Abstract
Tools for tuning transcription in mammalian cells have broad applications, from basic biological discovery to human gene therapy. While precise control over target gene transcription via dosing with small molecules (drugs) is highly sought, the design of such inducible systems that meet required performance metrics poses a great challenge in mammalian cell synthetic biology. Important characteristics include tight and tunable gene expression with low background, minimal drug toxicity, and orthogonality. Here we review small-molecule-inducible transcriptional control devices that have demonstrated success in mammalian cells and mouse models. Most of these systems employ natural or designed ligand-binding protein domains to directly or indirectly communicate with transcription machinery at a target sequence, via carefully constructed fusions. Example fusions include those to transcription activator like effectors (TALEs), DNA-targeting proteins (e.g. dCas systems) fused to transactivating domains, and recombinases. Similar to the architecture of Type I nuclear receptors, many of the systems are designed such that the transcriptional controller is excluded from the nuclease in the absence of an inducer. Techniques that use ligand-induced proteolysis and antibody-based chemically induced dimerizers are also described. Collectively, these transcriptional control devices take advantage of a variety of recently developed molecular biology tools and cell biology insights and represent both proof-of-concept (e.g. targeting reporter gene expression) and disease-targeting studies.
Keywords: CRISPRa/CRISPRi, TALEs, Hormone receptors, Bacterial transcriptional factors, CAR T cells
Introduction
Tools to regulate gene expression in mammalian cells are essential to biotechnology, with applications that range from fundamental genetic studies to therapeutics that target diseases such as diabetes, arthritis, and cancer[1, 2]. Quantifying the contribution of individual genes to the overall function and phenotype of cells has been classically accomplished in genetics by utilizing gene knockout and complementation studies[3, 4]. The study of lethality-inducing or essential genes, however, is complicated and necessitates the precise control of the timing, amount and often cell-type specific expression of these genes[5–7]. Also, since mammalian cells have interconnected networks with built-in redundancy, it is desirable to manipulate the timing and magnitude of gene-expression exogenously in a defined manner[7, 8].
Synthetic receptors, when expressed in human cells, can alter the function and fate of these cells within human beings in a controlled manner. With the successful translation of chimeric antigen receptor (CAR) T cells, it is clear that cell-based therapies have the opportunity to alter medicine[9, 10]. The success of CAR T cells for the treatment of leukemias has spurred the development of other immune cells like NK cells as living drugs for the treatment of cancer and autoimmune diseases[11–13]. As the complexity and potency of these engineered cell therapies increases, the potential for on-target off-tumor toxicity also increases. Unpredictable destruction of unintended cells/tissue leading to toxicity and even death has been observed with immune cell fusions[14]. Moreover, one of the biggest advantages of cell-based therapies, the ability of the transfused cells to proliferate within the host, is also their biggest concern. The modified T cells or even a very small number of contaminating leukemic cells can grow unchecked after transfusion[15]. It is thus desirable to control the fate of these engineered cells after human administration.
There are at least five important considerations in the selection and design of small-molecule inducible regulators that make them challenging to implement. The first and most basic consideration is the choice of whether regulation is implemented by control of transcription, control of translation or post-translational localization. The second consideration is the size of the regulator. Smaller regulators impose a smaller metabolic demand within the cells, and for protein-based regulators of non-human origin, smaller proteins are less likely to be immunogenic. Third, for human-derived regulators, the crosstalk of the regulator in altering non-targeted protein expression by regulating endogenous loci within the transferred cells is an important consideration. An associated concern is the ability of the ligand to alter protein expression at endogenous loci in non-targeted cells and tissue. Fourth, the small molecule used for regulation must be bioavailable and non-toxic with defined pharmacokinetics. Fifth, the temporal dynamics (time taken to achieve a desired on/off ratio), and the magnitude of inducibility are essential considerations for these response systems. Reversibility, the ability of the system to reset in the absence of the small molecule, can also be an essential criterion.
In this review, we describe recent reports (over approximately the last five years) on the design and implementation of circuits to control gene expression in mammalian cells.. These systems are broadly categorized as synthetic protein fusions between a ligand receptor and a DNA-targeting domain, regulators based on bacterial transcription factors, and a handful of “other” systems less easily classified. The gene regulation tools described include both those which primarily facilitate in vitro studies, and those with applications in tissue engineering or potential human therapies. While such breadth makes it difficult to provide direct comparisons between most of these systems, attention is paid to the operational characteristics listed above, with key advantages/disadvantages pointed out. Aptamer-based inducible gene expression systems are beyond the scope of this review, though interested readers may refer to other recent reviews[16–18].
Receptor-based systems
The use of a synthetic transcriptional regulator in which a fused, small-molecule-inducible receptor acts as the sensor domain in mammalian cells was first described in the 1990s, when the yeast transcription factor, Gal4, was fused to the estrogen receptor protein (Gal4-ER). Gal4-ER was shown to regulate endogenous genes in rat fibroblasts, in an estrogen-dependent manner[19]. Since then, the modular nature of human and insect hormone receptors has been exploited to induce the expression of user-defined gene targets. Popular approaches to achieving specific control over target genes involve either fusing the small-molecule ligand-binding domain (LBD) (Box 1) of a receptor to a customized DNA-binding protein or rewiring the signal relayed by an induced receptor protein to activate specific downstream targets. Below we highlight recent progress in the design and application of such exogenously controlled, ligand-inducible, mammalian gene expression systems. The core design elements comprise a DNA binding module (DBD) that determines specificity for the target loci, an activator module that dramatically alters the rate of transcription by RNA polymerase, and a sensor module that ensures that the system is ligand-responsive (LBD). Based on the mode of gene targeting (DBD), we classify the receptor-based inducible systems into the three broad categories of those involving: (a) transcription activator-like effectors (TALEs), (b) RNA-guided CRISPR/dCas9 (dead Cas9) proteins, and (c) other DNA targeting proteins.
Box 1: Useful terms.
AD - Activation domain. A protein domain that recruits RNA polymerase II holoenzyme and eukaryotic transcription factors, to initiate transcription. Examples of some commonly used ADs include VP16, VP64, VPR, and VPH (all defined below).
Aptamer - Small (20 to 60 nucleotides) single-stranded RNA or DNA oligonucleotides able to bind target molecules with high affinity and specificity. Many synthetically generated aptamers can bind various targets, ranging from simple inorganic molecules to large protein complexes, and entire cells
CID - Chemical induced dimerization. Dimerization of interacting protein partners in the presence of a small-molecule inducer. Interacting proteins may undergo homodimerization (e.g. DmrB domains homodimerize in the presence of rapamycin) or heterodimerization (e.g. DmrA and DmrC domains heterodimerize in the presence of rapamycin) in the presence of an inducer. The DmrD domain undergoes reverse dimerization by dissociating in the presence of rapamycin.
CreER - inducible Cre/lox system consisting of cyclization recombinase (Cre) enzyme fused to the synthetic estrogen receptor (ER) protein. The ER fusion sequesters Cre in the cytoplasm through the ER and heat shock protein (HSP90) interaction in the cytoplasm. Induction with 4-hydroxytamoxifen (4-OHT) induces a conformational change in the ER, releasing the ER-Cre fusion from HSP90 interaction and allowing nuclear transport of the recombinase enzyme. The CreERT2 recombinase which has three mutations in the receptor’s LBD is currently the most efficient CreER system[20].
CRISPRa - CRISPR activation. Programmable gene activation brought about by coexpression of dCas9 protein fused to an activator domain and a customizable sgRNA. The dCas9-sgRNA complex is usually directed to promoter regions, by sgRNAs, to recruit additional transcription factors and upregulate expression of the target gene.
CRISPRi - CRISPR interference. Programmable gene silencing accomplished by the coexpression of dCas9 protein fused to a silencing domain and a customizable single guide RNA (sgRNA). The dCas9-sgRNA complex binds to DNA elements complementary to the sgRNA and causes a steric block that halts transcript elongation by RNA polymerase, resulting in the repression of the target gene.
DBD - DNA binding domain. Protein domain which recognizes and binds to specific DNA target sequences.
GPCR - G protein-coupled receptor. GPCRs are cell surface proteins that mediate cellular responses to a variety of extracellular signals. GPCR signaling is often re-purposed to allow inducible control over target gene expression. Details are described in this review.
LBD - Ligand binding domain. Protein domain which recognizes or binds a specific ligand (proteins, surface antigens, hormones, and small molecules) and undergoes conformational change leading to subsequent changes in neighboring domain.
NLS - Nuclear localization signal sequence. Short peptide motifs mainly composed of basic amino acid residues that mediate the nuclear import of proteins by binding to importin receptors (karyopherins) on the nuclear membrane. Refer to[21] for further information.
RXR - Retinoid X receptor. A member of thyroid/steroid hormone nuclear receptor that gets activated by 9-cis-retinoic acid.
SAM - Synergistic activation mediator. A three-component system used for effective transcriptional activation by recruiting multiple ADs at the target site. SAM is usually used in dCas-based transcriptional regulation. Refer[22] for details.
sgRNA - Single guide RNA. Single RNA molecule containing the crispr RNA (crRNA), that is complementary to target sequence, fused to the scaffold tracr RNA.
SunTag - Signal amplification system that recruits multiple ADs to target DNA via protein-protein interactions between a polypeptide sequence and its interacting antibody. Refer[23] for more information.
TEVp - Tobacco Etch Virus protease. TEVp cleaves proteins containing the amino acid sequence ENLYFQ|S/G also called the TEVp cleavage site (TCS).
TF - Transcriptional factor. Proteins that bind to DNA regulatory sequences and modulate the rate of gene expression. In this review, we focus on metabolite sensing TFs which modulate gene expression by changing the availability of the specific metabolite inputs.
VP16 - Herpes simplex viral (HSV) protein 16. VP16 is involved in the activation of the viral immediate-early genes in HSV. It is often used as an AD in synthetic transcriptional factors.
VP64 - Strong transcriptional AD derived by fusing four copies of VP16.
VPH- the combination of VP64, p65 activation domain from transcriptional factor NF-kappa-B, and heat shock factor (HSF-1). HSF-1 activates gene expression in response to a variety of stresses, including heat shock, oxidative stress, as well as inflammation and infection in humans.
VPR - combination of VP64, p65, and Rta activator from Epstein-Barr virus
Receptor-TALE fusions for inducible gene expression
Transcription activator-like effectors (TALEs) are bacterial DNA binding proteins that offer modular DNA recognition with single-nucleotide resolution and therefore have great potential as tools for programmable gene regulation. TALE proteins consist of a modular DNA-binding domain (DBD) along with a C-terminal, endogenous nuclear localization signal (NLS), and an activation domain (AD). A typical TALE DBD consists of a series of 33- to 35-amino acid repeats. Within each repeat are two adjacent amino acid residues (usually at residues 12 and 13), termed repeat variable diresidues (RVDs), which recognize a single, specific base pair (bp) of DNA. The most abundant, naturally occurring RVDs, His-Asp, Asn-Asn, Asn-Ile, and Asn-Gly, enable specific recognition of cytosine, guanine or adenine, adenine, and thymine, respectively. Carefully assembled TALE repeats are used as custom DBDs capable of recognizing user-defined DNA sequences[24]. By replacing the naturally occurring AD with other executer domains, TALEs have been converted into artificial sequence-specific nucleases, transcription repressors or activators, methylases, and recombinases for use in mammalian cells[25].
