Personalized cancer therapy matches the plan of treatment with specific molecular alterations. This is the result of pivotal advances in our knowledge of tumor biology focused on the mechanisms underpinning uncontrolled tumor growth, evasion from host immune—surveillance, cell survival, migration, and the ever-increasing number of available targeted and immune therapies.
Nowadays, next-generation sequencing (NGS) is widely used for the molecular profiling of solid tumors. Over the past few years, this multigene approach has proved to be winning over a single gene sequential strategy. In many academic centers, NGS can offer a comprehensive genomic profile (CGP) to identify all four classes of alterations (base-pair substitutions, copy number variations, insertions/deletions, and rearrangements) in hundreds of genes. The NGS approach is particularly useful in NSCLC, where the number of actionable gene alterations is high [1,2] and the available histological or cytological samples can be scarce [3,4]. The National Comprehensive Cancer Network (NCCN) guidelines recommended broad molecular profiling in metastatic NSCLC, including BRAF, ERBB2 (HER2), MET, RET, NTRK and ROS1 in addition to EGFR and ALK analysis [5]. Moreover, predictive biomarkers of response to immunotherapy are essential, beyond testing for tumor programmed death-ligand 1 (PD-L1) expression [6]. Recently, tumor mutational burden (TMB), defined as the total number of mutations per coding area of the tumor genome, has been evaluated in different solid tumors [7]. However, TMB has technical and clinical limitations, and despite an initial success from clinical trials, its role as a biomarker for immunotherapy is still uncertain [8,9].
In this continuously evolving perspective, the implementation of large NGS panels is the most pragmatic way to investigate clinically useful molecular markers simultaneously. CGP can detect concurrent or uncommon alterations in frequently mutated genes, reflecting tumor genomic heterogeneity and complexity. Studies from different centers reported successful implementation of CGP [10,11,12]. In the Netherlands, the DRUP study (NCT02925234) and the TAPUR study in the United States (NCT 0269353) evaluated the outcome of patients treated with targeted therapies based on specific molecular alterations. The TAPUR study stemmed from the recognition that the rapid dissemination of genomic profiling offers an opportunity to learn from the application of precision cancer medicine in practice providing a framework for clinical decision support [13]. Indeed, CGP may produce a volume of molecular data that is tricky to manage and difficult to transpose into clinical indications. Clinical decision-making can be particularly complex for patients with advanced NSCLC, considering the number of actionable molecular aberrations and available targeted therapies. Different thorny questions could emerge: how to face tumor heterogeneity, which alterations might induce tumor resistance, or which mutation is the driver genomic aberration to be targeted first.
In this scenario, a multidisciplinary molecular tumor board (MTB) represents a valuable tool to support patient’s physicians to foster precision medicine strategies. MTB is a dynamic team, gathering professionals from different specializations, including oncologists, pathologists, molecular biologists, geneticists, radiotherapists, surgeons, translational scientists, pharmacologists, and bioinformaticians. In these meetings, all the information regarding the patient’s history, imaging, histopathology, laboratory results, and CGP are summarized and reviewed. CGP results can also be evaluated by experts using cancer-related databases [14,15,16]. However, the large-scale sequencing of tumors and the exponential growth of bioinformatics data make their interpretation complicated, prefiguring a gap between the knowledge and the implications of genomics in therapeutic planning. Moreover, more than three million peer-reviewed publications are related to cancer and more than 250 US FDA-approved oncology drugs make keeping up to date an insurmountable task.
