Abstract
The integrity and regulation of the nuclear lamina is essential for nuclear organization and chromatin stability, with its dysregulation being linked to laminopathy diseases and cancer. Although numerous posttranslational modifications have been identified on lamins, few have been ascribed a regulatory function. Here, we establish that lamin B1 (LMNB1) acetylation at K134 is a molecular toggle that controls nuclear periphery stability, cell cycle progression, and DNA repair. LMNB1 acetylation prevents lamina disruption during herpesvirus type 1 (HSV-1) infection, thereby inhibiting virus production. We also demonstrate the broad impact of this site on laminar processes in uninfected cells. LMNB1 acetylation negatively regulates canonical nonhomologous end joining by impairing the recruitment of 53BP1 to damaged DNA. This defect causes a delay in DNA damage resolution and a persistent activation of the G1/S checkpoint. Altogether, we reveal LMNB1 acetylation as a mechanism for controlling DNA repair pathway choice and stabilizing the nuclear periphery.
INTRODUCTION
The nuclear lamina (NL) is the proteinaceous network located on the inner side of the nuclear membrane that modulates core nuclear functions, including defining nuclear shape, providing spatial organization within the nucleus, and tethering and stabilizing chromatin (1–5). In humans, the NL is composed of four type V intermediate filament proteins- the separately encoded lamin B1 (LMNB1) and lamin B2 (LMNB2) and the splice variants lamin A (LMNA) and lamin C (LMNC) (5). All of these proteins have a similar structure, and each of the nuclear lamins assembles into higher order filaments with multifunctional roles.
Given the importance of lamins in maintaining nuclear integrity and functions, it is not surprising that the NL has been linked to regulating multiple cellular processes. For example, phosphorylation leads to disruption of the nuclear lamina preceding mitosis (6,7). In particular, CDK1 phosphorylates the head domain of lamins to disrupt the laminar polymers (8). The lamina also regulates gene expression through interactions with lamin associated domains (LADs) on both heterochromatin and euchromatin (1,2). Additionally, the lamins help maintain genomic integrity and participate in the repair of damaged DNA. The decision to utilize either nonhomologous end joining (NHEJ) or homologous recombination (HR) to repair double stranded DNA breaks (DSBs) is modulated by proximity to the nuclear lamina, although the mechanism underlying this decision remains to be fully defined (9,10). In agreement with their impact on several cellular processes, mutations in nuclear lamins manifest in the development of a wide variety of rare but devastating diseases known as laminopathies, which include muscular disorders and progeria (4). Lamin functions and dysregulation are also linked to cancer biology, as their rearrangement is needed for maintaining nuclear integrity and the balance between nuclear rigidity and pliability when cells migrate through small and stiff spaces (11–13).
A key aspect of lamin biology comes from work that has demonstrated that the lamins A/C and B1 form interacting but distinct networks, indicating that each of these lamins likely has individual functions (14,15). This observation is supported by findings that the knockout or mutation of individual lamins results in distinct nuclear phenotypes and disruptions (15–17). While differences in lamin regulation and function have been probed by studying alterations in their protein–protein and protein–DNA interactions (1,18–21), accumulating evidence has shown that post-translational modifications (PTMs) provide another layer of regulation of NL functions (22). Mass spectrometry-based studies have identified the presence of several PTMs on human lamins, including phosphorylation, acetylation, farnesylation, methylation, ubiquitination and SUMOylation (22–24). While the functions of a few site-specific PTMs on particular lamins have been determined (5,25), most of these modifications remain uncharacterized, including those on LMNB1, the most ancient and ubiquitously expressed lamin (3).
Our lab identified site-specific alterations in LMNB1 acetylation (26) during infection with human cytomegalovirus (HCMV), a nuclear-replicating dsDNA virus that causes congenital diseases and morbidity and mortality in immunocompromised individuals (27,28). We found that LMNB1 K134 acetylation acts as a host response to suppress HCMV production. K134 acetylation increases over the course of infection and functionally stabilizes the nuclear periphery, which is otherwise disrupted by HCMV infection (26). Increasing nuclear integrity impairs viral capsid nuclear egress and attenuates virus production. Although elevated LMNB1 K134 acetylation impairs viral replication late in infection, we found this modification to also be present in uninfected cells. This suggests that this acetylation could also serve as a regulatory point outside the context of infection. Given that herpesviruses induce cell cycle arrest in G1 to facilitate viral genome replication (29–31) and that lamins can modify the cell cycle, we propose that the K134 acetylation may be broadly relevant in protecting nuclear stability and be tied to the progression of the cell cycle.
Here, we establish that the lamin B1 acetylation status acts as a regulatory toggle that controls DNA repair, cell cycle progression, and the stability of the nuclear periphery. First, we use a nuclear-replicating herpesvirus to demonstrate the broader impact of the LMNB1 K134 acetylation for maintaining nuclear periphery integrity during viral infections. Next, combining microscopy, flow cytometry, and proximity-based proteomics, we discover a function for LMNB1 acetylation in uninfected cells. We demonstrate that LMNB1 acetylation impedes the localization of 53BP1 to DNA damage foci, thereby inhibiting canonical nonhomologous end joining (cNHEJ), delaying DNA damage resolution, and inducing persistent activation of the G1/S checkpoint. Together, these results highlight LMNB1 acetylation as a regulatory hub during viral infections and DNA damage response.
MATERIALS AND METHODS
Cell lines
MRC5 primary human lung fibroblasts (ATCC CCL-171, male, passage number 20–28), U2OS human bone osteosarcoma cells (ATCC HTB-96, female), and U2OS DNA repair reporter cell lines (EJ5 and EJ7, a kind gift from Jeremy Stark (32,33)) were cultured in Dulbecco's modified Eagle's medium (DMEM) (Life Technologies) supplemented with 10% fetal bovine serum (v/v) (Gemini Bio-Products) and 1% penicillin–streptomycin (P/S) (v/v) (GIBCO) at 37°C and 5% CO2. The original MRC5 cell line, as well as the mCherry-LMNB1 and 3HA-BirA(miniTurbo)-LMNB1 cell lines, were authenticated by morphology checking by microscopy. Phoenix cells (ATCC CRL-3213, embryonic) were cultured under the same conditions but without the addition of antibiotic. Mycoplasma detection assays were performed by ATCC by agar culture, PCR-assay, and Hoechst DNA staining.
Virus strain and viral infection
WT HSV-1 (17+ strain, gift from Beate Sodeik) was produced and propagated as previously described (34). Briefly, wild-type HSV-1 was produced by electroporating pBAC-HSV-1 into U2OS cells and harvesting P0 stocks. To generate P1 stocks, P0 stocks were used to infect U2OS cells at a low MOI. Virus was collected when cells displayed 100% cytopathic effect and was concentrated by ultracentrifugation. Viral stock titer was determined by plaque assay (see below). MRC5 cells (2.5 × 105/well of a six-well tissue culture plate) that were transfected with the LMNB1 constructs were infected with WT HSV-1 in half volume DMEM plus 2% FBS (v/v) and 1% P/S (v/v) at a MOI of 5 (microscopy) or MOI of 1 (qPCR). Plates were incubated at 37°C and 5% CO2 and were rocked every 10 min for 1 h. Infection media was replaced with complete media, and infection progressed until collection at desired time points. HSV-1 titers were calculated by plaque assay. Briefly, supernatants were collected at 24 hpi. The supernatants were spun down at 2000 × g for 5 min to pellet cellular debris, and the clarified supernatants were stored at –80°C. A reporter plate of U2OS cells was cultured until 90–95% confluent. Collected viral supernatants were thawed and diluted in a serial manner into DMEM supplemented with 2% FBS (v/v) and 1% P/S (v/v) and added to the reporter plate. Plates were rocked every 10 min for 1 h. At this time, media was replaced with methocel media (1% methocel w/v), and cells were incubated at 37°C and 5% CO2 for 72 h. Methocel was aspirated, and cells were washed with 1× PBS. Cells were incubated with crystal violet (1% crystal violet w/v, 50% methanol v/v) for 15 min at room temperature before rinsing and plaque quantification.
Sample collection for HSV-1 viral genome quantification
MRC5 cells expressing mCherry-LMNB1 WT, K134Q or K134R were plated at 1.25 × 105 cells/ml the day before infection. Cells were infected with HSV-1 at a MOI of 1 as described above. At the indicated time points, the cells were washed and scraped into PBS, pelleted at 9000 rpm for 5 min, and flash frozen in liquid nitrogen. For nuclear/cytoplasmic fractionation, the cells were pelleted and immediately fractionated using the NE-PER nuclear/cytoplasmic fractionation kit (Life Technologies Corporation). Briefly, a packed cell volume of 10 μl was resuspended in CERI supplemented with 1:100 HALT Protease and Phosphatase Inhibitor by vortexing for 15 s and then incubated on ice for 10 min. CERII was added to the tube, which was then vortexed for 5 s, incubated on ice for 1 min, vortexed for 5 s and centrifuged at 4°C for 5 min at 16 000 × g. The supernatant (cytoplasmic fraction) was transferred to a new tube, flash frozen in liquid nitrogen, and stored at –80°C until ready for further sample preparation. The pellet (nuclear fraction) was resuspended in NER by vortexing for 15 s. The sample was then incubated on ice and was vortexed for 15 s every 10 min for a total of 40 min. The sample was centrifuged for 10 min at 4°C at 16 000 × g, and the supernatant (nuclear fraction) was transferred to a new tube, flash frozen, and stored at –80°C until ready for DNA extraction.
DNA extraction for qPCR
For whole cell lysate samples, the cell pellet was resuspended in 500 μl media and sonicated 5 times for 10 s each time, icing the sample between each round of sonication. The sample was then centrifuged at 5000 rpm for 1 min, and the supernatant was transferred to a new tube. For each sample type (whole cell lysate, nuclear fraction, cytoplasmic fraction), the sample was diluted into qPCR DNA Resuspension Buffer (400 mM NaCl, 10 mM Tris pH 7.5, 10 mM EDTA) at a 1:4 sample to qPCR DNA Resuspension Buffer ratio. Proteinase K (40 μg/ml) and SDS (0.16%) were added to the sample, and the sample was incubated overnight at 37°C. The next day, a phenol:chloroform extraction was performed by adding 1:1 phenol:chloroform (v/v) to the sample. The sample was vortexed for 5 s, mixed by rotating at 4°C for 1 min, and centrifuged for 1 min at 13 000 rpm. The supernatant was then transferred to a new tube. This extraction was repeated a total of three times with phenol:chloroform and one time with chloroform. Four microliters linear acrylamide, 1:10 (v/v) 3 M NaOAc, and 2 volumes ice cold 100% ethanol were added to the sample, and the sample was incubated overnight at –20°C. The next day, the sample was centrifuged at 13 000 rpm for 10 min at 4°C. The supernatant was decanted, and the pellet was washed with 1 ml 70% ice cold ethanol and centrifuged at 13 000 rpm for 2 min at 4°C. The supernatant was decanted, and the pellet was dried via speedvac. The DNA pellet was resuspended in water.
qPCR of viral genomes
Quantification of viral genomes was by qPCR targeting the UL30 locus of the viral genome (F: 5′-GCGAAAAGACGTTCACCAAG-3′; R: 5′-CGGAGACGGTATCGTCGTAA-3′) and endogenous GAPDH for normalization (F:5′-TTCGACAGTCAGCCGCATCTTCTT-3′; R: 5′-CAGGCGCCCAATACGACCAAATC-3′). Primers were used at 5 μM in SYBR green PCR master mix (Life Technologies). Template DNA was diluted 1:50–1:250-fold, depending upon the application. Amplification was on the ViiA 7 real-time PCR systems (Applied Biosystems) in the Princeton Genomics Core facility. The UL30 amplicon sequence was cloned into pCR2.1 using the topo-TA cloning system in order to generate a standard for absolute quantification of viral genomes. Relative viral DNA was determined using the ΔΔCT method with GAPDH as an internal control. Relative standards were preformed to calculate amplification efficiency by five-fold serial dilution of pooled samples for each reaction. Efficiencies were between 80% and 110%.
Transient DNA transfection
To complete the transfection, 300 ng of mini-prepped mCherry-LMNB1 WT, K134Q or K134R [previously generated in (26)] were incubated with 1 μl Xtremegene (Sigma-Aldrich) diluted in 200 μl OMEM (Life Technologies) for 20 min at room temperature. This mixture was added to a dish of previously seeded 1.4 × 105 MRC5 cells in OMEM for a total volume of 2 ml (six-well tissue culture plate), and the cells were incubated with the mixture for 5–6 h. The cells were washed with complete media and then incubated with complete media for ∼40 h before infection.
Cloning of mCherry-LMNB1, miniTurbo-LMNB1 and CRISPR-resistant mCherry-LMNB1
The mCherry-Lamin B1–10 construct (Addgene, Plasmid #55069) that was previously altered via site-directed mutagenesis to generate K134Q and K134R (26) was used as the parent construct for MRC5 and U2OS mCherry-LMNB1 WT, K134Q and K134R stable lines. The 3xHA-miniTurbo-NLS_pCDNA3 construct (Addgene, Plasmid #107172) was used as the parent plasmid for miniTurbo stable cell lines. mCherry-LMNB1 (WT, K134Q or K134R) was amplified by PCR using the following primers: F: 5′-GGCGAATTCATGGTGAGCAAGGGCG-3′ and R: 5′-GCCGGATCCTACATAATTGCACAGCTTCTATTGGATGC-3′. 3xHA-miniTurbo was amplified by PCR using the following primers: F: 5′-GGCGAATTCATGTACCCGTATGATGTTCCGGATTAC-3′ and R: 5′-GCCCTCGAGACTTTTCGGCAGACC-3′. For mCherry-LMNB1, the purified PCR product and the pLXSN retroviral plasmid (Clontech) were digested with EcoRI (New England Biolabs (NEB)) and BamHI (NEB). The insert and vector were ligated together to generate the plasmid pLXSN-mCherry-LMNB1 (WT, K134Q or K134R). Plasmids were confirmed by sequencing (Genewiz). For 3xHA-miniTurbo, the insert and the pLXSN-mCherry-LMNB1 (WT, K134Q or K134R) vector were digested with EcoRI (NEB) and XhoI (NEB). The insert and vector were ligated together to generate the plasmid pLXSN-3xHA-BirA(miniTurbo)-LMNB1 (WT, K134Q or K134R). Plasmids were confirmed by sequencing (Genewiz). Cas9 (CRISPR)-resistant pLXSN-LMNB1 was generated by site directed mutagenesis to generate the following sequence in LMNB1 5′-GCACGAGACCCGTTTAGTAG-3′. This mutagenesis produces silent mutations that are codon optimized in Homo sapiens from the WT sequence of 5′-GCATGAAACGCGCTTGGTA-3′ that is targeted by the guide RNA discussed below. We amplified the pLXSN-LMNB1 plasmid (with associated K134 mutations) with primers facing opposing directions and containing mutations in 5′ overhangs in the following 5′-phoshorylated primers: F: 5′-CGTTTAGTAGAGGTGGATTCTGGGC-3′ and R: 5′-GGTCTCGTGCTTCCTTCTGGTCTCGTTAATC-3′. Briefly, the entire plasmid was amplified using the high-fidelity Phusion polymerase. The resulting amplicon was verified by gel electrophoresis, treated with DpnI to digest the parental plasmid, and ligated to re-circularize the amplicon. These plasmids were then transfected into E. coli and the resulting mutants were sequence verified as described above.
