Abstract
The 70% ethanolic extracts from eight neglected fruits; Muntingia calabura, Leucaena leucocephala, Spondias dulcis, Syzygium jambos, Mangifera caesia, Ardisia elliptica, Cynometra cauliflora and Ficus auriculata were evaluated for their 2,2-diphenyl-1-picrylhydrazyl (DPPH) free radical scavenging, α-glucosidase inhibitory activities as well as total phenolic content. The results of this study revealed that M. caesia fruit extract demonstrated the most potent radical scavenging activity. Among the fruits examined for α-glucosidase inhibitory activity, M. calabura and F. auriculata exhibited strong activity with no significant difference. The Pearson correlation indicated that the activities of M. caesia and F. auriculata contributed by phenolic compounds. A total of 65 metabolites were tentatively identified by using ultra-high-performance liquid chromatography tandem mass spectrometry (UHLPC-MS/MS). These findings suggested that the possible application of M. caesia and F. auriculata as a functional food with antioxidant and α-glucosidase inhibitory properties.
Keywords: TPC, DPPH radical scavenging, α-Glucosidase inhibitory, UHPLC-MS/MS
Introduction
Diabetes mellitus (DM) is a group of non-communicable disease that can be categorized into two types, type 1 and type 2. In both types, the body has difficulty transporting blood sugar to the cells, which causes glucose level in the blood to remain elevated while the cells begin to starve (Akhtar et al., 2018). According to the World Health Organization (WHO) in (2018), approximately 150 million people are currently suffering from this disease, and this number might double by the year 2025.
Many factors can contribute to the increasing incidence of DM, mostly are due to the oxidative stress induced by free radical formation that can cause β-cells of the pancreas to malfunction, insulin resistance and impaired glucose tolerance (Akhtar et al., 2018). Moreover, the digestion of dietary carbohydrates that release glucose also leads to postprandial hyperglycemia (Anjum and Tripathi, 2019). α-Amylase and α-glucosidase are crucial enzymes that breakdown carbohydrates and help intestinal absorption (Akhtar et al., 2018). Apart from oxidative stress and carbohydrates digestion lead to DM, the aging process, consuming unhealthy diets and a sedentary lifestyle are also risk factors that lead to these diabetes-related illnesses. Antioxidants are preventative treatments that can reduce the complications associated oxidative stress (Lawal et al., 2017). Antioxidants are substances that stabilized free radicals and may protect cells from undesirable changes or cellular structure damages. Many previous studies suggest that antioxidants and α-glucosidase inhibitors from natural sources especially fruits may exert antidiabetic effects, hence these natural compounds have gained the attention of researchers worldwide (Putri et al., 2017; Muniyandi et al., 2019). Several of the reported classes of compound that displayed various biological effects are including phenolics, tannins and anthocyanins, which are the most important groups of secondary metabolites (Muniyandi et al., 2019). Hence, this has led to an influx interest to study and identify antioxidant and antidiabetic agents from natural sources including fruits and plants (Akhtar et al., 2018).
Malaysia is recognized as a country with a vast diversity of flora and fauna. The various species of plants, animals and microorganisms offer a great source of nutritious food and medicines. In addition to the commonly consumed local plants and fruits, there are also neglected fruits species that have the potential to act as alternative sources of micronutrients, and bioactive plant metabolites. In traditional medicine, many of these neglected fruits have been used to treat various diseases, such as treating wounds, hemorrhage, dysentery, gastrointestinal problems, alleviate cold and fever, as well as diabetes and several cancers (El-Fishawy et al. 2011; Puangpradab et al., 2018). These fruits including “ceri hutan” (Muntingia calabura L. (Muntingiaceae)), “petai belalang” (Leucaena leucocephala (Lam.) de Wit (Fabaceae)), “kedondong” (Spondias dulcis Parkinson (Anacardiaceae)), “jambu mawar” (Syzygium jambos L. (Myrtaceae)), “binjai” (Mangifera caesia Jack (Anacardiaceae)), “mata itik” (Ardisia elliptica Thunb. (Primulaceae)), “katak puru” (Cynometra cauliflora L. (Fabaceae)), “ara” (Ficus auriculata Lour. (Moraceae)) and others. Nevertheless, biological activity and detail metabolite characterization of these neglected fruits are still lacking and has yet to be determined. Due to this reason, the present study aimed to determine DPPH radical scavenging and α-glucosidase inhibitory activities as well as the total phenolic content (TPC) of the selected local neglected fruits. In addition, the active extracts were profiled using ultra-high-performance liquid chromatography tandem mass spectroscopy (UHPLC-MS/MS) to obtain a better insight into the chemical constituents that could be contributing to the activity.
Materials and methods
Chemical reagents
Absolute ethanol, Folin-Ciocalteu (FC) reagent, LCMS grade (water, methanol, acetonitrile), formic acid (FA) and gallic acid (GA) were purchased from Merck Millipore International (Darmstadt, Germany). Sodium carbonates, quercetin, 2,2-diphenyl-1-picrylhydrazyl (DPPH), p-nitro-phenyl-α-D-glucopyranose (PNPG), sodium phosphate monobasic monohydrate, sodium phosphate dibasic, glycine and α-glucosidase enzyme were obtained from Sigma (Aldrich, Germany). Water was purified by a MiliO system (Millipore Bedford, USA).
Fruit materials
The fruit materials (ripen stage) of M. calabura (SK 3345/18), L. leucocephala (MFI 0079/19), S. dulcis (MFI 0065/19), S. jambodpfys (MFI 0053/19), A. elliptica (MFI 0054/19), C. cauliflora (SK 1757/11) and F. auriculata (MFI 0146/19) were obtained from Jengka, Pahang, meanwhile M. caesia (MFI 0148/19) were collected from Segamat, Johor in February 2018. All of the fruits sample were authenticated by Dr. Mohd Firdaus Ismail, an in-house botanist at Biodiversity Unit, Institute of Bioscience, Universiti Putra Malaysia. The fruits (including peels) were washed and sliced into small pieces then were dried in oven (Smoke Master, Technical Cooperation, Japan) under controlled temperature at 40 °C and left until the constant weight achieved. The samples were then ground into fine powder, sieved using a 250 µm and stored in the chiller at 4 °C until further use.
Fruit extraction
The extraction process was followed method by Mediani et al. (2014) with some modification. The extraction was carried out by weighing 10 g of the dried fruits powder and adding 100 mL of 70% ethanol. The mixture was subjected to sonication at controlled temperature (40 °C) using ultrasonic bath cleaner Thermo–10D Ultrasonic Cleaner, (Fisher scientific, USA) for 1 h and filtered using Whatman filter paper No.1 before subjected to rotary vacuum evaporator (Buchi Laboratoriums Technik, Flawil, Switzerland) to obtain the concentrated crude extract. Then, the same procedure was repeated twice later for 30 min to obtain the maximum yield of extraction. The crude extracts were weighed and subjected to freeze-drying ScanVac CoolSafe Freeze DryerTM (Labogene, Lynge, Denmark) for 3 days, then stored in the chiller at 4 °C until further analysis.
Total phenolic content assay
This assay was performed by following to Mediani et al. (2014) with some modifications. GA was used as a standard and a standard curve was obtained in determination the TPC of the fruit extracts. Briefly, 20 µL of each sample (three replicates) using six serial dilutions starting with 100 µg/mL of a stock was mixed with 100 µL of FC reagent in a 96-well plate. The mixture was left at room temperature for 5 min incubation to react. After that, 80 µL of 7.5% sodium carbonate was added to each well. Then, the plate was incubated in the dark for 30 min before the absorbance was measured at 750 nm using microplate reader Tecan Infinite F200 Pro plate reader (Tecan Group Ltd, Männedorf, Switzerland). The results were expressed in mg of gallic acid equivalents per gram of crude extract (mg GAE/g crude extract).
Free radical scavenging assay
The assay was performed as described by Mediani et al. (2014). Then, 100 µL of DPPH (80 mg/L) was added to 50 µL of test extract (330-80 µg/mL) or quercetin (positive control) in 96-well microplate and the mixture has been left to stand in the dark for 30 min at room temperature. The absorbance was recorded by using Infinite F200 Pro microplate reader (Tecan, Männedorf, Switzerland) at 517 nm. All tests were performed in triplicates. The result was expressed in IC50 value as µg/mL of crude extract, which indicates the concentration of sample required to scavenge 50% of DPPH free radicals.
