Abstract
Altered expression of long noncoding RNA (lncRNA), longer than 200 nucleotides without potential for coding protein, has been observed in diverse human diseases including viral diseases. It is largely unknown whether lncRNA would deregulate in SARS-CoV-2 infection, causing ongoing pandemic COVID-19. To identify, if lncRNA was deregulated in SARS-CoV-2 infected cells, we analyzed in silico the data in GSE147507. It was revealed that expression of 20 lncRNA like MALAT1, NEAT1 was increased and 4 lncRNA like PART1, TP53TG1 was decreased in at least two independent cell lines infected with SARS-CoV-2. Expression of NEAT1 was also increased in lungs tissue of COVID-19 patients. The deregulated lncRNA could interact with more than 2800 genes/proteins and 422 microRNAs as revealed from the database that catalogs experimentally determined interactions. Analysis with the interacting gene/protein partners of deregulated lncRNAs revealed that these genes/proteins were associated with many pathways related to viral infection, inflammation and immune functions. To find out whether these lncRNAs could be regulated by STATs and interferon regulatory factors (IRFs), we used ChIPBase v2.0 that catalogs experimentally determined binding from ChIP-seq data. It was revealed that any one of the transcription factors IRF1, IRF4, STAT1, STAT3 and STAT5A had experimentally determined binding at regions within -5kb to +1kb of the deregulated lncRNAs in at least 2 independent cell lines/conditions. Our analysis revealed that several lncRNAs could be regulated by IRF1, IRF4 STAT1 and STAT3 in response to SARS-CoV-2 infection and lncRNAs might be involved in antiviral response. However, these in silico observations are necessary to be validated experimentally.
Keywords: Long non-coding RNA, NEAT1, MALAT1, Interferon regulatory factors, SARS-CoV-2, COVID-19
Long non-coding RNA; NEAT1; MALAT1; Interferon regulatory factors; SARS-CoV-2; COVID-19
1. Introduction
Long noncoding RNA (lncRNA) is functional RNA, longer than 200 nucleotides without potential for coding protein. Like protein-coding genes, lncRNAs are regulated by transcription factors and other regulators. LncRNA interacts with genomic regions, mRNA, protein, microRNA and modulates the levels of protein-coding genes at transcription, post-transcription or post-translation levels. DNA-lncRNA interaction forms DNA-RNA triple helix at genomic region facilitating or inhibiting recruitment of transcription factors and regulates expression of nearby genes. Interaction of lncRNA with mRNA or protein may stabilize or destabilize mRNA or protein [1, 2]. By altering protein level, lncRNA modulates many biological processes, functions and pathways including, immunity, inflammation [3, 4, 5].
1.1. Immunological defects in COVID-19
Ongoing pandemic coronavirus disease 19 (COVID-19) is caused by infection with severe acute respiratory coronavirus 2 (SARS-CoV-2). Major symptoms of COVID-19 include fever, dry cough, breathing difficulty, diarrhoea, and throat sore. Small proportion of infected individuals progresses to severe conditions with multi-organ failure and may be fatal [6]. Hyperinflammatory response, mediated through dysregulated macrophages, innate and adaptive immunity have been considered as pathological conditions in severe COVID-19. Increased levels of cytokines like IL-6, IL-7, TNF, inflammatory chemokines like CCL2, CCL3, chemokine ligand CXCL10 and α-chain of IL-2 receptor has been observed and reviewed [7]. Increased chemokine expression in the absence or reduced levels of Type I and III interferons in different tissues from COVID-19 patients and infected cells with SARS-CoV-2 have been observed [8, 9, 10]. Based on this data, it has been proposed that use of exogenous IFN to stimulate antiviral immunity might be successful for treating SARS-CoV-2 infection [11]. In contrast, increased levels of type I IFN genes in COVID-19 patients [12] and SARS-CoV-2 infected organoid [13] have been observed. Enhanced expression of IFN-stimulated genes (ISGs) has been reported in different cells from patients [14, 15] and in mouse model of COVID-19 [16]. Reduced IFN-1 response and higher levels ISGs observed in PBMC was resolved by identifying transient low plasma IFNA levels in PBMC originated from lungs [17]. Differences in results published by different investigators, particularly role IFN-1 in COVID-19 might also be attributed to use of different platforms for single-cell sequencing, unsupervised analysis of highly multi-dimensional data, use of different cell types or mixture of cell types and definition of the severity including sampling time points [15]. However molecular mechanisms of altered expression of cytokine, chemokine, interferon and other inflammatory molecules remain unclear.
1.2. Altered expression of long non-coding RNA in viral infection
Deregulation of lncRNA has been observed in many infectious diseases, especially in diseases caused by viral infection and reviewed [18]. Infections with influenza virus (IAV), HIV, herpes simplex virus (HSV), hepatitis C virus (HCV), hepatitis B virus (HBV), severe acute respiratory syndrome coronavirus (SARS-CoV) and others alter the expression of host lncRNA [19, 20]. Expression of lncRNA EGOT, NEAT1, BISPR/LncBST2, LINC01191/VIN/lnc-ACTR3, ISR (IFN-stimulated lncRNA) 2 and ISR8, LOC100506319/LINC01988/LncRNA-PAAN, LOC102637961/lncRNA-ACOD1, MIR155HG, TSPOAP1-AS1, PSMB8-AS1, PSORS1C3, IVRPIE was increased and expression of NRAV/DYNLL1AS1 was decreased in IAV infected cells [21, 22, 23, 24, 25, 26, 27, 28]. In HIV infected cells expression of ISR2, NEAT1, GOMAFU/MIAT, LUST/RBM5-AS1, BIC/MIR155HG, MALAT1, NRIR/lncCMPK2, MIR3945HG, FIRRE, LINC02574, LINC01426 is increased while expression of NRON, PANDAR/PANDA, CDKN2B-AS1/ANRIL DD3/PCA, DANCR, CMPD/LINC01152, NCRMS/RMST, GAS5, LINC00173, lincRNA-p21/TP53COR1 and SAF/FAS-AS1 is decreased [29, 30, 31]. Increased expression of NEAT1, MAMDC2-AS1, PRINS, ISR2 and LOC102637961/lncRNA-ACOD1 is observed in HSV infected cells [24, 27, 32, 33, 34]. Infection with HCV increases expression of lncRNA BISPR, negative regulator of interferon response (NRIR), EGOT, ISR2 (GBP1 pseudogene 1), ISR8 (AC116366.6), GAS5, CFAP58-DT/lncITPRIP-1, lncRNA32/LUARIS. Expression of lncRNA32/LUARIS is increased in EMCV and HBV infected cells. Expression of NEAT1 is increased in Semliki Forest virus infected cells. Increased expression of NeST/IFNG-AS1 is observed in Theiler's virus-infected cells (Reviewed in [33]). In respiratory syncytial virus (RSV) infected cells expression of NRAV/ DYNLL1-AS1 and MEG3 is decreased [35, 36]. Differential expression of hundreds of annotated and un-annotated lncRNAs has been observed in mice infected with SARS-CoV and IAV; expression of some was dependent on STAT1 or IFNAR. Result obtained from this study shows that altered lncRNAs were similar in both the infection and they had characteristics kinetic expression profiles in IFNAR and STAT1 knockout mice during SARS-CoV infection [37].
To summarize, expression of NEAT1 is increased in IAV, HIV, HSV, Japanese encephalitis virus, rabies virus and Hantaan orthohantavirus (HTNV) infected cells. Increased expression of EGOT is observed in IAV, HIV, HCV and Semliki Forest virus. Expression of LOC102637961/lncRNA-ACOD1, a lncRNA identified by its nearest coding gene aconitate decarboxylase 1 (Acod1), is increased in IAV, HSV and vascular stomatitis virus (VSV) infected cells while expression of NRAV/DYNLL1AS1 is inhibited by IAV, HSV and Sendai virus (SeV) infections. These results indicate that lncRNAs were differentially expressed in viral infection, and may be involved in viral replication and pathogenesis [25, 33]. Possible involvement of representative lncRNAs in diverse viral and host functions in response to infection is shown in Supplementary Text Table STT1.1.
1.3. Mechanism(s) of actions of lncRNAs in virus-infected cells
Several current reviews are available to show that deregulated lncRNAs are involved in antiviral activity [25, 33, 38]. LncRNAs are observed to modulate (i) viral replication and growth, and (ii) expression of IFNs, and ISGs, possibly by interacting with transcription factors like STAT1, STAT3, NFκB and IRFs. Expressions of several lncRNAs are modulated by treatment with IFNs.
