Abstract
In arthropods, zinc finger-associated domains (ZADs) are found at the N-termini of many DNA-binding proteins with tandem arrays of Cys2-His2 zinc fingers (ZAD-C2H2 proteins). ZAD-C2H2 proteins undergo fast evolutionary lineage-specific expansion and functional diversification. Here, we show that all ZADs from Drosophila melanogaster form homodimers, but only certain ZADs with high homology can also heterodimerize. CG2712, for example, is unable to heterodimerize with its paralog, the previously characterized insulator protein Zw5, with which it shares 46% homology. We obtained a crystal structure of CG2712 protein's ZAD domain that, in spite of a low sequence homology, has similar spatial organization with the only known ZAD structure (from Grauzone protein). Steric clashes prevented the formation of heterodimers between Grauzone and CG2712 ZADs. Using detailed structural analysis, site-directed mutagenesis, and molecular dynamics simulations, we demonstrated that rapid evolutionary acquisition of interaction specificity was mediated by the more energy-favorable formation of homodimers in comparison to heterodimers, and that this specificity was achieved by multiple amino acid substitutions resulting in the formation or breaking of stabilizing interactions. We speculate that specific homodimerization of ZAD-C2H2 proteins is important for their architectural role in genome organization.
INTRODUCTION
Proteins with tandem arrays of Cys2-His2 zinc fingers (C2H2 proteins) comprise the largest family of transcription factors in higher eukaryotes (1). A unique feature of C2H2 proteins is their specific binding to long (12–40 bp) DNA sequences, which distinguishes this class of proteins from other transcription factors (2,3). A zinc finger-associated domain (ZAD) is found at the N-termini of many C2H2 zinc-finger transcription factors in arthropods (4). In Drosophila melanogaster, 98 of a putative 326 C2H2 proteins have an N-terminal ZAD (5).
Expansion of ZADs in arthropods is a result of gene duplications persisting through positive evolutionary selection; therefore, domains can be clustered on the basis of their amino acid sequence similarity (4). Seven paralogous groups were identified in Drosophila and grouped into four larger clades, with the similarity within paralogous groups varying from 85% to only 20% in distant clusters (4,5).
Most ZAD-C2H2 proteins are involved in regulation of gene expression. The Zipic, Sry δ, Pita, Piragua and Zw5 proteins are important during development, and most of their binding sites are in promoter regions (6–10). Among the ZAD-C2H2 proteins, the most well studied is motif 1 binding protein (M1BP), which binds to the core promoter sequences of >2000 Drosophila genes (11). In particular, M1BP was shown to recruit TATA-binding protein-related factor 2 (TRF2) to promoters of genes encoding ribosomal proteins (12). Some other ZAD-C2H2 proteins have more specific or redundant roles in transcription regulation. For example, ZAD and Architectural Function 1 protein (ZAF1) binds to nearly 90 promoters in embryos, but has redundant functions in gene regulation (13). Grauzone is expressed at all stages of Drosophila development, but is only strongly required for activity of the promoter of the cortex gene involved in meiosis in oocytes (14,15). The proteins Quib, Séance, and Molten defective are responsible for the activity of promoters of the genes involved in steroid hormone biosynthesis (16,17).
The ability to organize chromatin loops and display insulator/architectural properties was demonstrated for several ZAD-C2H2 proteins. The Zw5 protein was originally identified as a key factor of the scs insulator at the boundary of the hsp70 cluster (18,19) and can effectively support long-range interaction between its binding sites (20). The Pita protein functions in cooperation with dCTCF and Su(Hw) in organization of insulator/boundary elements in the bithorax complex (21,22). In transgenic lines, the ZAF1 protein can form a chromatin loop that isolates the eye enhancer from the white gene (13). In all cases, the homodimerization activity of ZADs was critical for chromatin loop formation that either blocks or supports long-distance enhancer–promoter interactions (10,13).
To date, the only structurally characterized ZAD, from the Grauzone protein, was shown to be a homodimer that adopted a unique fold with four conserved cysteines coordinating zinc ions in each monomer and contained a large amphipathic α-helix involved in a specific interaction with another monomer (23). Several other ZADs have been confirmed to also form homodimers (7,10,24,25). The ZADs of three proteins (Pita, Zw5 and ZIPIC) are able to form homodimers, but fail to effectively heterodimerize. As a consequence, hetero-pairs of these proteins do not support long-distance interactions in transgenic model systems (10). These results allow us to assume that most of the ZAD-C2H2 proteins have similar architectural properties that are based on the predominant ability of ZADs to form homodimers, but not heterodimers.
In this study, we investigated the prevalence of cognate ZADs being able to heterodimerize. We performed analyses of dimerization between ZAD paralogs within clusters with the highest sequence homology. Finally, to investigate structural determinants of dimerization specificity of ZADs that evolved after gene duplication events, we obtained a crystal structure of the ZAD from the CG2712 protein, a paralog of the well-studied Zw5 protein, together with molecular modeling and subsequent analysis of other dimers of ZADs.
MATERIALS AND METHODS
Phylogenetic analysis
Primary processing and multiple sequence alignment were performed in ClustalW (26). After multiple alignments of all 98 ZAD sequences from D. melanogaster, the sequences were manually trimmed on both sides (Supplementary Figure S1). Evolutionary analyses were conducted with MEGA X (27). The evolutionary distances were computed using the Dayhoff matrix-based method (28). All positions with <20% site coverage were eliminated; thus, there were a total of 83 positions remained in the final dataset. The variation rate among sites was modeled with a gamma distribution (shape parameter = 2.6, determined using the maximum likelihood approach). The phylogenetic tree was reconstructed using the neighbor-joining method. The percentage of replicate trees, in which the associated sequences are clustered together, was determined using the bootstrap test (1000 replicates). The tree was visualized and annotated with the number of exons in ZADs using the ggtree R package (29). Sequence logo was created using WebLogo (30).
Plasmids and cloning
CG2712 [1–90] was cloned in frame with a TEV-cleavable GST-tag in the modified vector pGEX-4T1 (GE Healthcare). For in vitro experiments, protein fragments were PCR-amplified using corresponding primers (Supplementary Table S1) from fly cDNA and subcloned into modified pGEX-4T1 (GE Healthcare), pMAL-C5X (New England Biolabs), or a vector derived from pACYC and pET28a(+) (Novagen) bearing a p15A replication origin, kanamycin resistance gene, and pET28a(+) MCS. For yeast two-hybrid assays, cDNAs encoding ZADs were amplified using the corresponding primers (see Supplementary Table S1) and fused with the DNA-binding or activation domain of GAL4 in the corresponding pGBT9 and pGAD424 vectors (Clontech). We also used a modified pGBT9 vector in which ZADs were cloned at the N-terminus of the GAL4 DNA-binding domain. PCR-directed mutagenesis was used to generate constructs with mutant ZADs using mutagenic primers (Supplementary Table S1).
Yeast two-hybrid assay
The yeast two-hybrid assay was performed as previously described (10). Briefly, for growth assays, plasmids were transformed into yeast strain pJ69–4A by the lithium acetate method, following standard Clontech protocol, and plated on media without tryptophan and leucine. After 2 days of growth at 30°C, the cells were plated on selective media without tryptophan, leucine, histidine, and adenine, and their growth was compared after 2–3 days. Each assay was repeated three times.
Protein expression and purification
E.coli BL21(DE)3 containing plasmid expressing CG2712 [1–90] with TEV-cleavable GST were grown at 37°C in 3 l of LB media containing 0.2 mM ZnSO4 until an optical density of 0.6 was reached, then cooled to 18°C and induced with 1 mM IPTG overnight at +18°C. Cells were pelleted, resuspended in degassed lysis buffer A [20 mM Tris (pH 7.4), 150 mM NaCl, 20 mM KCl, 5 mM MgSO4, 0.1 mM ZnCl2, 10% w/w glycerol, 0.1% NP40, 1 mM dithiothreitol (DTT)] containing protease inhibitors, sonicated, centrifuged at 20 000 × g for 1 h and applied to 4 ml of pre-equilibrated glutathione-resin (Pierce). The resin was washed with lysis buffer containing 500 mM NaCl and subjected to TEV cleavage overnight at 4°C at constant rotation in degassed buffer containing 20 mM Tris (pH 8.0), 200 mM NaCl, 5 mM sodium citrate, 0.01 mM ZnCl2 and 1 mM DTT. Flowthrough containing cleaved protein was collected and the buffer was changed to degassed 20 mM Tris (pH 7.4), 50 mM NaCl, 0.1 mM ZnCl2 and 1 mM DTT using a HiPrep DeSalting column (GE Healthcare) applied to SOURCE15Q resin (GE Healthcare). Flowthrough was collected, adjusted to 100 mM NaCl, and concentrated using Amicon concentrators. MBP-pulldown was performed with Immobilized Amylose resin (New England Biolabs) in buffer A containing 5 mM DTT. BL21 cells co-transformed with plasmids expressing MBP-fused and 6xHis-Thioredoxin-fused ZADs were grown in LB media to an A600 of 1.0 at 37°C and then induced with 1 mM IPTG at 18°C overnight. ZnCl2 was added to final concentration 100 μM before induction. Cells were disrupted by sonication, centrifuged, applied to resin for 10 min at +4°C, after that resin was washed four times with buffer A containing 500 mM NaCl and bound proteins were eluted with 50 mM maltose, 100 mM Tris, pH 8.0, 100 mM NaCl for 15 min. 6xHis-pulldown was performed similarly with Zn-IDA resin (Cube Biotech) in buffer B (50 mM HEPES–KOH, pH 7.6, with 500 mM NaCl, 5 mM MgCl2, 0.1 mM ZnCl2, 20 mM imidazole, 5% glycerol, 0.1% NP-40, and 5 mM β-mercaptoethanol) containing 1 mM PMSF and Calbiochem Complete Protease Inhibitor Cocktail VII (5 μl/ml), washed four times with buffer B containing 30 mM imidazole and proteins were eluted with buffer C (50 mM HEPES–KOH, pH 7.6, with 500 mM NaCl, 250 mM imidazole and 5 mM β-mercaptoethanol) (20 min at +4°C). Chemical cross-linking was carried out for 10 min at room temperature in buffer B containing 20 mM HEPES–KOH, pH 7.7; 150 mM NaCl, 20 mM imidazole, 1 mM β-mercaptoethanol. Prior to cross-linking, protein concentration was adjusted to 10 μM for at least 1 h. Reaction was quenched with 50 mM Tris–HCl (pH 6.8), and samples were resolved using SDS-PAGE followed by silver-staining.