Generation of stable cell lines
To generate stable cell lines, each plasmid was transfected into Phoenix cells. Briefly, 7 μg plasmid and 435 μl OMEM were incubated in one tube while 35 μl Lipofectamine 2000 (Thermo Fisher Scientific) and 435 μl OMEM were incubated in a separate tube for 1 min. The two tubes were combined and incubated at room temperature for 5 min. The mixture was dropped onto Phoenix cells and incubated overnight. The following day, the media was changed to DMEM supplemented with 10% FBS. Retrovirus was collected at 48 and 72 h post-transfection, filtered through a 0.45 μm Acrodisc® Syringe Filter with Supor® Membrane (VWR), mixed one to one (v/v) with complete media, and used to transduce MRC5, U2OS or EJ5 Reporter cells. Cells stably expressing the constructs of interest were selected with 400 μg/ml G418 (Thermo Fisher Scientific) until the cells on a control plate with no G418 resistance were dead. Stable cell lines were maintained in half selection media (200 μg/ml G418) until ready to use for an experiment. When cells were seeded for an experiment, G418 was not included in the media. Endogenous LMNB1 knockouts were generated in MRC5 cells using the TrueCut system (Invitrogen). Briefly, cells were seeded the evening prior to transfection at 0.8 × 105 cells/ml. LMNB1 sequence specific TrueGuide sgRNA (Invitrogen #A35533) targeting the sequence 5′-GCACGAGACCCGTTTAGTAG-3′ (Invitrogen #A35526) was used. The sgRNA was combined in equal molar ratios to TrueCut Cas9 Protein V2 (Invitrogen #A36499) in OMEM (ThermoFisher #31985062) and 2:1 CRISPRMAX transfection reagent (ThermoFisher #CMAX0001). Transfection and editing efficiency were >80%.
LMNB1 sequence alignment and phylogenetic analysis
Protein amino acid sequences were retrieved from Uniprot for LMNB1 from 18 different species and for all four human lamins (LMNB1, LMNB2, LMNA, LMNC). These sequences were aligned by MultAlin and annotated by ESPript3. LMNB1 phylogenetic analysis was performed using MEGAX v10.1.8 using the Neighbor-Joining method with evolutionary distances computed using the Poisson correction method.
Bleomycin and hydrogen peroxide treatment
Stable MRC5 or U2OS mCherry-LMNB1 (WT, K134Q or K134R) cells were treated with 10 μg/ml bleomycin (ApexBIO) diluted in media for 1 h or with 35 or 200 μM hydrogen peroxide diluted in media for 30 min. For flow cytometry, media was replaced with fresh complete media plus 10 μM EdU (Fisher Scientific) and incubated for 1 h prior to collection. For microscopy, media was replaced with complete media. The 0 h post-treatment sample was collected immediately, and samples were subsequently collected every hour for 6 h post-treatment. Flow cytometry and immunofluorescence microscopy are described below.
Cell cycle flow cytometry
Untreated or bleomycin-treated 70% confluent MRC5 cells were incubated with 10 μM EdU for 1 h prior to collection. Cells were washed with 1× PBS, trypsinized, centrifuged for 5 min at 400 × g, and fixed in 4% paraformaldehyde (PFA) (Electron Microscopy Sciences) for 20 min at room temperature. Samples were washed in wash buffer (1× PBS, 2% FBS) two times by centrifuging at 800 × g for 8 min. Samples were permeabilized in 0.25% Triton X-100 in wash buffer on ice for 15 min, then washed in wash buffer two times by centrifuging at 800 × g for 8 min. S phase cells were labelled by conjugating azide-AlexaFluor-488 (ClickChemistryTools) to EdU by incubating with Click-IT staining solution (10 mM sodium ascorbate, 2 mM CuSO4, and 10 μM Azide-AlexaFluor 488 in 1x PBS) for 1 h. Samples were washed twice with wash buffer at 1200 × g for 8 min. M phase cells were stained for 1 h with anti-pH3 S10 (histone H3 phosphorylated at S10) pre-conjugated to AlexaFluor 647 (1:100, Cell Signaling #3458S) diluted in wash buffer. To stain for DNA content (for G1 and G2 phases), cells were incubated with DAPI (1:500, ThermoFisher Scientific) in wash buffer prior to flow cytometry. Cells were passed through a cell strainer, and flow cytometry analysis was performed in the Princeton Flow Cytometry Resource Facility on a BD LSRII Multi-Laser Analyzer w/ HTS with FACSDIVA software and then analyzed using FCSExpress v7.01.
Immunofluorescence microscopy and analysis
MRC5 or U2OS cells grown on 22 × 22 mm glass coverslips (VWR) were fixed in 4% PFA for 15 min at room temperature. Cells were washed three times with 1× PBS and then permeabilized in 0.2% Triton X-100 in PBS for 10 min at room temperature. Cells were washed once with 1x PBS and then blocked in 2.5% human serum, 2.5% goat serum in 1× PBT (0.1% Triton) for 30 min. Samples were incubated for 1 h with the following primary antibodies diluted into block: ICP4 (Ms, 1:300, Abcam, Ab6514) γH2AX (Ms, 1:500, BioLegend, #613402), 53BP1 (Rb, 1:100, Cell Signaling, #4937), HA (Ms, 1:1000, VWR, clone 16B12). Secondary staining was performed by incubating for 1 h with the appropriate secondary antibody or streptavidin conjugated to AlexaFluor488 diluted in block: goat anti-Ms IgG highly cross-adsorbed AlexaFluor488 (1:2000, Life Technologies A-11001), goat anti-Rb IgG cross-adsorbed AlexaFluor488 (1:2000, Thermo Fisher Scientific, A11008), goat anti-Ms IgG cross-adsorbed AlexaFluor568 (1:2000, Invitrogen A11019), streptavidin-AlexaFluor488 (1:1000, Thermo Fisher Scientific, S32354) and DAPI (1:1000, Thermo Fisher Scientific). Slides were mounted using Prolong Diamond and were imaged in the Princeton Confocal Imaging Core using an inverted fluorescence confocal microscope (Nikon Ti-E) equipped with a Yokogawa spinning disc (CSU-21) and digital CMOS camera (Hamamatsu ORCA-Flash TuCam) using a Nikon 100× Plan Apo objective with a 100× magnification. Image analysis was performed using ImageJ. Average fluorescence intensity within a nucleus was quantified following definition of the nucleus as an ROI and background subtraction in the channel to be quantified. Foci were quantified within nuclear ROIs after background subtraction in the channel to be quantified using the 3D Objects Counter plugin in ImageJ. Laminar breaks were manually quantified after background subtraction in the channel to be quantified, straightening, and binarizing each lamina. Average laminar area was measured as the area of the linearized nuclear lamina. Average laminar width was calculated by measuring the width of the nuclear lamina at four points across the lamina and taking the average of those four measurements. Levels of γH2AX in stable cell lines of mCherry-LMNB1 WT and K134Q expressed in an endogenous LMNB1 KO background were measured and stained for as described above except that a Nikon Plan Apo λ 20× objective at a 20× magnification was used.
EU labeling and microscopy
Cells were seeded on 15 mm coverslips at a density of 0.5 × 105 cells/ml the day before the experiment. Cells were treated with bleomycin (10 μg/ml) for 1 h. The bleomycin was then washed out, and the cells were labelled with a final concentration of 1 mM EU for 1 h. The cells were then washed with PBS and fixed in 4% paraformaldehyde for 15 min. Cells were permeabilized in 0.2% PBT (Triton in PBS) for 10 min at room temperature. A Click-IT reaction was performed to enable visualization of the EU labeling. Specifically, slides were stained for 1 h with 10 μM Azide-AlexaFluor-488, 2 mM CuSO4, 100 mM sodium ascorbate, in 1× PBS. Slides were blocked in 2.5% human serum, 2.5% goat serum for 30 min. Slides were incubated with primary antibody (γH2AX, Ms, 1:500, Biolegend, #613402) for 1 h, then incubated with secondary antibody (goat anti-mouse IgG (H+L) highly cross-adsorbed secondary antibody, Alexa Fluor 633, 1:2000, Thermo Fisher Scientific) and DAPI (1:1000, Thermo Fisher Scientific) for 1 h. Slides were mounted using Prolong Diamond and were imaged in the Princeton Confocal Imaging Core as described above. Image analysis was performed in ImageJ. Z-stacks were converted to maximum projection images, and nuclear ROIs were defined based on DAPI staining. The integrated density of the EU staining was measured for each nucleus.
Proximity ligation assay (PLA)
A proximity ligation assay was performed using the Duolink In Situ Detection Reagents Green (Millipore Sigma) following the manufacturer's protocol. Briefly, 2 × 105 cells were seeded per 35 mm scope dish the day before the experiment. The next day, cells were either untreated or treated with 10 μg/ml bleomycin for 1 h. The media was changed, and cells recovered for 1 h before being washed with 1× PBS and fixed in 4% paraformaldehyde for 15 min. Cells were permeabilized with 0.2% PBT for 10 min at room temperature. Cells were blocked in 2.5% human serum, 2.5% goat serum for 30 min at room temperature. Cells were then incubated with the correct pair of primary antibodies including LMNB1 (1:500, Ms, Proteintech, #66095-1-Ig) and histone H3 tri methyl K9 (H3K9me3) (1:500, Rb, abcam, ab8898), histone H3 tri methyl K4 (H3K4me3) (1:500, Rb, abcam, ab8580) or 53BP1 (Rb, 1:100, Cell Signaling #4937) for 1 h at room temperature. Samples were washed 2 times for 5 min with Wash Buffer A and then incubated with the Duolink PLA Probes (1:5 Duolink In Situ PLA Probe Anti-Mouse MINUS and 1:5 Duolink In Situ PLA Probe Anti-Rabbit PLUS) for 1 h in a humidified chamber at 37°C. Samples were washed 2 times for 5 min with Wash Buffer A and then incubated with Ligation Solution for 30 min in a humidified chamber at 37°C. Samples were washed 2 times for 5 min with Wash Buffer A and then incubated with the Amplification Solution for 100 min in a humidified chamber at 37°C. Then, samples were washed 2 times for 5 min with 1× Wash Buffer B and 1 time for 2 min with 0.01× Wash Buffer B. Nuclei were stained by incubating the samples with DAPI (1:1000) diluted in 0.01× Wash Buffer B for 10 min. Samples were then washed 2 times for 5 min with 0.01× Wash Buffer B. Samples were imaged in the Princeton Confocal Imaging Core as described above. Image analysis was performed in ImageJ. Nuclear ROIs were defined from a maximum projection of the DAPI channel. The PLA channel (488) was background-subtracted using a rolling ball radius of 200. The 3D-object counter was used to count the number of PLA foci.
Comet assay
To assess the persistence of DNA damage in untreated cells and 3 h post-bleomycin treatment, an Alkaline Comet Assay (Trevigen) was performed as per the manufacturer's instructions. The comets were stained with 1× SYBR Gold (Thermo Fisher Scientific), and images of comets were acquired as described above except that a Nikon Plan Apo λ 20× objective at a 20× magnification was used. At least 175 comets across two replicates were analyzed. Comet tail length and total length were measured manually and the percent tail DNA was calculated in Microsoft Excel.
Western blot analysis
Samples were lysed in a buffer containing 50 mM Tris–HCl pH 8.0, 100 mM NaCl, 0.5 mM EDTA, and 4% SDS by heating at 95°C for 5 min and then sonicating for ten 1-s pulses in a cup horn sonicator. This process was repeated until lysis was complete. Protein concentration was determined by BCA assay, and equal amounts (μg) of sample were loaded on a 10% tris-glycine SDS-polyacrylamide gel. Proteins were transferred onto a polyvinylidene fluoride (PVDF) membrane overnight at 4°C at 30 V. Membranes were cut and blocked with 5% milk in 1× TBS for 1 h. All primary antibodies were diluted into 5% milk in 1× TBST (0.1% Tween-20) and incubated overnight at 4°C. The following primary antibodies were used: ICP27 (1:1000, Ms, abcam, ab31631), ICP8 (1:1000, Ms, abcam, ab20194), gC (1:1000, Ms, abcam, ab6509), pRb-S780 (1:1000, Rb, clone D59B7, Cell Signaling Technology, #8180), LMNB1 (1:1000, Rb, abcam, ab16048), RFP (reactive against variants of RFP, 1:1000, Rb, Rockland, #600-401-379) and tubulin (1:5000, Ms, clone DM1A, Sigma, #T6199). The next day, membranes were incubated for 1 h at room temperature with the following secondary antibodies diluted in 5% milk in 1× TBST: mouse anti-rabbit IgG HRP-conjugated (1:5000, Jackson Immuno Research), goat anti-mouse HRP-conjugated (1:5000, Jackson Immuno Research) or Alexa 680 anti-mouse or anti-rabbit IgG (H+L) (Invitrogen, 1:10 000). If multiple proteins needed to be blotted for at similar molecular weights, the blots were stripped and re-probed using the following method: wash for 10 min with 1× TBST two times, wash for 20 min with 1× Reblot solution (EMD Millipore ReBlot Plus), wash for 10 min with 1× PBS two times, continue to block, primary and secondary as described above. Densitometry analysis was performed using ImageJ or ImageStudioLite for Odyssey software. All samples were normalized to the loading control (tubulin).
