Abstract
Objectives:
Erosive hand osteoarthritis (EOA) is a severe and rapidly progressing subset of hand osteoarthritis. Its etiology remains largely unknown, which has hindered development of successful treatments. Our goals were to test the hypothesis that EOA would demonstrate familial clustering in a large statewide population linked to genealogical records, and to determine the association of potential risk factors with EOA.
Methods:
Patients diagnosed with EOA were identified by searching 4,741,840 unique medical records from a comprehensive statewide database, the Utah Population Database (UPDB). Affected individuals were mapped to pedigrees to identify high-risk families with excess clustering of EOA as defined by a Familial Standardized Incidence Ratio (FSIR) of ≥ 2.0. The magnitude of familial risk of EOA in related individuals was calculated using Cox regression models. Association of potential EOA risk factors was analyzed using multivariate conditional logistic regression and logistic regression models.
Results:
We identified 703 affected individuals linked to 240 unrelated high-risk pedigrees with excess clustering of EOA (FSIR ≥ 2.0). The relative risk of developing EOA was significantly elevated in first-degree relatives. There was a significant association with the diagnosis of EOA and age, sex, diabetes, and obesity.
Conclusions:
Familial clustering of EOA observed in a statewide database indicates a potential genetic contribution to the etiology of the disease. Age, sex, diabetes, and obesity are risk factors for EOA. Identification of causal gene variants in these high-risk families may provide insight into the genes and pathways that contribute to EOA onset and progression.
Keywords: Erosive hand osteoarthritis, osteoarthritis, genetics, osteoarthritis cohort, osteoarthritis risk factors
Introduction:
Hand osteoarthritis (OA) is the most prevalent form of OA and is a major cause of disability.1–7 It is a heterogeneous disorder with a substantial genetic contribution.8 Despite significant heritability of hand OA, very few genes and pathways have been discovered that modify the course of the disease.6, 7 Erosive hand osteoarthritis (EOA) is often considered a more severe form of hand OA that affects the distal and proximal interphalangeal joints, although it is unclear if EOA represents a distinct form of general hand OA or simply a more severe stage of the disease.9–13 EOA is defined by its sudden onset, rapid progression, and radiological evidence of central subchondral erosions that have a ‘gull-wing’ or ‘saw-tooth’ appearance, collapse of the subchondral bone, and marginal osteophyte formation.9, 14, 15 Despite the prevalence and severity of EOA16, 17, there are no therapeutics that are effective in preventing the onset or limiting the progression of the disease.18
The main obstacle to the development of disease-modifying therapies is limited understanding of the disease process.19, 20 We have limited knowledge of the genes that confer susceptibility to EOA.21–23 EOA is suggested to have a familial contribution24, but this analysis was limited to sibling pairs. The genetic studies of EOA to date have been limited in size and scope, which has hindered to identification of targets for development of therapeutic intervention.
There have been several described risk factors associated with EOA including sex, alcohol consumption, and obesity, although there has been some discrepancy in risk factors between cohorts.16, 17, 25, 26 Determining the contribution of risk factors in different cohorts allows for a more representative view of patient characteristics associated with the pathogenesis of EOA, and may provide clinically useful information to identify groups at an increased risk of disease development.
Our goal was to utilize the Utah Population Database (UPDB), a large statewide population database linked to comprehensive genealogical records,27–34 to perform a retrospective population-based study to i) test our hypothesis that EOA clusters in large families, ii) define the magnitude of familial risk of EOA, and iii) evaluate our cohort for potential risk factors associated with EOA.
Methods:
Study Approval:
This study was approved by the Institutional Review Boards of the University of Utah (IRB # 79442) and Intermountain Healthcare (IRB # 1050554) and by the Resource for Genetic and Epidemiologic Research.
The Utah Population Database (UPDB):
Our study utilized data drawn from UPDB (https://uofuhealth.utah.edu/huntsman/utah-population-database/). The UPDB is one of the world’s largest and most comprehensive sources of linked population-based information for demographic and genetic studies. The UPDB contains data on over 11 million individuals from the late 18th century to the present. Data are updated as they become available from statewide birth and death certificates, hospitalizations, ambulatory surgeries, and drivers licenses. UPDB creates and maintains links between the database and the medical records held by the two largest healthcare providers in Utah as well as Medicare claims. The multigenerational pedigrees representing Utah’s founders and their descendants were constructed based on data provided by the Genealogical Society of Utah (GSU). Pedigrees spanning the past century have been expanded extensively based on vital records and, together with the GSU data, form the basis of the deep genealogical structure of the UPDB. The UPDB has been used in the early investigational stages to demonstrate familial clustering of diseases35–37, and has been instrumental to the discovery of many disease causing genes, including breast and ovarian cancer31, 34, colon cancer28, and prostate cancer.38
Selection of Cases:
We identified individuals diagnosed with erosive hand osteoarthritis (EOA) between October 1st, 2015 - December 31st, 2019 in the UPDB using the International Classification of Diseases (ICD) Tenth revisions code: ICD-10 M15.4 form 4,741,840 unique medical records. Individuals were excluded if they were also diagnosed with rheumatoid arthritis (ICD-9 714.0, ICD-10 M05.xx), other rheumatoid arthritis subtypes (ICD-9 714.2, ICD-10 M06.xx), or juvenile rheumatoid arthritis (ICD-9 714.3, ICD-10 M08.xx). The ICD codes are provider entered based on patient visits with the possibility of the billing team reviewing and modifying for improved accuracy (https://uofuhealth.utah.edu/huntsman/utah-population-database/data/). Affected individuals were required to have relatives in the UPDB to be included in our study cohort so we could link them to pedigrees. The selection of controls for familial risk analysis and unaffected individuals for age-standardized sex-specific EOA incidence rate analysis are described below.