Crystallization and data collection
An initial 96-well format crystallization screening of CG2712 [1–90] was performed with a robotic crystallization system (Rigaku, USA) and commercially available crystallization screens (Hampton Research, USA) using the sitting drop vapor diffusion method at 15°C. The protein concentration was 7 mg/ml in the following buffer: 20 mM Tris pH 7.4, 100 mM NaCl, 1 mM DTT and 1 mM ZnCl2. The drop volume was 0.02 μl with 50:50 protein to precipitant ratio. Optimization of initial conditions was made by the hanging drop vapor diffusion method in a 24-well plate with 3 μl drop volume (50:50 ratio). The best crystals were obtained in a crystallization condition containing 0.1 M 2-(N-morpholino)ethanesulfonic acid (pH 6.5) and 1.3 M magnesium sulfate. Crystals suitable for data collection were grown within 10 days.
Immediately before data collection, crystals of CG2712 were briefly soaked in mother liquor containing 25% glycerol as a cryoprotectant. Crystals were then flash-cooled to 100 K in liquid nitrogen. The X-ray diffraction data were collected at the BL41XU beamline of the Spring8 synchrotron (Harima Science Garden, Japan) equipped with a Pilatus detector. The data were indexed, integrated, and scaled using iMosflm (31). Based on the L-test (32) the data was not twinned. The program Pointless (33) suggested the P41212 space group. The data collection and processing statistics are summarized in Supplementary Table S2.
Structure solution and refinement
Despite the presence of a homologous Grauzone ZAD structure, all attempts to use molecular replacement failed. Thus, the structure of 2712 was solved by the SAD method using the Zn2+ ions as anomalous scatterers. The location of the Zn2+ ions was deduced with Phenix.hyss (34). Phaser (35) was used to phase the data. Subsequently, Parrot (36) was used for density modification and to solve phase ambiguity. The initial model was automatically built with Buccaneer (37). Some structure features, such as loops connecting secondary structure elements and the four cysteine residues coordinating each zinc ion, were built manually during the refinement.
The data set collected at remote wavelength (Supplementary Table S2) was used for structure refinement. Refinement was carried out with the REFMAC5 program of the CCP4 suite (38). TLS was introduced together with isotropic B-factor refinement. The visual inspection of electron density maps and the manual rebuilding of the model were carried out with the COOT interactive graphics program (39). The resolution was successively increased to 2.0 Å. In the final model, the asymmetric unit contains two independent copies of the protein (chain A and B) as well as 145 water molecules, two zinc ions, and one molecule of glycerol from cryo-solution. Nine (seven for chain B) N-terminal and eight C-terminal amino acids of each chain have no electron density due to their high mobility. Region 39–42 of chain B has a very weak electron density, possibly for the same reason. The structure refinement statistics are provided in Supplementary Table S2.
Structure analysis and validation
The visual inspection of the structure model was carried out with the COOT (39), Pymol (The PyMOL Molecular Graphics System, Schrödinger, LLC) and Chimera (40) software. The structure comparison and superposition were made using the PDBeFOLD program (41). The contacts were analyzed using the PDBePISA (42) and WHATIF (43). The dimeric interfaces were visualized with LigPlot (44).
Molecular dynamics simulations
Molecular dynamics (MD) simulations were performed with NAMD 2.11 software (45) and CHARMM36 force field (46). For the preparation of the model systems and further analysis of MD trajectories, VMD software (47) was used.
The X-ray structure of the CG2712 monomer was used as a template for the Zw5 and dv2712 monomers, which were modeled with Modeller (48). The Rosetta online server (49) was used to build models of CG2712–DV2712, CG2712–Zw5 and Zw5–Zw5.
Molecular dynamics (MD) simulations were performed in two variants: unconstrained MD to analyze the main interaction, and steered MD (SMD) to estimate the work needed to disrupt the dimers, as a reflection of the free energy of dimerization. For all unconstrained MD simulations, a TIP3P water box was added with boundaries in at least 10 Å from protein atoms, and for steered molecular dynamics (SMD) simulations, to assure adequate space for separation of monomers, water molecules were added with boundaries of at least 30 Å. Sodium and chloride ions were added to a final ion concentration of 0.15 M. MD runs were performed with the following conditions: NPT ensemble, 298 K, 1 atm, periodical boundary conditions, 1 fs time-step. All systems were subjected to two 5000 step minimizations: with protein Cα atoms fixed, and full optimization. Then 5 ns MD simulations were performed to optimize water boxes. Unconstrained MD productive runs were 300 ns, extended to 500 ns for the Zw5–Zw5 dimer due to RMSD increase.
Following analysis of unconstrained MD, SMD simulations were preceded by 50 ns (500 ns for the Zw5–Zw5 dimer) unconstrained MD trajectories of corresponding systems. In SMD simulations, rupturing force was applied to the center of mass (COM) of Cα atoms of each monomer with a constant pulling velocity of 0.5 Å/ns. The force constant was adjusted in series of test runs and set to 1 kcal/mol·Å2. Test runs demonstrated that for CG2712–CG2712 and Zw5–Zw5 dimers, binding of monomers is so strong that COM separation was achieved at the cost of destruction of the main α-helix. To avoid this, additional constraints were applied to maintain the secondary structure of the helices. For further details, see Supplementary Data.
RESULTS
Only highly homologous ZADs can heterodimerize
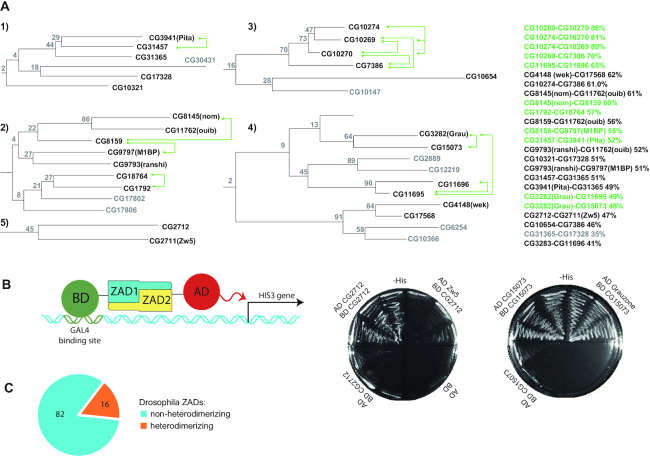
ZADs in arthropods underwent lineage-specific expansion through the process of gene duplication. To determine how common it is for ZADs to form homo- but not heterodimers, we studied the interaction between ZADs that have the maximum homology in their amino acid sequences. We used several phylogenetic approaches to search the paralogous clusters of ZAD-containing proteins. To ensure the most proper clustering, we monitored the integrity of some paralogous groups that are encoded by gene clusters (e.g. CG9797 [M1BP], CG9793, CG11762, CG8145 and CG8159) and the categorization of paralogous ZADs within one cluster to either the one-exon or the two-exon group (Supplementary Figure S2), which was proposed to be the important subdivision principle (4). Some paralogous ZADs that do not reside within the same genomic cluster (for example, Pita-CG31457-CG31365) are most likely retrocopies, which explains why only one of these proteins could have an intron within the ZAD. The best results were obtained using phylogenetic analysis based on the Dayhoff protein amino acid replacement matrices (PAM (50)), which suggested that all of the ZADs can be grouped into three clades originating from the most ancient duplications, in accordance with a previous report (5). We found five distinct paralogous clusters (Figure 1A, Supplementary Figure S2) containing ZADs with homology >45% (the percentage of identical and similar residues), while in other clusters sequence homology does not exceed 30%.
Figure 1.
(A) Clusters of zinc finger-associated domains of Drosophila melanogaster containing highly homologous paralogs tested for ability to heterodimerize using a yeast two-hybrid assay. Heterodimerizing pairs within clusters are shown by green connectors. Domains that were not tested are colored grey. Bootstrap values of confidence are shown at the branching points. The levels of homology between the most similar pairs are shown at the right. The complete phylogenetic tree is shown in Supplementary Figure S2, corresponding sequence alignment in Supplementary Figure S1. Complete results of testing the binary interactions are shown in Supplementary Table S3 and Supplementary Figure S3. Cluster numbering corresponds to the numbering in Supplementary Table S3. (B) The scheme of Yeast two-hybrid assay. BD stands for GAL4 DNA-binding domain, AD– GAL4 activation domain. Interaction between tested proteins leads to activation of HIS3 gene, which permits yeast growth on -His media. The example –His plates of heterodimerizing and non-heterodimerizing ZAD pairs are shown in the right. (C) The diagram showing the number of Drosophila ZADs found to have/do not have heterodimerization partner.
