Abstract
Among the various types of breast cancer, the luminal B subtype is the most common in young women, and ESR1-CCDC170 (E:C) fusion is the most frequent oncogenic fusion driver of the luminal B subtype. Nevertheless, treatments targeting E:C fusion has not been well established yet. Hence, the aim of this study is to investigate potential therapies targeting E:C fusion based on systematic bioinformatical analysis of the Cancer Genome Atlas (TCGA) data. One thousand related genes were extracted using transcriptome analysis, and major signaling pathways associated with breast cancer were identified with over-representation analysis. Then, we conducted drug-target network analysis based on the OncoKB and CIViC databases, and finally selected potentially applicable drug candidates. Six major cancer-related signaling pathways (p53, ATR/ATM, FOXM1, hedgehog, cell cycle, and Aurora B) were significantly altered in E:C fusion-positive cases of breast cancer. Further investigation revealed that nine genes (AURKB, HDAC2, PLK1, CENPA, CHEK1, CHEK2, RB1, CCNA2, and MDM2) in coordination with E:C fusion were found to be common denominators in three or more of these pathways, thereby making them promising gene biomarkers for target therapy. Among the 21 putative actionable drugs inferred by drug-target network analysis, palbociclib, alpelisib, ribociclib, dexamethasone, checkpoint kinase inhibitor AXD 7762, irinotecan, milademetan tosylate, R05045337, cisplatin, prexasertib, and olaparib were considered promising drug candidates targeting genes involved in at least two E:C fusion-related pathways.
Keywords: ESR1, CCDC170, breast cancer, gene fusion, TCGA, bioinformatics, drug repositioning
1. Introduction
Breast cancer, aside from skin cancer, is the most commonly diagnosed cancer in women worldwide [1]. Recent statistics report the emergence of 250,000 new cases of breast cancer solely in 2017, contributing to the 12% of women diagnosed with breast cancer in the United States [2]. Molecular classification divides breast cancer into four major classes: luminal A, luminal B, human epidermal growth factor receptor 2 (HER2)-enriched (HER2-E), and basal-like subtype [2]. Among them, luminal B remains to be the most common subtype in young women, accounting for 15%–20% of the total breast cancer cases, and within luminal B, ESR1-CCDC170 fusion-positive subtype, constituting 6% to 8% of the luminal B class, persists to be the most dominant subtype [3,4,5,6,7,8,9].
ESR1-CCDC170 fusion causing chimeric mRNA is known to be formed by a tandem duplication at the 6q25.1 location on a coiled-coil domain containing 170 (CCDC170) adjacent to the ESR1 gene [8,10]. It has been reported that the polymorphism of the CCDC170 gene correlates with breast cancer susceptibility [11,12]. ESR1-CCDC170 fusion-positive cancers treated with endocrine therapy showed reduced treatment efficiency in mouse models [13]. Although its effect has been studied in relation to ovarian cancer, the molecular signaling involved in the induction of ESR1-CCDC170 fusion-positive breast cancer has yet to be elucidated [14].
Here, we systematically analyzed the molecular pathological features of ESR1-CCDC170 fusion-positive breast cancer through the data analysis of the Cancer Genome Atlas (TCGA) and identified the activated oncogenic pathways. In addition, putative target genes and actionable drugs were inferred and prioritized by performing network analysis using both transcriptomic signatures and drug-target databases, such as OncoKB and CIViC.
2. Experimental Section
2.1. Sample Acquisition and Quality Control
Gene level 3 (RNA-seq by expectation-maximization, RSEM) mRNA expression with normalized read count values of TCGA breast cancer carcinoma (BRCA) was obtained from the Broad GDAC Firehose website (https://gdac.broadinstitute.org). Related clinical feature data, including information about the samples’ mutation annotation format (MAF) files, molecular subtypes, and tumor-node-metastasis (TNM) stages, were obtained from the website mentioned above.
2.2. Case-Control Selection
A previous study confirmed 319 fusion genes in TCGA clinical breast cancer tumors [15]. Unlike other in-frame fusion genes, ESR1-CCDC170 is known as a breast cancer-specific oncogenic fusion gene. Using the TCGA fusion gene data portal (Jackson Laboratory, https://www.tumorfusions.org), we identified 11 samples of CCDC170 fusion, which were cross-checked with increased CCDC170 expression level. Furthermore, only tumor samples that had the barcode 01A (primary solid tumor) were selectively chosen by disregarding other types of tumor samples, 11A (normal) or 06A (metastasized). Among the remaining samples, 50 samples with the highest expression of CCDC170 were confirmed as upregulated controls for analyzing the network within the noncoding region of the fusion gene. From the upregulated control samples, 2 outlier samples were filtered out using the IQR (interquartile range) method. The same number of controls (n = 48) with the lowest expression of CCDC170 were then selected from the remaining samples.
2.3. Selection of Genes Affected by ESR1-CCDC170 Fusion
RNA expression data from TCGA were made into a two-dimensional matrix composed of the selected 11 fusion samples and 48 control samples. Each column represents the patient ID, while each row represents the gene name. Based on the RNA expression matrix, variance tests were conducted using independent two-sample t-tests. To select genes in coordination with CCDC170 in the RNA expression, t-tests were performed between E:C fusion-positive and fusion-negative cases. Mostly affected 1000 genes were selected (adjust p-value < 2.0 × 10−8).
