Abstract
Background:
Studies of germ-free (GF) mice demonstrate that gut microbiota can influence behaviour by modulating neurochemical pathways in the brain, and that bacterial colonization normalizes behavioural deficits in GF-mice. Since disrupted GABAergic and glutamatergic signaling are reported in mood disorders, this study investigated the effect of gut microbiota manipulations on EIB-relevant gene expression in the brain.
Methods:
GF Swiss-Webster mice were colonized with E. coli JM83, complex microbiota (specific-pathogen-free; SPF), or no microbiota, and compared with controls (n=6/group). 21 synaptic genes representing GABAergic, glutamatergic, BDNF, and astrocytic functions were measured in the hippocampus, amygdala, and prefrontal cortex using quantitative PCR. Gene co-expression analysis was used to identify gene modules related to colonization status, and compared by permutation analysis. Gene expression profiles were compared to existing post-mortem cohorts of depressed subjects (n=28 cases vs 28 controls).
Results:
Region-specific alterations in gene expression were observed in GF-mice compared to controls. 58% of all genes (14/24) altered in GF-mice were normalized following SPF-colonization. GF-mice displayed disorganization of gene co-expression networks in all three brain regions (hippocampus, p=0.0003; amygdala, p=0.0012; mPFC, p=0.0069), which was restored by SPF colonization in hippocampus (p v.s. GF =0.0003, p v.s. control = 0.60). The hippocampal gene expression profile in GF-mice was significantly correlated with that in human depression (ρ=0.51, p=0.027), and this correlation was not observed after colonization.
Conclusion:
Together, we show that the absence of gut microbiota disrupts the expression of EIB-relevant genes in mice, and colonization restores EIB-relevant expression, in ways that are relevant to human depression.
Keywords: Excitation-inhibition balance, gut-brain axis, germ-free mice, gut microbiota, brain
Introduction
Knowledge of the role of intestinal microbiota on human health is rapidly expanding. Microbiota show a profound influence on numerous host processes, including immune system development, brain function, and behaviour (Bercik et al., 2011; Collins et al., 2012; McVey Neufeld et al., 2019; Sharon et al., 2019; Valles-Colomer et al., 2019). Mental illness represents the leading global disease burden, but treatments for common illnesses such as anxiety disorders and depression show poor response rates of approximately 50% (Bandelow et al., 2014; Prins et al., 2011). Modulating intestinal microbiota represents a novel and unexploited therapeutic approach, although it requires a more refined understanding of how microbiota affect biological systems related to mental illness.
γ-amino butyric acid (GABA), the primary inhibitory neurotransmitter in the brain, is implicated in psychiatric and neurological disorders, including anxiety, major depressive disorder (MDD), bipolar disorder, schizophrenia, autism, Alzheimer’s disease, and epilepsy (Lin and Sibille, 2013; Schur et al., 2016; Treiman, 2001). Neuroimaging meta-analyses have shown significantly decreased GABA concentrations in cortical brain regions of depressed patients (Schur et al., 2016). Human and rodent studies have identified altered GABAergic and glutamatergic gene expression in the anterior cingulate cortex, amygdala, and hippocampus (Chang et al., 2014; Guilloux et al., 2012; Kohen et al., 2014; Lussier et al., 2013; Tripp et al., 2012). Coordinated activity of inhibitory GABAergic interneurons with excitatory glutamatergic pyramidal cells is required to maintain the excitation-inhibition balance (EIB) and proper information processing (Fee et al., 2017). EIB maintenance also requires neurotrophic and astrocytic support, both of which show deficits in depression and animal models of stress (Guilloux et al., 2012; Oh et al., 2012; Poskanzer and Yuste, 2016).
Disrupted communication along the gut-brain axis is implicated in gastrointestinal and psychiatric disorders (De Palma et al., 2015; De Palma et al., 2014). Dysbiosis, a maladaptive shift in the balance of microbiota, is observed in many brain disorders that demonstrate changes in EIB, such as epilepsy, MDD, bipolar disorder, schizophrenia, and autism (Dahlin and Prast-Nielsen, 2019; Fields et al., 2018; Grochowska et al., 2018; Lum et al., 2020). Oral administration of probiotic Bifidobacterium longum (NCC3001) to patients with IBS and co-morbid depression improved depression scores and altered coordinated activity of brain networks involved in mood control (Pinto-Sanchez et al., 2017). Dysbiosis is also observed in animal models relevant to such disorders (Lum et al., 2020). Microbiota transplantation from patients with irritable bowel syndrome (IBS) induced gut and brain dysfunction in recipient mice (De Palma et al., 2017). Germ-free (GF) mice, lacking gut microbiota from birth, display increased exploration and impaired social and fear-related behaviors (Bercik et al., 2011; Desbonnet et al., 2014; Diaz et al., 2011; Hoban et al., 2018; Stilling et al., 2018), increased blood brain barrier permeability (Braniste et al., 2014), increased hypothalamic-pituitary-adrenal axis responsivity (Sudo et al., 2004), and impaired microglial development (Erny et al., 2015). Strikingly, these changes are reversed by colonization with specific-pathogen-free (SPF) microbiota (Clarke et al., 2013; Diaz et al., 2011; Hoban et al., 2018; Neufeld et al., 2011; Sudo et al., 2004; Vodicka et al., 2018).
Although microbiota can affect the GABAergic and glutamatergic signaling underlying EIB, a system often dysregulated in mood and anxiety disorders, a broad analysis of EIB-relevant gene expression in critical brain regions has not been performed. Here, we investigated the effect of microbiota on the expression of a representative group of synaptic genes frequently implicated in depression, anxiety-like behaviour, and EIB regulation (Oh et al., 2019). We measured gene expression levels in the amygdala, hippocampus, and mPFC from GF-mice, ex-GF-mice colonized with SPF microbiota or mono-colonized with E. coli, and compared them to control mice. We decided to use E.coli JM83 (derivative of E.coli K 12 commensal strain) for mono-colonization for consistency with prior literature on GF mouse model (Hapfelmeier et al., 2010). E.coli JM83 is a good candidate to study the gut microbiota-brain interaction because it is easy to grow and resilient to harsh conditions within the GF facilities. Being a non-pathogenic bacterial strain, E.coli JM83, does not induce overt inflammation following mono-colonization and as a result cause minimal mortality in GF-mice.
Within the framework of our hypothesis stating that microbiota can regulate aspects of EIB, we predicted that GF-mice would show altered gene-expression in a brain region-dependent manner and that bacterial colonization would normalize these changes. We also predicted that the effect of microbiota manipulation would be reflected in the dynamic interrelationship of these genes, as measured by gene co-expression analysis. Finally, we tested whether microbiota-induced gene expression changes parallel those observed in depression.
Methods
Gnotobiotic mice
10–14 week old GF Swiss Webster mice (n=6/group) were maintained axenic in sterile isolators at the Farncombe Family Axenic Gnotobiotic Unit of Central Animal Facility, McMaster University, Canada. 12-week old conventional Swiss Webster mice (n=6) were used as controls (Taconic Farms, Hudson, NY, USA). Handling of GF and colonized mice was performed in gnotobiotic isolators under strict axenic conditions, as previously described (De Palma et al., 2015). Mice were housed 3–4/cage and maintained on a 12-hour day/night cycle with unrestricted access to autoclaved pelleted diet (Teklad S-2335 Mouse Breeder Sterilizable Diet, Envigo, product code 7004) and water. Microbiota status was assessed regularly by direct bacteriology and 16s rRNA PCR testing. Given technical and practical constraints of breeding GF-mice, uniform sex distributions were not possible. Control and mono-colonized groups contained two females, whereas GF and SPF-colonized contained one. All experiments were approved by the McMaster University Animal Care Committee.