DNA damage repair reporter cell assay
Double stranded DNA break repair analysis was performed as described in (32, 33). Briefly, reporter cells were incubated with 1 ml antibiotic free media for 2 h prior to transfection. For EJ5 cells, 3.6 μl Lipofectamine 2000 was diluted into 100 μl OMEM and incubated for 5 min at room temperature. This mixture was added to a mixture of 800 μg pCBASceI (Addgene, #26477) plus 400 μg mCherry-LMNB1 construct of interest (WT, K134Q, K134R). For EJ7 cells, 3.6 μl Lipofectamine 2000 was diluted into 100 μl OMEM and incubated for 5 min at room temperature. This mixture was added to a mixture of 400 μg 7a sgRNA for EJ7-GFP and 4-μHOM reporters (Addgene, #113620), 400 μg 7b sgRNA for EJ7-GFP reporter (Addgene, #113624) and 400 μg mCherry-LMNB1 construct of interest (WT, K134Q, K134R). The combined mixture was incubated at room temperature for 25 min. Transfection complexes were added to the reporter cells, and the cells were incubated for 3 h. The cells were then washed, and 4 ml complete media was added back. The cells were incubated for 72 h. After 3 days, the cells were trypsinized, filtered through a cell strainer and analyzed by flow cytometry at the Princeton Flow Cytometry Resource Facility on a BD LSRII Multi-Laser Analyzer w/ HTS with FACSDIVA software. Flow cytometry data was analyzed by FCSExpress v7.01 software. The gating approach used is shown in Supplementary Figure S4A.
Analysis of mechanism of DNA repair using EJ5 reporter cells
EJ5 mCherry-LMNB1 WT, K134Q or K134R stable cell lines with endogenous LMNB1 knocked out were transfected with I-SceI as described above. After 72 h, the cells were collected for flow cytometry as described above and sorted on FACSAria Fusion cell sorter (BD Biosciences, San Jose, CA) into single cell colonies in a 96-well plate containing 100 μl of complete growth media. Cells were gated on forward scatter area versus height (FSC-A versus FSC-H) signals to eliminate doublets and on mCherry versus GFP (mCH-A versus GFP-A) signals to identify the double positive population. An untransfected negative control was run for comparison and optimal sort gate setting. Clonal populations of cells were grown until confluent, transferred to a 24-well dish, grown until confluent and then harvested. DNA extraction was performed as described above for qPCR analysis. The region around the I-SceI restriction sites was amplified by PCR using the following primers: F: 5′-CTGCTAACCATGTTCATGCC-3′ and R: 5′-AAGTCGTGCTGCTTCATGTG-3′. The PCR reaction was as follows: 10 μl 5× HF Phusion Buffer, 2 μl forward primer, 2 μl reverse primer, 4 μl 2.5 mM dNTPs, 1 μl Phusion polymerase, 120 ng template DNA and autoclaved water were combined to a final volume of 50 μl per reaction. Then, the following PCR reaction was run: (i) 94°C for 3 min, (ii) 94°C for 45 s, (iii) 63°C for 45 s, (iv) 72°C for 1 min 45 s, (v) repeat steps 2–4 for a total of 35 cycles, (vi) 72°C for 7 min. The PCR product was column purified using a DNA Clean & Concentrator (ZymoResearch) and sent for sequencing. Sequences were aligned in SnapGene and the manner in which the DNA was repaired was analyzed by manual inspection.
miniTurbo sample preparation for mass spectrometry analysis
A 15 cm plate of MRC5 stable LMNB1 cells (WT, K134Q or K134R) was incubated with 10 μg/ml bleomycin for 1 h. After treatment, the media was replaced with fresh media. At 1 h post-treatment, cells were incubated with 500 μM biotin (Sigma Aldrich) for 10 min at 37°C and then washed 5 times with ice cold 1× PBS. Cells were collected by scraping, flash frozen in liquid nitrogen and stored at –80°C. Cells were lysed in RIPA buffer (50 mM Tris pH 8, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton-X 100) plus 1:100 HALT protease and phosphatase inhibitor (Fisher Scientific) by first rotating for 5 min at 4°C and then sonicating in a cup horn sonicator 10 times with 1 s pulses twice. Samples were incubated on ice for 30 s between sonication rounds. The lysate was clarified by centrifuging at 10 000 rpm for 10 min at 4°C, and a BCA assay was conducted to determine protein concentration. 50 μl streptavidin coated beads (Thermo Fisher Scientific)/IP were washed 2 times with RIPA buffer. 1000 μg protein lysate (2 mg/ml concentration) was incubated with the beads for 1 h at 4°C with rotation. The samples were washed in the following series: 2 times with 1 ml RIPA buffer, 1 time with 1 ml 1 M KCl, 1 time with 1 ml 0.1 M Na2CO3, 1 time with 1 ml 2 M urea in 10 mM Tris–HCl (pH 8), and 2 times with 1 ml RIPA buffer. Beads were resuspended in RIPA buffer and transferred to a new LoBind microcentrifuge tube (Amuza, Inc., Eicom USA). Beads were washed 2 times with 200 ul 50 mM Tris–HCl pH 7.5 and 2 times with 1 ml 2 M urea in 50 mM Tris pH 7.5. Beads were incubated in 80 μl of trypsin buffer (2 M urea, 1 mM DTT, 0.8 μg trypsin in 50 mM Tris pH 7.5) shaking at 600 rpm for 1 h at 25°C. The supernatant was transferred to a new tube, and the beads were washed twice with 60 ul 2 M urea in 50 mM Tris pH 7.5. The washes were combined with the supernatant. Samples were reduced and alkylated with 25 mM Tris(2-carboxyethyl)phosphine (TCEP) (Thermo Fisher) and 50 mM chloroacetamide (Fisher Scientific) by shaking for 1.5 h at 600 rpm at 25°C. Samples were then diluted 1:1 with 50 mM Tris pH 8 and digested with 1 μg of trypsin overnight, shaking at 600 rpm at 25°C. Samples were acidified to 1% TFA, incubated on ice for 15 min, and centrifuged at 4000 × g for 5 min at 4°C. Samples were desalted over SDB-RPS StageTips (3 M Analytical Bio-technologies), dried in a speedvac and resuspended in 1% formic acid/1% acetonitrile.
Mass spectrometry data acquisition and analysis for miniTurbo
Samples were run on a Q-Exactive HF mass spectrometer (ThermoFisher Scientific) equipped with an EASYSpray ion source (ThermoFisher Scientific). Peptides were separated on an EASYSpray C18 column (2 μm × 75 μm × 50 cm) over a 150 min gradient at a flow rate of 250 nl/min as follows: 1% B to 5% B over 14 min, 5% B to 30% B over 150 min, 30% B to 97% B over 1 min, hold at 97% B for 1 min and 70% B for 1 min. MS1 scans were collected with the following parameters: 12 000 resolution, 30 ms MIT, 3e6 AGC, scan range 350–1800 m/z, loop count 15 and data collected in profile. MS2 scans were collected with the following parameters: 30 000 resolution, 150 ms MIT, 1e5 AGC, 1.2 m/z isolation window, loop count of 15, NCE of 28, 100.0 m/z fixed first mass, peptide match set to preferred and data collected at a dynamic exclusion of 30 s in profile. MS/MS spectra were analyzed by Proteome Discoverer v2.4 (Thermo Fisher Scientific). Spectra were searched using Sequest HT against a combined Uniprot database containing human protein sequences (downloaded January 2020) and common contaminants. The Spectrum Files RC node was used to perform offline mass recalibration, and the Minora Feature Detector node was used for label-free MS1 quantification. Fully tryptic peptides with a maximum of two missed cleavages, a 4 ppm precursor mass tolerance, and a 0.02 Da fragment mass tolerance were used in the search. Post-translational modifications (PTMs) that were allowed included the static modification carbamidomethylation of cysteine and the dynamic modifications of oxidation of methionine, deamidation of asparagine, loss of methionine plus acetylation of the N-terminus of the protein, acetylation of lysine, acetylation of the N-terminus of the protein and phosphorylation of serine, threonine and tyrosine. The Percolator node was used for peptide spectrum match (PSM) validation and the ptmRS node was used for assigning PTM sites. PSMs were assembled into peptide and protein identifications with a false discovery rate of <1% for both the peptide and protein level with at least two unique peptides identified per protein. Protein abundances from each run were normalized to the average protein abundance. Nuclear proteins were annotated based on Uniprot assignments. Putative altered interactions due to the presence of the acetyl mimic were determined by calculating the average fold change compared to WT or charge mimic. Proteins that had at least a 2-fold change in abundance in at least one comparison were considered for further analysis. Cytoscape v7.2 and the STRING plugin were used for network generation and visualization.
Parallel reaction monitoring (PRM) quantification of mCherry-LMNB1 and pRb protein levels
Cells were lysed in lysis buffer (50 mM Tris–HCl pH 8.0, 100 mM NaCl, 0.5 mM EDTA and 4% SDS) by heating at 95°C for 5 min and then sonicating for ten 1-s pulses in a cup horn sonicator. This process was repeated until lysis was complete. A BCA assay was performed to determine protein concentration. Cell lysates were reduced and alkylated at 70°C for 20 min and then proteins were precipitated by methanol/chloroform extraction. The protein pellets were resuspended in 25 mM HEPES pH 8.2 at 0.5 μg/μl via cup horn sonication. Proteins were digested overnight at 37°C using a 1:50 trypsin:protein ratio. Digested peptides were acidified to 1% trifluoroacetic acid (TFA) and desalted using an SDB-RPS StageTip Desalting (3 M Analytical Biotechnologies). Samples were resuspended in 1% formic acid, 1% acetonitrile. Samples were run on a Q-Exactive HF mass spectrometer (ThermoFisher Scientific) equipped with a Nanospray Flex Ion Source (ThermoFisher Scientific). Peptides were separated on an in-house packed 50 cm column (360 μm od, 75 μm id, Fisher Scientific) packed with ReproSil-Pur C18 material (120 Å pore size, 1.9 μm particle size, ESI Source Solutions) equipped with a stainless steel emitter (ThermoFisher Scientific) over a flow gradient of 60 min at a flow rate of 250 nl/min as follows: 1% B for 12 min, 3% B for 60 min and 97% B for 18 min. Full MS1 scans were performed with the following parameters: 30 000 resolution, 3e6 AGC target, 15 ms maximum injection time, scan range of 350–1800 m/z, and spectrum data collected in profile. Targeted MS2 scans were performed with the following parameters: 30 000 resolution, 1e5 AGC target, 60 ms maximum injection time, isolation window of 0.8, fixed first mass 125, spectrum data collected in profile and retention time windows of 6 min. The signature peptides (shown in figure panels) used for PRM analysis of mCherry-LMNB1 and pRb were selected and analyzed using Skyline v20.1 (35). Peptide abundances were calculated using the summed area under the curve for four transition ions per peptide. Peak areas were normalized to loading control peptides from GAPDH (VGVNGFGR and AGAHLQGGAK), HIST1H3A (EIAQDFK), and TUB1A (FDLMYAK and EDMAALEK).
Quantification of endogenous LMNB1 acetylation
To quantify the abundance of the K134 endogenous acetylation in cells enriched in the G2 phase of the cell cycle, six 70% confluent plates of MRC5 cells were treated with 10 μM CDK1 inhibitor RO-3306 (Sigma Aldrich) for 48 h. Samples were then collected, and acetylated peptides were enriched and prepared for mass spectrometry analysis as previously described (26, 36). Briefly, cell pellets were lysed, proteins were reduced and alkylated, proteins were precipitated by methanol/chloroform extraction, and proteins were digested with trypsin overnight. The digested peptides were cleaned up using an Oasis Column (Waters) and lyophilized for 2 days. Next, a 50 μg protein aliquot was reserved for whole proteome analysis, and then the acetylated peptides were enriched by anti-acetyl lysine immunoaffinity purification (PTMscan kit, Cell Signaling Technology). The enriched peptides and whole proteome samples were desalted using SDB-RPS StageTip Desalting (3M Analytical Biotechnologies) and resuspended in 1% formic acid, 1% acetonitrile. Tryptic peptides were analyzed on a Q-Exactive HF mass spectrometer (ThermoFisher Scientific) equipped with a Nanospray Flex Ion Source (ThermoFisher Scientific). Peptides were separated on an in-house packed 50 cm column (360 μm od, 75 μm id, Fisher Scientific) packed with ReproSil-Pur C18 material (120 Å pore size, 1.9 μm particle size, ESI Source Solutions) equipped with a stainless steel emitter (ThermoFisher Scientific) over a 150 min linear gradient at a flow rate of 250 nl/min (3–35% B over 150 min). MS1 scans were collected with the following parameters: 12 000 resolution, 30 ms MIT, 3e6 AGC, scan range 350–1800 m/z, and data collected in profile. MS2 scans were collected with the following parameters: 30 000 resolution, 150 ms MIT, 1e5 AGC, 1.6 m/z isolation window, loop count of 10, NCE of 28, 100.0 m/z fixed first mass, peptide match set to preferred and data collected at a dynamic exclusion of 45 s in centroid. MS/MS spectra were analyzed by Proteome Discoverer v2.4 (PD2.4, Thermo Fisher Scientific) and MS1 spectra were analyzed by Skyline v20 (35). In Proteome Discoverer, spectra from the CDK1 inhibitor-treated (CDK1i) acetyl-IP and those from previously collected untreated samples (26) were searched using Sequest HT against a combined Uniprot database containing human protein sequences (downloaded January 2020) and common contaminants. The search was performed as above for the miniTurbo data. Acetyl peptides that had constant abundances across the samples (untreated and CDK1i) were selected as normalization peptides. Raw files were imported into Skyline for MS1-based quantification. MS1 scans containing either the LMNB1 K134 acetylated peptide (ESDLNGAQIK[+42]LR) or the loading control peptide (FAU/RS30 FVNVVPTFGK[+42]K peptide) were selected based on either the retention time assignments and MS/MS identifications made by PD2.4 or by parallel reaction monitoring (PRM) identification made in separate experiments (data not shown) with the same LC gradient. Furthermore, MS1 peak selection required less than 5 ppm mass difference and the presence of the M+1 and M+2 isotopes in the appropriate abundance in relation to the monoisotopic peak. Summed peak area was calculated from the M+0, M+1 and M+2 isotopic species, and the summed peak area for the LMNB1 K134 acetylated peptide was normalized to the summed peak area of the loading control peptide.
