Abstract
Objective.
Eicosanoids modulate inflammation via complex networks involving different pathways and downstream mediators, including oxylipins. Although altered eicosanoids are linked to rheumatoid arthritis (RA), suggesting that metabolization is enhanced, the role of oxylipins in disease stratification remains unexplored. This study was undertaken to characterize oxylipin networks during the earliest stages of RA and evaluate their associations with clinical features and treatment outcomes.
Methods.
In total, 60 patients with early RA (according to the American College of Rheumatology/European League Against Rheumatism 2010 criteria), 11 individuals with clinically suspect arthralgia (CSA), and 28 healthy control subjects were recruited. Serum samples were collected at the time of onset. In the early RA group, 50 patients who had not been exposed to disease-modifying antirheumatic drug (DMARD) or glucocorticoid treatment at the time of recruitment were prospectively followed up at 6 and 12 months after having received conventional synthetic DMARDs. A total of 75 oxylipins, mostly derived from arachidonic, eicosapentanoic, and linoleic acids, were identified in the serum by liquid chromatography tandem mass spectrometry.
Results.
Univariate analyses demonstrated differences in expression patterns of 14 oxylipins across the RA, CSA, and healthy control groups, with each exhibiting a different trajectory. Network analyses revealed a strong grouping pattern of oxylipins in RA patients, whereas in individuals with CSA, a fuzzy network of oxylipins with higher degree and closeness was found. Partial least-squares discriminant analyses yielded variable important projection scores of >1 for 22 oxylipins, which allowed the identification of 2 clusters. Cluster usage differed among the groups (P = 0.003), and showed associations with disease severity and low rates of remission at 6 and 12 months in RA patients who were initially treatment-naive. Pathway enrichment analyses revealed different precursors and pathways between the groups, highlighting the relevance of the arachidonic acid pathway in individuals with CSA and the lipooxygenase pathway in patients with early RA. In applying distinct oxylipin signatures, subsets of seropositive and seronegative RA could be identified.
Conclusion.
Oxylipin networks differ across stages during the earliest phases of RA. These distinct oxylipin networks could potentially elucidate pathways with clinical relevance for disease progression, clinical heterogeneity, and treatment response.
INTRODUCTION
Rheumatoid arthritis (RA) is an immune-mediated rheumatic condition characterized by chronic inflammation and joint destruction (1). Early diagnosis and prompt treatment guided by treat-to-target goals is crucial to ensure long-term disease control (2). Interventions during the early phase of RA are associated with higher rates of remission, probably due to pathogenic mechanisms occurring at this point (3). Therefore, research into the early disease phase is of utmost scientific relevance. The development of RA is a multistep process, in which different phases can be distinguished (4). The recognition of the symptomatic phase preceding clinical arthritis, referred to as clinically suspect arthralgia (CSA), represents the first opportunity to identify patients at risk for progression to RA (5). This stage serves not only to open a window for characterization of the changes underlying the shift from systemic autoimmunity to overt joint synovitis, but also to delineate potential targets for prevention of disease progression (5,6). Although the cellular and proteomic characterization of these stages have been extensively pursued, the metabolomics, and mainly the lipidomics, have received less attention.
In the “omics” era, lipids are emerging as pivotal mediators for several biologic processes (7). Polyunsaturated fatty acids (PUFAs) and eicosanoids form one of the most complex networks in biology, controlling many physiologic and pathologic processes, often in opposing directions (8). The role of eicosanoids in RA dates back to the 1970s and mid-1980s, with the description of the cyclooxygenase (COX) and lipooxygenase (LOX) pathways (9). However, the presence of these enzymes could not be used to fully account for the underlying pathologic processes, and despite profound improvements in the clinical management of RA, these pathways were observed to remain active even after treatment (8). Consequently, a knowledge plateau was reached, in part due to technical limitations. It was not until recently that the CYP450-derived lipid species were described (10), although their role is still poorly characterized.
Lipidomics and high-throughput approaches have started a new investigative period aimed at attaining a better understanding of the control of local inflammation in RA and its progression to either resolution or chronification (11). A number of novel lipid mediators, such as oxylipins, are now recognized as active players in either controlling the resolution of the disease or fueling inflammation (11–13). Oxylipins have been reported to promote migration of polymorphonuclear cells, enhance vascular permeability, control cytokine production, and modulate oxidative stress species (11). Thus, oxylipins may be considered attractive candidate biomarkers for further clarifying the mechanisms of tissue injury and disease aggravation.
Previous results from our group revealed altered levels of different PUFAs in patients with RA (14), which may reflect an accelerated metabolization toward their downstream mediators, including oxylipins. It is plausible that applying new analytical technologies will allow us to answer the question as to whether PUFA-derived oxylipin networks are altered in RA, and help us to comprehensively analyze how disturbed oxylipin networks may be associated with the clinical phenotype of the earliest stages of the disease. Therefore, the aims of the present study were to 1) analyze oxylipin levels and networks during the earliest stages of RA, including the preclinical CSA stage, 2) evaluate whether oxylipin expression patterns may help identify patients with specific clinical features and treatment responses, and 3) evaluate whether profiling of oxylipins may identify pathways related to disease heterogeneity.
PATIENTS AND METHODS
Study participants.
The study was approved by the local institutional review board (Comité de Ética de Investigación Clínica del Principado de Asturias) in compliance with the Declaration of Helsinki. All study subjects gave written informed consent.
Our study involved 60 patients with early RA, diagnosed according to the American College of Rheumatology (ACR)/European League Against Rheumatism (EULAR) 2010 classification criteria (15), from the early arthritic clinic in the Department of Rheumatology at Hospital Universitario Central de Asturias in Oviedo, Spain. Patients were recruited at the time of disease onset. A complete clinical examination (see Supplementary Methods, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41537/abstract) was performed on all patients during the recruitment appointment. Composite measures of disease activity were calculated, including the Disease Activity Score in 28 joints using the erythrocyte sedimentation rate (DAS28-ESR) (16) and the Simplified Disease Activity Index (SDAI) (17). Patients who had not been exposed to any disease-modifying antirheumatic drugs (DMARDs) or glucocorticoids at the time of recruitment were considered to be treatment-naive (n = 50), and this group was prospectively followed up with complete clinical examinations at 6 months (n = 46) and 12 months (n = 40) after having received treatment with conventional synthetic DMARDs (csDMARDs).
Clinical management was performed according to the EULAR recommendations for the management of RA with synthetic and biologic DMARDs (18). Clinical response was evaluated using the EULAR response criteria (19) at 6 and 12 months of follow-up, and patients exhibiting a good response were considered to be responders, whereas those with moderate or no response were classified as nonresponders. Patients switching to a different csDMARD during the first 12 months were also classified as nonresponders.
Individuals with CSA were recruited from the same clinic. These individuals were considered to have CSA if they met ≥4 of the criteria in the EULAR definition of arthralgia suspicious for progression to RA (5), which allows a sensitivity of 70% and specificity of 93.6%. Subjects without arthritis were recruited as healthy controls among age- and sex-matched individuals from the same population of subjects who were without a diagnosis of inflammatory rheumatic disease.
Exclusion criteria applied to all of the groups included presence of a preexisting autoimmune or inflammatory condition, medical prescription for nonsteroidal antiinflammatory drugs or coxibs, usage of fish oil supplements, or history of cancer or recent infection (within 3 months of study recruitment). A fasting blood sample was collected from all individuals by venipuncture, and serum samples were transferred to the laboratory, processed under controlled, standardized procedures, and stored at −80°C within 2 hours of processing. Plasma samples from a subgroup of subjects (n = 6) were collected and processed in parallel.
Lipid extraction and liquid chromatography tandem mass spectrometry (LC-MS/MS) measurement of oxylipins.
All serum samples obtained at baseline were thawed once and immediately used for isolation of free fatty acids and oxylipins as described previously (20,21). In brief, 50 μl of serum was spiked with a cocktail of 26 deuterated internal standards that also included some selected PUFAs, brought to a volume of 1 ml with 10% methanol. Thereafter, the samples were purified by solid-phase extraction on a Strata-X column (see Supplementary Methods [http://onlinelibrary.wiley.com/doi/10.1002/art.41537/abstract]).
Oxylipins in the sera were analyzed and quantified by LC-MS/MS as described previously (20,21) (see Supplementary Methods [http://onlinelibrary.wiley.com/doi/10.1002/art.41537/abstract]). Oxylipins and free fatty acids were quantitated using the stable isotope dilution method. Identical amounts of deuterated internal standards were added to each sample and to all of the primary standards used to generate standard curves (see Supplementary Methods [http://onlinelibrary.wiley.com/doi/10.1002/art.41537/abstract]). The levels of oxylipins are expressed in pmoles/ml.
