Abstract
Glucocorticoid receptor (GR) is an essential transcription factor (TF), controlling metabolism, development and immune responses. SUMOylation regulates chromatin occupancy and target gene expression of GR in a locus-selective manner, but the mechanism of regulation has remained elusive. Here, we identify the protein network around chromatin-bound GR by using selective isolation of chromatin-associated proteins and show that the network is affected by receptor SUMOylation, with several nuclear receptor coregulators and chromatin modifiers preferring interaction with SUMOylation-deficient GR and proteins implicated in transcriptional repression preferring interaction with SUMOylation-competent GR. This difference is reflected in our chromatin binding, chromatin accessibility and gene expression data, showing that the SUMOylation-deficient GR is more potent in binding and opening chromatin at glucocorticoid-regulated enhancers and inducing expression of target loci. Blockage of SUMOylation by a SUMO-activating enzyme inhibitor (ML-792) phenocopied to a large extent the consequences of GR SUMOylation deficiency on chromatin binding and target gene expression. Our results thus show that SUMOylation modulates the specificity of GR by regulating its chromatin protein network and accessibility at GR-bound enhancers. We speculate that many other SUMOylated TFs utilize a similar regulatory mechanism.
INTRODUCTION
Glucocorticoid receptor (GR) belongs to the nuclear receptor (NR) family and it functions as key transcription factor (TF) by mediating the effects of glucocorticoids in metabolism, development and immune response throughout the human body (1,2). Synthetic GR agonists are widely used pharmaceuticals due to their potent anti-immune effects (3). Glucocorticoids are also a central component of the therapy regimen for patients with GR positive lymphoid cancers, such as B cell acute lymphoblastic leukemia (B-ALL) (4). Upon binding to its agonist, GR translocates to the nucleus where it binds to gene enhancers to modulate their transcriptional state. To regulate genes, GR needs to interact and cooperate with coregulator proteins that, based on in vitro assays, are classically categorized as coactivators and corepressors. Functionally they act by (i) bridging the enhancer-bound TF to the RNA polymerase II (Pol2) machinery, (ii) promoting chromatin remodeling by sliding, switching or evicting nucleosomes or (iii) covalently modifying amino acids of histones and other proteins (5,6) to regulate chromatin accessibility or TF activity. However, unbiased investigations of protein networks around chromatin-bound GR are scarce.
Small ubiquitin-related modifier (SUMO) is a bulky covalent modification that targets especially nuclear proteins, including a number of physiologically important TFs. Mammalian cells express three SUMO isoforms SUMO1, -2 and -3, ca. 100-amino-acid proteins that can be conjugated to specific lysine residues on target proteins. SUMO2 and SUMO3 are nearly (97%) identical (herein collectively referred to as SUMO2/3), whereas SUMO1 has only ∼50% identity with SUMO2/3 (7,8). SUMOs activated by SAE1/2 heterodimer are conjugated to target lysines by UBC9. SUMO ligases, such as PIAS proteins, can assist SUMOylation by guiding the target residue to the active site of the UBC9 (9). The modification is highly reversible: SUMO-specific proteases (SENPs) rapidly cleave SUMOs from target proteins, releasing free SUMO for a new cycle of conjugation (10). Components of SUMO pathway, such as PIAS proteins, are previously recognized coregulators for NRs and other TFs (11).
In vitro studies have linked SUMOylation to transcriptional repression through the recruitment of corepressors (12), but recent unbiased genome-wide studies have shown that SUMOylation regulates chromatin occupancy of many key TFs at enhancers, targeting gene expression in a more versatile and site-specific manner (13–17). However, the molecular mechanism(s) by which SUMOylation influences TF binding/occupancy on chromatin have remained uncovered. In this work, by using chromatin immunoprecipitation with selective isolation of chromatin-associated proteins (ChIP-SICAP) and mass spectrometry (MS) from cells stably isotopically-labeled with amino acids in culture (SILAC) (18), we firstly uncover the composition of the chromatin protein network around the hormone-bound GR, which markedly overlaps with the factors important for the growth of B-ALL cells (19). Secondly, we show that the protein network is modulated by the SUMOylation status of GR. The effect of SUMOylation is reflected in our ATAC-seq, ChIP-seq and RNA-seq data, showing that SUMOylation affects chromatin accessibility and GR binding at glucocorticoid-regulated enhancers and thereby expression of GR target genes. A similar mechanism is likely utilized by many other SUMOylated TFs.
MATERIALS AND METHODS
Plasmids
For generation of pcDNA5/TO/FRT-3x-FLAG-PIAS1 complementary cDNA from CMV-3x-FLAG-PIAS1 was inserted as BamHI fragments into pcDNA5/TO (Invitrogen) backbone. For generation of N-terminally BirA*-tagged GR3KR (pcDNA5-FRT-TO-HA-BirA-GR3KR), cDNA of the human GR isoform alpha mutant (K277,293,703R) (20) was transferred with Gateway-cloning (Invitrogen) to the destination vector pcDNA5-FRT-TO-HA-BirA-GW (gift from Dr. Maria Vartiainen, University of Helsinki, Finland) as previously described (21). For generation of the N-terminally EGFP-tagged NCOA1, cDNA of the human NCOA1 isoform SRC1a from pSG5-SRC1a (22) (gift from Dr. Parker, Imperial Cancer Research Fund, London, UK) was transferred with Gateway-cloning (Invitrogen) to the destination vector pDest-C1-EGFP-GW (gift from Dr. Maria Vartiainen, University of Helsinki, Finland). The cloning services of the Genome Biology Unit (GBU) at the University of Helsinki were used for generation of the N-terminally mCherry-tagged GRwt and GR3KR constructs. Briefly, cDNA of the human GR isoform alpha and its 3KR mutant were transferred with Gateway-cloning (Invitrogen) to the destination vector mCherry-GW. All plasmids were verified by sequencing.
Antibodies
Anti-GR (sc-1003), IgG (sc-2025, used as a ChIP-seq control) and anti-GAPDH (sc-25778) were from Santa Cruz Biotechnology (Santa Cruz, CA, USA); anti-SUMO2/3 (M114-3, used for ChIP-seq and western blotting) was from MBL International Corporation (Woburn, MA, USA); anti-PIAS (ab77231) (recognize PIAS1 and PIAS2) and anti-H3K4me2 (ab7766) were from Abcam (Cambridge, UK); anti-RNA polymerase II (Pol2) (8WG16, MMS-126R) from Covance Inc. (Princeton, NJ, USA; currently from BioLegend, 664906); anti-NCOA1 (A300–343A) and anti-NCOR1 (A301–145A) from Bethyl Laboratories Inc. (Montgomery, TX, USA); IgG (10400C, used as a SUMO2/3 IP control) from Life Technologies (Carlsbad, CA, USA).
Cell lines and culture
Culturing of isogenic HEK293 cells (Flp-In™ -293, Invitrogen) stably expressing GRwt (HEK293flpGR) or GR3KR (HEK293flpGR3KR) was done as previously described (13). Our previous study (13) confirmed the suitability of these cell lines to investigate GR SUMOylation. Briefly, the cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Gibco/Invitrogen) supplemented with 10% (v/v) fetal bovine serum (FBS), 25 μg/ml penicillin, 25 μg/ml streptomycin and 100 μg/ml hygromycin-B (Invitrogen). Isogenic HEK293 cells (Flp-In 293 T-REx™, Invitrogen) expressing in a tetracycline (tet)-inducible manner PIAS1 (HEK293flpPIAS1) were generated according to manufacturer's instructions and as previously described (13,23). HEK293flpPIAS1 cells were maintained as HEK293flpGR cells with additional supplementation of 15 μg/ml blasticidin (Invitrogen). Generation and culturing of HEK293 cells (Flp-In 293 T-REx™, Invitrogen) expressing in a tet-inducible manner BirA*-GR3KR was performed according to manufacturer's instructions and as previously described (21). The cells were maintained in DMEM supplemented with 10% (v/v) FBS, 25 μg/ml penicillin, 25 μg/ml streptomycin, 50 μg/ml hygromycin-B and 15 μg/ml blasticidin. The tet-inducible BirA*-GRwt -expressing HEK293 cells were maintained as the BirA*-GR3KR cells. Cells were routinely tested for mycoplasma contamination. In all experiments, cells were grown in DMEM supplemented with 2.5% (v/v) charcoal-treated FBS (steroid-depleted medium) before treatments with 100 nM dexamethasone (Dex, Sigma-Aldrich).
ChIP-SICAP sample preparation
HEK293flpGR or HEK293flpGR3KR cells were grown in 10-cm dishes in SILAC labeled growth DMEM, high glucose, no glutamine, no lysine, no arginine (LIFE Technologies/Thermo Fisher Scientific, A1443101) supplemented with dialyzed 10% FBS (LIFE Technologies, 26400044) containing standard Glutamax, penicillin/streptomycin and Na-pyruvate. The proper amino acids were added to the corresponding medium at the final concentration of 84 μg/ml (for Arg) and 146 μg/ml final concentration (for Lys). Mixture of vehicle (ethanol, EtOH)-treated HEK293flpGR and HEK293flpGR3KR cells were metabolic labeled with light SILAC (Arg 0 and Lys 0, Sigma-Aldrich, A8094 and L8662), Dex-treated HEK293flpGR cells were labeled with medium SILAC (Arg 6 and Lys 4, Sigma-Aldrich, 643440 and 616192), and Dex-treated HEK293flpGR3KR cells were labeled with heavy SILAC (Arg10 and Lys8, Sigma-Aldrich, 608033 and 608041). GR interactomes from vehicle-treated cells were assumed not to differ between the cell lines, because the GR is not chromatin-bound in the absence of ligand and the ChIP-SICAP primarily identifies chromatin-bound interactors. Cells were treated with 100 nM Dex or vehicle 2 h before performing ChIP. Cells were crosslinked for 10min with 1% formaldehyde, crosslinking stopped with 120 mM glycine for 10min, and cells lysed in RIPA buffer (0.1% SDS, 0.5% Na-deoxycholate, 0.5% NP-40 in PBS 1×) for 10min on ice. Chromatin preparation and ChIP were performed as described for ChIP-seq with 2 μg of anti-GR antibodies followed by capture of on-chromatin GR interactors as described in (18). Briefly, 3′-ends of sheared DNA were in vitro biotinylated through terminal deoxynucleotidyl transferase (ThermoFisher Scientific EP0161) and used as bait in streptavidin pull down. Tryptic protein digestion was performed on streptavidin beads and the resulting peptides were subjected to peptide SP3 de-salting (24,25) before MS analysis.
MS analysis of ChIP-SICAP samples
Peptides were eluted in trifluoroacetic acid (TFA) 0.1% and then loaded on a trap column (PepMap100 C18 Nano-Trap 2 cm × 100 μm) followed by separation over a 50cm analytical column (Acclaim PepMap RSLC, 75 μm × 2 μm) using a 70 min linear gradient of acetonitrile from 6 to 40% (Thermo EasynLC 1200, Thermo Fisher Scientific). Peptides were analyzed on a Tri-Hybrid Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific) operated in data dependent acquisition mode with HCD fragmentation. The MS1 and MS2 scans were acquired in the Orbitrap and ion trap, respectively. Raw data were analyzed with MaxQuant version 1.5.1.2 based on the Andromeda search Engine (26,27) and peptide identification was performed using Uniprot database of Human (canonical). Protein and peptide FDR were set to 1%. Methionine oxidation and N-terminal acetylation were set as variable modifications, while carbamidomethylation of cysteine residues was set as fixed modification. Match between runs, second peptide and re-quantify options were active and both label-free quantification (LFQ) and intensity-based absolute quantification (iBAQ) scores were calculated. Only proteins with at least 1 unique peptide in both replicates were used for further processing steps. GR was retrieved among the top 15 proteins in each replicate according to the iBAQ scores. Proteins with iBAQ scores higher than GR included histones, abundant non-histone chromatin proteins (HMGN1, HMGN2, HMGB1), fc receptor-like protein 3 (FCRL3), ubiquitin and SUMO2 (Supplementary Table S1).
Statistical analyses of ChIP-SICAP MS data
Bioconductor package Linear Models for Microarray Data (limma, version 3.42.2) (28) within R (www.R-project.org) was used to determine differentially enriched proteins in ChIP-SICAP data by using log2(FC) SILAC-ratios from two biological replicates as input. The P-values were adjusted using Benjamini–Hochberg method. For determining Dex-induced interactors of the GRwt (M/L) or GR3KR (H/L), proteins with adjusted P-value <0.05 were considered as significantly Dex-induced. For proteins without light value the ratio was imputed with the constant 50, which corresponds to 5.64 upon log2 transformation. When interactor enrichment in GRwt was compared with GR3KR (H/M), distributions of the ratios were normalized by the median. Normalized ratios were used as input for statistical analysis with limma, and proteins with adjusted P-value <0.05, log2(H/M) <0 were considered GRwt-enriched and with adjusted P-value <0.05, log2(H/M) >0 GR3KR-enriched. For proteins without medium or high value the ratio was imputed with the constant 50 or 0.02, respectively, which correspond to 5.64 or –5.64 upon log2 transformation, respectively. Selected publications were used to determine composition of protein groups and complexes (29–35). Physical interactions between complex subunits were acquired from the STRING database (36) and complexes visualized with Cytoscape (37).
Analysis of SUMO2/3 targets
HEK293 cells expressing in a tet-inducible manner BirA*-GRwt were grown on 15-cm dishes (3 × 106 cells per dish) for 3 days after which the medium was replaced with steroid-depleted medium. After growing in steroid-depleted medium for 24 h, cells were treated with 30 ng/ml tet for the next 22 h, after which 100 nM Dex or vehicle (EtOH) was added for 2 h before harvesting. When harvesting, 20 mM final concentration of N-ethylmaleimide (NEM, E3876, Sigma-Aldrich) was added to the medium to inhibit de-SUMOylation. Cell lysis and immunoprecipitation with anti-SUMO2/3 were performed as described (38). Briefly, ∼5 × 108 cells were lysed, lysates sonicated with a probe sonicator and supplemented with 50 mM dithiothreitol. Lysates were boiled at 97 °C for 10 min, diluted with RIPA buffer and supplemented with 10 mM NEM, centrifuged at 16 000 × g for 15 min at 4 °C and supernatants filtrated through 0.45 μm membrane. SUMO2/3 antibody produced in-house as described (38) and normal mouse IgG (Life Technologies) were coupled to protein G agarose beads (5015952001, Roche Diagnostics) and lysates incubated with antibody-coupled beads 16 h at 4 °C. The beads were rigorously washed, and proteins eluted twice with 0.5 mg/ml SUMO2 epitope peptide (IRFRFDGQPI, Panatecs Biochemical Solutions, Germany). Eluates were combined and analyzed by MS.