α-Glucosidase inhibitory activity assay
The α-glucosidase inhibition assay was conducted with some modifications as described by Lawal et al. (2017). The fruit extracts are prepared at 100 µg/mL (stock) with six serial dilutions. The α-glucosidase enzyme (0.02 U/well) and PNPG substrate (1 mM) were prepared in 50 mM phosphate buffer (pH 6.5). Then, a 10 µL of the enzyme was mixed with 10 µL of the test sample and 130 µL of 30 mM phosphate buffer in 96-well plate. The negative control was prepared by substituting the test sample with solvent, meanwhile blank sample (140 µL of 30 mM phosphate buffer and 10 µL of sample) and blank solvent (140 µL of 30 mM phosphate buffer and 10 µL of solvent). The mixture was incubated at room temperature for 5 min. Then, 50 µL of PNPG was added into each well of test sample, negative and positive controls while the others were loaded with 50 µL of 30 mM phosphate buffer (pH 6.5). After a 15 min of the incubation at room temperature, the reaction was stopped by adding 50 μL of 2 M glycine (pH 10). The percentage inhibition was calculated as % = [(an − as)/an] × 100%, an is the absorbance difference value between negative control and the blank, whereas as is the absorbance difference value between sample and the blank. Quercetin was used as positive control and analyses were performed in triplicates. The results were expressed in IC50 value as µg/mL.
UHPLC-MS/MS analysis
The UHPLC-MS/MS analysis of the active extracts was acquired following the method previously described by Lawal et al. (2017). 100 mg of the extract was dissolved in 1 mL of LCMS grade absolute methanol following by sonication and filtration through a 0.22 µm PTFE membrane into a 2 mL screw-capped sample vial. The molecular ion identification is achieved using a ThermoFisher Scientific™ Model Q Exactive™ Hybrid Quadrupole-Orbitrap mass spectrometry (San Jose, CA, USA) equipped with electrospray ionization (ESI) source coupled to an UPLC binary pump, a diode array detector (DAD) (200–650 nm range, 5 nm bandwidth) and an auto-sampler. The column used for the reversed phase was ACQUITY UPLC HSS T3 (1.8 μm, 2.1 × 150 mm). The mobile phase used was consisted of LCMS grade water (solvent A) and acetonitrile (solvent B) contained 0.1% FA. The flow rate was 0.4 mL/min with the injection volume of 2 µL and the column temperature was maintained at 40 °C. The gradient program started with 5-100% solvent B from 0 to 35 min. The MS analytical conditions were as follows: spray volt-pressure—4.0 kV; equipment temperature—29 °C; capillary temperature—350 °C; auxiliary gas at 40 units; sheath gas at 80 units; scan range 150-1500 m/z and collision-induced dissociation (CID) energy was adjusted to 30%. The data recorded and processed using Thermo Xcalibur Qual Browser software 4.0.
Statistical analysis
The biological activity results were expressed as mean ± standard deviation of three replicates. The analysis of significant difference among the results were obtained by analysis of variance (ANOVA) and correlation analysis was done by Pearson’s correlation analysis using Minitab software (Version 16, Minitab Inc., State College, PA, USA).
Results and discussion
Total phenolic content
Phenolic compounds found in most plants and fruits are beneficial for human diets due to their potential biological activities. The TPC is an important indicator for determining the amounts of antioxidants present in samples (Sanchez-Rabaneda et al., 2003). The TPCs of the tested fruits extract are presented in Table 1; they varied from 8.97 to 147.99 mg GAE/g crude extracts. In this study, the highest TPC was noted for L. leucocephala followed by C. cauliflora and A. elliptica (122.04 and 113.73 mg GAE/g crude extract, respectively), meanwhile S. jambos had the lowest TPC with no significant different (p > 0.05) with S. dulcis (9.65 mg GAE/g crude extract). The TPCs of F. auriculata, M. caesia and M. calabura had a significant different (p < 0.05), with values of 65.72, 48.54, and 38.95 mg GAE/g crude extract, respectively. The study by Chew et al. (2011) also reported the presence of TPC in aqueous methanolic extract from L. leucocephala fruit. However, the ethanolic extract in this study able to extract better phenolic content from L. leucocephala than methanolic extract. Moreover, 80% methanolic and acetone extracts of S. jambos were also reported to have lower TPC compared with current study (Hainida et al., 2009; Saikia et al., 2016). The variation in the results with previous studies may be due to the differences in the types of the phenolic compounds present in the diverse plant materials and their solubility in various organic solvents (Santhirasegaram et al., 2015). Variations of the extraction method, temperature and time could also influence the extraction efficiency of phenolic compounds (Abd Ghafar et al., 2018).
Table 1.
Total phenolic content, DPPH free radical scavenging and α-glucosidase inhibitory activities of the fruits extract
| Samples | Total phenolic content (mg GAE/crude extract) | DPPH radical scavenging activity (IC50, µg/mL) | α-Glucosidase inhibitory activity (IC50, µg/mL) |
|---|---|---|---|
| M. calabura | 38. 95 ± 3.45d | 8.49 ± 0.24b | 0.10 ± 0.01a |
| L. leucocephala | 147.99 ± 2.60a | 13.66 ± 0.55d | 3.43 ± 0.05e |
| S. dulcis | 9.65 ± 0.51e | 27.14 ± 0.63f | 4.73 ± 0.22f |
| S. jambos | 8.97 ± 0.31e | 24.44 ± 0.61e | 0.67 ± 0.04b |
| M. caesia | 48.54 ± 1.75d | 4.55 ± 0.40a | 23.93 ± 1.28 h |
| A. elliptica | 113.73 ± 4.92b | 13.45 ± 0.92d | 1.17 ± 0.07c |
| C. cauliflora | 122.04 ± 3.17b | 11.33 ± 0.15c | 3.01 ± 0.19d |
| F. auriculata | 65.72 ± 2.18c | 7.74 ± 0.42b | 0.12 ± 0.01a |
| Quercetin | – | 7.77 ± 0.09b | 6.04 ± 1.08 g |
Values are the mean ± standard deviation of triplicates. Mean with different subscript letter indicates the samples are significantly different (p < 0.05)
DPPH free radical scavenging activity
The generation of free radicals in human biological systems is usually associated with the development of chronic diseases; these free radicals can be scavenged by antioxidant agents (Gomathi et al., 2013). The free radical scavenging activity of the eight neglected fruit extracts are presented in Table 1. Interestingly, M. caesia and F. auriculata extracts revealed strong antioxidant activity with IC50 values of 4.55 and 7.74 µg/mL, respectively. In contrast, Mirfat et al. (2016) and Puangpradab et al. (2018) reported that M. caesia and F. auriculata fruit extracts have lower radical scavenging activity. The differences in activity of these extracts might be due to the different solvent system and method of extraction used. The present study reveals that the efficiency of the sonication-assisted extraction compared with shaking which can affect the extractable bioactive compounds (Saifullah et al., 2020). M. calabura showed IC50 value of 8.49 µg/mL with no significant different (p > 0.05) with F. auriculata. Meanwhile, S. dulcis and S. jambos demonstrated the lowest activity with IC50 values of 27.14 and 24.44 µg/mL, respectively, with no significant different between each other (p > 0.05). Pearson correlation analysis was conducted to investigate the relationship between TPC and antioxidant activity of M. caesia and F. auriculata extracts. Both extracts showed strong positive correlation between TPC and free radical scavenging assay with R values of 0.70 and 1.00, respectively. This finding suggested that the phenolic compounds in both extracts might contribute to the antioxidant activity and was in agreement with previous studies (Maity et al., 2013; Mediani et al., 2014; Yao et al., 2004).
α-Glucosidase inhibitory activity
One of the enzymes involved in carbohydrates digestion is α-glucosidase, which breaks down oligosaccharides and disaccharides into absorbable monomers for intestinal absorption (Anjum and Tripathi, 2019). Inhibition of this enzyme can effectively reduce postprandial blood glucose levels, especially in type 2 diabetic patients. The α-glucosidase inhibitory activity of the extracts is presented in Table 1. M. calabura and F. auriculata extracts showed strong α-glucosidase inhibitory activity with IC50 values of 0.10 and 0.12 µg/mL, respectively, without a statistical difference (p > 0.05). Interestingly, the IC50 values shown by most of the fruit extracts were lower than the quercetin standard, with exception for M. caesia fruit (IC50 value of 23.93 µg/mL). The effectiveness of the inhibitory activity towards α-glucosidase enzyme were as follows: M. calabura > F. auriculata > S. jambos > A. elliptica > C. cauliflora > L. leucocephala > S. dulcis > M. caesia. The antidiabetic activity of M. calabura fruit extract has been supported by McCune et al. (2011) using a different assay. They also reported that this fruit extract has the potential to prevent diabetes, cancer, cardiovascular diseases and other inflammatory diseases. Gomathi et al. (2013) also concluded that M. calabura extract could be a potential source of bioactive compounds for anti-inflammatory-related diseases. The α-glucosidase inhibitory activity of different polarity fractions of F. auriculata fruit also have been reported by Anjum and Tripathi, (2019). The methanol fraction of the extract showed a strong inhibition towards α-glucosidase enzyme compared to the other fractions. Pearson correlation analysis showed negative correlation between TPC and α-glucosidase inhibitory activity of M. calabura extract with R value of -0.42. This indicates that another group of compounds could be the major contributor as α-glucosidase inhibitors of M. calabura or might attributed due to synergistic effects of various compounds (Nor-Azman et al., 2018). Meanwhile, F. auriculata showed strong positive correlation (R value of 1.00) to the α-glucosidase inhibitory activity, which implying the presence of phenolic compounds in the extract responsible for the bioactivity (Abd Ghafar et al., 2018; Anjum and Tripathi, 2019).
