions or deletions (indels), due to a loss of normal mismatch repair (MMR) ability (1). High levels of MSI are predictive for colorectal cancer (CRC) therapy outcome in chemotherapy and immunotherapy and has been associated with distinct characteristics and favorable results including better prognosis, a higher 5-year survival, and lesser metastasis (1,2). Although the MSI phenotype has been observed across tumor types (3), it is most common in colon adenocarcinoma, stomach adenocarcinoma , and uterine corpus endometrial carcinoma (4). Thus, clinical centers perform immunohistochemistry-based MSI testing or PCR-capillary electrophoresis (PCR-CE) testing based on five established tumor-specific MSI-markers (5), for these tumor types (6,7). MSI testing via next generation sequencing (NGS) has created expanded possibilities for pan-cancer MSI testing based on larger panels of markers (8). NGS enables quantification of ‘MSI-intensity’ as a biomarker for immunotherapy response (9), or identification of MSI at low levels in circulating DNA obtained from liquid biopsies (10,11). Along with MSI information, NGS tests can provide additional cancer biomarkers such as tumor mutational burden (TMB) or copy number variations (10). Despite these expanded possibilities, the complexities of NGS sample preparation can make it cumbersome compared to established multiplex-PCR capillary electrophoresis-based detection of MSI (5). Detection of MSI as part of exome sequencing or existing targeted re-sequencing panels (8) have high cost and can be wasteful when MSI-generated indels are the main endpoint of interest, especially for longitudinal MSI monitoring at low tumor fractions. MSI-focused panels, on the other hand, mostly rely on hybrid capture of target regions, which present challenges for microsatellites traced in circulating-DNA due to stutter, low inputs and low tumor fraction (11). Without exception, all current approaches require at least 5–10 ng input DNA (10–15) which may not be available in certain settings.
Here we present inter-Alu-PCR–based MSI detection, a new method for assessment of MSI which combines the speed and convenience of PCR-based approaches with the breadth of NGS methodologies. Alu is a ∼300 bp DNA stretch, dispersed throughout the genome with a copy number of more than 1 million, amounting to ∼11% of the human genome (6). It contains a consensus body sequence and a poly-adenine region at the 3′ end (A-tails). The A-tails are variable in length at each locus and are prone to accumulation of mutation and shrinkage, thereby forming variant microsatellite-like structures at the end of Alu elements (7). Given these Alu features, inter-Alu-PCR using primers that extend outward from the head of Alu and from the A-tail region, provides a rapid approach to capture Alu A-tails within amplicons generated from adjacent Alu elements, Figure 1A. A custom-made software analysis tool, MSI-Tracer, enables Alu-associated MSI tracing at low-levels from tissue biopsies or circulating-DNA. We demonstrate that due to the exceedingly high Alu copy number, a single PCR reaction for inter-Alu sequence amplification retrieves enough information to enable identification of MSI starting from just 100 pg circulating-DNA, reducing current NGS limits by ∼2-orders of magnitude and roughly equivalent to circulating-DNA obtained from finger-sticks (16), thereby expanding the possibilities for liquid-biopsy based detection of MSI.
Figure 1.
Concept and workflow of NGS-based MSI detection using inter-Alu-PCR. (A) Poly-A-tails at the 3′ end of Alu repeats form microsatellite-like structures. Inter-Alu-PCR amplifies the regions between two neighbor Alus and captures microsatellites at multiple loci simultaneously. (B) Schematic overview of MSI detection by inter-Alu-PCR and NGS. Following DNA extraction, inter-Alu-PCR is conducted with normal and interrogated DNA using modified Alu primers that extend outward from the Alu head and tail. As Alu primers contain part of the adaptor sequences, libraries are directly prepared via index PCR from amplified product after purification. Following sequencing, the sample MSI status or MSI-associated indels is analyzed.
MATERIALS AND METHODS
Cell line and clinical sample DNA preparation
Genomic DNA (gDNA) from cell lines HCT-15 (Sigma, Aldrich) and Human genomic DNA (Promega) were used as mutant and wildtype control respectively. Snap-frozen colon adenocarcinoma stage II/III and paired normal tissue biopsies from treatment-naïve patients were obtained from the Massachusetts General Hospital Tumor Bank and gDNA was extracted using the Blood and Tissue kit (Qiagen). For serial dilution samples, normal and tumor gDNA were randomly fragmented using dsDNA Shearase Plus (ZYMO Research, CA, USA) and tumor DNA were serially mixed into normal DNA. Plasma from healthy volunteers and from stage I/II colon adenocarcinoma treatment-naïve patients were obtained under consent and Institutional Review Board approval by the Dana Farber Cancer Institute GI Bank and cfDNA was isolated using QIAamp Circulating Nucleic Acids Kit (Qiagen). The concentration of isolated DNA was quantified on a Qubit 3.0 fluorometer using dsDNA HS assay kit (Thermo Fisher Scientific). The MSI status of clinical tumor was examined by Promega MSI Analysis kit v.1.2 (supplementary methods).