MS analysis of SUMO2/3 targets and GR3KR BioID samples
BioID samples of GR3KR were prepared as described (21) MS analyses were performed on Orbitrap Elite hybrid mass spectrometer coupled to EASY-nLC II -system using the Xcalibur version 2.7.0 SP1 (Thermo Fisher Scientific). Proteins were identified using Proteome Discoverer™ software with SEQUEST search engine (version 1.4, Thermo Scientific). Thermo .raw files were searched against the human component of the UniProt-database complemented with trypsin, BSA and tag sequences. Trypsin was used as the enzyme specificity. Search parameters specified a precursor ion tolerance of 15 ppm and fragment ion tolerance of 0.8 Da, with up to two missed cleavages allowed for trypsin. Carbamidomethylation of cysteine residues was used as static modification whereas oxidation of methionine and biotinylation (only for BioID samples) of lysine residues or N-terminus were used as dynamic modification. Peptide FDR was calculated using Percolator node of software and set to <0.01. Spectral counting was used to produce semiquantitative data.
Statistical analyses of SUMO2/3 and GR BioID MS data
Significance analysis of interactome (SAINTexpress) (39,40) was used to determine significantly enriched proteins. In SUMO2/3 IP experiments, significantly SUMOylated proteins were determined by setting the IgG sample as the control and Dex and vehicle samples as the experiment. To determine enrichment of SUMO2/3 targets in Dex, the vehicle samples were set as the control. Proteins with FDR <0.05 were considered as significantly enriched in each comparison. SAINTexpress output files from SUMO2/3 IP are in Supplementary Table S2. In BioID experiments, significantly Dex-induced GR3KR interactors were determined with SAINTexpress by setting the Dex samples as the experiment and vehicle samples as the control. Proteins with loose filtering FDR <0.3 were considered as significantly enriched (Supplementary Table S3). Western blotting showed that streptavidin affinity-purified eluates from BirA*-GRwt and BirA*-GR3KR cell lines contain comparable levels of GR and biotinylated proteins (Supplementary Figure S5). Keratins, tubulins, bovine serum albumin and trypsin were considered as contaminants.
ChIP-seq
Chromatin immunoprecipitation (ChIP) was performed as described (13). Briefly, HEK293flpGR, HEK293flpGR3KR and HEK293flpPIAS1 cells were seeded at ∼70% confluence into 10-cm plates and allowed to grow in steroid-depleted transfection medium for 72 h. HEK293flpGR and HEK293flpGR3KR cells were treated with vehicle, or Dex (100 nM) for 1 h and HEK293flpPIAS1 cells with 100 ng/ml of tet for 24 h prior ChIP. In ML-792 (MedKoo Biosciences Inc., 407886) experiments, HEK293flpGR and HEK293flpGR3KR cells were exposed to 1 μM ML-792 (or DMSO as control) for 24 h. Subsequently, the cells were treated with vehicle or Dex (100 nM) for the final 1 h. Chromatin was fragmented to an average size of 300–500 bp by sonication (Bioruptor, UCD-300, Diagenode). Antibodies were coupled to magnetic protein G beads (Dynabeads, Invitrogen) for 16 h, sonicated lysates were incubated with antibody-coupled beads for 16 h. Antibodies used per IP: GR, 2 μg; NCOR1, 1.4 μg; NCOA1, 3 μg; Pol2, 1 μg, SUMO2/3, 1 μg; H3K4me2, 1 μg; PIAS, 1.5 μl; IgG, 1 μg. Between 2 and 4 IP samples were pooled for one ChIP-seq sample. ChIP-seq sequencing libraries were generated according to manufacturer's protocol using NEBNext Ultra II DNA Library Prep Kit (E7645L, NEB). In general, two biological replicate samples were sequenced using Illumina HiSeq 2000 (50SE) or NextSeq 500 (75SE).
ATAC-seq
ATAC-seq was performed as described (41). Briefly, HEK293flpGR and HEK293flpGR3KR cells were seeded, grown and hormone treated as in ChIP-seq. The cells were detached from the plates using 2 ml of Accutase (Thermo Fisher Scientific, A6964) by incubating 5 min at RT. Dex-treated cells were detached with Accutase in the presence of 100 nM Dex. Accutase was inactivated with 3.5 ml of growth media. For nuclei isolation, the cell pellets were resuspended in a concentration of 5 million cells per ml in Buffer A [15 mM Tris–HCl (pH 8), 15 mM NaCl, 60 mM KCl, 1 mM EDTA, 0.5 mM EGTA, 0.5 mM spermidine (Sigma-Aldrich, S2626), protease inhibitor cocktail]. Subsequently, equal volume of Buffer A with 0.04% (w/v) IGEPAL (Sigma-Aldrich, I8896) was added, to obtain a concentration of 2.5 million cells per ml with 0.02% (w/v) IGEPAL. Samples were incubated on ice for 10 min and washed two times with Buffer A without IGEPAL. Isolation of nuclei was verified by Trypan Blue counting. Subsequently, the rest of ATAC followed published protocol (42). Briefly, 100 000 nuclei were subjected to Tn5 transposition reaction using 2.5 μl TDE1 from Nextera DNA Library Prep Kit (Illumina, FC-121-1030). After adding the transposition reaction mix, the samples were incubated 45 min at 37°C with 800 rpm shaking, and subsequently DNA was purified using MinElute PCR Purification kit (Qiagen, 28004). Transposed DNA was PCR amplified and barcoded using published primers (42), and NEBNext High-Fidelity 2× PCR Master Mix (New England Biolabs, M0551S). Appropriate number of PCR cycles was determined as described (42). After PCR, the samples were size selected using SPRIselect (Beckman Coulter #B23317) to remove <150 bp and >800 bp fragments according to manufacturer's instructions. Size selection verification, library concentration and size were analyzed using Agilent Bioanalyzer High Sensitivity DNA Analysis kit (Agilent, 5067-4626). Two biological replicate samples were sequenced using Illumina NextSeq 500 (40PE).
RNA-seq and data analysis
HEK293flpGR and HEK293flpGR3KR cells were seeded onto 6-well plates and grown for 48 h, after which the medium was replaced with steroid-depleted medium. After growing in steroid-depleted medium for 24h, the cells were exposed to 1 μM ML-792 (or DMSO control) for 24 h and treated with vehicle or Dex (100 nM) for the last 6 h. RNA was extracted with RNeasy® Plus Mini Kit (Qiagen) and mRNA isolated using NEBNext Poly(A) mRNA Magnetic Isolation Module (E7490, New England Biolabs) according to manufacturer's recommendations. RNA-seq libraries were prepared using NEBNext Ultra II Directional RNA Library Prep Kit (E7765, New England Biolabs) and two biological replicate samples were sequenced with Illumina NextSeq 500 (75SE). RNA-seq raw reads were quality controlled, and adapters and reads smaller than 36 nt were removed. Trimmed raw reads were aligned to hg19 genome using STAR (43) with default settings. Total count per gene was calculated using TPM normalization and differentially expressed genes were analyzed with DESeq2 using HOMER (44) for all comparisons. RNA-seq analysis was performed as described (45). Genes with TPM >0.5 in any treatment were considered as expressed and those with adjusted P-value of <0.01 and log2(fold change) of <–0.5 or >0.5 as differentially expressed. Differentially expressed gene sets were subjected to pathway analysis with DAVID functional annotation tool (GOTERM_BP_DIRECT) (46,47).
ChIP-seq and ATAC-seq data analysis
ChIP-seq data analysis was performed as previously described (13,48). For ATAC-seq data, after filtering low quality reads as with ChIP-seq data analysis, paired-end samples were aligned to hg19 genome using Bowtie2 (49). Alignment was performed with end-to-end sensitive mode allowing no mismatches. Around 20% of the reads were mapped to mitochondrial DNA. From the two biological replicate sample, at least 50 million unique non-mitochondrial reads were obtained for each condition. Downstream data analysis was performed using HOMER (44). Peaks in each dataset were called using findPeaks with style factor, FDR <0.01, > 25 tags, >4-fold over control sample and local background. IgG sample from the background HEK293flpFRT cells was used as control sample. DESeq2 (50) through getDiffrentialPeaksReplicates.pl was used to isolate differential binding peaks (FDR < 0.05, fold change [FC] > 2) between the GRwt and GR3KR (C1–C3). For determination of pre-bound, Dex-induced, and non-GR SUMO2/3 sites, SUMO2/3 peaks were called from each sample as indicated above. Dex-induced SUMO2/3 sites peaks showed tag FC > 4 Dex/EtOH, and Poisson P-value <0.0001. SUMO2/3 sites that did not meet these criteria were classified as non-induced SUMO2/3 sites. Overlap of non-induced SUMO2/3 and GR peaks defined pre-bound SUMO2/3 and non-GR SUMO2/3 sites. GR-binding sites changed upon ML-792 treatment were determined with the same criteria. For determination of Dex-independent and -dependent sites, indicated EtOH or Dex ChIP-seq samples from GRwt and GR3KR cells were combined and peaks called as indicated above. Dex-dependent peaks showed less than 1 tag per bp per site at EtOH samples and showed tag FC > 4 Dex/EtOH, with Poisson P-value <0.0001. Sites that did not meet these criteria were classified as Dex-independent sites. The following count of sites were determined as Dex-independent; 42 977 for NCOA1, 8 584 for NCOR1 and 89 260 for SUMO2/3. The following count of sites were determined as Dex-dependent; 67 458 for NCOA1, 11 848 for NCOR1 and 25 766 for SUMO2/3. Chromatin accessibility populations were defined on the basis of FC; Dex-induced sites had FC > 4 and Dex-decreased FC < 4 Dex/EtOH, with Poisson P-value <0.0001. The rest of the sites were classified as unresponsive to Dex treatment. Pre-accessible and de novo C2 sites were defined on the basis of ATAC-seq data. De novo sites had <1.5 log2 tags, while pre-accessible sites had >1.5 log2 tags in EtOH-treated ATAC-seq sample from GRwt cells. Aggregate plots and heatmaps were generated with 10 bp or 20 bp bins surrounding ±1 kb area around the center of the peak. All plots were normalized to 10 million mapped reads and further to local tag density, tags per bp per site. Box plots represented either log2 tag counts or log2 tag count comparison of Dex/EtOH. Log2 transformed tag counts were used in the scatter plots. AnnotatePeaks.pl was used to calculate the enrichment of sites to different genomic location; promoter, intron, intergenic, exon and miscellaneous (other) sites. Miscellaneous sites consist of UTRs and non-coding RNAs. De novo motif searches were performed using findMotifsGenome.pl with the following parameters; 200 bp peak size window, strings with 2 mismatches, binomial distribution to score motif P-values, and either 50 000 background regions (GR binding sites) or 250 000 background regions (ATAC sites) (Supplementary Table S4). Motif heatmap was generated using hierarchical clustering with Euclidean distance. Association of GR binding sites to Dex-regulated genes (peak-centric analysis) was performed on the basis of linear distance using AnnotatePeaks.pl. C1–C3 cluster sites were checked against the union of Dex up-regulated or Dex down-regulated genes in GRwt and GR3KR cells using RNA-seq data. The log2 Dex/EtOH induction was shown in box plots. Statistical significance was determined either with unpaired two-sample t-test (two samples), or with One-way ANOVA with Bonferroni post hoc test (three or more samples). Published datasets were analyzed as indicated above.
FRAP
HEK293 cells were seeded to μ-slide eight-well chambers (Ibidi) in DMEM supplemented with 2.5% (v/v) charcoal-treated FBS (steroid-depleted medium) and transfected with constructs expressing EGFP-tagged NCOA1 or NCOA2 and mCherry-tagged GRwt or GR3KR. Two days later, cells were induced with 100 nM dex for 1h, and the nucleus was scanned using 488 and 561 nm excitations at 250-ms intervals with Zeiss LSM 800 microscope. After 20 scans, a high intensity bleach pulse at 488 and at 561 nm were applied simultaneously to a 1.5-μm wide rectangular area spanning the nucleus and scanning of the nucleus was continued until equilibrium in fluorescence distribution was reached. The fluorescence recovery was analyzed from the bleached area. Measurements with NCOA1 or NCOA2 together with GR were performed in two individual experiments where a minimum of 10 nuclei were measured in each experiment.
Public datasets
The following publicly available sequencing datasets were used: GSE48379 for GR and SUMO-2/3 ChIP-seq in HEK293flpGR cells, GR and SUMO-2/3 ChIP-seq in HEK293flpGR3KR cells, and IgG ChIP-seq in HEK293flpFRT cells (13); GSE32970 for DNase-seq in HEK293T cells (51); GSE30263 for CTCF ChIP-seq in HEK293 cells (52); ENCSR000FCH for H3K27ac ChIP-seq in HEK293 cells (53); ENCSR000FCJ for H3K9me3 ChIP-seq in HEK293 cells (53); ENCSR000FCG for H3K4me1 ChIP-seq in HEK293 cells (53); GSE35583 for H3K4me3 ChIP-seq in HEK293 cells (52); GSE91583 for GFP-ATF2 ChIP-seq in HEK293flp cells (52).