Bacterial colonization of GF-mice
GF-mice (n=6/group) were colonized with SPF microbiota or mono-colonized with 109 CFU of E. coli JM83 via a single intragastric gavage as previously described (De Palma et al., 2015). After four weeks, mice were sacrificed and brains immediately snap frozen in isopentane over dry ice and stored at −80°C (Figure 1). E. coli JM83 was grown in LB broth and incubated at 37°C, and shaking at 160 rpm for 12 hours. Bacteria were collected by centrifugation (15 min, 3500g), washed in sterile PBS, and concentrated to a density of 109 CFU/ml in PBS, under a sterile laminar flow hood. Fresh cecal contents from two SPF Swiss Webster donors (1 male, 1 female, 12 weeks old) were pooled and suspended in sterile PBS for the ex-GF SPF colonization. Bacterial suspensions were sealed in sterile tubes and imported into sterile isolators for intragastric gavage.
Figure 1. Colonization timeline.
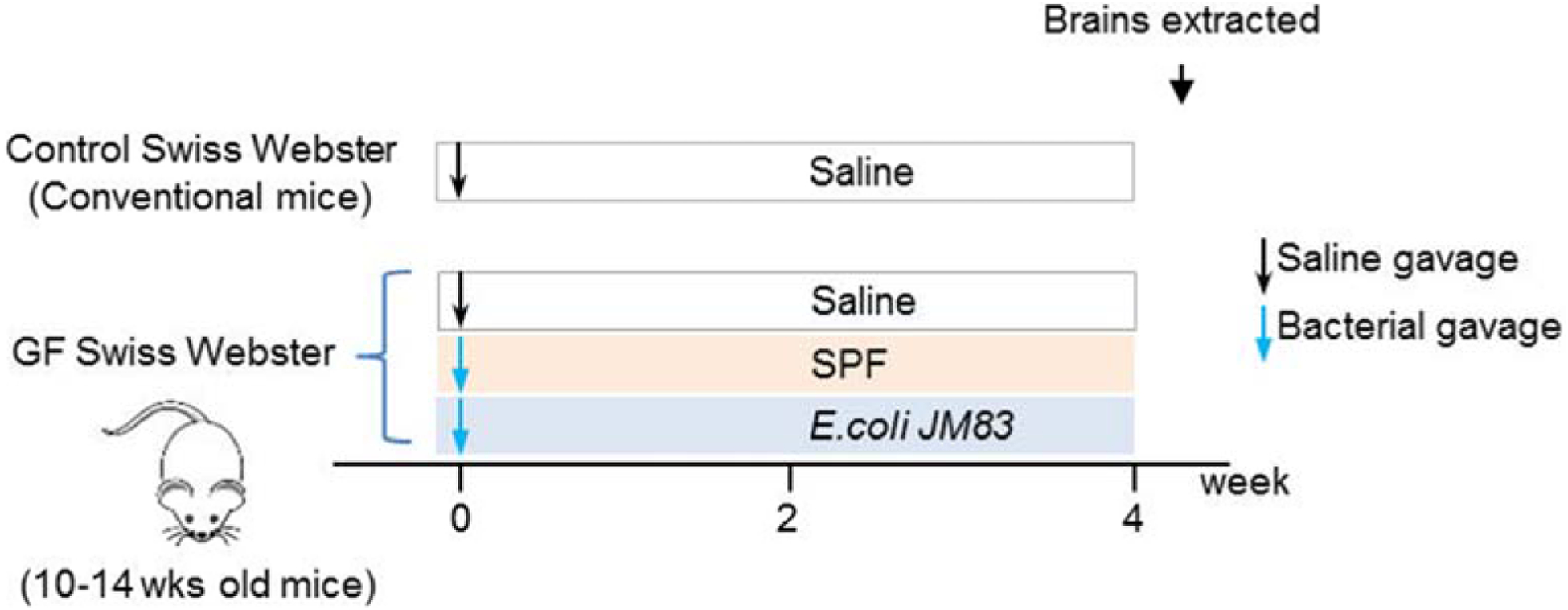
10–14 weeks old Swiss Webster GF-mice were colonized with SPF microbiota or mono-colonized with 109 CFU of E. coli JM83 via a single intragastric gavage. Control (conventional mice) and GF cohorts received saline via a single intragastric gavage. After four weeks, mice were sacrificed and brains collected for different assays. (n=6/group, 3–4/cage)
Sample Preparation
Under RNase-free conditions, brains were sectioned to the mPFC (bregma ~+1.7mm), amygdala (bregma ~−1.3mm), then hippocampus (bregma ~−2.7mm) using a cryostat (Leica), and grey matter collected via micropunch (George Paxinos, 2012). RNA extraction and cDNA synthesis were performed using PicoPure RNA isolation and Superscript VILO cDNA synthesis kits, respectively (ThermoFisher Scientific, Walton, MA).
Quantitative Polymerase Chain Reaction (qPCR)
qPCR was performed on a CFX96 Touch Thermocycler (Bio-Rad, Hercules, CA) using SYBR green Master Mix (Bio-Rad, Hercules, CA) using primers for Bdnf, Dlg4, Gabra1, Gabra2, Gabra5, Gad1, Gad2, Gfap, Gria1, Grin2a, Grin2b, Ntrk2.fl, Ntrk2.t1, Pvalb, Slc17a7, Slc1a2, Slc32a1, Slc6a1, Sst, Vgf, and Vip. Gapdh and Ppia were used as loading controls, were highly correlated (r>0.9), and showed less than a 5% change across groups in all brain regions. Primers were developed as previously described (Oh et al., 2016). Primer sequences are provided in Supplemental Table 1
Statistical Analysis
Differential Gene Expression:
Log2-based delta Ct values were transformed into non-log expression values (arbitrary units). Gene expression was compared across groups within each region using one-way ANOVA.
ANOVAs were corrected for multiple comparisons across the 21 genes × 3 regions studied using the Benjamini-Hochberg false discovery rate (FDR) method (Benjamini and Hochberg, 2000). ANOVA results were considered significant at 5% FDR, and pair-wise post-hoc t-tests were performed with Bonferroni correction. Statistical analyses were performed in R, version 3.5.1 (Team, 2018).
Co-expression Analysis:
Weighted gene co-expression network analysis (WGCNA) (Langfelder and Horvath, 2008) was used to assess changes in the co-ordinated expression of genes across experimental groups within each brain region. Briefly, WGCNA generates a correlation matrix within each group, by which genes are then hierarchically clustered. Modules of highly co-expressed genes are generated, such that a given gene may only be a member of one module. A soft-thresholding power of β=4 was used as this showed the greatest similarity in scale-free topology and mean connectivity across networks. Given the moderate sample size and number of genes in this study we used WGCNA primarily to group genes by rough similarity in gene expression, an approach suited to capture large effects. We opted not to examine eigengene correlations, hub genes, or differences in intra-module measures of connectivity which are more routinely reported in larger datasets.
For between-group comparisons, the proportion of genes present across modules was measured, and empirical p-values representing the significance of these differences were determined using permutation testing (n=10,000). Stouffer’s p-value meta-analysis (i.e. equal weighting) was used to combine the multiple between-group module comparisons (after Bonferroni correction) (Heard and Rubin-Delanchy, 2018). Co-expression networks were visualized using Cytoscape (Shannon et al., 2003).
Comparison to Large-Scale MDD Microarray Datasets
We compared experimental group fold-changes to published post-mortem transcriptomic studies in MDD subjects (Ding et al., 2015; Duric et al., 2010). Analyses were restricted to male subjects given the predominantly male group composition of the mouse studies. Mean log2 fold-changes (LFCs) in GF, mono-colonized, and SPF-colonized groups were compared gene-wise to LFCs from post-mortem MDD datasets from hippocampal region CA1 (Duric et al., 2010), amygdala, and anterior cingulate cortex (ACC) (Ding et al., 2015). Study selection was primarily driven by data availability. Other transcriptomic studies of MDD in the hippocampus, amygdala, and ACC were available through the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/), though incomplete annotations of Affymetrix probe IDs, incomplete datasets, and absence of raw data precluded their inclusion. The recent region-specific RNAseq dataset of (Labonte et al., 2017) did not contain hippocampal samples and was not used. The ACC is homologous to the murine mPFC regions examined here (George Paxinos, 2012). Similarity in LFC profile was assessed by Spearman correlation given the log-scale nature of the data, differences in expression measures, and our interest in comparing the profiles (i.e. rank-orders) of expression rather than absolute values.