2.4. Pathway Analysis via ConsensusPathDB (CPDB) and Over-Representation
The selected 1000 genes that correlated to the reference gene (CCDC170) from the aforementioned RNA expression data were used to perform over-representation analysis (ORA) via ConsensusPathDB (CPDB, http://cpdb.molgen.mpg.de/CPDB 11th September 2020). We inputted a gene list with the option of Entrez Gene using pathway-based sets with a minimum overlap input list (n = 2) and p-value cutoff (p-value < 0.01). A total of 113 biological pathways were merged and curated by CPDB from the following sources, according to data from BioCarta (https://maayanlab.cloud/Harmonizome/dataset/Biocarta+Pathways), INOH [16], KEGG [17], NetPath [18], PID [19], Reactome [20], and WikiPathways [21]. In consideration of the ontological characteristics and the proportion of duplicated genes, the pathways, which were enriched with selected 1000 genes (q-value < 0.05), were condensed into 15 cancer-related pathways, and their components were 184 genes.
2.5. Druggable Pathway Analysis via CIViC and OncoKB
The “Clinical Evidence Summaries” data, released on 1 October 2017, were downloaded from the Clinical Interpretations of Variants in Cancer (CIViC) website (https://civic.genome.wustl.edu/releases), and the “Actionable Variants” data were accessed and downloaded on 17 October 2017 from the OncoKB website (http://oncokb.org/). A total of 673 CIViC variants (181 genes) with expected therapy efficacy in 148 OncoKB actionable variants (53 genes) were integrated. A total of 113 CCDC170-correlated genes were matched to the CIViC and OncoKB variants.
2.6. Statistical Analysis and Data Visualization
The open software R version 3.4.3 was used to process all statistical analyses for selecting genes correlated to CCDC170, including the variance test and independent two-sample t-test. An RNA expression heatmap was also visualized using ComplexHeatmap, a package for R. A KEGG mapper (https://www.genome.jp/kegg/mapper.html 11th September 2020) was used to visualize target pathways related to DNA damage response. Cytoscape version 3.5.3 was used to analyze and express the complex network between targetable drugs and therapeutic agents. Our study defined statistical significance with a p-value of < 0.05 and false detection rate (FDR) with a q-value of <0.001.
3. Results
3.1. Clinico-Pathological Characteristics
We checked the clinico-pathological characteristics of 11 ESR1-CCDC170 fusion-positive and 48 fusion-negative patients among 1095 breast cancer patients in Broad GDAC Firehose (Figure 1, Table 1). Two significant differences were identified between fusion-positive and negative patients.
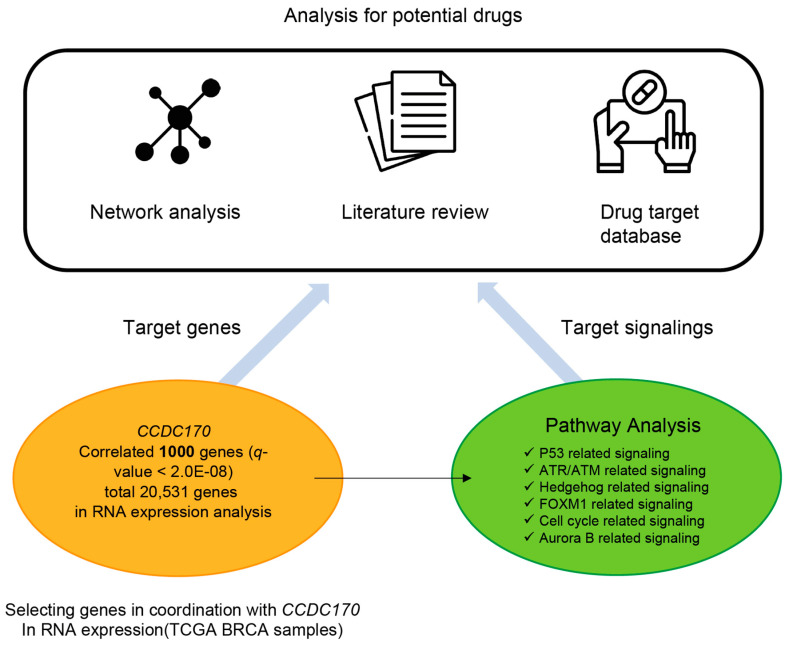
Figure 1.
Overall schematics. Transcriptome data for breast cancer (BRCA) were obtained from the Broad GDAC Firehose database. Following the RNA measurement analysis of a total of 20,531 genes, 1000 genes correlated with CCDC170 were selected (q < 2.0 × 10−8). Over-representation analysis of the 1000 genes demonstrated a significant relationship with six major cancer-related pathways (p53, ATR/ARM, hedgehog, FOXM1, cell cycle, and Aurora B). Potential gene targets and drug candidates were isolated via drug network analysis using a drug-target database on genes correlated to CCDC170 and the literature review.
Table 1.