1.4. Long non-coding RNA modulates viral replication
Interferon (IFN)-stimulated lncRNA ISR [24] and MIR155HG [22], IVRPIE [21] suppress IAV replication and growth while LOC100506319/LINC01988/LncRNA-PAAN [26], TSPOAP1-AS1 [28], NRAV/ DYNLL1-AS1 (Reviewed in [33]) and PSMB8-AS1 [23] enhance IAV replication and growth. EGOT and CFAP58-DT/lncITPRIP-1inhibit replication and growth of HCV (Reviewed in [33]); NEAT1 inhibits replication and growth of HIV [29] and HTNV [39]. LncRNA32/LUARIS inhibits the replication of EMCV, HBV and HCV [40]. NRIR/lncCMPK2 suppresses HCV replication [41].
1.5. Long non-coding RNA modulates interferon
Overexpression of NeST/IFNG-AS1 inhibits clearance of Theiler's virus by enhancing IFNG, thus regulating Type II interferon response (Reviewed in Liu and Ding [33]). Increased expression of MIR155HG could enhance the level of INFB (Maarouf et al., 2019). A novel lncRNA IVRPIE (Inhibiting IAV Replication by Promoting IFN and ISGs Expression) interacts with hnRNP U and regulates positively expression of INFB and several ISGs like IRF1, IFIT1, IFIT3, Mx1, ISG15, and IFI44L through histone modifications of target genes. Taken together; these findings suggest that IVRPIE is a critical regulator of host antiviral response [21].
1.6. LncRNA modulates interferon-stimulated genes (ISGs)
Several evidences are available to show that altered expression of the lncRNA in different viral infected cells might involve in modulation ISGs. For examples, CFAP58-DT/lncITPRIP-1 suppresses HCV replication by increasing ISGs [42]. Expressions of several ISGs like IFIT2, IFIT3, IFITM3, OASL, and MxA were reduced in cells overexpressing NRAV/DYNLL1-AS1 [43]. LncRNA32/LUARIS inhibits the replication of EMCV, HBV and HCV, associated with hnRNP U and activates transcription factor ATF2 modulates expression of ISGs [40]. LincRNA-cox2/Ptgs2os2, induced by TLR ligands depending on MyD88 and NF-κB, reduced the expression of some ISGs, like IRF7 and Rasd2. LincRNA-Cox-2/Ptgs2os2 interacts with HnRNP-A/B and hnRNP-A2/B1 to regulate expression of ISGs [44]. TLR ligands suppress the expression of TTC39A-AS1/lincRNA-EPS and enhance expression of ISGs like IFIT2, RASD2, OAS1 and GPB5 by recruiting hnRNPL [45]. In IFN-stimulated hepatocytes, knocked down of NRIR/lncCMPK2 resulted in the transcriptional up-regulation of many ISGs like CMPK2, RASD2, ISG15, CXCL10, IFIT3, and IFITM1 [41]. Expression of several ISGs, including GBP1, ISG15, MXA, BST2, ISG56, IFI6, and IFITM1 was increased in EGOT knockdown cells with or without HCV infection. This result shows that EGOT may promote viral replication by blocking the IFN antiviral response [46].
1.7. Modulation lncRNA by interferon
Many lncRNAs are differentially expressed following IFN stimulation in high throughput studies; specific role of the identified lncRNAs in viral infection has been identified only for a small fraction and reviewed [38]. Expression of BISPR/lncBST2 is increased by IFNA or IFNG treatment. HCV or HEV infected Huh7 cells and in liver of HCV-infected patients show similar result. Expression of both BISPR/lncBST2 and BST2 was STAT-dependent. Based on the inhibitory effect of BST2 on virion secretion, BISPR/ lncBST2 might be involved in regulating viral infection partially by increasing the expression of antiviral protein BST2 and reviewed [25]. NRIR/lncCMPK2 is stimulated by IFNA or IFNG and possibly regulated by STAT2. Expression of NRIR/ lncCMPK2 is increased in liver of chronic HCV infected patients [41]. IFNA treatment stimulates expression of EGOT. In response to HCV infection, EGOT is activated by NFκB [46]. IAV infection in mouse model enhances expression of interferon (IFN)-stimulated lncRNA (ISR) like ISR2, ISR8. Knockdown of the lncRNAs in cells resulted in increased IAV replication and over expression reduced the viral replication. IFNB treatment induces the lncRNA expression. Induction of the lncRNA by IAV infection was not observed in cells deficient of IFNAR1. LncRNA ISR is regulated by RIG-I-dependent signaling that regulates IFNB production during IAV infection, and has an inhibitory capacity in viral replication [24].
1.8. Long non-coding RNA targets proteins coded by HIV to modulate viral infection
Expression of MALAT1 is increased in HIV infected cells. MALAT1 interacts with chromatin modulator polycomb repressive complex PRC2, releases the core component enhancer of zeste homolog 2 (EZH2) from binding with HIV-1 LTR promoter and removes PRC2 complex-mediated methylation of histone H3 on lysine 27 (H3K27me3). Thus MALAT1 may relieve epigenetic silencing of HIV-1 transcription [47]. NRON, highly expressed in resting CD4+ T lymphocytes, involves in HIV latency inducing Tat protein degradation and potentially suppresses the viral transcription by decreasing the cellular abundance of viral transactivator protein Tat [48].
1.9. Modulation of viral infection by NEAT1 through paraspeckle formation
Increased expression of NEAT1 has been observed in HIV, IAV, HTNV, HSV, Japanese encephalitis and rabies virus-infected cells. Mechanism of actions of increased NEAT1 has been worked out in detail in HTNV infected cells. Silencing NEAT1 by siRNA in HTNV infected HUVEC human cells, enhances HTNV replication while exogenous over-expression of NEAT1 effectively inhibited the replication. NEAT1 overexpression increased IFNB production and inhibited HTNV replication. Similar inhibitory effect of NEAT1 on HTNV virus titers is also observed in animals [39]. This result shows that NEAT1 affects HTNV viral replication through IFNB. Besides, NEAT1 has been shown to regulate expression of IL8 by relocating SFPQ from its promoter and recruiting SFPQ into the paraspeckles [49]. Antiviral activity of NEAT1, observed in several experiments, could be mediated through formation of paraspeckles, a class of membraneless subnuclear bodies, observed in the interchromatin space of mammalian cells. Paraspeckles are RNA-protein structures formed by the interaction between NEAT1, an indispensable structural component, and different class of RNA and many RNA binding proteins. Paraspeckles could sequester paraspeckle-localizing proteins and RNA and modulates their functions outside the paraspeckles, thus acting as molecular sponges [50, 51]. It has been revealed that NEAT1 may promote IFN responses by acting as a positive feedback for RIG-I signaling. Increased NEAT1 by interacting with SFPQ, relocates SFPQ from the promoters of RIG-I and DDX60 to paraspeckles. Thus NEAT1 removes the transcriptional inhibitory effects of SFPQ on RIG-I and DDX60, resulting in increased expression of transcriptional factor IRF7, which in turn induced the expression of IFN and NEAT1 [39]. NEAT1 may also activate IRF3 through formation of multi-subunit complex with HEXIM1. This NEAT1-HEXIM1 complex interacts with cGAS sensor and its partner PQBP1, releases proteins from paraspeckle. Released proteins are recruited to STING and activates IRF3 producing type 1 IFN. These results indicate that NEAT1 has a critical role in the antiviral response of IFN through (i) RIG-I signaling and (ii) cGAS-STING-IRF3 pathway and reviewed [25]. Paraspeckles is induced by IAV, HSV [49] and HIV [29].
Altered expression of lncRNA in SARS-CoV-2 infected cells or tissues from COVID-19 has not been studied to the best of our knowledge. Reusing the RNA sequencing data in GSE147507, we observed that expression of several lncRNAs were altered in SARS-CoV-2 infected cell lines and lung tissue from COVID-19 patients. Interacting protein/gene partners of these deregulated lncRNAs were associated with pathways relevant for viral replication, inflammation and immune function indicating possible role of the lncRNA in SARS-CoV-2 infection.
2. Materials and methods
2.1. Data from GEO accession GSE147507
Signature of protein-coding genes in SARS-CoV-2 infected cells in vitro and lung tissue from COVID-19 patients has been reported [8]. In this study, cell lines A549 and Calu3 cells were infected separately with SARS-CoV-2 (USA-WA1/2020). A549 and Calu3 cells were derived from human alveolar basal epithelial adenocarcinoma and human lung epithelial tumor respectively. In some experiments, A549 cells were transfected with ACE2 and then infected with SARS-CoV-2. All together there were 4 experimental conditions for which we analyzed the differential lncRNA expression: (i) expression in A549 cells infected with SARS-CoV-2 compared to expression in mock-transfected A549 cells, (ii) expression in A549 cells expressing exogenous ACE2 and infected with SARS-CoV-2 compared with that of in mock-transfected A549 cells, (iii) expression in A549 cells expressing exogenous ACE2 and infected with SARS-CoV-2 compared with that of in exogenous expressing ACE2 in A549 cells and (iv) expression in Calu-3 cells infected with SARS-CoV-2 compared to expression in mock-transfected Calu-3 cells. There was a biological replicate of the condition in (iii) Besides, sequencing data for RNA samples from lungs tissues from COVID-19 patients and control was also analyzed.