RESULTS
Protein interaction network of the chromatin-bound GR
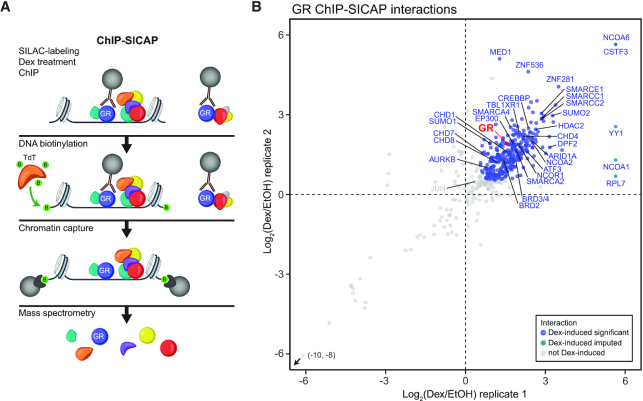
To better understand the mechanisms by which GR regulates gene transcription, we applied ChIP-SICAP, which purifies exclusively chromatin-bound proteins (18) to reveal the composition of the protein network around the chromatin-bound GR (Figure 1A). Medium SILAC-labelled isogenic HEK293 cells that stably express GR (HEK293flpGR) (13) were exposed to dexamethasone (Dex, a synthetic GR agonist), while light SILAC-labelled cells were treated with ethanol (EtOH, vehicle) before performing ChIP-SICAP experiment. All samples showed comparable levels of GR capture in ChIP, retrieving GR among the most abundantly identified proteins in each sample (Supplementary Figure S1A). Biological replicates also showed good correlation between the obtained networks (Pearson's r = 0.827) (Figure 1B). ChIP-SICAP identified 317 (significant with adjusted P-value < 0.05 or imputed) Dex-induced GR interactors (i.e. interacting with the receptor or binding at the same 200–300 bp chromatin fragment) as determined by applying moderated t-statistics with linear model analysis (limma) (28) (Figure 1B). The full list of Dex-induced GR interactors is reported in Supplementary Table S1. Dex-induced interactors included expected NR coactivators (e.g. NCOA2, EP300 and CREBBP), NR corepressors (e.g. NCOR1, TBL1XR1 and HDAC2), and AP-1 components (JUN and ATF3), as well as several other transcriptional coregulators and proteins involved in chromatin remodeling. The latter group includes members of the bromodomain and extra-terminal motif (BET) family (BRD2 and BRD3/4), chromodomain helicase DNA-binding (CHD) family (CHD1, CHD4, CHD7 and CHD8) and different subunits of the BAF (SWI/SNF) complex (ARID1A, SMARCC1, SMARCC2, SMARCE1, SMARCD2, DPF2, ACTL6A, SMARCA4 and SMARCA2).
Figure 1.
GR interacts with various coregulators on chromatin. (A) Schematic representation of the ChIP-SICAP protocol. (B) Scatter plot showing chromatin-associated proteins identified with GR ChIP-SICAP. Blue dots with blue rim represent significantly (adj. P-value < 0.05) Dex-induced interactions, blue dots with green rim represent imputed Dex-induced interactions and grey dots represent interactions that are not Dex-induced.
Next, we compared our GR ChIP-SICAP data of Dex-induced GR interactors with genes that in a genome-wide RNAi screen influenced the growth and Dex-sensitivity of NALM-6 cells that model B-ALL (19). Intriguingly, many of the identified GR chromatin partners were products of genes influencing the growth (116 genes) and Dex-sensitivity (47 genes) (Supplementary Figure S2) of NALM-6 cells. These results indicate that many of the GR chromatin partners in HEK293 cells have physiologically relevant functions also in B-ALL cells.
GR and many of its chromatin partners are SUMO2/3 targets
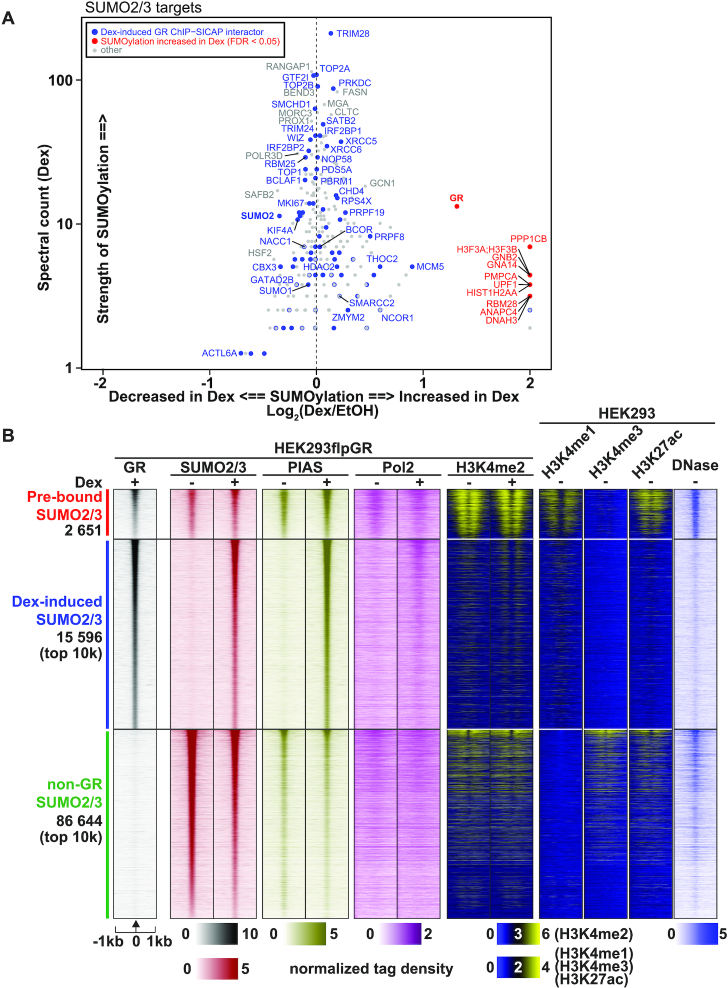
Several studies have shown that GR is modified by SUMO2/3 in various cell lines (54–63), including HEK293 cells (63) (Supplementary Table S2). To verify this hypothesis in our experimental setup, we employed the protocol by Barysch et al. (38) with a monoclonal SUMO2/3 antibody to identify SUMO2/3 targets in Dex- and vehicle-treated HEK293 cells that stably express GR. Significance analysis of interactome (SAINTexpress) (39,40) identified 450 proteins as significant (FDR < 0.05) SUMO2/3 targets when compared to the IgG control (Figure 2A). Most of them (91%) (including all our top 100 SUMO2/3 targets) have been identified as SUMO2/3 targets in other studies (54–67) (Supplementary Table S2). Interestingly, ca. 26% (82 of 317) of Dex-induced GR interactors were identified as SUMO2/3 targets (Figure 2A). Some of them, such as TRIM28, TOP2A and TOP2B, were among the most SUMOylated proteins. SUMOylation of 11 proteins, including the GR, was significantly (FDR < 0.05) increased in response to Dex treatment (Figure 2A, Supplementary Table S2), whereas no protein showed reduced modification in response to Dex. The GR was targeted by SUMO2/3 only in the presence of Dex, indicating that the binding of agonist triggers the SUMOylation of the receptor.
Figure 2.
GR and its chromatin partners are SUMO2/3 targets. (A) Scatterplot showing proteins identified as significantly SUMOylated (enriched over IgG, SAINTexpress FDR < 0.05) in SUMO2/3-pulldown in HEK293 cells expressing GR. X-axis shows the effect of Dex on the SUMOylation of each protein as Log2(Dex/EtOH). Spectral count (Dex) on the y-axis represents relative abundancy of the protein in the anti-SUMO2/3 immunoprecipitates. Spectral counts were normalized to the total spectral count sum of each sample. Significant (adj. P-value < 0.05) or imputed Dex-induced interactors identified with GR ChIP-SICAP are shown in blue. Proteins with significantly (FDR < 0.05) increased SUMOylation in Dex compared to vehicle are shown in red. There were no Dex-induced GR ChIP-SICAP interactors with significantly increased SUMOylation in Dex. (B) Heat maps representing GR, SUMO2/3, PIAS, Pol2 and H3K4me2 ChIP-seq data from HEK293flpGR (wt) in the presence and absence of Dex, and H3K4me1, H3K4me3 and H3K27ac ChIP-seq data from HEK293, and DNase-seq (DNase) data from HEK293T cells. Each heat map represents ±1kb around the center of the SUMO2/3 peak. Binding intensity (tags per bp per site) scale is noted below on a linear scale. All heat maps are normalized to a total of 10 million reads, and further to local tag density. Clusters represent from top to bottom; GR sites with pre-bound SUMO2/3, GR sites with Dex-induced SUMO2/3 (only top 10k shown), and non-GR SUMO2/3 sites (only top 10k shown).
Next, we performed ChIP-seq with SUMO2/3 antibody in vehicle and Dex treated HEK293flpGR cells and compared it to our previously published GR cistrome from HEK293 cells (13) to examine whether the SUMOylation of GR interactors is reflected on the genome-wide occupancy of SUMO2/3 at GR-binding sites (GRBs). We also performed ChIP-seq for PIAS SUMO ligases (11) to examine whether these components of the SUMOylation machinery are present at the binding sites. In parallel, we used antibodies against RNA polymerase II (Pol2) and the active histone mark H3K4me2. Clustering of ChIP-seq obtained from SUMO2/3 chromatin-binding sites yielded three clusters: a small cluster of pre-bound SUMO2/3 (GRBs that are already bound by SUMO2/3 prior to Dex exposure), a cluster of Dex-induced SUMO2/3 (GRBs that bind SUMO2/3 only after Dex exposure) and a larger cluster of non-GR SUMO2/3 (SUMO2/3-bound sites that do not bind GR) (Figure 2B). As expected, Dex-induced chromatin binding of PIAS proteins mirrored that of SUMO2/3 in HEK293flpGR cells (Figure 2B). In the absence of Dex, overexpression of PIAS1 (HEK293flpPIAS1 cells) did not force binding of SUMO2/3 or PIAS to these Dex-responsive sites, further confirming that their binding is highly dependent on the activation of GR (Supplementary Figure S3A). Based on publicly available chromatin accessibility (DNase-seq) and histone modification (ChIP-seq) data from HEK293 cells, we can speculate that chromatin sites where Dex induces the binding of SUMO2/3 are less accessible and harbor less of active histone modifications (H3K27ac and H3K4me3) compared to other SUMO2/3-bound sites (Figure 2B, Supplementary Figure S3B). However, enhancers, marked by H3K4me1, are enriched among Dex-induced sites compared to non-GR SUMO2/3 sites (Figure 2B, Supplementary Figure S3B). After Dex induction, an active histone modification H3K4me2 was redistributed (Figure 2B, Supplementary Figure S3C) and the occupancy of Pol2 was increased at the Dex-induced SUMO2/3 chromatin-binding sites (Figure 2B), both indicative of enhancer activation. Taken together, these findings indicate that a significant fraction of GR chromatin partners are SUMO2/3 targets and their recruitment to GRBs leads to redistribution of SUMO2/3 on chromatin. This notion is also supported by the identification of SUMO2 as a Dex-induced GR chromatin partner in ChIP-SICAP (Figure 1B).
SUMOylation of GR modulates the protein interaction network of the chromatin-bound GR
The main SUMOylated target lysines of the GR (K277 and K293) are located at ΨKxE (where Ψ is a hydrophobic residue) SUMO consensus motifs in the N-terminal domain of the receptor (63), while the C-terminal domain harbors a weaker SUMO consensus motif-embedded lysine (K703). We have previously shown that mutation of lysines at the three SUMO consensus motifs leads to a SUMOylation-defective GR (GR3KR) that shows enhanced activity in reporter gene assays (20), that binds more prevalently to chromatin and that harbors a distinct chromatin-binding pattern from the wild-type receptor (GRwt) (13).
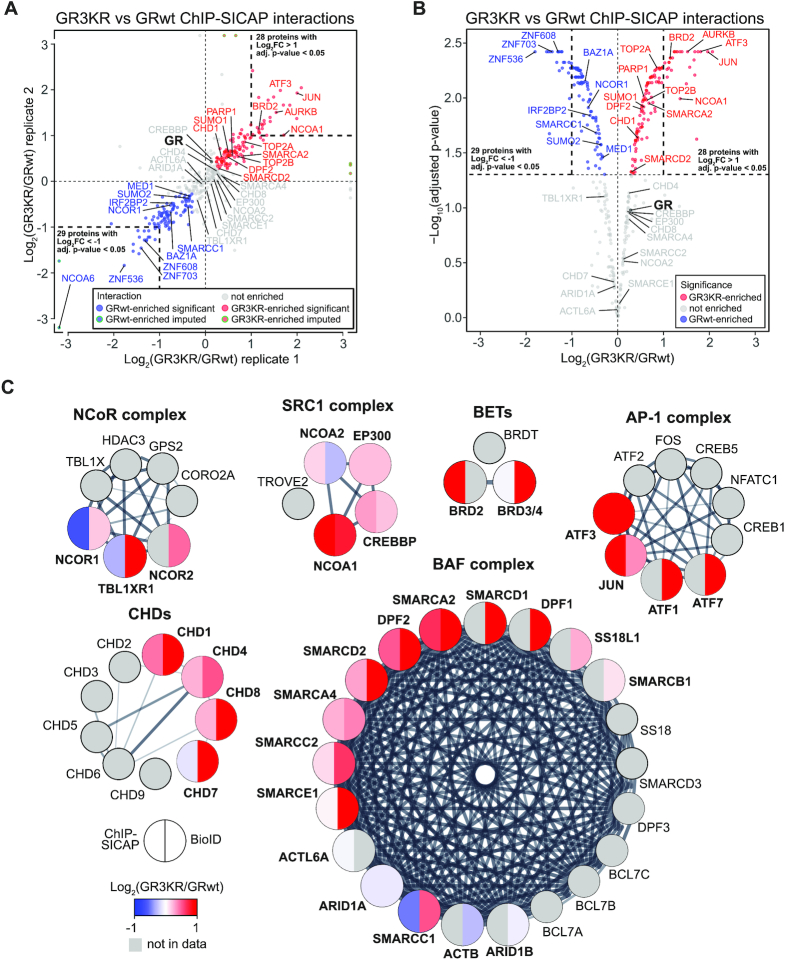
To investigate the role of the SUMO target lysines of GR on the receptor's on-chromatin interactome, we applied ChIP-SICAP in heavy-labeled HEK293flp cells expressing GR3KR (HEK293flpGR3KR) and compared the interactome to that of GRwt. Western blotting showed that the cellular expression of GR3KR protein is slightly lower than that of GRwt (Supplementary Figure S4A and B). The GR3KR ChIP-SICAP samples were processed in parallel with the (above described) GRwt samples in a triple SILAC experimental setting. Western blotting analysis confirmed that GRwt and GR3KR inputs, flow-throughs and eluates from ChIP contained comparable levels of GR protein (Supplementary Figure S4C–E). Both GR3KR replicates showed comparable levels of GR capture in ChIP-SICAP, retrieving GR among the most abundantly identified proteins (Supplementary Figure S4F). Biological replicates also showed concordant interactomes (Pearson's r = 0.845) (Supplementary Figure S4G). ChIP-SICAP identified 342 (significant with adjusted P-value < 0.05 or imputed) Dex-induced GR3KR interactors as determined with limma (Supplementary Figure S4G and H, Supplementary Table S1).