Comparisons in clinical and pathological characteristics of ESR1-CCDC170 fusion-positive and negative BRCA patients and control cohorts. The clinical and pathological characteristics between ESR1-CCDC170 fusion-positive, negative BRCA, and control cohorts were compared.
| Total (n = 1095) |
Control (n = 48) |
Fusion (n = 11) |
p-Values | |
|---|---|---|---|---|
| age | 46–72 | 44–68 | 49–71 | 0.001 |
| sex | ||||
| - female | 1082 (98.8%) | 48 (100.0%) | 11 (100.0%) | |
| Vital status | 1 | |||
| - alive | 991 (90.5%) | 42 (87.5%) | 10 (90.9%) | |
| - dead | 104 (9.5%) | 6 (13.6%) | 1 (9.1%) | |
| stage | NS | |||
| - stage I | 90 (8.3%) | 2 (4.2%) | 1 (9.1%) | |
| - stage Ia | 85 (7.8%) | 3 (6.2%) | 1 (9.1%) | |
| - stage Ib | 6 (0.6%) | 0 (0.0%) | 0 (0.0%) | |
| - stage II | 6 (0.6%) | 0 (0.0%) | 0 (0.0%) | |
| - stage IIa | 359 (33.0%) | 24 (50.0%) | 3 (27.3%) | |
| - stage IIb | 257 (23.6%) | 9 (18.8%) | 3 (27.3%) | |
| - stage III | 2 (0.2%) | 0 (0.0%) | 0 (0.0%) | |
| - stage IIIa | 156 (14.4%) | 4 (8.3%) | 2 (18.2%) | |
| - stage IIIb | 27 (2.5%) | 0 (0.0%) | 0 (0.0%) | |
| - stage IIIc | 65 (6.0%) | 3 (6.2%) | 1 (9.1%) | |
| - stage Iv | 20 (1.8%) | 2 (4.2%) | 0 (0.0%) | |
| - stage x | 14 (1.3%) | 1 (2.1%) | 0 (0.0%) | |
| ER status | 0 | |||
| - positive | 808 (77.2%) | 10 (20.8%) | 10 (90.9%) | |
| - negative | 237 (22.5%) | 38 (79.2%) | 0 (0.0%) | |
| - indeterminate | 2 (0.2%) | 0 (0.0%) | 1 (9.1%) | |
| PR status | 0 | |||
| - positive | 700 (66.9%) | 2 (4.3%) | 7 (63.6%) | |
| - negative | 342 (32.7%) | 45 (95.7%) | 4 (36.4%) | |
| - indeterminate | 4 (0.4%) | 0 (0.0%) | 0 (0.0%) | |
| HER2 IHC | 0 | |||
| 0 | 61 (9.8%) | 7 (21.9%) | 0 (0.0%) | |
| - 1+ | 270 (43.4%) | 12 (37.5%) | 4 (44.4%) | |
| - 2+ | 199 (32.1%) | 10 (31.2%) | 1 (11.1%) | |
| - 3+ | 90 (14.5%) | 3 (9.4%) | 4 (44.4%) | |
| subtype | 0 | |||
| - Basal | 190 (18.0%) | 42 (89.4%) | 0 (0.0%) | |
| - Her2 | 82 (7.8%) | 5 (10.6%) | 1 (9.1%) | |
| - LumA | 566 (53.6%) | 0 (0.0%) | 5 (45.5%) | |
| - LumB | 217 (20.6%) | 0 (0.0%) | 5 (45.5%) | |
| PIK3CA mutation | 301 (31.2%) | 3 (6.8%) | 1 (10.0%) | NS |
| CDH1 mutation | 96 (10.0%) | 0 (0.0%) | 1 (10.0%) | NS |
| TP53 mutation | 264 (27.4%) | 29 (65.9%) | 3 (30.0%) | NS |
| BRCA1 mutation | 11 (1.1%) | 0 (0.0%) | 0 (0.0%) | NS |
| BRCA2 mutation | 12 (1.2%) | 1 (2.7%) | 1 (10.0%) | NS |
First, CCDC170 fusion-positive patients had a high rate of ER-positive (90.9%) and PR-positive (63.6%), whereas fusion-negative patients displayed significantly lower rates of 20.8% and 4.3%, respectively (p < 0.05). Additionally, HER2 immunohistochemistry (IHC) results showed a significantly higher rate of 3+ for fusion-positive patients than for patients with fusion-negative (44.4% vs. 9.4%, p < 0.05, Table 1). According to the findings above, CCDC170 fusion-positive BRCA appears to closely resemble characteristics typical of triple-positive breast cancer in this cohort.
Second, the pathological subtype of fusion-positive patients was found to have a high proportion of the luminal A (45.5%) and luminal B subtypes (45.5%), while 90.9% of the 48 cases with the lowest CCDC170 expression were found to be basal type (p < 0.05, Table 1). Taken together, the CCDC170 fusion-positive BRCA showed a mutually exclusive relationship with the basal-type breast cancer cells.
On the other hand, no significant differences in age, sex, vital status, and TNM stage were observed between the two groups. In addition, the five gene variants (PIK3CA, CDH1, TP53, BRCA1, and BRCA2) frequently found in BRCA showed no significant difference between the two groups. Based on these results, CCDC170 fusion-positive BRCA patients have distinct pathological characteristics in terms of tumor subtype and triple positive tendency.
3.2. Key Pathways and Genes Altered in ESR1-CCDC170 Fusion-Positive Breast Cancer
One thousand genes were obtained by an independent t-test (q < 2.0 × 10−8) and inputted for performing the over-representation analysis (ORA) of the ConsensusPathDB website to select cancer-related pathways. As a result, a total of six cancer-related pathways (p53, ATR/ATM, FOXM1, hedgehog, Cell cycle, and Aurora B-related signaling pathways) were discerned.
In the six major cancer-related pathways, 137 genes were significantly over- or under-expressed in the CCDC170 fusion-positive cases compared with the CCDC170 fusion-negative controls (Figure 2, Figure S1).