Statistical analysis.
Continuous variables are presented as the mean ± SD or median with interquartile range, whereas categorical variables are summarized as the number (percentage) of subjects. For each oxylipin, the fatty acid precursor and pathway (first enzyme acting on the precursors) were retrieved from the literature. If a nonenzymatic mechanism was described in the literature, regardless of the experimental setting, it was included as well, in a conservative approach.
Nonparametric tests were used to evaluate differences in oxylipin profiles across the groups, and the P values obtained were adjusted for multiple testing using the Benjamini-Hochberg procedure (22). Linear regression models were used to control comparisons for potential confounders. Oxylipin levels were log-transformed, normalized to the median value, and scaled using the range-scaling method (23).
Correlograms and network analyses were built to analyze the correlations among the oxylipins across conditions. Centrality measures were calculated for each metabolite and condition (24). Partial least-squares discriminant analysis (PLS-DA) was used to identify discriminant metabolites, with the analyses controlled for multicollinearity, and cross-validation accuracy and permutation model statistics were retrieved. Oxylipins contributing to group discrimination in the PLS-DA were selected on the basis of having a variable important projection (VIP) score of >1 (24). VIP-selected metabolites were used in unsupervised cluster analyses.
For comparisons between 2 groups, orthogonal PLS-DA (OPLS-DA) models were constructed to evaluate group discrimination. These models enhance the interpretation and discrimination based on intra- and interclass information, without a significant effect on prediction power (25). Cross-validation of the OPLS-DA models was performed by permutation analyses, and Q2 P values were computed. Correlation analyses against prespecified patterns were assessed, and correlation coefficients (determined using Spearman’s rank correlation test), P values, and false discovery rates were computed.
Pathway enrichment analyses were performed using the KEGG human genome library, with a global test for pathway enrichment. Pathway topology analysis was used to assess betweenness centrality. Raw P values (adjusted), Holm’s P values, and pathway impact were obtained for each pathway. Statistical analyses were carried out using R version 3.6.3 and MetaboAnalyst version 4.0.
RESULTS
Serum oxylipin levels during the earliest stages of RA.
Serum oxylipins were measured in 60 patients with early RA (including 50 treatment-naive patients), 11 individuals with CSA, and 28 matched healthy controls (characteristics of the subjects are listed in Supplementary Table 1, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41537/abstract). A total of 74 oxylipins derived from arachidonic acid (AA) (n = 39), docosahexanoic acid (DHA) (n = 13), linoleic acid (LA) (n = 7), eicosapentanoic acid (EPA) (n = 7), dihomo-γ-linolenic acid (DHGLA) (n = 3), α-linolenic acid (ALA) (n = 2), and oleic acid (OA) (n = 2) were identified by LC-MS/MS in the serum from patients with early RA (a complete list of the identified oxylipins is shown in Supplementary Table 2, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41537/abstract). In subgroup analyses, a good correlation was observed between the oxylipin levels measured in the serum and oxylipin levels measured in the plasma (median Spearman’s R = 0.870, range 0.750–1.000).
Moreover, the peaks corresponding to leukotriene B4 (LTB4) and 5S,12S-dihydroxyeicosatetraenoic acid (5S,12S-diHETE) were compared in the serum samples from patients with RA. Both metabolites showed similar fragmentation patterns by MS, but showed baseline separation under the LC conditions employed, thus excluding the possibility of a potential overlap in the chromatogram. Furthermore, a comparative analysis of both analytes allowed us to exclude the possibility of a significant platelet—neutrophil activation matrix during sample collection and preparation (see Supplementary Figure 1, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41537/abstract), as 5S, 12S-diHETE is considered a product of activated platelets and neutrophils.
A total of 14 oxylipins exhibited different serum levels across the groups (see Supplementary Table 3, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41537/abstract). Oxylipins exhibited different trajectories, with some showing peaking levels in patients with RA, whereas others showed peaking levels in individuals with CSA or healthy controls (see Supplementary Figure 2, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41537/abstract). The change was not always gradual across the groups, thus pointing to the existence of complex, individual patterns requiring a global approach. No associations with age, sex, body mass index, or traditional cardiovascular risk factors were observed in any of the study groups (all P > 0.05). Similarly, exclusion of patients who were receiving medications at the time of recruitment (n = 10) did not change these results. Oxylipin levels were not observed to parallel the extent of disease activity in RA patients (all P > 0.05). It must be noted that most of the patients had a status of high disease activity.
Correlograms showing the associations among the oxylipins (Figure 1A) provided evidence of more defined oxylipin groupings in healthy controls and patients with RA, in contrast to a more widespread pattern in individuals with CSA. Network graphs were generated to analyze the interactions among the oxylipins (Figure 1B). These analyses confirmed that the different clinical stages were hallmarked by distinct oxylipin profiles. Healthy controls exhibited a well-defined group of oxylipins that were closely associated with each other, comprising mostly AA-derived and DHA-derived oxylipins, the latter being in a central location. Overall, a relatively clear grouping pattern by precursors could be distinguished.
Figure 1.
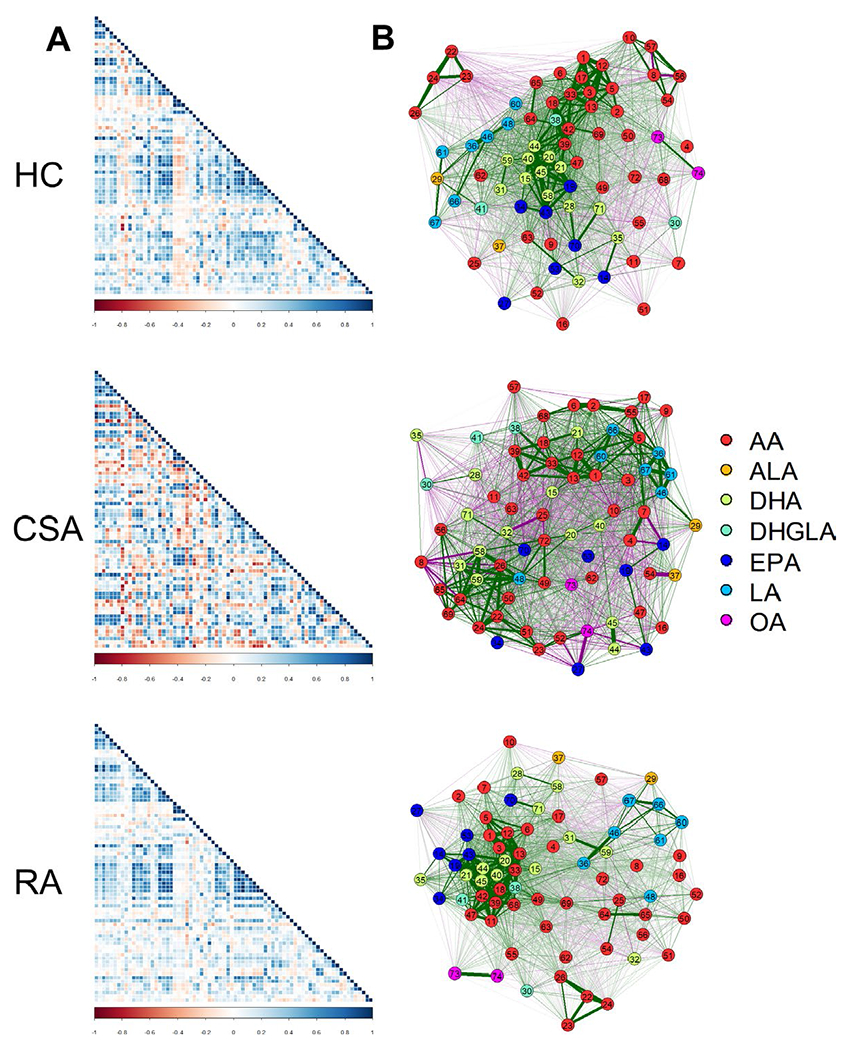