Inter-Alu-PCR and NGS
Inter-Alu-PCR was performed using 0.1-1ng DNA input in a 25 μl reaction mixture on a CFX Connect™ real-time PCR machine (Biorad) per protocol provided in Supplementary Table S1.
The Alu tail primer (5′- CGCTCTTCCGATCTCTGGAGCGAGACTCCGTCTCA-3′) and the Alu head primer (5′- TGCTCTTCCGATCTGACTGGTCTCGATCTCCTGACCTC-3′) were adapted from AluY consensus primers AluY278T18 and AluY66H21 employed previously for inter-Alu-PCR (17) with modifications for library construction as described in supplementary methods.
MSI bioinformatic analysis
Sequencing data were mapped to human reference genome (hg38) using BWA-MEM alignment software. Bam files were processed to retain only sequences with mapping quality >60. Bam files were further processed with SamJdk to remove reads where clipping exceeds 5% (18). The insert size distribution was calculated using Qualimap 2.2.1 (19). MSI evaluation was performed via MSIsensor or MSI-tracer on experimental data and TCGA data (supplementary methods).
MSIsensor
MSIsensor is a publicly-available software that identifies somatic variants at microsatellite regions via a two-step process (20). First, MSIsensor scans reference genome and generates a list of microsatellite sites, which includes homopolymers of at least 5 bp and repeat units with maximum length of 5 bp. Second, it interrogates microsatellite sites containing more than 20 sequence reads and generates a distribution file that compares microsatellite length in the tumor and normal samples. Significantly varied loci are identified via Chi-Squared Test and the percent of unstable loci is used as MSI score. Unmatched analysis was conducted by comparing interrogated samples to HMC human male control DNA.
MSI-tracer
As an alternative to MSIsensor, a custom-made python-based MSI status calling algorithm, MSI-tracer, was developed to assess MSI status. MSI-tracer is designed specifically for detecting MSI-caused deletions at low tumor purities and compares the interrogated samples versus a normal sample. The software utilizes the MS distribution files derived from MSIsensor as input and compares the distribution of MS size from interrogated vs. normal samples at Alu-tail-adjacent homopolymers at least 10 bp long, containing pure poly-A/T runs and with sequencing coverage ≥ X (default X = 20). Deletion-containing sites at each locus are scored when (a) at least two distinct poly-A/T in the interrogated sample are ≥2 bp shorter compared to the shortest poly-A/T in the normal sample, and (b) these deletions are supported by at least N sequencing reads (default threshold N = 5). The MSI-tracer score is calculated by dividing the number of deletion-containing MS sites to the total number of MS sites. Alternatively, the absolute number of deletion-containing MS under the same overall sequencing reads for interrogated and normal samples, is computed. If the sequencing reads between interrogated and normal DNA are substantially (>10%) different, in-silico down-sampling is applied to perform the comparison under equal sequence reads. Experiments with low-tumor purity samples are usually designed to yield 15–20 × 106 paired end sequence reads per sample. The Alu MSI-tracer code is available on Github https://github.com/Amakri1020/MSI-Tracer.
Statistical analysis
The Student t test was employed to analyze the statistical differences of the MSI-tracer score or the number of MSI events per sample. The Bonferroni-corrected P value (Paltered = 0.05/number of experimental repeats) was used to determine statistical significance.
RESULTS
Inter-Alu-PCR
Inter-Alu-PCR is performed by employing a pair of Alu primers AluY278T18 and AluY66H21 containing adaptor oligonucleotides towards their 5′ ends. This primer modification renders amplicons compatible with library preparation with no need for adaptor ligation. Next, inter-Alu-PCR products are directly subjected to library index PCR to complete a sequencing sample preparation protocol over 2–3 h, Figure 1B. This is followed by overnight sequencing and analysis to evaluate MSI status. To profile inter-Alu-PCR amplicon sizes, we examined NGS-sequence inserts generated when starting from 1ng intact gDNA, sheared DNA or cfDNA. Under our PCR amplification conditions, the most frequent library inserts were between 110 and 135 bp irrespective of DNA type used. Sheared DNA and cfDNA share similar patterns, while gDNA displays a broader fragment distribution extending to about 375 bp (Supplementary Figure S1A). While the average distance of Alu residues across the genome is ∼ 2.4 kb (17,21), the 30 sec PCR extension time applied coupled with paired-end 150 bp sequencing favor amplification and sequencing of successive Alu elements within ∼100–150 bp. This is also compatible with the anticipated fragment sizes in circulating-DNA. Inter-Alu-PCR from intact gDNA enriches more MS sites than fragmented DNA when 1 million sequencing reads are applied (Supplementary Figure S1B). The number of distinct MS sites sequenced increases with sequencing reads and tends to saturate at higher depth. Using fragmented DNA, ∼1000 distinct MS sites are obtained with just 30 000 reads, while 16 355 MS sites, corresponding to ∼1.6% of overall Alu elements, are obtained with 50 × 106 sequencing reads (Supplementary Figure S1C). The relative distribution of all MS types, and the size of poly-A/T MS amplified with inter-Alu-PCR are depicted in Supplementary Figures S1D and E, respectively. The impact of DNA input 10 pg–1 ng using cfDNA and ∼15 × 106 reads per sample is depicted in Supplementary Figure S1F. cfDNA input of 0.1–1 ng captures 6000–14 000 distinct MS, of which 2000–4000 have a coverage >20 and can be analyzed. Moreover, the coverage of inter-Alu MS sites, the reproducibility between different runs of the same sample and the length distribution of obtained MS were assessed (Supplementary Figures S2A–D). In summary, using traces of intact or fragmented DNA, inter-Alu-PCR samples contiguous Alu elements within <150 bp of each other containing thousands MS sites for analysis.