Identification of compounds in the active fruit extracts by UHPLC-MS/MS
M. caesia and F. auriculata fruit extracts exhibited the most significant activity towards free radical scavenging and α-glucosidase inhibitory, respectively as compared to the other fruit extracts. Hence, M. caesia, and F. auriculata fruit extracts were subjected to UHPLC-MS/MS analysis to profile the metabolites present in the extracts in order to obtain a better insight into the chemical constituents that could be contributing to the activity. M. calabura fruit extract was not analyzed due to inadequate amount of sample.
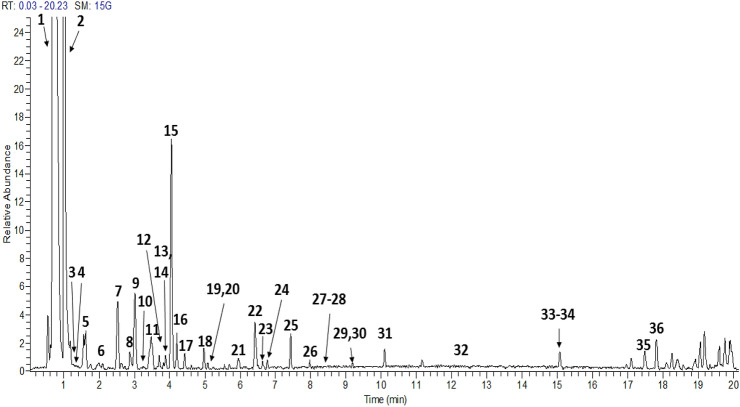
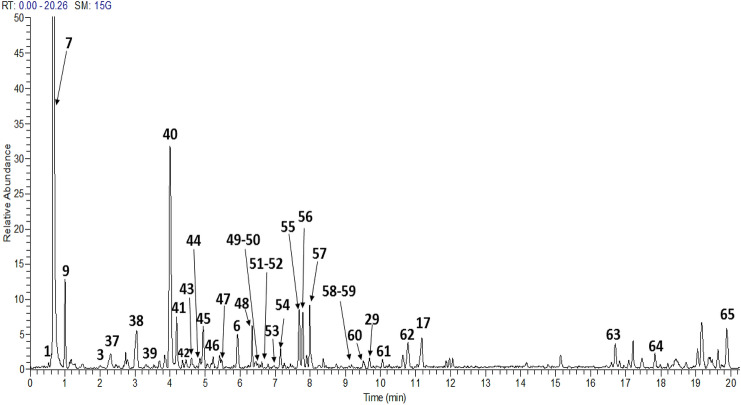
In the MS analysis, generally the glycoside linkage gets cleaved, resulting in the loss of several mass fragments at m/z 176 amu (glucuronic acid), 162 amu (hexose, glucose, galactose), 146 amu (deoxyhexose, rhamnose) and 132 amu (pentose, xylose, arabinose) (Maity et al., 2013). The total ion chromatograms (TIC) of both fruit extracts are presented in Fig. 1 (M. caesia) and Fig. 2 (F. auriculata), while Table 2 (M. caesia) and Table 3 (F. auriculata) summarizing the retention time (RT), MS/MS data and the identified metabolites. For convenience, the peak numbers were assigned to the respective compounds. A total of 65 metabolites from both fruit extracts were tentatively identified based on the mass fragmentation data in comparison with literature and online databases. All the metabolites that were detected and identified were in the negative ion mode [M-H]−.
Fig. 1.
Total ion chromatogram profile of 70% ethanol of M. caesia fruit extract. For peak assignments, see Table 2
Fig. 2.
Total ion chromatogram profile of 70% ethanol of F. auriculata fruit extract. For peak assignments, see Table 3
Table 2.
Tentative identification of metabolites presents in M. caesia fruit extract
| Peak No. | RT (min) | MF | MW (g/mol) | Theoretical mass (m/z) | Measured mass [M-H]− | Delta | UV (nm) | MS/MS Fragment ions | Tentative identification | References |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.66 | C15H18O9 | 342.29 | 341.0878 | 341.1073 | 0.0195 | 266 | 179, 89, 59 | Caffeoyl glucose | Santhirasegaram et al. (2015) |
| 2 | 0.97 | C15H18O9 | 342.29 | 341.0878 | 341.1079 | 0.0201 | 206 | 179, 89, 59 | Caffeoyl glucose isomer | Santhirasegaram et al. (2015) |
| 3 | 1.19 | C13H16O10 | 332.26 | 331.0670 | 331.0660 | 0.0010 | 278 | 169, 125, 107 | Monogalloyl glucose | Santhirasegaram et al. (2015) |
| 4 | 1.56 | C7H6O5 | 170.11 | 169.0143 | 169.0129 | 0.0014 | 214, 272 | 12, 69, 67 | Gallic acid | Maity et al. (2013); Santhirasegaram et al. (2015) |
| 5 | 1.62 | C7H6O5 | 170.11 | 169.0143 | 169.0128 | 0.0015 | 214, 272 | 125, 69, 67 | Gallic acid isomer | Maity et al. (2013); Santhirasegaram et al. (2015) |
| 6 | 2.28 | C6H8O6 | 176.12 | 175.0397 | 174.9549 | 0.0848 | 210 | 146, 118, 115, 87, 59 | 4-hydroxy-6-methylcoumarin | Abd Ghafar et al. (2018) |
| 7 | 2.57 | C7H12O6 | 192.16 | 191.0561 | 191.0184 | 0.0377 | 212 | 111, 87, 85 | Quinic acid | Santhirasegaram et al. (2015) |
| 8 | 2.87 | C22H18O14 | 506.37 | 505.2595 | 505.1158 | 0.1437 | 216 | 331, 169 | Galloyl hexose derivative | Abu-Reidah et al. (2015) |
| 9 | 3.08 | C7H12O6 | 192.16 | 191.0561 | 191.0185 | 0.0376 | 214 | 111, 87, 85 | Quinic acid isomer | Santhirasegaram et al. (2015) |
| 10 | 3.20 | C76H52O46 | 1701.19 | 1699.1657 | 183.0286 | ND | 214, 276 | 140, 124, 78 | Tannic acid | Santhirasegaram et al. (2015); Ertas et al. (2014) |
| 11 | 3.49 | C14H6O8 | 302.19 | 300.9990 | 300.0389 | 0.9601 | 214, 276 | 282, 256, 230, 185 | Ellagic acid | Santhirasegaram et al. (2015) |
| 12 | 3.72 | C10H8O3 | 176.16 | 175.0401 | 175.0598 | 0.0197 | 216, 270 | 146, 131, 118, 115, 113, 85 | Herniarin/7-methoxycoumarin | Ahmad & Misra (1997) |
| 13 | 3.88 | C6H8O6 | 176.12 | 175.0397 | 174.9547 | 0.0846 | 214, 278 | 146, 118, 115, 87, 59 | 4-hydroxy-6-methylcoumarin isomer | Abd Ghafar et al. (2018) |
| 14 | 3.89 | C21H22O12 | 466.39 | 465.1039 | 465.1022 | 0.0017 | 214, 276 | 303, 285 | 3,4′,5,6,7-Pentahyroxyflavanone-O-hexoside | Dias et al. (2010) |
| 15 | 4.05 | C44H46O22 | 926.82 | 925.2336 | 925.3477 | 0.1141 | 214, 274 | 609, 293 | Peonidin 3-O-galactoside (dimer) | Berardini et al. (2005) |
| 16 | 4.19 | C15H12O7 | 304.25 | 303.0510 | 303.0500 | 0.0010 | 214, 282 | 285, 259, 193, 151 | Taxifolin/Dihydroquercetin | Ye et al. (2012) |
| 17 | 4.45 | C16H12O6 | 300.26 | 299.0561 | 299.1126 | 0.0565 | 216, 276 | 299, 256, 229, 175, 89, 59 | Gliricidin | Ye et al. (2012) |
| 18 | 4.99 | C21H22O11 | 450.39 | 449.1087 | 449.1074 | 0.0013 | 214, 274 | 287, 269, 151, 135, 125 | Flavanomarein | Dias et al. (2010) |
| 19 | 5.08 | C21H22O11 | 450.39 | 449.1087 | 449.1077 | 0.0010 | 216, 274 | 287, 269, 151, 125 | Flavanomarein isomer | Dias et al. (2010) |
| 20 | 5.09 | C22H20O10 | 444.38 | 443.0984 | 443.0794 | 0.0190 | 216, 276 | 281, 237, 167, 112 | Rothindin/Psedobaptigenin-7-O-glucoside | Lin et al. (2000) |
| 21 | 6.10 | C21H20O12 | 464.37 | 463.0882 | 463.0877 | 0.0005 | 218, 276 | 300, 271, 255, 151 | Quercetin-O-hexoside | Kumar et al. (2015) |
| 22 | 6.45 | C21H20O12 | 464.37 | 463.0882 | 463.0865 | 0.0017 | 218, 270 | 316, 287, 271, 178, 151 | Myricitrin | Kumar et al. (2015) |
| 23 | 6.65 | C21H20O12 | 464.37 | 463.0882 | 463.2535 | 0.1653 | 218, 276 | 301, 300, 271, 255 | Quercetin-O-hexoside isomer | Kumar et al. (2015) |
| 24 | 6.78 | C15H12O6 | 288.25 | 287.0561 | 287.1279 | 0.0718 | 218, 276 | 269, 243, 225, 161, 107 | 3,7,3′,4′-Tetrahydroxyflavanone | Ye et al. (2012) |
| 25 | 7.40 | C21H20O11 | 448.37 | 447.0933 | 447.0921 | 0.0012 | 218, 270 | 301, 300, 271, 255, 178, 151 | Quercetin-3-O-rhamnoside | Kumar et al. (2015) |
| 26 | 7.72 | C15H12O5 | 272.25 | 271.0612 | 271.0494 | 0.0118 | 220, 276 | 241, 227, | Rubrofusarin | Fathalla et al. (2018) |
| 27 | 8.27 | C21H20O10 | 432.37 | 431.0981 | 431.0972 | 0.0009 | 220, 276 | 285, 284, 255, 227 | Kaempferol-3-O-rhamnoside | Zhou et al. (2018) |
| 28 | 8.32 | C21H20O10 | 432.37 | 431.0981 | 431.0972 | 0.0009 | 220, 276 | 285, 284, 255, 227, 178 | Kaempferol-3-O-rhamnoside isomer | Zhou et al. (2018) |