We downloaded the raw RNA sequencing data from Gene Expression Omnibus (GSE147507). The data contained the raw read counts obtained for different experimental conditions, divided into distinct series. Each series had two or more replicates in which mock-treated cells served as control. We carried out differential expression analysis between the mock-treated samples and the infected samples as contained in the respective series using Bioconductor package edgeR. Low abundant genes were filtered out from the dataset before carrying out the analysis. EdgeR uses TMM (Trimmed mean of M-values) to normalize for library composition by computing a set of scale factors with which the effective library sizes are calculated. It thereby estimates dispersion and fits generalized linear models based on negative binomial distribution. The correction for false discovery rates (FDR) was done by Benjamini-Hochberg method. Genes which had false discovery rate (FDR) ≤ 0.05 were considered to be significant. Other important parameters we looked into were magnitude of the fold changes with respect to the controls, given in the column logFC and the average counts per million of the genes in the samples, as provided in the log(cpm) column in the output files. Based on these values the candidate genes were selected for further analysis (Supplementary Text Table STT2.1).
The heatmaps depicting the expression of the deregulated lncRNAs in A549 and Calu cells, in control and experimental conditions, were generated using the heatmap.2 function of “gplots” package in R. The normalized counts of the lncRNAs (log(cpm) values) were used as inputs to construct the heatmaps. We have used the ‘Ward.D2’ method of hierarchical clustering for the lncRNAs, and z-scores were calculated for each lncRNA. The clustering has been done only on the genes, while the samples have been arranged in a sequential manner as per their experimental set, with the mock-treated samples and the experimental samples grouped separately.
2.2. Interacting partners of long non-coding RNA
We downloaded experimentally determined and curated physical interaction data of noncoding RNA with DNA, mRNA, microRNA, proteins and others from NPInter v4.0 (http://bigdata.ibp.ac.cn/npinter4) [52]. The database catalogues experimentally derived interactions, collected manually from publications in peer-reviewed journals and annotated using other databases like NONCODE, miRBase and UniProt. Annotation of genes in NPInter database was further annotated using NCBI database (ftp://ftp.ncbi.nih.gov/gene/DATA/GENE_INFO/Mammalia/).
2.3. Association of genes/proteins with biological pathways
To identify possible functional implication of the deregulated lncRNAs, we analyzed the interacting protein and mRNA partners of the deregulated lncRNA by enrichment analysis. Enrichment analysis provides information about the overrepresentation of the given genes in particular pathway. We carried out enrichment analysis using online facility at Enrichr at https://amp.pharm.mssm.edu/Enrichr/ [53]. Enrichr is an integrative web-based software application for analysis of a gene-set comparing with various gene-set libraries. Given an input list of genes, it provides enrichment for different libraries like BioPlanet pathways and Gene Ontology (GO) terms for biological process. The online facility uses pathways catalogued in various databases; we have chosen the BioPlanet pathways for comprehensive coverage of different disease conditions including infection disease [54].
2.4. Binding of transcription factors at the putative promoters of lncRNAs
To find binding abilities of transcription factors at the putative promoters (-5Kb upstream to +1kb downstream of transcription start sites of the lncRNAs), we utilized the searchable ChIPBase v2.0 (http://rna.sysu.edu.cn/chipbase/) database. This database catalogues curated experimental data from chromatin immune-precipitation followed by sequencing (ChIP-seq) datasets [55]. Binding sites for transcription factors (TFs) IRF1-IRF5, IRF8, IRF9 and STAT1-STA4, STAT5A, STAT6, known to initiate the antiviral responses, were obtained from the database. We have also included MYC as MALAT1 and NEAT1 interacts with active chromatin site of the MYC gene [56] and activation of MYC has been observed in IAV infection. Activation of MYC has also been shown to modulate IAV-induced changes in metabolism; inhibition of MYC activation reversed infection-induced changes in metabolism [57]. For several transcription factors, more than one cell line or different conditions were used to determine binding of TF by ChIP-seq. For example, binding sites of MYC were observed in different cell lines like Foreskin fibroblast cells, B-cell lymphoma cell lines (RAMOS, Raji and Blue1), HEK293T cells expressing wild type MYC and mutant DNA binding domain of MYC. Binding sites of STAT1 were observed in un-stimulated and IFNG stimulated HeLa cells, K562 cells treated with IFNG and IFNA separately. Binding sites of IRF1 were observed in LoVo cells and K562 cells treated with IFNG and IFNA separately. Detail of the cell lines and the conditions obtained from ChIPBase is shown in the Supplementary text Table STT2.2. We have considered only those TFs that were obtained in two independent cell lines and/or different conditions.
To find out the whether binding of these TFs at the putative promoters of the deregulated lncRNAs over the randomly chosen TFs, we have tested randomly generated 4 sets of TFs, each containing 30 TFs and identified their binding sites within -5Kb to +1 Kb of the transcription start sites of the deregulated lncRNAs. Out of the 120 randomly chosen TFs, ASCL1, CAMTA2, ETV4, E2F2, HEY1, HEYL, HSF4, IRF1, IRF9, IRX5_l, KLF1, KLF6, LHX3, MEOX2, NFATC3, OTX2, RELA, SETDB1, SIX5, TP53, VDR, ZBTB2, and TGIF1 had ChIP-seq data in ChiPBase database. Since IRF1 was also among the TFs we used for our targeted analysis, we omit the data for IRF1 from these sets. For each of 22 TFs, we collected the binding sites within -5Kb to +1 Kb of the transcription start sites. Total number of binding sites for each of the deregulated lncRNAs were compared with that of obtained with MYC, IRF1-IRF5 and STAT1-STAT5A.
For comparison of different set of genes/proteins, we used online facility at http://bioinformatics.psb.ugent.be/webtools/Venn/.
2.5. Statistical analysis
For analysis of raw sequencing data form GSE147507, we used Bioconductor package edgeR. Low abundant genes were filtered out from the dataset before carrying out analysis. Genes which had false discovery rate (FDR) ≤ 0.05 were considered to be significant.
For association of genes/protein with different pathway, we used online facility Enrichr at https://amp.pharm.mssm.edu/Enrichr/. Given a set of genes as input, this database returns results of enriched pathways with significant levels (p-values and adjusted p-values with multiple testing corrections) as shown in the Supplementary Tables SXT3A. For over representation of binding sites at the different regions within -30Kb to +10Kb of the transcription start sites and preferences of binding of MYC, STATs and IRFs over the randomly chosen TFs, we used ANOVA (one way or 2 way). All statistical analyses were performed using Graph pad prism software (Version 8, Sandiego, CA, USA).
3. Result
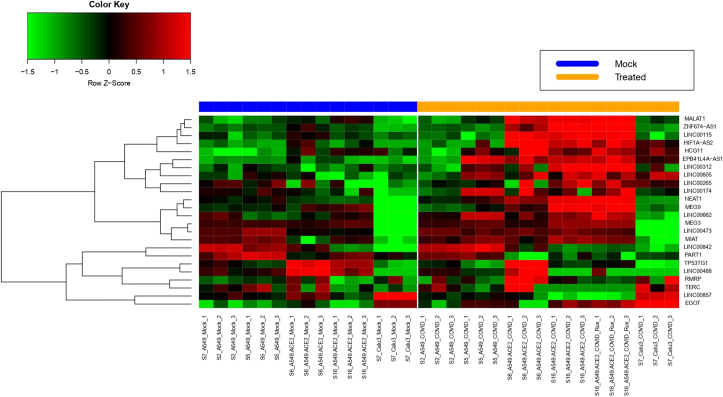
Expression of several lncRNA was deregulated in SARS-CoV-2 infected cells. Among the significantly (FDR≤ 0.05) altered expression, increased expressions of EGOT, EPB41L4A-AS1, HCG11, HIF1A-AS2, LINC00115, LINC00174, LINC00265, LINC00312, LINC00473, LINC00605, LINC00662, LINC00842, MALAT1, MEG3, MEG9, MIAT, NEAT1, RMRP, TERC and ZNF674-AS1 and decreased expressions of LINC00488, LINC00857, PART1, TP53TG1 were observed in more than one cell lines (Figure 1). For example, expression of EGOT was increased 3.0, 4.6 and 3.4 fold in A549 cells transfected with ACE2 were infected with SARS-CoV-2 and compared with either mock-infected A549 and mock A549 transfected with ACE2 and Calu cells infected with the virus respectively. Expressions of several lncRNAs were altered only in one cell line or increased in one cell line and decreased in another cell line (Supplementary Text Table STT2A). Besides, expressions few pseudogenes or noncoding RNA other than lncRNA were also altered (Supplementary Text Table STT2B). In our subsequent analysis, we have used only those lincRNAs that were altered consistently in more than one experiment. Detailed result is shown in the Supplementary Tables SXT1A- SXT1E.
Figure 1.