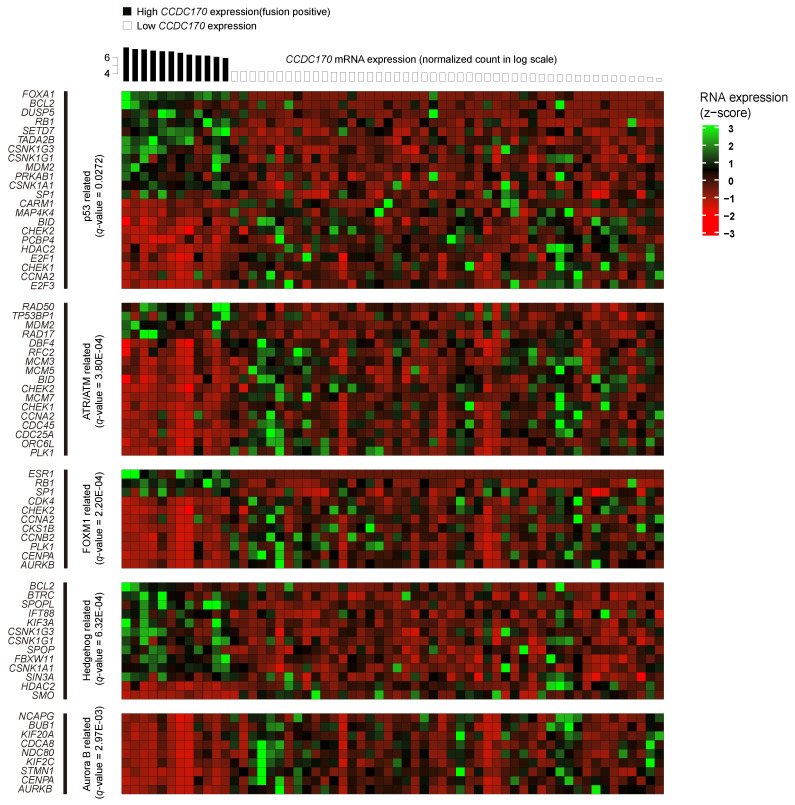
Figure 2.
Gene expression heatmap of cancer-related pathways correlated with CCDC170 RNA expression. Of the analyzed genes, 72 of the genes associated with P53, ATR/ATM, FOXM1, hedgehog, and Aurora B demonstrated significant differences in expression in CCDC170 fusion-positive BRCA samples when compared with the control group. Over-representation analysis using ConsensusPathDB (CPDB) yielded statistically significant pathways related to cancer (q < 0.05). The x-axis is indicative of the sample, while the y-axis is indicative of its respective RNA expression. The RNA expression was converted into z-score prior to representation on the heatmap.
Of the six pathways, two concerning p53- and ATR/ATM-related signaling pathways were associated with DNA damage response. Mapping with the KEGG pathway revealed 22 genes that are involved in the p53-related pathway and 17 genes in the ATR/ATM-related pathway. Both pathways are highly relevant to the promotion and maintenance of the cell cycle (Figure 3). Genes with multiple hits of more than two that coincide for both p53- and ATR/ATM-related signaling pathways are CCNA2(CycA), MDM2, CHEK1(Chk1), and CHEK2(Chk2).
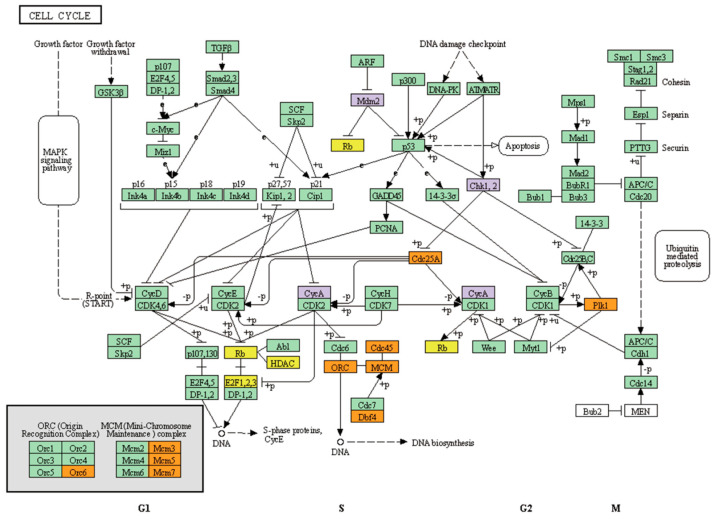
Figure 3.
Over- and under-expressed genes are enriched in the human cell cycle pathway. The KEGG pathway map for the human cell cycle signaling pathway, has04110, was visualized using the KEGG mapper. Among the pathways, p53 and ATR/ATM shared a significant correlation with the identified genes. Genes associated with the p53 signaling pathway are boxed in yellow, ATR/ATM in orange, and common denominators for both pathways in purple.
There were 39 genes involved in multiple pathways (Figure 4, Table S2), of which AURKB, HDAC2, PLK1, CENPA, CHEK1, CHEK2, RB1, CCNA2, and MDM2 were included in at least three pathways that are important for tumor proliferation and maintenance specific to ESR1-CCDC170 fusion-positive BRCA patients.