| 29 | 9.51 | C15H10O7 | 302.23 | 301.0354 | 301.0344 | 0.0010 | 222 | 273., 178, 151,121, 108 | Quercetin | Kumar et al. (2015) |
| 30 | 9.57 | C15H10O7 | 302.23 | 301.0354 | 301.0343 | 0.0011 | 222 | 273, 178, 151,121, 108 | Quercetin isomer | Kumar et al. (2015) |
| 31 | 10.66 | C15H12O5 | 272.25 | 271.0612 | 271.0602 | 0.0010 | 222 | 151, 135, 119, 107 | Butin | Jin et al. (2015) |
| 32 | 12.43 | C22H23O11 | 463.41 | 462.1168 | 461.2166 | 0.9002 | 222 | 299, 283, 255, 245, 193, 164. | Gliricidin-O-hexoside | Ye et al. (2012) |
| 33 | 15.04 | ND | ND | ND | 304.9128 | ND | 222 | 174, 146 | Cinnamic acid derivative | Hofmann et al. (2016) |
| 34 | 15.09 | ND | ND | ND | 304.9126 | ND | 222 | 174, 146 | Cinnamic acid derivative isomer | Hofmann et al. (2016) |
| 35 | 17.47 | C26H26O16 | 594.47 | 593.4678 | 593.2713 | 0.1965 | 224 | 315, 277, 241 | Isorhamnetin hexose-malic acid | Abu-Reidah et al. (2015) |
| 36 | 17.79 | C16H12O4 | 268.26 | 267.0663 | 267.1956 | 0.1293 | 224 | 252, 223, 133 | Formononetin | Ye et al. (2012) |
Delta is defined as the absolute difference between theoretical mass and measured mass of compound. ND is not determined
Table 3.
Tentative identification of metabolites presents in F. auriculata fruit extract
| Peak No. | RT (min) | MF | MW (g/mol) | Theoretical mass (m/z) | Measured mass [M-H]− | Delta | UV (nm) | MS/MS Fragment ions | Tentative identification | References |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.67 | C15H18O9 | 342.29 | 341.0878 | 341.1074 | 0.0196 | ND | 179, 119, 89, 85, 71, 59 | Caffeoyl glucose | Santhirasegaram et al. (2015) |
| 7 | 0.74 | C7H12O6 | 192.16 | 191.0561 | 191.0544 | 0.0017 | 208 | 111, 87, 85 | Quinic acid | Santhirasegaram et al. (2015) |
| 9 | 0.99 | C7H12O6 | 192.16 | 191.0561 | 191.0547 | 0.0014 | 206 | 111, 87, 85 | Quinic acid isomer | Santhirasegaram et al. (2015) |
| 3 | 2.23 | C13H16O10 | 332.26 | 331.0670 | 331.0660 | 0.0010 | 210 | 169, 151, 125, 107 | Monogalloyl glucose | Santhirasegaram et al. (2015) |
| 37 | 2.29 | C13H16O10 | 332.26 | 331.0670 | 331.0659 | 0.0011 | 210 | 169, 151, 125, 107 | Monogalloyl glucose isomer | Santhirasegaram et al. (2015) |
| 38 | 3.03 | C16H18O9 | 354.30 | 353.0878 | 353.0863 | 0.0015 | 216, 324 | 191, 179, 135 | Chlorogenic acid/Caffeoylquinic acid | Hofmann et al. (2016) |
| 39 | 3.12 | C9H18O4 | 180.15 | 179.0350 | 179.0336 | 0.0014 | 218, 284 | 179, 150, 135, 134, 89 | Caffeic acid | Sanchez-Rabaneda et al. (2003) |
| 40 | 3.99 | C16H18O9 | 354.31 | 353.0878 | 353.06555 | 0.0223 | 218, 326 | 191 | 5-caffeoylquinic acid | Dou et al. (2007) |
| 41 | 4.11 | C27H30O15 | 594.51 | 593.1507 | 593.1494 | 0.0013 | 218, 284 | 593, 285, 284, 256, 255 | Kaempferol-3-O-rutinoside | Kumar et al. (2015) |
| 42 | 4.23 | C27H30O17 | 626.51 | 625.1410 | 625.1388 | 0.0021 | 218, 290, 324 | 625, 463, 462, 301, 300, 299 | Quercetin 3,4′-diglucoside | Lawal et al. (2017) |
| 43 | 4.30 | C27H30O15 | 594.51 | 593.1507 | 593.1499 | 0.0008 | 218, 284 | 593, 285, 284, 256, 255 | Kaempferol-3-O-rutinoside isomer | Kumar et al. (2015) |
| 44 | 4.87 | C15H14O6 | 290.26 | 289.0718 | 289.0707 | 0.0011 | 218, 270 | 245, 203, 151, 125, 123, 109 | Catechin | Sanchez-Rabaneda et al. (2003) |
| 45 | 4.93 | C15H14O6 | 290.26 | 289.0718 | 289.0706 | 0.0012 | 218, 278 | 245, 203, 151, 125, 123, 109 | Epicatechin | Sanchez-Rabaneda et al. (2003) |
| 46 | 5.61 | C21H20O11 | 448.37 | 447.0933 | 447.0921 | 0.0012 | 218, 282 | 339, 327, 299, 285, 151 | Luteolin-6-C- β-D-glucoside (Isoorientin) | Sanchez-Rabaneda et al. (2003) |
| 47 | 5.67 | C21H20O11 | 448.37 | 447.0933 | 447.0919 | 0.0014 | 218, 278 | 357, 339, 327, 299, 285, 151 | Luteolin-8-C- glucoside (Orientin) | Sanchez-Rabaneda et al. (2003) |
| 6 | 5.94 | C6H8O6 | 176.12 | 175.0397 | 174.9545 | 0.0852 | 220, 282 | 146, 118 | 4-hydroxy-6-methylcoumarin | Abd Ghafar et al. (2018) |
| 48 | 6.35 | C21H20O10 | 432.37 | 431.0984 | 431.0969 | 0.0015 | 220, 268 | 341, 311, 283, 271, 269 | Apigenin-6-C-glucoside (Vitexin) | Sanchez-Rabaneda et al. (2003) |
| 49 | 6.47 | C21H20O10 | 432.37 | 431.0984 | 431.0968 | 0.0016 | 220, 278 | 353, 341, 311, 283, 269 | Apigenin-8-C-glucoside (Isovitexin) | Sanchez-Rabaneda et al. (2003) |
| 50 | 6.55 | C21H8O13 | 478.35 | 477.0675 | 477.0662 | 0.0013 | 220, 278 | 477, 301, 255, 178, 151, 121 | Quercetin-3-O-glucuronide | Barros et al. (2014) |
| 51 | 6.61 | C21H20O12 | 464.37 | 463.0882 | 463.0870 | 0.0012 | 220, 282 | 300, 271, 255, 151 | Quercetin-3-O-glucoside | Kumar et al. (2015) |
| 52 | 6.89 | C21H20O10 | 432.37 | 431.0984 | 431.1671 | 0.0687 | 218 | 431, 269, 268 | Apigenin-7-O-glucoside | Plazonic et al. (2009) |
| 53 | 6.91 | C20H18O11 | 434.35 | 433.0776 | 433.0766 | 0.0010 | 220, 278 | 301, 300, 271, 255 | Quercetin-3-O-arabinoside | Kumar et al. (2015) |
| 54 | 7.05 | C20H18O11 | 434.35 | 433.0776 | 433.0764 | 0.0012 | 220, 282 | 301, 300, 271, 255 | Quercetin-3-O-arabinoside isomer | Kumar et al. (2015) |
| 55 | 7.69 | C25H24O12 | 516.45 | 515.1195 | 515.1174 | 0.0021 | 220, 328 | 353, 191, 179.0336, 173 | Dicaffeoylquinic acid | Dias et al. (2010) |
| 56 | 7.78 | C20H18O12 | 450.34 | 449.0726 | 449.1076 | 0.0347 | 220, 288 | 287, 151, 135 | Myricetin-O-pentoside | Hofmann et al. (2016) |
| 57 | 7.97 | C21H22O11 | 450.39 | 449.1089 | 449.2020 | 0.0931 | 222, 284 | 287, 286, 199 | 3,5′,5′,7-Tetrahydroxyflavanone-O-hexoside | Dias et al. (2010) |
| 58 | 9.48 | C15H10O6 | 286.23 | 285.0405 | 285.0396 | 0.0009 | 222 | 217, 199, 175, 151, 133, 107 | Luteolin | Sanchez-Rabaneda et al. (2003) |
| 59 | 9.48 | C15H10O6 | 286.23 | 285.0404 | 285.0405 | 0.0000 | 222 | 217, 15, 133 | Kaempferol | Sanchez-Rabaneda et al. (2003) |
| 60 | 9.54 | C15H10O6 | 286.23 | 285.0405 | 285.0394 | 0.0011 | 222 | 217, 199, 175, 151, 133, 107 | Luteolin isomer | Sanchez-Rabaneda et al. (2003) |
| 29 | 9.56 | C15H10O7 | 302.23 | 301.0354 | 301.0351 | 0.0003 | 220 | 273, 178, 151, 121, 108 | Quercetin | Kumar et al. (2015) |
| 61 | 10.00 | C22H18O11 | 458.37 | 457.0776 | 457.1686 | 0.0910 | 220 | 305, 317, 225, 217, 167, 109 | Epigallocatechin gallate | Maity et al. (2013) |
| 62 | 10.79 | C15H10O5 | 270.23 | 269.0456 | 269.0446 | 0.0010 | 222 | 225, 151, 117, 107 | Apigenin | Sanchez-Rabaneda et al. (2003) |