Statistical analysis
GraphPad Prism v.9.0 was used for all statistical analysis. Bar graphs represent means, and error bars indicate standard error of the mean (SEM), unless otherwise noted. The number and type of replicates are indicated in the figure legends. Significance tests were performed using a one-way ANOVA with a post-hoc test, as indicated in the figure legends, unless otherwise specified. Statistical significance is indicated as asterisks in figures: * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001 and **** P ≤ 0.0001.
RESULTS
Lamin B1 acetylation stabilizes the nuclear periphery during HSV-1 infection
Despite many broad similarities, the replication cycles of alpha and beta herpesviruses are strikingly distinct (Figure 1A). For example, the alphaherpesvirus HSV-1 replicates within 18 h, while the viral replication cycle of HCMV, a betaherpesvirus, progresses over 4–5 days. Notably, while HCMV induces kidney-bean shaped nuclear curvature and ER and Golgi remodeling into the nuclear-adjacent viral assembly complex (37, 38), these structures are not present during HSV-1 infection. Despite these differences, these herpesviruses share a common focal point in their replication cycles: viral DNA is packaged and capsids are assembled within the nucleus. Since the herpesvirus capsid is too large to fit through a nuclear pore, it must egress from the nucleus by budding through the nuclear membrane. Therefore, herpesviruses must interact with and disrupt the nuclear lamina to gain access to the nuclear membrane to allow capsids to leave the nucleus for final maturation and cellular egress (39–41).
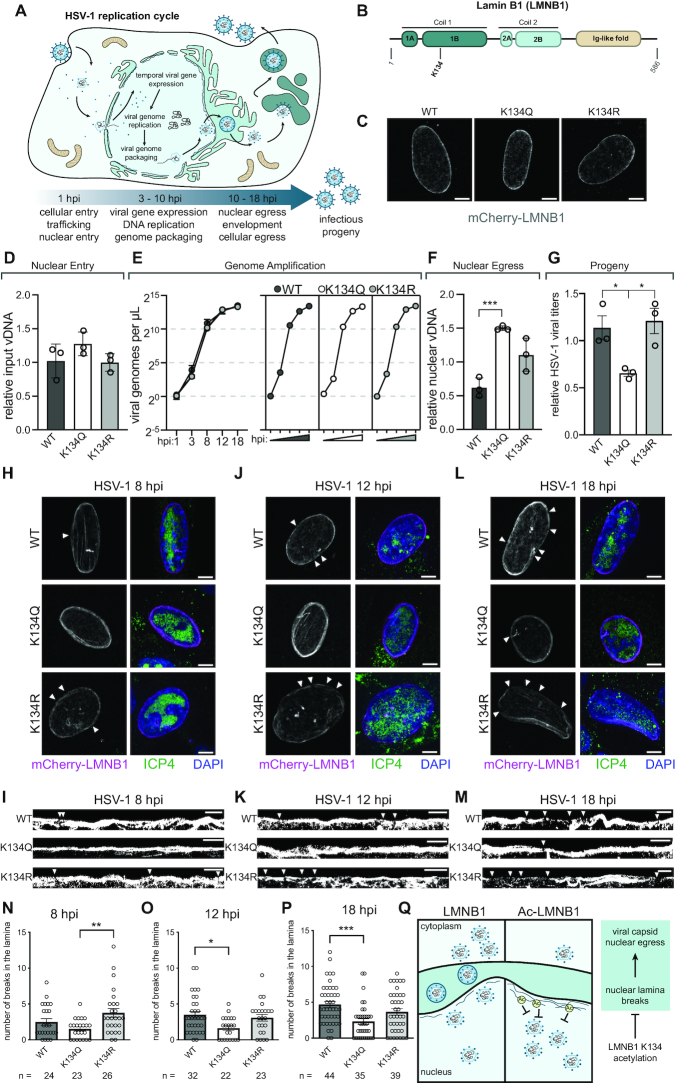
Figure 1.
LMNB1 acetyl mimic preserves laminar integrity during HSV-1 infection. (A) HSV-1 replication cycle demonstrating the temporal progression of viral infection and highlighting nuclear events. (B) Lamin B1 (LMNB1) has five domains, and lysine 134 (K134) falls in the Coil 1B domain. (C) mCherry-LMNB1 morphology in uninfected cells expressing WT, K134Q or K134R LMNB1. Representative immunofluorescence (IF) images are shown, scale bar represents 5 μm. (D) Relative input vDNA (viral genomes) ± SD, determined using the ΔΔCT method with GAPDH as an internal control, present at 1 hpi in WT, K134Q and K134R expressing cells, n = 3 biological replicates. (E) Viral genomes/μl at time points throughout the HSV-1 replication cycle. Individual genome amplification plots for WT, K134Q and K134R expressing cells are shown on the right. n = at least two biological replicates. (F) Relative viral DNA (viral genomes) ± SD present in the nuclear fraction of the 18 hpi WT, K134Q and K134R expressing cells. Unpaired t-test, n = 3 biological replicates. (G) Effect of LMNB1 mutants on HSV-1 viral titers, average ± SEM, one-way ANOVA, Tukey post-hoc test, n = 3 biological replicates. (H, J, L) Representative IF images of MRC5 cells expressing WT, K134Q or K134R mCherry-LMNB1 and infected with HSV-1 at (H) 8 hpi, (J) 12 hpi and (L) 18 hpi. Scale bar represents 5 μm. Left: mCherry-LMNB1, white arrows indicate breaks. Right: merged image of mCherry-LMNB1 (magenta), ICP4 (marker of viral infection, green) and DAPI (blue). (I, K, M) Representative IF images of linearized and binarized mCherry-LMNB1 at (I) 8 hpi, (K) 12 hpi and (M) 18 hpi, breaks indicated by white arrows. Scale bar represents 5 μm. (N, O, P) Quantification of number of breaks in the nuclear lamina in MRC5 cells expressing WT, K134Q or K134R mCherry-LMNB1 at (N) 8 hpi, (O) 12 hpi and (P) 18 hpi. Average ± SEM, one-way ANOVA, Bonferroni post-hoc test, n (number of cells) is indicated in the figure. (Q) Proposed model for the function of LMNB1 K134 acetylation during HSV-1 infection. K134 acetylation may stabilize LMNB1, causing it to incur fewer breaks and thereby inhibit viral capsid nuclear egress.
We have previously shown that acetylation at lysine 134 (K134) in the Coil 1B region of LMNB1 (Figure 1B) decreases HCMV titers and stabilizes the nuclear periphery during HCMV infection (26). We hypothesized that LMNB1 acetylation could be a conserved mechanism for maintaining laminar integrity during infection with nuclear-replicating viruses. To test this hypothesis, we transfected human fibroblasts with WT, acetyl mimic (K134Q) or charge mimic (K134R) versions of LMNB1 tagged with mCherry. All three LMNB1 constructs were expressed at similar levels (Supplementary Figure S1A), and we did not observe any differences in the number of breaks in LMNB1 between the WT and the two mutants in uninfected cells (Figure 1C, Supplementary Figure S1B). To confirm that acetylation-driven laminar stability can inhibit infectious virus production during HSV-1 infection, we performed a titer assay of virions produced from infected fibroblasts expressing WT or mutant LMNB1. Infection of acetyl mimic cells produced 50% fewer infectious virions than in either WT or charge mimic cells (Figure 1G, Supplementary Figure S1C), demonstrating that LMNB1 acetylation is antiviral against herpesvirus infections.
To pinpoint at which stage LMNB1 acetylation affects viral replication, we assessed the impact of the acetyl mimic on delivery of viral genomes to the nucleus, viral gene expression, viral genome replication and viral capsid egress from the nucleus. By measuring viral genome abundance by qPCR over the time course of infection, we observed that LMNB1 K134 acetylation status did not affect viral entry or genome replication (Figure 1D, E). We further found that expression of viral immediate early genes was not impacted by the LMNB1 mutants by staining for the HSV-1 protein ICP4, which had similar levels in both the WT and mutant cells (Supplementary Figure S1D). Western blot analysis of representative immediate early, early and late viral proteins confirmed that the LMNB1 mutants do not impact viral gene expression (Supplementary Figure S1E). Given that our findings did not suggest that LMNB1 acetylation impaired early events in the viral cycle, we predicted that the presence of the acetyl mimic was likely impacting viral capsid egress from the nucleus. To test this hypothesis, we performed nuclear/cytoplasmic fractionation of infected cells at 18 hpi (Supplementary Figure S1F) followed by qPCR for viral genomes in each of these subcellular compartments. In line with our predicted model that LMNB1 acetylation impedes viral capsid nuclear egress, we observed a significant accumulation of viral genomes in the nucleus of the acetyl mimic cells compared to the wild type cells and a reduction in the number of viral genomes in the cytoplasm of the acetyl mimic cells (Figure 1F, Supplementary Figure S1G).
To investigate if the acetyl mimic was leading to an accumulation of viral capsids by stabilizing the nuclear periphery, we infected these cells with HSV-1 and collected samples at mid (8 h post-infection (hpi), Figure 1H, I, N) and late stages of viral infection (12 and 18 hpi, Figure 1J–M, O–P). Already at 8 hpi there were significantly more disruptions in the LMNB1 network in the presence of the charge mimic (K134R) in comparison to the acetyl mimic (K134Q) (Figure 1H, I, N). As infection progressed through 12 hpi (Figure 1J, K, O) and 18 hpi (Figure 1L, M, P, Supplementary Figure S2A), both WT and the charge mimic accumulated similar numbers of breaks, while the acetyl mimic remained stable and consistently displayed fewer breaks than WT or K134R. These analyses suggest that acetylation of LMNB1 inhibits breaks in the nuclear lamina that are otherwise induced during later stages of HSV-1 infection. Consistent with these conclusions, we observed no significant differences in either the overall laminar thickness or laminar area in the presence of the acetyl mimic (Supplementary Figure S2B, C), indicating that the acetyl mimic stabilizes the lamina but does not cause any additional accumulation of laminar proteins.
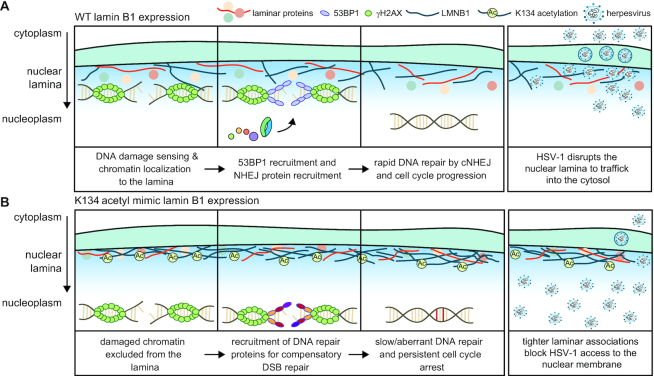
Together, these data lead us to a model in which LMNB1 K134 acetylation stabilizes the nuclear lamina, thereby inhibiting the ability of the viral capsid to egress from the nucleus (Figure 1Q). These findings show that LMNB1 acetylation can stabilize the nuclear lamina during distinct viral infections and on shorter time scales (<24 h) than previously observed (26). Given this finding, as well as our previous identification of this acetylation in uninfected cells and the knowledge that lamins have functional roles throughout the cell cycle (42, 43), we asked whether LMNB1 acetylation is poised to have a functional role on the time scales of cell cycle progression, thereby playing a broader role in regulating nuclear stability in uninfected cells. Further supporting this prediction is that a hallmark of herpesviruses infections is their ability to arrest the cell cycle in G1 phase so that viral replication can occur (29, 30, 44, 45).
Lamin B1 lysine 134 acetylation impacts the G1 to S phase cell cycle transition
Although we and others have detected numerous LMNB1 acetylation sites outside the context of infection, including K134, nothing is known about the timing and physiological relevance of these acetylations (26, 46–48). Given that acetyl-regulated sites are often conserved (49–51), as a first step to understand if there is a broader role for LMNB1 K134 as a regulator of a cellular process outside the context of viral infection, we assessed the evolutionary history of this site among 18 different species (Figure 2A, Supplementary Figure S3A). We found that the LMNB1 lysine 134 is conserved in mammalian and some avian species but is an unmodifiable positively charged arginine in frogs and fish. Thus, it appears that this lysine may have evolved more recently based on species phylogeny and could serve as a regulatory toggle in LMNB1 function. This idea is further supported when comparing the sequences of the three human lamins (LMNB1, LMNB2 and LMNA/C). While these three proteins share at least 40% amino acid sequence conservation, the site that corresponds with K134 is an arginine in both LMNB2 and LMNA/C (Figure 2B, Supplementary Figure S3B). This site-specificity of K134 in LMNB1 further suggests that it may have a regulatory function. As we observed an increase in K134 acetylation starting at early stages of HCMV infection (26) that aligns with the timing of virally-induced cell cycle regulation, we asked if LMNB1 acetylation affects cell cycle progression in uninfected cells.
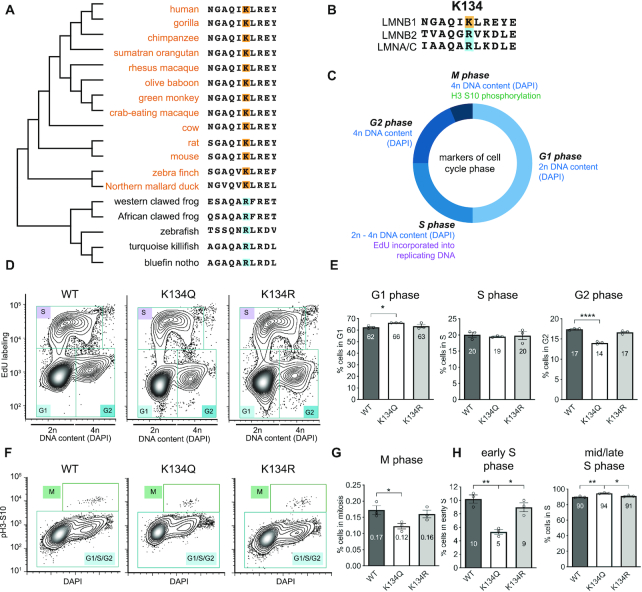
Figure 2.