Next, we compared the Dex-induced chromatin partners of GR3KR to GRwt by using the heavy/medium SILAC ratios. Biological replicates showed concordant interactomes (Pearson's r = 0.871). Analysis with limma revealed that, from Dex-induced GR interactors, 102 proteins significantly preferred GRwt (adjusted P-value < 0.05 or imputed, and log2[3KR/wt] < 0), whereas 122 proteins preferred GR3KR (adjusted P-value < 0.05 or imputed, and log2[3KR/wt] > 0) (Supplementary Table S1). GRwt-preferred interactors encompassed coactivators NCOA6 and MED1, but this group of proteins was interestingly more enriched with corepressors or proteins implicated in transcriptional repression, including NCOR1, IRF2BP2, ZNF536, ZNF608 and ZNF703 (NLZ1) and BAZ1A (ACF1) (Figure 3A and B, Supplementary Table S1). GR3KR-preferred interactors in turn included SMARCA2 ATPase subunit, SMARCD2 and DPF2 from the BAF chromatin remodeling complex, chromatin remodeling factor CHD1, BET family chromatin reader BRD2, topoisomerase II homologues TOP2A and TOP2B, Aurora kinase B (AURKB), coactivator NCOA1, and JUN and ATF3 from the AP-1 TF complex (Figure 3A and B, Supplementary Table S1). We next complemented the comparison of the on-chromatin interactome of GRwt and that of GR3KR by using proximity-dependent biotin identification (BioID) which maps protein-protein interactions of Dex-activated GR in the nuclear space (21). BioID employs fusion of GR to a mutated Escherichia coli biotin ligase (BirA*) that covalently attaches biotin to primary amines within 10 nm range (68). BioID revealed 138 Dex-induced GR3KR interactors as determined with SAINTexpress (FDR < 0.3) (39,40) (Supplementary Table S3). Comparison to GRwt BioID data (21) showed that 79 proteins preferred GR3KR over GRwt (FDR < 0.3), whereas only one protein preferred GRwt (Supplementary Table S3). Although the ChIP-SICAP and the BioID yielded differences, notably, the GR3KR-preferred interactions with the BAF chromatin remodeling complex subunits SMARCA2 and DPF2, CHD1 and ATF3 were also found as GR3KR-preferred interactions in BioID. Additionally, chromatin remodelers CHD7 and CHD8, and ATF1 and ATF7 of the AP-1 TF complex were GR3KR-preferred interactors in BioID (Figure 3C, Supplementary Table S3).
Figure 3.
A number of transcriptional regulators prefer SUMOylation-deficient GR to SUMOylation-competent GR. (A) Scatter plot showing correlation of on-chromatin GR interactors enriched in GR3KR over GRwt cells in the biological replicates. Only Dex-induced interactions are shown. Blue dots represent GRwt-enriched interactions (adjusted P-value < 0.05 or imputed, and log2[3KR/wt] < 0), red dots GR3KR-enriched interactions (adjusted P-value < 0.05 or imputed, and log2[3KR/wt] > 0) and gray dots interactions that are not enriched. (B) Volcano plot representing significance of interactions that are differentially enriched with GR3KR compared to GRwt. Only Dex-induced interactions are shown. Dot colors are the same as in (A), but imputed values are not shown. (C) GR interactor enrichment in nuclear protein complexes. Color of each protein represents enrichment in GRwt (blue) compared with GR3KR (red). Left side of the circle represents interactor enrichment in ChIP-SICAP and right side in BioID data. Proteins in grey belong to the protein complex but were not found in proteomics data. Interactions between complex subunits were acquired from the STRING database. Edge thickness represents confidence of interaction.
Inhibition of SUMOylation using a selective SAE inhibitor enhances GR chromatin binding and target gene expression
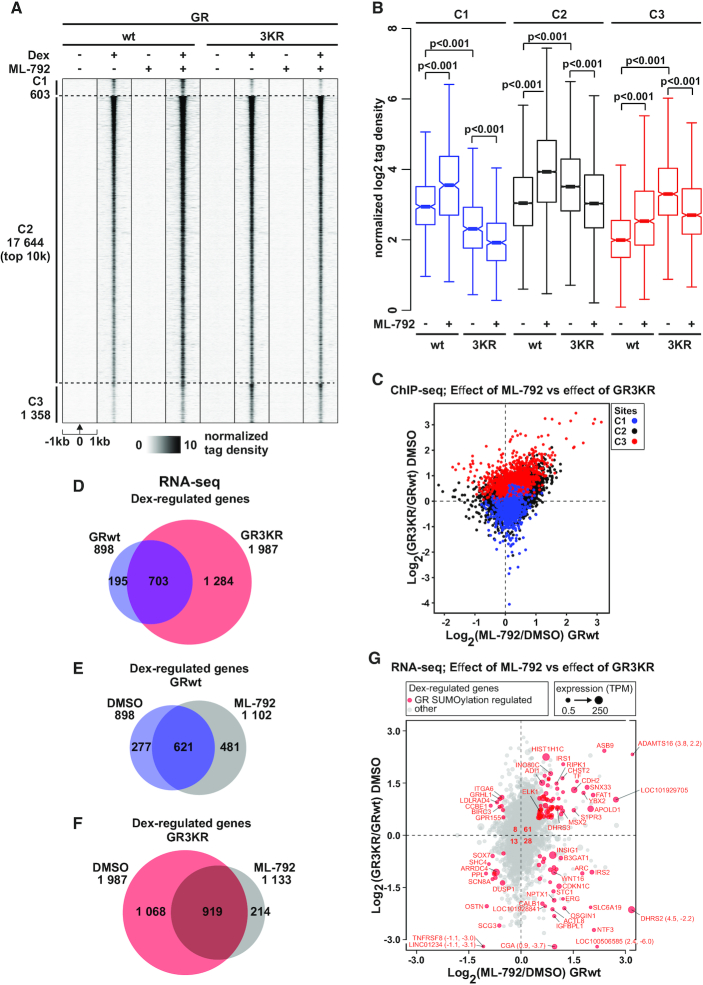
We have previously shown that SUMOylation of GR enhances chromatin occupancy and target gene expression of GR, not at all loci, but in a locus-selective manner (13). To confirm the results obtained with SUMOylation site-mutated receptor and that the mutation principally compromised the SUMOylation, not other lysine modifications in the receptor, we utilized a novel potent SUMO-activating enzyme (SAE) inhibitor ML-792 (69) in ChIP-seq and RNA-seq experiments. We first re-determined and clustered the chromatin GR-binding sites (GRBs) from our previous study (13) (see Materials and Methods for details) as sites preferred by GRwt (cluster [C]1, blue color), sites shared by GRwt and GR3KR (C2, black color), and sites preferred by GR3KR (C3, red color) (Figure 4A, Supplementary Figure S6A-B). We first confirmed by western blotting with SUMO2/3 antibody that exposure of HEK293flpGR cells to ML-792 indeed results in a robust inhibition of protein SUMOylation (Supplementary Figure S6C). HEK293flpGR and HEK293flpGR3KR cells were exposed to ML-792 (or DMSO) for 23 h before 1 h treatment with Dex (or vehicle), and ChIP-seq was performed with GR antibody. In the absence of SUMOylation inhibitor, GR and GR3KR showed their preferred binding to C1–C3 GRBs as described previously (Figure 4A and B). In line with the enhanced chromatin occupancy of the SUMOylation-deficient GR, inhibition of SUMOylation significantly enhanced the chromatin occupancy of SUMOylation-competent GR at C1, C2 and C3, resulting also in > 3 500 novel GRwt-binding sites (Figure 4A and B) (Supplementary Figure S6D and E). As expected, ML-792 was not able augment the chromatin occupancy of GR3KR, but the inhibitor interestingly weakened the binding of SUMOylation-deficient receptor at all clusters. The latter finding suggest an interplay between the SUMOylation of the receptor and other chromatin proteins in the regulation of chromatin GR. Cross-comparison of the effect of GR SUMOylation sites and that of ML-792 on the chromatin occupancy of the receptor indicated a large number of the C3 GRBs with enhanced occupancy of GRwt upon inhibition of SUMOylation (Figure 4C). Examples of genome browser tracks of GRBs unchanged and induced by ML-792 are shown in Supplementary Figure S7A–D.
Figure 4.
SUMOylation inhibitor ML-792 blunts the differences in GR chromatin binding and target gene expression between GRwt and GR3KR cells. (A) GR ChIP-seq profiles from DMSO- or ML-792-treated HEK293flpGRwt (wt) and HEK293flpGR3KR (3KR) cells in the presence or absence of Dex. Each heat map represents ±1 kb around the center of the GR peak. C1 represents GRwt-preferred, C2-shared, and C3 GR3KR-preferred binding sites. Only top 10 000 sites (top 10k) are shown for C2. Binding intensity (tags per bp per site) scale is noted below on a linear scale. All heat maps are normalized to a total of 10 million reads, and further to local tag density. (B) Box plots representing normalized log2 tag density of ML-792 GR ChIP-seq data at C1 (blue), C2 (black) and C3 (red) sites. P-values were calculated with one-way ANOVA with Bonferroni post hoc test. All box plots are normalized to total of 10 million reads. Blue color represents GRwt (wt), red color GR3KR (3KR). (C) Scatter plot representing GR ChIP-seq log2 tag density of ML-792/DMSO from GRwt cells (x-axis) and GR3KR/GRwt from DMSO treated cells (y-axis). GR-binding clusters are color coded, C1 (blue), C2 (black) and C3 (red). (D–F) Comparison of Dex-regulated genes from RNA-seq between (D) DMSO-treated GRwt and GR3KR cells, (E) ML-792- and DMSO-treated GRwt cells, and (F) ML-792- and DMSO-treated GR3KR cells. (G) Scatter plot representing RNA-seq log2 fold change of ML-792/DMSO from GRwt cells (x-axis) and GR3KR/GRwt from DMSO treated cells (y-axis). Circle size depicts TPM values. Genes that are significantly regulated by both ML-792 (adjusted P-value < 0.01 and log2[ML-792/DMSO] > 0.5 or < –0.5) and GR3KR (adjusted P-value < 0.01 and log2[GR3KR/GRwt] > 0.5 or < –0.5) are highlighted in red (GR SUMOylation regulated genes). Number of GR SUMOylation regulated genes belonging to each sector is shown in the middle. Figure shows only genes that are Dex-regulated (adjusted P-value < 0.01 and log2[Dex/EtOH] > 0.5 or < –0.5).
Since inhibition of SUMOylation in GRwt cells blunted the differences in chromatin binding between GRwt and GR3KR cells, we next analyzed the effect of ML-792 also on Dex-regulated gene expression by RNA-seq as described in Methods. Firstly, RNA-seq analyses recapitulated the main differences in the Dex-regulated gene expression between GRwt and GR3KR cells previously measured by gene arrays (13) (Figure 4D, Supplementary Figure S8A). For example, FKBP5 and TSC22D3 showed similar Dex regulation, while CDKN1C and MAFB displayed significantly stronger Dex induction by GRwt. ELK1 and CXXC4 in turn showed significantly more robust Dex induction by GR3KR (13) (Supplementary Figure S8B). In agreement with the ChIP-seq data, inhibition of SUMOylation increased the number of Dex-regulated genes in GRwt cells (Figure 4E, Supplementary Figure S8B), whereas the inhibition decreased their number in GR3KR cells (Figure 4F). Cross-comparison of the effect of GR SUMOylation sites and that of ML-792 on the target gene expression indicated that over half of the GR3KR target genes that are also regulated by ML-792 show enhanced Dex regulation in GRwt cells upon ML-792 treatment (Figure 4G). Pathway analyses indicated that both GR SUMOylation sites and ML-792 influence expression of genes associated with cell growth and proliferation processes (Supplementary Figure S8C), which is in line with the effect of GR SUMOylation sites on cell growth characteristic and growth response to Dex (13). Finally, we associated C1–C3 sites (ChIP-seq data) to Dex-regulated genes (RNA-seq data) (Supplementary Figure S8D and E). At the C1 sites, the Dex up-regulation did not significantly differ between the receptor forms, and ML-792 did not yield any significant effect, whereas the Dex up-regulation differed at the C2 and C3 sites, with GR3KR showing stronger Dex induction (Supplementary Figure S8D). ML-792 enhanced Dex up-regulation at C2 sites in GRwt (P < 0.001) and GR3KR cells (P < 0.05) and resulted in an increasing trend of Dex induction at C3 sites in GRwt cells but not in GR3KR cells. Overall, our results with the SUMOylation inhibitor ML-792 are in line with the consequences of the GR SUMOylation site mutations, thus confirming the role of GR SUMOylation in the regulation of receptor chromatin occupancy and target gene regulation.