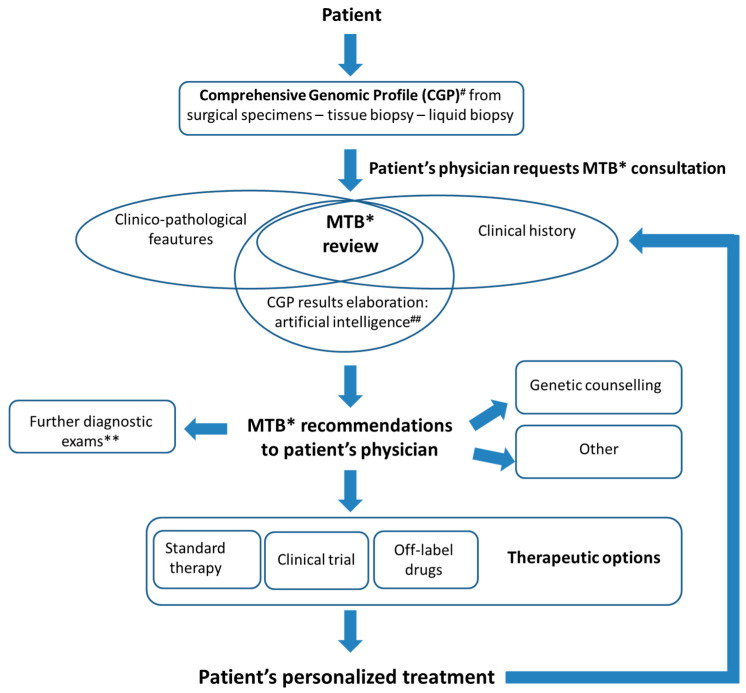
To overcome these gaps, dedicated artificial intelligence (AI) tools are necessary [17]. AI offers a review of relevant data and evidence, matches patients’ clinical and molecular data to clinical trials avoiding a manual screening that can result in a low percentage of eligibility [18]. These systems allow a strict collaboration among specialists and increases workflow efficiency guaranteeing a comprehensive patient overview and tracking MTB decisions and follow-up. In addition, a cloud-based virtual molecular tumor board (VMTB) platform has been recently developed, with scoring models for ranking cancer treatments, allowing specialists to discuss selected cases in a time- and location-independent way [19]. Moreover, a molecular tumor board portal (MTBP) with automated NGS data interpretation and reporting, and virtual meeting, has been introduced for unifying the CGP result management across seven European cancer centers within the Cancer Core Europe (CCE) network [20]. With these supports, MTB can critically discuss the results exposed by a coordinator/navigator with the final aim to find the best match among clinico-pathological characteristics, genomic alterations and available targeted therapies. Other issues to be addressed could be the need for additional medical examinations or recommendations of genetic counseling (Figure 1).
Figure 1.
The Molecular Tumor Board (MTB) at European Institute of Oncology: the way to precision medicine. The MTB plays a pivotal role to integrate patient’s clinico-pathological features with tumor molecular data derived from comprehensive genomic profiling, taking advantage of artificial intelligence systems. The main goal is to offer to patient’s physician recommendations regarding the best—patient-tailored—therapeutic options. # Comprehensive genomic profiling performed with Oncomine Comprehensive Assay v.3 (Thermo Fisher Scientific, Waltham, MA, USA) or FoundationOne CDX (Foundation medicine—Roche) [31]. ## Artificial intelligence support: Navify (Roche Diagnostics). * MTB: Molecular Tumor Board. Multidisciplinary team of specialists including oncologists, surgeons, pathologists, biologists, bioinformatics experts, geneticists, basic and translational scientists, radiologists. ** Diagnostic exams including radiologic exam, re-biopsy, histopathological revision or additional molecular tests.