To study interactions between Drosophila ZADs, we used the yeast two-hybrid system that previously showed effectiveness in testing dimeric interactions between the ZADs of Pita, Zw5 and ZIPIC (10). Most of possible pairs of ZADs from five distant clusters mentioned above were selected and tested for homo- and heterodimerization (Figure 1A and B, Supplementary Table S3 and Supplementary Figure S3). In total 25 ZADs were included in the analysis. As shown in Figure 1A, most of the tested pairs of ZADs with higher than 51% homology interact with each other (the only exceptions are CG17568-Wek, CG8159-CG11762 and CG10274-CG7386, which have 55–62% homology), whereas CG3282(Grau)-CG11695 and CG3282(Grau)-CG15073 are the only domains with sequence similarity below 51% (49% and 48%, respectively) that are able to heterodimerize. Thus, using a threshold of 45%, we found most of the potentially heterodimerizing domains. The analysis demonstrated that 16 of the 98 Drosophila ZADs belonging to 4 out of 5 paralogous clusters (Figure 1A) have heterodimerization partners comprising 13 heterodimerizing pairs. Another 31 tested pairs of ZADs from these clusters with sequence similarity in range 30–62% were found to form only homodimers (Figure 1C, Supplementary Figure S3). We did not observe formation of heterodimers between ZADs from different clusters.
We performed analysis of conservation between DNA-binding residues within zinc-fingers of ZAD-C2H2 proteins from paralogous clusters containing heterodimerizing ZADs. No obvious correlation was found between the ability of ZADs to heterodimerize and conservation of DNA-binding position within zinc-fingers of corresponding proteins (Supplementary Figure S4). Only proteins CG10270-CG10269-CG10274-CG7386 from highly homologous cluster have almost identical zinc-fingers and probably functionally substitute each other. Thus, much likely most proteins with heterodimerizing ZADs have different DNA-recognition patterns.
Selective homodimerization of ZADs of the Zw5 and CG2712 proteins is preserved in cognate Drosophila species
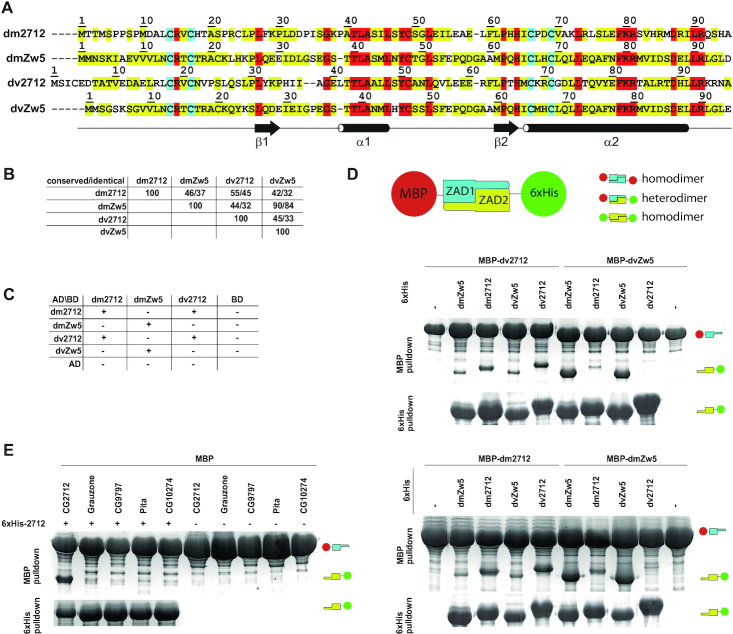
For further analysis of the mechanisms underlying the loss of heterodimerization ability between highly homologous ZADs of paralogs, we focused on the ZADs of the CG2712 and Zw5 proteins, which are encoded by genes organized in one cluster. CG2712 and Zw5 are conserved among Drosophila species, suggesting that their duplication occurred before the evolution of the genus Drosophila. The ZADs of the Zw5 orthologs from D. melanogaster and D. virilis (GJ18900, hereafter dvZw5) share 84% identical residues (90% homology). The ZADs of the CG2712 orthologs (GJ19263 in D. virilis, hereafter dv2712) are less conserved, with only 45% identical and 55% homologous residues (Figure 2A and B). Overall sequence identity among these four domains is only 24%. We studied ability of these ZADs to heterodimerize using pulldown and yeast two-hybrid assays. Results shown in Figure 2C revealed that both CG2712 and Zw5 from D. melanogaster specifically interact with corresponding D. virilis orthologues in a yeast two-hybrid assay, but do not display formation of heterodimers. In pulldown assays, which were performed at much higher protein concentrations, some heterodimers were formed, but a strong preference for homodimers was clear (Figure 2D). These results suggested the potential for heterodimer formation at high protein concentrations, but a preference for homodimers. In general, MBP-pulldown assays were found to be less specific than yeast two-hybrid assays due to the much higher protein concentration in bacteria cells compared to yeast cells, in which less-efficient interactions with higher dissociation rates were observed. 6xHis pulldowns were found to be nonspecific, since many ZADs tend to bind to metal-chelating resins; thus, 6xHis pulldowns were used only as protein expression controls. We tested the ability of CG2712 ZAD to heterodimerize with ZADs from other paralogous clusters in a MBP-pulldown assay, which revealed a very low presence of heterodimers (Figure 2E).
Figure 2.
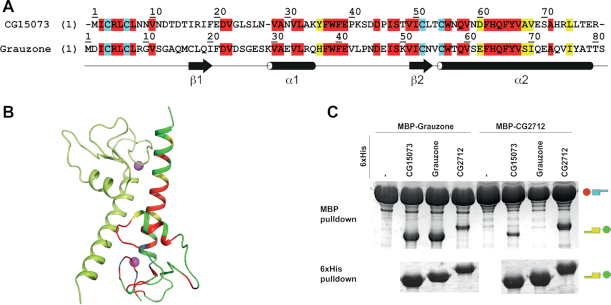
Specificity of homodimerization retained between CG2712 and Zw5 orthologs. (A) Multiple sequence alignment of CG2712 and Zw5 zinc finger-associated domains (ZADs) from Drosophila melanogaster (dm) and D. virilis (dv). Identical residues are shown in red; conserved are in yellow, invariant cysteines are in cyan. (B) Percentage of conserved/identical residues in ZADs of CG2712 and Zw5 from D. melanogaster and D. virilis. (C) Testing of heterodimerization ability of dm/dv CG2712 and Zw5 ZADs in a yeast two-hybrid assay. The dvZw5 ZAD demonstrated strong self-activation properties when fused to Gal4 DNA-binding domain and was omitted for clarity. AD/BD denotes activation and DNA-binding domains of GAL4. (D) Testing of heterodimerization ability of dm/dv CG2712 and Zw5 ZADs in MBP or 6xHis-pulldown assay after co-expression in bacteria. The experiment scheme is shown on top. Small cartoons on the right show the positions of MBP-fused ZADs (MBP is 46kDa) and Thioredoxin-6xHis-fused ZADs (Thioredoxin-6xHis is 17kDa). On the scheme thioredoxin was not shown for clarity. Upper panels show results of interaction for MBP-fused D. virilis 2712 and Zw5 ZADs, bottom panels – for MBP-fused ZADs from D. melanogaster 2712 and Zw5. (E) Results of testing of heterodimerization ability between ZADs of CG2712 and proteins from other paralogous clusters in MBP or 6xHis-pulldown assay after co-expression in bacteria. Designations are as in panel D.
Thus, CG2712 ZAD can effectively dimerize with its ortholog dv2712, but not with the paralogs Zw5 and dvZw5, despite the fact that these pairs have comparable sequence similarity.
Spatial structure of the CG2712 ZAD is highly similar to that of the Grauzone ZAD despite a low sequence similarity
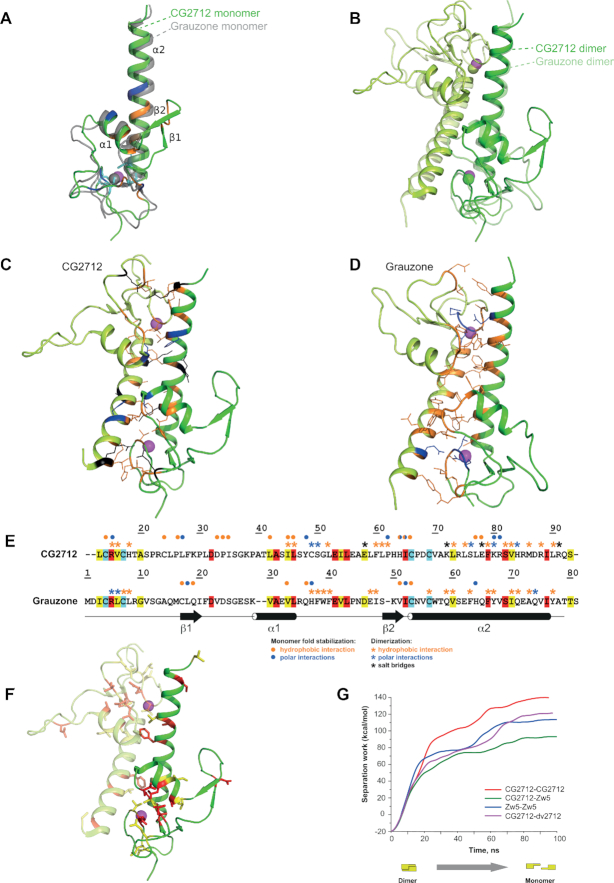
To understand the mechanisms underlying the predominance of homodimerization of ZADs from Zw5 and CG2712, we obtained a crystal structure of CG2712 ZAD at 2.0 Å resolution (Figure 3A). An asymmetric unit of the crystal contained two monomers of CG2712 that form a dumbbell-like homodimer (Figure 3B). The monomer possesses two α-helices—a short α1 (residues 42–51) and a long C-terminal α2 (66–91)—as well as a β-sheet composed of two strands, β1 (residues 31–33) and β2 (residues 37-40). The zinc ion, which plays a structural role knotting the N-terminal loop (residues 10–22), the loop region between two α-helices, and C-terminal α2, is coordinated via four cysteine residues invariant among all the ZADs (Supplementary Figure S1). In addition to zinc coordination, the CG2712 monomer fold is strengthened by a number of interactions, which can be classified as: (a) between the α1 and α2 helices, (b) between the looped regions near zinc ions and (c) between the C-terminal α2 helix and the looped region preceding the short α1 helix. Interface (a) is formed by hydrogen bonding between residues Y48, K80, and R81 and is strengthened by hydrophobic interactions of L42, L73, I45, L46 and L77. Interface (b) is strengthened by hydrogen bonding of R15, L25, P62, H64 and I65 and by hydrophobic interactions via C14, C24, P26, H64, I65, and C66. Finally, interface (c) contains only hydrophobic interactions between D33, P34, I35, L77 and E78 (Figures 3A and 5A). Noteworthy, the last interface exists only in case of one monomer from the asymmetric unit, which makes doubtful its significance for proper domain folding.