Heatmap representing the expression of the 24 lncRNAs found to be upregulated (20) or downregulated (4) in SARS-CoV-2 infected cells wrt to mock-treated cells, for more than one experimental condition. The rows represent the lncRNAs while the columns are the experimental samples grouped according to mock-treatment or SARS-CoV-2 treatment, in A549 and Calu cell lines. The samples are named according to the experimental set they have been taken from (Series2, Series5, Series6, Series7 and Series16), followed by the cell line (A549/ Calu) and the treatment (Mock/ SARS-CoV-2).
3.1. Interacting partners of lncRNAs
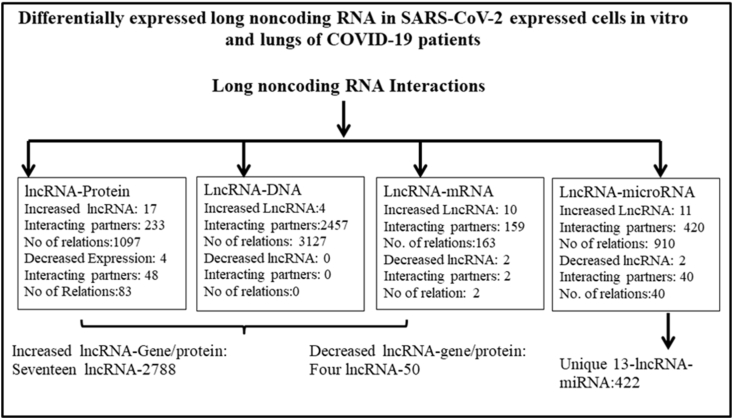
Interacting partners of lncRNAs may provide functional information of the deregulated lncRNAs. Different experimentally determined interacting partners of the lncRNAs were taken from NPInter database (http://bigdata.ibp.ac.cn/npinter4) as described in the material and methods. These interacting partners were determined mainly in high throughput assays, curetted and provided in the database. Summary of the result is shown in Figure 2. All together 2788 unique genes/proteins interact with 17 lncRNAs that have been shown to increase in SARS-CoV-2 infected cells. It was evident that NEAT1 interacts with maximum number of genes/proteins, followed by MALAT1. NEAT1 and MALAT1 together target more than 95% of the genes/proteins; the majority of the interactions were at DNA level (Supplementary Tables SXT2A and SXT2B). Similarly, 50 genes/proteins were target of four lncRNAs that are decreased in SARS-CoV-2 infected cells. Altered lncRNAs could also interact with 422 miRNAs (Supplementary Tables SXT2C and SXT2D).
Figure 2.
Summary of the interaction of lncRNA with DNA, mRNA, microRNA (miRNA) and proteins taken from NPInter v4.0 (http://bigdata.ibp.ac.cn/npinter4). This experimental data was obtained in high throughput assays in different cell lines and catalogued in the database; whether similar interactions could also be obtained in cell lines infected with SARS-CoV-2 remain unknown.
3.2. NCATS BioPlanet (https://tripod.nih.gov/bioplanet/) pathways associated with lncRNA interacting proteins/genes
To obtain the functional implications of the deregulated lncRNA, we used their interacting partners for the association of the genes/protein with different pathways described in NCATS BioPlanet (https://tripod.nih.gov/bioplanet/). Result of such analysis using Enrichr (Supplementary Tables S3) showed that 1378 BioPlanet pathways were associated with lncRNA interacting genes/proteins (Supplementary Table SXT3A) and summarized in Supplementary Text Table STT3.1A. We categorized the pathways related to (i) virus life cycle like Influenza viral RNA transcription and replication pathway and others, (ii) interleukin, interferon, and inflammation like Interleukin-6 signaling pathway, interleukin-2 signaling pathway, chemokine signaling pathway, Antiviral mechanism by interferon-stimulated genes and others, (iii) related to immunity and immune cell dysfunctions like B cell survival pathway, Natural killer cell receptor signaling pathway and others and (iv) other pathways, mainly related to signaling pathways like messenger RNA processing, IGF1 pathway, mTOR signaling pathway and many others. Representative result is shown in Table 1 and detail result is shown in Supplementary Table STT3B. Immune cell dysfunctions observed in COVID-19 is shown in Supplementary Table STT3C. In these 3 categories together, 473 lncRNA interacting genes/proteins are associated with 80 different BioPlanet pathways related to viral life cycles, interleukin, interferon, inflammation, immunity and immune cell dysfunctions. Detailed result of association of genes with each category is shown in Supplementary Tables SXT3B- SXT3D.
Table 1.
Representative result of the association of deregulated lncRNA interacting partners with pathways related to infection, inflammation and immune functions.
| Pathway group | Pathways (total no of pathways) | Total unique genes/proteins |
|---|---|---|
| Virus replication, transcription, life cycle-related pathways | HIV factor interactions with host, HIV genome transcription, Human cytomegalovirus and MAP kinase pathways, Influenza factor interactions with host, Influenza infection, Influenza viral RNA transcription and replication, SARS coronavirus protease, Viral messenger RNA biosynthesis and others (15) | 111, 253 Gene-pathway relations |
| Interleukin, interferon, inflammation-related pathways | Antiviral mechanism by interferon-stimulated genes, Chemokine signaling pathway, Inflammatory response pathway, Interferon alpha/beta signaling, Interferon gamma signaling regulation, Interleukin-2 receptor beta chain in T cell activation, Interleukin-6 signaling pathway, Type I interferon (interferon-alpha/beta) pathway and Type II interferon signaling (interferon-gamma) and others (36) | 288, 505 Gene-pathway relations |
| Immune cell functions, immunity-related pathways | Adaptive immune system, Antigen processing: cross presentation, B cell survival pathway, MHC class II antigen presentation, Natural killer cell-mediated cytotoxicity, Platelet activation, signaling and aggregation, T cell receptor signaling in naive CD4+ T cells T cell receptor signaling in naive CD8+ T cells, T cell signal transduction and others (29) | 239, 699 Gene-pathway relations |
3.3. Possible regulation of long non-coding RNA by IFN-regulatory (IFR) proteins, STATs and MYC
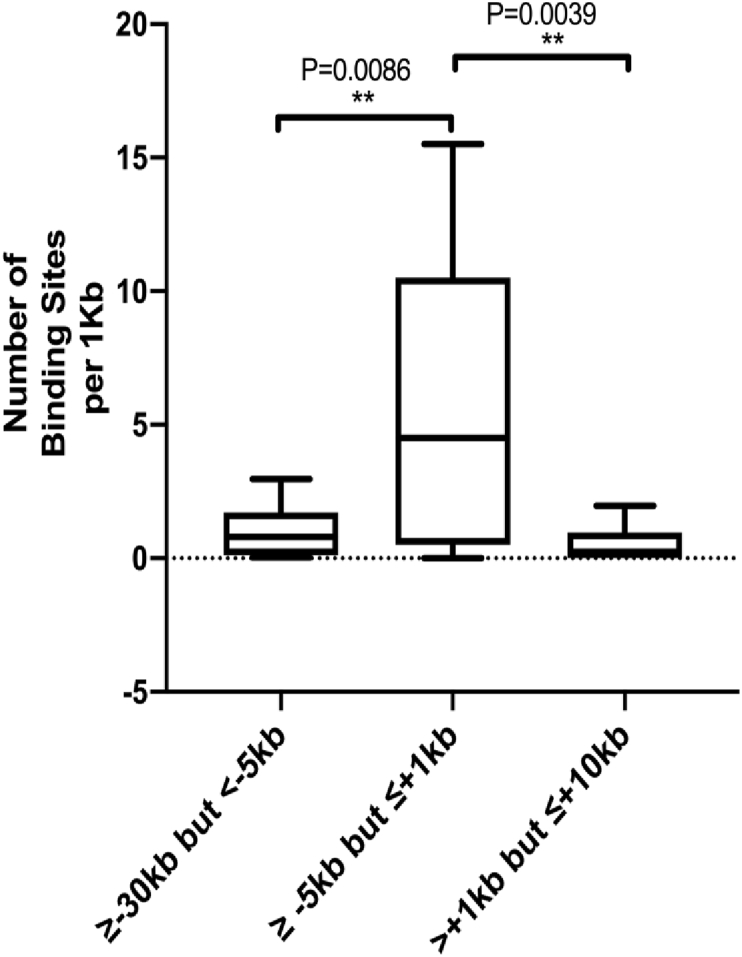
Given that transcription factors like IFN-regulatory proteins, NFκB, and others to initiate antiviral response by the host [58] and secreted interleukins through interleukin receptors activate the JAK-STAT signaling cascade to induce expression of antiviral interferon-stimulated genes (ISGs) [59], we searched whether deregulated lncRNAs could be targets of transcription factors IFR1-IFR5, IRF8, and IRF9, STAT1-STAT4, STA5A and STAT6 in ChIPBase v2.0 (http://rna.sysu.edu.cn/chipbase/) database. This database catalogs ChIP-seq data in different cell lines at different conditions (Table STT2.2). We also include MYC as MALAT1 and NEAT1 interacts with this TF [56] and known to involve in IAV infection [57]. Number of binding sites per 1Kb of all these TRFs was significantly higher at regions -5Kb to +1Kb of transcription start sites (TSS) of the lncRNA in the region between -30Kb to +10Kb of the TSS (Figure 3). Total number of binding sites in the regions are shown in the Supplementary Table SXT4. This result indicates that these TFs might have preference for binding at -5Kb to +1Kb of TSS, the putative promoters of the lncRNA.