LMNB1 K134 is highly conserved and K134 acetyl mimic impedes the G1 to S cell cycle transition. (A) LMNB1 K134 conservation among 18 species arranged by phylogenetic similarity. Lysines are highlighted in orange and arginines are highlighted in blue. (B) LMNB1 K134 conservation with the other human lamins (LMNA/C and LMNB2). Lysines are highlighted in orange and arginines are highlighted in blue. (C) Cell cycle phase was determined by flow cytometry using the combination of three distinct markers (DAPI, EdU incorporation and histone 3 serine 10 phosphorylation). (D, F) To determine which stage of the cell cycle LMNB1 K134 acetylation may affect, cell cycle analysis was performed by flow cytometry of MRC5 cells stably expressing WT, K134Q or K134R mCherry-LMNB1. Representative contour plots are shown with each cell cycle stage indicated. (E, G) Average percentage of cells ± SEM in each phase of the cell cycle. (E) left: G1 phase, middle: S phase, right: G2 phase (G) M phase. (H) Average percentage of cells ± SEM in (left) early S phase and (right) mid/late S phase, n = 3 biological replicates, one-way ANOVA, Bonferroni post-hoc test, average is indicated in each bar.
To test if LMNB1 K134 acetylation impacted cell growth, we established stable fibroblast cell lines expressing mCherry-LMNB1 WT, acetyl mimic (K134Q) or charge mimic (K134R), confirmed similar expression of the mCherry-LMNB1 constructs in these cell lines (Supplementary Figure S3C–E), and measured the cell cycle profile of each of these cell lines by flow cytometry (Supplementary Figure S4A). To distinguish G1 and G2 phases, we stained DNA with DAPI (Figure 2C). S phase cells were measured via incorporation of a fluorophore-taggable nucleotide analogue, EdU, to mark DNA replication. Finally, mitotic cells were quantified by histone 3 phosphorylation at Ser10 (Figure 2C), which labels mitotic entry (52). We observed that expression of acetyl mimic LMNB1 caused cells to accumulate in G1 phase and concomitantly decreased the proportion of cells in G2 and M phases relative to WT LMNB1 expression (Figure 2D–G, Supplementary Figure S4B, D). Expression of the charge mimic restores the percentages of cells in these phases to those in WT cells (Figure 2D–G). Because cell cycle progression is a cyclical process with entry into S phase as the rate limiting step, an increase in G1 levels coupled to a decrease in mitotic entry suggests that there is a defect in S phase entry. We did not observe a gross defect in S phase levels (Figure 2D, E); however, subdividing S phase into early S phase and mid/late S phase revealed a sharp decrease in S phase entry, but not S phase progression (Figure 2H, Supplementary Figure S4C). These sub-divided S phase data suggest that acetylation of LMNB1 slows the transition from G1 to S phase, which is regulated by the G1/S DNA damage checkpoint. Therefore, we predict that LMNB1 acetylation may impact the assessment or resolution of the G1/S checkpoint in uninfected cells.
LMNB1 acetyl mimic induces persistent activation of the G1/S DNA damage checkpoint
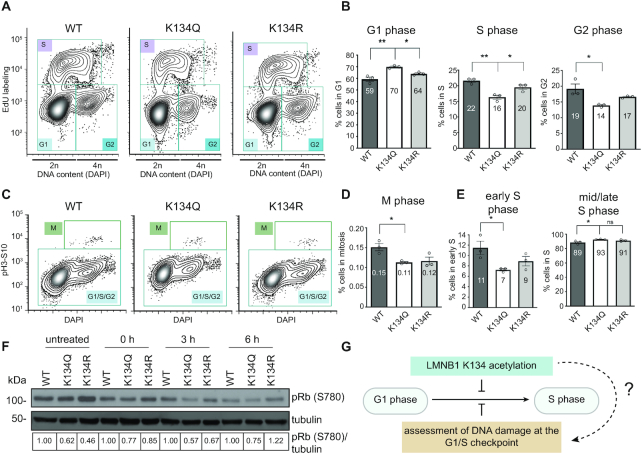
Based on our findings that cells expressing the LMNB1 K134 acetyl mimic accumulate in G1 phase of the cell cycle and enter S phase at a slower rate, we hypothesized that this acetylation may cause activation of the G1/S cell cycle checkpoint. An important trigger for activating this checkpoint is the presence of DNA damage in the cell. DNA damage, such as DSBs, occurs at a low level in any population of cells and must be repaired to license cell cycle progression (53, 54). To determine if K134 acetylation regulates the G1/S checkpoint through assessment of DNA damage, we asked if stimulating the DNA damage checkpoint exacerbated the cell cycle defect that we observed in homeostatic cells. Accordingly, we performed cell cycle profiling after inducing DNA damage in WT, K134Q and K134R mCherry-LMNB1 cells by either an optimized concentration of bleomycin (specific induction of DSBs) (Supplementary Figure S4H) (55) or hydrogen peroxide (nonspecific induction of DSBs). Supporting that LMNB1 K134 acetylation impacts the G1/S DNA damage checkpoint, we found that LMNB1 acetyl mimic expressing cells accumulate in G1 phase following both bleomycin treatment (Figure 3A–D, Supplementary Figure S4E, G) and treatment with both low (35 μM) and high (200 μM) doses of hydrogen peroxide treatment; we observed concomitant decreases in the percentage of acetyl mimic cells in S, G2 and M phases (Figure 3A–D, Supplementary Figure S4I–N). As expected, the percentages of LMNB1 charge mimic cells in each of these phases were similar to WT LMNB1 levels (Figure 3A–D, Supplementary Figure S4J–N). Upon bleomycin treatment, we also observed a decrease in the percentage of K134Q cells in early S phase (Figure 3E, Supplementary Figure S4F). To test the activation of the G1/S checkpoint, we performed western blot analysis for phosphorylation of pRb at S780 (Figure 3F). It is well-established that pRb S780 phosphorylation by cyclin D-CDK4 is required for cells to progress through the G1/S checkpoint as this phosphorylation inactivates pRb, freeing E2F transcription factors to induce expression of genes needed to enter S phase (56, 57). Over a time course of recovery from bleomycin treatment, we observed that the acetyl mimic cells consistently expressed less phosphorylated pRb (pRb S780) than the WT cells, indicating that less pRb is inactivated in these cells. In contrast, the levels of total pRb remained unchanged, as assessed by targeted mass spectrometry (Supplementary Figure S4O). This observation confirms our flow cytometry data that the expression of the acetyl mimic induces an accumulation of cells in G1 phase due to activation of the G1/S checkpoint in response to DNA damage (Figure 3G).
Figure 3.
K134 acetyl mimic expression reduces G1/S checkpoint resolution. (A, C) To determine which stage of the cell cycle LMNB1 K134 acetylation may affect in the presence of DSBs, cell cycle analysis was performed by flow cytometry of MRC5 cells stably expressing WT, K134Q or K134R mCherry-LMNB1 that were treated with bleomycin to induce DSBs. Representative contour plots are shown with each cell cycle stage indicated. (B, D) Average percentage of cells ± SEM in each phase of the cell cycle. (B) Left: G1 phase, middle: S phase, right: G2 phase (D) M phase. (E) Average percentage of cells ± SEM in (left) early S phase and (right) mid/late S phase, n = 3 biological replicates, one-way ANOVA, Bonferroni post-hoc test, average is indicated in each bar. (F) Representative western blot showing pRb (S780) phosphorylation levels in cells stably expressing WT, K134Q or K134R mCherry-LMNB1 across a time course of recovery from bleomycin treatment. (G) Proposed hypothesis that LMNB1 acetylation may affect progression through the G1/S checkpoint. As the G1/S checkpoint is regulated by assessing DNA damage, we propose that LMNB1 K134 acetylation triggers a stall in this checkpoint.
LMNB1 acetyl mimic reduces laminar association with chromatin
Because S phase entry was delayed in cells expressing acetyl mimic LMNB1 after experiencing DNA damage, we next sought to understand how LMNB1 acetylation affects DNA repair. A key factor regulating the response to DNA damage at the G1/S checkpoint is the recruitment of proteins to resolve DNA damage. In particular, certain DNA repair processes occur through associations with the nuclear lamina. To investigate LMNB1 interacting proteins following DNA damage, we performed proximity labeling (miniTurbo) (58) followed by mass spectrometry. We generated fibroblast cell lines stably expressing WT, acetyl mimic (K134Q) and charge mimic (K134R) LMNB1 fused to an engineered and promiscuous version of the biotin ligase BirA that can label nearby proteins (10–15 nm) after the addition of exogenous biotin. After confirming the correct localization of the tagged protein to the nuclear periphery and that the promiscuous biotin ligase was active (Supplementary Figure S5A, B), we treated the cells with bleomycin to induce DNA damage and then with biotin for proximity labeling (Figure 4A). Streptavidin immunoaffinity purification (IP) followed by mass spectrometry identified 110 nuclear proteins, including the previously established LMNB1 interactors LMNA, LMNB2, LBR and H2AFY (19) (Supplementary Figure S5C and Supplementary Table S1).
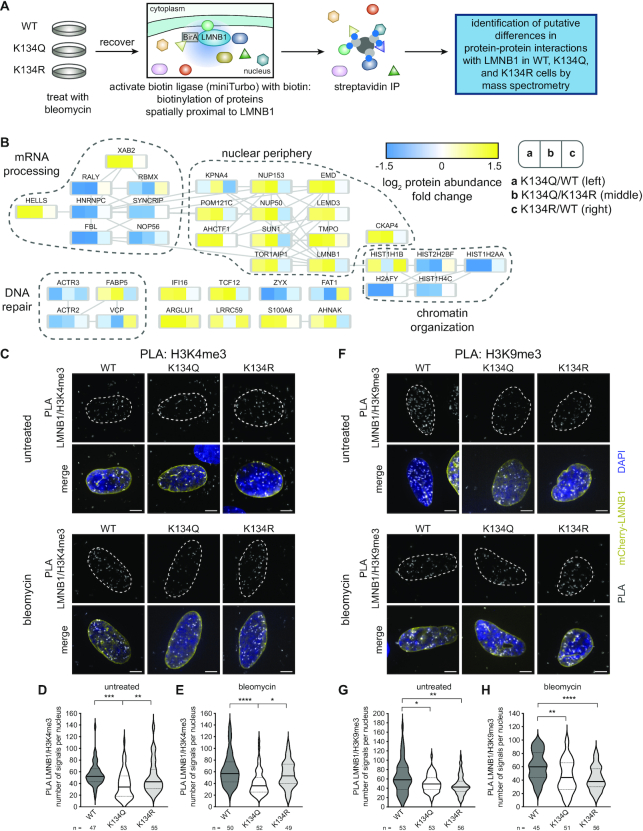
Figure 4.
K134 acetyl mimic expression alters LMNB1 protein associations and decreases association with chromatin. (A) Workflow for miniTurbo followed by mass spectrometry identification of changes in protein-protein associations with WT, K134Q or K134R LMNB1 during DNA repair after DNA damage induced by bleomycin treatment. (B) Nuclear proteins that exhibited a two-fold change in at least one comparison (K134Q/WT, K134Q/K134R or K134R/WT). Increased abundance is shown in yellow, while decreased abundance is shown in blue. (C, F) Representative IF images of (C) LMNB1-H3K4me3 (euchromatin marker) and (F) LMNB1-H3K9me3 (heterochromatin marker) proximity ligation assays (PLA) before (upper) or after (lower) bleomycin treatment. In the merged image, mCherry-LMNB1 (yellow), PLA (grey), DAPI (blue), scale bar represents 5 μm. (D, E, G, H) Quantification of the number of PLA puncta present (D, G) before or (E, H) after bleomycin treatment. Violin plot showing median (solid line) and interquartile range (doted lines), one-way ANOVA, Tukey post-hoc test, n (number of cells) is indicated in the figure.
To identify LMNB1 interactions that were disrupted by K134 acetylation, we focused on the nuclear proteins that had at least a two-fold change in abundance in the acetyl mimic compared to either the WT or the charge mimic (Figure 4B). In line with our previous analysis that LMNB1 acetylation stabilizes the nuclear lamina, we observed a 2–3-fold increased abundance of nuclear periphery proteins, including emerin (EMD), TMPO, LEMD3 and SUN1, in the acetyl mimic IP compared to either the WT or charge mimic IP. We also found a 2–5-fold decrease in four histone proteins in the K134 acetyl mimic IP compared to the WT or charge mimic LMNB1 IPs, suggesting that LMNB1 acetylation induces a defect in chromatin localization to the nuclear periphery after DNA damage, which is a necessary step in certain DNA repair pathways (9). To test the effect of the expression of the acetyl mimic on the association between LMNB1 and chromatin, we performed a proximity ligation assay (PLA) between LMNB1 and heterochromatin (H3K9me3) or euchromatin (H3K4me3) marks in both untreated and bleomycin treated conditions (Figure 4C–H, Supplementary Figure S5D). We observed a significant reduction in the number of PLA signals in the presence of the LMNB1 acetyl mimic for both euchromatin (Figure 4C–E) and heterochromatin (Figure 4F–H) in both untreated and DNA-damage-induced conditions. To assess whether this decrease in association with chromatin impacts gene expression, we labelled actively transcribed RNA with EU, and did not observe any effect on global transcription rates (Supplementary Figure S5E,F). These observations align with our miniTurbo data to indicate that LMNB1 acetylation reduces the association between the nuclear periphery and chromatin. This acetylation-driven decrease in LMNB1 proximal histones is likely due to defective chromatin tethering, which is an essential step in cNHEJ (9). Corroborating this hypothesis, we further found decreased association with a cluster of proteins that are important for DNA repair. Together, our findings that LMNB1 K134 acetylation both delays S phase entry and decreases the association of chromatin with and the recruitment of DNA repair proteins to the nuclear periphery suggest that acetylation at this residue may stall the G1/S checkpoint by impacting the assessment or repair of DNA damage.