Analyses of correlations among oxylipins. A, Correlation matrices among oxylipins were plotted in correlograms by group (healthy controls [HC], individuals with clinically suspect arthralgia [CSA], and patients with rheumatoid arthritis [RA]). Tile colors represent the proportional strength of the correlation between each pair of oxylipins. B, Network analyses based on oxylipin concentrations were performed across study groups. Each node corresponds to a single oxylipin (numbered 1–74). Node colors represent each precursor, including arachidonic acid (AA), docosahexanoic acid (DHA), eicosapentanoic acid (EPA), linoleic acid (LA), dihomo-γ-linolenic acid (DHGLA), α-linolenic acid (ALA), and oleic acid (OA). Lines between nodes illustrate the strength (width) and type (positive [green] versus negative [red]) of the correlations between each pair of oxylipins. The relative position of the nodes parallels its correlation: nodes more closely correlated are located closer to each other. 1 = thromboxane B2; 2 = prostaglandin F2±; 3 = prostaglandin E2; 4 = prostaglandin D2; 5 = thromboxane B1; 6 = prostaglandin E1; 7 = thromboxane B3; 8 = prostaglandin E3; 9 = 20-hydroxy-prostaglandin E2; 10 = 13, 14-dihydro-15-keto-prostaglandin E2; 11 = tetranor 12-hydroxyeicosatetraenoic acid; 12 = 12-hydroxy-5,8,10-heptadecatrienoic acid; 13 = 11-hydroxyeicosatetraenoic acid; 14 = 11-hydroxyeicosapentaenoic acid; 15 = 13-hydroxy-docosahexaenoic acid; 16 = prostaglandin B2; 17 = prostaglandin J2; 18 = 9-hydroxyeicosatetraenoic acid; 19 = 9-hydroxyeicosa-pentaenoic acid; 20 = 16-hydroxy-docosahexaenoic acid; 21 = 20-hydroxy-docosahexaenoic acid; 22 = leukotriene B1; 23 = 20-OH-leukotriene B4; 24 = 12-oxo-leukotriene B4; 25 = leukotriene E4; 26 = 5-hydroxyeicosatetraenoic acid; 27 = 5- hydroxyeicosa-pentaenoic acid; 28 = 4-hydroxyoctadeca-4,7,10,12,16-pentaenoic acid; 29 = 9-hydroxy-10,12,15-octadecatrienoic acid; 30 = 5-hydroxyicosatrienoic acid; 31 = 7,17-dihydroxy-5,8,10,13,15,19-docosahexaenoic acid/resolvin D5; 32 = 10,17-dihydroxydocosahexaenoic acid/neuroprotectin D1; 33 = 15-hydroxyeicosatetraenoic acid; 34 = 15-hydroxy-5,8,11,13,17-eicosapentaenoicacid; 35 = 17-hydroxy-docosahexaenoic acid; 36 = 13-hydroxy-9,11-octadecadienoic acid; 37 = 13-hydroxy-10,12,15-octadecatrienoic acid; 38 = 15-hydroxyicosatrienoic acid; 39 = 8-hydroxyeicosatetraenoic acid; 40 = 10-hydroxy-docosahexaenoic acid; 41 = 8-hydroxyicosatrienoic acid; 42 = 12-hydroxyeicosatetraenoic acid; 43 = 12-hydroxyeicosa-pentaenoic acid; 44 = 14-hydroxy-docosahexaenoic acid; 45 = 11-hydroxy-docosahexaenoic acid; 46 = 9-hydroxy-9,11-octadecadienoic acid; 47 = 12-oxoeicosa-5,8,10,14-tetraenoic acid; 48 = 9-oxooctadeca-10,12-dienoic acid; 49 = 20-hydroxyeicosatetraenoic acid; 50 = 18-hydroxyeicosatetraenoic acid; 51 = 17-hydroxyeicosatetraenoic acid; 52 = 16-hydroxyeicosatetraenoic acid; 53 = 18-hydroxyeicosa-pentaenoic acid; 54 = 5,6-epoxyeicosa-8,11,14-trienoic acid; 55 = 8,9-epoxyeicosa-8,11,14-trienoic acid; 56 = 11,12-epoxyeicosa-8,11,14-trienoic acid; 57 = 14,15-epoxyeicosa-8,11,14-trienoic acid; 58 = 16(17)-epoxy-4,7,10,13,19-docosapentaenoic acid; 59 = 19,20-dihydroxy-4,7,10,13,16-docosapentaenoic acid; 60 = 9,10-epoxyoctadec-12-enoic acid; 61 = 12,13-epoxyoctadec-12-enoic acid; 62 = 5,6-dihydroxyicosatrienoic acid; 63 = 8,9-dihydroxyicosatrienoic acid; 64 = 11,12-dihydroxyicosatrienoic acid; 65 = 14,15-dihydroxyicosatrienoic acid; 66 = 9,10-dihydroxy-12-octadecenoic acid; 67 = 12,13-dihydroxy-12-octadecenoic acid; 68 = arachidonic acid; 69 = adrenic acid; 70 = eicosapentanoic acid; 71 = docosahexanoic acid; 72 = 20-carboxy-arachidonic acid; 73 = 9-nitrooleate; 74 = 10-nitrooleate.
In patients with RA, a smaller group of oxylipins could be observed. These oxylipins were strongly correlated with each other, were located in an eccentric location, and included EPA-, DHA-, and AA-derived species, although a more diverse grouping was noted. Moreover, some nodes served as links between this group and smaller groups within the rest of the network.
Finally, the oxylipin network in individuals with CSA exhibited a fuzzy pattern, with less clear groupings, a more heterogeneous distribution of nodes, and a higher number of connections among them. Values for each centrality measure (degree, expected influence, betweenness, and closeness) supported these findings, since higher degree and closeness was observed in individuals with CSA, whereas specific compounds exhibited higher betweenness in patients with RA (see Supplementary Figure 3, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41537/abstract). These results confirm that quantitative and qualitative differences in oxylipin levels were present during the earliest phases of RA, even at the preclinical stage of arthralgia.
Identification of clinically relevant clusters by oxylipin profiling.
Multivariate approaches were conducted to capture the global picture of oxylipin disturbances. A PLS-DA with all identified metabolites (12.1% of the total variance explained, R2 = 0.461, 71.0% cross-validation accuracy, and empirical permutation P = 5 × 10−4) achieved a partial discrimination among the groups (Figure 2A), although a certain overlap existed. Interestingly, the PLS-DA findings in the CSA group revealed an intermediate oxylipin profile, falling between the profiles of the healthy control and RA groups. These findings indicate that oxylipin profiles may not be useful for accurate prediction of group classification, but do suggest that oxylip-in-based group similarities may exist.
Figure 2.
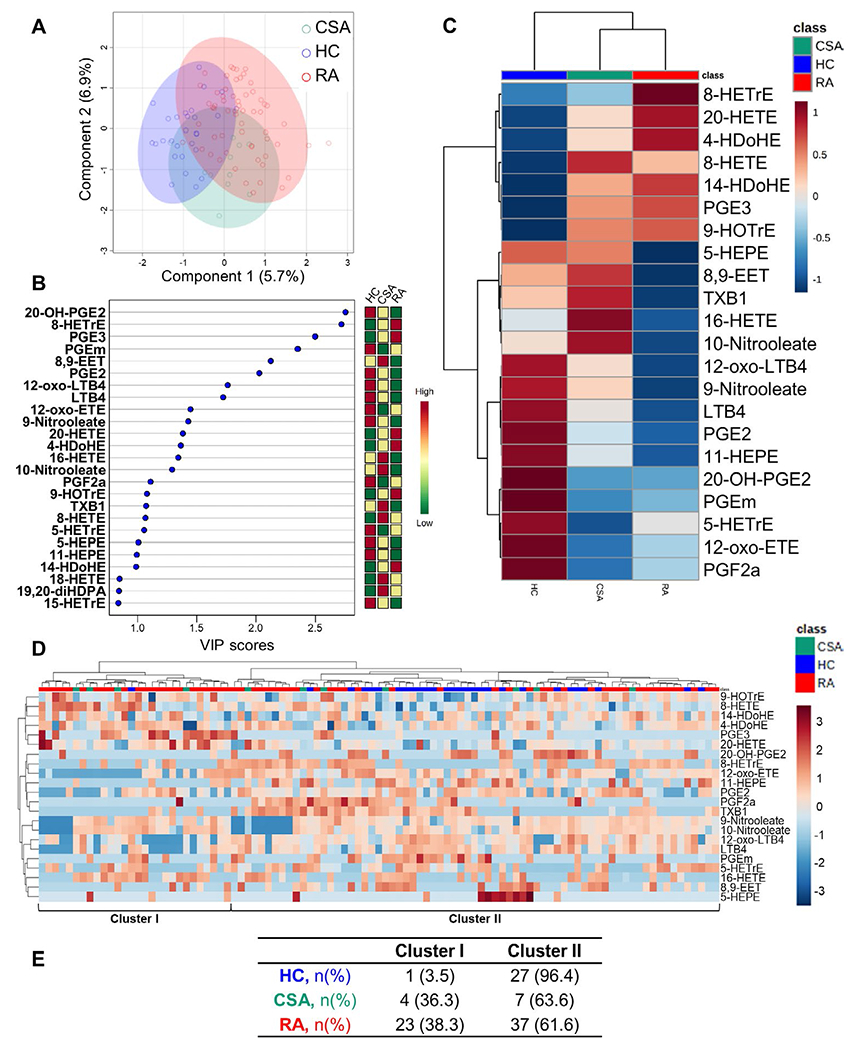
Oxylipin profiling across study groups. A, Partial least-squares discriminant analysis (PLS-DA) was used to assess the discriminant capacity of all identified oxylipins, based on the amount of variance explained by the first 2 components. B, The top 25 oxylipins were ranked based on variable important projection (VIP) scores from the PLS-DA model (left). The heatmap indicates the concentration ranks across the different groups (right). C, A group-averaged heatmap was constructed based on the 22 oxylipins with a VIP score >1. D, A heatmap based on the 22 oxylipins with a VIP score >1 was used to identify 2 oxylipin clusters according to levels (ranging from high [shades of red] to low [shades of blue]). In C and D, the upper key indicates group classes. E, The number (%) of individuals in each oxylipin cluster is shown by study group. See Figure 1 for other definitions.