Validation of Cases:
We reviewed the medical charts of 57 random cases to determine if the EOA diagnosis based on coding was correct. Of the 57 cases, we were able to chart review 48 (84.2%). All 48 individuals had a verifiable EOA diagnosis confirmed by radiographic evidence (44/48) or diagnosis by a rheumatologist (33/48) or orthopaedic surgeon. No individuals (0/48) had gout, rheumatoid arthritis, or psoriatic arthritis. Three individuals (6.31%) had psoriasis noted on a skin exam. One individual (2.1%) had evidence of erosive OA in another joint (the 1st metatarsophalangeal joint). We were unable to verify the EOA diagnosis of 9 (15.8%) individuals because we did not have access to their medical records. These individuals were likely diagnosed outside of the U of U Healthcare System and Intermountain Healthcare Hospital and Clinics. These two health systems serve ~75–85% of the state and this number correlates well with the number of individuals (84.2%) we were able to chart review.
High Risk Pedigree Identification:
To determine if there was excess familial clustering of EOA in each pedigree, we utilized the Familial Standardized Incidence Ratio (FSIR).39 FSIR allows for the quantification of familial risk of a disease by comparing the incidence of a disease in a family to its expected incidence in the general population. FSIR is a statistical method that accounts for the number of biological relatives in a pedigree, the degree of relatedness, and the age at which an individual was diagnosed. Exact one-sided Poisson probabilities were calculated under the null hypothesis of no familial enrichment of EOA. Individuals were grouped into fourteen categories based on age (0–30, 31–40, 41–50, 51–60, 61–70, 71–80, and 81–120) and sex. To determine the incidence ratio, the number of years prior to and after diagnosis was calculated for all affected and unaffected individuals, and then the number of living diagnosed years was divided by the number of living undiagnosed years. To determine the pedigree incidence ratio, the UPDB was analyzed to identify the founders of pedigrees containing an affected individual, the affection status of every individual biological relative in each pedigree was determined, and incidence ratio was calculated as described above. The pedigree’s incidence ratio/whole population incidence ratio was used to determine the FSIR. High-risk pedigrees were selected if they had two or more living affected individuals, and if the FSIR was ≥ 2 and significant (p < 0.05) using a chi-squared test as described by Kerber.39
Familial Risk Analysis:
Controls with no history of EOA were randomly selected from the Utah population and matched 10:1 to corresponding EOA cases on sex, birth year, and whether born in Utah. Additionally, we imposed the restrictions that controls must be alive in the matched cases’ diagnosis year. Cases and controls were followed from birth until death, or 2019, or diagnosis year, whichever occurred first. Estimates of familial risk were based on a hazard rate ratio (HR) of familial recurrence, which represents the ratio of the hazard rate for the occurrence of EOA among relatives of the cases with the comparable hazard rate among the relatives of the matching controls. The HRs were calculated using Cox regression models, additionally adjusting for sex and birth year. Because observations within families are non-independent, a Huber-White sandwich estimator of variance for clustered data was used to correct for any families that were analyzed multiple times because of the multiple EOA cases within the family.40 Analyses were performed separately in which specific groups of relatives of the cases were compared to the comparable relatives of the matched controls as follows: first-degree relatives, second-degree relatives, first cousins and second cousins.
Age-Standardized Sex-Specific EOA Incidence Rates:
We selected all individuals with birth year and informative sex who resided in Utah from 2015 until 2018 or died in Utah, whichever happened first. This resulted in identification of 606 individuals with EOA. In contrast to the EOA cohort used to determine familial risk and identification of high-risk pedigrees, we chose to exclude the patients diagnosed with EOA in 2019 because 2018 is the last year the UPDB received death certificates. Demographic characteristics of the EOA cases and non-EOA population were compared using t-tests for continuous variables and chi-square tests for categorical variables (no adjustments) (Supplemental Table 1).
Age-standardized incidence rates by sex were calculated using the direct method. Person-years were calculated for affected individuals (cases) and unaffected individuals. Cases contributed one person-year for every year lived in Utah from 2015 until diagnosed with EOA. Person-years contributed from each unaffected individual was one additional year for every year lived in Utah from 2015 until death or 2018, whichever occurred first. The female-to-male incidence ratios were calculated by dividing the rate in males by that in females for each age group, and the corresponding 95% CI was estimated assuming log-normal distribution. Logistic regression models were used to assess the association between EOA and sex, additionally adjusting for birth year and whether the subjects were Caucasian and Hispanic.
Risk Factor Analysis:
Specific ICD-9 and ICD-10 codes were used to identify risk factors among study patients (Supplemental Table 2). Relative risk of EOA were calculated using gender-specific Cox proportional hazards models with adjustments for sex, birth year, race and ethnicity, and clustering for common mothers. We adjusted independently for obesity and diabetes since obesity is a risk factor for type 2 diabetes. Follow up time as defined by year from 2015 to date of death, date of last reside in Utah, or date of first EOA diagnosis, whichever happened first. Odds ratios and 95% CI were calculated.
Results:
Identification and demographic detail of the erosive hand osteoarthritis cohort:
To identify individuals diagnosed with EOA, we searched the UPDB for individuals with the ICD-10 code ICD-10 M15.4 from October 2015 – December 2019 and excluded patients with a rheumatoid arthritis diagnosis. This query identified 703 individuals for analysis with a mean age at time of diagnosis was 67 years (± 11.54), 80.23% were female, and 90.9% of individuals were white (Table 1). We performed manual chart review on a random subset of individuals to verify the EOA diagnosis (see Methods and Supplemental Figure 1).
Table 1 -.