Figure 3.
Total binding sites of the TFs IFR1-IFR5, IRF8, IRF9, STAT1-STAT4, STA5A, STAT6 and MYC at -30Kb to +10Kb of the TSS of the deregulated lncRNA obtained from ChIPBase in different cell lines and conditions by ChIP-seq.
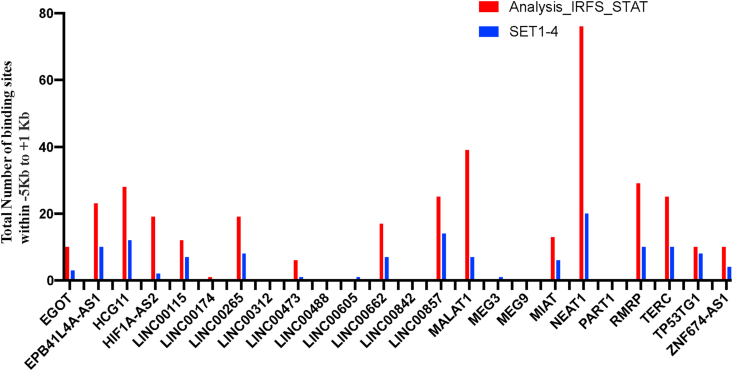
To find out the specificity and/or preference of IRFs, STATs and MYC for binding to the putative promoters of the deregulated lncRNAs, we have tested randomly chosen 22 TFs for their binding to -5Kb to +1Kb of the TSS. Number of binding sites within -5Kb to +1 Kb of the 22 randomly chosen TFs at the putative promoters of the deregulated lncRNAs were significantly (p = 0.0053) lower compared to that of obtained with MYC, IRF1-IRF5, IRF8, IRF9, STAT1-STAT4, STA5A and STAT6. The result (set 1–4) is shown in the Figure 4 and the detail result is shown Supplementary Table SXT5 This result shows that the MYC, IRFs and STATs have preference for binding to the putative promoters of the deregulated lncRNAs in different cell lines and conditions over randomly chosen TFs.
Figure 4.
Overrepresentation of binding sites of the transcription factors IRFs, STATs and MYC at the putative promoters of the deregulated lncRNA in comparison of the randomly chosen 22 TFs denoted by Set 1–4. For details see the supplementary Text Supplementary Text ST2.2.
For stringency, we considered only those TF-lncRNA relations that are observed in more than one independent cell line/condition in ChiPBase. Using this criteria it was observed that IRF1, STAT1 and MYC1 could bind to majority of the lncRNA. For example binding of IRF1 was observed within -5Kb and +1Kb sequences from the TSS of EPB41L4A-AS1, HCG11, HIF1A-AS2, LINC00115, LINC00662, MALAT1 and NEAT1 in more than one cell line and treated with IFNA and IFNG, LINC00265 in IFNG treated and untreated cells, MIAT, TERC, ZNF674-AS1 in cells treated with IFNA or IFNG or untreated cell. IRF1 could also bind at the putative promoters of LINC00857 and TP53TG1 in cells treated with IFNA or IFNG and untreated cells; expression of these lncRNAs was decreased. Increased expression of LINC00265 and decreased expression of TP53TG1 could be mediated by IRF3; IRF4 binds to putative promoters of EGOT, EPB41L4A-AS1, HCG11, HIF1A-AS2, LINC00265, MALAT1, NEAT1, RMRP and TERC that were increased in SARS-CoV-2 infected cells. IRF2, IRF5, IRF8, IRF9 did not have any binding with the deregulated lncRNA following our criteria.
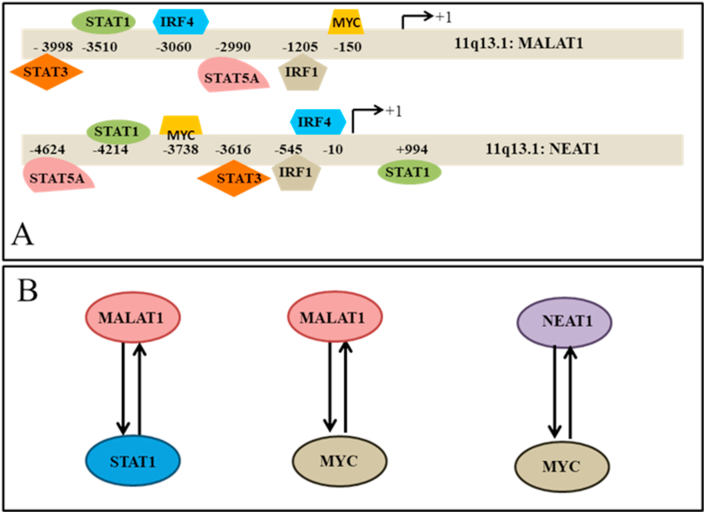
MYC could bind at -5Kb to +1Kb upstream regions of EPB41L4A-AS1, HCG11, HIF1A-AS2, LINC00115, LINC00265, LINC00662, MALAT1, NEAT1, RMRP, TERC and ZNF674-AS1; expression of these gene was increased, LINC00857 and TP53TG1. Expression of the latter two lncRNA was decreased. Interestingly, in HEK293T cells expressing mutant MYC that could not bind to DNA, binding of MYC to the putative promoters of these lncRNAs was not observed. This indicates that binding of MYC could regulate the expression of these lincRNAs. STAT1 binds to the promoters of EPB41L4A-AS1, HCG11, LINC00265, LINC00473, LINC00662, MALAT1, MIAT, NEAT1, TERC, ZNF674-AS1 in IFNG stimulated and un-stimulated cells; RMRP in un-stimulated cells; expression of these lncRNAs was increased in SARS-CoV-2 infected cells. STAT1 also binds to the putative promoter of LINC00857 in IFNG stimulated and un-stimulated cells. No binding site was observed for IRF2, IRF5, IRF8, IRF9. STAT2, STAT4 and STAT6 in more than one independent samples. Summary of the result is shown in Table 2. Binding of different TFs at the putative promoters of MALAT1 and NEAT1 in more than one independent experiment is shown in Figure 5A. Altered expression of the lncRNAs and their possible regulation by transcription factors associated with viral infections [58, 59] indicates that these lncRNAs might be involved in pathogenesis of SARS-CoV-2 infection.
Table 2.
Summary of binding of different transcription factors in more than one cell line at the putative promoters of deregulated lncRNA in SARS-CoV-2 infected cells.
| TFs | LncRNAs (conditions) |
|---|---|
| IRF1 | EPB41L4A-AS1, HCG11, HIF1A-AS2, LINC00115, LINC00662, MALAT1, NEAT1, (Binding observed in IFNA and IFNG treated cells), LINC00265 (Binding observed in IFNG treated cells and untreated cells), MIAT, TERC, ZNF674-AS1 (Binding observed in IFNA, IFNG treated cells and untreated cells)[Increased expression] LINC00857, TP53TG1 (Binding observed in IFNA, IFNG treated cells and untreated cells)[Decreased expression] |
| IRF3 | LINC00265 (Increased expression) TP53TG1 (Decreased expression) |
| IRF4 | EGOT, EPB41L4A-AS1, HCG11, HIF1A-AS2,LINC00265, MALAT1, NEAT1, RMRP, TERC (Increased expression) |
| STAT1 | EPB41L4A-AS1, HCG11, LINC00265, LINC00473, LINC00662, MALAT1, MIAT, NEAT1, TERC, ZNF674-AS1 (Observed in IFNG stimulated and un-stimulated cells), RMRP (un-stimulated) [Increased expression] LINC00857 (Observed in IFNG stimulated and un-stimulated cells) [Decreased expression] |
| STAT3 | EPB41L4A-AS1, HCG11, LINC00265, LINC00662, MALAT1, MIAT, NEAT1, RMRP, TERC, ZNF667-AS1 (Increased expression) LINC00857, TP53TG1 (Decreased expression) |
| STAT5A | EPB41L4A-AS1, MALAT1, NEAT1 |
| MYC | EPB41L4A-AS1, HCG11, HIF1A-AS2, LINC00115, LINC00265, LINC00662, MALAT1, NEAT1, RMRP, TERC, ZNF674-AS1 (Increased expression) LINC00857, TP53TG1 (Decreased expression) |
Figure 5.