A total of 22 oxylipins had a VIP score of >1 (Figure 2B). This finding was used for heatmap visualization and cluster analysis (including all study subjects). A group-averaged heatmap (Figure 2C) confirmed the previous global differences in oxylipin levels, with an intermediate, “transitional” profile observed in individuals with CSA, although some CSA-specific disturbances were also noted. Interestingly, the RA and CSA groups showed a close similarity of profiles.
Results of the cluster analysis (Figure 2D) allowed the identification of 2 oxylipin clusters (cluster I and cluster II). Cluster usage differed among the groups (P = 0.003) (Figure 2E), thus confirming the differences observed in network analyses and the partial overlap observed in the PLS-DA model. These findings point to a potential clinical relevance of the oxylipin profiles.
Whereas the healthy controls mostly had groupings in cluster I, individuals with CSA and patients with RAwere observed to have groupings in both cluster I and cluster II. Therefore, we analyzed whether the different oxylipin clusters were related to specific clinical features in RA. Patients with RA exhibiting cluster I had higher scores on visual analog scales (VAS) for patient global assessment (P = 0.016) and patient assessment of pain (P = 0.003) as compared to the scores from their cluster II counterparts, whereas no between-cluster differences in other RA features were noted (Table 1).
Table 1.
Clinical features of the rheumatoid arthritis patients at the time of recruitment, by oxylipin cluster*
| Cluster I (n = 23) |
Cluster II (n = 37) |
|
|---|---|---|
| Clinical features | ||
| Duration of symptoms, weeks | 24.00 (11.50–37.00) | 20.00 (8.50–30.00) |
| Morning stiffness, minutes | 60.00 (15.00–90.00) | 30.00 (12.50–120.00) |
| Tender joint count (of 28 joints) | 9.00 (6.00–14.00) | 8.00 (4.50–13.00) |
| Swollen joint count (of 28 joints) | 6.00 (3.00–10.00) | 5.00 (3.00–8.50) |
| ESR, mm/hour | 19.00 (11.00–37.00) | 20.00 (7.50–34.00) |
| CRP, mg/dl | 0.80 (0.30–3.20) | 0.60 (0.20–1.65) |
| Patient global assessment (0–100 VAS) | 70.00 (60.00–90.00) | 50.00 (40.00–72.50)† |
| Patient pain assessment (0–10 VAS) | 8.00 (7.00–8.00) | 6.00 (5.00–8.00)‡ |
| DAS28 (scale 0–10) | 5.66 (4.68–6.45) | 5.05 (3.86–6.07) |
| SDAI (scale 0–86) | 29.60 (23.12–39.37) | 26.30 (18.15–35.05) |
| HAQ (scale 0–3) | 1.50 (1.66–0.65) | 1.10 (0.60–1.65) |
| Fatigue (0–10 VAS) | 5.00 (0.00–8.00) | 6.00 (1.50–8.00) |
| RF+, no. (%) | 16 (69.5) | 21 (56.7) |
| ACPA+, no. (%) | 15 (65.2) | 20 (54.0) |
| RF−/ACPA−, no. (%) | 5 (21.7) | 14 (37.8) |
| Traditional CV risk factors, no. (%) | ||
| Hypertension | 9 (39.1) | 12 (32.4) |
| Diabetes | 3 (13.0) | 4 (10.8) |
| Dyslipidemia | 8 (34.7) | 11 (29.7) |
| Smoking | 6 (26.0) | 17 (45.9) |
| Treatments, no. (%) | ||
| None§ | 18 (78.2) | 32 (86.4) |
| Glucocorticoids | 4 (17.3) | 4 (10.8) |
| Methotrexate | 2 (8.6) | 3 (8.1) |
Except where indicated otherwise, values are the median (interquartile range). Differences were assessed by Mann-Whitney U or chi-square test (or Fisher’s exact test, as appropriate), according to the distribution of the variables; other than those indicated by footnotes, all P values between cluster I and cluster II were nonsignificant at P < 0.05. ESR = erythrocyte sedimentation rate; CRP = C-reactive protein; VAS = visual analog scale; DAS28 = Disease Activity Score in 28 joints; SDAI = Simplified Disease Activity Index; HAQ = Health Assessment Questionnaire; RF+ = rheumatoid factor-positive; ACPA+ = anti–citrullinated protein antibody–positive; CV = cardiovascular.
P = 0.016 versus cluster I.
P = 0.003 versus cluster I.
Among the patients with early rheumatoid arthritis, 50 were recruited before being exposed to any treatment (designated treatment-naive).
The clinical response to csDMARD treatment (low-dose glucocorticoids and methotrexate) was compared between clusters among the RA patients who were initially treatment-naive. Interestingly, at 6 months following csDMARD treatment, cluster I patients were less likely to achieve a EULAR good response compared to their cluster II counterparts (5 [31.2%] of 16 patients in cluster I versus 21 [70.0%] of 30 patients in cluster II classified as responders; P = 0012). Equivalent results were observed when the response at 6 months was assessed as achievement of DAS28 remission (4 of 16 patients in cluster I versus 20 of 30 patients in cluster II; P = 0.007) and SDAI remission (3 of 13 patients in cluster I versus 14 of 29 patients in cluster II; P = 0.05). Furthermore, differences in response between the 2 clusters were also seen at 12 months, according to the frequency of a EULAR good response (4 of 13 patients in cluster I versus 17 of 27 patients in cluster II; P = 0.056) and achievement of remission based on the DAS28 criteria (4 of 13 patients in cluster I versus 17 of 27 patients in cluster II; P = 0.056). Two patients in cluster I and 3 patients in cluster II were switched to a different csDMARD at 12 months. No patients were switched to a biologic DMARD.
All of these results suggest that oxylipin profiling can delineate disease clusters that could be differentiated across conditions, with CSA showing an intermediate profile. Moreover, these profiles are associated with clinical features and early treatment response in RA. Thus, our findings support the role of oxylipins in shaping the early RA clinical phenotype.
Identification of pathways with clinical relevance for arthritis using oxylipin signatures.
We next aimed to identify whether oxylipins could delineate metabolic pathways related to disease progression or clinical heterogeneity at the onset of RA. First, an OPLS-DA method was carried out to evaluate whether oxylipins can discriminate between healthy controls and individuals with CSA (Figure 3A) (permutation empiric Q2 P = 0.014). Since a discrimination was achieved, a correlation analysis against the prespecified transition pattern of healthy control→CSA was performed to identify those oxylipins showing a linear increase in absolute levels in individuals with CSA relative to healthy controls (Figure 3B). A group of 8 oxylipins showing this pattern was identified, deriving from AA (4 oxylipins), LA (2 oxylipins), DHA (1 oxylipin), and DHGL4 (1 oxylipin) precursors, and from the LOX (4 oxylipins), CYP (3 oxylipins), and COX (1 oxylipin) pathways (see Supplementary Table 4, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41537/abstract).
Figure 3.