One-component TALE-based systems consist of a single polypeptide that is a linear fusion between the DBD, activator and sensor modules. The monomeric receptor undergoes conformational changes upon binding a small-molecule, leading to exposure of the AD and subsequent transcriptional activation. Mercer et al. designed a tripartite TALE-based transcription factor (TALE-TF) by fusing the heterodimeric single-chain retinoid X-α/ecdysone (RXE) LBD to the synthetic TALE protein (Arv15, DBD) linked to four copies of the herpes simplex virus transactivation domain, VP16[26]. The RXE-LBD undergoes an intramolecular rearrangement in response to the binding of the agonist molecule ponasterone A (PonA). This rearrangement exposes the previously sequestered AD and results in transcriptional activation upon the binding of Arv15 to its cognate DNA-binding site, AvrXa7. This TALE-TF, designated RXE-Avr15-VP64, acted as a PonA inducible activator of transcription. To track the ligand-responsiveness of this system, a standard luciferase-based reporter assay was utilized. For comparison, the authors also constructed the ER-Avr15-VP64, and the homodimerization of ER mediated by its small-molecule ligand 4-hydroxytamoxifen (4-OHT) enables activation of this TALE-TF. Upon activation with PonA, the monomeric RXE-Avr15-VP64 activator increased luciferase gene expression ∼1400-fold and displayed tunable activation in response to different ligand concentrations. By comparison, ER-Avr15-VP64 activated with 4-OHT demonstrated lower fold-induction (~350-fold) and displayed a more switch-like behavior in response to increasing ligand concentrations[26]. Collectively, these results established the RXE-Avr15-VP64 as a sensitive and regulatable activator with a large dynamic range. Further, the one-component design of this system makes it simple to integrate the RXE-Avr15-VP64 transactivator within gene networks. A single polypeptide system also potentially offers faster response times as compared to dimerizing activators. It will be important to determine off-target effects induced by the RXE-TFs, as TALEs can potentially tolerate mismatches of several nucleotides[26].
Lonzaric et al. attempted to minimize background expression by circularly locking the TF protein, thus preventing its interaction with DNA. The circularly locked, ‘rapa-TALE’ is a single polypeptide chain formed by linking two copies of the rapamycin binding domain DmrD to each end of a TALE-VP16 fusion protein[27]. In the absence of rapamycin inducer, DmrD forms homodimers and circularizes the rapa-TALE TF, while the addition of rapamycin or its analogs disrupts the formation of the dimer and enables activation of gene expression. When using the rapa-TALE system in HEK cells, the addition of rapamycin led to 17-fold activation of the luciferase gene, which was brought down to background expression levels after subsequent removal of rapamycin from the cells. The monomeric rapa-TALE design represents a novel approach to reversible gene expression, but still demonstrated leaky activation of the luciferase gene in the absence of rapamycin inducer. As posited by the authors, the leaky expression of the rapa-TALE system arises probably due to incomplete or late locking, with a fraction of the protein molecules remaining in a linear conformation even without rapamycin[27].
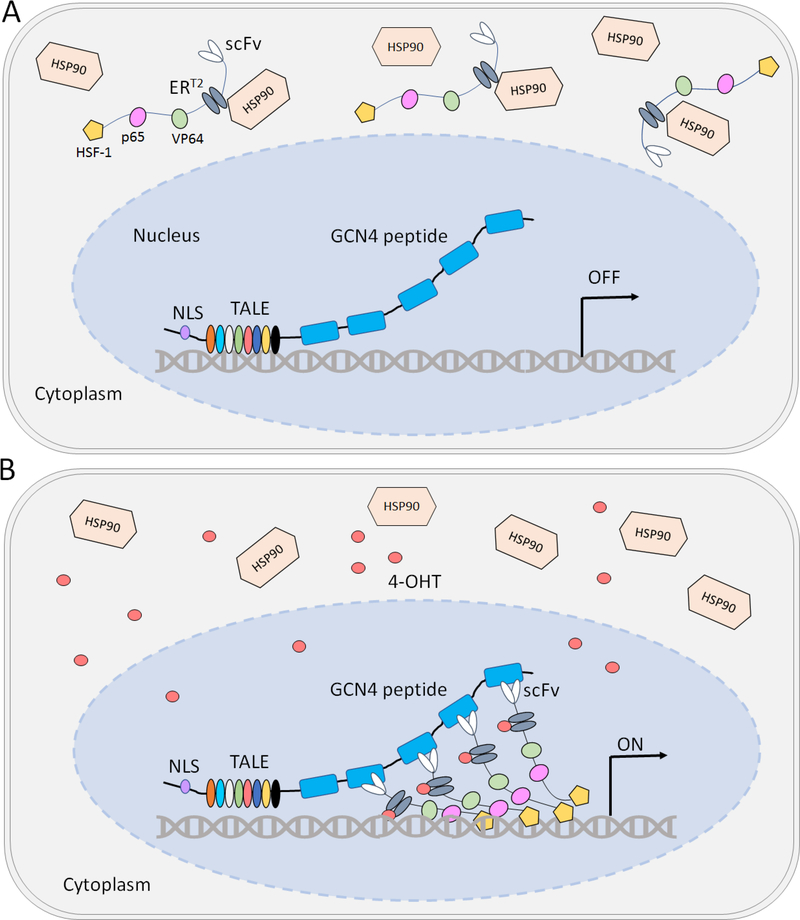
A more common approach to reduce background expression is to separate the DBD and the AD such that they are expressed as separate polypeptides that only come together in the presence of the ligand. In the case of the TALE designs, these multi-component designs improve TALE’s accessibility to the target DNA due to separation of the bulky AD and lower the background transgene expression by enabling sequestration of the AD in the absence of a small-molecule inducer. Zhao et al. designed the ‘HIT-TALE SunTag’ (short for Hybrid inducible technology) consisting of a TALE array fused to 10 tandem repeats of the GCN4 peptide (Fig. 1). The peptide sequence can interact with a single-chain variable antibody fragment (scFv) and recruit an AD fused to the scFv at the target site[28]. For inducible control, the scFv is further fused to the estrogen receptor (scFv-ER-AD) which dimerizes and mediates nuclear transport upon binding 4-OHT. When compared with the direct ER-TALE-VPH fusion, the HIT-TALE-SunTag system brought about a 275% increase in mRNA expression of the target gene in HEK293T cells with background expression reduced to undetectable levels[28]. The HIT-TALE SunTag system thus enabled tight, tunable, and reversible gene activation with notably no background signal in the absence of 4-OHT. The system can also be applied for other applications including transcriptional repression, and epigenetic modulation.
Fig. 1. Inducible gene expression using the HIT-TALE-SunTag system.
A. In the absence of the inducer (4-OHT), the fusion protein containing estrogen receptor (ERT2) and the activator domains (VP64, p65, and HSF-1) remain sequestered in the cytoplasm by interactions with the HSP90 chaperone. The nuclear-tagged TALE-GCN4 complex is localized at the target locus but without the activator domains, TALE-GCN4 complex cannot turn ON transcription of the target gene. B. Upon addition of 4-OHT, the ERT2 fusion complex dissociates from HSP90 and translocates in the nucleus where the single-chain antibody (scFv) interacts with the GCN4 peptide and recruits the multiple activator domains to the target site to activate gene transcription. The DNA binding TALE array determines the target site. The same design can be adapted to dCas9-sgRNA based systems wherein DNA-targeting specificity is determined by the sgRNA.
As stated above, the modular nature of TALE scaffolds and the ability to assemble custom recognition motifs with single-nucleotide resolution implies that the TALE-based TFs can be used to target endogenous genomic sites in any species of interest. However, the extensive protein engineering required to construct a TALE and the requirement of co-delivery of multiple engineered factors to achieve robust transcriptional activation has limited TALEs application in large-scale genomic perturbation[25].
Receptor-dCas fusions for inducible gene expression
CRISPR–Cas systems have been repurposed for programmable and targeted gene regulation by using “nuclease dead” Cas (dCas or dead Cas) proteins that are constructed by mutating the active site residue(s), in Cas9 protein, to alanine (D10A and H840A in Cas9 from Streptococcus pyogenes) and thus abolishing Cas9 nuclease activity[29, 30]. dCas tools have advanced from simple fusions with single activators or repressors (CRISPRi/a tools, or first generation dCas9 tools), to recruiters of multiple copies of activators for efficient gene activation (signal amplifiers, or second generation dCas9 tools)[31]. More recently, ligand-inducible controllers of gene expression have been developed (inducible CRISPRi/a, or third generation dCas9 tools)[31]. Inducible CRISPRi/a can be achieved by regulating the expression of sgRNA or dCas proteins[32, 33] or by regulating the availability of the dCas proteins for gene targeting. Though regulating the expression of sgRNA or dCas protein has enabled efficient CRISPR activity with minimal background[32], such a regulatory system often has slower dynamics than regulating dCas availability, due to the rate-limiting transcription, translation, and protein folding steps[32]. In this section, we discuss inducible gene switches where the DNA targeting activity of dCas variants is regulated by the availability of an inducer.
Similar to the TALE designs, dual polypeptide, inducible CRISPRi/a can be achieved by separately fusing the dCas protein and the AD to single chains of a homodimeric or heterodimeric nuclear receptors such that the receptor undergoes dimerization only in presence of a small-molecule inducer. Gao et al. designed an inducible dCas9-based transcriptional regulation system in which the S. pyogenes (Sp) dCas9 and an activator (VPR activator) or repressor (KRAB repressor) were fused to complementary pairs of six previously reported chemical-and light-inducible heterodimerization domains and measured the activation and repression efficiencies of each inducible dCas9 system using a GFP-based fluorescent reporter in HEK293T cells[34]. The abscisic acid (ABA)-inducible (ABI–PYL1[35]) and the gibberellin (GA)-inducible (GID1–GAI[36]) systems were noted as the most potent systems which activated protein expression 165- and 94-fold respectively in presence of their inducers, with no background activation in the absence of the inducer.
Chen et al. used a similar two-component design that relied on the heterodimerization of the PYL1 and ABI domains in the presence of ABA. In place of a VPR activator, Chen et al. utilized P300 histone acetyltransferase (HAT) domain. In the presence of ABA, the dCas9-P300 HAT protein complex was used for engineering the epigenome of HEK293T cells at the Interleukin 1 Receptor Antagonist (IL1RN) gene locus. The IL1RN gene has a natural low expression level in HEK293T cells[37]. In presence of ABA inducer and IL1RN specific sgRNA, HEK cells expressing the ABA-inducible dCas9-P300 HAT protein complex demonstrated a 30-fold increase in the IL1RN mRNA level while cells not induced with ABA did not show this increase in IL1RN mRNA. Chen et al., further designed sgRNA targeting other genes in HEK293T cells, thereby demonstrating the modularity of their ABA-inducible dCas9-P300 HAT system[38]. Notably, the ABA-inducible and GA-inducible systems activated protein expression with dCas9 fused to ABI or GID1 protein domains (respectively), but not with dCas9 fused to the PYL1 or GAI domains, suggesting the spatial orientation of the fused protein can interfere with chemical-induced dimerization[38].
To reduce background expression and increase the efficiency of CRISPRa by recruiting multiple ADs to the target site, Lu et al. designed the ‘HIT-dCas9-SunTag’ system. As with the HIT-TALE-SunTag system (Figure 1), key features of the HIT-dCas9-SunTag system are nuclear transport of the AD component only upon addition of the inducer, and inducible recruitment of multiple ADs to the target gene to bring about efficient gene activation[39]. Using the HIT-dCas9-SunTag system, Lu et al. reported a 600-fold increase in the expression of the luciferase reporter, while eliminating background luciferase expression in absence of the inducer, as compared to a 150-fold increase in luciferase expression when using a direct dCas9-ER-VPH fusion[39]. The authors thus demonstrated reduced background expression by using drug inducible AD recruitment in their HIT-dCas9-SunTag system. As with the TALE systems, the HIT-dCas9-SunTag system is limited by its requirement of delivering multiple engineered components to the target locus to effect gene expression.