LMNB1 acetyl mimic delays the repair, but not recognition, of DNA damage
To determine if delayed S phase entry and decreased levels of DNA repair proteins at the nuclear periphery in LMNB1 acetyl mimic expressing cells were the result of defective DNA repair, we measured recovery following DNA damage. Specifically, we performed a comet assay for untreated cells and for cells 3 h post-bleomycin treatment. As expected, we observed an increase in percent tail DNA, an indication of the extent of DNA damage, in treated cells in comparison to untreated cells (Figure 5A, Supplementary Figure S6A). Notably, we observed an elevated percentage of tail DNA in the presence of the LMNB1 acetyl mimic compared to either the WT or the charge mimic at 3 h post-treatment, indicating that DNA damage is persisting in the presence of the acetyl mimic and that DNA repair is delayed (Figure 5A, Supplementary Figure S6A). In agreement with this observation, we noticed that a lower percentage of acetyl mimic comets had no tails (no damage) than either the WT or the charge mimic at 3 hpt (Supplementary Figure S6B).
Figure 5.
LMNB1 acetyl mimic impedes DNA repair and dissipation of γH2AX DNA damage foci and impedes cNHEJ. (A) Assessment of recovery from DNA damage by alkaline comet assay. Average ±95% confidence interval. Total number of comets analyzed (n, combined from two biological replicates) is indicated. Kruskal–Wallis test was used to determine significance. Representative IF images of different amounts of DNA damage (% tail DNA) are shown. Scale bar represents 10 μm. (B) Workflow for experimental setup. Untreated cells were collected prior to the start of the treatment. All other samples were treated with bleomycin for 1 h to induce DSBs. Afterward, the media was replaced and the 0 h post-treatment (hpt) sample was collected. Samples were collected every hour for 6 hpt. (C) Representative IF images of γH2AX foci (green) in MRC5 cells stably expressing WT, K134Q or K134R mCherry-LMNB1 (magenta) at the indicated time points post-bleomycin treatment. Scale bar represents 5 μm. (D) Average γH2AX fluorescence intensity ± SEM at each time point post-bleomycin treatment. The number of cells (n) is indicated, one-way ANOVA, Bonferroni post-hoc test. (E) Left: Average γH2AX fluorescence intensity ± SEM (shaded) over the course of recovery from bleomycin treatment. Right: Average number of γH2AX foci ± SEM (shaded) over the course of recovery from bleomycin treatment. (F) Left: Schematic of EJ7 reporter strategy for assessing cNHEJ activity. Cells are transfected with CRISPR gRNAs and Cas9 that induce blunt end double strand breaks on either side of a spacer inserted at the coding sequence for G67 in GFP. If repaired through cNHEJ, the coding sequence of GFP is repaired and the cells will fluoresce green. Right: Average percentage of mCherry-LMNB1 WT, K134Q or K134R expressing cells that have successfully repaired ± SEM, n = 3 biological replicates, two-sided Student's t-test. (G) Left: Schematic of EJ5 reporter strategy for assessing all end joining (EJ). Cells are transfected with a plasmid for the I-SceI rare-cutting endonuclease. If repaired through EJ, the promoter and the coding region of GFP will be in frame and the cells will fluoresce green. Right: Average percentage of mCherry-LMNB1 WT, K134Q or K134R expressing cells that have successfully repaired ± SEM, n = 3 biological replicates, two-sided Student's t-test. (H) Schematic depicting some examples of how the I-SceI DSBs in the EJ5 reporter cells may be repaired. These include full repair, trimming of the sticky ends (indicated by blue and magenta) or end resection further into the I-SceI region beyond the sticky ends. (I) Quantification of the percentage of DSBs that were repaired by various types of repair in the mCherry-LMNB1 WT, K134Q and K134R EJ5 stable cell lines. (J) Relative abundance of the endogenous LMNB1 K134 acetylation in cells synchronized in G2 phase (CDK1i) compared to unsynchronized (untreated) cells. Mean ± SD.
To assess if this persistence in DNA damage was due to a delay in the recognition or the resolution of DNA damage, we measured histone H2AX phosphorylation kinetics (γH2AX, marker of DNA damage). We treated fibroblasts expressing WT, acetyl mimic or charge mimic mCherry-LMNB1 with bleomycin for 1 h, then measured recovery from and repair of DSBs in 1 h increments over a span of 6 h (Figure 5B). As expected, bleomycin treatment induced similar accumulation of punctate γH2AX after just 1 h of exposure in all three cell lines (Figure 5C, D), indicating that this early step of the DNA repair signaling pathway is not impaired by LMNB1 K134 acetylation. The media was changed after the initial 1 h bleomycin treatment, causing γH2AX levels to gradually decrease in both LMNB1 WT and K134R expressing cells as DNA repair occurred (Figure 5C, D). However, γH2AX levels and numbers of foci were consistently elevated in fibroblasts expressing K134Q, indicating that DNA repair was delayed (Figure 5C–E, Supplementary Figure S6C). We reproduced these findings with regard to the acetyl mimic in another cell type, U2OS (Supplementary Figure S6D–F) and in fibroblasts expressing CRISPR-resistant mCherry-LMNB1 WT and K134Q in an endogenous LMNB1 knockout background (Supplementary Figure S7A, D).
Double stranded DNA break repair is necessary for genomic stability because this form of DNA damage can cause chromosomal rearrangements that lead to the loss of genetic information. Given that expression of the acetyl mimic LMNB1 delayed both cell cycle progression through the G1/S checkpoint and recovery from DNA damage (delayed γH2AX dissipation), we predicted that LMNB1 K134 acetylation may affect DNA DSB repair. As cNHEJ is the predominant form of DSB repair that occurs during G1 phase of the cell cycle, we hypothesized that cNHEJ is impaired by LMNB1 K134 acetylation. To test this hypothesis, we employed U2OS-based DNA repair reporter lines (32, 33) that express GFP when a targeted DSB is repaired by end joining (EJ) (Figure 5F, G). Briefly, we co-transfected the required plasmids to induce a targeted DSB in combination with WT, acetyl mimic or charge mimic LMNB1 expression plasmids and measured the relative rate of DNA repair (percentage of GFP positive cells) in LMNB1 expressing cells (mCherry positive). We found that cNHEJ, measured specifically by the EJ7 reporter line, was significantly reduced in the presence of the acetyl mimic compared to WT and charge mimic LMNB1 (Figure 5F, Supplementary Figure S7B). These findings agree with our observation that expression of K134Q also results in decreased association of LMNB1 with proximal histones (Figure 4B), as chromatin proximity to the nuclear lamina promotes cNHEJ (9). Importantly, our data show that LMNB1 acetylation specifically impedes cNHEJ, and not DNA repair in general, since γH2AX levels eventually did decrease (Figure 5C-E). Furthermore, we found that total EJ was not decreased by expression of the LMNB1 acetyl mimic in the EJ5 reporter cell line, which measures total repair by both cNHEJ and its compensatory pathway, alternative EJ (alt-EJ) (Figure 5G, Supplementary Figure S7C). These data suggest that the presence of the acetyl mimic may bias the repair towards alt-EJ.
To test this hypothesis, we generated stable cells expressing WT or mutant LMNB1 in U2OS EJ5 cells that lack endogenous LMNB1. We accomplished this by generating CRISPR-resistant LMNB1 cell lines expressing mCherry-tagged WT, K134Q or K134R LMNB1. We then optimized and implemented a recombinant CRISPR/Cas9-system to KO endogenous LMNB1 (Supplementary Figure S7A). Next, we induced DSBs with the restriction enzyme I-SceI and employed flow cytometry to sort the double positive (mCherry and GFP) cells into single cell colonies. We expanded these colonies and performed PCR followed by sequencing to determine how the DSBs induced by I-SceI were repaired (Figure 5H). We determined that some of the clones from the WT and the K134R cells (25% and 46%, respectively) had faithful repair of the I-SceI site, while none of the K134Q clones had faithful repair (Figure 5H, I, Supplementary Figure S7E). Moreover, 92% of the breaks in the K134Q clones were repaired either by end resection or trimming of the broken ends, while a smaller percentage of the breaks in the WT and K134R clones (58% and 38%, respectively) were repaired in this manner that results in a loss of genetic information. These data show that expression of the acetyl mimic biases DNA repair towards alt-EJ, especially since the charge mimic (which cannot be acetylated) had a higher proportion of clones that were repaired faithfully than the WT. Together, these data indicate that LMNB1 K134 acetylation impedes DNA repair through the highly efficient cNHEJ pathway and biases DNA repair towards hypermutagenic DNA repair pathways.
Given that we found that cNHEJ is negatively regulated by LMNB1 K134 acetylation and that cNHEJ is the predominant form of DNA repair that occurs in G1 phase, we predicted that increased K134 acetylation may be linked to duplication of the genome in S phase, at which point there may be a preference for DNA repair pathways other than cNHEJ, such as HR. Therefore, we expected to observe increased LMNB1 K134 acetylation after S phase entry. To enrich for cells that had entered S phase and duplicated their genome, we treated MRC5 cells with a CDK1 inhibitor, RO-3306, that prevents mitotic entry and enriches for cells in S and G2 phases (52). Using anti-acetyl-lysine immunoaffinity purification followed by mass spectrometry, we found an increased abundance of acetylated LMNB1 K134 after CDK1 inhibition (Figure 5J). Given that cNHEJ proteins are highly expressed throughout the cell cycle, these findings point to a potential cellular mechanism to regulate DNA repair pathway fate (NHEJ versus HR) via LMNB1 acetylation regulation based on cell cycle stage.
LMNB1 acetyl mimic delays recruitment of cNHEJ repair factor 53BP1 to DNA repair foci
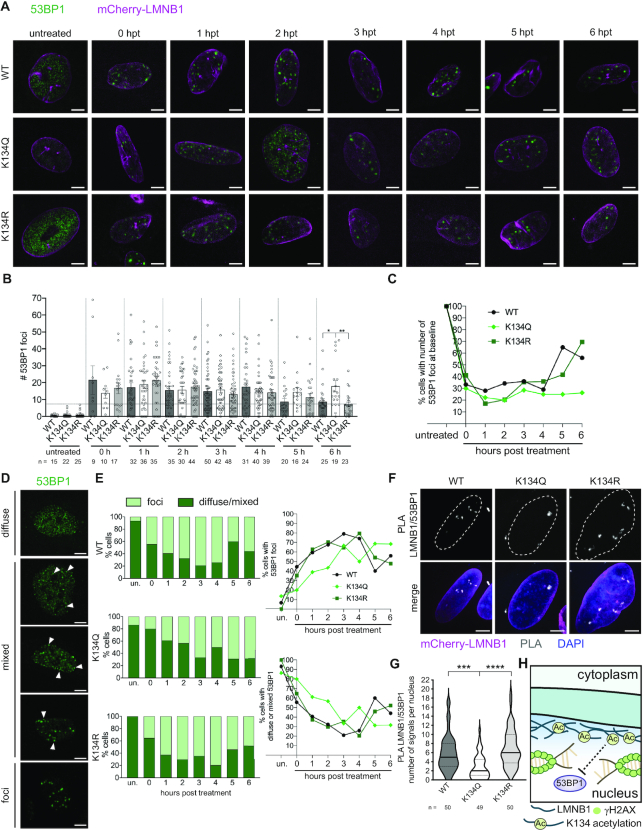
In order to understand how LMNB1 acetylation biased DNA repair against cNHEJ, we considered that K134Q expression was linked to increased end resection. A key step in committing a DSB to repair by the cNHEJ pathway is assembly of 53BP1 into discrete foci to suppress DNA end resection and promote DNA repair (59). To determine if LMNB1 acetylation leads to defective cNHEJ due to impaired 53BP1 recruitment, we treated WT, acetyl mimic and charge mimic mCherry-LMNB1 cells with bleomycin and assessed 53BP1 localization by microscopy. In untreated fibroblasts and U2OS cells, we found that 53BP1 rarely formed foci and diffusely stained the nucleoplasm (Figure 6A, B, Supplementary Figure S8A, B). Upon treatment with bleomycin to induce DNA damage, 53BP1 assembled into distinct foci in LMNB1 K134 WT and charge mimic cells (Figure 6A, B, Supplementary Figure S8A, B). These foci gradually resolved and returned to numbers found in untreated cells (baseline numbers) over a 6 h period after bleomycin was removed from the media (Figure 6C, Supplementary Figure S8C). 53BP1 foci also formed following bleomycin treatment in K134 acetyl mimic LMNB1 cells; however, these foci failed to resolve over a 6 h time course following bleomycin wash out.
Figure 6.
LMNB1 acetyl mimic delays 53BP1 foci formation after DNA damage. (A) Representative IF images of 53BP1 foci (green) in MRC5 cells stably expressing WT, K134Q or K134R mCherry-LMNB1 (magenta) at the indicated time points post-bleomycin treatment. Scale bar represents 5 μm. (B) Average number of 53BP1 foci ± SEM at each time point post-bleomycin treatment. The number of cells (n) is indicated, one-way ANOVA, Bonferroni post-hoc test. Baseline number of foci is indicated by the horizontal dashed line. (C) Average percentage of 53BP1 foci at or below baseline numbers (number of foci present in untreated cells) over the course of recovery from bleomycin treatment. (D) Representative IF images of the three 53BP1 phenotypes (diffuse, mixed, foci). Scale bar represents 5 μm. (E) (left) Percentage of WT, K134Q or K134R cells with distinct 53BP1 foci or diffuse/mixed 53BP1 over the course of recovery from bleomycin treatment (shown as percentage out of total). (right) Percentage of cells with (top) distinct 53BP1 foci or (bottom) diffuse/mixed 53BP1 over the course of recovery from bleomycin treatment. (F) Representative IF images of LMNB1–53BP1 proximity ligation assay 1 hpt after bleomycin treatment. Nuclei are outlined. In the merged image, mCherry-LMNB1 (magenta), PLA (grey), DAPI (blue), scale bar represents 5 μm. (G) Quantification of the number of PLA puncta. Violin plot showing median (solid line) and interquartile range (doted lines), one-way ANOVA, Tukey post-hoc test, n (number of cells) is indicated in the figure. (H) Proposed model for the effect of LMNB1 acetylation on impeding the recruitment of the NHEJ repair factor 53BP1 to sites of DSBs.