| 17 | 11.22 | C16H12O6 | 300.26 | 299.0561 | 299.0550 | 0.0010 | 222, 288 | 284, 270, 256 | Gliricidin | Ye et al. (2012) |
| 63 | 16.71 | C13H12O9 | 312.22 | 311.0409 | 311.0550 | 0.0141 | 224, 306 | 179, 149, 133, 115 | Caftaric acid | Abu-Reidah et al. (2015); Barros et al. (2014) |
| 64 | 17.82 | C15H16O9 | 340.28 | 339.0722 | 339.0706 | 0.0016 | 224, 316 | 339, 177 | Aesculin | Li et al. (2013) |
| 65 | 19.94 | C15H18O8 | 326.29 | 325.0929 | 325.0916 | 0.0013 | 224, 282 | 269, 189, 183, 145 | p-coumaroylhexose | Kajdzanoska et al. (2010) |
Note: Delta is defined as the absolute difference between theoretical mass and measured mass of compound. ND is not determined
Metabolites profiling of M. caesia fruit extract
Peaks 1 and 2 were tentatively identified as caffeoyl glucose and its isomer, respectively, with deprotonated molecular ions both at m/z 341, which also provided the same fragment ions at m/z 179, 89 and 59. These metabolites exhibited fragmentation patterns with the loss of the glucose moiety at m/z 179 (loss of m/z 162) and were consistent with previous research (Santhirasegaram et al., 2015). Peak 3 was assigned as monogalloyl glucose, gave a deprotonated molecular ion at m/z 331 and yielded fragment ions at m/z 169 (loss of glucose), which originated from gallic acid, 125 (loss of glucose + CO2) and 107 (Santhirasegaram et al., 2015). Another galloyl derivative is galloyl hexose, represented by peak 8 with a deprotonated molecular ion at m/z 505 and characterized by fragment ions at m/z 331 and 169 (Abu-Reidah et al., 2015). Peaks 4 and 5 were assigned as gallic acid and its isomer, respectively, based on the molecular ions at m/z 169 that further showed fragmentation at m/z 125 (loss of COOH), 69 and 67, which was similar with previously described (Abd Ghafar et al., 2018). Peaks 7 and 9 were identified as quinic acid and its isomer, respectively, as they gave an identical deprotonated molecular ion at m/z 191 and fragment ions at m/z 111, 87 and 85 (Santhirasegaram et al., 2015). For peak 10 was identified as tannic acid, the deprotonated molecular ion was found at m/z 183 and displayed the MS/MS data at m/z 140, 124, and 78 (Ertas et al., 2014). Peak 11 was tentatively identified as ellagic acid characterized by having a molecular ion at m/z 300. This metabolite showed similar fragment ions as previously reported at m/z 255 [M-H–CO2], 230 [M-H-CO2-CO], and 185 [M-H-2CO2-CO] (Santhirasegaram et al., 2015). The deprotonated molecular ion for peak 26 was observed at m/z 271 and was identified as rubrofusarin. It gave the fragment ions at m/z 241, suggesting the losses of carbonyl and further cleavage at m/z 227 and m/z 225 to lose methylene and phenolic hydroxy, respectively (Fathalla et al., 2018). Peaks 33 and 34 were identified as an unknown cinnamic acid derivative and a cinnamic acid isomer, respectively. Both showed a precursor ion at m/z 304 and a product ion at m/z 146 (cinnamic acid moiety) (Hofmann et al., 2016). Peak 35 was identified as isorhamnetin hexose-malic acid, which showed a product ion at m/z 315, corresponding to the neutral loss of hexose-malic acid moiety [M-H-278], that provided the isorhamnetin aglycone (Abu-Reidah et al., 2015).
Furthermore, 11 metabolites (peaks 14, 15, 17, 18, 19, 20, 22, 24, 31, 32 and 36) present in the extracts were classified as flavonoids derivatives. For peak 14, the precursor ion was observed at m/z 465 and further fragmentation at m/z 303 (loss of hexose) and 285 (loss of hexose + H2O) (Dias et al., 2010), hence, was tentatively identified as 3,4′,5,6,7-pentahydroxyflavone-O-hexoside. The molecular ion of peak 15 was observed at m/z 925 and identified as a dimer for peonidin-3-O-galactoside (m/z 463). Further fragment ions of this metabolite were congruent with reported data at m/z 609 and 293 (Berardini et al., 2005). Peaks 17 and 32 were assigned as gliricidin and gliricidin-O-hexoside, respectively, with a deprotonated molecular ion at m/z 299 and 461, respectively. Peak 17 presented a molecular ion at m/z 299 and showed fragment ions at m/z 256 and 175, similar to that previously reported by Ye et al. (2012) and was confirmed by comparing with the reference standard in their study. Meanwhile, peak 32 with m/z 461 as the precursor ion, showed the MS/MS data corresponding to the loss of hexose moiety at m/z 299 and the subsequent fragmentation pattern of gliricidin (Ye et al., 2012). Peaks 18 and 19 gave a molecular ion at m/z 449 and identical fragment ions at 287 (loss of glucose) and 269 (loss of glucose + H2O) (Dias et al., 2010). Therefore, these peaks were assigned as flavanomarein and its isomer, respectively. Peak 20 was tentatively identified as rothindin (psedopatigenin-7-O-glucoside), gave a deprotonated molecular ion at m/z 443 and fragment ions at m/z 281 (loss of glucose moiety) as reported previously (Lin et al., 2000). For peak 22, the precursor ion was observed at m/z 463 and characterized by product ions at m/z 316 (loss m/z 146) that corresponded to the loss of sugar moiety through cleavage of the C-O bond and followed by m/z 271 due to the loss of HCO2 (from m/z 316) (Kumar et al., 2015). This peak was identified as myricitrin. In addition, peaks 24 and 31 were tentatively assigned as 3,7,3′,4′-tetrahydroxyflavanone and butin, respectively. For peak 24, it gave a deprotonated molecular ion at m/z 287 and yielded the fragment ion due to the natural losses at m/z 269 [M-H-H2O], m/z 243 [M-H-CO2] and m/z 161 (Ye et al., 2012). Meanwhile, peak 31 presented a deprotonated molecular ion at m/z 271 and exhibited a loss of sugar moiety at m/z 135. A similar fragment ion was observed in a previous result reported by Jin et al. (2015), wherein this compound was identified in comparison with butin standard. Furthermore, peak 36 presented a molecular ion at m/z 267 and yielded MS/MS data at m/z 252 [M-H-CH3], which suggesting that the presence of the methoxyl group, at m/z 223 (loss of CO2) and the ion at m/z 163 indicated that the methoxyl group should be located at the ring B and subsequently identified as formononetin (Ye et al., 2012). This metabolite and its fragment ions were identified in comparison with the refence standard.