Figure 3.
Crystal structure of CG2712 ZAD is highly similar to that of Grauzone ZAD. (A) The crystal structure of CG2712 ZAD monomer. Residues stabilizing the monomer are colored blue for hydrogen bonds, orange for hydrophobic interactions and cyan for invariant cysteine residues. The zinc ion is colored in magenta. The superposed Grauzone monomer is colored in gray and is semi-transparent for clarity. (B) Overlay of CG2712 and Grauzone ZAD homodimers. Superposition was made using one of the monomers. Dimers are colored by chain, with the Grauzone dimer depicted as semi-transparent. Zinc ions are in magenta and green for CG2712 and Grauzone ZADs, accordingly. (C) Dimeric interface of the CG2712 ZAD homodimer. Side chains of the residues involved in the interface are shown as wire and are colored as blue for hydrogen bonds, black for salt bridges, and orange for hydrophobic interactions. (D) Dimeric interface of Grauzone ZAD homodimer. Color scheme and orientation of the molecule is similar to panel C. (E) Amino acid alignment of Grauzone and CG2712 ZADs (color scheme is similar to Figure 2A). Blue asterisks show residues involved in polar interactions between monomers, black – residues involved in salt bridges, while orange asterisks mark residues forming hydrophobic contacts according to molecular dynamics simulation (CG2712) or crystal structure analysis (Grauzone). Circles show interactions stabilizing the corresponding monomer. Color scheme is the same as for asterisks. (F) Residues conserved between CG2712 and Grauzone ZADs depicted on the crystal structure of CG2712 ZAD. Color scheme is similar to Figure 2A. (G) Results of steered molecular dynamics simulation showing the energy required for dimer dissociation as a function of time.
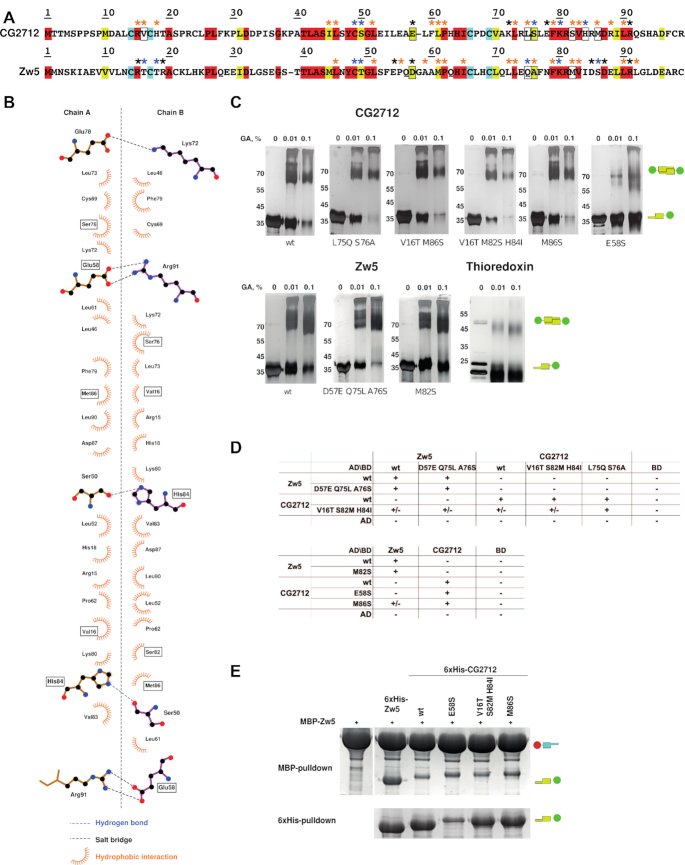
Figure 5.
Site-directed mutagenesis suggested the cumulative effect of stabilizing interactions in the specificity of dimer formation. (A) Sequence alignment of CG2712 and Zw5 ZADs. Identical residues are shown in red, conserved – in yellow, invariant cysteines are in cyan. Blue asterisks show residues involved in hydrogen bonding, black – residues involved in salt bridges, while orange asterisks mark residues forming hydrophobic contacts according to molecular dynamics simulation (for further details see Supplementary Tables S5 and S6). Residues subjected to mutagenesis are shown in frame. (B) The plot of the CG2712 ZAD dimerization interface made for crystal structure. Point mutations introduced in CG2712 ZAD are shown in frame. (C) Results of testing the impact of substitutions on homodimerization using a chemical cross-linking assay with increasing concentrations of glutaraldehyde (concentrations are shown on the top). Thioredoxin was used as negative control. Positions of the molecular weight markers are shown on the left rows. Designations are as on Figure 2D. (D) Results of testing the effect of substitutions in a yeast two-hybrid assay. AD/BD denotes activation and DNA-binding domains of GAL4; +/- denotes weak growth. (E) Testing of the impact of point mutations on the heterodimerization ability of CG2712/Zw5 ZADs in a pulldown assay after co-expression in bacteria. Designations are as on Figure 2D. For complete results, see Supplementary Figure S6.
Comparison with the only known structure of ZAD, Grauzone (PDB ID: 1PZW), revealed that in spite of a very low sequence homology between these ZADs (25%), their monomeric structures are similar (Figure 3A), with a corresponding RMSD between 1.6 and 3.0 Å2 (the range corresponds to differences between the two CG2712 monomers from the asymmetric unit of the crystal). The relatively high RMSD is a consequence of the different conformations of the looped regions, as the positions and orientations of all α-helices almost coincide. In contrast to the CG2712 crystal structure, a β-sheet in the Grauzone ZAD is located in a different region preceding the α2 helix. The distribution of the residues involved in the monomer folding and dimer formation of Grauzone and CG2712 ZADs is shown in the Supplementary Figure S5.
CG2712 has an extensive dimerization interface (Figure 3C), which covers approximately 18% of the total solvent accessible area of each monomer and mostly involves residues from both α-helices. The interface is strengthened by hydrophobic interactions via residues V16, H18, L46, L52, L61, P62, K72, L73, L75, F79, V83, M86 and L90 of each monomer. The corresponding energy gain upon dimer formation (–18.2 kcal/mol) is comparable to that of Grauzone (–19.5 kcal/mol). However, the CG2712 interface is additionally fastened by five salt bridges and two hydrogen bonds, versus only four hydrogen bonds for Grauzone (Supplementary Table S4). Detailed analysis of the interfaces of both ZADs revealed that they differ significantly by location and type of interacting amino acids (Figure 3C and D) and are primarily composed of non-conserved residues (Figure 3E and F). Only the three residues forming the hydrophobic interface are conserved or semi-conserved between CG2712 and Grauzone, namely V16 (L6 in Grauzone), F79 (F66) and V83 (I70) (Figure 3E). Additionally, in contrast to the R5 residue of Grauzone, which forms a direct hydrogen bond to Q74 of the adjacent monomer, the corresponding R15 of CG2712 is not hydrogen-bonded to D87 (Q74 in Grauzone) directly. However, the latter bond is formed via an intermediate water molecule found in the crystal structure of CG2712 ZAD.
Thus, in spite of the similar monomeric structures of CG2712 and Grauzone ZADs, discrepancies in the dimerization mechanism, involving almost completely non-conserved amino acids forming the interface, lead to the inability of these domains to form heterodimers due, at least in part, to steric hindrances. Accordingly, the presence of heterodimers between CG2712 and Grauzone or ZADs from distant clusters is negligible even in pulldown assays (Figure 2E).
Structural basis for the dimerization specificity of highly homologous ZADs
The CG2712 ZAD crystal structure demonstrates no significant steric hindrances preventing CG2712-Zw5 heterodimerization
Taking into account the results of the structural comparison between Grauzone and CG2712 dimers, we aimed to examine the heterodimerization potential of particular ZADs with high homology: CG2712 and Zw5 from D. melanogaster as well as the CG2712 ortholog from D. virilis. Sequence homologies between these domains are 46% for CG2712 and Zw5 and 55% for CG2712 and dv2712. The Zw5 ortholog from D. virilis (dvZw5) has an almost identical sequence to Zw5, with a few substitutions in loops; thus, this domain thus provides no extra information on structural aspects of dimerization specificity and was excluded from further analysis. In sum, we focused on a comparative analysis of CG2712-CG2712, Zw5-Zw5, CG2712-Zw5 and CG2712-dv2712 ZAD dimers.