Baseline Patient Characteristics of the Erosive Hand Osteoarthritis Cohort.
| Number of Individuals | 703 |
|---|---|
| Age (years) | 67 ± 11.54 (Range 12–94) |
| Race | |
| White | 639 (90.9%) |
| Non-white | 64 (9.1%) |
| Sex | |
| Female | 564 (80.23%) |
| Male | 139 (19.77%) |
Identification of High-Risk Pedigrees:
To test if there is significant familial clustering of EOA in our cohort, we analyzed individuals diagnosed with EOA that linked to a pedigree using the Familial Standardized Incidence Ratio (FSIR) calculation.39 We identified 240 unrelated, multigenerational, high-risk pedigrees that had at least two living members in the UPDB and an increased clustering of EOA, defined by a FSIR ≥ 2.0 (p-value < 0.05). Of the 240 high-risk pedigrees, the FSIR ranged from 2.0 – 210.8 (mean 11.7 ± SD = 11.7 ± 210.2; 1st and 3rd quartiles = 4.8 and 11.7, respectively). Founder birth year, number of descendants, number of affected individuals, and FSIR values are indicated for 10 representative high-risk pedigrees in Table 2. Figure 1 is an example of a multigenerational high-risk pedigree with at least 15 known affected individuals and a FSIR of 2.06 (affected individuals are represented by black circles/squares and individuals with an unknown affection status are represented by white circles/squares). The identification of high-risk pedigrees indicates significant familial clustering of EOA in our cohort.
Table 2 -.
High-Risk Pedigrees with Excess Familial Clustering of Erosive Hand Osteoarthritis. FSIR and p-values were calculated according to Kerber39.
| Founder Birth Year | Number of Descendants | Number of Affected Individuals | FSIR# |
|---|---|---|---|
| 1682* | 109,720 | 15 | 2.1 |
| 1789 | 81,204 | 12 | 2.1 |
| 1758 | 34,750 | 8 | 3.5 |
| 1795 | 300,10 | 8 | 3.0 |
| 1715 | 38,586 | 8 | 2.4 |
| 1780 | 34,156 | 7 | 2.8 |
| 1794 | 7,262 | 5 | 9.7 |
| 1779 | 13,015 | 5 | 4.7 |
| 1762 | 6677 | 4 | 11.8 |
| 1805 | 5,182 | 3 | 15.5 |
Abbreviations: FSIR = familial standardized incidence ratio.
indicates pedigree represented in Figure 1.
indicates that all FSIR p-values are p < 0.05
Figure 1 – A.
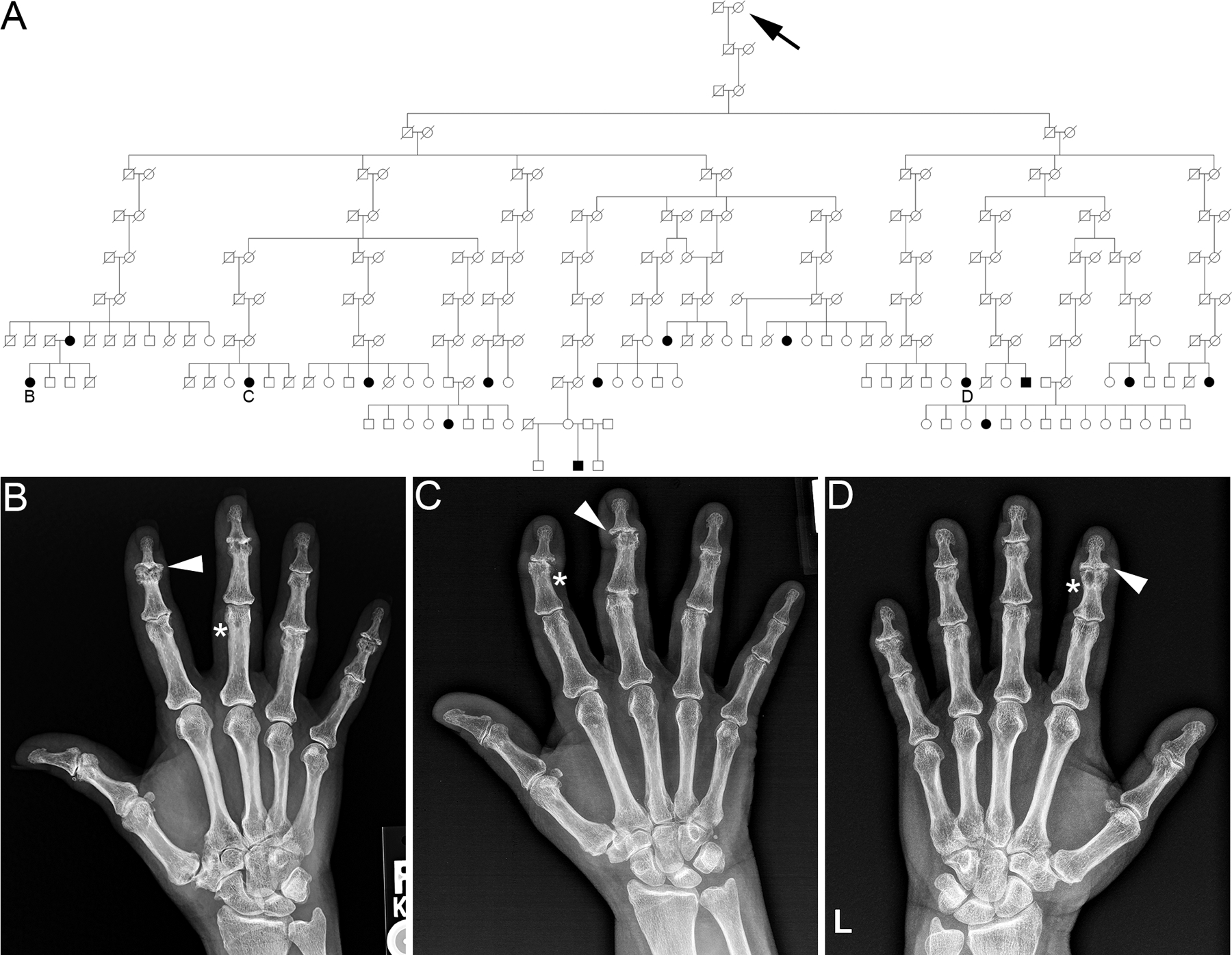
, Example of a high-risk erosive hand osteoarthritis pedigree identified in the Utah Population Database. Circles = females, squares = males, arrow = family founder, slashes = deceased. White circles/squares = affection status unknown. Black circles/squares = individuals affected with erosive hand osteoarthritis. B, C, D indicate individuals with hand radiographs. B-D, Hand radiographs of individuals (B-D) in the high-risk EOA pedigree (A). Asterisks indicate central subchondral erosions and arrowheads indicate a ‘gull-wing’ appearance of the joint.