Quercetin derivatives have also been identified in M. caesia fruit extracts. All these compounds displayed a common fragmentation ion at m/z 271, corresponding to the loss of CHO or H2CO (Kumar et al., 2015). They were also identified by their characteristics fragment ion at m/z 151 through heterolytic cleavage. These compounds generally displayed fragmentation patterns due to the loss of sugar moiety. For authentication, the molecular ion for peak 16 was observed at m/z 303, which belong to dihydroquercetin, displayed fragment ions similar to those described by Ye et al. (2012), and its product ion at m/z 285 indicated its flavonol type due to the neutral loss of 18 amu from the parent ion. Other quercetin derivatives were also observed at peaks 21, 23, 25, 29 and 30. For peaks 21, 23 and 25, there was a quercetin fragment ion from the loss of glucuronyl (m/z 176), glucosyl (m/z 162) and rhamnosyl (m/z 146) moieties, respectively. Consequently, they were tentatively identified as quercetin-O-hexoside, quercetin-O-hexoside isomer and quercetin-3-O-rhamnoside, respectively. Furthermore, both the observed peaks 29 and 30, had a molecular ion at m/z 301 and exhibited a similar MS/MS spectrum as reported by Kumar et al. (2015). Therefore, these peaks were assigned as quercetin and quercetin isomer, respectively.
In addition, coumarin derivatives were observed in M. caesia fruit extracts at peaks 6, 12 and 13. Peaks 6 and 13 presented a deprotonated molecular ion at m/z 174, which were tentatively identified as 4-hydroxy-6-methylcoumarin and its isomer, respectively. Further fragmentation patterns of the compound at m/z 147 and 119 were observed, that corresponded to the loss of CO and C2O2, respectively, which was consistent with a previous reported (Abd Ghafar et al., 2018). In addition, the deprotonated molecular ion at m/z 175 for peak 12 displayed further product ions at m/z 148 [M-H-CO] and m/z 133, consequently assigned as 7-methoxycoumarin (herniarin), and the fragment ions were confirmed in comparison with the study of Ahmad and Misra, (1997) regarding the isolation of this compound from Matricaria chamomilla flowers. In addition, kaempferol derivatives were identified in the extracts at peaks 27 and 28. Both of these metabolites presented a precursor ion at m/z 431 and displayed identical common fragment ions of kaempferol at m/z 285 (loss of rhamnose), m/z 255 (loss of glucose moiety) and m/z 178. These fragmentations were consistent with previously reported data (Zhou et al. 2018) and were identified as kaempferol-3-O-rhamnoside and its isomer, respectively.
Metabolites profiling of F. auriculata fruit extracts
A total of 36 metabolites were tentatively identified in F. auriculata fruit extract. Peaks 1, 7, 9, 3, 37, 6, 29 and 17 were assigned as caffeoyl glucose, quinic acid, quinic acid isomer, monogalloyl glucose, monogalloyl glucose isomer, 4-hydroxy-6-methylcoumarin, quercetin and gliricidin, respectively. Similarly, these compounds were identified in M. caesia fruit extract and their MS/MS data were consistent as previously described.
Peak 38 provided the molecular ion at m/z 353, which was tentatively identified as chlorogenic acid (caffeoylquinic acid). This compound revealed the fragmentation ions at m/z 191 [quinic acid-H], 179 [caffeic acid-H] and subsequently the loss of CO2 from the transition of m/z 179 to m/z 135 (Hofmann et al., 2016). The deprotonated molecular ion of peak 39 was observed at m/z 179, which belonged to caffeic acid, and loss of CO2 was observed as a characteristic ion at m/z 135 (Sanchez-Rabaneda et al., 2003). In addition, 5-caffeoylquinic acid was assigned for peak 40, which gave a precursor ion at m/z 353 and displayed similar MS/MS data reported at m/z 191, corresponding to the deprotonated quinic acid (Dou et al., 2007). For peak 55, the molecular ion at m/z 515 was identified as dicaffeoylquinic acid, which has a characteristic ion of caffeoylquinic acid (m/z 353), and further fragmented at m/z 191(quinic acid), and 179 (caffeic acid) (Dias et al., 2010). Peak 63 was suggested as caftaric acid as the deprotonated molecular ion at m/z 311 and fragment ions at m/z 179 [M-H-tartaric], 149 [M-H-caffeoyl], 133 and 115 were consistent with reported data (Abu-Reidah et al., 2015). Peaks 64 and 65 were tentatively identified as aesculin and p-coumaroylhexose, respectively. The MS/MS patterns were observed at m/z 339 as the precursor ion and the product ion at m/z 177 for aesculin, suggesting the loss of glucose moiety (m/z 162). Meanwhile, for p-coumaroylhexose, the precursor ion was observed at m/z 325 and further fragmented at m/z 189 and 145, resulting from the loss of glucose unit and m/z 163 belong to p-coumaric acid, which were similarly to those described by Kajdzanoska et al. (2010).
Six metabolites (peaks 42, 50, 51, 53, 54 and 29) were identified as quercetin derivatives based on the presence of aglycone fragment ions at m/z 301 and the characteristic fragment ions at m/z 271 and 151 in their MS/MS spectra (Kumar et al., 2015). Peak 42 presented a deprotonated molecular ion at m/z 625 and fragment ions at m/z 463 (loss of glucose) and 301 (quercetin), which was identified as quercetin-3,4′-diglucoside. This metabolite exhibited the loss of the two glucose moieties at m/z 324 and m/z 162 (Lawal et al., 2017). The molecular ion of peak 50 was obtained at m/z 477 and displayed a fragment ion at m/z 301, indicating the loss of the glucuronic unit (176 amu). Therefore, on the basis of this information, this peak was assigned as quercetin-3-O-glucuronide, which was in agreement with previously reported data (Barros et al., 2014). Peaks 51, 53 and 54 were tentatively identified and characterized as quercetin-3-O-glucoside, quercetin-3-O-arabinoside and quercetin-3-O-arabinoside isomer, with a deprotonated molecular ion at m/z 463 and 433, respectively. These compounds exhibited similar fragment ions at m/z 300, 271 and 255, which was consistent with a previous study by Kumar et al. (2015), corresponding to the losses of glucuronyl (m/z 176), glucosyl (162) and rhamnosyl (146) moieties.
Regarding apigenin derivatives, altogether four compounds (peaks 48, 49, 52 and 62) were found in F. auriculata fruit extracts. These metabolites showed the loss of m/z at 120 and 90 in their MS/MS data, which is the common fragmentation patterns of C-glycoside that corresponded to the cross-ring cleavages in the sugar moiety. Peaks 48 and 49 were assigned as apigenin-8-C-glucoside (vitexin) and apigenin-6-C-glucoside (isovitexin), respectively. Both metabolites showed the same deprotonated molecular ion at m/z 431 and fragmentation ions at m/z 341 (loss of m/z 90) and 311 (loss of m/z 120), which provides evidence of the cross ring cleavage on the glucose moiety of the molecules that produced 1,3 and 1,2 cross-ring glucose attached to the apigenin aglycone, respectively. These fragmentation ions were similar to those reported in a previous study (Sanchez-Rabaneda et al., 2003). The fragment ion that differentiate isovitexin and vitexin was the ion observed at m/z 268 as reported by Sanchez-Rabaneda et al. (2003). Peak 52 with a molecular ion at m/z 431 was identified as apigenin-7-O-glucoside. The fragmentation ion of this compound was observed at m/z 269, resulting in the loss of glucose unit (162 amu) (Plazonic et al., 2009). Peak 62 with a deprotonated molecular ion at m/z 269 was identified as apigenin. Further fragmentation of this molecular ion gave two fragment ions at m/z 225 and 151, which were reported as the base peak for apigenin (Sanchez-Rabaneda et al., 2003).