Recent interest in the Cas12a (previously called Cpf1) CRISPR associated nuclease has led to the development of inducible CRISPR/dCas12a transcriptional activators. Employing dCas12a for gene regulation offers advantages such as the use of shorter length CRISPR RNAs (crRNAs) for guiding dCas12a to targets (no tracrRNA required), the ability to target T-rich PAMs, and ease of multiplexed regulation through the use of a single transcript coding multiple crRNAs, which are processed by dCas12a’s inherent RNase activity. Like the dCas9 based transcriptional activators described above, Tak et al. developed CRISPR/Cas12a activator by fusing the Lachnospiraceae bacterium dCas12a (Lb-dCas12a) and the p65 transactivator separately to the DmrA and DmrC interacting proteins, which dimerize in presence of rapamycin[40]. Chemical induced heterodimerization of the nuclear tagged- DmrA and DmrC domains enabled 9-fold to 40-fold simultaneous upregulation of the three endogenous genes target genes in HEK293 and human U20S cells (derived from bone tissue) in presence of constitutively expressed guide RNA[40]. The study thus paved the way for future efforts to develop regulatory domains for dCas12a as those previously designed for Sp-dCas9 proteins. Though the use of a single transcript encoding multiple crRNAs reduces the cloning effort, the authors noted that the efficiency of target gene activation is 2-fold lower compared to expressing each crRNA separately.
In another dCas12a application, Liu et al. combined the biosensing abilities of riboswitches with the DNA targeting activity of dCas12a[41]. They re-designed the crRNA to incorporate a theophylline binding aptamer at its 3’ end such that, in the absence of theophylline the guide region of the crRNA pairs with an anti-sense stem in the crRNA-riboswitch. In the presence of theophylline, a conformational change in the RNA structure exposes the guide region to enable the recruitment of the dCas12a-regulator (activator or silencer) to the target gene[41]. The dCas12a-riboswitch activator brought about theophylline dose-dependent activation of DNA (cytosine-5)-methyltransferase-1 (DNMT1) expression (involved in DNA methylation), with ~90-fold upregulation in 1 mM theophylline while the dCas12a-riboswitch silencer completely turned off the DNMT1 expression at 1 mM theophylline[41].
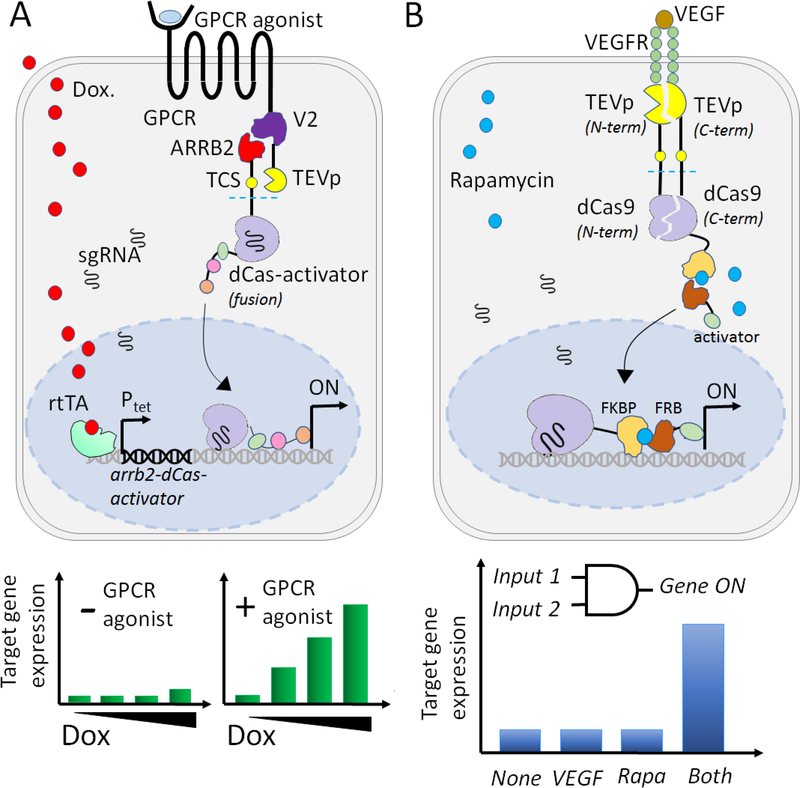
The single-input inducible gene control systems described above have simple design principles and lead to predictable response performance. However, in some cases, the single input may not be sufficient to recognize complex cellular environments, and additional layers of control may be desired[42]. To incorporate an additional layer of regulation and thus enable stringent control of target expression, dual-input inducible systems have recently been described[43, 44]. The dual-input systems are uniquely responsive to the presence of both ligands to activate/repress gene expression. Kipniss et al. utilized various ligand-sensing G-protein coupled receptors (GPCRs), interacting with the dCas9 protein (termed “ChaCha” design), allowing for modulation of gene expression in response to diverse ligands including synthetic compounds (clozapine-N-oxide (CNO), salvinorin B, and isoproterenol), hormones (vasopressin, thyrotropin-releasing hormone), mitogens (neuromedin B), chemokines (stromal-derived factor 1), and fatty acids (lysophosphatidic acid)[43]. The ChaCha design for inducible CRISPRa consists of two distinct polypeptides that work together to sense dual inputs. The first polypeptide is a tripartite fusion comprising a small-molecule inducible sensor protein (evolved human GPCRs), a dimerizer protein (V2 tail sequence from arginine vasopressin receptor 2 (AVPR2)), and an effector (TEVp) (Fig 2A). The second polypeptide, synthesized only in the presence of doxycycline, comprises of the adaptor protein, Beta-Arrestin-2 (ARRB2) fused to an activator module comprising of dCas9-ADs via a TEV protease sensitive linker (TCS). In the presence of the GPCR agonist, the V2 sequence heterodimerizes with the ARBB2 protein and this enables TEVp to cleave the TCS, leading to the proteolytic release of the dCas9-VPR transactivator. The dCas9-VPR translocates to the nucleus and subsequently activates the expression of the genes of interest. The group demonstrated that the CNO-activated ChaCha system had a lower leakiness and resulted in 3.7-fold higher GFP activation in HEK293T cells, as compared to a previously reported CRISPR Tango design[45]. Kipniss et al. also generated a toolkit of eight CRISPR ChaCha systems, by simply varying the GPCR used in the system, thereby expanding the applicability of ligand-inducible gene control by CRISPR-dCas9[43]. Designing dual-input or multi-input gene control is often a time-consuming process and is generally only sought when single-input systems cannot provide the desired level of specificity[42].
Fig. 2. Dual input transcriptional regulation.
A. Two layers of inducibility are achieved with the ChaCha system. In the first layer, doxycycline (Dox) induces the expression of ARRB2-dCas-activator fusion protein through a Tet-ON system (see text for details) while a second layer of control is achieved when a GPCR agonist leads to GPCR signal activation. Without the appropriate GPCR agonist, increasing dox concentration does not alter target gene expression. The binding of the GPCR agonist induces a conformational change in the V2 domain and allows the interaction between ARRB2 and V2. This ARRB2-V2 interaction further leads to the proteolytic release of the dCas9-activator. The nuclear-tagged dCas9-VPR localizes in the nucleus and upregulates target gene expression in the presence of constitutively expressed sgRNAs. B. Multilayered transcriptional activation is induced by VEGF and rapamycin inducers. VEGF binds to VEGFR and induces the proteolytic release of the membrane-tethered split-dCas9 fragments. Rapamycin induces heterodimerization of rapamycin-binding proteins (FKBP-FRB), leading to the recruitment of the FRB-fused activator to the target gene by dCas9-sgRNA complex. Once at the target gene, the dCas9-VP64 transactivator turns ON gene transcription.
Several recent studies describe the use of a split-dCas9 system for better control of inducible gene regulation. The initial report by Zetsche et al. utilized split-dCas9 fragments attached to two separate heterodimerizing nuclear receptors, with the intent of using inducer-mediated heterodimerization to reconstitute the split-dCas9[46]. The system brought about significant target gene activation, but with a high background activity due to sgRNA-mediated dimerization of the split fragments in the absence of inducer. To reduce background, Baeumler et al. designed a three-polypeptide dual inducer system (AND gate). The circuit comprises of tethered the N- and C-terminal split dCas9 fragments to the plasma membrane via the heterodimeric vascular endothelial growth factor receptors (VEGFR1 and VEGFR2), each carrying a complementary fragment of the split TEVp, and TCS cleavage sites (Fig. 2B). In the presence of VEGF, the receptors dimerize and bring together the split TEVp fragments. The assembled and functional TEVp cleaves the paired dCas9 fragments that self-assemble and translocate to the nucleus. When VEGF was available, the split-dCas9/VEGFR system mediated 48.5-fold target (ASCL1) gene expression, in the presence of a single constitutive ASCL1 targeting sgRNA, as compared to the no VEGF control[44]. Multilayered inducibility was added to this design by further separating the VP64 transactivator from the dCas9 fragments. In the modified design, the membrane-tethered C-terminal dCas9 fragment was fused to the FKBP interacting domain while the VP64 activator was fused to the FRB domain[47] (Fig. 2B). Rapamycin induces the heterodimerization of the FRB and FKBP domains enabling fusion of the targeting (dCas9) and the activator modules (VP64). In the presence of VEGF but the absence of rapamycin, the membrane-released split-dCas9 fragments can assemble and translocate to the nucleus but cannot activate target genes due to lack of VP64 association. This concurrent stimulation using two inducers resulted in extremely tight control over the target gene, with 95-fold induced activation relative to the no inducer control[44].
In another example of multi-drug-input control linked to gene expression, Foight et al. developed chemically induced dimerization (CID) systems based on clinically approved drugs that bind protease NS3a from hepatitis C virus (HCV)[48]. They started with de novo-designed protein scaffolds and used computational design, followed by optimization through yeast surface display, to isolate “reader” proteins, which bind to specific drug-NS3a complexes with high affinity. For example, designed reader protein DNCR2 shows 36 pM affinity toward the danoprevir:NS3 complex. Their so-called PROCISiR system (Pleiotropic Response Outputs from a Chemically-Inducible Single Receiver) allows for programing diverse cellular responses based on a single receiver protein[48]. In one application of this system for drug-induced transcription in HEK293 cells, NS3a was fused to dCas9, while DNCR2 was fused to VPR. Addition of danoprevir induced expression of the target gene, while subsequent addition of competitive binder grazoprevir reversed expression and allowed for a graded response. In another dCas9-dependent design, NS3a was instead fused to VPR, while different reader proteins were fused to RNA binding proteins specific to different RNA hairpins. Guide RNAs designed to target specific genes also included the RNA hairpins that serve as ligands for the reader protein fusions. VPR and hence gene activation was thus directed to different target genes, depending on the drug added. The responsive nature of the PROCISiR architecture holds great promise in achieving diverse modes of transcriptional control at multiple loci.
Despite the ease of multiplexed gene regulation offered by CRISPRi/a tool, a variety of limitations of the inducible CRISPR/Cas systems remain to be addressed[49]. For example, binding of multiple sgRNAs and dCas9s to the chromatin may lead to changes in the genome organization[31]. Further, the desired use of many different guide RNAs simultaneously leads to scalability issues, due to the generation of unstable constructs prone to recombination in E.coli during clonal propagation. Moreover, the use of multiple controls to regulate sgRNAs expression causes undesired increases in plasmid size, reducing the efficiency transduction and chromosomal integration. Finally, off-target Cas9-binding events have also been widely reported, resulting in non-specific genomic effects[50, 51].
In a direct comparison between TALE-based and dCas9- based designer TFs, TALE-based activators showed stronger target gene activation than dCas9-based activators (over 5000-fold activation by TALE-VPR, vs. about 1000-fold activation by dCas9-VPR activators)[52]. A possible reason for the poorer performance of the dCas9 regulator is the requirement for sgRNAs targeting multiple sites to achieve significant activation[53]. The authors thus suggest the use of TALE-based transcriptional regulators for use in layered circuits to avoid crosstalk between the multiple targeting gRNAs[52].