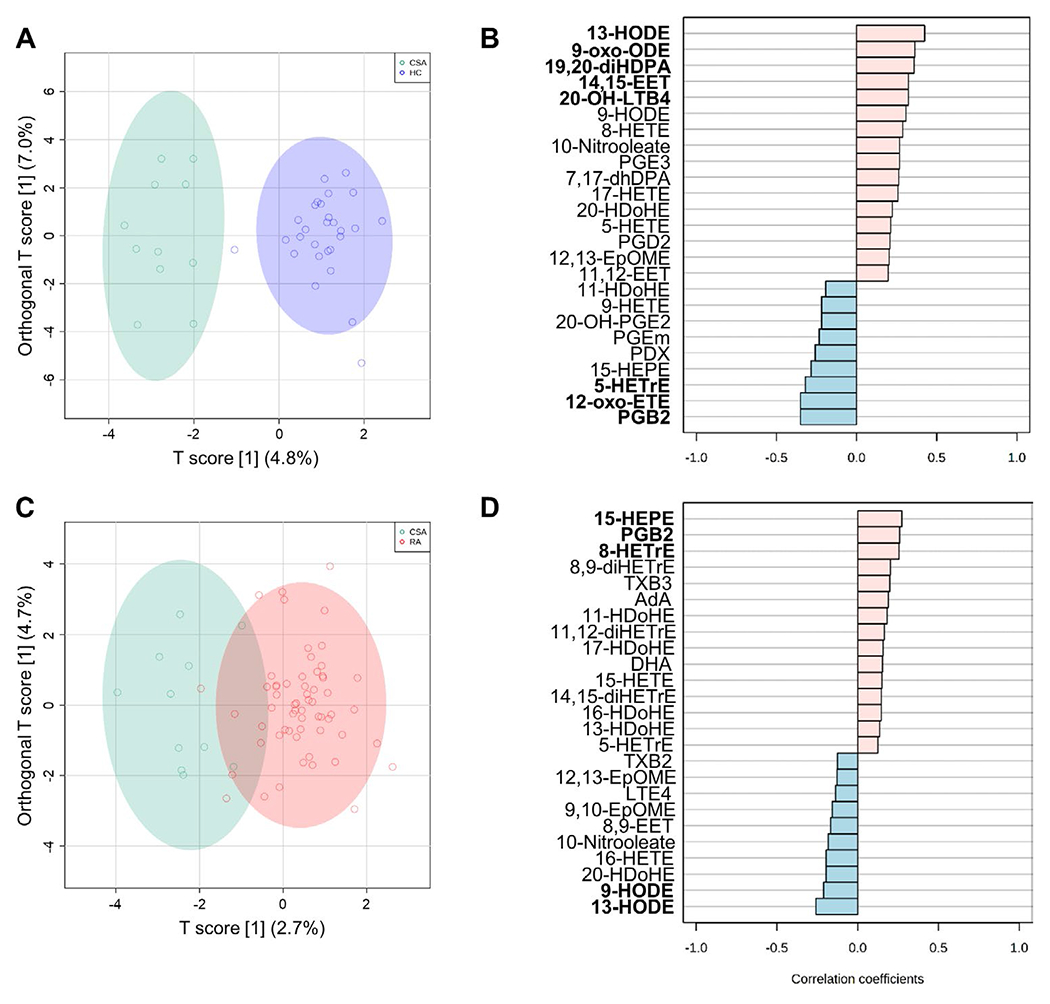
Oxylipin signatures associated with the earliest stages of RA. A and C, Orthagonal partial least-squares discriminant analysis models were constructed to evaluate the discriminant capacity of all oxylipins identified, showing comparisons between the healthy control and CSA groups (A) or the CSA and RA groups (C). B and D, Correlation patterns were determined for the top 25 oxylipins exhibiting the transition patterns healthy controlÂCSA (B) or CSAÂRA (D), based on Spearman’s rank correlation test as a distance measure. Oxylipins with statistically significant correlations are highlighted in boldface type. See Figure 1 for definitions.
A similar discriminant analysis for the pattern of CSA→RA (permutation empiric Q2 P = 0.054) was carried out, resulting in identification of 5 species of significance in discriminating patients with early RA, derived from LA (2 oxylipins), EPA (1 oxylipin), AA (1 oxylipin), and DHGLA (1 oxylipin), with almost all (4 of the 5) originating from the LOX pathway (Figures 3C and D) (see also Supplementary Table 5, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41537/abstract).
Furthermore, pathway enrichment analyses confirmed a higher impact of AA metabolism for the healthy control versus CSA comparison, whereas LA metabolism was ranked as the pathway with the highest impact for the CSA versus RA comparison (see Supplementary Tables 6 and 7, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41537/abstract). Interestingly, analysis of the healthy control→CSA→RA pattern identified a group of 18 species, mostly deriving from AA (12 species) and from the COX pathway (8 species) (see Supplementary Figures 4A and B, available on the Arthritis 6 Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41537/abstract).
In addition, we evaluated whether oxylipin expression patterns can reveal differences between the seropositive RA (rheumatoid factor [RF]–positive/anti–citrullinated protein antibody [ACPA]–positive) subset and the seronegative RA (RF-negative/ACPA-negative) subset. An OPLS-DA of the oxylipin profiles revealed a good discrimination between healthy controls and patients with seronegative RA (permutation empiric Q2 P = 0.05) (Figure 4A). Correlation analyses identified a group of 7 oxylipins that were differentially expressed (Figure 4B), mostly deriving from AA (3 oxylipins) and OA (2 oxylipins), with the COX pathway (3 oxylipins) and nitration pathway (2 oxylipins) being the most important (see Supplementary Table 8, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41537/abstract). An OPLS-DA of the oxylipin profiles also revealed that healthy controls could be discriminated from patients with seropositive RA (permutation Q2 empiric P = 0.054), although a partial overlap was noted (Figure 4C). In this case, a higher number of oxylipins (13 oxylipins) was found to be significant in the correlation analysis (Figure 4D), with AA (9 oxylipins) and 5-LOX (6 oxylipins) being the most common precursors and pathways retrieved, respectively (see Supplementary Table 9, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41537/abstract). Accordingly, pathway enrichment analyses revealed a higher relevance of the AA metabolism pathway in the seropositive RA subset than in the seronegative RA subset (see Supplementary Tables 10 and 11, available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41537/abstract).
Figure 4.
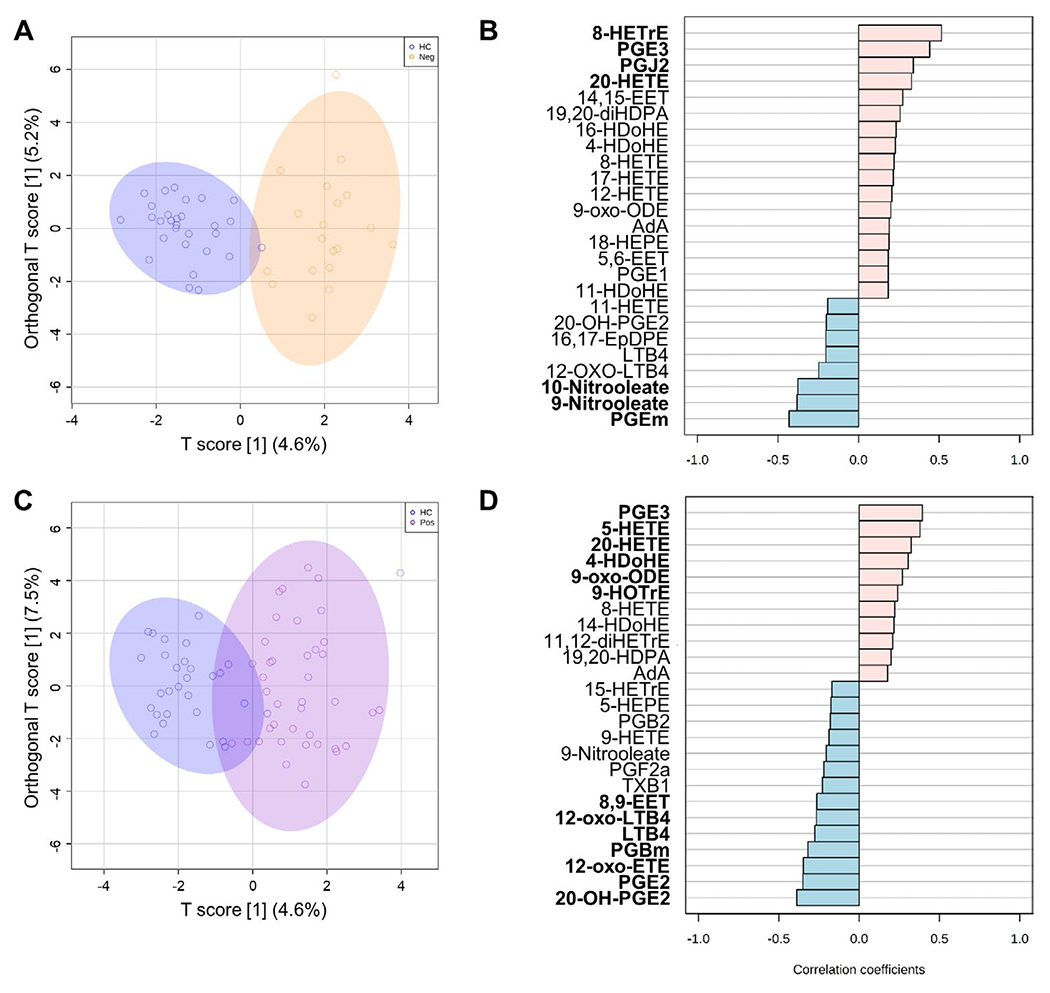
Oxylipin signatures associated with seropositivity in early RA. A and C, Orthagonal partial least-squares discriminant analysis models were constructed to evaluate the discriminant capacity of all oxylipins identified, showing comparisons between the healthy control and seronegative RA (rheumatoid factor-negative/anti-citrullinated protein antibody-negative) groups (A) or the healthy control and seropositive RA groups (C. B and D, Correlation patterns were determined for the top 25 oxylipins exhibiting the transition patterns healthy control→eronegative RA (B) or healthy control→seropositive RA (D), based on Spearman’s rank correlation test as a distance measure. Oxylipins with statistically significant correlations are highlighted in boldface type. See Figure 1 for definitions.