MSI status analysis
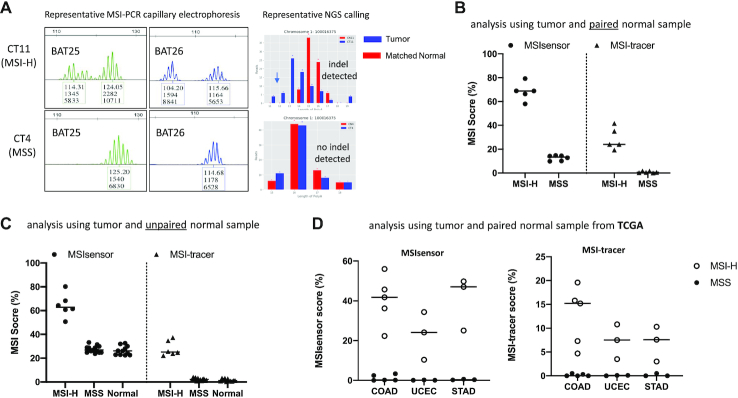
Analysis of MSI status using inter-Alu-PCR-captured MS was evaluated by employing tissue biopsy-derived DNA from 18 colon cancer patients. 11 of these tumor samples had DNA from matched normal tissue also available, while the remaining 7 tumor samples were unpaired. The MSI status for the 18 samples were first characterized via multiplexed PCR followed by capillary-based fragment length analysis using the standard 5-marker system. 5 MSI-H (28%) and 13 MSS patients were identified (supplementary Table S2). NGS analysis of inter-Alu-PCR products was then applied to the 11 tumors with matched normal tissues. Sequencing of individual microsatellites yield distributions of ‘stutter’ fragments for the interrogated tumor sample and corresponding normal tissue and indel sites are scored based on comparison of fragment sizes (Figure 2A). Data were analyzed by MSIsensor, an established bioinformatic tool for MSI status classification (20), and by MSI-tracer, a new algorithm for calling MSI-caused indels, developed by us, directed specifically to the detection of low-level indels. Both algorithms show distinct clustering of MSI-H and MSS tumors (Figure 2B) regardless of sequencing depth, down to 40 000 sequencing reads per sample (Supplementary Figures S3A and S3B). Agreement in classification is demonstrated with multiplexed-PCR-electrophoresis in 11/11 cases. Since matched normal tissue is not always available in practice, we also evaluated use of DNA from unpaired normal samples. We replaced DNA from matched normal tissue with DNA from a mix of normal individuals (Promega human male control DNA, HMC) and repeated the analysis, this time including also samples from 7 colon cancer patients with unpaired normal tissue. This approach retained the clustering between MSI-H and MSS samples, albeit with a reduced discrimination for both MSIsensor and MSI-tracer analyses (Figure 2C). The increased MSI score in un-paired samples is potentially due to germline polymorphisms scored as somatic indels. Overall, the data indicate that inter-Alu-PCR in combination with NGS can accurately identify MSI status using ultra-low-pass sequencing without the need for matched normal tissues.
Figure 2.
MSI status analysis by inter-Alu-PCR using Microseq ultra-low pass sequencing (∼0.1–0.2 × 106 reads per sample). (A) Representative PCR-CE and NGS results from MSI-H and MSS tumor DNA. (B) Analysis of tumor and matched normal tissue samples. Inter-Alu-PCR was conducted on DNA from 11 colon cancer patients (five MSI-H and six MSS as assessed by the 5-plex PCR-CE assay). MSI status was evaluated via MSIsensor and MSI-tracer software. (C) Analysis of unpaired samples (tumor samples with un-matched normal tissue). Inter-Alu-PCR was applied on DNA from 18 tumors (5 MSI-H and 13 MSS) plus 11 normal tissues obtained from colon cancer patients, and were compared against human male control (HMC) DNA. (D) MSI analysis based on inter-Alu-PCR-obtained MS in different cancer types. Whole genome sequencing data for three MSI-prone cancer colon adenocarcinomas (COAD, n = 10), corpus endometrial carcinoma (UCEC, n = 6) and Stomach adenocarcinoma (STAD, n = 6) were obtained from TCGA database. Inter-Alu regions were extracted using bed files derived from inter-Alu-PCR Hiseq data. MSI status of MSS and MSI-H tumors were examined via MSIsensor and MSI-tracer.