While analyzing the number of 53BP1 foci in cells expressing WT and K134 mutant LMNB1, we also observed a population of cells that failed to form discrete 53BP1 foci following bleomycin treatment (Figure 6D, E). While discrete 53BP1 foci formed rapidly in LMNB1 K134 WT and charge mimic cells, in cells expressing acetyl mimic LMNB1, 53BP1 aggregation into distinct DNA repair foci was delayed by several hours (Figure 6D, E). To directly assess the association between LMNB1 and 53BP1, we performed a PLA assay at 1 h post-bleomycin treatment (Figure 6F, G). We observed significantly less association between 53BP1 and acetyl mimic LMNB1 compared to either WT or charge mimic, while we saw no difference in 53BP1 association between the WT and the charge mimic. These data show that LMNB1 acetylation may affect DNA repair by impacting both the assembly of 53BP1 into DNA repair foci and the resolution of these foci after they have formed (Figure 6H). Altogether, these findings provide a mechanism linking LMNB1 acetylation to impaired cell cycle progression beyond the G1/S boundary as a result of slower, less effective cNHEJ due to delayed 53BP1 foci formation and resolution.
DISCUSSION
The nuclear lamina is a dense fibrillar network of proteins that regulate nuclear processes including chromatin remodeling, transcription, mitosis, DNA replication, and DNA repair (1–5). While we and others have shown that lamins A/C, B1 and B2 undergo extensive post-translational modification, only a few of these modifications have known functions (5, 22, 25). Our study defines previously uncharacterized functions for LMNB1 K134 acetylation. We demonstrate that LMNB1 K134 acetylation during herpesvirus infections stabilizes the nuclear lamina. By proximity labeling integrated with mass spectrometry, we show that this acetylation increased LMNB1 association with nuclear periphery proteins, in agreement with our findings that this modification conferred stability to the lamina during HSV-1 infection. Conversely, after DNA damage, laminar interactions with several histones and with proteins related to DNA repair were diminished in the presence of the acetyl mimic. Using microscopy and cell reporter assays, we found that LMNB1 K134 acetylation inhibited the repair of DSBs by the cNHEJ pathway, which caused γH2AX foci to be repaired more slowly and delayed recovery from the G1/S checkpoint. We further identified that cNHEJ was impaired at the 53BP1 recruitment step, which is an essential platform to recruit DNA repair proteins (60) and that acetylation of LMNB1 biases DNA repair towards alternative pathways that increased end resection. These findings lead us to propose a model in which LMNB1 K134 acetylation stabilizes the laminar network and inhibits association of nucleoplasmic factors, such as DNA repair enzymes and chromatin, with the lamina (Figure 7). We further propose that this same stabilization mechanism prevents DNA replicating viruses, such as HSV-1 or HCMV, from disrupting the nuclear lamina for viral capsid nuclear egress.
Figure 7.
Proposed model for how LMNB1 K134 acetylation can stabilize the nuclear lamina and act as a molecular toggle for DNA DSB repair at the nuclear periphery. (A) In the presence of WT LMNB1, the lamina is accessible and DNA damage repair factors can be recruited to the nuclear periphery. 53BP1 foci can activate the cNHEJ pathway for rapid DNA repair and subsequently, cell cycle progression. During herpesvirus infection, the nuclear lamina can be disrupted to facilitate viral capsid nuclear egress. (B) In cells expressing LMNB1 K134 acetyl mimic, intra-laminar associations are favored and the lamina is inaccessible to damaged chromatin. Because 53BP1 recruitment to damaged DNA is impaired, the G1/S checkpoint is persistently activated, and DNA repair may be slowly accomplished by compensatory pathways. During herpesvirus infection, the nuclear lamina is stabilized, impeding viral capsid nuclear egress.
In addition to exogenous sources of genotoxicity, all cells experience a high level of endogenous DNA damage caused by processes like metabolism, transcription and DNA replication. This DNA damage must be repaired to maintain genomic integrity. The DNA damage response has evolved in all living organisms to initiate DNA repair and to halt the cell cycle to prevent damaged, unrepaired DNA from being passed to daughter cells. The nuclear lamina has been previously implicated in cell cycle control (6, 7) and as an organizing space for DNA repair and repair pathway fate (9, 10). Moreover, defects in DNA repair have been linked to laminopathies (4). For example, silencing LMNB1 decreases the expression of DNA repair proteins (42, 61), knockout of 53BP1 delays cell and animal growth (62), and an interaction between 53BP1 and LMNA/C is essential for cNHEJ (63). Here, we expand on the current literature by showing that LMNB1 acetylation at K134 leads to activation of the G1/S checkpoint due to defective cNHEJ. To the best of our knowledge, no prior study has established a connection between LMNB1 PTMs or mutations, the formation of 53BP1 DNA repair foci and successful DNA repair by cNHEJ.
The nuclear lamins form distinct, but interacting networks (14, 15). Chromatin anchoring to the nuclear lamina is an essential step for productive cNHEJ, while chromatin mobility is necessary for the strand invasion step of HR (9, 64–66). Given the well-established link between LMNA/C and 53BP1 (63) and that silencing LMNB1 impedes HR, but not cNHEJ (42, 61), we propose that LMNB1 K134 acetylation status serves as a regulatory toggle to locally control laminar network accessibility, thus allowing for the precise spatial organization of nuclear processes (Figure 7). Supporting this model, our miniTurbo analysis revealed that, following DNA damage, the K134 acetyl mimic increased LMNB1 association with nuclear periphery proteins but reduced association of LMNB1 with several histone proteins. We also observed a decreased association with transitional endoplasmic reticulum ATPase (VCP), a protein which has been implicated in the recruitment of 53BP1 to sites of DSBs (67, 68) and, through our PLA analysis, a decreased association with 53BP1 itself. Therefore, LMNB1 acetylation represents a possible mechanism to control DNA repair pathway choice in S phase to promote error-free HR over NHEJ despite both pathways being active in S phase (60, 69). Because we found that LMNB1 K134 acetylation is elevated by CDK1 inhibition, which synchronizes cells that have 4N DNA content, we expect that K134 acetylation is linked to S phase entry. This is further supported by the knowledge that herpesviruses replicate in cells that are in a G1/S cell cycle transition where cellular replication proteins are expressed but host DNA replication is blocked (70, 71). We have seen that LMNB1 acetylation increases throughout HCMV infection (26), which may be a consequence of this infection induced pseudo-G1/S environment. During DNA damage, our data show that LMNB1 acetylation impedes DNA repair, which is linked to slower S phase entry. By contrast, herpesvirus infection bypasses the G1/S DNA damage checkpoint (70). Therefore, we expect that the defect in herpesvirus replication is due to a broader mechanism that regulates the accessibility of the laminar microenvironment to host and viral proteins (Figure 7). Moreover, it is interesting to conjecture that the presence of this regulatory lysine may have evolved, in part, due to selective pressure from mammalian herpesviruses, which coincides with the diversification of the Alloherpesviridae (amphibians and fish) and Herpesviridae (birds and mammals) families (72, 73).
The model that LMNB1 acetylation could alter the homeostatic nuclear periphery landscape may point to a mechanism to regulate nuclear processes that require tethering to or interaction with other laminar proteins and may shed light on molecular decisions that influence DSB repair pathway choice, as discussed above. The observations that several deacetylases localize to DNA damage foci and are required for repair by cNHEJ (74, 75) further support a model in which laminar deacetylation facilitates chromatin tethering and provide a mechanism to regulate LMNB1 acetylation. While no known LMNB1 deacetylases have been identified, it has been established that inhibition of all HDACs except for class III HDACs (sirtuins) has no effect on LMNB1 acetylation (76). Consequently, it is likely that one of the nuclear class III HDACs (sirtuins 1 and 6) deacetylates LMNB1, and previous work has shown that sirtuin 6 localizes to the nuclear periphery (77). Furthermore, analysis of the entire p300/CBP acetylome uncovered a modest effect of p300 inhibition or overexpression on LMNB1 K134 acetylation levels, hinting that this may be the acetyltransferase responsible for this modification (46). Future work to define the temporality of LMNB1 K134 acetylation and which acetyltransferases and deacetylases regulate this modification will allow us to better understand the role of this modification as a molecular toggle of the balance between laminar integrity and accessibility that helps to regulate nuclear periphery processes. As it is known that LMNB1 can be dynamically modified by multiple PTMs (22–24), including ubiquitination at K134 (78), further studies to address how crosstalk among these PTMs impacts laminar function are needed, especially since alterations to nuclear laminar structure impacting downstream laminar-associated events, such as DNA repair, are key factors in laminopathies (4). Our work here underscores a regulatory role for LMNB1 acetylation that contributes to laminar stability and DNA repair, processes affected by laminopathies and viral infections.
DATA AVAILABILITY
The mass spectrometry miniTurbo and acetylome proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD022974. The flow cytometry data have been deposited to the FlowRepository with the repository ID: FR-FCM-Z2XN.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jeremy Stark (City of Hope) for the gift of the DNA repair reporter cells. At Princeton University, we thank Gary Laevsky and Claudia Kaestner from the Princeton Confocal Imaging Core, Tina DeCoste and Katherine Rittenbach from the Flow Cytometry Resource Facility and Jessica Wiggins from the Genomics Core Facility.
Contributor Information
Laura A Murray-Nerger, Department of Molecular Biology, Princeton University, Lewis Thomas Laboratory, Washington Road, Princeton, NJ 08544, USA.
Joshua L Justice, Department of Molecular Biology, Princeton University, Lewis Thomas Laboratory, Washington Road, Princeton, NJ 08544, USA.
Pranav Rekapalli, Department of Molecular Biology, Princeton University, Lewis Thomas Laboratory, Washington Road, Princeton, NJ 08544, USA.
Josiah E Hutton, Department of Molecular Biology, Princeton University, Lewis Thomas Laboratory, Washington Road, Princeton, NJ 08544, USA.