The implementation of MTB has been recently reported as a valuable clinical tool in different scenarios [21,22,23,24,25,26]. In particular, according to the 4-year experience of West Cancer Center and Research Institute (Memphis, TN, USA), MTB expressed 837 recommendations, including the administration of standard therapy (37%), clinical trial participation (31%) and off-label therapy use (10%) [21]. In a timeframe of 3 years, the MTB at the Institute Curie in Paris (France) enabled the inclusion of 10% of patients into a clinical trial with matched therapy [23]. According to the Johns Hopkins-MTB recommendations (Baltimore, Maryland, United States), 43% of patients receiving a genomically matched therapy derived a clinical benefit lasting at least 6 months [25]. The University Medical Center in Groningen (the Netherlands) reported that MTB indications resulted in a positive clinical response in the majority (81%) of patients affected by metastatic NSCLC with rare or complex mutational profiles, achieving an objective response rate of 67%, with a median progression-free survival of 6.3 months and overall survival of 10.4 months [22]. However, drug accessibility may represent a crucial barrier, thus, clinical and hospital pharmacists should be involved in MTB to facilitate drug administration, even in expanded access programs or in compassionate use. Indeed, only by receiving personalized treatments can the improvement of patient survival associated with the CGP approach be achieved [27,28,29].
In conclusion, objective data are consistent with the indication that patients with advanced/metastatic disease often have distinct molecular alterations that may require matched treatments rather than standard therapies derived from non-biomarker-based populations [29,30]. In this setting, molecular tumor boards can be valuable tools to support a patient’s tailored treatment.
Funding
This research received no external funding.
Conflicts of Interest
E.G.-R. received honoraria from Roche; C.F. and M.B. declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ettinger D.S., Aisner D.L., Wood D.E., Akerley W., Bauman J., Chang J.Y., Chirieac L.R., D’Amico T.A., Dilling T.J., Dobelbower M., et al. NCCN Guidelines Insights: Non-Small Cell Lung Cancer, Version 5. 2018. Natl. Compr. Canc. Netw. 2018;16:807–821. doi: 10.6004/jnccn.2018.0062. [DOI] [PubMed] [Google Scholar]
- 2.Kalemkerian G.P., Narula N., Kennedy E.B., Biermann W.A., Donington J., Leighl N.B., Lew M., Pantelas J., Ramalingam S.S., Reck M., et al. Molecular Testing Guideline for the Selection of Patients With Lung Cancer for Treatment With Targeted Tyrosine Kinase Inhibitors: American Society of Clinical Oncology Endorsement of the College of American Pathologists/International Association for the Study of Lung Cancer/Association for Molecular Pathology Clinical Practice Guideline Update. J. Clin. Oncol. 2018;36:911–919. doi: 10.1200/JCO.2017.76.7293. [DOI] [PubMed] [Google Scholar]
- 3.Stoy S.P., Segal J.P., Mueller J., Furtado L.V., Vokes E.E., Patel J.D., Murgu S. Feasibility of Endobronchial Ultrasound-guided Transbronchial Needle Aspiration Cytology Specimens for Next Generation Sequencing in Non-small-cell Lung Cancer. Clin. Lung Cancer. 2018;19:230–238. doi: 10.1016/j.cllc.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 4.Fumagalli C., Casadio C., Barberis M., Guarize J., Guerini-Rocco E. Letter to the Editor. Clin. Lung Cancer. 2018;19:e439–e440. doi: 10.1016/j.cllc.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 5.NCCN Clinical Practise Guidelines in Oncology (NCCN Guidelines) [(accessed on 30 December 2020)]; Available online: www2.tri-kobe.org/nccn/guideline/lung/english/non_small.pdf.