For MSI-tracer analysis, the software counts the number of sequencing reads that present distinct shorter poly-As relative to a similarly treated normal sample, and then applies a threshold-based filter for a binary determination of deletion-positive or negative sites. Supplementary Figure S3C shows that the determination of MSI status does not depend significantly on the threshold chosen for this filter; for example, setting a threshold of 2, 5 or 10 sequencing reads containing shorter poly-As in the interrogated sample relative to a compared normal sample does not significantly affect MSI classification.
The application of inter-Alu-PCR for MSI identification was further studied in different cancer types that underwent whole genome sequencing by the TCGA consortium (22). Samples with pre-defined MSI status were selected from MSI-prone tumors COAD (colon adenocarcinoma), UCEC (corpus endometrial carcinoma) and STAD (Stomach adenocarcinoma), as listed in Supplementary Table S3. Inter-Alu regions were extracted from whole-genome sequencing data and MSI status identification was performed. Distinct clustering between MSI-H and MSS samples was evident in all cases (Figure 2D), indicating the wide applicability of inter-Alu-PCR for MSI analysis across cancer types.
Limit of detection
To assess the lowest limit of detection (LOD) of MSI-caused indels, serial dilutions of MSI-H tumor DNA into matched normal DNA were tested. The accuracy of our dilution approach was orthogonally validated on tumor-specific somatic mutations, such as KRAS for a subset of the samples using digital droplet PCR (23,24), Supplementary Figure S4. DNA from MSI-H colon cancer specimens with high original tumor purity, (CT18 and CT11, ∼50–70% tumor) was fragmented and serially diluted, then 1ng was used as input for inter-Alu-PCR–NGS.
First, we addressed a scenario where a low-tumor purity clinical sample from a patient, such as circulating-DNA, is interrogated in the absence of paired normal sample. Application of MSIsensor analysis had limited power to determine indels at tumor purities <5% (not shown), as also reported by other groups (25). We therefore developed MSI-tracer for detecting low-level deletions in Alu-element poly-A/T-rich tails, when interrogated versus unpaired normal DNA. Using MSI-tracer, CT18 or CT11 DNA dilutions of 3% or higher had MSI-tracer score higher than any of 19 normal and MSS samples interrogated in the same manner (Figure 3A, and Supplementary Figure S5) corresponding to an LOD ∼1.5% for somatic indels given a ∼50% tumor purity. This analysis was performed via MSI-tracer using 20 × 106 sequencing reads for tumor dilutions and normal DNA mixture from five healthy individuals (Promega HMC DNA), which yielded ∼5000 Alu-MS sites per sample having adequate coverage (>20) for indel assessment. Supplementary Figure S6A presents representative poly-A distributions obtained in serial dilution experiments.
Figure 3.
Assessment of limits of detection. (A) Serial dilution of MSI-H samples CT18 and CT11 analyzed against non-matched normal tissue via MSI-tracer for MSI classification. (B) Analysis for MSI classification based on 115 MS loci commonly altered in the examined MSI-H tissue samples. (C) Serial dilution of MSI-H samples CT18 and CT11 analyzed against corresponding matched normal tissue via MSI-tracer for MSI classification. (D) Tracing deletions that are clonally present in the primary tumor, in serial dilutions of DNA from MSI-H samples CT18 and CT11 into DNA from matched normal tissue. An average of 15 × 106 sequencing reads/sample (Hiseq) was used. Statistics are based on two or three independent experimental repeats per sample. Error bars represent standard deviation from two to three independent repeats. Asterisks in 3C and 3D represent serial dilutions with significantly different MSI-tracer score as compared to matched normal samples (CN18, CN11). Bonferroni-adjusted P-values were used to assess significance
To improve the lowest limits of detection when no information from the primary tumor is available, we sought to identify ‘pre-determined’ subsets of Alu-poly-A/T sites that harbor recurrent somatic indels at all MSI-H tumor samples analyzed in this work. A group of 115 informative Alu-poly-A/T sites was thus identified, Supplementary Table S4. Focusing MSI-tracer analysis solely to this group of 115 informative MS loci improved the LOD to 0.3–1% for CT18 and CT11, respectively (Figure 3B), corresponding to an LOD of 0.15–0.5% for somatic indels. Therefore, in the absence of paired normal, the subset of recurrent somatic indels can be used to filter the data and reduce random noise.
Next, we also examined the scenario where matched normal is available. The dilution samples were interrogated with their corresponding normal. Dilutions exceeding 1% of CT18 and 3% of CT11 showed significantly higher MSI-tracer score, corresponding to an LOD of ∼0.5–1.5% given a ∼50% tumor purity (Figure 3C). Finally, inter-Alu-PCR was also applied for tracing tumor-specific microsatellite deletions (tumor fingerprint) in liquid biopsies. Thereby potentially employing inter-Alu-PCR for minimal residual disease detection in patients with MSI-H primary tumors. To this end, MSI-tracer was adapted to follow only deletions present in the primary tumor at high clonality (>30%). Presence of deletions were detectable down to 0.1% and 1% for CT18 and CT11 respectively (Figure 3D) corresponding to a somatic indel LOD 0.05–0.5% for detecting traces of tumor DNA within excess normal DNA.