Familial Risk:
To determine whether there is an increased risk of EOA among closely related individuals, we examined the relative risk of developing EOA in first- and second-degree relatives and first and second cousins in our cohort. The risk of developing EOA was approximately 5.5-fold greater in first-degree relatives of EOA cases compared to controls (Relative Risk, 5.53 [95% CI, 2.1 – 14.58], p < 0.001) (Table 3). We were unable to detect a significant elevated risk of EOA in second-degree relatives or first and second cousins of EOA cases. Together with the familial clustering of EOA, these data suggests a potential underlying genetic contribution to EOA.
Table 3 -.
Increased Familial Risk of Erosive Hand Osteoarthritis.
| Relationship | Cases | Controls | Relative Risk (Coefficient and 95% CI) | p-value | ||
|---|---|---|---|---|---|---|
| Affected | Unaffected | Affected | Unaffected | |||
| First-degree relative | 8 | 2654 | 15 | 27902 | 5.53 (2.1 – 14.58) | 0.001 |
| Second-degree relative | 2 | 5661 | 10 | 57601 | 2.14 (0.46 – 10.06) | 0.334 |
| First cousins | 6 | 6835 | 34 | 73694 | 11.08 (1.09 – 112.58) | 0.166 |
| Second cousins | 18 | 49275 | 289 | 520441 | 0.66 (0.4 – 1.06) | 0.087 |
Abbreviations: CI - 95% confidence interval
Age-Standardized Sex-Specific Incidence Rates of EOA:
Hand OA affects females more than males and this trend appears to be valid for EOA.6, 7, 13, 24, 25 To determine if there is an age and sex bias associated with EOA, we examined age-standardized sex-specific incidence rates of EOA in our statewide cohort from October 2015 - December 2018. We found a significant association between sex and age with EOA. Out of 2,065,277 unaffected individuals and 606 EOA cases, 80% of EOA cases were female (51.5% female in the unaffected group) and older (birth year, EOA cases- 1950.7 ± 11.5 and unaffected individuals - 1979.8 ± 23) when compared to the unaffected group (p < 0.001) (Supplemental Table 1). We also determined that females have a significantly higher rate of EOA from the ages of 40–89 compared to males, with the highest female-to-male incidence ratios being 4.730 (95% CI, 3.956 – 5.655) in the 60–69 age group (Table 4). Logistic regression analysis indicated that females have a 3.48-fold increased risk of EOA diagnosis compared to males even after adjusting for birth year, race and ethnicity (Relative Risk, 3.48 [95% CI, 2.85 – 4.25). Our results indicate that being female is a significant risk factor for EOA.
Table 4 -.
Age-Specific Incidence Rates of Erosive Hand Osteoarthritis by Sex and Female-to-Male Incidence Ratios.
| Age (years) | Male | Female | Female-to-male ratio (95% CI) |
||
|---|---|---|---|---|---|
| N cases | Rate Per 1000 | N cases | Rate Per 1000 | ||
| < 20 | 2 | 0.002 | 8 | 0.007 | 4.135 (0.878, 19.472) |
| 20–29 | 11 | 0.02 | 10 | 0.016 | 0.790 (0.336, 1.860) |
| 30–39 | 7 | 0.012 | 16 | 0.026 | 2.073 (0.853, 5.038) |
| 40–49 | 34 | 0.07 | 76 | 0.149 | 2.129 (1.421, 3.189) |
| 50–59 | 79 | 0.182 | 388 | 0.851 | 4.676 (3.671, 5.956) |
| 60–69 | 144 | 0.37 | 728 | 1.753 | 4.730 (3.956, 5.655) |
| 70–79 | 151 | 0.646 | 519 | 1.993 | 3.080 (2.570, 3.693) |
| 80–89 | 49 | 0.436 | 178 | 1.281 | 2.936 (2.140, 4.028) |
| 90+ | 7 | 0.252 | 17 | 0.378 | 1.475 (0.612, 3.558) |
Risk Factors Associated with EOA:
Knowledge of risk factors that may contribute to EOA remains incomplete. We analyzed the association of several risk factors with EOA that have been previously associated with general hand OA and EOA (see Supplemental Table 2 for risk factor diagnostic codes).16, 17, 25, 26 We examined the association of tobacco use, alcohol use, diabetes, obesity, and having a first-degree relative with EOA in the same cohort used for the age- and sex-specific analysis. Because females were at a higher risk for EOA (Table 4), we examined the relative risk of the above risk factors independently in males and females while adjusting for demographic features (see Methods and Supplemental Table 1). A history of obesity (Relative Risk, 1.50 [95% CI, 1.18 – 1.90]) was significantly associated with EOA in females, while diabetes was significantly associated in males (Relative Risk, 1.65 [95% CI, 1.05 – 2.59]). In both the female and male EOA cohorts, independent of obesity and diabetes, having a first-degree was a significant risk factor for EOA with the risk being greater in males than females (Table 5). No significant associations were detected between EOA diagnosis and tobacco or alcohol use. These data indicate obesity, diabetes, and having a first-degree relative with EOA are all significant risk factors for EOA.