- 6.Bodor J.N., Boumber Y., Borghaei H. Biomarkers for Immune Checkpoint Inhibition in Non–Small Cell Lung Cancer (NSCLC) Cancer. 2020;126:260–270. doi: 10.1002/cncr.32468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yarchoan M., Hopkins A., Jafee E.M. Tumor mutational burden and response to PD-1 inhibition. N. Engl. J. Med. 2017;377:2500. doi: 10.1056/NEJMc1713444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hellmann M.D., Ciuleanu T.E., Pluzanski A., Lee G.A., Otterson C., Audigier-Valette E., Minenza H., Linardou S., Burgers P., Salman H., et al. Nivolumab plus ipili-mumab in lung cancer with a high tumor mutational burden. N. Engl. J. Med. 2018;378:2093–2104. doi: 10.1056/NEJMoa1801946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hellmann M.D., Rizvi N.A., Goldman J.W., Gettinger S.N., Borghaei H., Brahmer J.R., Ready N.E., Gerber D.E., Chow L.Q., Juergens R.A., et al. Nivolumab plus ipilimumab as first-line treatment for advanced non-small-cell lung cancer (CheckMate 012): Results of an open-label, phase 1, multicohort study. Lancet Oncol. 2017;18:31–41. doi: 10.1016/S1470-2045(16)30624-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nesline M.K., DePietro P., Dy G.K., Early A., Papanicolau-Sengos A., Conroy J.M., Lenzo F.L., Glenn S.T., Chen H., Grand’Maison A., et al. Oncologist uptake of comprehensive genomic profile guided targeted therapy. Oncotarget. 2019;10:4616–4629. doi: 10.18632/oncotarget.27047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsimberidou A.M., Hong D.S., Ye Y., Cartwright C., Wheler J.J., Falchook G.S., Naing A., Fu S., Piha-Paul S., Janku F., et al. Initiative for Molecular Profiling and Advanced Cancer Therapy (IMPACT): An MD Anderson Precision Medicine Study. JCO Precis. Oncol. 2017;1:1–18. doi: 10.1200/PO.17.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh A.P., Shum E., Rajdev L., Cheng H., Goel S., Perez-Soler R., Halmos B. Impact and diagnostic gaps of comprehensive genomic profiling in real-world clinical practice. Cancers. 2020;12:1156. doi: 10.3390/cancers12051156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mangat P.K., Halabi S., Bruinooge S.S., GarrettMayer E., Alva A., Janeway K.A., Stella P.J., Voest E., Yost K.J., Perlmutter J., et al. Rationale and Design of the Targeted Agent and Profiling Utilization Registry (TAPUR) Study. JCO Precis. Oncol. 2018;2:1–14. doi: 10.1200/PO.18.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tate J.G., Bamford S., Jubb H.C., Bamford S., Jubb H.C., Sondka Z., Beare D.M., Bindal N., Boutselakis H., Cole C.G., et al. COSMIC: The catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47:D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor A.D., Micheel C.M., Anderson I.A., Levy M.A., Lovly C.M. The path(way) less traveled: A pathway-oriented approach to providing information about precision cancer medicine on my cancer genome. Transl. Oncol. 2016;9:163–165. doi: 10.1016/j.tranon.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chakravarty D., Gao J., Phillips S.M., Kundra R., Zhang H., Wang J., Rudolph J.E., Yaeger R., Soumerai T., Nissan M.H., et al. OncoKB: A precision oncology knowledge base. JCO Precis. Oncol. 2017;1:1–16. doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perakis S.O., Weber S., Zhou Q., Graf R., Hojas S., Riedl J.M., Gerger A., Dandachi N., Balic M., Hoefler G., et al. Comparison of three commercial decision support platforms for matching of next-generation sequencing results with therapies in patients with cancer. ESMO Open. 2020;5:e000872. doi: 10.1136/esmoopen-2020-000872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The Role of AI with Tumor Boards. [(accessed on 30 December 2020)]; Available online: https://www.idigitalhealth.com/news/the-role-of-ai-with-tumor-boards.