A common mechanism to gain specificity is the presence of steric clashes between residues on an interface, which hamper dimerization. Homology modeling of all heterodimers was conducted, with thorough inspection for possible steric hindrances. This revealed the following eventual clashes between side chains in the CG2712-Zw5 dimeric interface: M86 (CG2712)-T16 (Zw5), M16-S86, L75-Q75, S76-A76 and L52-I84. The latter clash also breaks two hydrogen bonds between the side chain of H84 and the main chain oxygen of S50 in the CG2712 homodimer. L75 resides at the center of α2 and clash with the Q75 of the adjacent subunit. However, some of these residues of CG2712 ZAD are conserved in dv2712, which is able to form heterodimers with CG2712. Moreover, in co-expression assays followed by pulldown, a small portion of CG2712-Zw5 and dv2712-Zw5 heterodimers was detected together with major portions of their corresponding homodimers (Figure 2D and Supplementary Figure S6A). These findings indicate that clashes found with homology modeling might not cause an inability to heterodimerize.
Molecular dynamics simulation revealed higher stability of homodimers compared to heterodimers
To further shed light on the occurrence of heterodimerization, we used protein–protein docking followed by MD simulation to model the structure of a Zw5 homodimer (Supplementary Figure S7), as well as CG2712-Zw5 and CG2712-dv2712 heterodimers. Additionally, the CG2712 homodimer was also treated with the same procedure based on its crystal structure in order to assess the accuracy of this approach and to allow proper comparison.
MD simulations of the CG2712 ZAD homodimer demonstrated its stability through the trajectory (Supplementary Figure S7), including all of the secondary structure elements. Dimerization interface analysis along the MD trajectory revealed no major discrepancies with the crystal structure of CG2712. The model inherited all the polar and hydrophobic interactions within the dimeric interface (Supplementary Table S5) and showed additional interactions along the MD trajectory, thus provides the applicability of such an approach to model other dimers.
Polar (salt bridges, hydrogen bonds) and non-polar (hydrophobic) interactions along MD trajectories were further compared for the dimers analyzed (Table 1 and Supplementary Figure S8). In all cases, salt bridges were more stable along the trajectories compared to hydrogen bonds. For the CG2712 homodimer, a salt bridge between residues R91 and E58 (Figure 5B, Table 1, and Supplementary Table S5) provides the major contribution to stability. This symmetrical interaction was also observed in the crystal structure and is maintained along the entire MD trajectory (Supplementary Figure S9A). The symmetric solvent-separated ion pairs D87-R15 is rather stable along the whole MD trajectory. The two other salt bridges, E78-K72 and E53-R88 exist only at short span of MD trajectory (Supplementary Figure S9B–D). The hydrogen bond between the side chains of both S76 residues is maintained over most of the trajectory. Less stable hydrogen bonds are listed in Supplementary Table S5. In comparison to the X-ray structure, more hydrophobic residues are participating in inter-monomer contacts along MD trajectory, these additional contributors are F60, R88 and I45 (Supplementary Table S5).
Table 1.
Results of dimerization interface analysis of zinc finger-associated domain dimers. Data averaged along the MD trajectory are shown. Detailed data are summarized in Supplementary Tables S5–S8.
| CG2712 homodimer* | Zw5 homodimer | CG2712 – Zw5 | CG2712- dv2712 | |
|---|---|---|---|---|
| Hydrogen bonds | S76-S76 | M16-T88 | ||
| S82-V16 | S86-T16 | S82-T16 | R85-V20 | |
| K80-S50 | S86-R15 | K80-T49 | K80-N53 | |
| H84-S50 | A59-L90 | S50-Q82 | ||
| Salt bridges | R91-E58** | R15-D87** | ||
| E78-K72** | D85-R19 | E58-R91 | E58-R93 | |
| D87-R15** | D57-R91 | R88-E54 | R91-E60 | |
| E53-R88 | E54-R91 | R91-E54 | E53-R95 | |
| Relative strength of hydrophobic interactions (%)* | 100 | 84 | 78 | 98 |
Residue numbering is in accordance to Figure 2A.
*Relative to a reference: CG2712 homodimer. Estimated by SMD simulations.
**This interaction is doubled in dimer due to symmetry.
In contrast to CG2712, the Zw5 ZAD homodimer has less stable salt bridges along the entire trajectory. A salt bridge R15-D87 (Supplementary Figure S10A) persists for only one pair of residues, while a bond formed by a vice versa pair breaks and is replaced by another salt bridge, D85-R19 (Supplementary Figure S10B), throughout the trajectory. Another salt bridge, D57-R91 (Supplementary Figure S10C), is formed along the trajectory and is disrupted later by formation of an E54-R91 salt bridge (Supplementary Figure S10D). A number of hydrogen bonds were detected along a significant part of trajectory, including mostly stable bonds S86-T16, S86-R15, and S86-T18 (Supplementary Table S6). The interface is strengthened by hydrophobic residues listed in Supplementary Table S6.
The CG2712-Zw5 heterodimer has only one salt bridge that is stable over the trajectory: E58-R91 (hereafter, the first amino acid corresponds to CG2712), similar to E58-R91 of the CG2712 homodimer (Supplementary Figure S11A). Other observed salt bridges were: the most stable R91-E54 as well as solvent separated ion pairs - R88-E54 and D87-R15 (Supplementary Figure S11B). Short-lived hydrogen bonds were observed during the MD, indicating their instability along the trajectory (Supplementary Table S7). Residues strengthening the heterodimeric hydrophobic core are listed in Supplementary Table S7.
Finally, the CG2712 ZAD residues involved in salt bridges in the CG2712–dv2712 dimeric interface are E53, E58 and R91 (Table 1). Hydrophobic residues of the interface are listed in Supplementary Table S8.
In summary, stability of the dimerization interfaces must be attributed first to salt bridges and hydrophobic interactions. The polar residues of CG2712 involved in salt bridges in either homo- or heterodimers include E53 (E54 in Zw5), E58, R88, and R91 (R91 in Zw5). This list is expanded by conserved L46 (L45 in Zw5), L52 (L51), P62 (P62), L73 (L73), F79 (F79), V83 (V83) and L90 (L90) stabilizing hydrophobic interface. The Zw5 residues P62 and I65 participate only in the corresponding homodimerization interface; their influence on the heterodimerization interface is much lower. I84 of Zw5 participates in hydrophobic interactions exclusively within the CG2712–Zw5 heterodimer, while its counterpart in CG2712, H84, is involved in interface formation within the homodimer only (Table 1 and Figure 5B).
To obtain a quantitative estimation of dimer stability, an SMD approach was used. SMD runs of 100 ns allow the dimer dissociation process to occur closer to equilibrium (Figure 3G). The results obtained confirmed that dimers can be ranked by their stability as follows: CG2712–CG2712, Zw5–Zw5 and CG2712–Zw5, with the latter dimer being the least stable, with an almost 40 kcal/mol difference compared to the former.
The structural results indicate that differences in amino acid sequences between Zw5 and CG2712 ZADs do not block the formation of heterodimers, but rather render it less stable through a loss of a significant number of stabilizing interactions. More stable hydrogen bonds in homodimers could also make heterodimerization less preferable.
Heterodimerizing CG15073 and Grauzone ZADs have similar dimerization interfaces
The paralogs CG15073 and Grauzone provide an opposite example to CG2712/Zw5, since the ZADs of these proteins efficiently form heterodimers, but do not interact with CG2712 (Figure 4C). These paralogs have 48% sequence homology (Figure 4A), which is comparable to the CG2712/Zw5 pair (46%). Since the crystal structure of the Grauzone ZAD is known, we analyzed the pattern of its dimerization interface and compared it with the corresponding residues in CG15073. Most of the hydrophobic residues at the dimerization interfaces of the Grauzone and CG15073 ZADs are identical or conserved (Figure 4A). The hydrogen bonds much likely are also retained (Q74 is substituted to histidine in CG15073), and no additional polar interactions stabilize Grauzone homodimers (Supplementary Table S4). Thus, in this case, homodimer seems not to have a significant advantage in energy efficiency compared to heterodimers, and heterodimers formation can also be explained by the mechanism proposed for the specificity of CG2712 and Zw5 homodimerization.
Figure 4.
Grauzone ZAD forms heterodimers with its paralog, CG15073 ZAD. (A) Sequence alignment of Grauzone and CG15073 ZAD sequences (identical residues are shown in red; conserved are in yellow). (B) Crystal structure of Grauzone dimer (PDB ID: 1PZW) colored by homology with CG15073 ZAD according to the sequence alignment shown in panel A. (C) Testing of heterodimerization ability of CG15073 and Grauzone ZADs upon co-expression followed by 6xHis-pulldown assay. Designations are as in the Figure 2D.
Mutagenesis of CG2712 and Zw5 ZADs proves energy advantages in the formation of homodimers compared to heterodimers
In order to confirm the results of in silico analysis and shift dimerization toward formation of heterodimers, we used site-directed mutagenesis. We aimed either to lower the stability of homodimers through removal of stable polar bonds or alteration of hydrophobic interactions (Tables 1 and 2, Figure 5A and B, and Supplementary Figure S12), or to fasten a possible heterodimer. The homo- and heterodimerization were studied using yeast two-hybrid and MBP-pulldown assays, while stability of dimers was assessed using a chemical cross-linking assay.
Table 2.
Summary of the impact of amino acid substitutions on homo- and heterodimerization
| Substitution | Description | Effect on homodimerization (cross-linking assay) | Effect on heterodimerization (Y2H) | Effect on heterodimerization (MBP pulldown) |
|---|---|---|---|---|
| CG2712 V16T S82M H84I | H-bond removal (H84I), alter homodimeric hydrophobic interface (V16T) | Weak | Weak | No |
| CG2712 E58S | Most stable salt bridge removal | Strong | No | Moderate |
| CG2712 L75Q S76A | H-bond removal (S76A). Alter homodimeric hydrophobic interface (L75Q) | No | No | No |
| CG2712 M86S | Alter homodimeric hydrophobic interface | Weak | Moderate | Moderate |
| Zw5 D57E Q75L A76S | engineered salt bridge (D57E) | Weak | No | No |
| Zw5 M82S | Decrease homodimeric hydrophobic interaction | No | No | No |
For details see Supplementary Figure S6.