Table 5 -.
Risk Factors Associated with Erosive Hand Osteoarthritis.
| Risk Factor/Clinical Diagnosis | Relative Risk with 95% Confidence Interval |
p-value | Risk Factor/Clinical Diagnosis | Relative Risk with 95% Confidence Interval |
p-value |
|---|---|---|---|---|---|
| (not adjusted for obesity) | (not adjusted for diabetes) | ||||
| Females | Females | ||||
| Tobacco Use | 1.3 (1.00 −1.68) | 0.051 | Tobacco Use | 1.27 (0.98–1.65) | 0.075 |
| Alcohol Use | 0.80 (0.26– 2.51) | 0.707 | Alcohol Use | 0.80 (0.26 − 2.50) | 0.704 |
| Diabetes | 1.23 (0.95–1.60) | 0.114 | Obesity | 1.50 (1.18 − 1.90) | 0.001 |
| FDR* with EOA | 6.40 (2.65 − 15.43) | < 0.001 | FDR with EOA | 6.42 (2.66 −15.49) | < 0.001 |
| Males | Males | ||||
| Tobacco Use | 0.79 (0.48 − 1.30) | 0.361 | Tobacco Use | 0.82 (0.49 − 1.35) | 0.426 |
| Alcohol Use | 2.48 (0.93 − 6.62) | 0.069 | Alcohol Use | 2.48 (0.93 − 6.64) | 0.069 |
| Diabetes | 1.65 (1.05 − 2.59) | 0.031 | Obesity | 1.22 (0.66 − 2.24) | 0.526 |
| FDR with EOA | 16.99 (5.37 − 53.75) | < 0.001 | FDR with EOA | 16.92 (5.35 − 53.54) | < 0.001 |
FDR = first-degree relative
Discussion:
We have used a unique statewide medical genetics resource, the Utah Population Database (UPDB), to identify a cohort of individuals diagnosed with erosive hand osteoarthritis (EOA). From this cohort we have i) identified 240 unrelated high-risk pedigrees demonstrating familial enrichment EOA, ii) determined that first-degree relatives of an individual with EOA is at approximately 5.5-fold increased risk of developing EOA, and iii) that sex, age, diabetes, obesity, and having a first-degree relative with EOA are significant risk factors associated with EOA. In sum, these data suggest that both genetic and physiological factors contribute to the development of EOA in a large population-based cohort.
Genetic Involvement in EOA:
Although hand OA is highly heritable8, few genes with large effects have been associated with the onset and progression of hand OA6, 7, and only three genes have been associated with the EOA phenotype.21–23 The predominant approach to discover hand OA gene variants has been genome-wide association studies (GWAS)41–46, which relies on large cohorts of cases and controls and well-defined phenotypes. The heterogeneous nature of hand OA has likely been a confounding factor in some GWAS. An alternative approach to GWAS is to study families with highly penetrant, severe or early-onset forms of OA.
The study of rare variants in affected families is a powerful way to identify gene variants with a determinate effect on disease development.47–50 Using the UPDB, we have identified 240 large multigenerational, high-risk pedigrees segregating EOA. Although a previous study described association of EOA in sibling pairs24, our study is unique because it is the first to identify a large number of multigenerational EOA pedigrees and determine relative risk among family members. Identification of causal gene variants in these families will inform us about genes and pathways that when disrupted contribute to EOA51.
Risk Factors Associated with EOA:
We examined the risk of developing EOA based on sex and age and found that females are 3.48-fold more likely to develop EOA than males with the highest female-to-male incidence ratios in the 60–69 age group. This suggests that EOA is similar to general hand OA in that females are disproportionately affected.6, 7, 13, 24, 25 When we subdivide risk factors based on sex and adjusted for a family history of EOA and other demographic factors, we found that obesity was a risk factor in females and diabetes a risk factors in males, while having a first-degree relative with EOA is a common risk factor to both sexes. Our data are consistent with other studies that have examined risk factors for EOA in other populations.16, 17, 25, 26 Awareness of these comorbidities observed to be significantly associated with EOA in the current study may help guide the clinical diagnosis of this condition in at-risk populations.
This study has several limitations. As for all database studies, it is unclear how errors in diagnostic coding would impact the study findings, and manual chart review was not possible for all individuals included in the analysis. We were able to chart review 48/57 (84.2%) randomly selected individuals. Of those 48 we were able to confirm that all individuals were correctly diagnosed (radiographic (44/48) and by a provider) with EOA. Our results are consistent with prior investigation, which has shown 93–97% rates of accuracy for UPDB diagnostic coding when compared to manual chart review.52–54 The relative risk and FSIR calculations are likely underestimates for EOA and is representative of symptomatic EOA, which is due to several factors. Our cohort was limited to individuals with an ICD-10 diagnosis for EOA, which has only been in use since October 2015, and our high-risk pedigree analysis can only identify individuals diagnosed in Utah. We are missing individuals who were diagnosed using different codes prior to October 2015, those diagnosed out of state, and affected individuals who have not sought out medical care. Because of these factors, our analyses are likely an underrepresentation of EOA, and in high-risk pedigrees we consider individuals without an EOA diagnosis as ‘affection status unknown’ until we can definitively determine if they are unaffected or affected.