Other receptor-based fusions for regulated gene expression
As discussed above, hormone receptors are alluring LBDs and have been widely used to tune gene expression in mammalian cells and in mouse models. Recently, tight control of the flippase (FLP) recombinase for overexpression and knockdown of proteins in hematopoietic and fibroblast cells was achieved through double induction control[54]. Briefly, the N-terminus of FLP was fused to a FKBP12 conditional destruction domain. The destruction domain is an engineered variant of the FKBP12 protein that is rapidly and constitutively degraded when expressed in mammalian cells[55]. The fusion of FLP to this FKBP12 destruction domain confers instability to FLP and leads to rapid clearance of the FLP protein from mammalian cells. The addition of the shield-1 ligand (a morpholine-containing, high-affinity ligand for FKBP) stabilizes the engineered FKBP12 variant and thus allows the fused FLP protein to be stably expressed in the cytoplasm of the host cells. A second layer of inducible regulation of FLP activity was added by fusing the C-terminal of the FLP enzyme to the estrogen receptor domain which sequesters the stabilized FLP protein within the cytoplasm until the availability of 4-OHT inducer. The fusion of FLP to engineered FKBP12 and ER domains allows for tight control of target gene regulation using shield-1 and 4-OHT ligands. This system resulted in a 5–10 fold reduction in leaky expression seen in the FLPs-ERT2 constructs. The inducible FLP recombinase activity has been used to knock down the tumor suppressor phosphatase and tensin homolog (PTEN) in human cell lines (SC-1 and K562) and mouse cell line (32D). A cell-dependent effect was observed in the recombination efficiency. For instance, maximum recombination happened in SC-1 fibroblasts following 4-OHT treatment possibly due to low levels of endogenous Hsp90, a chaperone protein. However, they only observed suboptimal recombination in hematopoietic (32D, K562, and HoxA9 bone marrow) cells. Hence, the percentage of recombined cells can be tuned by changing the concentration and combination of drugs as well as the application time. The main advantage of the inducible FKBP12-FLP-ERT2 fusion protein is the reduction of the leaky expression observed in the FLP-ERT2 fusion proteins.
The mammalian central nervous system is composed of heterogeneous subpopulations of several neuronal and glial cells. To understand the mechanisms governing the differentiation of these cell types, several mouse models have been developed that utilize the regulatory regions of retinal progenitor-specific genes to control the expression of Cre recombinase, thus allowing selective genetic modification in retinal progenitors. None of the existing models offer on-demand control for selectively studying gene function at different developmental time points. To overcome this issue, a tamoxifen-inducible hormone receptor system, Rax-CreERT2 mouse line was designed in which the CreERT2 recombinase is implanted into the endogenous Rax gene, followed by a pGK-neo sequence between Frt sites, for targeted selection in embryonic stem cells. Next, the Rax-CreERT2 mice were mated with R26-CAG-lox-stop-lox-tdTom (Ai9) which expresses the tdtomato reporter protein conditioned on Cre excision activity [56]. Addition of 4-OHT induces Cre activity, which in turn enables the expression of tdtomato in Rax-expressing progenitors. This method thus allows the tracking of all Rax-progenitors during different stages of development by tracking the expression of tdtomato, and is applicable in cell lineage experiments and for understanding gene functions in the development of retinal, hypothalamus and pituitary [56].
Pancreatic ductal adenocarcinoma (PDAC) is a disease developed by pancreatic expression of oncogenic KRAS mutations (Kras G12D or Kras G12V). KRAS gene is a subtype of RAS genes, the most frequently mutated oncogene family in human cancer, and mutations in KRAS are observed in nearly 100% of PDACs[57]. The current genetically engineered mouse models of PDAC are based on the Cre-loxp system that only allows for mutant Kras activation and does not address the critical feature of the disease which is sequential tumorigeneses and heterogeneity. Schönhuber et al. have addressed this issue by using a dual-recombinase system consisting of flippase and Cre recombinase to generate mouse models that allow for controlled independent or sequential gene expression of Kras-driven PDAC. In this model, the Pdx1-Flp line was used along with a tdTomato-EGFP-Cre reporter (R26mT-mG) that switched the gene expression from tdTomato to EGFP in the presence of tamoxifen-induced CreERT2 in the Flp-lineage. Thus, the dual-recombinase approach can advance our understanding of multistep genetic carcinogenesis and manipulation of tumor microenvironment[58].
Chimeric Antigen Receptor (CAR) T cells that are engineered to target antigens specific to tumor cells have shown promising responses in clinical trials for the treatment of leukemias/lymphomas. However, the response must be regulatable to avoid any potential off-targets and cytokine storm. To address this issue, a tamoxifen-inducible Flp recombinase-based flip excision (FLEx) system was used to regulate anti-Her2-CAR expression in T cells[59]. The FLEx switch was modified to work with the FlpO/frt recombinase system such that the nucleocytoplasmic shuttling of the FlpO recombinase fused to ERT2 estrogen receptor was dependent on the presence of 4-OHT. This gene circuit was further developed to allow for turn on (ON), turn off (OFF) or expression Level Switch (EXP) by altering the orientation of EF1α promoter to tune CAR expression. The results indicate significant changes in anti-Her2-CAR expression after one day of induction in all three designed circuits, with faster kinetics being observed in the ON switch. A low level of basal CAR expression was observed in a small population of cells (8%) with the ON switch system. This system demonstrated memory such that 15 days post-induction, the level of CAR expression was the same for the cells that were continuously induced for the entire 15-day period and the ones induced for only two days. This is particularly advantageous to avoid prolonged drug exposure that can cause side effects. Although this model has not been implemented in other CAR T-cells to modulate the gene expression, it can be potentially utilized to regulate the expression of any gene of interest that plays a key role in T cell function and behavior.
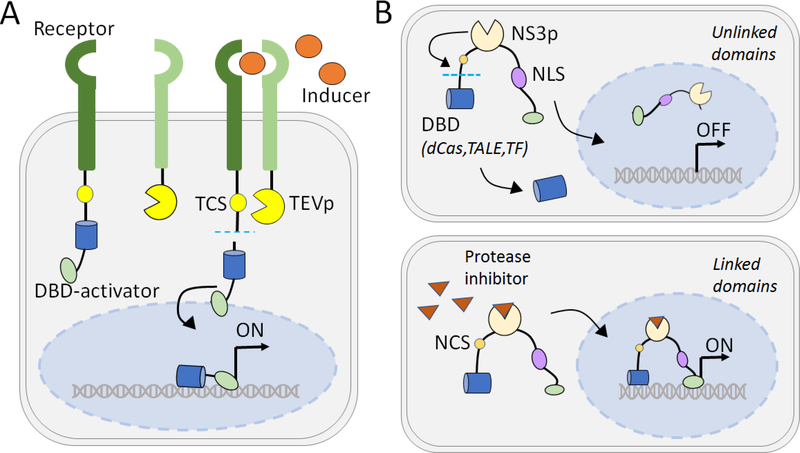
Biosensors developed in mammalian cells that rely on an extracellular signal also depend on host cell receptors or transduction pathways, which might be subject to cross talk and regulation by native cellular components[60–63]. Modular Extracellular Sensor Architecture (MESA) is a platform that is orthogonal to native cellular pathways and exerts independent, tunable gene regulation function in mammalian cells[64]. This system is designed such that ligand-inducible dimerization of a cell surface receptor brings a protease in proximity to its target sequence to release a transcription factor. The transcription factor then localizes to the nucleus to activate a transgene under the control of a minimal promoter (Fig. 3A). A non-transgenic MESA platform was developed in HEK293FT cells where the N- and C-termini of TEVp are fused to FKBP and FRB extracellular domains, which heterodimerize in the presence of small molecule rapamycin. Cleavage by TEVp releases the Tet transactivator (tTA) that targets the Tet operator and activates target gene expression. A 10.6-fold induction of YFP from a tTA-responsive reporter was achieved by reducing the quantity of both PCN and PCC transfected. This study provides a framework for a custom-designed MESA platform that can potentially respond to novel ligands.
Fig. 3. Small-molecule induced proteolytic release of synthetic transcriptional factor.
A. Modular Extracellular Sensor Architecture (MESA) platform induces receptor dimerization in the presence of small-molecule inducers leading to proteolytic release of a synthetic transcriptional factor (tet transactivator). The TF released upon proteolysis localizes to the nucleus to enable activation of the target gene expression. B. StaPL module for inducible transcriptional regulation. In the absence of an inducer, cis-cleaving NS3 protease releases the associated TF. The unlinked TF remains in the cytoplasm while the activator domains translocate to the nucleus but cannot activate the target gene without the associated TF. In presence of a small-molecule drug, the activity of the protease is inhibited thereby allowing nuclear localization of the linked TF and activator domains at the target site to turn ON gene expression.
Glioblastoma is an aggressive brain tumor and its tumor microenvironment is associated with immunosuppressive signals that prevent innate and adaptive immune system to recognize the tumor. One way to overcome immunosuppression is to increase the level of IL-12 in the tumor microenvironment (IL-12 plays a key role in linking innate and adaptive immune responses). To regulate expression of IL-12 in glioblastoma mouse models, a rheoswitch system that contains of a co-activation partner (fusion between a transcription activation domain and a nuclear factor domain), a ligand-inducible transcription factor (fusion between a DNA binding domain and a nuclear factor ligand binding domain), an inducible promoter, and an activator ligand was developed[65]. The addition of veledimex, a synthetic analog of the hormone ecdysone, leads to the formation of a stable complex between the two fusion proteins and formation of an active transcription factor complex resulting in transcriptional activation from the inducible promoter. Through the recruitment of other transcriptional co-activators and components of the transcriptional machinery, the expression of a gene that is under control of the Rheoswitch Therapeutic System (RTS) can be regulated. Using this approach, a 5–10-day GL-21 murine glioma tumor cell line was intratumorally injected with Ad-RTS-mIL-12, which harbors an adenoviral vector carrying the IL-12 gene. The expression of IL-12 was regulated by the oral administration of veledimex. The same group previously showed that Ad-RTS-mIL-12 decreased tumor growth rate in breast and melanoma cancers in mice[42]. This study showed that treatment with veledimex at 10 or 30 mg/m2/day led to the survival of the mice for 85 days, tumor-free. Note the advantage of administering veledimex to mice orally in this study. The promising results of the preclinical studies led to ongoing human studies of Ad-RTS-mIL-12+ veledimex in cancer subjects of glioblastoma[66].
One drawback of most hormone receptor-based gene switches is the requirement to deliver the effector and driver components on two separate plasmids, leading to imbalanced expression of the two plasmids and inducer-triggered induction of the transgenes. Lee et al. developed a single vector for inducible transgene expression in target cells, based on the ecdysone receptor (EcR)[67]. The construct contains the activation domain of VP16, GAL4 DBD, and a modified version of EcR that does not bind to RXR. When the ecdysone agonist tebufenozide is absent, the GAL4 DBD and modified EcR nuclear receptor (GvECR) form a homodimer in the cytosol. Upon tebufenozide addition, the protein enters the nucleus and functions as a transcription factor, activating gene expression in a dose-dependent manner and reaching ~170-fold induction after treatment with 10 μmol/l tebufenozide. This singular, adenoviral gene switch can thus regulate the spatiotemporal expression of transgenes. It is noted that prolonged gene therapy may be implausible with this system since stable genome integration is not possible.