We introduced a number of mutations. (i) E58S was introduced into CG2712 to break the most stable salt bridge with R91. (ii) To lower the free energy gain of homodimer formation, a non-conserved residue substitution (M86S) was introduced into CG2712, since this residue has significant impact on the estimated free energy of homodimer formation (Supplementary Figure S12B). A similar substitution, M82S, was introduced in Zw5, as it lowers free energy gain according to PDBePISA analysis (Supplementary Figure S12C). (iii) To fasten heterodimerization, a double mutation (L75Q/S76A) was introduced into CG2712 to remove the potential L75–Q75 clash in the heterodimer, and the reverse mutation, Q75L/A76S, was made in Zw5. From an evolutionary point of view, these substitutions are the most likely to confer specificity, since they simultaneously change interacting residues in both monomers. This also breaks a hydrogen bond between two S76 residues of the CG2712 homodimer that was shown to be highly stable according to the MD simulation (Supplementary Table S5). (iv) A triple mutation, V16T/S82M/H84I, was introduced into CG2712 to alter important polar and hydrophobic interactions in the corresponding homodimeric interface (Table 1 and Figure 5B). (v) The D57E mutation was introduced into Zw5 to allow salt-bridge formation with CG2712 in the heterodimer (Figure 5A).
The effects of all of the substitutions are summarized in Table 2. From chemical cross-linking assays, it is evident that the removal of a salt bridge in CG2712 (through the E58S mutation) had the strongest effect on homodimerization, as other substitutions did not significantly affect homodimerization of either CG2712 or Zw5 (Figure 5C) in vitro. Removal of the salt bridge increased the efficiency of heterodimerization in the MBP-pulldown assay in spite of a significantly lower expression level (Figure 5E), likely because it destabilizes both homo- and heterodimers (according to MD simulation), which results in a lower protein yield.
The most significant impact on heterodimerization was observed with the 2712M86S mutation in both the yeast two-hybrid (Figure 5D) and pulldown assays (Figure 5E). Besides E58S, M86S theoretically has the strongest influence on CG2712 homodimer stability, altering solvation free energy and making formation of heterodimers with Zw5 more energetically favorable (Supplementary Figure S6). M86S effect on removal of the potential clash of M86 (CG2712)–T16 (Zw5) seems improbable, since M86 is also absent in dv2712 (Figure 2A) and V16T has very little effect in combination with S82M/H84I substitutions.
Other substitutions did not significantly affect either the homodimerization or heterodimerization of CG2712 (Figure 5D): in the yeast two-hybrid assay, only a weak heterodimerization was observed between 2712V16TS82MH84I and Zw5 (and only when CG2712 was fused to the GAL4- activation domain, not to the DNA-binding domain), suggesting a cumulative effect of these substitutions. Since H84I substitution breaks two H-bonds, this could also slightly destabilize the homodimer, which is detectable in the two-hybrid assay.
In summary, salt-bridge removal has the strongest effect on homodimerization compared with H-bond removal or altering hydrophobic interactions. Meanwhile, heterodimerization was affected by mutations in residues involved in either hydrophobic interactions or salt-bridge formation.
Chimeric Zw5 ZAD with the α2-helix of CG2712 dimerizes with ZADs of both Zw5 and CG2712, but fails to form homodimers
Mutagenic analysis revealed the possibility of CG2712–Zw5 ZAD heterodimer formation and suggested that specificity is predominantly determined by the energetic favorability of the homodimeric form. To further validate this hypothesis, we designed chimeric ZADs, swapping the long α2-helices between CG2712 and Zw5. We predicted that such ZADs would not be able to efficiently form homodimers, since their α2-helices must interact with the zinc-coordinating module (including loops) from another protein. We investigated whether these chimeric ZADs would be able to heterodimerize with ZADs of both CG2712 and Zw5 proteins, which would indicate that there is no steric barrier.
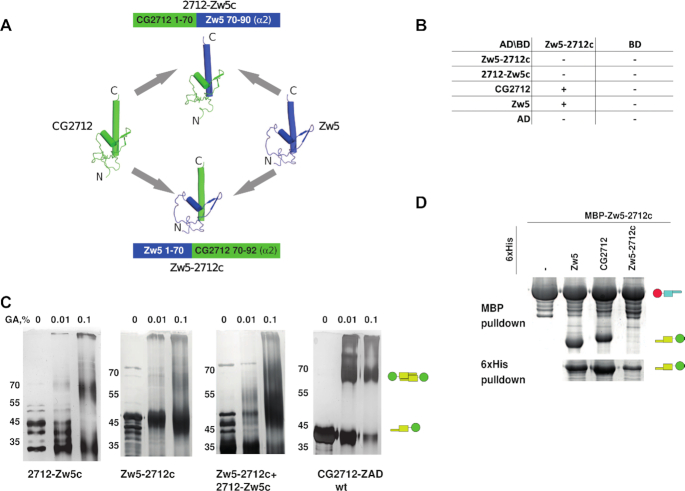
Chimeric ZADs consisting of CG2712 with its α2-helix replaced with that of Zw5 (2712-Zw5c) and vice versa (Zw5–2712c) were created (Figure 6A). Their dimerization specificity was studied using yeast two-hybrid and MBP-pulldown assays.
Figure 6.
Chimeric zinc finger-associated domain (ZAD) Zw5 with α2-helix from CG2712 can dimerize with both CG2712 and Zw5 ZADs. (A) Schematic representation of protein chimeras Zw5–2712c (Zw5 ZAD with α2-helix from CG2712 ZAD) and 2712-Zw5c (CG2712 ZAD with α2-helix from ZAD of Zw5). (B) Results of testing of the homo- and heterodimerization abilities of chimeric ZAD Zw5–2712c using a yeast two-hybrid assay. The 2712-Zw5c ZAD demonstrated strong self-activation properties only when fused to Gal4 DNA-binding domain. Designations are as in Figure 4C. (C) Testing of homodimerization ability of chimeras in a chemical crosslinking assay. (D) Results of testing of the homo- and heterodimerization abilities of chimeric ZADs Zw5–2712c and 2712–Zw5c using co-expression in bacteria followed by MBP-pulldown assay. Designations are as in the Figure 2D.
Both chimeric Zw5–2712c and 2712–Zw5c failed to form homodimers (Figure 6B and C), which is in accordance with the model of low free energy gain upon heterodimer formation between CG2712 and Zw5 and the absence of polar bonds that can stabilize this interaction. The 2712–Zw5c ZAD exhibits strong self-activation properties in yeasts when fused to GAL4 DNA-binding domain. This ZAD does not interact neither with Zw5/CG2712 nor with Zw5–2712c ZADs and is likely incorrectly folded. In accordance with the proposed model, the chimeric Zw5–2712c ZAD was able to interact with both CG2712 and Zw5 ZADs (Figure 6B and D). The fact that it can interact with both proteins further confirms that no steric hindrances prevented this interaction. Formation of homodimers by CG2712 and Zw5 is clearly more energy efficient, but since Zw5–2712c cannot form homodimers, the intermediate level of energy efficiency enforced by both polar and non-polar bonds seems to be sufficient for the detection of heterodimers between Zw5–2712c and both CG2712 and Zw5. This confirms the suggestion that dimerization specificity in ZADs is a cumulative effect of substitutions stabilizing homodimers via hydrophobic and polar interactions.
DISCUSSION
Only four zinc-coordinating cysteines and several amino acids involved in supporting the structure are conserved between Drosophila ZADs, with the most divergent ZADs in Drosophila having <18% homology (4). ZADs were classified as a subgroup of treble-clef zinc fingers, which are known to have high variation in amino acid sequence (23,51). According to this, the spatial structures of the Grauzone and CG2712 ZADs are very similar at the level of monomers and corresponding dimers, in spite of having only 25% identical residues. On the other hand, details of the dimerization interfaces of these domains differ significantly, blocking the formation of CG2712-Grauzone heterodimers and determining the specificity of their interactions. It seems a variety in primary sequence of ZAD domains, defines, from one hand, secondary structure elements, and provides, from the other hand, the versatility of conformations that can be adopted by the flexible linker regions. It affords within a commonly shared scaffold a large combinatorial space for selecting tunable residue-specific dimerization capabilities and provides an efficient and versatile platform for fast adaptations.
Our results showed that ZADs are a unique, diverse group of domains that preferentially homodimerize. We analyzed all pairs of Drosophila ZADs with homology over 45%, and heterodimerization was confirmed for only 13 pairs (16 ZADs, nearly 16% of Drosophila ZADs). We estimate that sequence similarity of 45% is a threshold for the potential ability of ZADs to heterodimerize. However, there are exceptions: some ZADs with relatively high similarity (like CG10274 and CG7386, with 61% similarity) lose the ability to heterodimerize despite being able to dimerize with another cognate proteins from the same cluster, suggesting that complex combinatorial interaction patterns could be formed. Thus, the evolutionary process directly affects the divergence of ZADs, resulting in a decrease and subsequent loss of their ability to form heterodimers.
Structure-based mutagenesis and MD simulation provided insight into the structural basis for the ability of closely related ZADs to predominantly form homodimers. We showed that ZADs of CG2712 or its paralog, Zw5, from D. melanogaster and their orthologs from D. virilis also preferentially form homodimers. However, CG2712 is unable to efficiently interact with Zw5, with which it shares 47% similarity, despite the absence of steric hindrances that could hamper the interaction between these two ZADs. Instead, most of amino acid differences accumulated in these paralogs led to stabilization of the homodimeric state through a higher free energy gain upon dimer formation, as well as through a relatively higher number of polar interactions compared to corresponding heterodimers. According to this mechanism, more evolutionary distant ZADs seem to exhibit amino acid substitutions that lead to structural restrictions preventing their heterodimerization.