Results
Absence of gut microbiota significantly altered glutamatergic/GABAergic synaptic gene expression
We sought to determine how microbiota influence EIB-relevant synaptic gene expression in mice. qPCR was performed on hippocampus, amygdala, and mPFC from conventional (control), GF, and ex-GF-mice colonized either with SPF microbiota or mono-colonized with E. coli (n=6/group) (Figure 1). We selected fifteen GABAergic/glutamatergic genes, four BDNF-related, and 2 astrocytic genes due to their role in EIB maintenance and their influence by BDNF (Ding et al., 2015; Oh et al., 2016; Oh et al., 2019).
Region-specific alterations in gene expression were observed in GF-mice compared to controls (Figure 2A, “GF” columns). Boxplot visualization of the data underlying this heatmap showing group differences and within-group low variability are shown in Figure 2B and Supplementary Figures 1. Broadly, the hippocampus was characterized by dysregulation across all functional groups of genes, the amygdala showed elevated expression of GABAergic genes, and the mPFC showed decreased expression of glutamatergic genes. The amygdala and mPFC showed generally opposing expression profiles in GABAergic and glutamatergic genes. Despite our modest sample size, scatterplots (Supplementary Figures 2–4) show robust group-level consistency in effects. In total, 24/63 (38%), of the 21 genes × 3 regions measured, were significantly differentially expressed in GF-mice, indicating that an absence of gut microbiota leads to region-specific alterations in EIB-relevant gene expression.
Figure 2. Region-specific effects of microbiota on EIB-relevant synaptic gene expression in mice.
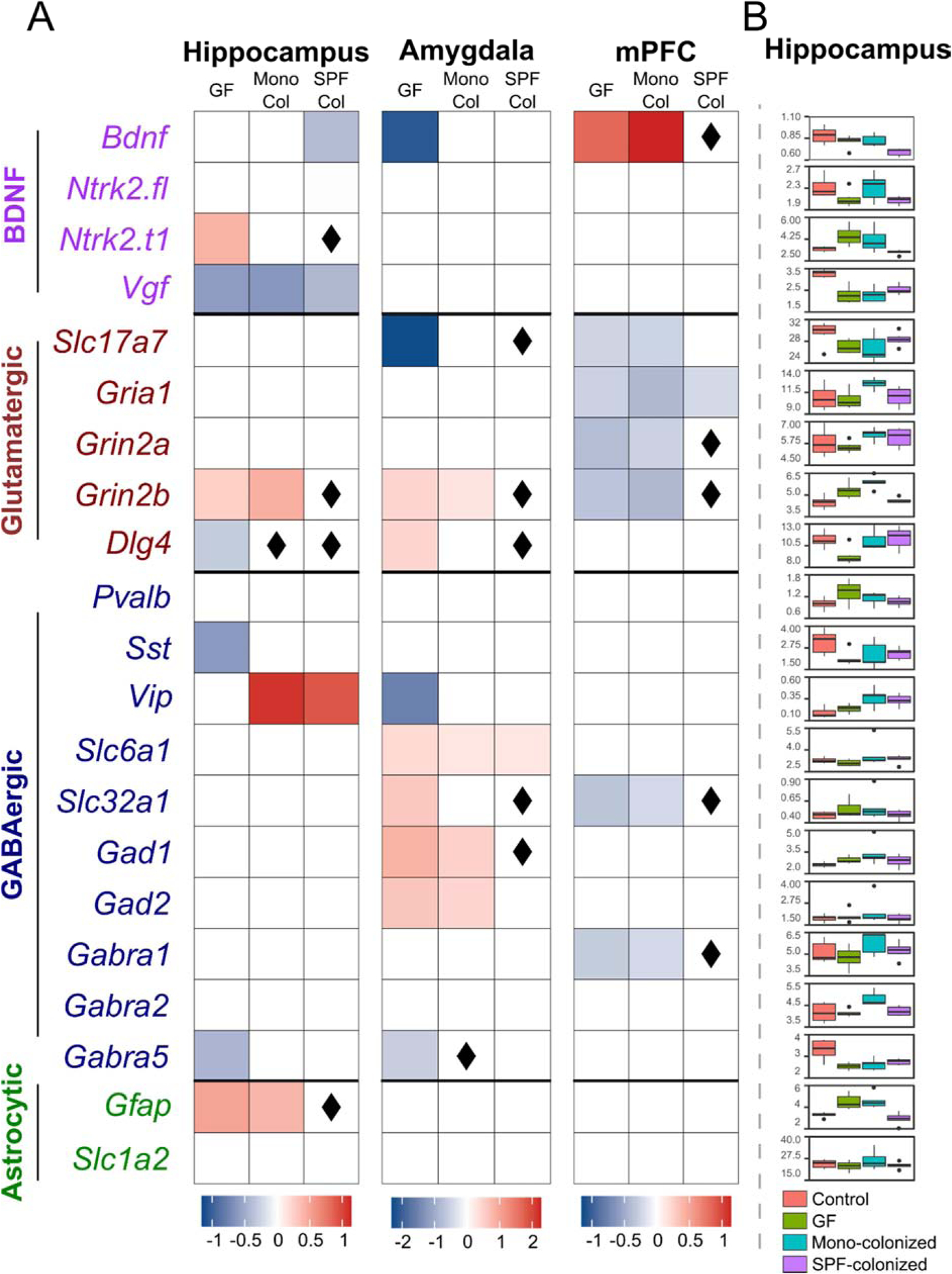
(A) Log2 fold-changes are visualized for genes showing a significant (p<0.05) effect of group in ANOVA and a significant difference (p<0.05) from controls (conventional mice) in post-hoc t-tests. Increased expression is visualized in red, and decreased expression in blue. ◊ indicates fully normalized genes, i.e. no significant difference (p>0.05) from controls and a significant difference (p<0.05) from germ-free (GF) mice in post-hoc t-tests. “Con” indicates conventional mice, “Mono Col” indicates mice colonized with E. coli JM83 alone, and “SPF Col” indicates mice colonized with SPF microbiota. (B) Tukey box-and-whisker plots showing raw gene expression data for the hippocampus. Controls are shown in red, GF mice in green, mono-colonized mice in blue, and SPF-colonized mice in violet.
Bacterial colonization of GF-mice with SPF microbiota normalized GABAergic/glutamatergic synaptic gene expression
In each brain region, we observed a striking normalization of gene expression in SPF-colonized mice, and a partial normalization in mono-colonized mice. We defined full normalization as genes showing both no significant difference between controls and colonized mice, and a significant difference between GF and colonized mice. Partial normalization indicates only a lack of a significant difference between controls and colonized mice. Notably, across all brain regions, 14/24 (58%) of the genes significantly altered in GF mice showed full normalization and 7/24 (29%) showed partial normalization after SPF colonization, resulting in 87% of genes being normalized to some degree by complex microbiota (Figure 2A, “SPF Col” columns). Conversely, only 2/24 (8%) genes altered in GF-mice showed full normalization and 8/24 (33%) showed partial normalization, resulting in 41% of genes affected by mono-colonization (Figure 2A, “Mono Col” columns).
SPF-colonization showed a similar magnitude of effect across all brain regions, with 4/8 differentially expressed genes in the hippocampus, 5/10 in the amygdala, and 5/7 in the mPFC being fully normalized. In the hippocampus, all GABAergic and glutamatergic genes altered in GF-mice were either fully or partially normalized in SPF-colonized mice. In the amygdala and mPFC, 8/9 and 5/6 of the GABAergic and glutamatergic genes were either fully or partially normalized, respectively. Mono-colonization fully normalized Dlg4 in the hippocampus, Gabra5 in the amygdala, and no gene in the mPFC. These results indicate that the composition of the gut microbiome has a dynamic and region-specific influence on EIB-relevant genes, and that normalization is dependent on the complexity of the microbiota.