Application on cfDNA samples
Detection of MSI-related indels in cfDNA obtained from cancer patients with MSI-H tumors is challenging due to the low DNA amount and low tumor fraction in the circulation. As a proof of principle using inter-Alu-PCR in detecting MSI-related poly-A deletions in cfDNA, 6 cfDNA samples obtained from colon cancer patients and four samples obtained from healthy volunteers were used. Two samples originated from patients with MSI-H tumors and contained BAT25/BAT26 indels detectable in cfDNA via multiplexed-PCR-CE, Supplementary Figure S7; and four cfDNA samples originated from patients with MSS tumors, showing no indels in cfDNA via multiplexed-PCR-CE.
These samples represent a scenario where, apart from cfDNA, there is no inter-Alu-PCR information either from the primary tumor, or from corresponding normal tissue. In the absence of matched normal DNA, cfDNA from cancer patients were interrogated against cfDNA from 4 arbitrary-chosen normal volunteers. Application of inter-Alu-PCR-NGS on 1ng cfDNA from MSI-H patients demonstrated a significantly higher MSI-tracer score, compared to MSS or normal volunteer samples (Figure 4A). This difference became more pronounced when analysis was targeted to the group of 115 informative Alu sites recurrently altered in MSI-H tumors, Figure 4B, consistent also with the serial dilution experiments in Figure 3B. Supplementary Figure S6B depicts representative poly-A distributions from cfDNA experiments, indicating presence of shorter poly-adenines for interrogated samples that are MSI-H in tumor and plasma, versus samples from any of the 4 normal volunteers.
Figure 4.
Detection of MSI-associated deletions in cfDNA from 10 individuals, including 4 healthy donors, 4 MSS colon cancer patients and 2 MSI-H colon cancer patients. No inter-Alu-PCR on primary tumor or matched normal tissue data were available for these samples. (A and C) inferred MSI status using inter-Alu-PCR and MSI-tracer at cfDNA input of 1 ng or 0.1 ng. (B and D) application of MSI-tracer analysis only on 115 informative inter-Alu-PCR sites commonly mutated in MSI-H tumors. (E) Venn diagram: triplicate independent determination of altered MS sites within set of 115 informative sites using 0.1 ng from cfDNA 3202 and 3400. Asterisks represent cfDNA samples with significantly different MSI-tracer score as compared to cfDNA from normal volunteers. Bonferroni-adjusted P-values were used to assess significance.
Finally, considering the limited availability of cfDNA in clinics, or the possibility of obtaining traces of cfDNA from finger-sticks (10) we also performed MSI analysis using lower cfDNA input. Repeating the analysis in the cfDNA samples described above, using 0.1ng cfDNA was again able to distinguish MSI-H from MSS patients and healthy donors (Figure 4C and D). Triplicate repeats indicated that inter-Alu-PCR from 0.1 ng cfDNA captures different groups of informative regions each time, but consistently shows significant increase of deletions in MSI-H patient cfDNA’ (Figure 4E). These data indicate that inter-Alu-PCR can perform MSI classification using cfDNA, and that the input DNA can be as low as 0.1 ng DNA.
DISCUSSION
Alu-PCR has been employed for detecting linkage disequilibrium (26), selection of human DNA from non-human samples (27), detection of copy number differences, mutator phenotypes and structural variation in cancer (17,28–29). In view of Alu structure which contains poly-adenine homopolymers near Alu ends, we hypothesized that inter-Alu-PCR should generate microsatellite-rich genomic fractions that enable efficient MSI detection, thereby expanding inter-Alu-PCR applications in cancer. Indeed, our data show that MSI status is predicted accurately for tissue samples, while application of MSI-tracer software enables tracing MSI-related poly-A deletions at low allelic frequencies with an LOD of 0.05–1.5%, depending also on the availability of matched tumor/normal samples and the degree of instability. This compares well to an LOD of 1–10% for PCR-CE (8,30) or ∼0.1% for indel-enriched PCR-CE (8,31–35) using mutation enrichment technologies (31,36–37); or to LOD ∼0.1–7% reported for NGS-based methods (10–15,38). Regarding bioinformatic analysis, while threshold-based statistical or machine-learning-based methods (MSIsensor. mSINGs, MANTIS, Cortes-Ciriano, MSI-ColonCore reviewed in (8)), may also determine MSI status from NGS data, the simpler MSI-tracer software developed here is specifically directed towards detection of low-level poly-A deletions using Alu-tails. Polynucleotide sequences are known to be prone to sequencing errors especially due to polymerase slippage events generating ‘stutter’ during sample preparation. MSI-tracer postulates that, in view of stutter-generated noise, at low-allelic levels only deletions generating distinctly shorter poly-A/Ts can be detected and focuses specifically on such alterations. The presence of stutter necessitates redundant coverage for each interrogated target, even for the larger deletions. Similar to other MSI-calling algorithms (24), for MSI-tracer a minimum sequence coverage of at least 20 for interrogated poly-A target was found necessary to provide reproducible statistics for reliable analysis. Application of experimental approaches to reduce PCR stutter (39,40) could be anticipated to further enhance the reported LODs for MSI detection using MSI-tracer.