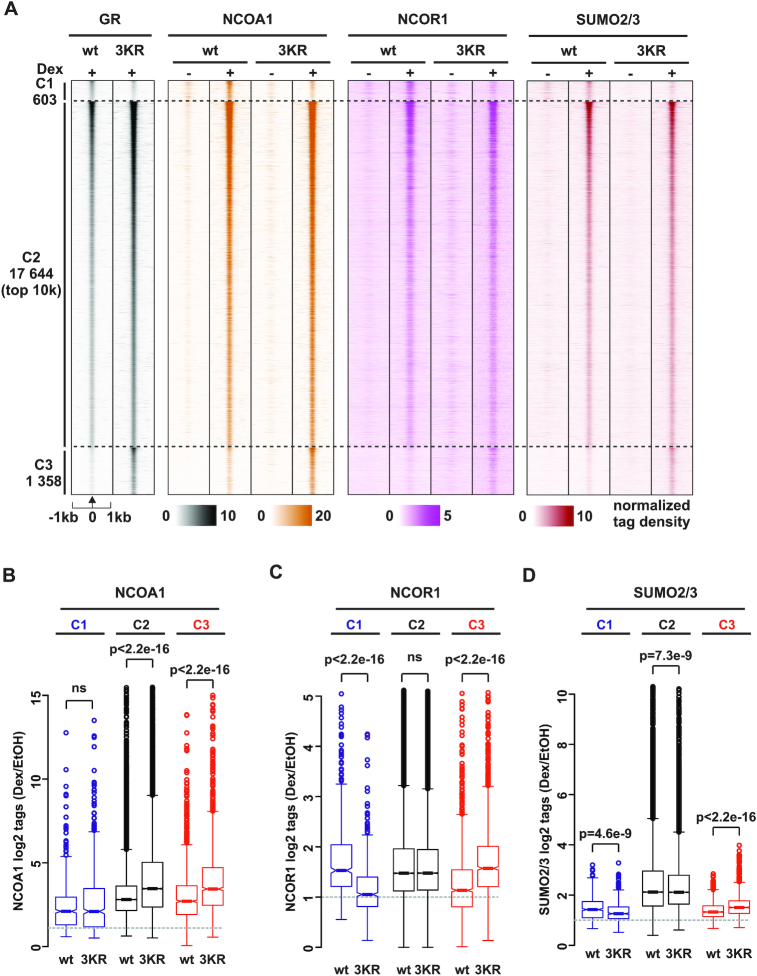
SUMOylation-deficient GR recruits more efficiently NCOA1, but not NCOR1, to chromatin
Next, we examined whether the differences in the interactomes of GRwt and GR3KR are reflected in the chromatin occupancy of the interactors. For these analyses, we focused on NCOA1, a GR coactivator that was significantly GR3KR-enriched in ChIP-SICAP and showed a marked GR3KR-enrichment in BioID (Figure 3C). We also determined the chromatin occupancy of corepressor NCOR1 that showed significant enrichment with GRwt in ChIP-SICAP, but a slight preference for GR3KR in BioID, and whose recruitment to certain GR-repressed chromatin loci has been reported to be regulated by the SUMOylation of the receptor (70,71) (Figure 3C). Lastly, since SUMO2 was significantly GRwt-enriched in ChIP-SICAP cells and we have observed differences in chromatin occupancy of SUMO2/3 between in Dex-treated GRwt and GR3KR cells (13), we determined the occupancy in vehicle-treated cells. We performed ChIP-seq experiments in HEK293flpGR and HEK293flpGR3KR cells with specific antibodies in the presence of Dex or vehicle and compared the obtained NCOA1-, NCOR1- and SUMO2/3-binding profiles to GRwt and GR3KR cistromes from the same cells (Figure 4A, Supplementary Figure S6A and B) (13). We first determined Dex-independent and Dex-dependent chromatin sites for NCOA1, NCOR1 and SUMO2/3 (see Materials and Methods for details). As shown in Supplementary Figure S9A, chromatin occupancy of these coregulators and that of SUMO2/3 do not differ between GRwt and GR3KR cells at the Dex-independent sites, whereas at the Dex-dependent sites, the NCOA1, but not the NCOR1, shows a stronger enrichment in GR3KR than in the GRwt cells. SUMO2/3 in turn shows a stronger enrichment in GRwt than in GR3KR cells at Dex-dependent sites (13). Subsequently, we investigated the occurrence of these proteins at individual GRB clusters. As shown in Figure 5A and B, the Dex-induced recruitment of NCOA1 is significantly stronger at C2 and C3 in GR3KR cells compared to GRwt cells, whereas there is no such difference at C1. In contrast, the occupancy of NCOR1 does not differ between the GRwt and GR3KR cells at C2, while at C1 and C3, the occupancy mirrored the preferred binding of GRwt and GR3KR, respectively (Figure 5A, C), with NCOR1 being more enriched at C1 in GRwt cells and at C3 in GR3KR cells. Also, the occurrence of SUMO2/3 at C1 and C3 mirrored the binding preference of GRwt and GR3KR. Thus, the enhanced interaction of GR3KR with NCOA1 in our proteomic assays is in line with increased Dex-induced recruitment and co-occupancy of NCOA1 at the GRBs in GR3KR-expressing cells in our genomic assays. Since the chromatin occupancy of GR3KR is higher than GRwt at most GRBs, this could contribute to the differential co-occupancy of NCOA1 at these sites. However, the NCOR1 in turn occupies the GRBs in GRwt- and GR3KR-expressing cells in a similar manner, suggesting that the GR-NCOR1 interaction at the genome-wide level is not generally influenced by the SUMOylation status of the GR.
Figure 5.
Recruitment of NCOA1, but not NCOR1, to chromatin is influenced by SUMOylation status of GR. (A) Heat maps showing GR, NCOA1, NCOR1 and SUMO2/3 ChIP-seq profiles from HEK293flpGRwt (wt) and HEK293flpGR3KR (3KR) cells in the presence and absence of Dex. Each heat map represents ±1 kb around the center of the GR peak. C1 represent GRwt preferred, C2 shared, and C3 GR3KR preferred binding sites. Only top 10 000 sites (top 10k) are shown for C2. Binding intensity (tags per bp per site) scale is noted below on a linear scale. All heat maps are normalized to a total of 10 million reads, and further to local tag density. (B–D) Box plots representing normalized log2 tag density of Dex/EtOH for (B) NCOA1 (C) NCOR1 and (D) SUMO2/3 at C1 (blue), C2 (black) and C3 (red) sites. P-values were calculated using unpaired two-sample t-test. All box plots are normalized to total of 10 million reads.
Since NCOA1 and SUMO2/3 at C2 sites show an opposing enrichment between GRwt and GR3KR cells, the higher chromatin occurrence of SUMO2/3 could potentially be associated with the lower occurrence of NCOA1 at the GRwt-occupied C2 sites. We therefore cross-compared the differences in the chromatin occupancy of SUMO2/3 to the differential binding of NCOA1 between the GRwt and GR3KR cells (Supplementary Figure S9B and C). However, the comparison indicated no simple link between the opposing enrichment of NCOA1 and SUMO2/3 at C2 GRBs (Supplementary Figure S9C). The above data suggest that the GR SUMOylation, not the occurrence of SUMO2/3 at GRBs per se, hampers the Dex-induced recruitment of NCOA1 onto chromatin. To complement this notion in live cells, we performed fluorescence recovery after photobleaching (FRAP) experiments. Longer residence times of GR on chromatin measured by FRAP correlate with greater transcriptional output (72), and activation of GR can increase the residence time of NCOA2 (73). However, depletion of endogenous NCOA2 does not impact the residence time of GR. We transfected HEK293 cells with mCherry-tagged GRwt or GR3KR and EGFP-tagged NCOA1 in the presence of Dex. In keeping with the interaction between the GR and the NCOA1, FRAP assays showed that the half-recovery times of both receptor forms significantly increased in the presence of an excess of NCOA1 (Supplementary Figure S9D and E). More interestingly, NCOA1 was significantly more potent in increasing the half recovery time of GR3KR than that of GRwt. On the other hand, the half-recovery time did not significantly change between the receptor forms in the absence of NCOA1, and NCOA1 showed no difference in its half recovery in the presence of GR3KR compared to that of GRwt (Supplementary Figure S9F and G). Our live cell data thus support our proteomic and ChIP-seq data showing enhanced association of NCOA1 with the SUMOylation-deficient GR.
Activated GR regulates chromatin accessibility
Our unbiased proteomic data indicate a prominent recruitment of different chromatin remodeling proteins by the GR including SMARCA2 (BRM) and DPF2 members of the BAF complex and other remodelers (e.g. BRD2 and CHD1) (Figure 3, Supplementary Table S1). Notably, the BAF complex has been previously associated with the action of GR on chromatin (74), where GR at many closed chromatin sites can recruit the BAF complex to GRBs to induce opening of chromatin (41,75,76). Other chromatin remodeling complexes have not been extensively studied for GR; however, the majority of enhancers harbor the activity of two or more chromatin remodeling complexes (77).
To assess the enhanced interaction of GR3KR with chromatin remodeling complexes, we investigated the chromatin accessibility by ATAC-seq (78) (see Methods for details). In both GRwt and GR3KR cells, we detected ∼300 000 open chromatin sites (Supplementary Figure S10A) which encompassed ∼270 000 sites unresponsive to Dex (DexUN). The DexUN chromatin sites were the most accessible regions (Supplementary Figure S10C), being highly enriched at promoters (Supplementary Figure S10B) and showing the presence of several histone marks associated with active transcription (Supplementary Figure S10H–J). These open chromatin sites most likely represent the cell type-specific chromatin landscape of the HEK293 cells (51,79). Interestingly, Dex exposure resulted in a varying degree of increase and decrease in the number of open chromatin sites (Supplementary Figure S10C), mostly at intergenic and intronic regions (Supplementary Figure S10B). The majority of Dex-induced open chromatin sites were shared between GRwt and GR3KR cells (DexUP-shared), but there was a clear cluster of sites where binding of GR3KR induced significantly more opening of chromatin than that of GRwt (DexUP-3KR) (Supplementary Figure S10C). At the DexUP-3KR sites, a robust redistribution of H3K4me2 enrichment was visible after Dex treatment only in GR3KR cells, whereas at the DexUP-shared sites, a robust redistribution of H3K4me2 enrichment was visible after Dex treatment in both cell lines (Supplementary Figure S10J). We also observed that Dex exposure closes a smaller number of open chromatin sites differently in GRwt cells (DexDN-wt) and GR3KR cells (DexDN-3KR) (Supplementary Figure S10C). Examples of genome browser tracks of different types of open chromatin sites are shown in Supplementary Figure S11A-E.
To gain insight into these different types of open chromatin sites, we performed de novo motif analyses (Supplementary Table S4). Analysis of enriched motifs indicated a clear separation of different open chromatin clusters (Supplementary Figure S10D). DexUN and DexDN open chromatin sites showed enrichment for bZIP motifs, such as AP-1, while their enrichment was low or absent at DexUP open chromatin sites (Supplementary Figure S10D). CTCF motifs were also highly enriched at DexUN sites. Binding of both CTCF and ATF2 (bZIP containing TF) was enriched at DexUN sites (Supplementary Figure S10E–F). DexDN sites also display enhanced binding of ATF2. DexUP open chromatin sites in turn showed a strong enrichment for NR3C-type motifs, such as GRE (Supplementary Figure S10D). These motifs were also enriched at DexUN sites, which is indicative of GR binding occurring at pre-accessible chromatin sites. The binding of GRwt and GR3KR can be observed at DexUP sites (Supplementary Figure S10G), clearly illustrating the active role of GR in remodeling of new open chromatin sites. Our chromatin accessibility data also suggest that the GR, especially its SUMOylation-deficient form, induces opening of compacted chromatin sites through the recruitment of chromatin remodeling factors.
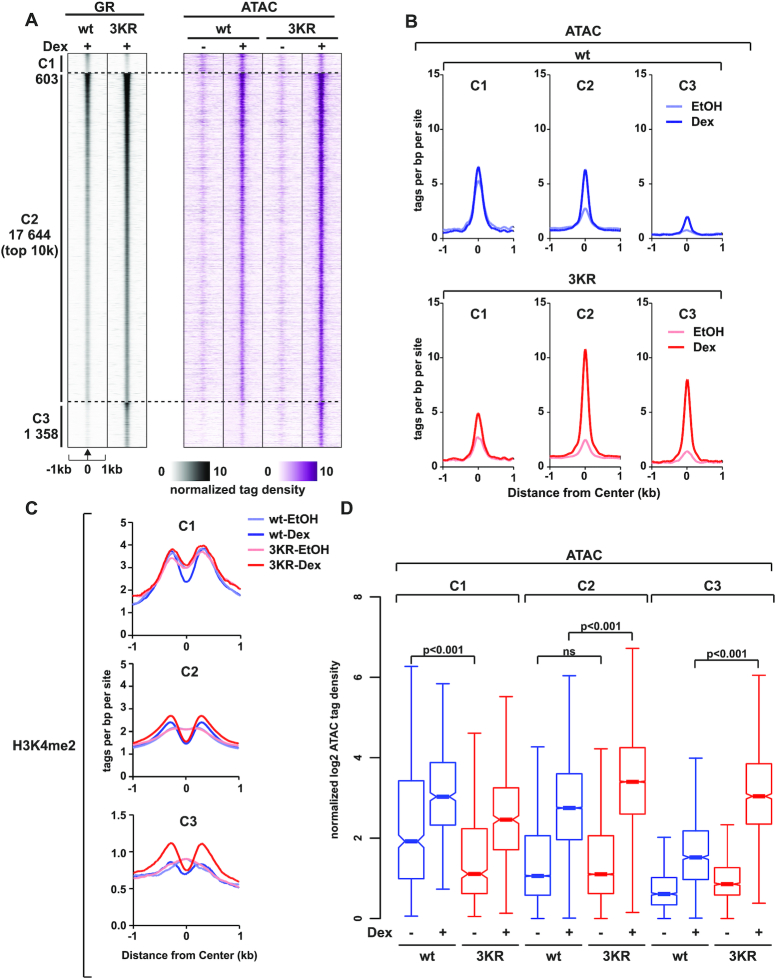
SUMOylation-deficient GR is more efficient at opening chromatin
The proteomics, ChIP-seq and ATAC-seq analyses suggest that the GR3KR is more efficient at promoting chromatin accessibility through its increased interaction with chromatin modifiers. To gain more insight into this, we focused on the chromatin accessibility changes at the GRB clusters defined in our ChIP-seq data, (compare Figure 4A, Supplementary Figure S6A and B). The C1 is the most accessible of the three clusters (C1–C3) before Dex exposure, which is reflected in the higher level of H3K4me2 in C1 than in C2 or C3 (Figure 6A–C). The C3 in turn shows lower enrichment of H3K27ac and higher enrichment of H3K9me3 than the C1 or C2 (Supplementary Figure S12). Furthermore, the C1 was more pre-accessible in GRwt cells than in GR3KR cells, possibly explaining the preferred binding of GRwt to this cluster (Figure 6B, D). This is likely, since ML-792 does not inhibit GRwt binding to these sites. The C3, in turn, shows very low pre-accessibility, but the binding of GR3KR efficiently increases the accessibility of this cluster (Figure 6A, B and D), whereas GRwt has only a very weak effect to the accessibility of these sites. These results are mirrored by the H3K4me2 enrichment, wherein a clear redistribution after Dex treatment is observed only in GR3KR cells (Figure 6C). Interestingly, at the C2, the pre-accessibility does not differ between the cell lines, but also in the C2, the binding of GR3KR increased chromatin accessibility significantly more than that of the GRwt (Figure 6B, D). These results further strengthen the notion that, although both receptor forms bind to the same sites, the GR3KR can interact more avidly than the GRwt with chromatin regulators to open chromatin.
Figure 6.