The diversity of ZADs generates a large group of C2H2 proteins that can specifically homodimerize using these domains. Heterodimeric interactions appear to transiently maintain the functions of recently emerged proteins, which exhibit complementary functions. Each protein's function then adapts to new requirements and acquires dimerization specificity, which in the case of ZADs could be achieved simply by accumulation of amino acid substitutions stabilizing homodimerization. This cumulative mechanism demonstrates possibly the fastest way of developing specificity through the many amino acid substitutions acquired by rapidly evolving proteins after gene duplications.
The highest number of ZAD proteins is present in insects (many species have 90–150 ZAD proteins), while the crustaceans studied thus far have less than 10 ZAD proteins (4,5). In vertebrates, only one gene (ZNF276) has been found that encodes a protein with a domain similar to ZAD (4). Currently, only one function of ZAD is evident - the dimerization of C2H2 proteins. This function of ZADs is necessary for effective binding of ZAD-C2H2 proteins to chromatin (13) and the formation of specific long-distance genomic interactions (10). Some ZADs are likely to be involved in the transport of ZAD-C2H2 proteins from the cytoplasm to the nucleus (52).
Interestingly, some mammalian C2H2 proteins have a domain called SCAN that could perform a homodimerization function similar to ZAD. The SCAN domain is found at the N-termini of 71, 38 and 28 C2H2 proteins in human, mice, and cows, respectively (53–55). The SCAN domain consists of five α-helices that can form an antiparallel homodimer (56). In contrast to ZADs, SCAN domains have more than 80% similarity in amino acid sequence, indicating that the potential amino acid variations in these domains are limited to a few structural elements. As a result, many SCAN domains are apparently capable of forming heterodimers, although this problem is not well studied (57). Another group of mammalian C2H2 proteins contains N-terminal BTB (bric-a-brac, tramtrack and broad complex)/POZ (poxvirus and zinc finger) domain that is a conserved among higher eukaryotes (58,59). The BTB domains usually form homodimers. However, the crystal structures of the heterodimers between the BTB-containing Bcl6, NAC1 and Miz-1 proteins are known. These domains share over 60% homology and most of contacts between the heterologous BTB domains are the same as in the case of their homo-dimers (60).
Most C2H2 proteins in higher eukaryotes do not contain N-terminal dimerization domains like ZAD, SCAN or BTB. For example, the main mammalian architectural protein CTCF has long been considered to lack a homodimerization domain (61). However, it has recently been shown that CTCF from different species has an unstructured N-terminal domain that is capable of homodimerization (62). This domain was shown to be important for the activity of Drosophila CTCF (63). The dimerization domains in CTCFs from different species lack secondary structure and sequence similarity, making it impossible to identify such domains using common bioinformatics approaches. Thus, there is a possibility that unstructured domains are widely distributed at the N-terminal ends of C2H2 proteins as an alternative to structural domains such as ZAD, SCAN or BTB. Interestingly, in Drosophila, the ZAD-C2H2 protein Pita and dCTCF have similar functions in organizing the boundaries in the bithorax complex and are able to functionally replace each other (21,22). Thus, different N-terminal dimerization domains might have similar roles in the activity of architectural proteins. Further studies are necessary to clarify the functional interchangeability of different N-terminal homodimerization domains and the role of ZAD-C2H2 proteins in the organization of chromosome architecture and the regulation of transcription.
DATA AVAILABILITY
Atomic coordinates and structure factors for the reported crystal structure have been deposited with the Protein Data Bank under accession number 6FP5.
Supplementary Material
ACKNOWLEDGEMENTS
Computer modeling was carried out using equipment from the shared research facilities of the HPC computing resources at Lomonosov Moscow State University (DOI:10.14529/jsfi190201). We acknowledge the Joint Supercomputer Center of the Russian Academy of Sciences for provision of computational time and Ministry of Science and Higher Education of the Russian Federation for analysis of protein–protein interactions and the Shared-Access Equipment Centre “Industrial Biotechnology” of Research Center of Biotechnology of the Russian Academy of Sciences for MALDI experiments. We are grateful to Irina Suvorova for help with figure design.
Contributor Information
Artem Bonchuk, Department of the Control of Genetic Processes, Institute of Gene Biology, Russian Academy of Sciences, Moscow 119334, Russia; Center for Precision Genome Editing and Genetic Technologies for Biomedicine, Institute of Gene Biology, Russian Academy of Sciences, Moscow 119334, Russia.
Konstantin Boyko, Department of the Control of Genetic Processes, Institute of Gene Biology, Russian Academy of Sciences, Moscow 119334, Russia; Bach Institute of Biochemistry, Research Center of Biotechnology of the Russian Academy of Sciences, Moscow 119071, Russia.
Anna Fedotova, Department of the Control of Genetic Processes, Institute of Gene Biology, Russian Academy of Sciences, Moscow 119334, Russia.
Alena Nikolaeva, Department of the Control of Genetic Processes, Institute of Gene Biology, Russian Academy of Sciences, Moscow 119334, Russia; National Research Center «Kurchatov Institute», Moscow 123182, Russia.
Sofya Lushchekina, Emanuel Institute of Biochemical Physics, Russian Academy of Sciences, Moscow 119334, Russia.
Anastasia Khrustaleva, Department of the Bioinformatics, Institute of Gene Biology, Russian Academy of Sciences, Moscow 119334, Russia.
Vladimir Popov, Bach Institute of Biochemistry, Research Center of Biotechnology of the Russian Academy of Sciences, Moscow 119071, Russia; National Research Center «Kurchatov Institute», Moscow 123182, Russia.
Pavel Georgiev, Department of the Control of Genetic Processes, Institute of Gene Biology, Russian Academy of Sciences, Moscow 119334, Russia.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Russian Science Foundation - project 19-74-10099 (expression and purification of ZAD domains and their mutants, protein crystallization, interaction examination and structural studies); Russian Foundation for Basic Research - grant 19-03-00043 (molecular modeling); Russian Federal Space Agency (diffraction data collection); Ministry of Science and Higher Education of the Russian Federation grant 075-15-2019-1661 (analysis of protein-protein interactions). Funding for open access charge: Russian Science Foundation; Ministry of Science and Higher Education of the Russian Federation.
Conflict of interest statement. None declared.
REFERENCES
- 1. Fedotova A.A., Bonchuk A.N., Mogila V.A., Georgiev P.G.. C2H2 zinc finger proteins: the largest but poorly explored family of higher eukaryotic transcription factors. Acta Naturae. 2017; 9:47–58. [PMC free article] [PubMed] [Google Scholar]
- 2. Lambert S.A., Jolma A., Campitelli L.F., Das P.K., Yin Y., Albu M., Chen X., Taipale J., Hughes T.R., Weirauch M.T.. The human transcription factors. Cell. 2018; 172:650–665. [DOI] [PubMed] [Google Scholar]
- 3. Guo Z., Qin J., Zhou X., Zhang Y.. Insect transcription factors: a landscape of their structures and biological functions in Drosophila and beyond. Int. J. Mol. Sci. 2018; 19:3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chung H.R., Lohr U., Jackle H.. Lineage-specific expansion of the zinc finger associated domain ZAD. Mol. Biol. Evol. 2007; 24:1934–1943. [DOI] [PubMed] [Google Scholar]
- 5. Chung H.R., Schafer U., Jackle H., Bohm S.. Genomic expansion and clustering of ZAD-containing C2H2 zinc-finger genes in Drosophila. EMBO Rep. 2002; 3:1158–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maksimenko O., Bartkuhn M., Stakhov V., Herold M., Zolotarev N., Jox T., Buxa M.K., Kirsch R., Bonchuk A., Fedotova A.et al.. Two new insulator proteins, Pita and ZIPIC, target CP190 to chromatin. Genome Res. 2015; 25:89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Crozatier M., Kongsuwan K., Ferrer P., Merriam J.R., Lengyel J.A., Vincent A.. Single amino acid exchanges in separate domains of the Drosophila serendipity delta zinc finger protein cause embryonic and sex biased lethality. Genetics. 1992; 131:905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Payre F., Noselli S., Lefrere V., Vincent A.. The closely related Drosophila sry beta and sry delta zinc finger proteins show differential embryonic expression and distinct patterns of binding sites on polytene chromosomes. Development. 1990; 110:141–149. [DOI] [PubMed] [Google Scholar]
- 9. Nazario-Yepiz N.O., Riesgo-Escovar J.R.. piragua encodes a zinc finger protein required for development in Drosophila. Mech. Dev. 2017; 144:171–181. [DOI] [PubMed] [Google Scholar]
- 10. Zolotarev N., Fedotova A., Kyrchanova O., Bonchuk A., Penin A.A., Lando A.S., Eliseeva I.A., Kulakovskiy I.V., Maksimenko O., Georgiev P.. Architectural proteins Pita, Zw5,and ZIPIC contain homodimerization domain and support specific long-range interactions in Drosophila. Nucleic Acids Res. 2016; 44:7228–7241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li J., Gilmour D.S.. Distinct mechanisms of transcriptional pausing orchestrated by GAGA factor and M1BP, a novel transcription factor. EMBO J. 2013; 32:1829–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baumann D.G., Gilmour D.S.. A sequence-specific core promoter-binding transcription factor recruits TRF2 to coordinately transcribe ribosomal protein genes. Nucleic Acids Res. 2017; 45:10481–10491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Maksimenko O., Kyrchanova O., Klimenko N., Zolotarev N., Elizarova A., Bonchuk A., Georgiev P.. Small Drosophila zinc finger C2H2 protein with an N-terminal zinc finger-associated domain demonstrates the architecture functions. Biochim. Biophys. Acta Gene Regul. Mech. 2020; 1863:194446. [DOI] [PubMed] [Google Scholar]