While NGS-based detection of MSI status is often done by using targeted re-sequencing panels developed for different purposes, such as detection of single point mutations (8), such panels have significantly lower density of microsatellites when compared to inter-Alu-PCR. This ultimately translates to higher requirements in starting DNA and sequencing depth for determination of MSI status at low tumor purities. As repeated measurements indicate in Figure 4E inter-Alu-PCR samples enough microsatellites to determine MSI status using just 100 pg of cfDNA and ∼15 × 106 sequencing reads. In contrast, the minimum amount of starting DNA for NGS platforms is 5–250 ng (10–15), i.e. ∼ 2 orders of magnitude higher than for inter-Alu-PCR. This enables the potential use of cfDNA obtained from finger-sticks (16), to enable minimally invasive, repeated testing for tumor load in MSI-positive patients undergoing treatment, including chemotherapy, standard radiation, brachytherapy or radiopharmaceutical treatments (41,42). Another potential advantage is the direct testing of MSI using inter-Alu-PCR on cfDNA from unpurified plasma (43), thereby addressing portions of circulating-DNA currently lost during purification (43). Additional practical advantages of inter-Alu-PCR are speed and cost, since sequencing sample preparation for 20–40 samples is complete within 4 h. If sequencing is performed overnight on Miseq platform, results can be obtained next day, bringing the cost-per-reaction and time-to-result to levels comparable to the established 5-plex PCR-CE test (5). Compared to PCR-CE inter-Alu-PCR requires 10–20-fold less DNA and provides additional potential such as tracing MSI at low-levels for minimal residual disease detection, copy-number variation and tumor-mutational-burden determination.
In summary, the combined practical and informational advantages of inter-Alu-PCR make it a powerful and practical tool for identifying tissue MSI-status or tracing MSI-associated indels in liquid biopsies using minute amounts of starting material.
Supplementary Material
Contributor Information
Fangyan Yu, Department of Radiation Oncology, Dana-Farber Cancer Institute and Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA.
Ka Wai Leong, Department of Radiation Oncology, Dana-Farber Cancer Institute and Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA.
Alexander Makrigiorgos, Department of Radiation Oncology, Dana-Farber Cancer Institute and Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA.
Viktor A Adalsteinsson, The Broad Institute of MIT and Harvard, Cambridge, MA, USA.
Ioannis Ladas, Department of Radiation Oncology, Dana-Farber Cancer Institute and Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA.
Kimmie Ng, Department of Medical Oncology, Dana-Farber Cancer Institute and Harvard Medicine School, Boston, MA, USA.
Harvey Mamon, Department of Radiation Oncology, Dana-Farber Cancer Institute and Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA.
G Mike Makrigiorgos, Department of Radiation Oncology, Dana-Farber Cancer Institute and Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01 CA221874 to G.M.M.]; the contents of this manuscript do not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. Funding for open access charge: Departmental funds.
Conflict of interest statement. None declared.
REFERENCES
- 1. Vilar E., Gruber S.B.. Microsatellite instability in colorectal cancer-the stable evidence. Nat. Rev. Clin. Oncol. 2010; 7:153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Le D.T., Uram J.N., Wang H., Bartlett B.R., Kemberling H., Eyring A.D., Skora A.D., Luber B.S., Azad N.S., Laheru D.et al.. PD-1 blockade in tumors with Mismatch-Repair deficiency. N. Engl. J. Med. 2015; 372:2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cortes-Ciriano I., Lee S., Park W.Y., Kim T.M., Park P.J.. A molecular portrait of microsatellite instability across multiple cancers. Nat. Commun. 2017; 8:15180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maruvka Y.E., Mouw K.W., Karlic R., Parasuraman P., Kamburov A., Polak P., Haradhvala N.J., Hess J.M., Rheinbay E., Brody Y.et al.. Analysis of somatic microsatellite indels identifies driver events in human tumors. Nat. Biotechnol. 2017; 35:951–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bacher J.W., Flanagan L.A., Smalley R.L., Nassif N.A., Burgart L.J., Halberg R.B., Megid W.M., Thibodeau S.N.. Development of a fluorescent multiplex assay for detection of MSI-High tumors. Dis. Markers. 2004; 20:237–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Watkins J.C., Yang E.J., Muto M.G., Feltmate C.M., Berkowitz R.S., Horowitz N.S., Syngal S., Yurgelun M.B., Chittenden A., Hornick J.L.et al.. Universal screening for mismatch-repair deficiency in endometrial cancers to identify patients with lynch syndrome and Lynch-like syndrome. Int. J. Gynecol. Pathol. 2017; 36:115–127. [DOI] [PubMed] [Google Scholar]