SUMOylation regulates the pioneer-like activity of GR in chromatin opening. (A) Heat maps showing GR ChIP-seq and ATAC-seq data from HEK293flpGRwt (wt) and HEK293flpGR3KR (3KR) cells in the presence and absence of Dex. Each heat map represents ±1 kb around the center of the GR peak. Binding intensity (tags per bp per site) scale is noted below on a linear scale. All heat maps are normalized to a total of 10 million reads, and further to local tag density. Sites have been sorted by GR signal, with the strongest GR signal on top. (B) Aggregate plots of ATAC-seq data for C1 (left), C2 (middle), and C3 (right) in GRwt (top, blue) and GR3KR (bottom, red) cells. Aggregate plots represent ±1kb around the center of the accessible region and are normalized to a total of 10 million reads, and further to local tag density. (C) Aggregate plots of H3K4me2 ChIP-seq data in the presence or absence of Dex for C1 (top), C2 (middle), and C3 (bottom) sites. Aggregate plots represent ±1kb around the center of the GR-binding site. Data are normalized to a total of 10 million reads, and further to local tag density. (D) Box plots representing normalized log2 tag density of ATAC-seq data at C1 (left), C2 (middle) and C3 (right) sites. P-values were calculated with One-way ANOVA with Bonferroni post hoc test. All box plots are normalized to total of 10 million reads. Blue color represents GRwt (wt), red color GR3KR (3KR).
Due to the above observations, we sub-divided C2 sites based on their pre-hormone accessibility to pre-accessible and de novo sites (Supplementary Figure S13A, E–F). Pre-accessible sites are open, while de novo sites remain closed (i.e. contain no Tn5 insertions) prior to hormone exposure. The stronger potential of GR3KR in increasing chromatin accessibility is seen both at the pre-accessible and at the de novo sites, with a clearer difference at the de novo sites (Supplementary Figure S13B, E–F). We also assessed the binding of NCOA1 at C2 pre-accessible and de novo sites, which showed that this coactivator was significantly more abundant with GR3KR than GRwt only at de novo sites (Supplementary Figure S13C, E–F). This was mirrored by the enrichment of H3K4me2, wherein a clear difference between GR3KR and GRwt was seen only at de novo sites (Supplementary Figure S13E and F). NCOR1, however, did not show a significant preference for either receptor form at C2 pre-accessible or de novo sites (Supplementary Figure S13D–F). These results reveal that in comparison to the GRwt, the GR3KR is especially efficient in recruiting NCOA1 and regulating chromatin accessibility at closed chromatin sites.
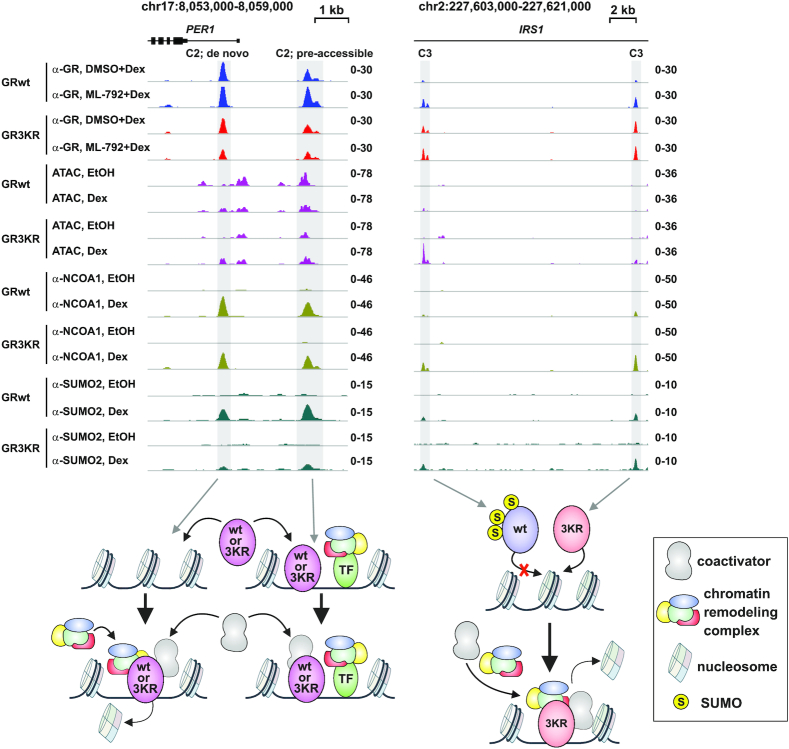
The target gene-selective effects of GR SUMOylation in the modulation of chromatin accessibility, coactivator NCOA1 recruitment and target gene expression are highlighted in two examples of GR target loci in Figure 7:PER1 locus is similarly Dex-regulated by GRwt and GR3KR, with both receptor forms binding to pre-accessible and de novo chromatin sites. Both receptor forms can recruit NCOA1 to the chromatin and induce the expression of PER1. In contrast, only GR3KR binds to enhancers at IRS1 locus, increasing its chromatin accessibility, recruiting NCOA1 to the enhancers, and resulting in Dex induction of the expression of the locus. Fittingly, the binding of GRwt to the IRS1 locus is rescued by the inhibition of SUMOylation, which also results in increased the expression of the locus (Figure 4G).
Figure 7.
Model examples of GR target loci differentially regulated by GR SUMOylation. Example of genome browser tracks of PER1 (left) and IRS1 (right) loci showing GR ChIP-seq, ATAC-seq, NCOA1 and SUMO2/3 ChIP-seq data from GRwt and GR3KR cells. PER1 is Dex induced in both cells. IRS1 is Dex induced only in GR3KR cells. Dex induction of IRS1 is increased in GRwt cells upon ML-792 treatment. PER1 loci harbor two GR binding sites, one at pre-accessible and one at de novo chromatin site. GRwt and GR3KR can bind to both sites and recruit NCOA1. At de novo site, both receptor forms induce chromatin accessibility. IRS1 locus harbors two GR3KR specific binding sites all de novo chromatin sites. GR3KR induces the recruitment of NCOA1 and chromatin accessibility at these sites. GRwt is able to bind to these sites upon ML-792 treatment.
DISCUSSION
Signal-activated TFs, such as NRs, adopt several modes of action for binding to chromatin (80). At the heart of these modes is the NR-mediated recruitment of a diverse group of proteins known as coregulators. However, systematic analyses of GR-centered chromatin protein networks have been scarce in the NR field. RIME (Rapid Immunoprecipitation Mass spectrometry of Endogenous proteins) with isobaric labelling has been successfully applied to identify protein interactions of estrogen receptor alpha and other TFs in breast cancer cells (81,82). Moreover, GR interactome has been recently investigated by ChIP coupled with MS (ChIP-MS) in mouse liver and mouse embryonic fibroblasts (83). Here, we utilized ChIP-SICAP (18), a novel proteomic methodology, to cast new light to the protein associations and chromatin function of GR, an important NR and drug target that has been widely studied for decades. We complemented our ChIP-SICAP data with biotin proximity mapping of the GR interactome, analysis of SUMO2/3-modified proteins as well as with genome-wide ChIP-seq and ATAC-seq data from the same cellular background to show that coregulator recruitment and chromatin openness induced by glucocorticoid-bound GR is influenced by the SUMOylation status of the receptor. Furthermore, ChIP-seq and RNA-seq analyses with SAE inhibitor ML-792 validate the role of GR SUMOylation in these events. Our data exemplify how a post-translational modification can fine-tune the response of a signal-activated TF.
Our ChIP-SICAP identifies proteins binding either to GR or to nearby (up to 200–300 bp) chromatin (18). The chromatin protein interactome of hormone-activated GR includes expected NR coregulators, e.g. coactivators EP300, NCOA2 and NCOA6, and corepressor NCOR1, but also proteins not previously linked to GR action. The latter group includes chromatin-looping associated factors YY1 and cohesin subunits SMC1A and SMC3. In keeping with the notion that most of the NR coregulators function as large multi-protein complexes (5,6), we also identify a number of BAF, CHD and NCOR complex components as chromatin partners of the GR (see Supplementary Table S1 for all hits). The biological relevance of our interactome data is highlighted by the fact that a marked number of the identified GR’s chromatin partners significantly affect the glucocorticoid sensitivity of B-ALL cells (19). Examples of these chromatin partners are CREBBP, EP300, ARID1A, CHD1, NCOA1, NCOR1 and SUMO2. Our GR chromatin interactome thus forms an important resource for the NR field in parallel with other recent GR interactome studies (21,84–86). Our work also shows that ChIP-SICAP is a powerful novel tool for identifying NR on-chromatin interactomes.
The two main SUMO2/3-modified lysine residues of GR reside at its N-terminal transactivation domain (20). Other studies have also shown that GR activity is regulated by synergy control motifs that overlap with the SUMOylation sites (87,88). These lysines may be targeted also by ubiquitination (89), but glucocorticoid has not been shown to promote the latter modification. Our analyses indicate that conjugation of endogenous SUMO2/3 to the GR is triggered by binding of glucocorticoid to the receptor. In addition, our previous re-ChIP analyses indicate the simultaneous presence of SUMO2/3 and GR and GRBs (13). We have shown that mutating these two SUMOylation sites (as well as a third site at the LBD) leads to a SUMOylation-defective GR that binds more prevalently to chromatin and harbors a distinct chromatin-binding pattern from the SUMOylation-competent receptor (13). Moreover, general inhibition of SUMOylation enables the SUMOylation-competent GR to bind to and regulate a striking number of chromatin sites and target genes otherwise bound and regulated merely by the SUMOylation-deficient GR, thus verifying that the behavior of the GR3KR in binding to chromatin and regulation of genes is causally linked to the GR SUMOylation. Although the SUMOylation inhibitor did not phenocopy all the differences between the SUMOylation-competent and -deficient GR, several of the remaining differences can be attributed to ML-792 inhibiting not only the SUMOylation of GR but also that of other proteins. This is likely, since SUMO1 and SUMO2 are associated with GR-occupied chromatin and many of the GR-interacting proteins are also modified by SUMOs. Interestingly, ChIP-SICAP uncovered a large group of protein associations influenced by the GR SUMOylation status. The SUMOylation sites of GR widely curbed the receptor's association with of a variety (∼100) of proteins, including the well-established coactivator NCOA1, chromatin remodelers (e.g. SMARCA2, BRD2, CHD1) and TFs (e.g. JUN and ATF3 from the AP-1 complex). Our genome-wide data showing that SUMOylation-deficient GR recruits NCOA1 to GRBs more efficiently than the wild-type receptor support our proteomic results. Furthermore, our live imaging data of GR–NCOA1 interaction are in line with our proteomic and genomic data; in the presence of NCOA1, the half recovery is slower with GR3KR than with GRwt. However, the depletion of endogenous NCOA2 does not impact the residence time or bound population of GR in single-molecule imaging experiments (73). Since single-molecule imaging mainly estimates stationary/bound molecules, while FRAP mostly assesses diffusing molecules, the differences between GR3KR and GRwt in the presence of an excess NCOA1 could be due to altered diffusion kinetics of the receptor. In addition, we cannot exclude the possibility that the observed difference in the chromatin interaction and recruitment of NCOA1 in part reflects increased association of SUMOylation-deficient GR with chromatin. Interestingly, aurora kinase B (AURKB), for which Poulard and colleagues have identified a non-canonical role in the regulation of glucocorticoid sensitivity of B-ALL cells (19), was also enriched with the SUMOylation-deficient GR. Moreover, SUMOylation pathway components were also among the top genes differentially expressed, overexpressed, in B-ALL patients at relapse versus their diagnosis stage (19), suggesting a link between enhanced SUMOylation and relapse or treatment resistance.
Somewhat unexpectedly, there were only a few well-recognized NR coregulators, such as coactivator NCOA6 (AIB3) and corepressor NCOR1, among the proteins preferring the SUMOylation-competent GR in ChIP-SICAP analysis. However, the latter group interestingly encompassed IRF2BP2, ZNF536, ZNF703 (NLZ1) and BAZ1A (ACF1) that are chromatin proteins implicated in NR-regulated transcriptional repression (90–93) and thus potentially capable of repressing GR at enhancers. Recent studies suggest that the SUMOylation of GR is required for the formation of NCOR1-containing repressive complex for tethered and negative GRE-mediated transrepression (70,71), or for repression of estrogen receptor signaling by GR (94). However, the genome-wide co-occupancy of NCOR1 at the GRBs is not affected by the SUMOylation status of GR. The inconsistencies between ChIP-SICAP, BioID, and ChIP-seq data with NCOR1 imply a complex nature of the interaction between the SUMOylation status of GR and the corepressor. Thus, the reported effect of GR SUMOylation on the NCOR1-containing complex might be restricted to a few specific loci.
Enrichment of chromatin remodeling factors was one of the differences between the chromatin interactome of SUMOylation competent and that of the modification deficient GR. The factors more avidly interacting with the SUMOylation-deficient GR included several subunits of the BAF complex and several CHD family members as well as BRD2. In addition, topoisomerase II homologues, TOP2A and TOP2B as well as poly(ADP ribose)polymerase 1 (PARP1), preferentially associated with non-SUMOylated GR. Interestingly, TOP2B and PARP1 have been shown to be required for BAF mediated transcriptional activation (95), while BRD2 influences glucocorticoid sensitivity of B-ALL cells (19,96). While some CHD proteins have been associated with GR-regulated gene expression only recently (97), the interplay between GR and BAF complex, especially its catalytic subunit SMARCA4 (BRG1) (74,98), as well as its importance at GR-regulated enhancers are well known (75,76). In addition, SMARCA2 (BRM) has been shown to be important for the regulation of GR target genes (99,100). Besides binding to pre-accessible, open chromatin sites, GR can bind to closed nucleosomal sites, recruiting BAF complex to evict nucleosomes and promote an open chromatin state (75). We also observe a similar pioneer factor-like mode of GR, when the receptor binds to closed chromatin sites and thereby increases their accessibility. The pioneer-like activity is particularly notable with the non-SUMOylated receptor, as it was able to access to a large subset of closed chromatin sites that are inaccessible to SUMOylated GR. Importantly, the effect of SUMOylation status on the interaction of the receptor with chromatin remodeling factors is reflected in our chromatin accessibility data, also on pre-accessible sites, and gene expression data, showing that the SUMOylation-deficient GR is more potent in opening of chromatin at glucocorticoid-regulated enhancers and inducing expression of their target loci. Moreover, the pioneer factor-like potential of non-SUMOylated GR might be potentiated by the enhanced interaction with topoisomerases, as TOP2 synergizes with BAF complex to transform facultative heterochromatin into more accessible chromatin (101). Finally, since SUMOylation on chromatin can act as a general stabilizer of chromatin states, controlling gene programs that need to be rewired during differentiation processes (102), SUMOylation of TFs directing differentiation processes could control their capacity to interact with the chromatin in a manner similar shown here for GR. In the case of a ‘classic’ pioneer TF, FOXA1, SUMOylation indeed has been demonstrated to restrict its pioneering activity on chromatin (103).