Absence of gut microbiota leads to profound dysregulations of gene co-expression networks in all three brain areas, which are restored by SPF colonization
We next applied weighted gene co-expression network analysis (WGCNA) to assess coordinated gene expression across areas and conditions. Gene co-expression networks were constructed for each brain region and assessed for degree of conservation across experimental groups, defined as the percentage of overlapping genes. Gene networks of control mice displayed highly organized co-expression structures in all three brain regions: nearly all GABA, glutamate, and astrocytic genes were co-expressed in a single module, with smaller modules containing BDNF genes (Figure 3, upper-most panels). See details in Supplementary Figures 5–7.
Figure 3. Region-specific effects of microbiota on gene co-expression networks.
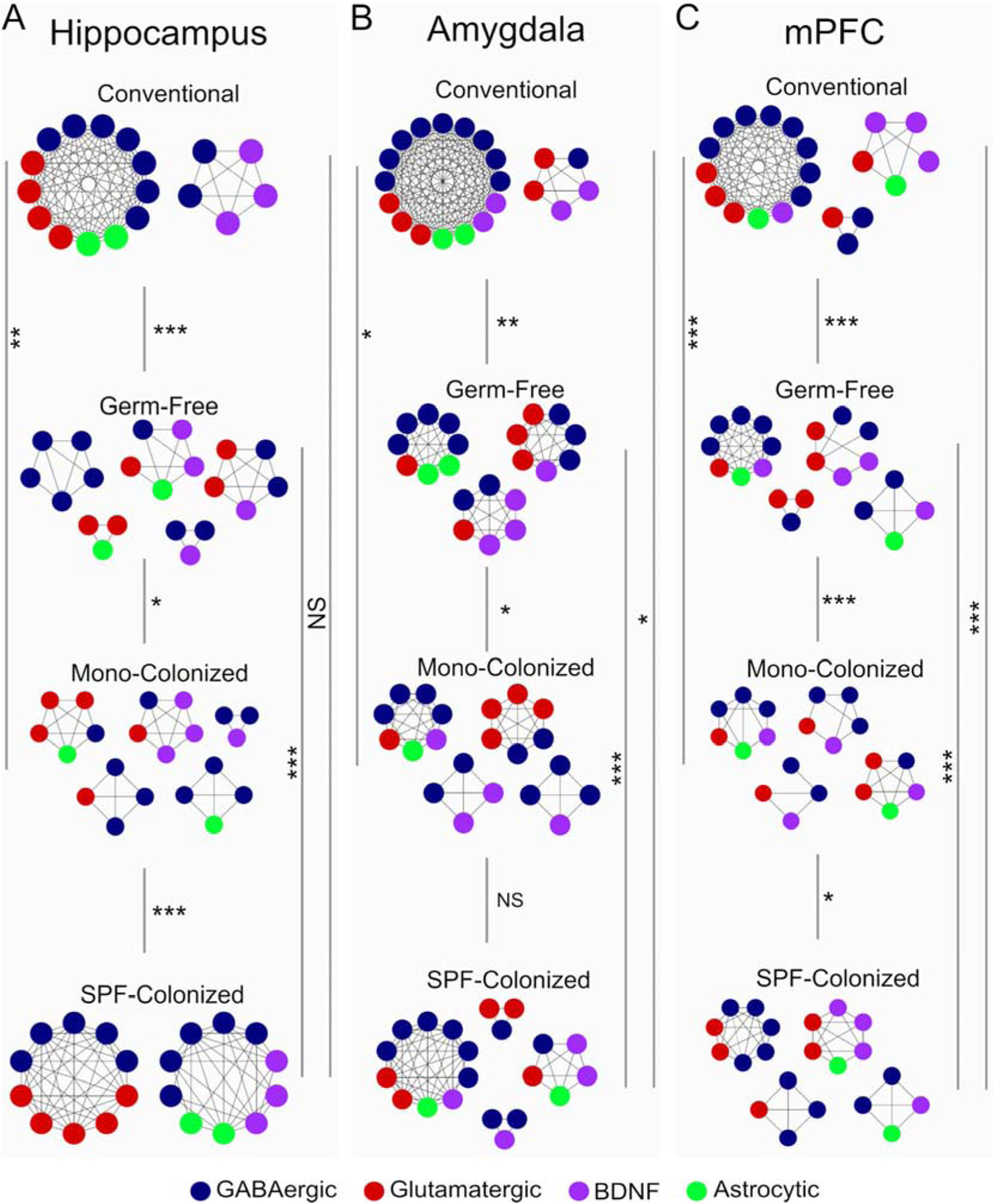
Nodes represent genes (gene names removed for visual clarity, see supplementary figures 1–3 for complete annotations), and edges represent correlated gene expression between genes. Similarity of modules across experimental groups was performed by permutation test, and statistical significance was determined by p-value meta-analysis of all module-wise comparisons across groups. *p<0.05, **p<0.01, ***p<0.001.
In contrast, GF-mice display significant disorganization of co-expression networks compared to control mice in all three brain regions (hippocampus, p=0.0003; amygdala, p=0.0012; mPFC, p=0.0069), manifested as increased numbers of smaller un-connected modules (Figure 3). Strikingly, colonization of GF-mice with SPF microbiota restored the overall gene co-expression structure in the hippocampus (p v.s. GF =0.0003, p v.s. control = 0.60). This normalization saw the majority of previously disorganized genes reorganize into a single module that was not significantly different from controls. This normalization was not observed in the amygdala (p v.s. GF = 0.010, p v.s. control = 0.011) or mPFC (p v.s. GF = 0.0062, p v.s. control = 0.0062) of SPF-colonized mice. Finally, gene modules in mono-colonized mice were different from the other groups with no apparent reorganization towards control modules in any region examined (Hippocampus, p v.s. GF = 0.012, p v.s. control = 0.0056; amygdala, p v.s. GF = 0.012, p v.s. control = 0.016; mPFC, p v.s. GF =0.0007, p v.s. control = 2.7×10−6).
Given our low sample size, we sought to support these co-expression findings with a parallel statistical approach, namely principal component analysis (PCA). PCA was performed on the omnibus gene expression within each brain region. We found the first 3 components consistently accounted for over 60% of the overall gene expression variance (hippocampus=60.6%, amygdala=75.6%, mPFC=73.2%). Moreover, plotting these 3 components revealed separation of the control and SPF-colonized groups from the GF and mono-colonized groups (Supplementary Figures 8–10). This separation was most robust in the hippocampus and weaker in the amygdala and mPFC, paralleling the co-expression results. These PCA findings support our co-expression results through independent analytical methods. Ultimately, these results indicate that gut microbiota have profound impacts on co-ordinated EIB-relevant gene expression in the brain, and that bacterial colonization appears to restore these patterns of co-expression in the hippocampus.
GF-mice and post-mortem MDD subjects have similar hippocampal gene expression profiles
Given the involvement of these genes in mood and anxiety disorders, we next determined the similarity of regional expression profiles to post-mortem MDD subjects. Gene expression changes for each experimental group was compared to the log2 fold change obtained from previously published large-scale gene expression studies of post-mortem samples from subjects with MDD (Table 1) (Duric et al., 2010; Joeyen-Waldorf et al., 2012). The gene expression profile of the hippocampus in GF-mice showed a significant correlation (ρ=0.51, p=0.027) with that of the hippocampus in MDD subjects (Figure 4). This correlation was not significant after mono-colonization (ρ=0.32, p=0.18) or SPF-colonization (ρ=0.28, p=0.24).
Table 1. GF-mice show a transcriptional profile significantly correlated to that of post-mortem MDD subjects.