Ileana M Cristea, Department of Molecular Biology, Princeton University, Lewis Thomas Laboratory, Washington Road, Princeton, NJ 08544, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health NIGMS [GM114141 to I.M.C.]; Edward Mallinckrodt Foundation [to I.M.C]; National Science Foundation Graduate Research Fellowship [NSF-GRFP DGE-1656466 to L.A.M.N.]; National Institutes of Health NIGMS [T32GM007388]. Funding for open access charge: NIGMS [GM114141]; Mallinckrodt Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1. Pascual-Reguant L., Blanco E., Galan S., Le Dily F., Cuartero Y., Serra-Bardenys G., Di Carlo V., Iturbide A., Cebrià-Costa J.P., Nonell L.et al.. Lamin B1 mapping reveals the existence of dynamic and functional euchromatin lamin B1 domains. Nat. Commun. 2018; 9:3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gesson K., Rescheneder P., Skoruppa M.P., Von Haeseler A., Dechat T., Foisner R.. A-type Lamins bind both hetero- and euchromatin, the latter being regulated by lamina-associated polypeptide 2 alpha. Genome Res. 2016; 26:462–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burke B., Stewart C.L.. The nuclear lamins: flexibility in function. Nat. Rev. Mol. Cell Biol. 2013; 14:13–24. [DOI] [PubMed] [Google Scholar]
- 4. Broers J.L.V., Ramaekers F.C.S., Bonne G., Ben Yaou R., Hutchison C.J.. Nuclear lamins: laminopathies and their role in premature ageing. Physiol. Rev. 2006; 86:967–1008. [DOI] [PubMed] [Google Scholar]
- 5. Dechat T., Pfleghaar K., Sengupta K., Shimi T., Shumaker D.K., Solimando L., Goldman R.D.. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008; 22:832–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Peter M., Nakagawa J., Doree M., Labbe J., Nigg E.. In vitro disassembly of the nuclear lamina and M phase-specific phosphorylation of lamins by cdc2 kinase. Cell. 1990; 61:591–602. [DOI] [PubMed] [Google Scholar]
- 7. Heald R., McKeon F.. Mutation of phosphorylation sites in lamin A that prevent nuclear lamina dissembly in mitosis. Cell. 1990; 61:579–589. [DOI] [PubMed] [Google Scholar]
- 8. Peter M., Heitlinger E., Haner M., Aebi U., Nigg E.A.. Disassembly of in vitro formed lamin head-to-tail polymers by CDC2 kinase. EMBO J. 1991; 10:1535–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lemaître C., Grabarz A., Tsouroula K., Andronov L., Furst A., Pankotai T., Heyer V., Rogier M., Attwood K.M., Kessler P.et al.. Nuclear position dictates DNA repair pathway choice. Genes Dev. 2014; 28:2450–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang K., Guo R., Xu D.. Non-homologous end joining: advances and frontiers. Acta Biochim. Biophys. Sin. (Shanghai). 2016; 48:632–640. [DOI] [PubMed] [Google Scholar]
- 11. Harada T., Swift J., Irianto J., Shin J.W., Spinler K.R., Athirasala A., Diegmiller R., Dingal P.C.D.P., Ivanovska I.L., Discher D.E.. Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. J. Cell Biol. 2014; 204:669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Irianto J., Xia Y., Pfeifer C.R., Athirasala A., Ji J., Alvey C., Tewari M., Bennett R.R., Harding S.M., Liu A.J.et al.. DNA damage follows repair factor depletion and portends genome variation in cancer cells after pore migration. Curr. Biol. 2017; 27:210–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ungricht R., Kutay U.. Mechanisms and functions of nuclear envelope remodelling. Nat. Rev. Mol. Cell Biol. 2017; 18:229–245. [DOI] [PubMed] [Google Scholar]
- 14. Nmezi B., Xu J., Fu R., Armiger T.J., Rodriguez-Bey G., Powell J.S., Ma H., Sullivan M., Tu Y., Chen N.Y.et al.. Concentric organization of A- and B-type lamins predicts their distinct roles in the spatial organization and stability of the nuclear lamina. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:4307–4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shimi T., Pfleghaar K., Kojima S.I., Pack C.G., Solovei I., Goldman A.E., Adam S.A., Shumaker D.K., Kinjo M., Cremer T.et al.. The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev. 2008; 22:3409–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sullivan T., Escalante-Alcalde D., Bhatt H., Anver M., Bhat N., Nagashima K., Stewart C.L., Burke B.. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 1999; 147:913–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen N.Y., Kim P., Weston T.A., Edillo L., Tu Y., Fong L.G., Young S.G.. Fibroblasts lacking nuclear lamins do not have nuclear blebs or protrusions but nevertheless have frequent nuclear membrane ruptures. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:10100–10105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guelen L., Pagie L., Brasset E., Meuleman W., Faza M.B., Talhout W., Eussen B.H., De Klein A., Wessels L., De Laat W.et al.. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008; 453:948–951. [DOI] [PubMed] [Google Scholar]
- 19. Fu Y., Lv P., Yan G., Fan H., Cheng L., Zhang F., Dang Y., Wu H., Wen B.. MacroH2A1 associates with nuclear lamina and maintains chromatin architecture in mouse liver cells. Sci. Rep. 2015; 5:17186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roux K.J., Kim D.I., Raida M., Burke B.. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012; 196:801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pickersgill H., Kalverda B., De Wit E., Talhout W., Fornerod M., Van Steensel B.. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat. Genet. 2006; 38:1005–1014. [DOI] [PubMed] [Google Scholar]
- 22. Simon D.N., Wilson K.L.. Partners and post-translational modifications of nuclear lamins. Chromosoma. 2013; 122:13–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Farnsworth C.C., Wolda S.L., Gelb M.H., Glomset J.A.. Human Lamin B contains a farnesylated cysteine residue. J. Biol. Chem. 1989; 264:20422–20429. [PMC free article] [PubMed] [Google Scholar]
- 24. Jung H.J., Nobumori C., Goulbourne C.N., Tu Y., Lee J.M., Tatar A., Wu D., Yoshinaga Y., De Jong P.J., Coffinier C.et al.. Farnesylation of lamin B1 is important for retention of nuclear chromatin during neuronal migration. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:1923–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karoutas A., Szymanski W., Rausch T., Guhathakurta S., Rog-Zielinska E.A., Peyronnet R., Seyfferth J., Chen H.R., de Leeuw R., Herquel B.et al.. The NSL complex maintains nuclear architecture stability via lamin A/C acetylation. Nat. Cell Biol. 2019; 21:1248–1260. [DOI] [PubMed] [Google Scholar]
- 26. Murray L.A., Sheng X., Cristea I.M.. Orchestration of protein acetylation as a toggle for cellular defense and virus replication. Nat. Commun. 2018; 9:4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rawlinson W.D., Boppana S.B., Fowler K.B., Kimberlin D.W., Lazzarotto T., Alain S., Daly K., Doutré S., Gibson L., Giles M.L.et al.. Congenital cytomegalovirus infection in pregnancy and the neonate: consensus recommendations for prevention, diagnosis, and therapy. Lancet Infect. Dis. 2017; 17:e177–e188. [DOI] [PubMed] [Google Scholar]
- 28. Freeman R.B. The ‘Indirect’ effects of cytomegalovirus infection: minireview. Am. J. Transplant. 2009; 9:2453–2458. [DOI] [PubMed] [Google Scholar]
- 29. Dittmer D., Mocarski E.S.. Human cytomegalovirus infection inhibits G1/S transition. J. Virol. 1997; 71:1629–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bresnahan W.A., Boldogh I., Thompson E.A., Albrecht T.. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology. 1996; 224:150–160. [DOI] [PubMed] [Google Scholar]
- 31. Flemington E.K. Herpesvirus lytic replication and the cell cycle: arresting new developments. J. Virol. 2001; 75:4475–4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bennardo N., Cheng A., Huang N., Stark J.M.. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008; 4:e1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bhargava R., Sandhu M., Muk S., Lee G., Vaidehi N., Stark J.M.. C-NHEJ without indels is robust and requires synergistic function of distinct XLF domains. Nat. Commun. 2018; 9:2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lum K.K., Song B., Federspiel J.D., Diner B.A., Howard T., Cristea I.M.. Interactome and proteome dynamics uncover immune modulatory associations of the pathogen sensing factor cGAS. Cell Syst. 2018; 7:627–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. MacLean B., Tomazela D.M., Shulman N., Chambers M., Finney G.L., Frewen B., Kern R., Tabb D.L., Liebler D.C., MacCoss M.J.. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010; 26:966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Murray L.A., Combs A.N., Rekapalli P., Cristea I.M.. Methods for characterizing protein acetylation during viral infection. Methods Enzym. 2019; 626:587–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jean Beltran P.M., Mathias R.A., Cristea I.M.. A portrait of the human organelle proteome in space and time during cytomegalovirus infection. Cell Syst. 2016; 3:361–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Buchkovich N.J., Maguire T.G., Alwine J.C.. Role of the Endoplasmic Reticulum chaperone BiP, SUN domain proteins, and dynein in altering nuclear morphology during human cytomegalovirus infection. J. Virol. 2010; 84:7005–7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Milbradt J., Webel R., Auerochs S., Sticht H., Marschall M.. Novel mode of phosphorylation-triggered reorganization of the nuclear lamina during nuclear egress of human cytomegalovirus. J. Biol. Chem. 2010; 285:13979–13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Reynolds A.E., Liang L., Baines J.D.. Conformational changes in the nuclear lamina induced by herpes simplex virus type 1 require genes UL31 and UL34. J. Virol. 2004; 78:5564–5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hamirally S., Kamil J.P., Ndassa-Colday Y.M., Lin A.J., Jahng W.J., Baek M.C., Noton S., Silva L.A., Simpson-Holley M., Knipe D.M.et al.. Viral mimicry of Cdc2/cyclin-dependent kinase 1 mediates disruption of nuclear lamina during human cytomegalovirus nuclear egress. PLoS Pathog. 2009; 5:e1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Butin-Israeli V., Adam S.A., Jain N., Otte G.L., Neems D., Wiesmüller L., Berger S.L., Goldman R.D.. Role of lamin B1 in chromatin instability. Mol. Cell. Biol. 2015; 35:884–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moir R.D., Spann T.P., Lopez-Soler R.I., Yoon M., Goldman A.E., Khuon S., Goldman R.D.. Review: the dynamics of the nuclear lamins during the cell cycle - Relationship between structure and function. J. Struct. Biol. 2000; 129:324–334. [DOI] [PubMed] [Google Scholar]
- 44. Lu M., Shenk T.. Human cytomegalovirus infection inhibits cell cycle progression at multiple points, including the transition from G1 to S. J. Virol. 1996; 70:8850–8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Paladino P., Marcon E., Greenblatt J., Frappier L.. Identification of herpesvirus proteins that contribute to G1/S arrest. J. Virol. 2014; 88:4480–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Weinert B.T., Narita T., Satpathy S., Srinivasan B., Hansen B.K., Schölz C., Hamilton W.B., Zucconi B.E., Wang W.W., Liu W.R.et al.. Time-resolved analysis reveals rapid dynamics and broad scope of the CBP/p300 acetylome. Cell. 2018; 174:231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lundby A., Lage K., Weinert B.T., Bekker-Jensen D.B., Secher A., Skovgaard T., Kelstrup C.D., Dmytriyev A., Choudhary C., Lundby C.et al.. Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep. 2012; 2:419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Weinert B.T., Schölz C., Wagner S.A., Iesmantavicius V., Su D., Daniel J.A., Choudhary C.. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 2013; 4:842–851. [DOI] [PubMed] [Google Scholar]
- 49. Choudhary C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T.C., Olsen J.V, Mann M.. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009; 325:834–840. [DOI] [PubMed] [Google Scholar]
- 50. Choudhary C., Weinert B.T., Nishida Y., Verdin E., Mann M.. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 2014; 15:536–550. [DOI] [PubMed] [Google Scholar]
- 51. Narasumani M., Harrison P.M.. Discerning evolutionary trends in post-translational modification and the effect of intrinsic disorder: analysis of methylation, acetylation and ubiquitination sites in human proteins. PLoS Comput. Biol. 2018; 14:e1006349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Justice J.L., Needham J.M., Thompson S.R.. BK Polyomavirus activates the DNA damage response to prolong S phase. J. Virol. 2019; 93:e00130-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hoeijmakers J.H.J. DNA damage, aging, and cancer. N. Engl. J. Med. 2009; 361:1475–1486. [DOI] [PubMed] [Google Scholar]
- 54. Mehta A., Haber J.E.. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 2014; 6:a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Povirk L.F., Han Y.H., Steighner R.J.. Structure of bleomycin-induced DNA double-strand breaks: predominance of blunt ends and single-base 5′ extensions. Biochemistry. 1989; 28:5808–5814. [DOI] [PubMed] [Google Scholar]
- 56. Kitagawa M., Higashi H., Jung H.K., Suzuki-Takahashi I., Ikeda M., Tamai K., Kato J.Y., Segawa K., Yoshida E., Nishimura S.et al.. The consensus motif for phosphorylation by cyclin D1-Cdk4 is different from that for phosphorylation by cyclin A/E-Cdk2. EMBO J. 1996; 15:7060–7069. [PMC free article] [PubMed] [Google Scholar]
- 57. Knudsen E.S., Wang J.Y.. Dual mechanisms for the inhibition of E2F binding to RB by cyclin-dependent kinase-mediated RB phosphorylation. Mol. Cell. Biol. 1997; 17:5771–5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Branon T.C., Bosch J.A., Sanchez A.D., Udeshi N.D., Svinkina T., Carr S.A., Feldman J.L., Perrimon N., Ting A.Y.. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 2018; 36:880–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Panier S., Boulton S.J.. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 2014; 15:7–18. [DOI] [PubMed] [Google Scholar]
- 60. Pannunzio N.R., Watanabe G., Lieber M.R.. Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J. Biol. Chem. 2018; 293:10512–10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Liu N.A., Sun J., Kono K., Horikoshi Y., Ikura T., Tong X., Haraguchi T., Tashiro S.. Regulation of homologous recombinational repair by lamin B1 in radiation-induced DNA damage. FASEB J. 2015; 29:2514–2525. [DOI] [PubMed] [Google Scholar]
- 62. Ward I.M., Minn K., van Deursen J., Chen J.. p53 binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol. Cell. Biol. 2003; 23:2556–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Redwood A.B., Perkins S.M., Vanderwaal R.P., Feng Z., Biehl K.J., Gonzalez-Suarez I., Morgado-Palacin L., Shi W., Sage J., Roti-Roti J.L.et al.. A dual role for A-type lamins in DNA double-strand break repair. Cell Cycle. 2011; 10:2549–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dion V., Kalck V., Horigome C., Towbin B.D., Gasser S.M.. Increased mobility of double-strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nat. Cell Biol. 2012; 14:502–509. [DOI] [PubMed] [Google Scholar]
- 65. Fontana G.A., Hess D., Reinert J.K., Mattarocci S., Falquet B., Klein D., Shore D., Thomä N.H., Rass U.. Rif1 S-acylation mediates DNA double-strand break repair at the inner nuclear membrane. Nat. Commun. 2019; 10:2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Horigome C., Oma Y., Konishi T., Schmid R., Marcomini I., Hauer M.H., Dion V., Harata M., Gasser S.M.. SWR1 and INO80 chromatin remodelers contribute to DNA double-strand break perinuclear anchorage site choice. Mol. Cell. 2014; 55:626–639. [DOI] [PubMed] [Google Scholar]
- 67. Acs K., Luijsterburg M.S., Ackermann L., Salomons F.A., Hoppe T., Dantuma N.P.. The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nat. Struct. Mol. Biol. 2011; 18:1345–1350. [DOI] [PubMed] [Google Scholar]
- 68. Meerang M., Ritz D., Paliwal S., Garajova Z., Bosshard M., Mailand N., Janscak P., Hübscher U., Meyer H., Ramadan K.. The ubiquitin-selective segregase VCP/p97 orchestrates the response to DNA double-strand breaks. Nat. Cell Biol. 2011; 13:1376–1382. [DOI] [PubMed] [Google Scholar]
- 69. Rothkamm K., Krüger I., Thompson L.H., Löbrich M.. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell. Biol. 2003; 23:5706–5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hume A.J., Kalejta R.F.. Regulation of the retinoblastoma proteins by the human herpesviruses. Cell Div. 2009; 4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Colao I., Pennisi R., Venuti A., Nygårdas M., Heikkilä O., Hukkanen V., Sciortino M.T.. The ERK-1 function is required for HSV-1-mediated G1/S progression in HEP-2 cells and contributes to virus growth. Sci. Rep. 2017; 7:9176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hanson L., Dishon A., Kotler M.. Herpesviruses that infect fish. Viruses. 2011; 3:2160–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Davison A.J., Eberle R., Ehlers B., Hayward G.S., McGeoch D.J., Minson A.C., Pellett P.E., Roizman B., Studdert M.J., Thiry E.. The order Herpesvirales. Arch. Virol. 2009; 154:171–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kciuk M., Bukowski K., Marciniak B., Kontek R.. Advances in DNA repair—emerging players in the arena of eukaryotic DNA repair. Int. J. Mol. Sci. 2020; 21:3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Roos W.P., Krumm A.. Survey and summary: the multifaceted influence of histone deacetylases on DNA damage signalling and DNA repair. Nucleic. Acids. Res. 2016; 44:10017–10030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Svinkina T., Gu H.B., Silva J.C., Mertins P., Qiao J., Fereshetian S., Jaffe J.D., Kuhn E., Udeshi N.D., Carr S.A.. Deep, quantitative coverage of the lysine acetylome using novel anti-acetyl-lysine antibodies and an optimized proteomic workflow. Mol. Cell. Proteomics. 2015; 14:2429–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Miteva Y.V, Cristea I.M.. A proteomic perspective of Sirtuin 6 (SIRT6) phosphorylation and interactions and their dependence on its catalytic activity. Mol. Cell. Proteomics. 2014; 13:168–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wagner S.A., Beli P., Weinert B.T., Nielsen M.L., Cox J., Mann M., Choudhary C.. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics. 2011; 10:doi:10.1074/mcp.M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry miniTurbo and acetylome proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD022974. The flow cytometry data have been deposited to the FlowRepository with the repository ID: FR-FCM-Z2XN.