- 7. Stadler Z.K. Diagnosis and management of DNA mismatch repair-deficient colorectal cancer. Hematol. Oncol. Clin. North Am. 2015; 29:29–41. [DOI] [PubMed] [Google Scholar]
- 8. Baudrin L.G., Deleuze J.F., How-Kit A.. Molecular and computational methods for the detection of microsatellite instability in cancer. Front. Oncol. 2018; 8:621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mandal R., Samstein R.M., Lee K.W., Havel J.J., Wang H., Krishna C., Sabio E.Y., Makarov V., Kuo F., Blecua P.et al.. Genetic diversity of tumors with mismatch repair deficiency influences anti-PD-1 immunotherapy response. Science. 2019; 364:485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Georgiadis A., Durham J.N., Keefer L.A., Bartlett B.R., Zielonka M., Murphy D., White J.R., Lu S., Verner E.L., Ruan F.et al.. Noninvasive detection of microsatellite instability and high tumor mutation burden in cancer patients treated with PD-1 blockade. Clin. Cancer Res. 2019; 25:7024–7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Willis J., Lefterova M.I., Artyomenko A., Kasi P.M., Nakamura Y., Mody K., Catenacci D.V.T., Fakih M., Barbacioru C., Zhao J.et al.. Validation of microsatellite instability detection using a comprehensive Plasma-Based genotyping panel. Clin. Cancer Res. 2019; 25:7035–7045. [DOI] [PubMed] [Google Scholar]
- 12. Waalkes A., Smith N., Penewit K., Hempelmann J., Konnick E.Q., Hause R.J., Pritchard C.C., Salipante S.J.. Accurate Pan-cancer molecular diagnosis of microsatellite instability by Single-Molecule molecular inversion probe capture and high-throughput sequencing. Clin. Chem. 2018; 64:950–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhu L., Huang Y., Fang X., Liu C., Deng W., Zhong C., Xu J., Xu D., Yuan Y.. A novel and reliable method to detect microsatellite instability in colorectal cancer by next-generation sequencing. J. Mol. Diagn. 2018; 20:225–231. [DOI] [PubMed] [Google Scholar]
- 14. Pabla S., Andreas J., Lenzo F.L., Burgher B., Hagen J., Giamo V., Nesline M.K., Wang Y., Gardner M., Conroy J.M.et al.. Development and analytical validation of a next-generation sequencing based microsatellite instability (MSI) assay. Oncotarget. 2019; 10:5181–5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gallon R., Sheth H., Hayes C., Redford L., Alhilal G., O’Brien O., Spiewak H., Waltham A., McAnulty C., Izuogu O.G.et al.. Sequencing-based microsatellite instability testing using as few as six markers for high-throughput clinical diagnostics. Hum. Mutat. 2020; 41:332–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gyanchandani R., Kvam E., Heller R., Finehout E., Smith N., Kota K., Nelson J.R., Griffin W., Puhalla S., Brufsky A.M.et al.. Whole genome amplification of cell-free DNA enables detection of circulating tumor DNA mutations from fingerstick capillary blood. Sci. Rep. 2018; 8:17313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mei L., Ding X., Tsang S.Y., Pun F.W., Ng S.K., Yang J., Zhao C., Li D., Wan W., Yu C.H.et al.. AluScan: a method for genome-wide scanning of sequence and structure variations in the human genome. BMC Genomics. 2011; 12:564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lindenbaum P., Redon R.. bioalcidae, samjs and vcffilterjs: object-oriented formatters and filters for bioinformatics files. Bioinformatics. 2018; 34:1224–1225. [DOI] [PubMed] [Google Scholar]
- 19. Okonechnikov K., Conesa A., Garcia-Alcalde F.. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics. 2016; 32:292–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Niu B., Ye K., Zhang Q., Lu C., Xie M., McLellan M.D., Wendl M.C., Ding L.. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics. 2014; 30:1015–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W.et al.. Initial sequencing and analysis of the human genome. Nature. 2001; 409:860–921. [DOI] [PubMed] [Google Scholar]
- 22. Cancer Genome Atlas Research, N. Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., Stuart J.M.. The cancer genome atlas Pan-cancer analysis project. Nat. Genet. 2013; 45:1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fitarelli-Kiehl M., Yu F., Ashtaputre R., Leong K.W., Ladas I., Supplee J., Paweletz C., Mitra D., Schoenfeld J.D., Parangi S.et al.. Denaturation-enhanced droplet digital PCR for liquid biopsies. Clin. Chem. 2018; 64:1762–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. dMIQE Group. Huggett J.F. The digital MIQE guidelines update: minimum information for publication of quantitative digital PCR experiments for 2020. Clin. Chem. 2020; 66:1012–1029. [DOI] [PubMed] [Google Scholar]