A: Binding of different transcription factors at the putative promoters of MALAT1 (hg38 chr11:65,497,688-65,506,431 Size: 8,744 Total Exon Count: 4 Strand: +) and NEAT1 (hg38 chr11:65,422,798-65,445,540 Size: 22,743 Total Exon Count: 1 Strand: +) in more than one independent experiment observed from ChiP-seq data in ChIPBase. For MALAT1 data for MYC (sample ID HUMHG01680), IRF1 (HUMHG05454, treated with IFNA), IRF4 (HUMHG04579), STAT1 (HUMHG00980, stimulated by IFNG), STAT3 (HUMHG04914) and STAT5A (HUMHG05298) is shown. For NEAT1 data for MYC (HUMHG01680), IRF1 (HUMHG05454, treated with IFNA), IRF4 (HUMHG03345), STAT1 (HUMHG00980 treated with IFNG), STAT3 (HUMHG02787) and STAT5A (HUMHG05298). Binding positions of these transcription factors at the putative promoters of MALAT1 and NEAT1 in other samples are shown in the Supplementary Table SXT6.
It has been shown earlier that MALAT1 and NEAT1 interact with regulatory region in the active chromatin sites of STAT1, MYC and many other active genes [56]. Thus such interaction may facilitate the transcription of the TFs cooperating with other TFs. We observed above that STAT1 and MYC occupied in the putative promoters of the two lncRNA in different cell lines. Combing the observations, we speculated that there might be a positive feedback loop between the lncRNAs and TFs (Figure 5B). Such relations may amplify the signal generated by STAT1 or MYC as have been observed for other regulators [60, 61]. Role of MYC in SARS-CoV-2 infection is not known. However, in IAV infection, metabolic deregulation has been observed to be controlled by MYC activation and inhibition of MYC activation reversed infection-induced changes in metabolism [57].
4. Discussion
In the present study, we analysed the RNA sequencing data obtained in different cell lines infected with SARS-CoV-2 and deposited in GEO database (GSE147507) and observed that expression of 20 lncRNAs was increased and 4 lncRNAs was decreased in SARS-CoV-2 infected cells in more than one experimental condition. Expression of NEAT1 was also increased significantly in lungs of COVID-19 patients. These lncRNAs, mainly NEAT1 and MALAT1 interact with thousands of genes/proteins as evident from the data in NPInter v4.0 database that catalogs experimentally determined interactions mainly in high-throughput assays and curated [52]. Association of the interacting partners of the deregulated lncRNA with pathways relevant for viral replication and growth, chemokine and cytokine and immune cell functions revealed that the deregulated lncRNA might play important role in cellular response to SARS-CoV-2 infection through their interacting partners. In addition, we observed from ChIPBase database, that transcription factors IRF1, IRF4, STAT1, STAT3, STAT5A and MYC could bind to the putative promoters of MALAT1 and NEAT1. Result of our analysis with different experimental data from various databases shows that NEAT1 and MALAT1 could contribute to antiviral response to SARS-CoV-2 infection by altering host gene expression.
Among the altered expression of lncRNAs identified in the manuscript, expression of MALAT1, MIAT and NEAT1 was common between HIV infected cells and SARS-CoV-2 infected cells and EGOT and NEAT1 was common between IAV and SARS-CoV-2 infected cells (Supplementary Text Table STT4.1). Most of the studies were carried out in cultured cells or animals infected with different viruses (Supplementary Text Table STT4.2). Recently, increased expression of MALAT1 was reported in bronchoalveolar lavage fluid of COVID-19 patients ([62], Figure 6 of the referred paper).
4.1. Interacting partners of deregulated lncRNAs and their association with COVID-19 relevant pathways
LncRNA interacting genes/proteins are associated with many pathways. Among these, 20 MALT1 and NEAT interacting gene/protein was associated and over represented with interleukin-6 signaling pathway. Excessive production of interleukin-6, popularly called “cytokine storm” has been associated with severe COVID-19 resulting in multi-organ failure [63]. Role of MALAT1 and NEAT1 in this processes, inferred from our in silico analysis, has yet to be validated. Mechanisms by which lncRNA including MALAT1 and NEAT1 can modulate IL-6 and NLRP3 inflammasome have recently been reviewed [64]. Interleukin-2 signaling pathway was significantly enriched with 148 interacting partners of 15 deregulated lncRNA. Decrease in CD8 + T cell and lymphocyte of COVID-19 patients has been shown to mediate through this pathway [65].
4.2. Possible role of deregulated lncRNA in SARS-CoV-2 in interferon responses
It is difficult to infer with high confidence the role of deregulated lncRNAs in IFN response in SARS-CoV-2 infected cells from the in silico analysis. Reduced levels of Type I and III interferon response have been observed in these cell lines, animal models and human lung tissue from the COVID-19 patients. However, a subset of ISGs is induced in these cells [8]. Enhanced expression of ISGs has also been reported in different cells from patients [14, 15] and in mouse model of COVID-19 [16]. Thus in this experimental condition, enhanced expression of the lncRNAs could be independent of Type I and III interferons, but some of the lncRNAs might modify the ISGs levels as observed in different virus infected cells [40, 41, 42, 43, 44] by different lncRNA. Interactions of hnRNP U with lncRNA TP53TG1, EGOT, EPB41L4A-AS1, HIF1A-AS2, LINC00174, LINC00473, LINC00662, MALAT1, MEG3, MEG9, RMRP, ZNF674-AS1 [66] LINC00842 [67], NEAT1 [56] in high throughput assays could contribute to the ISGs levels as has been observed with LncRNA32/LUARIS [40]. Similar interactions of PART1, TP53TG1, EPB41L4A-AS1, HIF1A-AS2, LINC00174, LINC00662, MALAT1, RMRP [66] and NEAT1 [56] may also modulate ISGs as has been observed with TTC39A-AS1/lincRNA-EPS [45]. It is unknown whether paraspeckles is induced in response to SARS-CoV-2 infection; although expression of NEAT1, the indispensible component of paraspeckles was increased in our analysis as well as various viral infections including IAV, HIV and HTNV. Thus similar mechanism might be operative in SARS-CoV-2 infected cells to increase ISGs.
4.3. Limitations of the study
In the present in silico study, we observed several deregulated lncRNAs in more than one experimental condition. Analysis with experimental databases for their interacting partners, association of pathways relevant for virus replication, transcription, life cycle-related, interleukin and interferon related functions, inflammation, immune cell functions and immunity indicates that lncRNA through their interacting partners might be involved in these pathways. Binding of several TFs like IRF1, STAT1 and STAT3 at the putative promoters of several deregulated lncRNAs from ChIP-seq databases indicates that these lncRNAs could be components of viral response. This in silico analysis generates several hypotheses for the role of lncRNAs in SARS-CoV-2 infection and needs to be validated.
5. Conclusion
Deregulated lncRNA might be involved in the regulation of ISGs and control viral replication. Interacting partners of the deregulated lncRNA are associated with interleukin-6 pathways, showing the possible role of lncRNA in SARS-CoV-2 infection. Some of the lncRNAs could be regulated by IFN-regulatory (IFR) proteins, STAT1 and STAT3, well established antiviral responsive regulators. This observation once validated experimentally in patient samples could provide a new therapeutic target combating SARS-CoV-2 infection.