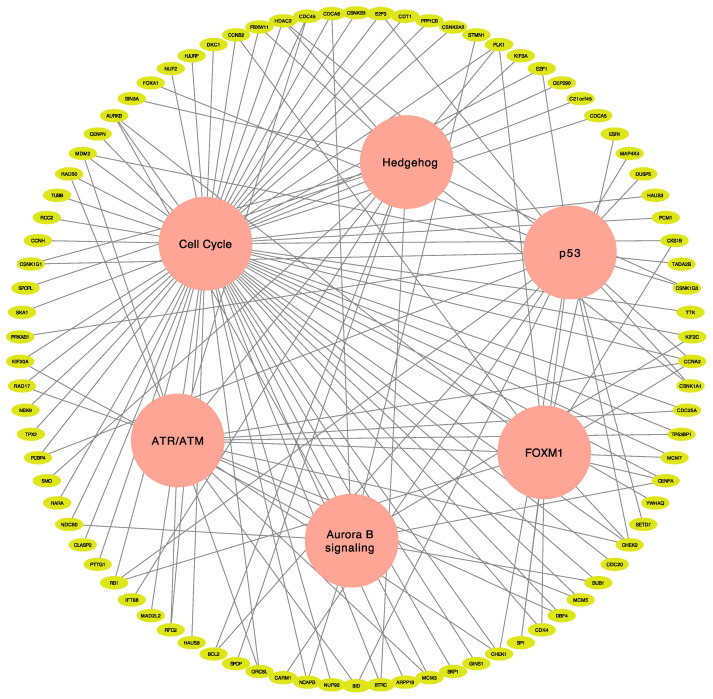
Figure 4.
Putative target genes involved in multiple pathways of ESR1-CCDC170 fusion-positive cancer. Six major cancer signaling pathways associated with p53, ATR/ATM, FOXM1, hedgehog, cell cycle, and Aurora B in accordance with their respective genes were visualized. Potential gene candidates involved in these pathways were discerned.
Further investigation of the 48 samples with the highest mRNA levels of CCDC170 with the differentially expressed gene (DEG) analysis showed similar patterns as the TCGA data obtained above in fusion-positive samples when compared with the control samples (Figure S2, Table S1). This suggests that a similar cell signaling is activated not just with fusion but with other possibilities in CCDC170 over-expression.
3.3. Identification of Actionable Targets and Potential Therapeutic Choice Using Network Analysis
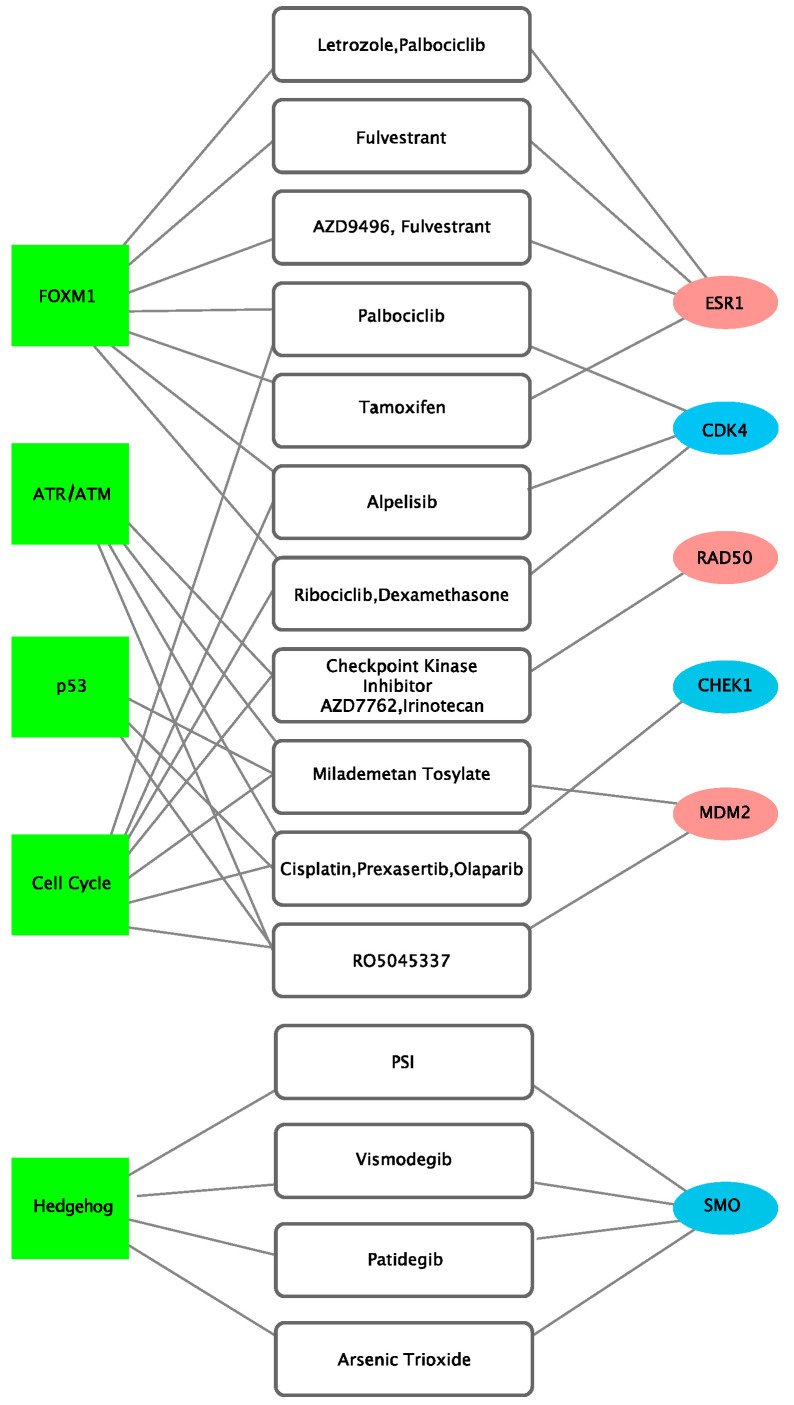
Actionable target genes and potentially available drugs were extracted by inputting the 137 genes in the following drug databases: CIViC (n = 673) and OncoKB (n = 262). ESR1, CDK4, RAD50, CHEK1, MDM2, and SMO were mapped as targetable genes. Results indicated the following drug-target relationships: letrozole, palbociclib, fulvestrant, AZD9496, and tamoxifen for ESR1; palbociclib, alpelisib, ribociclib, and dexamethasone for CDK4; checkpoint kinase inhibitor AZD7762 and irinotecan for RAD50; cisplatin, prexasertib, and olaparib for CHEK1; milademetan tosylate and RO5045337 for MDM2; and PSI, vismodegib, patidegib, and arsenic trioxide for SMO (Figure 5).