- 25. Jia P., Yang X., Guo L., Liu B., Lin J., Liang H., Sun J., Zhang C., Ye K.. MSIsensor-pro: Fast, accurate, and Matched-normal-sample-free detection of microsatellite instability. Genomics Proteomics Bioinformatics. 2020; 18:65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Labuda M., Labuda D., Korab-Laskowska M., Cole D.E., Zietkiewicz E., Weissenbach J., Popowska E., Pronicka E., Root A.W., Glorieux F.H.. Linkage disequilibrium analysis in young populations: pseudo-vitamin D-deficiency rickets and the founder effect in French Canadians. Am. J. Hum. Genet. 1996; 59:633–643. [PMC free article] [PubMed] [Google Scholar]
- 27. Nelson D.L., Ledbetter S.A., Corbo L., Victoria M.F., Ramirez-Solis R., Webster T.D., Ledbetter D.H., Caskey C.T.. Alu polymerase chain reaction: a method for rapid isolation of human-specific sequences from complex DNA sources. Proc. Natl Acad. Sci. U.S.A. 1989; 86:6686–6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Srivastava T., Seth A., Datta K., Chosdol K., Chattopadhyay P., Sinha S.. Inter-alu PCR detects high frequency of genetic alterations in glioma cells exposed to sub-lethal cisplatin. Int. J. Cancer. 2005; 117:683–689. [DOI] [PubMed] [Google Scholar]
- 29. Krajinovic M., Richer C., Labuda D., Sinnett D.. Detection of a mutator phenotype in cancer cells by inter-Alu polymerase chain reaction. Cancer Res. 1996; 56:2733–2737. [PubMed] [Google Scholar]
- 30. Berg K.D., Glaser C.L., Thompson R.E., Hamilton S.R., Griffin C.A., Eshleman J.R.. Detection of microsatellite instability by fluorescence multiplex polymerase chain reaction. J. Mol. Diagn. 2000; 2:20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li J., Wang L., Mamon H., Kulke M.H., Berbeco R., Makrigiorgos G.M.. Replacing PCR with COLD-PCR enriches variant DNA sequences and redefines the sensitivity of genetic testing. Nat. Med. 2008; 14:579–584. [DOI] [PubMed] [Google Scholar]
- 32. How-Kit A., Daunay A., Buhard O., Meiller C., Sahbatou M., Collura A., Duval A., Deleuze J.F.. Major improvement in the detection of microsatellite instability in colorectal cancer using HSP110 T17 E-ice-COLD-PCR. Hum. Mutat. 2017; 39:441–453. [DOI] [PubMed] [Google Scholar]
- 33. Song C., Liu Y., Fontana R., Makrigiorgos A., Mamon H., Kulke M.H., Makrigiorgos G.M.. Elimination of unaltered DNA in mixed clinical samples via nuclease-assisted minor-allele enrichment. Nucleic Acids Res. 2016; 44:e146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ladas I., Yu F., Leong K.W., Fitarelli-Kiehl M., Song C., Ashtaputre R., Kulke M., Mamon H., Makrigiorgos G.M.. Enhanced detection of microsatellite instability using pre-PCR elimination of wild-type DNA homo-polymers in tissue and liquid biopsies. Nucleic Acids Res. 2018; 46:e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Baudrin L.G., Duval A., Daunay A., Buhard O., Bui H., Deleuze J.F., How-Kit A.. Improved microsatellite instability detection and identification by Nuclease-Assisted microsatellite instability enrichment using HSP110 T17. Clin. Chem. 2018; 64:1252–1253. [DOI] [PubMed] [Google Scholar]
- 36. Murphy D.M., Bejar R., Stevenson K., Neuberg D., Shi Y., Cubrich C., Richardson K., Eastlake P., Garcia-Manero G., Kantarjian H.et al.. NRAS mutations with low allele burden have independent prognostic significance for patients with lower risk myelodysplastic syndromes. Leukemia. 2013; 27:2077–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Galbiati S., Brisci A., Lalatta F., Seia M., Makrigiorgos G.M., Ferrari M., Cremonesi L.. Full COLD-PCR protocol for noninvasive prenatal diagnosis of genetic diseases. Clin. Chem. 2011; 57:136–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Milbury C.A., Correll M., Quackenbush J., Rubio R., Makrigiorgos G.M.. COLD-PCR enrichment of rare cancer mutations prior to targeted amplicon resequencing. Clin. Chem. 2012; 58:580–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Daunay A., Duval A., Baudrin L.G., Buhard O., Renault V., Deleuze J.F., How-Kit A.. Low temperature isothermal amplification of microsatellites drastically reduces stutter artifact formation and improves microsatellite instability detection in cancer. Nucleic Acids Res. 2019; 47:e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fazekas A., Steeves R., Newmaster S.. Improving sequencing quality from PCR products containing long mononucleotide repeats. BioTechniques. 2010; 48:277–285. [DOI] [PubMed] [Google Scholar]
- 41. Lo Y.M., Leung S.F., Chan L.Y., Chan A.T., Lo K.W., Johnson P.J., Huang D.P.. Kinetics of plasma Epstein-Barr virus DNA during radiation therapy for nasopharyngeal carcinoma. Cancer Res. 2000; 60:2351–2355. [PubMed] [Google Scholar]
- 42. Kassis A.I., Wen P.Y., Van den Abbeele A.D., Baranowska-Kortylewicz J., Makrigiorgos G.M., Metz K.R., Matalka K.Z., Cook C.U., Sahu S.K., Black P.M.et al.. 5-[125I]Iodo-2'-Deoxyuridine in the radiotherapy of brain tumors in rats. J. Nucl. Med. 1998; 39:1148–1154. [PubMed] [Google Scholar]
- 43. Breitbach S., Tug S., Helmig S., Zahn D., Kubiak T., Michal M., Gori T., Ehlert T., Beiter T., Simon P.. Direct quantification of cell-free, circulating DNA from unpurified plasma. PLoS One. 2014; 9:e87838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.