Declarations
Author contribution statement
Nitai P. Bhattacharyya: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Sayantan Laha, Chinmay Saha: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Susmita Dutta, Madhurima Basu, Raghunath Chatterjee, Sujoy Ghosh: Analyzed and interpreted the data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Akhade V.S., Pal D., Kanduri C. Long noncoding RNA: genome organization and mechanism of action. Adv. Exp. Med. Biol. 2017;1008:47–74. doi: 10.1007/978-981-10-5203-3_2. [DOI] [PubMed] [Google Scholar]
- 2.Noh J.H., Kim K.M., McClusky W.G., Abdelmohsen K., Gorospe M. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscipl. Rev. RNA. 2018;9:e1471. doi: 10.1002/wrna.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yao R.W., Wang Y., Chen L.L. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 2019;21:542–551. doi: 10.1038/s41556-019-0311-8. [DOI] [PubMed] [Google Scholar]
- 4.Atianand M.K., Caffrey D.R., Fitzgerald K.A. Immunobiology of long noncoding RNAs. Annu. Rev. Immunol. 2017;35:177–198. doi: 10.1146/annurev-immunol-041015-055459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stojic L., Lun A.T.L., Mascalchi P., Ernst C., Redmond A.M., Mangei J., Barr A.R., Bousgouni V., Bakal C., Marioni J.C., Odom D.T., Gergely F. A high-content RNAi screen reveals multiple roles for long noncoding RNAs in cell division. Nat. Commun. 2020;11:1851. doi: 10.1038/s41467-020-14978-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Moller R., Jordan T.X., Oishi K., Panis M., Sachs D., Wang T.T., Schwartz R.E., Lim J.K., Albrecht R.A., tenOever B.R. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045. doi: 10.1016/j.cell.2020.04.026. e1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bost P., Giladi A., Liu Y., Bendjelal Y., Xu G., David E., Blecher-Gonen R., Cohen M., Medaglia C., Li H., Deczkowska A., Zhang S., Schwikowski B., Zhang Z., Amit I. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell. 2020;181:1475–1488. doi: 10.1016/j.cell.2020.05.006. e1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu H., Chan J.F., Wang Y., Yuen T.T., Chai Y., Hou Y., Shuai H., Yang D., Hu B., Huang X., Zhang X., Cai J.P., Zhou J., Yuan S., Kok K.H., To K.K., Chan I.H., Zhang A.J., Sit K.Y., Au W.K., Yuen K.Y. Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: an Ex Vivo study with implications for the pathogenesis of COVID-19. Clin. Infect. Dis. 2020;71:1400–1409. doi: 10.1093/cid/ciaa410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Brien T.R., Thomas D.L., Jackson S.S., Prokunina-Olsson L., Donnelly R.P., Hartmann R. Weak induction of interferon expression by severe acute respiratory syndrome coronavirus 2 supports clinical trials of interferon-lambda to treat early coronavirus disease 2019. Clin. Infect. Dis. 2020;71:1410–1412. doi: 10.1093/cid/ciaa453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antonelli G., Turriziani O., Pierangeli A., d'Ettorre G., Galardo G., Pugliese F., Mastroianni C.M., Scagnolari C. Type I interferons can be detected in respiratory swabs from SARS-Cov-2 infected patients. J. Clin. Virol. 2020;128:104450. doi: 10.1016/j.jcv.2020.104450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., Paul van Schayck J., Mykytyn A.Z., Duimel H.Q., van Donselaar E., Riesebosch S., Kuijpers H.J.H., Schipper D., van de Wetering W.J., de Graaf M., Koopmans M., Cuppen E., Peters P.J., Haagmans B.L., Clevers H. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Z., Ren L., Zhang L., Zhong J., Xiao Y., Jia Z., Guo L., Yang J., Wang C., Jiang S., Yang D., Zhang G., Li H., Chen F., Xu Y., Chen M., Gao Z., Dong J., Liu B., Zhang X., Wang W., He K., Jin Q., Li M., Wang J. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27:883–890. doi: 10.1016/j.chom.2020.04.017. e882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J.S., Park S., Jeong H.W., Ahn J.Y., Choi S.J., Lee H., Choi B., Nam S.K., Sa M., Kwon J.S., Jeong S.J., Lee H.K., Park S.H., Choi J.Y., Kim S.H., Jung I., Shin E.C. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Israelow B., Song E., Mao T., Lu P., Meir A., Liu F., Alfajaro M.M., Wei J., Dong H., Homer R.J., Ring A., Wilen C.B., Iwasaki A. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J. Exp. Med. 2020;217 doi: 10.1084/jem.20201241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arunachalam P.S., Wimmers F., Mok C.K.P., Perera R., Scott M., Hagan T., Sigal N., Feng Y., Bristow L., Tak-Yin Tsang O., Wagh D., Coller J., Pellegrini K.L., Kazmin D., Alaaeddine G., Leung W.S., Chan J.M.C., Chik T.S.H., Choi C.Y.C., Huerta C., Paine McCullough M., Lv H., Anderson E., Edupuganti S., Upadhyay A.A., Bosinger S.E., Maecker H.T., Khatri P., Rouphael N., Peiris M., Pulendran B. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yi K., Zhang Y., Wang Y., Wang Z., Xie M., Jin Z., Zhao T. Long noncoding RNA and its role in virus infection and pathogenesis. Front. Biosci. (Landmark Ed.) 2019;24:777–789. doi: 10.2741/4750. [DOI] [PubMed] [Google Scholar]
- 19.Wang J., Cen S. Roles of lncRNAs in influenza virus infection. Emerg. Microb. Infect. 2020;9:1407–1414. doi: 10.1080/22221751.2020.1778429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y., Yu T., Ding Y., Li Y., Lei J., Hu B., Zhou J. Analysis of expression profiles of long noncoding RNAs and mRNAs in A549 cells infected with H3N2 swine influenza virus by RNA sequencing. Virol. Sin. 2020;35:171–180. doi: 10.1007/s12250-019-00170-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao L., Xia M., Wang K., Lai C., Fan H., Gu H., Yang P., Wang X. A long non-coding RNA IVRPIE promotes host antiviral immune responses through regulating interferon beta1 and ISG expression. Front. Microbiol. 2020;11:260. doi: 10.3389/fmicb.2020.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maarouf M., Chen B., Chen Y., Wang X., Rai K.R., Zhao Z., Liu S., Li Y., Xiao M., Chen J.L. Identification of lncRNA-155 encoded by MIR155HG as a novel regulator of innate immunity against influenza A virus infection. Cell Microbiol. 2019;21 doi: 10.1111/cmi.13036. [DOI] [PubMed] [Google Scholar]
- 23.More S., Zhu Z., Lin K., Huang C., Pushparaj S., Liang Y., Sathiaseelan R., Yang X., Liu L. Long non-coding RNA PSMB8-AS1 regulates influenza virus replication. RNA Biol. 2019;16:340–353. doi: 10.1080/15476286.2019.1572448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan Q., Zhao Z., Liao Y., Chiu S.H., Wang S., Chen B., Chen N., Chen Y., Chen J.L. Identification of an interferon-stimulated long noncoding RNA (LncRNA ISR) involved in regulation of influenza A virus replication. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20205118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu L., Wang T., Tang Q., Li G., Wu P., Chen K. Long non-coding RNAs: regulators of viral infection and the interferon antiviral response. Front. Microbiol. 2018;9:1621. doi: 10.3389/fmicb.2018.01621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J., Wang Y., Zhou R., Zhao J., Zhang Y., Yi D., Li Q., Zhou J., Guo F., Liang C., Li X., Cen S. Host long noncoding RNA lncRNA-PAAN regulates the replication of influenza A virus. Viruses. 2018;10 doi: 10.3390/v10060330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang P., Xu J., Wang Y., Cao X. An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism. Science. 2017;358:1051–1055. doi: 10.1126/science.aao0409. [DOI] [PubMed] [Google Scholar]
- 28.Wang Q., Zhang D., Feng W., Guo Y., Sun X., Zhang M., Guan Z., Duan M. Long noncoding RNA TSPOAP1 antisense RNA 1 negatively modulates type I IFN signaling to facilitate influenza A virus replication. J. Med. Virol. 2019 doi: 10.1002/jmv.25483. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Q., Chen C.Y., Yedavalli V.S., Jeang K.T. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. mBio. 2013;4 doi: 10.1128/mBio.00596-12. 512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schynkel T., Szaniawski M.A., Spivak A.M., Bosque A., Planelles V., Vandekerckhove L., Trypsteen W. Interferon-mediated long non-coding RNA response in macrophages in the context of HIV. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21207741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ray R.M., Morris K.V. Long non-coding RNAs mechanisms of action in HIV-1 modulation and the identification of novel therapeutic targets. Noncoding RNA. 2020;6 doi: 10.3390/ncrna6010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y., Huang L., Luo W., Li F., Xiao J., Qin S., Wang Z., Song X., Jin F. Single-cell RNA-sequencing analysis identifies host long noncoding RNA MAMDC2-AS1 as a co-factor for HSV-1 nuclear transport. Int. J. Biol. Sci. 2020;16:1586–1603. doi: 10.7150/ijbs.42556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu W., Ding C. Roles of LncRNAs in viral infections. Front. Cell Infect. Microbiol. 2017;7:205. doi: 10.3389/fcimb.2017.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sonkoly E., Bata-Csorgo Z., Pivarcsi A., Polyanka H., Kenderessy-Szabo A., Molnar G., Szentpali K., Bari L., Megyeri K., Mandi Y., Dobozy A., Kemeny L., Szell M. Identification and characterization of a novel, psoriasis susceptibility-related noncoding RNA gene, PRINS. J. Biol. Chem. 2005;280:24159–24167. doi: 10.1074/jbc.M501704200. [DOI] [PubMed] [Google Scholar]