T cells engineered to target tumor cells by expression of tumor-specific T cell receptors (TCRs) are potential immunotherapy treatments that must be controlled to prevent side effects and overactivity. Current “safety switches” are often time-consuming and use a single drug to turn ON or OFF the T cell activity[68–71]. Tighter control of T cell activation can be achieved using two different drugs, one to turn the cell ON, and another OFF. Researchers, therefore, created a dual-gated gene switch by fusing an analog-sensitive zeta-chain-associated protein kinase 70 (ZAP70) allele to the ligand-binding domain of the ERT2 that turns on or off the activity in response to 3-MB-PP1 (an ATP analog) and 4-OHT molecules, respectively[72]. ZAP70 plays an important role in T cell activation by binding to phosphorylated CD3 immunoreceptor tyrosine-based activation motifs (ITAMs) upon T cell activation. This binding interaction results in kinase phosphorylation and activation of downstream proteins involved in a signal transduction pathway. Temporal control of signaling cascade in the TCR pathway through a switch of ZAP70 expression using this strategy led to T cell activation in less than 2 min, and inhibition within 1 min, as measured by intracellular calcium levels and expression of CD69. However, surprisingly this system was not compatible with CARs although it contains the CD3 domain that interacts with ZAP70. In these cells, T cell activation was turned off by the addition of the 3-MB-PP1, however, the addition of the 4-OHT ligand resulted in a leaky elevated CD69 expression. The switch also inhibits cytokine secretion, suggesting further optimization is needed before use in adoptive T cell therapy[72].
Bacterial TF-based systems
A vast array of bacterial transcription factors (TFs) consist of a single polypeptide that links a DNA binding domain (DBD) that recognizes DNA operator sequences around the promoter of a target gene (or operon), and a ligand-binding domain (LBD), or sensory domain. These TFs typically function as multimers. These TFs operate as simple yet effective gene switches whereby gene expression is regulated in response to a small molecule ligand, often in a dose-dependent manner. As a result, they are commonly repurposed for control of synthetic transgene expression in mammalian systems. This is accomplished through relatively simple modifications of the TF, and addition of cognate operator repeats to suitable mammalian promoter sequences[1]. Table 1 lists many bacterial-derived, inducible TF systems that have been demonstrated to operate in mammalian systems, along with their respective small molecule inducer(s). Here, we review the three most widely used and commercially available bacterial TF-based inducible systems, with a focus on more recent applications in mammalian gene control.
Table 1.
Prokaryotic transcriptional regulators used for inducible control over gene expression in mammalian cells
| Regulator | Source organism | Role in source organism | Inducer | Host organism/ cells | Reference |
|---|---|---|---|---|---|
| AlcR | Aspergillus nidulans | Activator | Acetaldehyde | Baby hamster kidney cells (BHK-21), CHO-K1, HeLa | [73] |
| AmtR | Corynebacterium Glutamicum | Repressor | GlnK protein | HEK-293, CHO | [74] |
| ArgR | Chlamydia pneumoniae | Repressor | L-arginine | CHO-K1 | [75] |
| BirA | E. coli | Repressor | Biotin | CHO-K1 | [76] |
| BmR1 | Bacillus megaterium | Repressor | Pentobarbital | K-293, CHO | [74] |
| CbaR | Comamonas testosteroni | Repressor | 3-Chlorobenzoate | HEK-293 cells, HeLa cells, telomerase-immortalized human mesenchymal stem cells | [77] |
| EmrR | Sinorhizobium meliloti | Repressor | Luteolin | HEK-293T, COS-7, human telomerase reverse transcriptase immortalized human mesenchymal stem cells (hMSC-hTERT) | [78] |
| EthR | Mycobacterium tuberculosis | Repressor | 2-Phenylethyl-butyrate | HEK-293 | [79] |
| FapR | Bacillus subtilis | Repressor | Malonyl-CoA | COS-1 cells | [80] |
| HdnoR | Arthrobacter nicotinovorans | Repressor | 6-Hydroxy-nicotine | CHO-K1 | [81] |
| HucR | Deinococcus radioduransR1 | Repressor | Uric acid | HeLa, HEK-293, human fibrosarcoma cells (HT-1080) | [82] |
| IcaR | Staphylococcus epidermidis | Repressor | Gentamycin | HEK-293, CHO | [74] |
| LmrA | Bacillus subtilis | Repressor | Lincomycin | HEK-293, CHO | [74] |
| LuxR | Vibrio fischeri | Activator | 3-Oxo-hexanoyl-homoserine lactone (3OC6HSL) | HEK-293 | [83] |
| McbR | Corynebacterium glutamicum | Repressor | L-methionine | HEK-293, CHO | [74] |
| MphR(A) | Escherichia coli | Repressor | Erythromycin | CHO-K1, NIH/3T3, and human HT-1080 cells | [84] |
| PcaV | Streptomyces coelicolor | Repressor | Green tea derived phenolic acid | HEK-293, Mouse and monkey models | [85] |
| PhlF | Pseudomonas fluorescensCHA0 | Repressor | 2,4-Diacetylphloroglucinol | HEK-293, CHO | [74] |
| PIP | Streptomyces coelicolor | Repressor | Pristinamycin I | CHO-K1 | [86] |
| PmeR | Pseudomonas syringae | Repressor | Phloretin, paraben, plant flavanoids | HEK-293, HeLa, HT-1080, hMSC-TERT,CHO-K1, BHK-21 | [87] |
| QacR | Staphylococcus aureus | Repressor | Several plant alkaloids | HEK-293, CHO | [74] |
| Rex | Streptomyces coelicolor | Repressor | NADH | CHO-K1 | [88] |
| ScbR | Streptomyces coelicolor | Repressor | 2-(1′-Hydroxy-6-methylheptyl)-3-(hydroxymethyl)-butanolide | HEK-293T, COS-7, BHK-21, human hepatocellular carcinoma cells (HepG2) | [89] |
| SimR | Streptomyces antibioticus Tü 6040 | Repressor | Simocyclinone D8 (SD8) | HEK-293T, COS-7, human telomerase reverse transcriptase immortalized human mesenchymal stem cells (hMSC-hTERT) | [78] |
| TraR | Agrobacterium tumefaciens | Activator | N-(3-oxo-octanoyl) homoserine lactone | HeLa, HEK-293, Huh7 (hepatoma), and RD (rhabdomyosarcoma) | [90] |
| TtgR | Pseudomonas putida | Repressor | Phloretin | CHO-K1, BHK-21, COS-7, HaCaT, HEK-293, HT-1080, and NIH/3T3 cell lines, as well as into primary human fibroblasts and keratinocytes | [91] |
| VanR | Caulobacter crescentus | Repressor | Vanillic acid | CHO-K1 | [92] |
TetR-based inducible systems
The tetracycline-controlled “Tet-Off” and “Tet-On” gene expression systems are the most widely used inducible systems to regulate the expression of mammalian genes[93]. A typical Tet-Off system, as first described by Gossen et al. [94] consists of the tetracycline repressor (TetR) fused to the VP16 activation domain of HSV. In the absence of inducer (tetracycline (Tc), or some Tc derivatives), this transactivating fusion protein (termed tTA) activates transcription of genes downstream of strategically placed tetO sequences. When the inducer is added the TetR component releases the operator, switching OFF expression. Isolation of a TetR variant that exhibits the opposite response to tetracycline (i.e., binds operator in the presence of Tc/Tc analogs) allowed the construction of a reverse transactivator (termed rtTA) which was used in the “Tet-On” system[93, 95].
TetR-based inducible systems have been extensively studied and optimized for mammalian gene control[74, 96]. Recently, to minimize the side effects associated with the use of chimeric antigen receptor T-cell therapy (CAR T therapy), Gu et al. developed inducible CD19CAR (iCAR19) T cells using the Tet-ON system for doxycycline (dox)-activated expression of the CD19-targeting CAR[97]. The iCAR19 cells were derived from human peripheral blood mononuclear cells (PBMCs) and engineered using GMP standards for potentially safe and controlled treatment of relapsed or refractory B-cell malignancies[97]. The group reported dox-induced CAR upregulation which was brought down to background levels within 24 h of removal of the dox inducer, thus producing an effective reversible switch. The iCAR19 cells further exhibited a dox-dependent, high cytotoxic activity (84% lysis) against Raji cells (Human Burkitt′s lymphoma cells expressing CD19) as compared to the cytotoxic activity in absence of dox (34% lysis) or against CD19-negative cells (<20% cell lysis)[97]. A notable drawback to this system is the “leaky” CAR expression in the uninduced state, leading to significant background activity of the CAR19-T cells. Furthermore, prolonged exposure to doxycycline may cause systemic toxicity as well as increased risk of generating antibiotic-resistant microorganisms in patients treated with the iCAR19 cells.
TetR-based inducible systems have also been used to build smart sensor devices which have the potential to detect and treat clinical disorders[98, 99]. By combining the small-molecule sensing properties of membrane receptors and TetR, Ye et al. engineered HEK-293 cells containing a synthetic insulin-sensor device for correcting insulin resistance in mice[98]. The device was designed using the human insulin receptor by re-wiring the receptor’s signaling pathway to phosphorylate a hybrid transcription factor, TetR-ELK1 where the TetR component serves as the DBD and recruits the ELK1 activator to the target site. In the basal state, when insulin is low, nuclear TetR-ELK1 stays bound to a chimeric promoter containing the heptameric operator module (tetO7) linked to a minimal version of the CMV promoter (PhCMV), while the ELK1 domain remains inactive. Insulin (from 2–20 ng/mL) triggers a signaling cascade from the insulin receptor, leading to the activation of mitogen-activated protein kinase (MAPK). Active MAPK phosphorylates and activates ELK1, thereby upregulating the expression of the target adiponectin gene. The addition of doxycycline disrupts operator binding, preventing transgene expression regardless of its phosphorylation state, thus providing dual-input regulation of transcription. HEK-293 cells containing this insulin sensor were implanted into hyper-insulinemic mice to bring about the adiponectin-mediated correction of insulin resistance through an insulin-induced reduction in blood glucose, free fatty acid, and cholesterol levels, along with an added layer of dox-repressed control of sensor activity to prevent adiponectin overdosing[98]. Repeated, subcutaneous insertion of engineered cells is notably a sub-optimal therapy option.
Based on a mechanism similar to the insulin-sensor device, Xu et al. used a G-protein coupled receptor (GPCR) and TetR combination to develop a treatment strategy for hepatogenous diabetes in mice [99]. Cells containing the GPCR GPBAR1 relay an activation signal for the upregulation of glucagon-like-peptide 1 (GLP-1), proven to have hepatoprotective effects in mice[100]. With this configuration, the GPBAR1 receptor activates protein kinase A (PKA) in the presence of inducer oleanolic acid (OA). Active PKA in turn phosphorylates the CREB1 activator of TetR-CREB1 in the nucleus of the engineered cells. In hepatogenous diabetes mouse models implanted with microencapsulated HEK293 cells containing the insulin-sensor device, administration of OA tablets (3 × 100 mg/kg/day), resulted in a 2.3-fold increase in bloodstream GLP-1 levels[99]. The microencapsulation of the engineered HEK293 cells ensures that communication between the host and graft cells occurs solely via diffusion of secretory metabolites across the semi-permeable capsule membrane. Administration of OA as inducer also offers potential therapeutic effects in treating hepatogenous diabetes [99]. Additionally, the dox-triggered safety switch enables precise and on-demand termination of gene circuit function during any unforeseeable scenarios in clinical applications.
The successful use of TetR-based gene regulators in mammalian cells has prompted studies into other TetR-like bacterial TFs, to develop new and orthogonal gene expression systems for mammalian cells. For example, Bojar and Fussenegger recently described the use of two TetR-like repressors, SimR (from Streptomyces antibioticus Tü 6040) and EmrR (from Sinorhizobium meliloti), to control gene expression in a variety of mammalian cell lines using the small-molecule inducers simocyclinone D8 (SD8) and luteolin, respectively[78]. The authors designed inducible ON-switches via fusion to KRAB domain, as well as OFF-switches via fusion to VP16. Such studies further demonstrate the versatility of TetR-like regulators for efficient and reversible control of mammalian gene expression, but also point to limitations that may arise due to inducer toxicity.