Thus, oxylipin profiling may help define the relevant pathways related to disease heterogeneity in patients with RA. We found that different species were related to the different disease stages underlying the early phase of RA. Similarly, the oxylipin profiles revealed divergent pathways between seronegative and seropositive RA.
DISCUSSION
A growing body of studies supports the use of lipidomics to gain a greater understanding of complex diseases and unveil pathogenic circuits linked to disease progression and new targets. The results herein support, for the first time, the occurrence of alterations in PUFA-derived oxylipins during the earliest phases of arthritis. Oxylipin profiling can identify subsets of patients with different clinical features and treatment responses, and may elucidate specific metabolic pathways that are differentially expressed between disease subsets. Based on our findings, oxylipin profiling could be considered an attractive source of biomarkers and potential targets for the early phase of inflammatory arthritis.
A remarkable finding from our study was the noticeable disturbance in oxylipin networks across the groups, which delineated distinct global patterns among the healthy controls, individuals with CSA, and patients with RA. Importantly, these patterns could not be attributed to class-specific (ω-3– or ω-6–derived) general impairments, but rather, might be attributed to distinct patterns for each individual compound. This notion has already been previously documented by our group (14) and by others (26) at the level of fatty acid precursors, thus underlining the need for complex, global approaches to account for the heterogeneity of these species and for a simultaneous assessment of the main pathways involved. Equivalent results were obtained in synovial fluid (27), thereby supporting this notion. Moreover, oxylipin levels were not related to demographic features or traditional cardiovascular risk factors in any of the study groups, consistent with previously reported findings. Furthermore, our findings in the cohort of treatment-naive patients with early RA rule out the possibility of the influence of disease duration and treatment exposure, thus pointing to oxylipin networks as playing an active role in the disease, as opposed to being innocent bystanders.
The most interesting result from our study was the identification of alterations in oxylipins during the earliest stages of RA, including early RA as well as CSA. Preventive interventions at the CSA stage (28) are limited because of the poor characterization of the underlying pathogenic mechanisms. Although previous studies have identified some metabolites (29), the lipid compartment has been largely neglected. Our results support the role of oxylipins as potential factors in this setting. These findings are in line with studies showing lower ω-3–derived PUFA levels in individuals at high risk of developing RA (30), and demonstrating changes in gene expression related to lipid metabolism at this stage (31). Moreover, we have recently reported that reduced circulating DHA, EPA, and AA levels can be found in patients with RA at the time of disease onset (14), suggesting that lipid metabolism is potentially disturbed in RA. The results herein reinforce this hypothesis, and go further by identifying the actual species altered in the arthralgia stage downstream of the main PUFA precursors.
Individuals with CSA exhibited a genuine widespread oxylipin network with higher degree and betweenness. This may be the result of a transitional status in which a high number of metabolic interactions are operating, or also a consequence of group heterogeneity. CSA itself is a very heterogeneous stage of arthritis, with a number of possible clinical outcomes reported (32), from resolution to chronification. The fact that individuals with CSA were equally represented in the 2 oxylipin clusters identified and that there was overlap in the PLS-DA findings between the healthy control and RA groups support this idea. More importantly, absolute levels of oxylipins also revealed specific alterations in the CSA group that were not present in patients with RA or healthy controls. Furthermore, an important number of oxylipins exhibited decreased levels in individuals with CSA, with a certain degree of recovery observed in patients with RA, although other trajectories were also noted (see Supplementary Figure 2, [http://onlinelibrary.wiley.com/doi/10.1002/art.41537/abstract]), and therefore further research is warranted.
Correlation analyses comparing healthy controls to individuals with CSA or individuals with CSA to patients with RA led to insights into the potential changes occurring in the multistep development of RA. First, the analysis comparing healthy controls to individuals with CSA identified 8 differentially expressed species originating from 4 different precursors and major pathways, whereas the comparison of individuals with CSA to patients with RA yielded fewer differentially expressed species, mostly derived from LAs and the LOX pathway. Pathway analyses supported these differences. Overall, these findings suggest that distinct oxylipin alterations are associated with the different stages along the course of RA. A more diverse picture is evident in the earlier preclinical arthralgia stage, which is consistent with the different risk factors and mechanisms associated with the first events in the triggering of RA (33), while a more convergent effect is evident when comparing individuals with CSA to patients with RA, in which mechanisms are thought to be shared among disease subsets (34,35). Importantly, these discrepancies may be the result of the natural regulation of eicosanoid pathways, hallmarked by an initial production of proinflammatory species (mostly AA-derived) that prompt a class-switch to an antiinflammatory, homeostatic response (36,37). However, antiinflammatory and proresolving functions are not equivalent (38), and it is plausible that a stronger shift toward proresolution may be needed to control the phase of CSA transitioning to RA.
Oxylipins showing a significant difference between individuals with CSA and patients with RA may be conceived as potential therapeutic targets to prevent disease progression. Actually, the LOX pathway has been recently described to be up-regulated at the synovial level in patients with RA in comparison to patients with osteoarthritis, while other enzymatic pathways remain unchanged (27). Due to the relevance of LOX-derived species in this setting, LOX inhibition may be an attractive therapeutic candidate. In fact, zileuton-mediated LOX inhibition has already been studied in patients with RA, although no clinical efficacy was demonstrated in those with established disease (39). However, in light of our results, further research on the effect of LOX inhibition on disease progression, rather than on management of the disease, must be considered. This is supported by studies in animal models in which treatment was administered very early (40). The fact that LOX expression is persistent along the disease course, remaining unchanged by conventional treatments (41), emphasizes the need to initiate earlier intervention. Alternatively, and due to the synergistic effects among the oxylipins, dual COX/LOX inhibitors may also be considered (42).
Importantly, differences were noted between the 2-step analyses (healthy controls versus individuals with CSA and individuals with CSA versus patients with RA) and the global analyses (healthy controls versus individuals with CSA versus patients with RA). Although the latter global approach mostly supported the role of AA and LOX as a whole, a compartmentalization was noted in the 2-step process, which aligns with the different oxylipin trajectories and allows for the identification of potential targets for tailored strategies, the main goal of personalized medicine (43), thus supporting the rationale of our analyses.