Figure 5.
Drug-target network of ESR1-CCDC170 fusion-positive BRCA cancer. Network visualization was demonstrated with Cytoscape, and drug-target relationship was identified with CIViC and OncoKB. Green boxes are representative of pathways, white boxes of drugs, and oval boxes of genes. Red oval boxes are genes that are over-expressed in fusion-positive cancer, whereas blue oval boxes are genes that are under-expressed in fusion-positive cancer.
Observing the druggable target genes associated with the main pathways of E:C fusion-positive BRCA, ESR1 and CDK4 genes were included in the FOXM1-related signaling pathway; RAD50 in the ATR/ATM-related signaling pathway; CHEK1 and MDM2 genes in the P53-related signaling pathway; CDK4, RAD50, and MDM2 genes in the cell cycle-related signaling pathway; and SMO genes in the hedgehog-related signaling pathway. Interestingly, four of the six targetable genes, CDK4, RAD50, CHEK1, and MDM2, were involved in two or more major cancer-related pathways. In the case of MDM2, three of the six pathways associated with E:C fusion-positive were identified to be involved.
4. Discussion
In this study, the characteristics of an ER-positive molecular subtype in CCDC170-subtype breast cancer were identified, and genes specifically regulated in E:C BRCA were identified and screened for BRCA-related signaling pathways. Additionally, information regarding optimal treatment targets and drugs for targeted therapy was provided. E:C fusion-positive BRCA requires a new therapeutic approach to overcome its relatively low response to hormone therapy [6,13,22,23,24].
A recent study on a potential targeted therapy for E:C BRCA performed by Li et al. was met with limitations with regard to a restricted number of cell line samples and proteins [13]. Our study has addressed this issue by performing analysis on a sufficient number of case-control TCGA human cancer samples and systematically testing the DEGs using more than 20,000 genes and cancer-specific pathways. Finally, we were able to propose a number of potential drugs with promising therapeutic effects.
The common early treatment options for breast cancer are generally divided into conventional chemotherapy (Adriamycin, cyclophosphamide, paclitaxel, and docetaxel), endocrine therapy (tamoxifen, letrozole, anastrozole, and exemestane), ERBB-targeted therapy (trastuzumab and pertuzumab), and combination treatment methods according to the pathological and molecular classification of breast cancer [2]. In the case of metastatic breast cancer, CDK4/6 inhibitor and PARP inhibitor are considered to be additional options [2]. On the other hand, many resistance mechanisms for drug therapy in breast cancer have been reported as follows: loss of estrogen receptor, deregulation of cell cycle for endocrine therapy, incomplete blockade of HER receptors, activation of the PI3K pathway, over-expression of estrogen receptor for HER2 inhibitors, polyclonal RB1 mutations for CDK 4/6 inhibitors, and so on [25,26]. Hence, finding novel therapeutic strategies using drug repositioning analysis is crucial for modern breast cancer treatment.
Among the repositioned drugs inferred in our study, CDK4/6 inhibitor (palbociclib), cisplatin, and PARP inhibitor are the drugs used as standard treatments for breast cancer patients with or without metastasis. On the other hand, AZD9496, alpelisib, dexamethasone, checkpoint kinase inhibitor AZD7762, irinotecan, cisplatin, prexasertib, milademetan tosylate, R05045337, PSI, vismodegib, patidegib, and arsenic trioxide are seen as putative actionable drugs that can be used for E:C fusion-positive BRCA proceeding in vitro and in vivo validation.
AURKB, HDAC2, PLK1, CENPA, CHEK1, CHEK2, RB1, and MDM2 genes, which were included in at least three pathways, are expected to play an important role in the promotion and maintenance of CCDC170-subtype breast cancer. For instance, PLK1 may act as a tumor suppressor gene that regulates estrogen receptor (ER)-regulated gene transcription in breast cancer [27]; RB1 gene, also a tumor suppressor gene, however, is frequently lost in triple-negative breast cancer [28]; CENPA is a significant prognostic marker for ER-positive patients [29]; and HDAC2 and CHEK2 genes have been significantly correlated to CCDC170 fusion subtype and have been reported to be associated with DDR functioning [30,31], which is also suggestive of the CCDC170 fusion subtype’s relationship with DDR.
In addition, we investigated whether there is a biological difference between the E:C fusion-positive group and the CCDC170 high-expression group without fusion. We found that there showed no major difference in cancer signaling except in several minor pathways, including the cilium assembly pathway and integrins in the angiogenesis pathway (Table S3).
In summary, this study presents core biomarkers and potentially actionable drugs specific to E:C fusion-positive breast cancer. Via in vitro experimentation, these candidates were confirmed to be strongly associated with this type of cancer, and their roles were verified by discerning their associated signaling pathways. We hope that our findings will be the steppingstone for future investigations, leading to the promotion of a targeted cancer therapy.