Recent ChIP-seq studies have addressed the role of SUMOylation of three human TFs, MITF (microphthalmia-associated transcription factor), GR, AR, and a yeast TF Sko1. In all cases, SUMOylation-deficient forms of these TFs displayed significantly enhanced interaction with chromatin by binding to numerous additional sites compared to their wild-type counterpart, thus suggesting a conserved role for the modification in restricting TF binding to chromatin (13–15,17). Most of the SUMO-targeted TFs, including the above listed ones, are endogenously modified only to a sub-stoichiometric level. The reason why a relatively small pool of SUMOylated TF can cause obvious effects on the transcriptional activity of TFs is likely to be due to the dynamic nature of the modification, i.e. rapid cycles of conjugation-deconjugation. Even though the equilibrium might lie on the side of the unmodified TF, a large portion of the pool of a given TF might be affected by SUMOylation in a short window of time. SUMOylation can lead to recruitment of repressive factors, and a repressive state can remain even after SUMO is deconjugated from the TF. Alternatively, SUMOylation can trigger a chain of events, leading to formation of a relatively stable complex. These complexes may remain even after deconjugation of SUMO. In both cases, the TF needs to be modified only for a short period of time. Based on the genome-wide chromatin-binding data of AR, GR, MITF and Sko1, Rosonina recently proposed a model for the role of TF SUMOylation in fine-tuning TF-binding site selection (16). According to this model, DNA binding triggers SUMOylation of TFs, which in turn alters their chromatin occupancy. However, the actual mechanism underlying the SUMOylation-altered chromatin occupancy of TFs has remained elusive. The DNA-TF interaction per se does not appear to be affected by the SUMOylation. The modification may in principle affect chromatin off-rate or nuclear mobility of its target TF, but these notions hardly explain the locus-selective effects of the SUMOylation on the TF. In the case of GR and AR, SUMOylation is induced by binding of their cognate hormones and promoted by PIAS proteins, such as PIAS1 and PIAS2 that are regulated by phosphorylation via other signaling pathways (11,20,104,105). Our data complement the model. We uncover that the hormone-triggered SUMOylation modulates the chromatin protein network of GR, curbing its protein-protein interactions with a number of prominent coregulators. Our live cell imaging data lend support to the notion that the recruited coactivator(s) can stabilize the association of GR with chromatin. The enhanced interaction of SUMOylation-deficient GR in particular with chromatin remodelers is likely to contribute to its promiscuous chromatin occupancy. We speculate that a similar mechanism might operate with many other SUMOylated TFs. Our study additionally foreshadows a concept that may apply more generally to TFs whose activity is regulated by covalent modifications that in turn modulate protein interaction networks on chromatin.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (106) via the PRIDE (107) partner repository with the following dataset identifiers: for SUMO2/3 IP PXD017714, for ChIP-SICAP PXD015183, for GR3KR BioID PXD017713. Newly generated ChIP-seq, RNA-seq and ATAC-seq datasets have been submitted to the NCBI Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/); accession code: GSE64373.
Supplementary Material
ACKNOWLEDGEMENTS
The hybridoma developed by Michael M. Matunis was obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA, USA. We thank Merja Räsänen for assistance in cell culture and SUMO2/3 antibody production, Eija Korhonen for assistance in cell culture and Sini Miettinen for assistance in MS sample preparation and MS runs. The EMBL GeneCore and BGI sequencing teams are greatly acknowledged for deep sequencing, and the UEF Bioinformatics Center for providing computational infrastructure. The UEF Cell and Tissue Imaging Unit is thanked for providing services and equipment for FRAP assays. The Genome Biology Unit (GBU) at the University of Helsinki is greatly thanked for cloning services.
Contributor Information
Ville Paakinaho, Institute of Biomedicine, University of Eastern Finland, Kuopio, Finland.
Joanna K Lempiäinen, Institute of Biomedicine, University of Eastern Finland, Kuopio, Finland.
Gianluca Sigismondo, German Cancer Research Center (DKFZ), Heidelberg, Germany.
Einari A Niskanen, Institute of Biomedicine, University of Eastern Finland, Kuopio, Finland.
Marjo Malinen, Institute of Biomedicine, University of Eastern Finland, Kuopio, Finland; Department of Environmental and Biological Sciences, University of Eastern Finland, Joensuu, Finland.
Tiina Jääskeläinen, Institute of Biomedicine, University of Eastern Finland, Kuopio, Finland.
Markku Varjosalo, Institute of Biotechnology, University of Helsinki, Helsinki, Finland.
Jeroen Krijgsveld, German Cancer Research Center (DKFZ), Heidelberg, Germany; Heidelberg University, Medical Faculty, Heidelberg, Germany.
Jorma J Palvimo, Institute of Biomedicine, University of Eastern Finland, Kuopio, Finland.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
V.P. was supported by the Academy of Finland; Cancer Foundation Finland; Sigrid Jusélius Foundation; J.K.L. was supported by the UEF Doctoral Programme in Molecular Medicine; Jalmari and Rauha Ahokas Foundation; Instrumentarium Foundation; Finnish Cultural Foundation; Emil Aaltonen Foundation; Orion Research Foundation; Kuopio University Foundation; Ida Montin Foundation; G.S. and J.K. acknowledge support by the Excellence Cluster CellNetworks (EXC81); M.V. and J.J.P. were supported by the Academy of Finland; Cancer Foundation Finland; Sigrid Jusélius Foundation. Funding for open access charge: The Academy of Finland.
Conflict of interest statement. None declared.
REFERENCES
- 1. Vandevyver S., Dejager L., Libert C.. Comprehensive overview of the structure and regulation of the glucocorticoid receptor. Endocr. Rev. 2014; 35:671–693. [DOI] [PubMed] [Google Scholar]
- 2. Weikum E.R., Knuesel M.T., Ortlund E.A., Yamamoto K.R.. Glucocorticoid receptor control of transcription: precision and plasticity via allostery. Nat. Rev. Mol. Cell Biol. 2017; 18:159–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cain D.W., Cidlowski J.A.. Immune regulation by glucocorticoids. Nat. Rev. Immunol. 2017; 17:233–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pui C.H., Evans W.E.. Treatment of acute lymphoblastic leukemia. N. Engl. J. Med. 2006; 354:166–178. [DOI] [PubMed] [Google Scholar]
- 5. Millard C.J., Watson P.J., Fairall L., Schwabe J.W.. An evolving understanding of nuclear receptor coregulator proteins. J. Mol. Endocrinol. 2013; 51:T23–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Meier K., Brehm A.. Chromatin regulation: how complex does it get. Epigenetics. 2014; 9:1485–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms. Genes Dev. 2004; 18:2046–2059. [DOI] [PubMed] [Google Scholar]
- 8. Tatham M.H., Jaffray E., Vaughan O.A., Desterro J.M., Botting C.H., Naismith J.H., Hay R.T.. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 2001; 276:35368–35374. [DOI] [PubMed] [Google Scholar]
- 9. van Wijk S.J., Timmers H.T.. The family of ubiquitin-conjugating enzymes (E2s): deciding between life and death of proteins. FASEB J. 2010; 24:981–993. [DOI] [PubMed] [Google Scholar]
- 10. Mukhopadhyay D., Dasso M.. Modification in reverse: the SUMO proteases. Trends Biochem. Sci. 2007; 32:286–295. [DOI] [PubMed] [Google Scholar]
- 11. Rytinki M.M., Kaikkonen S., Pehkonen P., Jaaskelainen T., Palvimo J.J.. PIAS proteins: pleiotropic interactors associated with SUMO. Cellular and Molecular Life Sciences: CMLS. 2009; 66:3029–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Treuter E., Venteclef N.. Transcriptional control of metabolic and inflammatory pathways by nuclear receptor SUMOylation. Biochim. Biophys. Acta. 2011; 1812:909–918. [DOI] [PubMed] [Google Scholar]
- 13. Paakinaho V., Kaikkonen S., Makkonen H., Benes V., Palvimo J.J.. SUMOylation regulates the chromatin occupancy and anti-proliferative gene programs of glucocorticoid receptor. Nucleic Acids Res. 2014; 42:1575–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sutinen P., Malinen M., Heikkinen S., Palvimo J.J.. SUMOylation modulates the transcriptional activity of androgen receptor in a target gene and pathway selective manner. Nucleic Acids Res. 2014; 42:8310–8319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bertolotto C., Lesueur F., Giuliano S., Strub T., de Lichy M., Bille K., Dessen P., d’Hayer B., Mohamdi H., Remenieras A.et al.. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature. 2011; 480:94–98. [DOI] [PubMed] [Google Scholar]
- 16. Rosonina E. A conserved role for transcription factor sumoylation in binding-site selection. Curr. Genet. 2019; 65:1307–1312. [DOI] [PubMed] [Google Scholar]
- 17. Sri Theivakadadcham V.S., Bergey B.G., Rosonina E.. Sumoylation of DNA-bound transcription factor Sko1 prevents its association with nontarget promoters. PLos Genet. 2019; 15:e1007991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rafiee M.R., Girardot C., Sigismondo G., Krijgsveld J.. Expanding the circuitry of pluripotency by selective isolation of chromatin-associated proteins. Mol. Cell. 2016; 64:624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Poulard C., Kim H.N., Fang M., Kruth K., Gagnieux C., Gerke D.S., Bhojwani D., Kim Y.M., Kampmann M., Stallcup M.R.et al.. Relapse-associated AURKB blunts the glucocorticoid sensitivity of B cell acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:3052–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tian S., Poukka H., Palvimo J.J., Janne O.A.. Small ubiquitin-related modifier-1 (SUMO-1) modification of the glucocorticoid receptor. Biochem. J. 2002; 367:907–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lempiainen J.K., Niskanen E.A., Vuoti K.M., Lampinen R.E., Goos H., Varjosalo M., Palvimo J.J.. Agonist-specific protein interactomes of glucocorticoid and androgen receptor as revealed by proximity mapping. Mol. Cell. Proteomics. 2017; 16:1462–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kalkhoven E., Valentine J.E., Heery D.M., Parker M.G.. Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J. 1998; 17:232–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rytinki M., Kaikkonen S., Sutinen P., Paakinaho V., Rahkama V., Palvimo J.J.. Dynamic SUMOylation is linked to the activity cycles of androgen receptor in the cell nucleus. Mol. Cell. Biol. 2012; 32:4195–4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hughes C.S., Moggridge S., Muller T., Sorensen P.H., Morin G.B., Krijgsveld J.. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat. Protoc. 2019; 14:68–85. [DOI] [PubMed] [Google Scholar]
- 25. Hughes C.S., Foehr S., Garfield D.A., Furlong E.E., Steinmetz L.M., Krijgsveld J.. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol. Syst. Biol. 2014; 10:757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M.. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics. 2014; 13:2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tyanova S., Temu T., Cox J.. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016; 11:2301–2319. [DOI] [PubMed] [Google Scholar]
- 28. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K.. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yao T.P., Ku G., Zhou N., Scully R., Livingston D.M.. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:10626–10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu R.C., Qin J., Hashimoto Y., Wong J., Xu J., Tsai S.Y., Tsai M.J., O’Malley B.W.. Regulation of SRC-3 (pCIP/ACTR/AIB-1/RAC-3/TRAM-1) coactivator activity by I kappa B kinase. Mol. Cell. Biol. 2002; 22:3549–3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yoon H.G., Chan D.W., Huang Z.Q., Li J., Fondell J.D., Qin J., Wong J.. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J. 2003; 22:1336–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sims J.K., Wade P.A.. SnapShot: chromatin remodeling: CHD. Cell. 2011; 144:626–626. [DOI] [PubMed] [Google Scholar]
- 33. Fujisawa T., Filippakopoulos P.. Functions of bromodomain-containing proteins and their roles in homeostasis and cancer. Nat. Rev. Mol. Cell Biol. 2017; 18:246–262. [DOI] [PubMed] [Google Scholar]
- 34. Li X., Wang W., Wang J., Malovannaya A., Xi Y., Li W., Guerra R., Hawke D.H., Qin J., Chen J.. Proteomic analyses reveal distinct chromatin-associated and soluble transcription factor complexes. Mol. Syst. Biol. 2015; 11:775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mashtalir N., D’Avino A.R., Michel B.C., Luo J., Pan J., Otto J.E., Zullow H.J., McKenzie Z.M., Kubiak R.L., St Pierre R.et al.. Modular organization and assembly of SWI/SNF family chromatin remodeling complexes. Cell. 2018; 175:1272–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P.et al.. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019; 47:D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T.. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Barysch S.V., Dittner C., Flotho A., Becker J., Melchior F.. Identification and analysis of endogenous SUMO1 and SUMO2/3 targets in mammalian cells and tissues using monoclonal antibodies. Nat. Protoc. 2014; 9:896–909. [DOI] [PubMed] [Google Scholar]
- 39. Teo G., Liu G., Zhang J., Nesvizhskii A.I., Gingras A.C., Choi H.. SAINTexpress: improvements and additional features in significance analysis of INTeractome software. J. Proteomics. 2014; 100:37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Choi H., Larsen B., Lin Z.Y., Breitkreutz A., Mellacheruvu D., Fermin D., Qin Z.S., Tyers M., Gingras A.C., Nesvizhskii A.I.. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat. Methods. 2011; 8:70–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Paakinaho V., Johnson T.A., Presman D.M., Hager G.L.. Glucocorticoid receptor quaternary structure drives chromatin occupancy and transcriptional outcome. Genome Res. 2019; 29:1223–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Buenrostro J.D., Wu B., Chang H.Y., Greenleaf W.J.. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 2015; 109:21.29.1–21.29.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lempiainen J.K., Manjur A.B.M.K., Malinen M., Ketola K., Niskanen E.A., Palvimo J.J. BCOR-coupled H2A monoubiquitination represses a subset of androgen receptor target genes regulating prostate cancer proliferation. Oncogene. 2020; 39:2391–2407. [DOI] [PubMed] [Google Scholar]