Our study does not evaluate the extent to which EOA is genetic. The enrichment of EOA in pedigrees is suggestive of a genetic contribution, especially in distant relatives, but we cannot rule out environmental or physiological influence on EOA particularly in light of the risk factor associations we observed. Although EOA segregates as an apparent dominant trait in many pedigrees, until we can phenotype all individuals, we cannot preclude the possibility EOA may be polygenic in some families.
To conclude, we demonstrated that EOA demonstrates familial enrichment, an increased relative risk among first-degree relatives, and identified significant EOA risk factors. Taken together, these findings suggest that EOA has a genetic and environmental component to its etiology. Genomic analysis of individuals within our high-risk pedigrees holds promise in identifying genetic variants associated with EOA. By identifying and studying gene variants that cause EOA, we may learn about the biological mechanisms that lead to other forms of OA, which may provide significant insight into surgical treatment or therapeutic intervention.
Supplementary Material
Supplemental Figure 1 – Hand radiographs of individuals in the EOA cohort. Each image represents a unique individual. Asterisks indicate central subchondral erosions and arrowheads indicate a ‘gull-wing’ appearance of the joint.
Supplemental Table 1 – Study Population Used for Age-Standardized Sex-Specific Incidence Rates of Erosive Hand Osteoarthritis.
Supplemental Table 1 – Identification of Risk Factors Using Diagnostic Coding.
Acknowledgments
This study was funded by the Skaggs Foundation for Research (MJJ and NHK), the Utah Genome Project (MJJ and NHK), and the Arthritis National Research Foundation (MJJ). We thank Dr. Rena D’Souza for critical feedback. The UPDB is supported by the Pedigree and Population Resource, the Program in Personalized Health and Center for Clinical and Translational Science, and NCI grant P30 CA2014.
This work was funded by the Skaggs Foundation for Research, the Utah Genome Project, and the Arthritis National Research Foundation.
Footnotes
Competing Interests: None
Data Sharing Statement: Data are available upon request.
Patient and Public Involvement: None.
References:
- 1.Goldring SR, Goldring MB. Clinical aspects, pathology and pathophysiology of osteoarthritis. J Musculoskelet Neuronal Interact. 2006. Oct-Dec; 6(4):376–378. [PubMed] [Google Scholar]
- 2.Neogi T The epidemiology and impact of pain in osteoarthritis. Osteoarthritis Cartilage. 2013. September; 21(9):1145–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pereira D, Peleteiro B, Araujo J, Branco J, Santos RA, Ramos E. The effect of osteoarthritis definition on prevalence and incidence estimates: a systematic review. Osteoarthritis Cartilage. 2011. November; 19(11):1270–1285. [DOI] [PubMed] [Google Scholar]
- 4.Snyder EA, Alvarez C, Golightly YM, Renner JB, Jordan JM, Nelson AE. Incidence and progression of hand osteoarthritis in a large community-based cohort: the Johnston County Osteoarthritis Project. Osteoarthritis Cartilage. 2020. February 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y, Niu J, Kelly-Hayes M, Chaisson CE, Aliabadi P, Felson DT. Prevalence of symptomatic hand osteoarthritis and its impact on functional status among the elderly: The Framingham Study. Am J Epidemiol. 2002. December 1; 156(11):1021–1027. [DOI] [PubMed] [Google Scholar]
- 6.Marshall M, Watt FE, Vincent TL, Dziedzic K. Hand osteoarthritis: clinical phenotypes, molecular mechanisms and disease management. Nat Rev Rheumatol. 2018. November; 14(11):641–656. [DOI] [PubMed] [Google Scholar]
- 7.Kloppenburg M, Kwok WY. Hand osteoarthritis--a heterogeneous disorder. Nat Rev Rheumatol. 2011. November 22; 8(1):22–31. [DOI] [PubMed] [Google Scholar]
- 8.Ishimori ML, Altman RD, Cohen MJ, Cui J, Guo X, Rotter JI, et al. Heritability patterns in hand osteoarthritis: the role of osteophytes. Arthritis Res Ther. 2010; 12(5):R180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banks SE. Erosive osteoarthritis: a current review of a clinical challenge. Clin Rheumatol. 2010. Jul; 29(7):697–706. [DOI] [PubMed] [Google Scholar]
- 10.Marshall M, Nicholls E, Kwok WY, Peat G, Kloppenburg M, van der Windt D, et al. Erosive osteoarthritis: a more severe form of radiographic hand osteoarthritis rather than a distinct entity? Ann Rheum Dis. 2015. January; 74(1):136–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Punzi L, Favero M, Frallonardo P, Ramonda R. Time to redefine erosive osteoarthritis. RMD Open. 2015; 1(1):e000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crain DC. Interphalangeal osteoarthritis. JAMA. 1961. March 25; 175:1049–1053. [DOI] [PubMed] [Google Scholar]
- 13.Peter JB, Pearson CM, Marmor L. Erosive osteoarthritis of the hands. Arthritis Rheum. 1966. June; 9(3):365–388. [DOI] [PubMed] [Google Scholar]
- 14.Addimanda O, Mancarella L, Dolzani P, Punzi L, Fioravanti A, Pignotti E, et al. Clinical and radiographic distribution of structural damage in erosive and nonerosive hand osteoarthritis. Arthritis Care Res (Hoboken). 2012. July; 64(7):1046–1053. [DOI] [PubMed] [Google Scholar]