LacI-based inducible systems
While LacI-based systems are used for regulating mammalian gene expression, a notable drawback is the leakiness of the LacI repressor. To overcome this limitation, Lee et al. employed a tighter-binding LacI repressor (LacIGY, identified in E. coli[101]. LacIGY showed the highest repression of a target gene when expressed in mouse NIH/3T3 embryonic cells, compared with other repressors that included WT-LacI, TetR, and TetR fused to a nuclear localization signal[101]. LacIGY was also able to repress the constitutively active dnmt1 encoding DNA methyltransferase 1 in intestine-specific lines of transgenic mice (here, the Villin promoter was targeted). Gene expression was mediated by addition of IPTG, and repression restored within 2–3 days of IPTG withdrawal[101]. This LacI “REMOTE-control” system (Reversible Manipulation of Transcription at Endogenous loci) enabled studying the effect of dnmt1 downregulation on development - a feat that had been difficult to achieve due to embryonic lethality of a dnmt1 knockout. This repressor thus demonstrated applicability in testing the reversibility of a phenotype and investigating gene function at different expression levels.
The LacI and TetR systems were recently developed and demonstrated to achieve tightly controlled CRISPR-Cas9-based genome editing in MC-38 cells, derived from C57BL6 murine colon adenocarcinoma cells[32]. The genome-wide screen incorporated the Brie sgRNA library (sgRNA library composed of 78,637 sgRNAs targeting 19,674 protein coding mouse genes and 1000 non-targeting control sgRNAs) into IPTG-inducible or dox-inducible constructs. Sun et al. were able to distinguish between essential and nonessential genes in a lethality screen by monitoring depletion of sgRNAs from the proliferating pool of cells. The researchers demonstrated application of the inducible sgRNA system in multiple human (L-363, A-498, LP-1, HEK293T, NCI-H1299, 786-O) and murine (MC-38, LL/2, 4T1, CT-26) cell lines, noting that the use of multiple operators (2xLacO and 2xTetO) significantly reduced background activity[32]. As with most bacterial and/or viral-derived recombinant systems, the LacI and TetR systems suffer from immunogenicity associated with their protein components; efforts are aimed at increasing their safety profiles. Despite this limitation, TetR-based systems are the most extensively studied and show the greatest potential for therapeutic applications, as demonstrated by treating metabolic disorders in diabetic mice.
CymR-based inducible systems
To complement the Tet-system, Mullick et al. developed a cumate-inducible system using the cymene repressor (CymR), derived from Pseudomonas putida[102]. In P. putida, CymR regulates expression from the p-cym and p-cmt operons, involved in cumate catabolism, by binding to operators (CuO) within the promoters of the two operons[103]. For use in mammalian transcription control, Mullick et al. placed a single CuO operator downstream of a minimal CMV promoter to generate a hybrid promoter regulated by CymR. CymR repressed target gene transcription from the minimal hybrid promoter, and addition of 10ug/ml cumate was able to fully activate gene expression. To demonstrate the modularity of the cumate-inducible system, Mullick et al. generated a cumate transactivator (cTA), by fusing a VP16 activation domain to CymR, as well as a cumate reverse transactivator (rcTA), by fusing VP16 to a CymR variant that instead binds operator in the presence of cumate[102]. While the use of rcTA is preferable, to enable inducible activation of transcription, its performance was sub-optimal due to high background expression. To improve rcTA activation, the authors combined CymR and rcTA to tightly regulate target gene expression in the absence of cumate, resulting in up to 700-fold activation in the presence of cumate[102]. The combined CymR and rcTA system was recently used to rapidly produce large amounts of therapeutic recombinant proteins (hCD200Fc and Rituximab) in CHO cells[104]. Notably, the hCD200Fc yield was up to 4-fold lower when a constitutive promoter was used instead of the cumate-inducible promoter, demonstrating the advantage of using a tunable and inducible promoter system to obtain high product yields in mammalian cell factories[104].
The use of bacterial TFs for mammalian gene regulation allows orthogonality in mammalian cells, thereby avoiding pleiotropic effects on host cell regulation. Further, the specificity of each TF for its inducer enables the use of multiple TFs within the same host cells without crosstalk. However, a notable limitation of these systems is the need to insert operator sequences, within the promoters, to enable binding of the TF regulators thus precluding the ability to control gene expression at endogenous loci within mammalian cells. This insertion may affect the functioning of a promoter, hence care must be taken in positioning the operators within mammalian promoter elements[1]. Additionally, the use of dox and other inducers, at their often relatively high concentrations needed to induce gene expression, may lead to cell-specific toxicity and cause undesired changes in cell function[105]. Finally, the use of prokaryotic and viral components in the rtTA system has been linked to potential immunogenicity, leading to adverse effects in clinical use of the Tet system[97].
Other small molecule-inducible systems
While synthetic, inducible control over mammalian genes has largely focused on the use of chimeric hormone receptors or bacterial transcription factors, other unique designs have been recently described. Two recent studies utilized small protein linkages or tags that enable drug-inducible stabilization[106] or proteolytic degradation[107] of a synthetic transcription factor, thereby mediating drug-inducible gene expression. As depicted in Fig. 3B, Jacobs et al. designed a stabilizable polypeptide linkage (StaPL) module to achieve drug-inducible protein stabilization. The StaPL module consists of a protease fused to an inducer binding sequence, wherein the proteolytic activity of the protease is inhibited in the presence of inducer[106]. Asunaprevir (ASV)-inhibited or telaprevir (TPV)-inhibited StaPL modules, derived from the HCV nonstructural protein 3 protease domain (NS3 protease), were linked to protease cleavage sites, yielding StaPLAI or StaPLTI, respectively[108]. These StaPLs were used to join an NLS sequence to a yellow fluorescent protein (YFP) sequence, such that nuclear localization of YFP occurred only in the presence of inducer. The orthogonal StaPL modules were further fused to a VEGFA-targeting zinc finger (ZF) domain (ZFVEGFA-StaPLAI-YFP-VPR or ZFVEGFA-StaPLTI-tdRFP-KRAB), resulting in ASV-mediated VEGF upregulation and TPV-mediated VEGF downregulation in HEK cells coexpressing both constructs[106]. The StaPL module was also used for inducible control of dCas9-based activation by inserting StaPLTI within a dCas9 activator, resulting in a 24-fold increase in target RFP fluorescence in the presence of TPV[106]. As many proteins tolerate the insertion of protein domains at exposed loops[109], the internal StaPL modules provide a versatile platform to create drug-stabilized protein variants. As with many other recombinant systems, the immunogenic nature of the protease used in the StaPL module limits its clinical applications.
Degron tags are peptide domains that greatly increase the rate of proteasomal degradation of a fused partner protein. By fusing the auxin-inducible degron (derived from the Arabidopsis thaliana IAA17 protein)[110] to a dCas9-PR transcription factor, Kleinjan et al. generated an auxin-controllable transcriptional activator which upregulated expression of gene targets in the absence of auxin, and markedly reduced target gene expression to background levels in the presence of auxin[107]. The AID-dCas9-PR protein was able to rapidly switch between expression states, with clearance of AID-dCas9-PR taking 1–2 hours after addition of auxin, and re-activation of target gene expression taking about 30 mins when replaced with medium lacking auxin[107]. The rapid action of the AID-dCas9-PR system allows for fast switching between different functionalities in the cell.
In attempts to overcome challenges of inducer toxicity and/or immunogenicity of proteins used in inducible gene regulation, Hill et al. developed antibody-based chemically induced dimerizers (AbCIDs) comprised of synthetically designed, single-chain human antibodies that selectively target a small molecule-protein complex. BCL-xL is a member of the anti-apoptotic BCL-2 family of proteins which binds to the commercially available ABT-737 drug, while exposing a large portion of the bound ABT-737 as a potential epitope for antibody binding[111]. Hill and coworkers isolated three AbCIDs (scAZ1, scAZ2, and scAZI3) that showed high selectivity for the BCL-xL/ABT-737 complex over BCL-xL alone and further demonstrated the use of the highly selective scAZ1 antibody in inducible CRISPRa[112]. For this purpose, Hill et al. fused scAZ1 to VPR, while dCas9 was fused to the BCL-xl domain. Constitutive sgRNAs expressed in HEK293T cells directed the dCas9-Bcl-xL fusion to a luciferase reporter gene which remained downregulated in the absence of ABT-737. Addition of 20nM ABT-737 upregulated luciferase expression by 20%[112]. The authors suggest their work paves a way for developing new tools using alternative binding domains like DARPins, nucleic acid-aptamer libraries, and knottins[112].
Bojar et al. designed another AbCID using the single-domain VHH camelid antibody (aCaffVHH), which undergoes homodimerization in the presence of caffeine. To regulate mammalian gene expression, the authors co-expressed in HEK293T cells aCaffVHH fused to the DBD of TetR, along with aCaffVHH fused to the VP activator. Caffeine addition upregulated expression of the reporter gene (human placental-secreted alkaline phosphatase (SEAP)), through a minimal promoter containing seven TetR binding operator sites[113]. The group also used the caffeine-stimulated AbCID system to trigger the expression of the human glucagon-like peptide (shGLP-1), for treating type-2-diabetes in mouse models with impaired insulin sensitivity[113]. This caffeine inducible AbCID makes it possible to fine-tune therapeutic transgene expression in response to routine intake of beverages such as tea and coffee. AbCID systems have great potential in therapeutic applications due to their non-immunogenic nature and use of safe small-molecule drug inducers.
In an exciting synthetic biology development toward genetically treating type-1 diabetes mellitus (T1DM), whereby insulin-producing pancreatic β cells are destroyed by the immune system, Saxena et al. designed a vanillic acid (VA)-responsive “band-pass filter” to coordinate the expression of three transcription factors (Ngn3, Pdx1, and MafA) involved in the differentiation of pancreatic progenitor stem cells into insulin-producing β-like cells[114]. They employed a VA-sensitive olfactory GPCR to sense extracellular VA levels and trigger a synthetic signaling cascade. Expression of a VA-dependent transactivator (VanA1), which subsequently activates other genes, is induced at medium VA concentrations. Meanwhile VanA1 action is inhibited at high VA levels. The synthetic orchestration of Ngn3, Pdx1 and MafA expression resulted in a lineage-controlled network that differentiates into glucose-sensitive insulin-secreting beta-like cells, with dynamics similar to that of human pancreatic islets. Synthetic, tunable gene circuits hold promise for programming somatic cells into autologous phenotypes for regenerative medicine.
Conclusion
Engineering control over gene expression in mammalian cells is met with many challenges. Small-molecule inducible systems provide a means of exogenous and often tunable control over transcription of target genes. In recent years, researchers have engineered, assembled, and repurposed a variety of molecular biology components to build such synthetic gene regulation systems. Developing systems with high specificity (minimal off-target effects and orthogonality) and tight regulation (low background and wide dynamic range) is essential to realizing their utility, in addition to a myriad of other important features such as stability, reversibility, and low toxicity. A common theme in many studies reviewed here is sequestration of key transcriptional machinery from the nucleus in the absence of inducer, as a means of minimizing background expression. Designing systems that require multiple inputs to fully actuate the transcriptional machinery is also thematic. Not surprisingly, many advances in synthetic control over mammalian gene expression take advantage of recently developed tools that offer improved specificity toward target genes (e.g. TALE and CRISPR devices), as well as continued insights into protein biochemistry, heterologous gene expression, and protein engineering capabilities. These reviewed studies collectively describe a variety of inducible gene expression platforms, from which further advances and applications are anticipated.