Our results shed new light on the potential role of oxylipins in early RA. Decreased EPA and DHA levels, which were linked to the altered levels of their derived species, were observed at the time of disease onset, thereby strengthening our previous findings (14). Cluster analyses revealed that 2 oxylipin profiles could be distinguished among RA patients. One of the clusters was predominantly present in healthy controls, which may be a more homeostatic profile, whereas the other cluster identified a group of RA patients with more severe clinical features, including higher VAS pain scores. This is aligned with previous evidence from clinical trials assessing fish oils and omega-3 supplements, in which a protective effect on pain was demonstrated in patients with RA (44,45). Importantly, eicosanoid metabolites are known to activate nociceptive pathways (46), but the actual mediators are unknown. Moreover, this cluster was also associated with csDMARD treatment outcomes. Since early remission is an important aim in treat-to-target strategies (47,48), oxylipin networks should be further studied either for their role as biomarkers or for their actionable mechanisms, to facilitate clinical management.
Finally, oxylipin profiling led us to identify differences between seronegative and seropositive RA. Although previous metabolomics studies have shown distinct metabolomic signatures in seronegative RA patients, the exact compounds have not been elucidated (49). Our results confirm that whereas both subsets could be distinguished from controls based on oxylipin signatures, the precursors and pathways greatly differed between them. These results underscore the differences between these 2 RA subsets and add another layer of complexity by identifying oxylipin networks as potential contributors. Whereas AA metabolism clearly dominated the oxylipin signature in seropositive RA patients, less impact was observed in seronegative RA patients, as demonstrated in the correlation and pathway analyses. Importantly, OA-derived nitrooleates, which are strong antiinflammatory lipids (50), and DHA- and EPA-derived species, in addition to COX products, were associated with seronegative RA. Due to the complexity of the seronegative subset of the disease, these findings warrant further research into these pathways.
In summary, the results of this study demonstrate that serum oxylipin levels were altered during the earliest stages of RA, and specific alterations were found even at the arthralgia stage, which may reflect an altered PUFA metabolism. Oxylipin networks at the time of onset of RA were related to the clinical phenotype of the disease and can be predictive of the response to treatment. More importantly, oxylipin profiling helped to identify metabolic pathways relevant to the heterogeneity of the disease.
Our study has key strengths, such as the comprehensive recruitment of the study subjects as well as characterization of the subjects from a clinical point of view, the use of a robust targeted metabolomics platform, and the use of a robust and well-adjusted multiparametric statistical approach. Yet, this study has some limitations that must be noted, including the cross-sectional design, which did not allow for prospective follow-up of the subjects with CSA, and lack of information on other lipid species. Although our approach included cross-validation and permutation tests, an important limitation of our findings is the lack of an external validation cohort. Under these circumstances, it is unclear whether the results could be generalized beyond the patient cohort recruited.
The potential effect of diet may also be a factor that should be considered, although our previous results failed to demonstrate a significant effect of diet on PUFA levels (51,52). Moreover, the effect of the sample type (serum versus plasma) must be taken into account to ensure comparability with other studies.
Finally, pathway assignment was based on information from the existing literature. Whether a promiscuous oxylipin production by other enzymes or by nonenzymatic reactions exists in pathologic conditions cannot be totally ruled out. However, data on the involvement of nonenzymatic pathways were extracted from the broad literature, and it has not been proven that these pathways were the main mechanisms in the setting of RA; therefore, its relevance needs to be evaluated with caution. Nevertheless, the use of pathway enrichment analyses and a well-recognized genome library confers some degree of validation to our results.
Taken together, these findings should notably improve our understanding of the eicosanoid networks in very early RA and should pave the ground for future, larger, multicentric and prospective studies to address this topic. However, studies in larger external cohorts of patients are needed to ensure the generalizability of our findings. In addition, to assess underlying metabolic changes in the inflamed joint, future research should also focus on the synovial membrane, as has been the focus in studies by other groups (27), including analysis of paired serum and synovial samples from RA patients. Expanding the lipid spectra to be investigated in patients with RA and conducting a global integration of the lipid layer with the rest of the clinical and biologic data are key steps that should be implemented as part of the research agenda.
Supplementary Material
Acknowledgments
Supported in part by the European Regional Development Funds, Instituto de Salud Carlos III Fondo de Investigación Sanitaria PI16/00113, Plan de Ciencia, Tecnología e Innovación FICYT 2018–2020 del Principado de Asturias IDI/2018/000152, and by the NIH (grant R01-AR-073324 to Dr. Guma, grant P30-DK-063491 to Dr. Quehenberger, and grant 5T32 AR064194-07 to Dr. Coras). Dr. Rodríguez-Carrio’s work was supported by a Ministry of Science, Innovation, and Universities postdoctoral contract from the Juan de la Cierva program (IJCI-2017-32070) and the Instituto de Salud Carlos III Sara Borrell program (CD19/00120).
Footnotes
No potential conflicts of interest relevant to this article were reported.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Smolen JS, Aletaha D, Barton A, Burmester GR, Emery P, Firestein GS, et al. Rheumatoid arthritis [review]. Nat Rev Dis Prim 2018; 4:18001. [DOI] [PubMed] [Google Scholar]
- 2.Smolen JS, Breedveld FC, Burmester GR, Bykerk V, Dougados M, Emery P, et al. Treating rheumatoid arthritis to target: 2014 update of the recommendations of an international task force. Ann Rheum Dis 2016;75:3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raza K, Buckley CE, Salmon M, Buckley CD. Treating very early rheumatoid arthritis. Best Pract Res Clin Rheumatol 2006;20:849–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerlag DM, Raza K, van Baarsen LG, Brouwer E, Buckley CD, Burmester GR, et al. EULAR recommendations for terminology and research in individuals at risk of rheumatoid arthritis: report from the Study Group for Risk Factors for Rheumatoid Arthritis. Ann Rheum Dis 2012;71:638–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Steenbergen HW, Aletaha D, Beaart-van de Voorde LJ, Brouwer E, Codreanu C, Combe B, et al. EULAR definition of arthralgia suspicious for progression to rheumatoid arthritis. Ann Rheum Dis 2017;76:491–6. [DOI] [PubMed] [Google Scholar]
- 6.Mankia K, Emery P. Preclinical rheumatoid arthritis: progress toward prevention [review]. Arthritis Rheumatol 2016;68:779–88. [DOI] [PubMed] [Google Scholar]
- 7.Guma M, Tiziani S, Firestein GS. Metabolomics in rheumatic diseases: desperately seeking biomarkers [review]. Nat Rev Rheumatol 2016;12:269–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korotkova M, Jakobsson PJ. Persisting eicosanoid pathways in rheumatic diseases [review]. Nat Rev Rheumatol 2014;10:229–41. [DOI] [PubMed] [Google Scholar]
- 9.Bombardieri S, Cattani P, Ciabattoni G, di Munno O, Pasero G, Patrono C, et al. The synovial prostaglandin system in chronic inflammatory arthritis: differential effects of steroidal and nonsteroidal anti-inflammatory drugs. Br J Pharmacol 1981;73:893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spector AA, Kim HY. Cytochrome P450 epoxygenase pathway of polyunsaturated fatty acid metabolism. Biochim Biophys Acta 2015;1851:356–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yacoubian S, Serhan CN. New endogenous anti-inflammatory and proresolving lipid mediators: implications for rheumatic diseases. Nat Clin Pract Rheumatol 2007;3:570–9. [DOI] [PubMed] [Google Scholar]
- 12.Serhan CN, Dalli J, Colas RA, Winkler JW, Chiang N. Protectins and maresins: new pro-resolving families of mediators in acute inflammation and resolution bioactive metabolome. Biochim Biophys Acta 2015;1851:397–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buczynski MW, Dumlao DS, Dennis EA. Thematic Review Series: proteomics—an integrated omics analysis of eicosanoid biology. J Lipid Res 2009;50:1015–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodríguez-Carrio J, Alperi-López M, López P, Ballina-García FJ, Suárez A. Non-esterified fatty acids profiling in rheumatoid arthritis: associations with clinical features and Th1 response. PLoS One 2016;11:e0159573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, Bingham CO, et al. 2010 rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum 2010;62:2569–81. [DOI] [PubMed] [Google Scholar]