- 14. Chen B., Harms E., Chu T., Henrion G., Strickland S.. Completion of meiosis in Drosophila oocytes requires transcriptional control by grauzone, a new zinc finger protein. Development. 2000; 127:1243–1251. [DOI] [PubMed] [Google Scholar]
- 15. Chu T., Henrion G., Haegeli V., Strickland S.. Cortex, a Drosophila gene required to complete oocyte meiosis, is a member of the Cdc20/fizzy protein family. Genesis. 2001; 29:141–152. [DOI] [PubMed] [Google Scholar]
- 16. Komura-Kawa T., Hirota K., Shimada-Niwa Y., Yamauchi R., Shimell M., Shinoda T., Fukamizu A., O’Connor M.B., Niwa R.. The drosophila zinc finger transcription factor ouija board controls ecdysteroid biosynthesis through specific regulation of spookier. PLoS Genet. 2015; 11:e1005712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Uryu O., Ou Q., Komura-Kawa T., Kamiyama T., Iga M., Syrzycka M., Hirota K., Kataoka H., Honda B.M., King-Jones K.et al.. Cooperative control of ecdysone biosynthesis in drosophila by transcription factors seance, ouija board, and molting defective. Genetics. 2018; 208:605–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Blanton J., Gaszner M., Schedl P.. Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev. 2003; 17:664–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gaszner M., Vazquez J., Schedl P.. The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes Dev. 1999; 13:2098–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kyrchanova O., Chetverina D., Maksimenko O., Kullyev A., Georgiev P.. Orientation-dependent interaction between Drosophila insulators is a property of this class of regulatory elements. Nucleic. Acids. Res. 2008; 36:7019–7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kyrchanova O., Maksimenko O., Ibragimov A., Sokolov V., Postika N., Lukyanova M., Schedl P., Georgiev P.. The insulator functions of the Drosophila polydactyl C2H2 zinc finger protein CTCF: Necessity versus sufficiency. Sci Adv. 2020; 6:eaaz3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kyrchanova O., Zolotarev N., Mogila V., Maksimenko O., Schedl P., Georgiev P.. Architectural protein Pita cooperates with dCTCF in organization of functional boundaries in Bithorax complex. Development. 2017; 144:2663–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jauch R., Bourenkov G.P., Chung H.R., Urlaub H., Reidt U., Jackle H., Wahl M.C.. The zinc finger-associated domain of the Drosophila transcription factor grauzone is a novel zinc-coordinating protein-protein interaction module. Structure. 2003; 11:1393–1402. [DOI] [PubMed] [Google Scholar]
- 24. Gibert J.M., Marcellini S., David J.R., Schlotterer C., Simpson P.. A major bristle QTL from a selected population of Drosophila uncovers the zinc-finger transcription factor poils-au-dos, a repressor of achaete-scute. Dev. Biol. 2005; 288:194–205. [DOI] [PubMed] [Google Scholar]
- 25. Chen L.Y., Wang J.C., Hyvert Y., Lin H.P., Perrimon N., Imler J.L., Hsu J.C.. Weckle is a zinc finger adaptor of the toll pathway in dorsoventral patterning of the Drosophila embryo. Curr. Biol. 2006; 16:1183–1193. [DOI] [PubMed] [Google Scholar]
- 26. Thompson J.D., Gibson T.J., Higgins D.G.. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics. 2002; Chapter 2:Unit 2 3. [DOI] [PubMed] [Google Scholar]
- 27. Kumar S., Stecher G., Li M., Knyaz C., Tamura K.. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018; 35:1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schwartz R.M., Dayhoff M.O.. Protein and nucleic Acid sequence data and phylogeny. Science. 1979; 205:1038–1039. [DOI] [PubMed] [Google Scholar]
- 29. Yu G. Using ggtree to visualize data on tree-like structures. Curr. Protoc. Bioinformatics. 2020; 69:e96. [DOI] [PubMed] [Google Scholar]
- 30. Crooks G.E., Hon G., Chandonia J.M., Brenner S.E.. WebLogo: a sequence logo generator. Genome Res. 2004; 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Battye T.G., Kontogiannis L., Johnson O., Powell H.R., Leslie A.G.. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D. Biol. Crystallogr. 2011; 67:271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Padilla J.E., Yeates T.O.. A statistic for local intensity differences: robustness to anisotropy and pseudo-centering and utility for detecting twinning. Acta Crystallogr. D. Biol. Crystallogr. 2003; 59:1124–1130. [DOI] [PubMed] [Google Scholar]
- 33. Evans P. Scaling and assessment of data quality. Acta Crystallogr. D. Biol. Crystallogr. 2006; 62:72–82. [DOI] [PubMed] [Google Scholar]
- 34. Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C.. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D. Biol. Crystallogr. 2002; 58:1948–1954. [DOI] [PubMed] [Google Scholar]
- 35. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cowtan K. Recent developments in classical density modification. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D. Biol. Crystallogr. 2006; 62:1002–1011. [DOI] [PubMed] [Google Scholar]
- 38. Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A.et al.. Overview of the CCP4 suite and current developments. Acta Crystallogr. D. Biol. Crystallogr. 2011; 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 41. Krissinel E., Henrick K.. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D. Biol. Crystallogr. 2004; 60:2256–2268. [DOI] [PubMed] [Google Scholar]
- 42. Krissinel E., Henrick K.. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007; 372:774–797. [DOI] [PubMed] [Google Scholar]
- 43. Hooft R.W., Sander C., Vriend G.. Positioning hydrogen atoms by optimizing hydrogen-bond networks in protein structures. Proteins. 1996; 26:363–376. [DOI] [PubMed] [Google Scholar]
- 44. Laskowski R.A., Swindells M.B.. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011; 51:2778–2786. [DOI] [PubMed] [Google Scholar]
- 45. Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kalé L., Schulten K.. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005; 26:1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Best R.B., Zhu X., Shim J., Lopes P.E., Mittal J., Feig M., Mackerell A.D. Jr.. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. J. Chem. Theory Comput. 2012; 8:3257–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Humphrey W., Dalke A., Schulten K.. VMD: visual molecular dynamics. J. Mol. Graph. 1996; 14:33–38. [DOI] [PubMed] [Google Scholar]
- 48. Webb B., Sali A.. Comparative protein structure modeling using MODELLER. Curr Protoc Protein Sci. 2016; 54:5.6.1–5.6.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lyskov S., Gray J.J.. The RosettaDock server for local protein-protein docking. Nucleic Acids Res. 2008; 36:W233–W238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kosiol C., Goldman N.. Different versions of the Dayhoff rate matrix. Mol. Biol. Evol. 2005; 22:193–199. [DOI] [PubMed] [Google Scholar]
- 51. Grishin N.V. Treble clef finger–a functionally diverse zinc-binding structural motif. Nucleic Acids Res. 2001; 29:1703–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zolotarev N.A., Maksimenko O.G., Georgiev P.G., Bonchuk A.N.. ZAD-domain is essential for nuclear localization of insulator proteins in Drosophila melanogaster. Acta Naturae. 2016; 8:97–102. [PMC free article] [PubMed] [Google Scholar]
- 53. Sander T.L., Haas A.L., Peterson M.J., Morris J.F.. Identification of a novel SCAN box-related protein that interacts with MZF1B. The leucine-rich SCAN box mediates hetero- and homoprotein associations. J. Biol. Chem. 2000; 275:12857–12867. [DOI] [PubMed] [Google Scholar]
- 54. Thomas J.H., Emerson R.O., Shendure J.. Extraordinary molecular evolution in the PRDM9 fertility gene. PLoS One. 2009; 4:e8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sander T.L., Stringer K.F., Maki J.L., Szauter P., Stone J.R., Collins T.. The SCAN domain defines a large family of zinc finger transcription factors. Gene. 2003; 310:29–38. [DOI] [PubMed] [Google Scholar]
- 56. Liang Y., Choo S.H., Rossbach M., Baburajendran N., Palasingam P., Kolatkar P.R.. Crystal optimization and preliminary diffraction data analysis of the SCAN domain of Zfp206. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2012; 68:443–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Liang Y., Huimei Hong F., Ganesan P., Jiang S., Jauch R., Stanton L.W., Kolatkar P.R.. Structural analysis and dimerization profile of the SCAN domain of the pluripotency factor Zfp206. Nucleic Acids Res. 2012; 40:8721–8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Stogios P.J., Downs G.S., Jauhal J.J., Nandra S.K., Prive G.G.. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005; 6:R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ahmad K.F., Engel C.K., Prive G.G.. Crystal structure of the BTB domain from PLZF. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:12123–12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stead M.A., Wright S.C.. Structures of heterodimeric POZ domains of Miz1/BCL6 and Miz1/NAC1. Acta Crystallogr. F Struct. Biol. Commun. 2014; 70:1591–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Arzate-Mejia R.G., Recillas-Targa F., Corces V.G.. Developing in 3D: the role of CTCF in cell differentiation. Development. 2018; 145:dev137729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bonchuk A., Kamalyan S., Mariasina S., Boyko K., Popov V., Maksimenko O., Georgiev P.. N-terminal domain of the architectural protein CTCF has similar structural organization and ability to self-association in bilaterian organisms. Sci. Rep. 2020; 10:2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bonchuk A., Maksimenko O., Kyrchanova O., Ivlieva T., Mogila V., Deshpande G., Wolle D., Schedl P., Georgiev P.. Functional role of dimerization and CP190 interacting domains of CTCF protein in Drosophila melanogaster. BMC Biol. 2015; 13:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the reported crystal structure have been deposited with the Protein Data Bank under accession number 6FP5.