Acknowledgments
Funding
This publication was supported by the NIH (U01AI148118), CPRIT (RP180466), MRA Established Investigator Award (509800), NSF (1705464, CBET-1511425), CDMRP (CA160591), and Owens foundation.
Footnotes
Declarations of Interest
The authors report no declarations of interest.
References
- [1].Auslander S, Fussenegger M, Engineering Gene Circuits for Mammalian Cell-Based Applications. Cold Spring Harbor Perspectives in Biology 2016, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wurm FM, Gwinn KA, Kingston RE, Inducible overproduction of the mouse c-myc protein in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America 1986, 83, 5414–5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, et al. , Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell 1993, 73, 585–596. [DOI] [PubMed] [Google Scholar]
- [4].Wu S, Huang J, Dong J, Pan D, hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 2003, 114, 445–456. [DOI] [PubMed] [Google Scholar]
- [5].Kim H, Kim M, Im SK, Fang S, Mouse Cre-LoxP system: general principles to determine tissue-specific roles of target genes. Lab Anim Res 2018, 34, 147–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jonkers J, Berns A, Conditional mouse models of sporadic cancer. Nat Rev Cancer 2002, 2, 251–265. [DOI] [PubMed] [Google Scholar]
- [7].Lampreht Tratar U, Horvat S, Cemazar M, Transgenic Mouse Models in Cancer Research. Front Oncol 2018, 8, 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kersten K, de Visser KE, van Miltenburg MH, Jonkers J, Genetically engineered mouse models in oncology research and cancer medicine. EMBO Mol Med 2017, 9, 137–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].June CH, O’Connor RS, Kawalekar OU, Ghassemi S, Milone MC, CAR T cell immunotherapy for human cancer. Science 2018, 359, 1361–1365. [DOI] [PubMed] [Google Scholar]
- [10].Majzner RG, Mackall CL, Clinical lessons learned from the first leg of the CAR T cell journey. Nature Medicine 2019, 25, 1341–1355. [DOI] [PubMed] [Google Scholar]
- [11].Rezvani K, Adoptive cell therapy using engineered natural killer cells. Bone Marrow Transplant 2019, 54, 785–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Tenspolde M, Zimmermann K, Weber LC, Hapke M, et al. , Regulatory T cells engineered with a novel insulin-specific chimeric antigen receptor as a candidate immunotherapy for type 1 diabetes. J Autoimmun 2019, 103, 102289. [DOI] [PubMed] [Google Scholar]
- [13].Kansal R, Richardson N, Neeli I, Khawaja S, et al. , Sustained B cell depletion by CD19-targeted CAR T cells is a highly effective treatment for murine lupus. Science Translational Medicine 2019, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Linette GP, Stadtmauer EA, Maus MV, Rapoport AP, et al. , Cardiovascular toxicity and titin cross-reactivity of affinity-enhanced T cells in myeloma and melanoma. Blood 2013, 122, 863–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ruella M, Xu J, Barrett DM, Fraietta JA, et al. , Induction of resistance to chimeric antigen receptor T cell therapy by transduction of a single leukemic B cell. Nature Medicine 2018, 24, 1499–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mustafina K, Fukunaga K, Yokobayashi Y, Design of Mammalian ON-Riboswitches Based on Tandemly Fused Aptamer and Ribozyme. ACS Synthetic Biology 2020, 9, 19–25. [DOI] [PubMed] [Google Scholar]
- [17].Berens C, Groher F, Suess B, RNA aptamers as genetic control devices: the potential of riboswitches as synthetic elements for regulating gene expression. Biotechnology Journal 2015, 10, 246–257. [DOI] [PubMed] [Google Scholar]
- [18].Yokobayashi Y, Aptamer-based and aptazyme-based riboswitches in mammalian cells. Current Opinion in Chemical Biology 2019, 52, 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Braselmann S, Graninger P, Busslinger M, A selective transcriptional induction system for mammalian cells based on Gal4-estrogen receptor fusion proteins. Proceedings of the National Academy of Sciences of the United States of America 1993, 90, 1657–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Indra AK, Warot X, Brocard J, Bornert JM, et al. , Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Research 1999, 27, 4324–4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kosugi S, Hasebe M, Matsumura N, Takashima H, et al. , Six classes of nuclear localization signals specific to different binding grooves of importin alpha. The Journal of Biological Chemistry 2009, 284, 478–485. [DOI] [PubMed] [Google Scholar]
- [22].Konermann S, Brigham MD, Trevino AE, Joung J, et al. , Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 2015, 517, 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chavez A, Tuttle M, Pruitt BW, Ewen-Campen B, et al. , Comparison of Cas9 activators in multiple species. Nature Methods 2016, 13, 563–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Scott JN, Kupinski AP, Kirkham CM, Tuma R, Boyes J, TALE proteins bind to both active and inactive chromatin. The Biochemical Journal 2014, 458, 153–158. [DOI] [PubMed] [Google Scholar]
- [25].Black JB, Perez-Pinera P, Gersbach CA, Mammalian Synthetic Biology: Engineering Biological Systems. Annual Review of Biomedical Engineering 2017, 19, 249–277. [DOI] [PubMed] [Google Scholar]
- [26].Mercer AC, Gaj T, Sirk SJ, Lamb BM, Barbas CF 3rd, Regulation of endogenous human gene expression by ligand-inducible TALE transcription factors. ACS Synthetic Biology 2014, 3, 723–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lonzaric J, Lebar T, Majerle A, Mancek-Keber M, Jerala R, Locked and proteolysis-based transcription activator-like effector (TALE) regulation. Nucleic Acids Research 2016, 44, 1471–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhao C, Zhang Y, Zhao Y, Ying Y, et al. , Multiple Chemical Inducible Tal Effectors for Genome Editing and Transcription Activation. ACS Chemical Biology 2018, 13, 609–617. [DOI] [PubMed] [Google Scholar]
- [29].Bikard D, Jiang W, Samai P, Hochschild A, et al. , Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Research 2013, 41, 7429–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhang X, Wang J, Cheng Q, Zheng X, et al. , Multiplex gene regulation by CRISPR-ddCpf1. Cell Discovery 2017, 3, 17018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Xu X, Qi LS, A CRISPR-dCas Toolbox for Genetic Engineering and Synthetic Biology. Journal of Molecular Biology 2019, 431, 34–47. [DOI] [PubMed] [Google Scholar]
- [32].Sun N, Petiwala S, Wang R, Lu C, et al. , Development of drug-inducible CRISPR-Cas9 systems for large-scale functional screening. BMC Genomics 2019, 20, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Cao J, Wu L, Zhang SM, Lu M, et al. , An easy and efficient inducible CRISPR/Cas9 platform with improved specificity for multiple gene targeting. Nucleic Acids Research 2016, 44, e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gao Y, Xiong X, Wong S, Charles EJ, et al. , Complex transcriptional modulation with orthogonal and inducible dCas9 regulators. Nature Methods 2016, 13, 1043–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Liang FS, Ho WQ, Crabtree GR, Engineering the ABA plant stress pathway for regulation of induced proximity. Science Signaling 2011, 4, rs2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Miyamoto T, DeRose R, Suarez A, Ueno T, et al. , Rapid and orthogonal logic gating with a gibberellin-induced dimerization system. Nature Chemical Biology 2012, 8, 465–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI, et al. , Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nature Biotechnology 2015, 33, 510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Chen T, Gao D, Zhang R, Zeng G, et al. , Chemically Controlled Epigenome Editing through an Inducible dCas9 System. Journal of the American Chemical Society 2017, 139, 11337–11340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lu J, Zhao C, Zhao Y, Zhang J, et al. , Multimode drug inducible CRISPR/Cas9 devices for transcriptional activation and genome editing. Nucleic Acids Research 2018, 46, e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Tak YE, Kleinstiver BP, Nunez JK, Hsu JY, et al. , Inducible and multiplex gene regulation using CRISPR-Cpf1-based transcription factors. Nature Methods 2017, 14, 1163–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Liu Y, Han J, Chen Z, Wu H, et al. , Engineering cell signaling using tunable CRISPR-Cpf1-based transcription factors. Nature Communications 2017, 8, 2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Chen X, Li T, Wang X, Du Z, et al. , Synthetic dual-input mammalian genetic circuits enable tunable and stringent transcription control by chemical and light. Nucleic Acids Research 2016, 44, 2677–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kipniss NH, Dingal P, Abbott TR, Gao Y, et al. , Engineering cell sensing and responses using a GPCR-coupled CRISPR-Cas system. Nature Communications 2017, 8, 2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Baeumler TA, Ahmed AA, Fulga TA, Engineering Synthetic Signaling Pathways with Programmable dCas9-Based Chimeric Receptors. Cell Reports 2017, 20, 2639–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kroeze WK, Sassano MF, Huang XP, Lansu K, et al. , PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nature Structural & Molecular Biology 2015, 22, 362–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Zetsche B, Volz SE, Zhang F, A split-Cas9 architecture for inducible genome editing and transcription modulation. Nature Biotechnology 2015, 33, 139–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Rivera VM, Berk L, Clackson T, Dimerizer-mediated regulation of gene expression in vitro. Cold Spring Harbor Protocols 2012, 2012, 815–820. [DOI] [PubMed] [Google Scholar]
- [48].Foight GW, Wang Z, Wei CT Jr, Greisen P, et al. , Multi-input chemical control of protein dimerization for programming graded cellular responses. Nat Biotechnol 2019, 37, 1209–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Dai X, Chen X, Fang Q, Li J, Bai Z, Inducible CRISPR genome-editing tool: classifications and future trends. Critical Reviews in Biotechnology 2018, 38, 573–586. [DOI] [PubMed] [Google Scholar]
- [50].Ricci CG, Chen JS, Miao Y, Jinek M, et al. , Deciphering Off-Target Effects in CRISPR-Cas9 through Accelerated Molecular Dynamics. ACS Central Science 2019, 5, 651–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH, Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Molecular Therapy. Nucleic Acids 2015, 4, e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lebar T, Jerala R, Benchmarking of TALE- and CRISPR/dCas9-Based Transcriptional Regulators in Mammalian Cells for the Construction of Synthetic Genetic Circuits. ACS Synthetic Biology 2016, 5, 1050–1058. [DOI] [PubMed] [Google Scholar]
- [53].Cheng AW, Wang H, Yang H, Shi L, et al. , Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Research 2013, 23, 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Maetzig T, Kuehle J, Schwarzer A, Turan S, et al. , All-in-One inducible lentiviral vector systems based on drug controlled FLP recombinase. Biomaterials 2014, 35, 4345–4356. [DOI] [PubMed] [Google Scholar]
- [55].Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ, A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell 2006, 126, 995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Pak T, Yoo S, Miranda-Angulo AL, Wang H, Blackshaw S, Rax-CreERT2 knock-in mice: a tool for selective and conditional gene deletion in progenitor cells and radial glia of the retina and hypothalamus. PloS One 2014, 9, e90381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Waters AM, Der CJ, KRAS: The Critical Driver and Therapeutic Target for Pancreatic Cancer. Cold Spring Harbor Perspectives in Medicine 2018, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Schonhuber N, Seidler B, Schuck K, Veltkamp C, et al. , A next-generation dual-recombinase system for time- and host-specific targeting of pancreatic cancer. Nature Medicine 2014, 20, 1340–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Chakravarti D, Caraballo LD, Weinberg BH, Wong WW, Inducible Gene Switches with Memory in Human T Cells for Cellular Immunotherapy. ACS Synthetic Biology 2019, 8, 1744–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Weber W, Bacchus W, Daoud-El Baba M, Fussenegger M, Vitamin H-regulated transgene expression in mammalian cells. Nucleic Acids Research 2007, 35, e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Fung E, Wong WW, Suen JK, Bulter T, et al. , A synthetic gene-metabolic oscillator. Nature 2005, 435, 118–122. [DOI] [PubMed] [Google Scholar]