- 16.Van der Heijde DM, van ’t Hof MA, van Riel PL, Theunisse LM, Lubberts EW, van Leeuwen MA, et al. Judging disease activity in clinical practice in rheumatoid arthritis: first step in the development of a disease activity score. Ann Rheum Dis 1990;49:916–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smolen JS, Breedveld FC, Schiff MH, Kalden JR, Emery P, Eberl G, et al. A Simplified Disease Activity Index for rheumatoid arthritis for use in clinical practice. Rheumatology (Oxford) 2003;42:244–57. [DOI] [PubMed] [Google Scholar]
- 18.Smolen JS, Landewé R, Bijlsma J, Burmester G, Chatzidionysiou K, Dougados M, et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2016 update. Ann Rheum Dis 2017;76:960–77. [DOI] [PubMed] [Google Scholar]
- 19.Van Gestel AM, Prevoo ML, van ť Hof MA, van Rijswijk MH, van de Putte LB, van Riel PL. Development and validation of the European League Against Rheumatism response criteria for rheumatoid arthritis: comparison with the preliminary American College of Rheumatology and the World Health Organization/International League Against Rheumatism criteria. Arthritis Rheum 1996;39:34–40. [DOI] [PubMed] [Google Scholar]
- 20.Quehenberger O, Armando AM, Brown AH, Milne SB, Myers DS, Merrill AH, et al. Lipidomics reveals a remarkable diversity of lipids in human plasma. J Lipid Res 2010;51:3299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Armando AM, Quehenberger O, Yan C, Dennis EA. Comprehensive ultra-performance liquid chromatographic separation and mass spectrometric analysis of eicosanoid metabolites in human samples. J Chromatogr A 2014;1359:60–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 1995;57:289–300. [Google Scholar]
- 23.Van den Berg RA, Hoefsloot HC, Westerhuis JA, Smilde AK, van der Werf MJ. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics 2006;7:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Opsahl T, Agneessens F, Skvoretz J. Node centrality in weighted networks: generalizing degree and shortest paths. Soc Networks 2010;32:245–51. [Google Scholar]
- 25.Trygg J, Wold S. Orthogonal projections to latent structures (O-PLS). J Chemom 2002;16:119–28. [Google Scholar]
- 26.Perreault M, Roke K, Badawi A, Nielsen DE, Abdelmagid SA, El-Sohemy A, et al. Plasma levels of 14:0, 16:0, 16:1n–7, and 20:3n–6 are positively associated, but 18:0 and 18:2n–6 are inversely associated with markers of inflammation in young healthy adults. Lipids 2014;49:255–63. [DOI] [PubMed] [Google Scholar]
- 27.Jónasdóttir HS, Brouwers H, Kwekkeboom JC, van der Linden HM, Huizinga T, Kloppenburg M, et al. Targeted lipidomics reveals activation of resolution pathways in knee osteoarthritis in humans. Osteoarthritis Cartilage 2017;25:1150–60. [DOI] [PubMed] [Google Scholar]
- 28.Gerlag DM, Norris JM, Tak PP. Towards prevention of autoantibody-positive rheumatoid arthritis: from lifestyle modification to preventive treatment. Rheumatology (Oxford) 2016;55:607–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Young SP, Kapoor SR, Viant MR, Byrne JJ, Filer A, Buckley CD, et al. The impact of inflammation on metabolomic profiles in patients with arthritis. Arthritis Rheum 2013;65:2015–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gan RW, Young KA, Zerbe GO, Demoruelle MK, Weisman MH, Buckner JH, et al. Lower ω-3 fatty acids are associated with the presence of anti-cyclic citrullinated peptide autoantibodies in a population at risk for future rheumatoid arthritis: a nested case-control study. Rheumatology (Oxford) 2016;55:367–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Baarsen L, de Hair M, Semmelink J, Choi I, Gerlag D, Tak P. A7.7 Synovial tissue profiling in autoantibody positive individuals without arthritis reveals gene signatures associated with subsequent development of rheumatoid arthritis. Ann Rheum Dis 2015;74:A77. [Google Scholar]
- 32.Boeters DM, Raza K, van der Helm-van Mil AH. Which patients presenting with arthralgia eventually develop rheumatoid arthritis? The current state of the art. RMD Open 2017;3:e000479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Catrina AI, Joshua V, Klareskog L, Malmström V. Mechanisms involved in triggering rheumatoid arthritis. Immunol Rev 2016;269:162–74. [DOI] [PubMed] [Google Scholar]
- 34.Catrina AI, Svensson CI, Malmström V, Schett G, Klareskog L. Mechanisms leading from systemic autoimmunity to joint-specific disease in rheumatoid arthritis [review]. Nat Rev Rheumatol 2017;13:79–86. [DOI] [PubMed] [Google Scholar]
- 35.McInnes IB, Schett G. The pathogenesis of rheumatoid arthritis. N Engl J Med 2011. ;365:2205–19. [DOI] [PubMed] [Google Scholar]
- 36.Serhan CN, Clish CB, Brannon J, Colgan SP, Chiang N, Gronert K. Novel functional sets of lipid-derived mediators with antiinflammatory actions generated from ω-3 fatty acids via cyclooxygenase 2-nonsteroidal antiinflammatory drugs and transcellular processing. J Exp Med 2000;192:1197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serhan CN, Savill J. Resolution of inflammation: the beginning programs the end. Nat Immunol 2005;6:1191–7. [DOI] [PubMed] [Google Scholar]
- 38.Serhan CN, Brain SD, Buckley CD, Gilroy DW, Haslett C, O’Neill L4J, et al. Resolution of inflammation: state of the art, definitions and terms. FASEB J 2007;21:325–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weinblatt ME, Kremer JM, Coblyn JS, Helfgott S, Maier AL, Petrillo G, et al. Zileuton, a 5-lipoxygenase inhibitor in rheumatoid arthritis. J Rheumatol 1992;19:1537–41. [PubMed] [Google Scholar]
- 40.Yang W, Wang X, Xu L, Li H, Wang R. LOX inhibitor HOEC interfered arachidonic acid metabolic flux in collagen-induced arthritis rats. Am J Transl Res 2018;10:2542–54. [PMC free article] [PubMed] [Google Scholar]
- 41.Gheorghe KR, Korotkova M, Catrina AI, Backman L, af Klint E, Claesson HE, et al. Expression of 5-lipoxygenase and 15-lipoxygenase in rheumatoid arthritis synovium and effects of intraarticular glucocorticoids. Arthritis Res Ther 2009;11:R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martel-Pelletier J, Lajeunesse D, Reboul P, Pelletier JP. Therapeutic role of dual inhibitors of 5-LOX and COX, selective and non-selective non-steroidal anti-inflammatory drugs. Ann Rheum Dis 2003;62:501–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coras R, Narasimhan R, Guma M. Liquid biopsies to guide therapeutic decisions in rheumatoid arthritis. Transl Res 2018;201:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Senftleber N, Nielsen S, Andersen J, Bliddal H, Tarp S, Lauritzen L, et al. Marine oil supplements for arthritis pain: a systematic review and meta-analysis of randomized trials [review]. Nutrients 2017;9:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miles EA, Calder PC. Influence of marine n-3 polyunsaturated fatty acids on immune function and a systematic review of their effects on clinical outcomes in rheumatoid arthritis. Br J Nutr 2012;107:S171–84. [DOI] [PubMed] [Google Scholar]
- 46.Svensson Cl, Zattoni M, Serhan CN. Lipoxins and aspirin-triggered lipoxin inhibit inflammatory pain processing. J Exp Med 2007;204:245–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bosello S, Fedele AL, Peluso G, Gremese E, Tolusso B, Ferraccioli G. Very early rheumatoid arthritis is the major predictor of major outcomes: clinical ACR remission and radiographic non-progression. Ann Rheum Dis 2011. ;70:1292–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aletaha D, Funovits J, Breedveld FC, Sharp J, Segurado O, Smolen JS. Rheumatoid arthritis joint progression in sustained remission is determined by disease activity levels preceding the period of radiographic assessment. Arthritis Rheum 2009;60:1242–9. [DOI] [PubMed] [Google Scholar]
- 49.Souto-Carneiro M, Tóth L, Behnisch R, Urbach K, Klika KD, Carvalho RA, et al. Differences in the serum metabolome and lipidome identify potential biomarkers for seronegative rheumatoid arthritis versus psoriatic arthritis. Ann Rheum Dis 2020;79:499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cui T, Schopfer FJ, Zhang J, Chen K, Ichikawa T, Baker PR, et al. Nitrated fatty acids: endogenous anti-inflammatory signaling mediators. J Biol Chem 2006;281:35686–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodríguez-Carrio J, Salazar N, Margolles A, González S, Gueimonde M, de los Reyes-Gavilán CG, et al. Free fatty acids profiles are related to gut microbiota signatures and short-chain fatty acids. Front Immunol 2017;8:823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodríguez-Carrio J, López P, Sánchez B, González S, Gueimonde M, Margolles A, et al. Intestinal dysbiosis is associated with altered short-chain fatty acids and serum-free fatty acids in systemic lupus erythematosus. Front Immunol 2017;8:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