- 35.Li J., Li M., Wang X., Sun M., Ma C., Liang W., Gao X., Wei L. Long noncoding RNA NRAV promotes respiratory syncytial virus replication by targeting the MicroRNA miR-509-3p/Rab5c Axis to regulate vesicle transportation. J. Virol. 2020;94 doi: 10.1128/JVI.00113-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tao X.W., Zeng L.K., Wang H.Z., Liu H.C. LncRNA MEG3 ameliorates respiratory syncytial virus infection by suppressing TLR4 signaling. Mol. Med. Rep. 2018;17:4138–4144. doi: 10.3892/mmr.2017.8303. [DOI] [PubMed] [Google Scholar]
- 37.Peng X., Gralinski L., Armour C.D., Ferris M.T., Thomas M.J., Proll S., Bradel-Tretheway B.G., Korth M.J., Castle J.C., Biery M.C., Bouzek H.K., Haynor D.R., Frieman M.B., Heise M., Raymond C.K., Baric R.S., Katze M.G. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. mBio. 2010;1 doi: 10.1128/mBio.00206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valadkhan S., Plasek L.M. Long non-coding RNA-mediated regulation of the interferon response: a new perspective on a familiar theme. Pathogens Immun. 2018;3:126–148. doi: 10.20411/pai.v3i1.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma H., Han P., Ye W., Chen H., Zheng X., Cheng L., Zhang L., Yu L., Wu X., Xu Z., Lei Y., Zhang F. The long noncoding RNA NEAT1 exerts antihantaviral effects by acting as positive feedback for RIG-I signaling. J. Virol. 2017;91 doi: 10.1128/JVI.02250-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishitsuji H., Ujino S., Yoshio S., Sugiyama M., Mizokami M., Kanto T., Shimotohno K. Long noncoding RNA #32 contributes to antiviral responses by controlling interferon-stimulated gene expression. Proc. Natl. Acad. Sci. U.S.A. 2016;113:10388–10393. doi: 10.1073/pnas.1525022113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kambara H., Niazi F., Kostadinova L., Moonka D.K., Siegel C.T., Post A.B., Carnero E., Barriocanal M., Fortes P., Anthony D.D., Valadkhan S. Negative regulation of the interferon response by an interferon-induced long non-coding RNA. Nucleic Acids Res. 2014;42:10668–10680. doi: 10.1093/nar/gku713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie Q., Chen S., Tian R., Huang X., Deng R., Xue B., Qin Y., Xu Y., Wang J., Guo M., Chen J., Tang S., Li G., Zhu H. Long noncoding RNA ITPRIP-1 positively regulates the innate immune response through promotion of oligomerization and activation of MDA5. J. Virol. 2018;92 doi: 10.1128/JVI.00507-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ouyang J., Zhu X., Chen Y., Wei H., Chen Q., Chi X., Qi B., Zhang L., Zhao Y., Gao G.F., Wang G., Chen J.L. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe. 2014;16:616–626. doi: 10.1016/j.chom.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carpenter S., Aiello D., Atianand M.K., Ricci E.P., Gandhi P., Hall L.L., Byron M., Monks B., Henry-Bezy M., Lawrence J.B., O'Neill L.A., Moore M.J., Caffrey D.R., Fitzgerald K.A. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341:789–792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Atianand M.K., Hu W., Satpathy A.T., Shen Y., Ricci E.P., Alvarez-Dominguez J.R., Bhatta A., Schattgen S.A., McGowan J.D., Blin J., Braun J.E., Gandhi P., Moore M.J., Chang H.Y., Lodish H.F., Caffrey D.R., Fitzgerald K.A. A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell. 2016;165:1672–1685. doi: 10.1016/j.cell.2016.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carnero E., Barriocanal M., Prior C., Pablo Unfried J., Segura V., Guruceaga E., Enguita M., Smerdou C., Gastaminza P., Fortes P. Long noncoding RNA EGOT negatively affects the antiviral response and favors HCV replication. EMBO Rep. 2016;17:1013–1028. doi: 10.15252/embr.201541763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qu D., Sun W.W., Li L., Ma L., Sun L., Jin X., Li T., Hou W., Wang J.H. Long noncoding RNA MALAT1 releases epigenetic silencing of HIV-1 replication by displacing the polycomb repressive complex 2 from binding to the LTR promoter. Nucleic Acids Res. 2019;47:3013–3027. doi: 10.1093/nar/gkz117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J., Chen C., Ma X., Geng G., Liu B., Zhang Y., Zhang S., Zhong F., Liu C., Yin Y., Cai W., Zhang H. Long noncoding RNA NRON contributes to HIV-1 latency by specifically inducing tat protein degradation. Nat. Commun. 2016;7:11730. doi: 10.1038/ncomms11730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Imamura K., Imamachi N., Akizuki G., Kumakura M., Kawaguchi A., Nagata K., Kato A., Kawaguchi Y., Sato H., Yoneda M., Kai C., Yada T., Suzuki Y., Yamada T., Ozawa T., Kaneki K., Inoue T., Kobayashi M., Kodama T., Wada Y., Sekimizu K., Akimitsu N. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell. 2014;53:393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 50.Nakagawa S., Yamazaki T., Hirose T. Molecular dissection of nuclear paraspeckles: towards understanding the emerging world of the RNP milieu. Open Biol. 2018;8 doi: 10.1098/rsob.180150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fox A.H., Lamond A.I. Paraspeckles. Cold Spring Harb. Perspect. Biol. 2010;2:a000687. doi: 10.1101/cshperspect.a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teng X., Chen X., Xue H., Tang Y., Zhang P., Kang Q., Hao Y., Chen R., Zhao Y., He S. NPInter v4.0: an integrated database of ncRNA interactions. Nucleic Acids Res. 2020;48:D160–D165. doi: 10.1093/nar/gkz969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., McDermott M.G., Monteiro C.D., Gundersen G.W., Ma'ayan A., Enrichr A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang R., Grishagin I., Wang Y., Zhao T., Greene J., Obenauer J.C., Ngan D., Nguyen D.T., Guha R., Jadhav A., Southall N., Simeonov A., Austin C.P. The NCATS BioPlanet - an integrated platform for exploring the universe of cellular signaling pathways for toxicology, systems biology, and chemical genomics. Front. Pharmacol. 2019;10:445. doi: 10.3389/fphar.2019.00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou K.R., Liu S., Sun W.J., Zheng L.L., Zhou H., Yang J.H., Qu L.H. ChIPBase v2.0: decoding transcriptional regulatory networks of non-coding RNAs and protein-coding genes from ChIP-seq data. Nucleic Acids Res. 2017;45:D43–D50. doi: 10.1093/nar/gkw965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.West J.A., Davis C.P., Sunwoo H., Simon M.D., Sadreyev R.I., Wang P.I., Tolstorukov M.Y., Kingston R.E. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell. 2014;55:791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smallwood H.S., Duan S., Morfouace M., Rezinciuc S., Shulkin B.L., Shelat A., Zink E.E., Milasta S., Bajracharya R., Oluwaseum A.J., Roussel M.F., Green D.R., Pasa-Tolic L., Thomas P.G. Targeting metabolic reprogramming by influenza infection for therapeutic intervention. Cell Rep. 2017;19:1640–1653. doi: 10.1016/j.celrep.2017.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hur S. Double-stranded RNA sensors and modulators in innate immunity. Annu. Rev. Immunol. 2019;37:349–375. doi: 10.1146/annurev-immunol-042718-041356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lazear H.M., Schoggins J.W., Diamond M.S. Shared and distinct functions of type I and type III interferons. Immunity. 2019;50:907–923. doi: 10.1016/j.immuni.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou J., Sun T., Jin S., Guo Z., Cui J. Dual feedforward loops modulate type I interferon responses and induce selective gene expression during TLR4 activation. iScience. 2020;23:100881. doi: 10.1016/j.isci.2020.100881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Michalska A., Blaszczyk K., Wesoly J., Bluyssen H.A.R. A positive feedback amplifier circuit that regulates interferon (IFN)-Stimulated gene expression and controls type I and type II IFN responses. Front. Immunol. 2018;9:1135. doi: 10.3389/fimmu.2018.01135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A., Guo D., Hu W., Yang J., Tang Z., Wu H., Lin Y., Zhang M., Zhang Q., Shi M., Zhou Y., Lan K., Chen Y. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microb. Infect. 2020;9:761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ragab D., Salah Eldin H., Taeimah M., Khattab R., Salem R. The COVID-19 cytokine storm; what we know so far. Front. Immunol. 2020;11:1446. doi: 10.3389/fimmu.2020.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paniri A., Akhavan-Niaki H. Emerging role of IL-6 and NLRP3 inflammasome as potential therapeutic targets to combat COVID-19: role of lncRNAs in cytokine storm modulation. Life Sci. 2020;257:118114. doi: 10.1016/j.lfs.2020.118114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shi H., Wang W., Yin J., Ouyang Y., Pang L., Feng Y., Qiao L., Guo X., Jin R., Chen D. The inhibition of IL-2/IL-2R gives rise to CD8(+) T cell and lymphocyte decrease through JAK1-STAT5 in critical patients with COVID-19 pneumonia. Cell Death Dis. 2020;11:429. doi: 10.1038/s41419-020-2636-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. ENCODE project. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xiao R., Tang P., Yang B., Huang J., Zhou Y., Shao C., Li H., Sun H., Zhang Y., Fu X.D. Nuclear matrix factor hnRNP U/SAF-A exerts a global control of alternative splicing by regulating U2 snRNP maturation. Mol. Cell. 2012;45:656–668. doi: 10.1016/j.molcel.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supplementary material/referenced in article.