- 15.Punzi L, Ramonda R, Sfriso P. Erosive osteoarthritis. Best Pract Res Clin Rheumatol. 2004. October; 18(5):739–758. [DOI] [PubMed] [Google Scholar]
- 16.Kwok WY, Kloppenburg M, Rosendaal FR, van Meurs JB, Hofman A, Bierma-Zeinstra SM. Erosive hand osteoarthritis: its prevalence and clinical impact in the general population and symptomatic hand osteoarthritis. Ann Rheum Dis. 2011. July; 70(7):1238–1242. [DOI] [PubMed] [Google Scholar]
- 17.Marshall M, Peat G, Nicholls E, van der Windt D, Myers H, Dziedzic K. Subsets of symptomatic hand osteoarthritis in community-dwelling older adults in the United Kingdom: prevalence, inter-relationships, risk factor profiles and clinical characteristics at baseline and 3-years. Osteoarthritis Cartilage. 2013. November; 21(11):1674–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kloppenburg M, Kroon FP, Blanco FJ, Doherty M, Dziedzic KS, Greibrokk E, et al. 2018 update of the EULAR recommendations for the management of hand osteoarthritis. Ann Rheum Dis. 2019. January; 78(1):16–24. [DOI] [PubMed] [Google Scholar]
- 19.Ramos YF, Meulenbelt I. Implementation of Functional Genomics for Bench-to-Bedside Transition in Osteoarthritis. Curr Rheumatol Rep. 2015. August; 17(8):53. [DOI] [PubMed] [Google Scholar]
- 20.Thysen S, Luyten FP, Lories RJ. Targets, models and challenges in osteoarthritis research. Dis Model Mech. 2015. January; 8(1):17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pattrick M, Manhire A, Ward AM, Doherty M. HLA-A, B antigens and alpha 1-antitrypsin phenotypes in nodal generalised osteoarthritis and erosive osteoarthritis. Ann Rheum Dis. 1989. June; 48(6):470–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramonda R, Musacchio E, Campana C, Frigato M, Frallonardo P, Barbieri V, et al. Immunogenetic aspects of erosive osteoarthritis of the hand in patients from northern Italy. Scand J Rheumatol. 2011. March; 40(2):139–144. [DOI] [PubMed] [Google Scholar]
- 23.Stern AG, de Carvalho MRC, Buck GA, Adler RA, Rao TPS, Disler D, et al. Association of erosive hand osteoarthritis with a single nucleotide polymorphism on the gene encoding interleukin-1 beta. Osteoarthritis and Cartilage. 2003; 11(6):394–402. [DOI] [PubMed] [Google Scholar]
- 24.Bijsterbosch J, van Bemmel JM, Watt I, Meulenbelt I, Rosendaal FR, Huizinga TW, et al. Systemic and local factors are involved in the evolution of erosions in hand osteoarthritis. Ann Rheum Dis. 2011. February; 70(2):326–330. [DOI] [PubMed] [Google Scholar]
- 25.Haugen IK, Englund M, Aliabadi P, Niu J, Clancy M, Kvien TK, et al. Prevalence, incidence and progression of hand osteoarthritis in the general population: the Framingham Osteoarthritis Study. Ann Rheum Dis. 2011. September; 70(9):1581–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haugen IK, Magnusson K, Turkiewicz A, Englund M. The Prevalence, Incidence, and Progression of Hand Osteoarthritis in Relation to Body Mass Index, Smoking, and Alcohol Consumption. J Rheumatol. 2017. September; 44(9):1402–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cannon-Albright LA, Goldgar DE, Neuhausen S, Gruis NA, Anderson DE, Lewis CM, et al. Localization of the 9p melanoma susceptibility locus (MLM) to a 2-cM region between D9S736 and D9S171. Genomics. 1994. September 1; 23(1):265–268. [DOI] [PubMed] [Google Scholar]
- 28.Cannon-Albright LA, Thomas TC, Bishop DT, Skolnick MH, Burt RW. Characteristics of familial colon cancer in a large population data base. Cancer. 1989. November 1; 64(9):1971–1975. [DOI] [PubMed] [Google Scholar]
- 29.Goldgar DE, Cannon-Albright LA, Oliphant A, Ward JH, Linker G, Swensen J, et al. Chromosome 17q linkage studies of 18 Utah breast cancer kindreds. Am J Hum Genet. 1993. April; 52(4):743–748. [PMC free article] [PubMed] [Google Scholar]
- 30.Lakhani SR, Gusterson BA, Jacquemier J, Sloane JP, Anderson TJ, van de Vijver MJ, et al. The pathology of familial breast cancer: histological features of cancers in families not attributable to mutations in BRCA1 or BRCA2. Clin Cancer Res. 2000. March; 6(3):782–789. [PubMed] [Google Scholar]
- 31.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994. October 7; 266(5182):66–71. [DOI] [PubMed] [Google Scholar]
- 32.Slattery ML, Kerber RA. A comprehensive evaluation of family history and breast cancer risk. The Utah Population Database. JAMA. 1993. October 6; 270(13):1563–1568. [PubMed] [Google Scholar]
- 33.Slattery ML, Kerber RA. Family history of cancer and colon cancer risk: the Utah Population Database. J Natl Cancer Inst. 1994. November 2; 86(21):1618–1626. [DOI] [PubMed] [Google Scholar]
- 34.Tavtigian SV, Simard J, Rommens J, Couch F, Shattuck-Eidens D, Neuhausen S, et al. The complete BRCA2 gene and mutations in chromosome 13q-linked kindreds. Nat Genet. 1996. March; 12(3):333–337. [DOI] [PubMed] [Google Scholar]
- 35.Abbott D, Brockmeyer D, Neklason DW, Teerlink C, Cannon-Albright LA. Population-based description of familial clustering of Chiari malformation Type I. J Neurosurg. 2018. February; 128(2):460–465. [DOI] [PubMed] [Google Scholar]