Spearman correlations, and p-values, comparing transcriptional changes observed to post-mortem cohorts of MDD subjects.
| Human MOD gene expression | |||||||
|---|---|---|---|---|---|---|---|
| Experimental Group | Hippocampus (Duric et al., 2010) | Amygdala (Ding et al., 2015) | mPFC (Ding et al., 2015) | ||||
| Rho | p | Rho | p | Rho | p | ||
| Mouse gene expression | GF | 0.51* | 0.027* | 0.19 | 0.43 | 0.43 | 0.059# |
| Mono-Colonized | 0.32 | 0.18 | 0.36 | 0.13 | 0.42 | 0.058# | |
| SPF-Colonized | 0.28 | 0.24 | 0.36 | 0.16 | 0.31 | 0.19 | |
p<0.05,
p<0.1.
Figure 4. Comparison of GF mouse and human MDD gene expression profiles in the hippocampus.
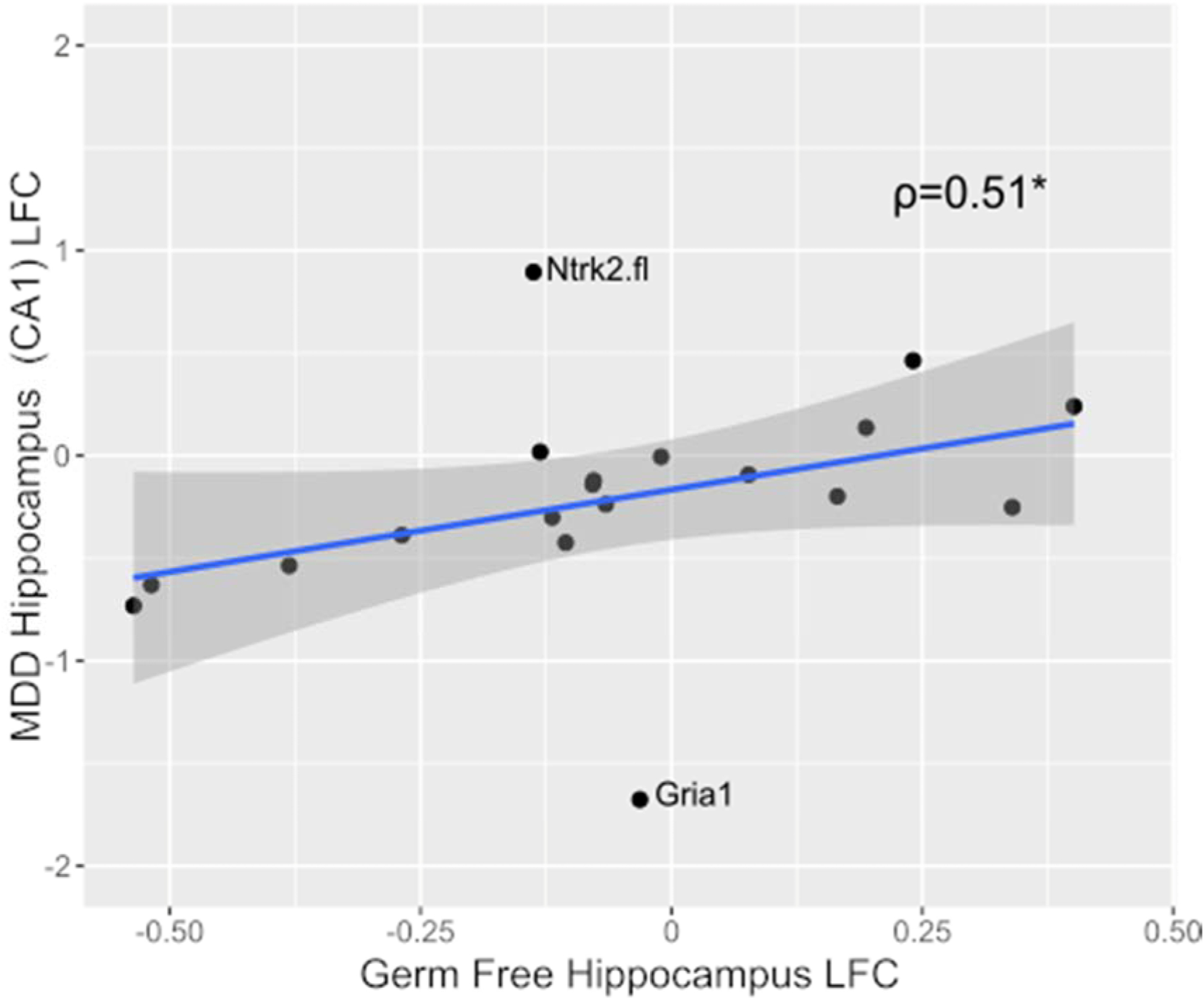
Log2 fold-change of the gene expression in GF-mice versus controls (conventional mice), compared to those observed in individuals with MDD versus controls. Spearman correlation revealed a significant positive association (ρ=0.51, p=0.027) between the two gene expression profiles. The shaded area shows the 95% confidence interval. This association remains significant after removal of Ntrk2.fl and Gria1 (ρ=0.742, p=9.48×10–4). *p<0.05.
A non-significant, but trend-level, correlation was observed between the expression profile of the ACC in MDD and the mPFC in both GF-mice (ρ=0.43, p=0.059) and mono-colonized mice (ρ=0.42, p=0.058). This correlation was also non-significant in SPF-colonized mice (ρ=0.31, p=0.19). No significant correlations were observed in the amygdala.
These results indicate that GF-mice have an EIB-relevant gene expression profile reminiscent of MDD in the hippocampus, and that this profile is responsive to changes in gut microbiota.
Discussion
In this study, we sought to determine the effect of gut microbiota on the expression of EIB-relevant genes, and the ability of different bacterial colonization schemes to reverse these effects. First, we found that GF-mice show region-specific changes in EIB-relevant gene expression in the hippocampus, amygdala, and mPFC. Second, colonization of GF-mice with complex SPF microbiota reversed the majority of these changes, whereas colonization with a single bacterial strain, E. coli JM83, showed a negligible effect. Third, GF-mice show significant disruptions in the robust co-expression patterns found in conventional (control) mice, across all brain regions. Fourth, colonization with SPF microbiota, but not mono-colonization, restores co-expression patterns in the hippocampus and marginally in the amygdala. Lastly, the profile of EIB-relevant gene expression in the hippocampus of GF-mice, but not SPF colonized mice, replicates that of the hippocampus in MDD subjects.
GF-mice show region-specific disruption in EIB-relevant genes and rescue by colonization
We observed diverse transcriptional changes in the hippocampus, up-regulation of glutamatergic and GABAergic genes in the amygdala and down-regulation of these genes in the mPFC of GF-mice. These three regions are highly relevant to psychiatric disorders given the mood-regulating role of the mPFC and amygdala, and the role of the hippocampus in learning and memory. Maintenance of EIB is critical to proper brain function, and EIB disruptions can corrupt information processing within and across brain regions, contributing to behavioural and cognitive deficits (Fee et al., 2017). GABA and glutamate genes showed concordant directionality within the amygdala and mPFC, suggesting maintenance of EIB within these brain regions but at an elevated level of activity in the amygdala, and reduced level in the mPFC. This mirrors a substantial body of literature in MDD and animal models of stress reporting increased amygdala activity and volume, with reciprocal activity and structural changes in the mPFC (Duric et al., 2013; Hill et al., 2011; Nikolova et al., 2018; Swartz et al., 2015; Victor et al., 2010). Convergent neuro-structural changes occur in GF-mice where aspiny and pyramidal neurons of the amygdala (GABAergic and glutamatergic cell-types, respectively) show dendritic hypertrophy (Luczynski et al., 2016). Reduced BDNF expression in the amygdala of GF-mice replicates previous observations in mice treated with antimicrobial agents (Bercik et al., 2011). A similar EIB-maintenance phenomenon appears to occur in the hippocampus, where disruptions in each functional group of genes were observed, and GABA and glutamate-related genes were altered in a balanced manner.