Acknowledgments
This study was supported by a VHS Medical Center Research Grant, Republic of Korea (VHSMC 20053), grant no. 18-2018-023, from the SNUBH Research Fund, and the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (grant no. NRF-2018R1D1A1A02086141).
Abbreviations
TCGA: Cancer Genome Atlas; IQR, interquartile range; E:C fusion-positive, ESR1-CCDC170 fusion-positive; HER2, human epidermal growth factor receptor 2; HER2-E, human epidermal growth factor receptor 2-enriched; ER, estrogen receptor; DDR, DNA damage response; CPDB, Consensus Pathway Database; ORA, over-representation analysis; FDR, false detection rate; DEG, differentially expressed gene; IHC, immunohistochemistry; NS, not significant; NA, not available.
Supplementary Materials
The following are available online at https://www.mdpi.com/2077-0383/10/4/582/s1: Figure S1: Heatmap of cell cycle-related and other miscellaneous genes with altered expression correlating to CCDC170 expression, Figure S2: Heatmap of cancer-related and other miscellaneous genes with altered expression correlating to CCDC170 expression in DEG analysis, Table S1: Comparisons in clinical and pathological characteristics of CCDC170 high expression group including E:C fusion-positive cases and CCDC170 high expression group without E:C fusion, Table S2: Thirty-nine genes with hits of multiple pathways, Table S3: Putative target genes involved in three major pathways of CCDC170 high expression group without fusion.
Author Contributions
Conceptualization, S.L., J.W.Y., and C.Y.H.; methodology, S.L., J.W.Y., and J.H.J.; formal analysis, S.L., J.W.Y., and J.H.J.; investigation, J.H.J., H.Y.K., S.L., and J.W.Y.; data curation, J.H.J.; writing—original draft preparation, J.H.J., H.Y.K., S.L., and J.W.Y.; writing—review and editing, J.H.J., H.Y.K., S.L., C.Y.H., and J.W.Y.; visualization, J.H.J., S.L., and J.W.Y.; supervision, S.L., C.Y.H., and J.W.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data is available in the following website: https://gdac.broadinstitute.org.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Waks A.G., Winer E.P. Breast cancer treatment: A review. JAMA. 2019;321:288–300. doi: 10.1001/jama.2018.19323. [DOI] [PubMed] [Google Scholar]
- 3.Fimereli D., Fumagalli D., Brown D., Gacquer D., Rothe F., Salgado R., Larsimont D., Sotiriou C., Detours V. Genomic hotspots but few recurrent fusion genes in breast cancer. Genes Chromosomes Cancer. 2018;57:331–338. doi: 10.1002/gcc.22533. [DOI] [PubMed] [Google Scholar]
- 4.Giltnane J.M., Hutchinson K.E., Stricker T.P., Formisano L., Young C.D., Estrada M.V., Nixon M.J., Du L., Sanchez V., Ericsson P.G., et al. Genomic profiling of ER(+) breast cancers after short-term estrogen suppression reveals alterations associated with endocrine resistance. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aai7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goksu S.S., Tastekin D., Arslan D., Gunduz S., Tatli A.M., Unal D., Salim D., Guler T., Coskun H.S. Clinicopathologic features and molecular subtypes of breast cancer in young women (age ≤ 35) Asian Pac. J. Cancer Prev. 2014;15:6665–6668. doi: 10.7314/APJCP.2014.15.16.6665. [DOI] [PubMed] [Google Scholar]
- 6.Hartmaier R.J., Trabucco S.E., Priedigkeit N., Chung J.H., Parachoniak C.A., Vanden Borre P., Morley S., Rosenzweig M., Gay L.M., Goldberg M.E., et al. Recurrent hyperactive ESR1 fusion proteins in endocrine therapy-resistant breast cancer. Ann. Oncol. 2018;29:872–880. doi: 10.1093/annonc/mdy025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matissek K.J., Onozato M.L., Sun S., Zheng Z., Schultz A., Lee J., Patel K., Jerevall P.L., Saladi S.V., Macleay A., et al. Expressed gene fusions as frequent drivers of poor outcomes in hormone receptor-positive breast cancer. Cancer Discov. 2018;8:336–353. doi: 10.1158/2159-8290.CD-17-0535. [DOI] [PubMed] [Google Scholar]
- 8.Veeraraghavan J., Tan Y., Cao X.X., Kim J.A., Wang X., Chamness G.C., Maiti S.N., Cooper L.J., Edwards D.P., Contreras A., et al. Recurrent ESR1-CCDC170 rearrangements in an aggressive subset of oestrogen receptor-positive breast cancers. Nat. Commun. 2014;5:4577. doi: 10.1038/ncomms5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yersal O., Barutca S. Biological subtypes of breast cancer: Prognostic and therapeutic implications. World J. Clin. Oncol. 2014;5:412–424. doi: 10.5306/wjco.v5.i3.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veeraraghavan J., Ma J., Hu Y., Wang X.S. Recurrent and pathological gene fusions in breast cancer: Current advances in genomic discovery and clinical implications. Breast Cancer Res. Treat. 2016;158:219–232. doi: 10.1007/s10549-016-3876-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turnbull C., Ahmed S., Morrison J., Pernet D., Renwick A., Maranian M., Seal S., Ghoussaini M., Hines S., Healey C.S., et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat. Genet. 