- 36.Coon H, Darlington TM, DiBlasi E, Callor WB, Ferris E, Fraser A, et al. Genome-wide significant regions in 43 Utah high-risk families implicate multiple genes involved in risk for completed suicide. Mol Psychiatry. 2018. October 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kazmers NH, Yu Z, Barker T, Abraham T, Romero R, Jurynec MJ. Evaluation for Kienbock Disease Familial Clustering: A Population-Based Cohort Study. J Hand Surg Am. 2020. January; 45(1):1–8 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neuhausen SL, Farnham JM, Kort E, Tavtigian SV, Skolnick MH, Cannon-Albright LA. Prostate cancer susceptibility locus HPC1 in Utah high-risk pedigrees. Hum Mol Genet. 1999. December; 8(13):2437–2442. [DOI] [PubMed] [Google Scholar]
- 39.Kerber RA. Method for calculating risk associated with family history of a disease. Genet Epidemiol. 1995; 12(3):291–301. [DOI] [PubMed] [Google Scholar]
- 40.Williams RL. A note on robust variance estimation for cluster-correlated data. Biometrics. 2000. June; 56(2):645–646. [DOI] [PubMed] [Google Scholar]
- 41.den Hollander W, Boer CG, Hart DJ, Yau MS, Ramos YFM, Metrustry S, et al. Genome-wide association and functional studies identify a role for matrix Gla protein in osteoarthritis of the hand. Ann Rheum Dis. 2017. December; 76(12):2046–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamalainen S, Solovieva S, Vehmas T, Luoma K, Leino-Arjas P, Hirvonen A. Genetic influences on hand osteoarthritis in Finnish women--a replication study of candidate genes. PLoS One. 2014; 9(5):e97417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kerkhof HJ, Lories RJ, Meulenbelt I, Jonsdottir I, Valdes AM, Arp P, et al. A genome-wide association study identifies an osteoarthritis susceptibility locus on chromosome 7q22. Arthritis Rheum. 2010. February; 62(2):499–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stefansson SE, Jonsson H, Ingvarsson T, Manolescu I, Jonsson HH, Olafsdottir G, et al. Genomewide scan for hand osteoarthritis: a novel mutation in matrilin-3. Am J Hum Genet. 2003. June; 72(6):1448–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Styrkarsdottir U, Thorleifsson G, Helgadottir HT, Bomer N, Metrustry S, Bierma-Zeinstra S, et al. Severe osteoarthritis of the hand associates with common variants within the ALDH1A2 gene and with rare variants at 1p31. Nat Genet. 2014. May; 46(5):498–502. [DOI] [PubMed] [Google Scholar]
- 46.Zhai G, van Meurs JB, Livshits G, Meulenbelt I, Valdes AM, Soranzo N, et al. A genome-wide association study suggests that a locus within the ataxin 2 binding protein 1 gene is associated with hand osteoarthritis: the Treat-OA consortium. J Med Genet. 2009. September; 46(9):614–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chakravarti A, Turner TN. Revealing rate-limiting steps in complex disease biology: The crucial importance of studying rare, extreme-phenotype families. Bioessays. 2016. June; 38(6):578–586. [DOI] [PubMed] [Google Scholar]
- 48.Jurynec MJ, Sawitzke AD, Beals TC, Redd MJ, Stevens J, Otterud B, et al. A hyperactivating proinflammatory RIPK2 allele associated with early-onset osteoarthritis. Hum Mol Genet. 2018. July 1; 27(13):2383–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramos YF, Bos SD, van der Breggen R, Kloppenburg M, Ye K, Lameijer EW, et al. A gain of function mutation in TNFRSF11B encoding osteoprotegerin causes osteoarthritis with chondrocalcinosis. Ann Rheum Dis. 2015. September; 74(9):1756–1762. [DOI] [PubMed] [Google Scholar]
- 50.Sliz E, Taipale M, Welling M, Skarp S, Alaraudanjoki V, Ignatius J, et al. TUFT1, a novel candidate gene for metatarsophalangeal osteoarthritis, plays a role in chondrogenesis on a calcium-related pathway. PLoS One. 2017; 12(4):e0175474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aury-Landas J, Marcelli C, Leclercq S, Boumediene K, Bauge C. Genetic Determinism of Primary Early-Onset Osteoarthritis. Trends Mol Med. 2016. January; 22(1):38–52. [DOI] [PubMed] [Google Scholar]
- 52.Peterson K, Firszt R, Fang J, Wong J, Smith KR, Brady KA. Risk of Autoimmunity in EoE and Families: A Population-Based Cohort Study. Am J Gastroenterol. 2016. July; 111(7):926–932. [DOI] [PubMed] [Google Scholar]
- 53.Samadder NJ, Curtin K, Tuohy TM, Pappas L, Boucher K, Provenzale D, et al. Characteristics of missed or interval colorectal cancer and patient survival: a population-based study. Gastroenterology. 2014. April; 146(4):950–960. [DOI] [PubMed] [Google Scholar]
- 54.Oakley GM, Curtin K, Orb Q, Schaefer C, Orlandi RR, Alt JA. Familial risk of chronic rhinosinusitis with and without nasal polyposis: genetics or environment. Int Forum Allergy Rhinol. 2015. April; 5(4):276–282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1 – Hand radiographs of individuals in the EOA cohort. Each image represents a unique individual. Asterisks indicate central subchondral erosions and arrowheads indicate a ‘gull-wing’ appearance of the joint.
Supplemental Table 1 – Study Population Used for Age-Standardized Sex-Specific Incidence Rates of Erosive Hand Osteoarthritis.
Supplemental Table 1 – Identification of Risk Factors Using Diagnostic Coding.