Our primary finding was a striking normalization of these differential expression profiles in GF-mice to control-like levels in all brain regions after colonization with SPF microbiota. Indeed, 58% of all genes altered in GF-mice were fully normalized, and 29% were partially normalized, to control levels. This was not observed with mono-colonization with E. coli JM83, where only 8% of genes were fully normalized (33% partially). This indicates that the complexity of the microbiota is proportional to its ability to normalize the expression of these EIB-relevant genes. There is evidence that manipulation of gut microbiota can modulate behaviour and brain chemistry both early (Chu et al., 2019; Desbonnet et al., 2015; Stilling et al., 2015) and later in life (Bercik et al., 2011; De Palma et al., 2015). Here, we found that the altered gene expression profile in 10–14 weeks old GF-mice normalized after SPF colonization. Future studies are needed to address the role of age in gut microbiota-induced alterations in EIB-relevant synaptic gene expression in the brain. Importantly, our co-expression results suggest additional region-specific critical periods.
Absence of gut microbiota disrupts gene co-expression networks, and microbiota colonization selectively reorganizes hippocampal networks
Gene co-expression networks typically reflect common metabolic, signaling, or regulatory pathways within cells and tissues (Langfelder and Horvath, 2008). Here, within a restricted set of synaptic genes, we observed highly organized co-expression in control mice, with GABAergic and glutamatergic genes clustering into a single co-expression module and residual genes in smaller modules. This highly organized expression is consistent with, and may reflect, the balance between excitatory and inhibitory tone present in controls and the dynamic interrelationship of these two neurotransmitter systems (Haider et al., 2006). Similar to our differential expression findings, GF-mice showed a significant disorganization of co-expression across all brain regions, characterized by dissolution of the large GABAergic/glutamatergic module into several small modules. Though speculative, such co-expression patterns may reflect a loss of proper synchrony in excitatory and inhibitory function in GF-mice. Interestingly, this disorganization was not reversed by mono-colonization in any group but was largely reversed in the hippocampus, but not the amygdala or mPFC, of SPF microbiota colonized mice. These findings were supported by our PCA results showing separation of control + SPF-colonized expression profiles from those of GF + mono-colonized mice. Given the importance of these genes in contributing to EIB (Oh et al., 2019), it is possible that residual co-expression differences reflect residual EIB deficits in the amygdala and mPFC. This may increase stress vulnerability in these mice, given the critical role of these two regions in the behavioural response to stress (Ghosal et al., 2017; Gilabert-Juan et al., 2013; Guilloux et al., 2012; Roozendaal et al., 2009). Moreover, these results further identify the insufficiency of mono-colonization with JM83 to restore control-like neurobiology. Overall, despite the limited number of samples and genes in our study, we consistently observed that synaptic genes group together in controls, segregate in GF-mice, and re-group only in the hippocampus of SPF-colonized mice.
GF-mice, but not SPF-colonized mice, recapitulate region-specific MDD expression profiles
Through comparisons of MDD-correlated, and gut microbiome-induced gene expression profiles, we showed that gut microbiota modulate similar neurobiological substrates to those affected in psychiatric disorders. Despite observations that GF-mice do not generally show a purely anxiety- or anhedonia-like behavioural profile, cross-species differences in changes in other brain regions, environmental, and genetic factors may explain the seemingly discordant behavioural profiles and concordant gene expression profiles (De Palma et al., 2015; Neufeld et al., 2011). Nevertheless, we found that GF-mice show a gene expression profile in the hippocampus that was significantly correlated to that of MDD subjects. However, this correlation was not observed in SPF-colonized mice. GF-mice and individuals with MDD show hippocampus-dependent cognitive deficits, suggesting that similar neurobiological changes may underlie these deficits. To our knowledge, this is the first comparison of brain region-specific gene expression in GF-mice and post-mortem psychiatric disorder cohorts. Clinical studies demonstrate altered gut microbiome profiles in MDD, and machine learning algorithms can, within datasets, distinguish symptomatic MDD subjects and healthy controls by microbiome composition (Simpson et al., 2020). Taken together, these findings indicate that there may be cross-species similarities in the relationship between gut microbiota and the brain, despite species differences in the specific composition of the gut microbiome (Hugenholtz and de Vos, 2018), and that altered gut microbiome may represent a contributing etiological factor leading to brain EIB gene expression changes in MDD. Further studies are needed to identify specific microbiome compositions which are associated with neurobiological and behavioural deficits, and the degrees of reversal that is achievable with different microbiome-based strategies.
In summary, these results demonstrate the possibility of modulating brain gene expression, relevant to information processing in cortical and sub-cortical regions, by modulating the composition of the gut microbiome. These changes appear to be relevant to systems involved in psychiatric disorders, and warrant further investigation to systematically characterize large-scale transcriptomic changes and associated behavioural changes.
Limitations, Future Directions, and Conclusions
Our results must be viewed in the context of several limitations. First, low sample size and varied sex composition limited our power to detect small effects and precluded inferences regarding sex-specificity, in addition to accessing behavioural consequences. Breeding difficulties and availability of GF facilities constrained the number of GF-mice available for use and thus imposed limitations on sample size and sex composition. Despite our limited sample size, our differential expression findings are relatively consistent and minimally influenced by outlying data. Second, acknowledging the limitations of co-expression analyses using few subjects and a restricted gene-list, we opted for a conservative analytical approach assessing only module composition (i.e. cross-tabulation based preservation), rather than intra- or inter-module connectivity which demand large samples and broader gene lists. Third, though the genes examined here are involved in EIB maintenance, they are not an exact EIB proxy. Changes in the expression of individual genes do not necessarily imply altered electrophysiological properties of neurons. However, diverse changes in these genes would be suggestive of regional or systems-level changes that may affect EIB regulation.
Future studies should endeavour to replicate our RNA findings in both sexes at proteomic, neurophysiological, behavioural and cognitive levels before definitive functional conclusions can be drawn. Though we draw comparisons between GF-mice and post-mortem MDD gene expression profiles, we do not anticipate these similarities being wholly specific to MDD. Future studies should also examine transdiagnostic and diagnosis-specific similarities to GF-mice, and determine if these changes represent a generalized association between microbiota and central gene expression.
In conclusion, these findings identify a robust influence of microbiota composition on EIB-relevant synaptic gene expression in multiple brain regions and provide evidence to further investigate the role of gut microbiota in the pathology of mood disorders.
Supplementary Material
Highlights.
Previous studies in mice show that gut microbiota can influence behaviour by modulating neurochemical pathways in the brain
In this study we report:
Absence of gut microbiota disrupts the expression and synchronization of excitation-inhibition relevant genes in the brain of mice
These changes parallel those observed in the hippocampus of human subjects with depression
Gut bacterial recolonization normalized the expression and synchronization of the same genes suggesting a role for the microbiome in disease and recovery
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare no competing financial conflicts of interest.