Furthermore, three kaempferol and one myricetin derivatives were also tentatively identified for peaks 41, 43, 59 and 56, respectively. Both the peaks 41 and 43 showed a deprotonated molecular ion at m/z 593 and were assigned as kaempferol-3-O-rutinoside and its isomer, respectively. For authentication, these compounds were matched with an authentic standard previously reported by Kumar et al. (2015), which displayed a characteristic fragment at m/z 285 corresponding to the loss of sugar moiety. Peak 59 was tentatively identified as kaempferol with a precursor ion at m/z 285 and further fragmented at m/z 217, 151 and 133 which consistent with those described by Sanchez-Rabaneda et al. (2003). Myricetin derivative was observed at peak 56, which was identified as myricetin-O-pentoside. The mass spectra of this metabolite contain all m/z fragments of the aglycone myricetin (m/z 287, m/z 151 and m/z 135), which corresponded to the loss of sugar units (Hofmann et al., 2016).
Other flavonoid compounds were also found in this fruit extract. Peak 44 showed a deprotonated molecular ion at m/z 289 and yielded fragments ion at 245, which was consistent with those reported for catechin (Sanchez-Rabaneda et al., 2003). Meanwhile, peak 45 gave a similar precursor ion and fragmentation patterns as those of catechin and was identified as epicatechin (catechin isomer). These two metabolites showed a loss of the CH2CHOH-group (at m/z 245). Peaks 46 and 47 were identified as isoorientin and orientin, respectively, due to the similar molecular ions at m/z 447 and the similar fragmentation ions at m/z 357, 327, 297 and 285 (Sanchez-Rabaneda et al., 2003). Both metabolites were characterized as C-glycosides of luteolin based on the loss of the glucose unit (m/z 90 and 120). The precursor ion for peak 57 was presented at m/z 449 and the fragmentation ions at m/z 287 [M-H-hexose], as similarly described by Dias et al. (2010). Hence, this peak was assigned as 3,5′,5′,7-tetrahydroxyflavanone-O-hexoside. Peaks 58 and 60 were identified as luteolin and its isomer, respectively, due to the similar deprotonated molecular ion at m/z 285 and the similar fragmentation ions at m/z 175, 151, 133 and 107 as reported previously (Sanchez-Rabaneda et al., 2003). Characterization of the luteolin was compared to the authentic standard compound, which showed product ions at m/z 151 and 133 in the earlier study. Peak 61 was assigned as epigallocatechin gallate as it gave a precursor ion at m/z 457 and the fragmentation ions at m/z 169 and 305, corresponding to gallic acid and epigallocatechin, respectively (Maity et al., 2013).
All the compounds identified in the fruit extracts of both M. caesia and F. auriculata might fulfil the structure–activity relationships for both activities. The differences in the chemical structure of the compounds were closely related to the various arrangement of the hydroxylation, alkylation, methoxylation and glycosylation sites (Abd Ghafar et al., 2018). In addition, substitutions, conjugations, and the degree of polymerization play important roles in determining the nature of the metabolites (Yao et al., 2004). Today, a greater number of studies are focusing on the health aspects of phenolic compounds, especially flavonoids constituents from plants. Numerous epidemiological studies have suggested that consumption of foods rich in phenolic compounds exhibited various biological benefits, including antioxidants, anti-inflammatory, antimicrobial and antiallergic and several other properties as well (Santhirasegaram et al., 2015; Yao et al., 2004). Therefore, the present study has demonstrated that the metabolites identified in the extracts of M. caesia and F. auriculata possess antioxidant activity and α-glucosidase inhibitory activities, respectively.
Overall, this study showed that the eight selected fruits demonstrated potential as natural sources of antioxidants and α-glucosidase inhibitors. Among the fruits tested, M. caesia showed a strong free radical scavenging activity, while M. calabura and F. auriculata possessed strong α-glucosidase inhibitory activity. In this report, 36 compounds were tentatively identified from the M. caesia and 36 compounds were identified from F. auriculata, resulting in a total of 65 different metabolites. The compounds identified comprising a derivative of quercetin, apigenin, kaempferol, myricetin, coumarin and phenolic acids. However, more research needs to be conducted to show the potential uses of these fruits as a natural antidiabetic agent, as they can be incorporated into functional foods and nutraceutical products.
Acknowledgements
This research was supported by grant from Universiti Putra Malaysia (UPM/700/2/1/GPB/2017/9597400) under Putra High Impact Grant Scheme.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Siti Norhamimah Mohamed Yunus, Email: cikmiemayunus@gmail.com.
Faridah Abas, Email: faridah_abas@upm.edu.my.
Ahmad Haniff Jaafar, Email: a_hanif@upm.edu.my.
Awanis Azizan, Email: awanis_azizan@yahoo.com.
Nur Khaleeda Zulaikha Zolkeflee, Email: khaleeda_zulaikha@yahoo.com.
Siti Zulaikha Abd Ghafar, Email: ctzue.agb@gmail.com.
References
- Abd Ghafar SZ, Mediani A, Maulidiani Ramli AS, Abas F. Antioxidant, α-glucosidase and nitric oxide inhibitory activities of Phyllanthus acidus and LC-MS/MS profile of the active extract. Food Biosci. 2018;25:134–140. doi: 10.1016/j.fbio.2018.08.009. [DOI] [Google Scholar]
- Abu-Reidah IM, Ali-Shtayeh MS, Jamous RM, Arraez-Roman D, Swgura-Carretero A. HPLC-DAD-ESI-MS/MS screening of bioactive components from Rhus coriaria L. (Suman) fruits. Food Chem. 2015;166:179–191. doi: 10.1016/j.foodchem.2014.06.011. [DOI] [PubMed] [Google Scholar]
- Ahmad A, Misra LN. Isolation of herniarin and other constituents from Matricaria chamomilla flowers. Int. J. Pharmacog. Phytochem. 1997;35:121–125. [Google Scholar]
- Akhtar N, Jafri L, Green BD, Kalsoom S, Mirza B. A multi-mode bioactive agent isolated from Ficus microcarpa L. Fill. with therapeutic potential for type 2 diabetes mellitus. Front. Pharmacol. 2018;9:1376. doi: 10.3389/fphar.2018.01376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjum N, Tripathi YC. In vitro alpha-amylase and alpha-glucosidase inhibitory activities of fruits of Ficus auriculata Lour. Int. J Pharm. Bio. Sci. 2019;10:134–141. [Google Scholar]
- Nor-Azman NII, Hashim N, Ahmad R. In-vitro α-Glucosidase inhibitory activities of Muntingia calabura Linn. Int. J. Eng. Tech. 2018;7:183–185. [Google Scholar]
- Barros A, Girones-Vilaplana A, Teixeira A, Collado-Gonzalez J, Moreno DA, Gil-Izqueierdo A, Rosa E, Dominguez-Perles R. Evaluation of grape (Vitis vinifera L.) stems from Portuguese varieties as a resource of (poly)phenolic compounds: A comparative study. Food Res. Int. 2014;65:375–384. doi: 10.1016/j.foodres.2014.07.021. [DOI] [Google Scholar]
- Berardini N, Schieber A, Klaiber I, Beifuss U, Carle R, Conrad J. 7-O-methylcyanidin 3-O-β-D-galactopyranoside, a novel anthocyanin from Mango (Mangifera indica L. cv. ‘Tommy Atkins’) peels. Z. Naturforsch. 2005;60:801–804. doi: 10.1515/znb-2005-0718. [DOI] [Google Scholar]
- Chew YL, Chan WWL, Tan PL, Stanslas J, Goh JK. Assessment of phytochemical content, polyphenolic composition, antioxidant and antibacterial activities of Leguminosae medical plants in Peninsular Malaysia. BMC Complement. Altern. Med. 2011;11:2–10. doi: 10.1186/1472-6882-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias T, Bronze MR, Houghton PJ, Mota-Filipe H, Paulo A. The flavonoid-rich fraction of Coreopsis tinctoria promotes glucose tolerance regain through pancreatic function recovery in streptozotocin-induced glucose-intolerant rats. J. Ethnopharmacol. 2010;132:483–490. doi: 10.1016/j.jep.2010.08.048. [DOI] [PubMed] [Google Scholar]