2010;42:504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y., He Y., Qin Z., Jiang Y., Jin G., Ma H., Dai J., Chen J., Hu Z., Guan X., et al. Evaluation of functional genetic variants at 6q25.1 and risk of breast cancer in a Chinese population. Breast Cancer Res. 2014;16:422. doi: 10.1186/s13058-014-0422-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L., Lin L., Veeraraghavan J., Hu Y., Wang X., Lee S., Tan Y., Schiff R., Wang X.S. Therapeutic role of recurrent ESR1-CCDC170 gene fusions in breast cancer endocrine resistance. Breast Cancer Res. 2020;22:84. doi: 10.1186/s13058-020-01325-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang S.Y.C., Lheureux S., Karakasis K., Burnier J.V., Bruce J.P., Clouthier D.L., Danesh A., Quevedo R., Dowar M., Hanna Y., et al. Landscape of genomic alterations in high-grade serous ovarian cancer from exceptional long- and short-term survivors. Genome Med. 2018;10:81. doi: 10.1186/s13073-018-0590-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshihara K., Wang Q., Torres-Garcia W., Zheng S., Vegesna R., Kim H., Verhaak R.G. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene. 2015;34:4845–4854. doi: 10.1038/onc.2014.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson R.G., Smith A.N., Bird C.C. Immunohistochemical detection of abnormal cell proliferation in colonic mucosa of subjects with polyps. J. Clin. Pathol. 1990;43:744–747. doi: 10.1136/jcp.43.9.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kandasamy K., Mohan S.S., Raju R., Keerthikumar S., Kumar G.S., Venugopal A.K., Telikicherla D., Navarro J.D., Mathivanan S., Pecquet C., et al. NetPath: A public resource of curated signal transduction pathways. Genome Biol. 2010;11:R3. doi: 10.1186/gb-2010-11-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaefer C.F., Anthony K., Krupa S., Buchoff J., Day M., Hannay T., Buetow K.H. PID: The pathway interaction database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fabregat A., Sidiropoulos K., Garapati P., Gillespie M., Hausmann K., Haw R., Jassal B., Jupe S., Korninger F., McKay S., et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2016;44:D481–D487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kutmon M., Riutta A., Nunes N., Hanspers K., Willighagen E.L., Bohler A., Melius J., Waagmeester A., Sinha S.R., Miller R., et al. WikiPathways: Capturing the full diversity of pathway knowledge. Nucleic Acids Res. 2016;44:D488–D494. doi: 10.1093/nar/gkv1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeselsohn R., Buchwalter G., de Angelis C., Brown M., Schiff R. ESR1 mutations—A mechanism for acquired endocrine resistance in breast cancer. Nat. Rev. Clin. Oncol. 2015;12:573–583. doi: 10.1038/nrclinonc.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lei J.T., Gou X., Seker S., Ellis M.J. ESR1 alterations and metastasis in estrogen receptor positive breast cancer. J. Cancer Metastasis Treat. 2019;5 doi: 10.20517/2394-4722.2019.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma C.X., Reinert T., Chmielewska I., Ellis M.J. Mechanisms of aromatase inhibitor resistance. Nat. Rev. Cancer. 2015;15:261–275. doi: 10.1038/nrc3920. [DOI] [PubMed] [Google Scholar]
- 25.Chun K.H., Park J.H., Fan S. Predicting and overcoming chemotherapeutic resistance in breast cancer. Adv. Exp. Med. Biol. 2017;1026:59–104. doi: 10.1007/978-981-10-6020-5_4. [DOI] [PubMed] [Google Scholar]
- 26.Condorelli R., Spring L., O’Shaughnessy J., Lacroix L., Bailleux C., Scott V., Dubois J., Nagy R.J., Lanman R.B., Iafrate A.J., et al. Polyclonal RB1 mutations and acquired resistance to CDK 4/6 inhibitors in patients with metastatic breast cancer. Ann. Oncol. 2018;29:640–645. doi: 10.1093/annonc/mdx784. [DOI] [PubMed] [Google Scholar]
- 27.Wierer M., Verde G., Pisano P., Molina H., Font-Mateu J., Di Croce L., Beato M. PLK1 signaling in breast cancer cells cooperates with estrogen receptor-dependent gene transcription. Cell Rep. 2013;3:2021–2032. doi: 10.1016/j.celrep.2013.05.024. [DOI] [PubMed] [Google Scholar]
- 28.Robinson T.J., Liu J.C., Vizeacoumar F., Sun T., Maclean N., Egan S.E., Schimmer A.D., Datti A., Zacksenhaus E. RB1 status in triple negative breast cancer cells dictates response to radiation treatment and selective therapeutic drugs. PLoS ONE. 2013;8:e78641. doi: 10.1371/journal.pone.0078641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGovern S.L., Qi Y., Pusztai L., Symmans W.F., Buchholz T.A. Centromere protein-A, an essential centromere protein, is a prognostic marker for relapse in estrogen receptor-positive breast cancer. Breast Cancer Res. 2012;14:R72. doi: 10.1186/bcr3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nevanlinna H., Bartek J. The CHEK2 gene and inherited breast cancer susceptibility. Oncogene. 2006;25:5912–5919. doi: 10.1038/sj.onc.1209877. [DOI] [PubMed] [Google Scholar]
- 31.Shan W., Jiang Y., Yu H., Huang Q., Liu L., Guo X., Li L., Mi Q., Zhang K., Yang Z. HDAC2 overexpression correlates with aggressive clinicopathological features and DNA-damage response pathway of breast cancer. Am. J. Cancer Res. 2017;7:1213–1226. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data is available in the following website: https://gdac.broadinstitute.org.