References
- Bandelow B, Lichte T, Rudolf S, Wiltink J, Beutel EM, 2014. The diagnosis of and treatment recommendations for anxiety disorders. Deutsches Ärzteblatt International 111(27–28), 473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y, 2000. On the Adaptive Control of the False Discovery Rate in Multiple Testing With Independent Statistics. Journal of Educational and Behavioral Statistics 25(1), 60–83. [Google Scholar]
- Bercik P, Denou E, Collins J, Jackson W, Lu J, Jury J, Deng Y, Blennerhassett P, Macri J, McCoy KD, Verdu EF, Collins SM, 2011. The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology 141(2), 599–609, 609. [DOI] [PubMed] [Google Scholar]
- Braniste V, Al-Asmakh M, Kowal C, Anuar F, Abbaspour A, Toth M, Korecka A, Bakocevic N, Ng LG, Kundu P, Gulyas B, Halldin C, Hultenby K, Nilsson H, Hebert H, Volpe BT, Diamond B, Pettersson S, 2014. The gut microbiota influences blood-brain barrier permeability in mice. Sci. Transl. Med 6(263), 263ra158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang LC, Jamain S, Lin CW, Rujescu D, Tseng GC, Sibille E, 2014. A conserved BDNF, glutamate- and GABA-enriched gene module related to human depression identified by coexpression meta-analysis and DNA variant genome-wide association studies. PLoS One 9(3), e90980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Murdock MH, Jing D, Won TH, Chung H, Kressel AM, Tsaava T, Addorisio ME, Putzel GG, Zhou L, Bessman NJ, Yang R, Moriyama S, Parkhurst CN, Li A, Meyer HC, Teng F, Chavan SS, Tracey KJ, Regev A, Schroeder FC, Lee FS, Liston C, Artis D, 2019. The microbiota regulate neuronal function and fear extinction learning. Nature 574(7779), 543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke G, Grenham S, Scully P, Fitzgerald P, Moloney RD, Shanahan F, Dinan TG, Cryan JF, 2013. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol. Psychiatry 18(6), 666–673. [DOI] [PubMed] [Google Scholar]
- Collins SM, Surette M, Bercik P, 2012. The interplay between the intestinal microbiota and the brain. Nat. Rev. Microbiol 10(11), 735–742. [DOI] [PubMed] [Google Scholar]
- Dahlin M, Prast-Nielsen S, 2019. The gut microbiome and epilepsy. EBioMedicine 44, 741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Palma G, Blennerhassett P, Lu J, Deng Y, Park AJ, Green W, Denou E, Silva MA, Santacruz A, Sanz Y, Surette MG, Verdu EF, Collins SM, Bercik P, 2015. Microbiota and host determinants of behavioural phenotype in maternally separated mice. Nat Commun 6, 7735. [DOI] [PubMed] [Google Scholar]
- De Palma G, Collins SM, Bercik P, Verdu EF, 2014. The microbiota-gut-brain axis in gastrointestinal disorders: stressed bugs, stressed brain or both? J Physiol 592(14), 2989–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Palma G, Lynch MD, Lu J, Dang VT, Deng Y, Jury J, Umeh G, Miranda PM, Pigrau Pastor M, Sidani S, Pinto-Sanchez MI, Philip V, McLean PG, Hagelsieb MG, Surette MG, Bergonzelli GE, Verdu EF, Britz-McKibbin P, Neufeld JD, Collins SM, Bercik P, 2017. Transplantation of fecal microbiota from patients with irritable bowel syndrome alters gut function and behavior in recipient mice. Sci Transl Med 9(379). [DOI] [PubMed] [Google Scholar]
- Desbonnet L, Clarke G, Shanahan F, Dinan TG, Cryan JF, 2014. Microbiota is essential for social development in the mouse. Mol Psychiatry 19(2), 146–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbonnet L, Clarke G, Traplin A, O’Sullivan O, Crispie F, Moloney RD, Cotter PD, Dinan TG, Cryan JF, 2015. Gut microbiota depletion from early adolescence in mice: Implications for brain and behaviour. Brain Behav Immun 48, 165–173. [DOI] [PubMed] [Google Scholar]
- Diaz HR, Wang S, Anuar F, Qian Y, Bjorkholm B, Samuelsson A, Hibberd ML, Forssberg H, Pettersson S, 2011. Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. U. S. A 108(7), 3047–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Chang LC, Wang X, Guilloux JP, Parrish J, Oh H, French BJ, Lewis DA, Tseng GC, Sibille E, 2015. Molecular and Genetic Characterization of Depression: Overlap with other Psychiatric Disorders and Aging. Mol Neuropsychiatry 1(1), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duric V, Banasr M, Licznerski P, Schmidt HD, Stockmeier CA, Simen AA, Newton SS, Duman RS, 2010. A negative regulator of MAP kinase causes depressive behavior. Nature medicine 16(11), 1328–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duric V, Banasr M, Stockmeier CA, Simen AA, Newton SS, Overholser JC, Jurjus GJ, Dieter L, Duman RS, 2013. Altered expression of synapse and glutamate related genes in post-mortem hippocampus of depressed subjects. The international journal of neuropsychopharmacology 16(1), 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erny D, Hrabe de Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E, Keren-Shaul H, Mahlakoiv T, Jakobshagen K, Buch T, Schwierzeck V, Utermohlen O, Chun E, Garrett WS, McCoy KD, Diefenbach A, Staeheli P, Stecher B, Amit I, Prinz M, 2015. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci 18(7), 965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fee C, Banasr M, Sibille E, 2017. Somatostatin-Positive Gamma-Aminobutyric Acid Interneuron Deficits in Depression: Cortical Microcircuit and Therapeutic Perspectives. Biol Psychiatry 82(8), 549–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields CT, Sampson TR, Bruce-Keller AJ, Kiraly DD, Hsiao EY, de Vries GJ, 2018. Defining Dysbiosis in Disorders of Movement and Motivation. The Journal of neuroscience : the official journal of the Society for Neuroscience 38(44), 9414–9422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George Paxinos KF, 2012. The Mouse Brain in Stereotaxic Coordinates, 4th Edition ed. Academic Press. [Google Scholar]
- Ghosal S, Hare BD, Duman RS, 2017. Prefrontal cortex GABAergic deficits and circuit dysfunction in the pathophysiology and treatment of chronic stress and depression. Current Opinion in Behavioral Sciences 14, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilabert-Juan J, Castillo-Gomez E, Guirado R, Moltó MD, Nacher J, 2013. Chronic stress alters inhibitory networks in the medial prefrontal cortex of adult mice. Brain Structure and Function 218(6), 1591–1605. [DOI] [PubMed] [Google Scholar]
- Grochowska M, Wojnar M, Radkowski M, 2018. The gut microbiota in neuropsychiatric disorders. Acta neurobiologiae experimentalis 78(2), 69–81. [PubMed] [Google Scholar]
- Guilloux J. p., Douillard-guilloux G, Kota R, Wang X, Martinowich K, Tseng GC, Lewis D.a., Sibille E, 2012. Molecular evidence for BDNF- and GABA-related dysfunctions in the amygdala of female subjects with Major Depression. Molecular Psychiatry 17(11), 1130–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider B, Duque A, Hasenstaub AR, McCormick DA, 2006. Neocortical Network Activity In Vivo Is Generated through a Dynamic Balance of Excitation and Inhibition. Journal of Neuroscience 26(17), 4535–4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapfelmeier S, Lawson MA, Slack E, Kirundi JK, Stoel M, Heikenwalder M, Cahenzli J, Velykoredko Y, Balmer ML, Endt K, Geuking MB, Curtiss R III, McCoy KD, Macpherson AJ, 2010. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 328(5986), 1705–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard NA, Rubin-Delanchy P, 2018. Choosing between methods of combining-values. Biometrika 105(1), 239–246. [Google Scholar]