- Dou J, Lee VSY, Tzen JTC, Lee M-C. Identification and comparison of phenolic compounds in the preparation of Oolong tea manufactured by semifermentation and drying processes. J. Agric. Food Chem. 2007;55:7462–7468. doi: 10.1021/jf0718603. [DOI] [PubMed] [Google Scholar]
- El-Fishawy A, Zayed R, Afifi S. Phytochemical and pharmacological studies of Ficus auriculata Lour. J. Nat. Prod. 2011;4:184–195. [Google Scholar]
- Ertas A, Yilmaz MA, Firat M. Chemical profile by LC-MS/MS, GC-MS and antioxidant activities of the essential oils and crude extracts of two Euphorbia species. Nat. Prod. Res. 2014;29:529–534. doi: 10.1080/14786419.2014.954113. [DOI] [PubMed] [Google Scholar]
- Fathalla N, Bishr M, Singab AN, Salama O. GC-MS and LC-MS identification of the phenolic compounds present in the ethyl acetate fraction obtained from Senna tora L. Roxb. seeds. Nat. Prod. Res. 2018;16:1–4. doi: 10.1080/14786419.2018.1508138. [DOI] [PubMed] [Google Scholar]
- Gomathi R, Anusuya N, Manian S. A dietary antioxidant supplementation of Jamaican cherries (Muntingia calabura L.). attenuates inflammatory related disorders. Food Sci. Biotechnol. 2013;22:787–794. doi: 10.1007/s10068-013-0146-1. [DOI] [Google Scholar]
- Hainida E, Ikram K, Hock K, Maleyki A, Jalil M. Antioxidant capacity and total phenolic content of Malaysian underutilized fruits. J. Food Compost. Anal. 2009;22:388–393. doi: 10.1016/j.jfca.2009.04.001. [DOI] [Google Scholar]
- Hofmann T, Nebehaj E, Albert L. Antioxidant properties and detailed polyphenol profiling of European hornbeam (Carpinus betulus L.) leaves by multiple antioxidant capacity assays and high-performance liquid chromatography/multistage electrospray mass spectrometry. Ind. Crops Prod. 2016;87:340–349. doi: 10.1016/j.indcrop.2016.04.037. [DOI] [Google Scholar]
- Jin MJ, Kim IS, Rehman SU, Dong M-S, Na C-S, Yoo HH. A liquid chromatograph-tandem mass spectrometry method for simultaneous quantitation of 10 bioactive components in Rhus verniciflua extracts. J. Chromatogr. Sci. 2015;4:390–396. doi: 10.1093/chromsci/bmv152. [DOI] [PubMed] [Google Scholar]
- Kajdzanoska M, Gjamovski V, Stefova M. HPLC-DAD-ESI-MSn identification of phenolic compounds in cultivated strawberries from Macedonia. Maced. J. Chem. Chem. En. 2010;29:181–194. doi: 10.20450/mjcce.2010.165. [DOI] [Google Scholar]
- Kumar S, Chandra P, Bajpai V, Singh A. Rapid qualitative and quantitative analysis of bioactive compounds from Phyllanthus amarus using LC/MS/MS techniques. Ind. Crops Prod. 2015;69:143–152. doi: 10.1016/j.indcrop.2015.02.012. [DOI] [Google Scholar]
- Lawal U, Leong SW, Shaari K, Ismail IS, Khatib A, Abas F. α-glucosidase inhibitory and antioxidant activities of different Ipomoea aquatica cultivars and LCMS/MS profiling of the active cultivar. J. Food Biochem. 2017;41(2):1–8. doi: 10.1111/jfbc.12303. [DOI] [Google Scholar]
- Li Y, Guo H, Wu Y, Geng Q, Dong D, Wu H, Li E. A sensitive and selective method for determination of Aesculin in Cortex fraxini by liquid chromatography quadrupole time-off light tandem mass spectrometry and application in pharmacokinetic study. J. Anal. Methods Chem. 2013: 1-6 (2013) [DOI] [PMC free article] [PubMed]
- Lin L-Z, He X-G, Lindenmaier M, Yang J, Cleary M, Qiu S-X, Cordell GA. LC-ESI-MS study of the flavonoid glycoside malonates of red clover (Trifolium pratense) J. Agric. Food Chem. 2000;48:354–365. doi: 10.1021/jf991002+. [DOI] [PubMed] [Google Scholar]
- Maity S, Chatterjee S, Variyar PS, Sharma A, Adhikari S, Mazumder S. Evaluation of antioxidant activity and characterization of phenolic constituents of Phyllantus amarus root. J. Agric. Food Chem. 2013;61:3443–3450. doi: 10.1021/jf3046686. [DOI] [PubMed] [Google Scholar]
- McCune LM, Kubota C, Stendell-Hollis NR, Thomson CA. Cherries and Health: a review. Crit. Rev. Food Sci. Nutr. 2011;51:1–12. doi: 10.1080/10408390903001719. [DOI] [PubMed] [Google Scholar]
- Mediani A, Abas F, Tan CP, Khatib A. Effects of different drying methods and storage time on free radical scavenging activity and total phenolic content of Cosmos caudatus. Antioxidants. 2014;3:358–370. doi: 10.3390/antiox3020358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirfat AHS, Salma I, Razali M. Natural antioxidant properties of selected wild Mangifera species in Malaysia. J. Trop. Agric. Food Sci. 2016;44:63–72. [Google Scholar]
- Muniyandi K, George E, Sathyanarayanan S, George BP, Abrahamse H, Thamburaj S, Thangaraj P. Phenolics, tannins, flavonoids and anthocyanins contents influenced antioxidant and anticancer activities of Rubus fruits from Western Ghats. India. Food Sci Hum Well. 2019;8:73–81. doi: 10.1016/j.fshw.2019.03.005. [DOI] [Google Scholar]
- Plazonic A, Bucar F, Males Z, Mornar A, Nigovic B, Kujundzic N. Identification and quantification of flavonoids and phenolic acids in Burr Parsley (Caucalis platycarpos L.), using high-performance liquid chromatography with diode array detection and electrospray ionization mass spectrometry. Molecules. 2009;14:2466–2490. doi: 10.3390/molecules14072466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puangpradab R, Suksathan R, Saratan N, Puangsombat P. Antioxidant properties and nutritive values of native figs consumed in northern Thailand. Acta Hortic. 2018;1210:281–288. doi: 10.17660/ActaHortic.2018.1210.39. [DOI] [Google Scholar]
- Putri ND, Nursyamsi KS, Prayogo YH, Sari DR-L, Budiarti E, Batubara I. Exploration of Mango fruits (Mangifera indica) as α-glucosidase inhibitors. Biosaintifika. 2017;9:554–559. doi: 10.15294/biosaintifika.v9i3.10516. [DOI] [Google Scholar]
- Saifullah M, Mc-Cullum R, McCluskey A, Vuong Q. Comparison of conventional extraction technique with ultrasound assisted extraction on recovery of phenolic compounds from lemon scented tea tree (Leptospermum petersonii) leaves. Heliyon. 2020;6(4):1–12. doi: 10.1016/j.heliyon.2020.e03666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia S, Mahnot NK, Lata C. Phytochemical content and antioxidant activities of thirteen fruits of Assam. India. Food Biosci. 2016;13:15–20. doi: 10.1016/j.fbio.2015.11.003. [DOI] [Google Scholar]
- Sanchez-Rabaneda F, Jauregui O, Casals I, Andres-Lacueva C, Izqueierdo-Pulido M, Lamuela-Raventos M. Liquid chromatographic/electrospray ionization tandem mass spectrometric study of the phenolic composition of cocoa (Theobroma cacao) J. Mass Spectrom. 2003;38:35–42. doi: 10.1002/jms.395. [DOI] [PubMed] [Google Scholar]
- Santhirasegaram V, Razali Z, George DS, Somasundram C. Effects of thermal and non-thermal processing on phenolic compounds, antioxidant activity and sensory attributes of Chokonan Mango (Mangifera indica L.) juice. Food Bioproc. Tech. 2015;8:2256–2267. doi: 10.1007/s11947-015-1576-y. [DOI] [Google Scholar]
- World Health Organization. Diabetes Country Profiles-Malaysia. (2018)
- Yao LH, Jiang YM, Shi J, Tomas-Barberan FA, Datta N, Singanusong R, Chen SS. Flavonoids in food and their health benefits. Plant Foods Hum. Nutr. 2004;59:113–122. doi: 10.1007/s11130-004-0049-7. [DOI] [PubMed] [Google Scholar]
- Ye M, Yang W-Z, Liu K-D, Qiao X, Li B-J, Cheng J, Feng J, Guo D-A, Zhao Y-Y. Characterization of flavonoids in Millettia nitida var. hirsutissima by HPLC/DAD/ESI-MS. J. Pharm. Anal. 2012;2:35–42. doi: 10.1016/j.jpha.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Shao Y, Fu J, Xiang L, Zheng Y, Li W. Characterization and quantification of taxifolin related flavonoids in Larix olgensis Henry Var. Koreana Nakai extract analysis and its antioxidant activity assay. Int. J. Pharmacol. 2018;14:534–545. doi: 10.3923/ijp.2018.534.545. [DOI] [Google Scholar]