- [62].Weber W, Daoud-El Baba M, Fussenegger M, Synthetic ecosystems based on airborne inter- and intrakingdom communication. Proceedings of the National Academy of Sciences of the United States of America 2007, 104, 10435–10440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Culler SJ, Hoff KG, Smolke CD, Reprogramming cellular behavior with RNA controllers responsive to endogenous proteins. Science 2010, 330, 1251–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Daringer NM, Dudek RM, Schwarz KA, Leonard JN, Modular extracellular sensor architecture for engineering mammalian cell-based devices. ACS Synthetic Biology 2014, 3, 892–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Chan T, Lewis J, Herberman RB, Chapter 25 - Therapeutic Efficacy and Systemic Antitumor T Cell Immunity Induced by RheoSwitch-Regulated IL-12 Expression after Intratumoral Injection of Adenovirus Vector or Vector-Transduced Dendritic Cells, in: Lattime EC, Gerson SL (Eds.), Gene Therapy of Cancer (Third Edition), Academic Press, San Diego: 2014, pp. 363–376. [Google Scholar]
- [66].Barrett JA, Cai H, Miao J, Khare PD, et al. , Regulated intratumoral expression of IL-12 using a RheoSwitch Therapeutic System((R)) (RTS((R))) gene switch as gene therapy for the treatment of glioma. Cancer Gene Therapy 2018, 25, 106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Lee S, Sohn KC, Choi DK, Won M, et al. , Ecdysone Receptor-based Singular Gene Switches for Regulated Transgene Expression in Cells and Adult Rodent Tissues. Molecular Therapy. Nucleic Acids 2016, 5, e367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Bonifant CL, Jackson HJ, Brentjens RJ, Curran KJ, Toxicity and management in CAR T-cell therapy. Mol Ther Oncolytics 2016, 3, 16011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Brudno JN, Kochenderfer JN, Toxicities of chimeric antigen receptor T cells: recognition and management. Blood 2016, 127, 3321–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Maude SL, Barrett D, Teachey DT, Grupp SA, Managing cytokine release syndrome associated with novel T cell-engaging therapies. Cancer J 2014, 20, 119–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].van den Berg JH, Gomez-Eerland R, van de Wiel B, Hulshoff L, et al. , Case Report of a Fatal Serious Adverse Event Upon Administration of T Cells Transduced With a MART-1-specific T-cell Receptor. Molecular therapy : the journal of the American Society of Gene Therapy 2015, 23, 1541–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Wong NML, Wong WW, Engineering a Dual Small Molecule Gated ZAP70 Switch in T Cells. ACS Synthetic Biology 2018, 7, 969–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Weber W, Rimann M, Spielmann M, Keller B, et al. , Gas-inducible transgene expression in mammalian cells and mice. Nat Biotechnol 2004, 22, 1440–1444. [DOI] [PubMed] [Google Scholar]
- [74].Stanton BC, Siciliano V, Ghodasara A, Wroblewska L, et al. , Systematic transfer of prokaryotic sensors and circuits to mammalian cells. ACS Synthetic Biology 2014, 3, 880–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Hartenbach S, Daoud-El Baba M, Weber W, Fussenegger M, An engineered L-arginine sensor of Chlamydia pneumoniae enables arginine-adjustable transcription control in mammalian cells and mice. Nucleic Acids Research 2007, 35, e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Weber W, Stelling J, Rimann M, Keller B, et al. , A synthetic time-delay circuit in mammalian cells and mice. Proc Natl Acad Sci U S A 2007, 104, 2643–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Xie M, Ye H, Hamri GC, Fussenegger M, Antagonistic control of a dual-input mammalian gene switch by food additives. Nucleic Acids Research 2014, 42, e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Bojar D, Fussenegger M, Programming mammalian gene expression with the antibiotic simocyclinone D8 and the flavonoid luteolin. Aiche Journal 2018, 64, 4237–4246. [Google Scholar]
- [79].Weber W, Schoenmakers R, Keller B, Gitzinger M, et al. , A synthetic mammalian gene circuit reveals antituberculosis compounds. Proc Natl Acad Sci U S A 2008, 105, 9994–9998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Ellis JM, Wolfgang MJ, A genetically encoded metabolite sensor for malonyl-CoA. Chemistry & Biology 2012, 19, 1333–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Malphettes L, Weber CC, El-Baba MD, Schoenmakers RG, et al. , A novel mammalian expression system derived from components coordinating nicotine degradation in arthrobacter nicotinovorans pAO1. Nucleic Acids Research 2005, 33, e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Kemmer C, Gitzinger M, Daoud-El Baba M, Djonov V, et al. , Self-sufficient control of urate homeostasis in mice by a synthetic circuit. Nat Biotechnol 2010, 28, 355–360. [DOI] [PubMed] [Google Scholar]
- [83].Miller M, Hafner M, Sontag E, Davidsohn N, et al. , Modular design of artificial tissue homeostasis: robust control through synthetic cellular heterogeneity. PLoS Comput Biol 2012, 8, e1002579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Weber W, Fux C, Daoud-el Baba M, Keller B, et al. , Macrolide-based transgene control in mammalian cells and mice. Nature Biotechnology 2002, 20, 901–907. [DOI] [PubMed] [Google Scholar]
- [85].Yin J, Yang L, Mou L, Dong K, et al. , A green tea-triggered genetic control system for treating diabetes in mice and monkeys. Science Translational Medicine 2019, 11. [DOI] [PubMed] [Google Scholar]
- [86].Fussenegger M, Morris RP, Fux C, Rimann M, et al. , Streptogramin-based gene regulation systems for mammalian cells. Nat Biotechnol 2000, 18, 1203–1208. [DOI] [PubMed] [Google Scholar]
- [87].Wang H, Ye H, Xie M, Daoud El-Baba M, Fussenegger M, Cosmetics-triggered percutaneous remote control of transgene expression in mice. Nucleic Acids Research 2015, 43, e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Weber W, Link N, Fussenegger M, A genetic redox sensor for mammalian cells. Metab Eng 2006, 8, 273–280. [DOI] [PubMed] [Google Scholar]
- [89].Weber W, Schoenmakers R, Spielmann M, El-Baba MD, et al. , Streptomyces-derived quorum-sensing systems engineered for adjustable transgene expression in mammalian cells and mice. Nucleic Acids Res 2003, 31, e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Neddermann P, Gargioli C, Muraglia E, Sambucini S, et al. , A novel, inducible, eukaryotic gene expression system based on the quorum-sensing transcription factor TraR. EMBO Reports 2003, 4, 159–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Gitzinger M, Kemmer C, El-Baba MD, Weber W, Fussenegger M, Controlling transgene expression in subcutaneous implants using a skin lotion containing the apple metabolite phloretin. Proceedings of the National Academy of Sciences of the United States of America 2009, 106, 10638–10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Gitzinger M, Kemmer C, Fluri DA, El-Baba MD, et al. , The food additive vanillic acid controls transgene expression in mammalian cells and mice. Nucleic Acids Research 2012, 40, e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Das AT, Tenenbaum L, Berkhout B, Tet-On Systems For Doxycycline-inducible Gene Expression. Current Gene Therapy 2016, 16, 156–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Gossen M, Bujard H, Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proceedings of the National Academy of Sciences of the United States of America 1992, 89, 5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Krueger C, Pfleiderer K, Hillen W, Berens C, Tetracycline derivatives: alternative effectors for Tet transregulators. BioTechniques 2004, 37, 546, 548, 550. [DOI] [PubMed] [Google Scholar]
- [96].Nevozhay D, Zal T, Balazsi G, Transferring a synthetic gene circuit from yeast to mammalian cells. Nature Communications 2013, 4, 1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Gu X, He D, Li C, Wang H, Yang G, Development of Inducible CD19-CAR T Cells with a Tet-On System for Controlled Activity and Enhanced Clinical Safety. International Journal of Molecular Sciences 2018, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Ye H, Xie M, Xue S, Charpin-El Hamri G, et al. , Self-adjusting synthetic gene circuit for correcting insulin resistance. Nature Biomedical Engineering 2017, 1, 0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Xue S, Yin J, Shao J, Yu Y, et al. , A Synthetic-Biology-Inspired Therapeutic Strategy for Targeting and Treating Hepatogenous Diabetes. Molecular Therapy : The Journal of the American Society of Gene Therapy 2017, 25, 443–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Trevaskis JL, Griffin PS, Wittmer C, Neuschwander-Tetri BA, et al. , Glucagon-like peptide-1 receptor agonism improves metabolic, biochemical, and histopathological indices of nonalcoholic steatohepatitis in mice. American Journal of Physiology. Gastrointestinal and Liver Physiology 2012, 302, G762–772. [DOI] [PubMed] [Google Scholar]
- [101].Lee KH, Oghamian S, Park JA, Kang L, Laird PW, The REMOTE-control system: a system for reversible and tunable control of endogenous gene expression in mice. Nucleic Acids Research 2017, 45, 12256–12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Mullick A, Xu Y, Warren R, Koutroumanis M, et al. , The cumate gene-switch: a system for regulated expression in mammalian cells. BMC Biotechnology 2006, 6, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Eaton RW, p-Cymene catabolic pathway in Pseudomonas putida F1: cloning and characterization of DNA encoding conversion of p-cymene to p-cumate. Journal of Bacteriology 1997, 179, 3171–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Poulain A, Perret S, Malenfant F, Mullick A, et al. , Rapid protein production from stable CHO cell pools using plasmid vector and the cumate gene-switch. Journal of Biotechnology 2017, 255, 16–27. [DOI] [PubMed] [Google Scholar]
- [105].Luger AL, Sauer B, Lorenz NI, Engel AL, et al. , Doxycycline Impairs Mitochondrial Function and Protects Human Glioma Cells from Hypoxia-Induced Cell Death: Implications of Using Tet-Inducible Systems. International Journal of Molecular Sciences 2018, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Jacobs CL, Badiee RK, Lin MZ, StaPLs: versatile genetically encoded modules for engineering drug-inducible proteins. Nature Methods 2018, 15, 523–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Kleinjan DA, Wardrope C, Nga Sou S, Rosser SJ, Drug-tunable multidimensional synthetic gene control using inducible degron-tagged dCas9 effectors. Nature Communications 2017, 8, 1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Chung HK, Jacobs CL, Huo Y, Yang J, et al. , Tunable and reversible drug control of protein production via a self-excising degron. Nature Chemical Biology 2015, 11, 713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Heinis C, Johnsson K, Using peptide loop insertion mutagenesis for the evolution of proteins. Methods in Molecular Biology 2010, 634, 217–232. [DOI] [PubMed] [Google Scholar]
- [110].Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M, An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nature Methods 2009, 6, 917–922. [DOI] [PubMed] [Google Scholar]
- [111].Lee EF, Czabotar PE, Smith BJ, Deshayes K, et al. , Crystal structure of ABT-737 complexed with Bcl-xL: implications for selectivity of antagonists of the Bcl-2 family. Cell Death and Differentiation 2007, 14, 1711–1713. [DOI] [PubMed] [Google Scholar]
- [112].Hill ZB, Martinko AJ, Nguyen DP, Wells JA, Human antibody-based chemically induced dimerizers for cell therapeutic applications. Nature Chemical Biology 2018, 14, 112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Bojar D, Scheller L, Hamri GC, Xie M, Fussenegger M, Caffeine-inducible gene switches controlling experimental diabetes. Nature Communications 2018, 9, 2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Saxena P, Heng BC, Bai P, Folcher M, et al. , A programmable synthetic lineage-control network that differentiates human IPSCs into glucose-sensitive insulin-secreting beta-like cells. Nat Commun 2016, 7, 11247. [DOI] [PMC free article] [PubMed] [Google Scholar]