- 19.Pishvaian M.J., Blais E.M., Bender R.J., Rao S., Boca S.M., Chung V., Hendifar A., Mikhail S., Sohal D., Pohlmann P., et al. A virtual molecular tumor board to improve efficiency and scalability of delivering precision oncology to physicians and their patients. JAMIA Open. 2019;2:505–515. doi: 10.1093/jamiaopen/ooz045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamborero D., Dienstmann R., Rachid M.H. Support systems to guide clinical decision-making in precision oncology: The Cancer Core Europe Molecular Tumor Board Portal. Nat. Med. 2020;26:992–994. doi: 10.1038/s41591-020-0969-2. [DOI] [PubMed] [Google Scholar]
- 21.VanderWalde A., Grothey A., Vaena D., Vidal G., ElNaggar A., Bufalino G., Schwartzberg L. Establishment of a Molecular Tumor Board (MTB) and Uptake of Recommendations in a Community Setting. J. Pers. Med. 2020;10:252. doi: 10.3390/jpm10040252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koopman B., van der Wekken A.J., ter Elst A., Hiltermann T.J.N., Vilacha J.F., Groves M.R., van den Berg A., Hiddinga B.I., Hijmering-Kappelle L.B.M., Stigt J.A., et al. Relevance and effectiveness of molecular tumor board recommendations for patients with non–small-cell lung cancer with rare or complex mutational profiles. JCO Precis. Oncol. 2020;4:393–410. doi: 10.1200/PO.20.00008. [DOI] [PubMed] [Google Scholar]
- 23.Basse C., Morel C., Alt M., Sablin M.P., Franck C., Pierron G., Callens C., Melaabi S., Masliah-Planchon J., Bataillon G., et al. Relevance of a molecular tumour board (MTB) for patients’ enrolment in clinical trials: Experience of the Institut Curie. ESMO Open. 2018;3:e000339. doi: 10.1136/esmoopen-2018-000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koopman B., Groen H.J.M., Ligtenberg M.J.L., Grunberg K., Monkhorst K., De langen A.J., Boelens M.C., Paats M.S., von der Thusen J.H., Dinjens N.M. Multicenter Comparison of Molecular Tumor Boards in The Netherlands: Definition, Composition, Methods, and Targeted Therapy Recommendations. Oncologist. 2020;25:1–12. doi: 10.1002/onco.13580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dalton W.B., Forde P.M., Kang H., Connolly R.M., Stearns V., Gocke C.D., Eshleman J.R., Axilbund J., Petry D., Geoghegan C., et al. Personalized Medicine in the Oncology Clinic: Implementation and Outcomes of the Johns Hopkins Molecular Tumor Board. JCO Precis. Oncol. 2017;1:1–19. doi: 10.1200/PO.16.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rolfo C., Manca P., Salgado R., Van Dam P., Dendooven A., Coelho A.M., Gandia J.F., Rutten A., Lybaert W., Vermeij J., et al. Multidisciplinary molecular tumour board: A tool to improve clinical practice and selection accrual for clinical trials in patients with cancer. ESMO Open. 2018;3:e000398. doi: 10.1136/esmoopen-2018-000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Normanno N., Barberis M., De Marinis F., Gridelli C. On The Behalf Of The Aiot Expert Panel. Molecular and Genomic Profiling of Lung Cancer in the Era of Precision Medicine: A Position Paper from the Italian Association of Thoracic Oncology (AIOT) Cancers. 2020;12:1627. doi: 10.3390/cancers12061627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fumagalli C., Guerini-Rocco E., Barberis M. Broad-based genomic sequencing in advanced non-small cell lung cancer in the dock. Transl. Lung Cancer Res. 2019;8:S360–S363. doi: 10.21037/tlcr.2019.04.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato S., Kim K.H., Lim H.J., Boichard A., Nikanjam M., Weihe E., Kuo D.J., Eskander R.N., Goodman A., Galanina N., et al. Real-world data from a molecular tumor board demonstrates improved outcomes with a precision N-of-One strategy. Nat. Commun. 2020;11:4965. doi: 10.1038/s41467-020-18613-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kato S., Kurasaki K., Ikeda S., Kurzrock R. Rare tumor clinic: The University of California San Diego Moores Cancer Center experience with a precision therapy approach. Oncologist. 2018;23:171–178. doi: 10.1634/theoncologist.2017-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.FoundationOneCDX Genomic Testing. [(accessed on 30 December 2020)]; Available online: https://www.foundationmedicine.com/genomictesting/foundation-one-cdx.