- 46. Huang da W., Sherman B.T., Lempicki R.A.. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009; 37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huang da W., Sherman B.T., Lempicki R.A.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 48. Toropainen S., Malinen M., Kaikkonen S., Rytinki M., Jaaskelainen T., Sahu B., Janne O.A., Palvimo J.J.. SUMO ligase PIAS1 functions as a target gene selective androgen receptor coregulator on prostate cancer cell chromatin. Nucleic Acids Res. 2015; 43:848–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Langmead B., Salzberg S.L.. Fast gapped-read alignment with bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Thurman R.E., Rynes E., Humbert R., Vierstra J., Maurano M.T., Haugen E., Sheffield N.C., Stergachis A.B., Wang H., Vernot B.et al.. The accessible chromatin landscape of the human genome. Nature. 2012; 489:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang J., Lee D., Dhiman V., Jiang P., Xu J., McGillivray P., Yang H., Liu J., Meyerson W., Clarke D.et al.. An integrative ENCODE resource for cancer genomics. Nat. Commun. 2020; 11:3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Golebiowski F., Matic I., Tatham M.H., Cole C., Yin Y., Nakamura A., Cox J., Barton G.J., Mann M., Hay R.T.. System-wide changes to SUMO modifications in response to heat shock. Sci. Signal. 2009; 2:ra24. [DOI] [PubMed] [Google Scholar]
- 55. Schimmel J., Eifler K., Sigurethsson J.O., Cuijpers S.A., Hendriks I.A., Verlaan-de Vries M., Kelstrup C.D., Francavilla C., Medema R.H., Olsen J.V.et al.. Uncovering SUMOylation dynamics during cell-cycle progression reveals FoxM1 as a key mitotic SUMO target protein. Mol. Cell. 2014; 53:1053–1066. [DOI] [PubMed] [Google Scholar]
- 56. Tammsalu T., Matic I., Jaffray E.G., Ibrahim A.F.M., Tatham M.H., Hay R.T. Proteome-wide identification of SUMO2 modification sites. Sci. Signal. 2014; 7:rs2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Impens F., Radoshevich L., Cossart P., Ribet D. Mapping of SUMO sites and analysis of SUMOylation changes induced by external stimuli. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:12432–12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hendriks I.A., D'Souza R.C., Yang B., Verlaan-de Vries M., Mann M., Vertegaal A.C. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat. Struct. Mol. Biol. 2014; 21:927–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hendriks I.A., D'Souza R.C., Chang J.G., Mann M., Vertegaal A.C. System-wide identification of wild-type SUMO-2 conjugation sites. Nat. Commun. 2015; 6:7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lamoliatte F., McManus F.P., Maarifi G., Chelbi-Alix M.K., Thibault P.. Uncovering the SUMOylation and ubiquitylation crosstalk in human cells using sequential peptide immunopurification. Nat. Commun. 2017; 8:14109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hendriks I.A., Lyon D., Young C., Jensen L.J., Vertegaal A.C., Nielsen M.L.. Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation. Nat. Struct. Mol. Biol. 2017; 24:325–336. [DOI] [PubMed] [Google Scholar]
- 62. Lumpkin R.J., Gu H., Zhu Y., Leonard M., Ahmad A.S., Clauser K.R., Meyer J.G., Bennett E.J., Komives E.A.. Site-specific identification and quantitation of endogenous SUMO modifications under native conditions. Nat. Commun. 2017; 8:1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hendriks I.A., Lyon D., Su D., Skotte N.H., Daniel J.A., Jensen L.J., Nielsen M.L.. Site-specific characterization of endogenous SUMOylation across species and organs. Nat. Commun. 2018; 9:2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lamoliatte F., Caron D., Durette C., Mahrouche L., Maroui M.A., Caron-Lizotte O., Bonneil E., Chelbi-Alix M.K., Thibault P.. Large-scale analysis of lysine SUMOylation by SUMO remnant immunoaffinity profiling. Nat. Commun. 2014; 5:5409. [DOI] [PubMed] [Google Scholar]
- 65. Becker J., Barysch S.V., Karaca S., Dittner C., Hsiao H.H., Berriel Diaz M., Herzig S., Urlaub H., Melchior F.. Detecting endogenous SUMO targets in mammalian cells and tissues. Nat. Struct. Mol. Biol. 2013; 20:525–531. [DOI] [PubMed] [Google Scholar]
- 66. Xiao Z., Chang J.G., Hendriks I.A., Sigurethsson J.O., Olsen J.V., Vertegaal A.C.. System-wide analysis of SUMOylation dynamics in response to replication stress reveals novel small ubiquitin-like modified target proteins and acceptor lysines relevant for genome stability. Mol. Cell. Proteomics. 2015; 14:1419–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hendriks I.A., Treffers L.W., Verlaan-de Vries M., Olsen J.V., Vertegaal A.C.O. SUMO-2 orchestrates chromatin modifiers in response to DNA damage. Cell. Rep. 2015; 10:1778–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Roux K.J., Kim D.I., Raida M., Burke B.. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012; 196:801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. He X., Riceberg J., Soucy T., Koenig E., Minissale J., Gallery M., Bernard H., Yang X., Liao H., Rabino C.et al.. Probing the roles of SUMOylation in cancer cell biology by using a selective SAE inhibitor. Nat. Chem. Biol. 2017; 13:1164–1171. [DOI] [PubMed] [Google Scholar]
- 70. Hua G., Paulen L., Chambon P.. GR SUMOylation and formation of an SUMO-SMRT/NCoR1-HDAC3 repressing complex is mandatory for GC-induced IR nGRE-mediated transrepression. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E626–E634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hua G., Ganti K.P., Chambon P.. Glucocorticoid-induced tethered transrepression requires SUMOylation of GR and formation of a SUMO-SMRT/NCoR1-HDAC3 repressing complex. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E635–E643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Stavreva D.A., Muller W.G., Hager G.L., Smith C.L., McNally J.G.. Rapid glucocorticoid receptor exchange at a promoter is coupled to transcription and regulated by chaperones and proteasomes. Mol. Cell. Biol. 2004; 24:2682–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Paakinaho V., Presman D.M., Ball D.A., Johnson T.A., Schiltz R.L., Levitt P., Mazza D., Morisaki T., Karpova T.S., Hager G.L.. Single-molecule analysis of steroid receptor and cofactor action in living cells. Nat. Commun. 2017; 8:15896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Swinstead E.E., Paakinaho V., Presman D.M., Hager G.L.. Pioneer factors and ATP-dependent chromatin remodeling factors interact dynamically: a new perspective: multiple transcription factors can effect chromatin pioneer functions through dynamic interactions with ATP-dependent chromatin remodeling factors. Bioessays. 2016; 38:1150–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Johnson T.A., Chereji R.V., Stavreva D.A., Morris S.A., Hager G.L., Clark D.J.. Conventional and pioneer modes of glucocorticoid receptor interaction with enhancer chromatin in vivo. Nucleic Acids Res. 2018; 46:203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hoffman J.A., Trotter K.W., Ward J.M., Archer T.K.. BRG1 governs glucocorticoid receptor interactions with chromatin and pioneer factors across the genome. Elife. 2018; 7:e35073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Morris S.A., Baek S., Sung M.H., John S., Wiench M., Johnson T.A., Schiltz R.L., Hager G.L.. Overlapping chromatin-remodeling systems collaborate genome wide at dynamic chromatin transitions. Nat. Struct. Mol. Biol. 2014; 21:73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J.. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013; 10:1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Heinz S., Romanoski C.E., Benner C., Glass C.K.. The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol. 2015; 16:144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Swinstead E.E., Paakinaho V., Hager G.L.. Chromatin reprogramming in breast cancer. Endocr. Relat. Cancer. 2018; 25:R385–R404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Siersbaek R., Scabia V., Nagarajan S., Chernukhin I., Papachristou E.K., Broome R., Johnston S.J., Joosten S.E.P., Green A.R., Kumar S.et al.. IL6/STAT3 signaling hijacks estrogen receptor alpha enhancers to drive breast cancer metastasis. Cancer. Cell. 2020; 38:412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Papachristou E.K., Kishore K., Holding A.N., Harvey K., Roumeliotis T.I., Chilamakuri C.S.R., Omarjee S., Chia K.M., Swarbrick A., Lim E.et al.. A quantitative mass spectrometry-based approach to monitor the dynamics of endogenous chromatin-associated protein complexes. Nat. Commun. 2018; 9:2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Escoter-Torres L., Greulich F., Quagliarini F., Wierer M., Uhlenhaut N.H.. Anti-inflammatory functions of the glucocorticoid receptor require DNA binding. Nucleic Acids Res. 2020; 48:8393–8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Clarisse D., Thommis J., Van Wesemael K., Houtman R., Ratman D., Tavernier J., Offner F., Beck I., De Bosscher K.. Coregulator profiling of the glucocorticoid receptor in lymphoid malignancies. Oncotarget. 2017; 8:109675–109691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hemmer M.C., Wierer M., Schachtrup K., Downes M., Hubner N., Evans R.M., Uhlenhaut N.H.. E47 modulates hepatic glucocorticoid action. Nat. Commun. 2019; 10:306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Dendoncker K., Timmermans S., Vandewalle J., Eggermont M., Lempiainen J., Paakinaho V., Van Hamme E., Dewaele S., Vandevyver S., Ballegeer M.et al.. TNF-alpha inhibits glucocorticoid receptor-induced gene expression by reshaping the GR nuclear cofactor profile. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:12942–12951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Holmstrom S.R., Chupreta S., So A.Y., Iniguez-Lluhi J.A.. SUMO-mediated inhibition of glucocorticoid receptor synergistic activity depends on stable assembly at the promoter but not on DAXX. Mol. Endocrinol. 2008; 22:2061–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Iniguez-Lluhi J.A., Pearce D. A common motif within the negative regulatory regions of multiple factors inhibits their transcriptional synergy. Mol. Cell. Biol. 2000; 20:6040–6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Akimov V., Barrio-Hernandez I., Hansen S.V.F., Hallenborg P., Pedersen A.K., Bekker-Jensen D.B., Puglia M., Christensen S.D.K., Vanselow J.T., Nielsen M.M.et al.. UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat. Struct. Mol. Biol. 2018; 25:631–640. [DOI] [PubMed] [Google Scholar]
- 90. Manjur A.B.M.K., Lempiainen J.K., Malinen M., Palvimo J.J., Niskanen E.A. IRF2BP2 modulates the crosstalk between glucocorticoid and TNF signaling. J. Steroid Biochem. Mol. Biol. 2019; 192:105382. [DOI] [PubMed] [Google Scholar]
- 91. Qin Z., Ren F., Xu X., Ren Y., Li H., Wang Y., Zhai Y., Chang Z.. ZNF536, a novel zinc finger protein specifically expressed in the brain, negatively regulates neuron differentiation by repressing retinoic acid-induced gene transcription. Mol. Cell. Biol. 2009; 29:3633–3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Sircoulomb F., Nicolas N., Ferrari A., Finetti P., Bekhouche I., Rousselet E., Lonigro A., Adelaide J., Baudelet E., Esteyries S.et al.. ZNF703 gene amplification at 8p12 specifies luminal B breast cancer. EMBO Mol. Med. 2011; 3:153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ewing A.K., Attner M., Chakravarti D. Novel regulatory role for human Acf1 in transcriptional repression of vitamin D3 receptor-regulated genes. Mol. Endocrinol. 2007; 21:1791–1806. [DOI] [PubMed] [Google Scholar]
- 94. Yang F., Ma Q., Liu Z., Li W., Tan Y., Jin C., Ma W., Hu Y., Shen J., Ohgi K.A.et al.. Glucocorticoid receptor:MegaTrans switching mediates the repression of an ERalpha-regulated transcriptional program. Mol. Cell. 2017; 66:321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Trotter K.W., King H.A., Archer T.K.. Glucocorticoid receptor transcriptional activation via the BRG1-dependent recruitment of TOP2beta and Ku70/86. Mol. Cell. Biol. 2015; 35:2799–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Kruth K.A., Fang M., Shelton D.N., Abu-Halawa O., Mahling R., Yang H., Weissman J.S., Loh M.L., Muschen M., Tasian S.K.et al.. Suppression of B-cell development genes is key to glucocorticoid efficacy in treatment of acute lymphoblastic leukemia. Blood. 2017; 129:3000–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Lee B.H., Stallcup M.R.. Glucocorticoid receptor binding to chromatin is selectively controlled by the coregulator hic-5 and chromatin remodeling enzymes. J. Biol. Chem. 2017; 292:9320–9334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. John S., Sabo P.J., Johnson T.A., Sung M.H., Biddie S.C., Lightman S.L., Voss T.C., Davis S.R., Meltzer P.S., Stamatoyannopoulos J.A.et al.. Interaction of the glucocorticoid receptor with the chromatin landscape. Mol. Cell. 2008; 29:611–624. [DOI] [PubMed] [Google Scholar]
- 99. Engel K.B., Yamamoto K.R.. The glucocorticoid receptor and the coregulator brm selectively modulate each other's occupancy and activity in a gene-specific manner. Mol. Cell. Biol. 2011; 31:3267–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Paakinaho V., Makkonen H., Jaaskelainen T., Palvimo J.J.. Glucocorticoid receptor activates poised FKBP51 locus through long-distance interactions. Mol. Endocrinol. 2010; 24:511–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Miller E.L., Hargreaves D.C., Kadoch C., Chang C.Y., Calarco J.P., Hodges C., Buenrostro J.D., Cui K., Greenleaf W.J., Zhao K.et al.. TOP2 synergizes with BAF chromatin remodeling for both resolution and formation of facultative heterochromatin. Nat. Struct. Mol. Biol. 2017; 24:344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Cossec J.C., Theurillat I., Chica C., Bua Aguin S., Gaume X., Andrieux A., Iturbide A., Jouvion G., Li H., Bossis G.et al.. SUMO safeguards somatic and pluripotent cell identities by enforcing distinct chromatin states. Cell. Stem Cell. 2018; 23:742–757. [DOI] [PubMed] [Google Scholar]
- 103. Sutinen P., Rahkama V., Rytinki M., Palvimo J.J.. Nuclear mobility and activity of FOXA1 with androgen receptor are regulated by SUMOylation. Mol. Endocrinol. 2014; 28:1719–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Poukka H., Karvonen U., Janne O.A., Palvimo J.J.. Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1). Proc. Natl. Acad. Sci. U.S.A. 2000; 97:14145–14150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Kotaja N., Karvonen U., Janne O.A., Palvimo J.J.. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol. Cell. Biol. 2002; 22:5222–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Deutsch E.W., Csordas A., Sun Z., Jarnuczak A., Perez-Riverol Y., Ternent T., Campbell D.S., Bernal-Llinares M., Okuda S., Kawano S.et al.. The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 2017; 45:D1100–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M.et al.. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019; 47:D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (106) via the PRIDE (107) partner repository with the following dataset identifiers: for SUMO2/3 IP PXD017714, for ChIP-SICAP PXD015183, for GR3KR BioID PXD017713. Newly generated ChIP-seq, RNA-seq and ATAC-seq datasets have been submitted to the NCBI Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/); accession code: GSE64373.