- Hill MN, Hillard CJ, McEwen BS, 2011. Alterations in corticolimbic dendritic morphology and emotional behavior in cannabinoid CB1 receptor-deficient mice parallel the effects of chronic stress. Cerebral cortex (New York, N.Y. : 1991) 21(9), 2056–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoban AE, Stilling RM, Moloney G, Shanahan F, Dinan TG, Clarke G, Cryan JF, 2018. The microbiome regulates amygdala-dependent fear recall. Mol Psychiatry 23(5), 1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz F, de Vos WM, 2018. Mouse models for human intestinal microbiota research: a critical evaluation. Cell Mol Life Sci 75(1), 149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joeyen-Waldorf J, Nikolova YS, Edgar N, Walsh C, Kota R, Lewis DA, Ferrell R, Manuck SB, Hariri AR, Sibille E, 2012. Adenylate cyclase 7 is implicated in the biology of depression and modulation of affective neural circuitry. Biol Psychiatry 71(7), 627–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohen R, Dobra A, Tracy JH, Haugen E, 2014. Transcriptome profiling of human hippocampus dentate gyrus granule cells in mental illness. Transl Psychiatry 4, e366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonte B, Engmann O, Purushothaman I, Menard C, Wang J, Tan C, Scarpa JR, Moy G, Loh YE, Cahill M, Lorsch ZS, Hamilton PJ, Calipari ES, Hodes GE, Issler O, Kronman H, Pfau M, Obradovic ALJ, Dong Y, Neve RL, Russo S, Kazarskis A, Tamminga C, Mechawar N, Turecki G, Zhang B, Shen L, Nestler EJ, 2017. Sex-specific transcriptional signatures in human depression. Nature medicine 23(9), 1102–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S, 2008. WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics 9, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin LC, Sibille E, 2013. Reduced brain somatostatin in mood disorders: a common pathophysiological substrate and drug target? Front Pharmacol 4, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luczynski P, Whelan SO, O’Sullivan C, Clarke G, Shanahan F, Dinan TG, Cryan JF, 2016. Adult microbiota-deficient mice have distinct dendritic morphological changes: differential effects in the amygdala and hippocampus. Eur J Neurosci 44(9), 2654–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum GR, Olson CA, Hsiao EY, 2020. Emerging roles for the intestinal microbiome in epilepsy. Neurobiology of Disease 135, 104576. [DOI] [PubMed] [Google Scholar]
- Lussier AL, Romay-Tallón R, Caruncho HJ, Kalynchuk LE, 2013. Altered GABAergic and glutamatergic activity within the rat hippocampus and amygdala in rats subjected to repeated corticosterone administration but not restraint stress. Neuroscience 231, 38–48. [DOI] [PubMed] [Google Scholar]
- McVey Neufeld KA, Bienenstock J, Bharwani A, Champagne-Jorgensen K, Mao Y, West C, Liu Y, Surette MG, Kunze W, Forsythe P, 2019. Oral selective serotonin reuptake inhibitors activate vagus nerve dependent gut-brain signalling. Sci Rep 9(1), 14290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld KM, Kang N, Bienenstock J, Foster JA, 2011. Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterol. Motil 23(3), 255–264, e119. [DOI] [PubMed] [Google Scholar]
- Nikolova YS, Misquitta KA, Rocco BR, Prevot TD, Knodt AR, Ellegood J, Voineskos AN, Lerch JP, Hariri AR, Sibille E, Banasr M, 2018. Shifting priorities: highly conserved behavioral and brain network adaptations to chronic stress across species. Translational Psychiatry 8(1), 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh DH, Son H, Hwang S, Kim SH, 2012. Neuropathological abnormalities of astrocytes, GABAergic neurons, and pyramidal neurons in the dorsolateral prefrontal cortices of patients with major depressive disorder. European Neuropsychopharmacology 22(5), 330–338. [DOI] [PubMed] [Google Scholar]
- Oh H, Lewis DA, Sibille E, 2016. The Role of BDNF in Age-Dependent Changes of Excitatory and Inhibitory Synaptic Markers in the Human Prefrontal Cortex. Neuropsychopharmacology 41(13), 3080–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Piantadosi SC, Rocco BR, Lewis DA, Watkins SC, Sibille E, 2019. The Role of Dendritic Brain-Derived Neurotrophic Factor Transcripts on Altered Inhibitory Circuitry in Depression. Biol Psychiatry 85(6), 517–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto-Sanchez MI, Hall GB, Ghajar K, Nardelli A, Bolino C, Lau JT, Martin FP, Cominetti O, Welsh C, Rieder A, Traynor J, Gregory C, De Palma G, Pigrau M, Ford AC, Macri J, Berger B, Bergonzelli G, Surette MG, Collins SM, Moayyedi P, Bercik P, 2017. Probiotic Bifidobacterium longum NCC3001 Reduces Depression Scores and Alters Brain Activity: A Pilot Study in Patients With Irritable Bowel Syndrome. Gastroenterology 153(2), 448–459.e448. [DOI] [PubMed] [Google Scholar]
- Poskanzer KE, Yuste R, 2016. Astrocytes regulate cortical state switching in vivo. Proceedings of the National Academy of Sciences of the United States of America 113(19), E2675–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins J, Olivier B, Korte SM, 2011. Triple reuptake inhibitors for treating subtypes of major depressive disorder: the monoamine hypothesis revisited. Expert opinion on investigational drugs 20(8), 1107–1130. [DOI] [PubMed] [Google Scholar]
- Roozendaal B, McEwen BS, Chattarji S, 2009. Stress, memory and the amygdala. Nature Reviews Neuroscience 10(6), 423–433. [DOI] [PubMed] [Google Scholar]
- Schur RR, Draisma LW, Wijnen JP, Boks MP, Koevoets MG, Joels M, Klomp DW, Kahn RS, Vinkers CH, 2016. Brain GABA levels across psychiatric disorders: A systematic literature review and meta-analysis of (1) H-MRS studies. Human brain mapping 37(9), 3337–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T, 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research 13(11), 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon G, Cruz NJ, Kang DW, Gandal MJ, Wang B, Kim YM, Zink EM, Casey CP, Taylor BC, Lane CJ, Bramer LM, Isern NG, Hoyt DW, Noecker C, Sweredoski MJ, Moradian A, Borenstein E, Jansson JK, Knight R, Metz TO, Lois C, Geschwind DH, Krajmalnik-Brown R, Mazmanian SK, 2019. Human Gut Microbiota from Autism Spectrum Disorder Promote Behavioral Symptoms in Mice. Cell 177(6), 1600–1618 e1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson CA, Mu A, Haslam N, Schwartz OS, Simmons JG, 2020. Feeling down? A systematic review of the gut microbiota in anxiety/depression and irritable bowel syndrome. J Affect Disord 266, 429–446. [DOI] [PubMed] [Google Scholar]
- Stilling RM, Moloney GM, Ryan FJ, Hoban AE, Bastiaanssen TF, Shanahan F, Clarke G, Claesson MJ, Dinan TG, Cryan JF, 2018. Social interaction-induced activation of RNA splicing in the amygdala of microbiome-deficient mice. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stilling RM, Ryan FJ, Hoban AE, Shanahan F, Clarke G, Claesson MJ, Dinan TG, Cryan JF, 2015. Microbes & neurodevelopment--Absence of microbiota during early life increases activity-related transcriptional pathways in the amygdala. Brain Behav Immun 50, 209–220. [DOI] [PubMed] [Google Scholar]
- Sudo N, Chida Y, Aiba Y, Sonoda J, Oyama N, Yu X-N, Kubo C, Koga Y, 2004. Postnatal microbial colonization programs the hypothalamic–pituitary–adrenal system for stress response in mice. The Journal of Physiology 558(1), 263–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz JR, Knodt AR, Radtke SR, Hariri AR, 2015. A neural biomarker of psychological vulnerability to future life stress. Neuron 85(3), 505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team, R.C., 2018. R: A Language and Environment for Statistical Computing. Vienna, Austria. [Google Scholar]
- Treiman DM, 2001. GABAergic mechanisms in epilepsy. Epilepsia 42 Suppl 3, 8–12. [DOI] [PubMed] [Google Scholar]
- Tripp A, Oh H, Guilloux JP, Martinowich K, Lewis DA, Sibille E, 2012. Brain-derived neurotrophic factor signaling and subgenual anterior cingulate cortex dysfunction in major depressive disorder. The American journal of psychiatry 169(11), 1194–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valles-Colomer M, Falony G, Darzi Y, Tigchelaar EF, Wang J, Tito RY, Schiweck C, Kurilshikov A, Joossens M, Wijmenga C, Claes S, Van Oudenhove L, Zhernakova A, Vieira-Silva S, Raes J, 2019. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat Microbiol 4(4), 623–632. [DOI] [PubMed] [Google Scholar]
- Victor TA, Furey ML, Fromm SJ, Ohman A, Drevets WC, 2010. Relationship between amygdala responses to masked faces and mood state and treatment in major depressive disorder. Archives of general psychiatry 67(11), 1128–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodicka M, Ergang P, Hrncir T, Mikulecka A, Kvapilova P, Vagnerova K, Sestakova B, Fajstova A, Hermanova P, Hudcovic T, Kozakova H, Pacha J, 2018. Microbiota affects the expression of genes involved in HPA axis regulation and local metabolism of glucocorticoids in chronic psychosocial stress. Brain Behav Immun 73, 615–624. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


