Abstract
The COVID-19 pandemic has become one of the biggest threats to the global healthcare system, creating an unprecedented condition worldwide. The necessity of rapid diagnosis calls for alternative methods to predict the condition of the patient, for which disease severity estimation on the basis of Lung Ultrasound (LUS) can be a safe, radiation-free, flexible, and favorable option. In this paper, a frame-based 4-score disease severity prediction architecture is proposed with the integration of deep convolutional and recurrent neural networks to consider both spatial and temporal features of the LUS frames. The proposed convolutional neural network (CNN) architecture implements an autoencoder network and separable convolutional branches fused with a modified DenseNet-201 network to build a vigorous, noise-free classification model. A five-fold cross-validation scheme is performed to affirm the efficacy of the proposed network. In-depth result analysis shows a promising improvement in the classification performance by introducing the Long Short-Term Memory (LSTM) layers after the proposed CNN architecture by an average of , which is approximately more than the traditional DenseNet architecture alone. From an extensive analysis, it is found that the proposed end-to-end scheme is very effective in detecting COVID-19 severity scores from LUS images.
Keywords: COVID-19, Lung ultrasound, Neural network, Severity score, Deep learning
1. Introduction
Since the beginning of the Coronavirus Disease (COVID-19) outbreak, researchers are attempting to find an alternative method to detect COVID-19 rapidly apart from the reverse transcription–polymerase chain reaction (RT-PCR) test, which is considered to be a gold-standard. The RT-PCR test is highly contingent upon the testing environment and sample collection procedures, and its testing capacity is limited [1,2]. Rapid testing is proved to be the most effective method to circumscribe the spread of this easily transmissible disease [3,4], which led researchers to search for a rapid diagnostic method. In this case, most of the past studies utilized mainly three types of radiological imaging techniques for COVID-19 detection, namely computed tomography (CT) scan, X-Ray, and lung ultrasound (LUS). Apart from these three techniques, there are some other works, where a combination of wearable medical sensors for extracting physiological signals [5] is employed. Among the radiological imaging methods, the CT scan provides a three-dimensional view of the lungs and is capable of detecting the manifestations of COVID-19 at various stages of the disease progression [2,6]. However, the CT screening is costly and exposes patients to radiation, which could be deleterious for them to some extent in the future [7]. The X-Ray is another attractive method because of its flexibility, low cost, and comparatively quicker approach [8,9]. But the characteristics of the disease and its pulmonary consolidations at various stages are not clearly visible in the X-ray images, since they are low-resolution by nature and contain overlapping projections [2,10]. The ultrasound imaging provides clear and real-time views of lungs with no future health hazards and is more pragmatic due to its effective functionality in bedside treatment and day-to-day checkup. A comparative performance analysis of COVID-19 detection by using the existing datasets of CT scan, X-ray, and ultrasound showed a superior detection performance in the case of LUS than that is obtained using CT scan or X-ray [11].
In literature, LUS imaging problems are mainly handled by the segmentation approach, frame-based and video-grading methods to classify into desired categories. The LUS has long been used to detect respiratory syndromes, with a better result for pneumonia diagnosis than that is obtained by X-Rays as per visual inspection by the experts on the respective fields [12]. After the outbreak of the COVID-19 pandemic, LUS has been appreciated as an effective visual-inspection based technique [[13], [14], [15]]. In particular, compared to the LUS based inspection, other techniques require larger use of tools and devices, which can lead to possible contamination and spread the disease [16]. Since the ultrasound dataset related to COVID-19 is severely limited and a considerable amount of annotated dataset is still publicly unavailable, very few research results have been reported so far on LUS considering the contemporary studies. Among them, in Refs. [7,17], LUS has been employed to detect specific patterns of the disease as well as the disease severity in terms of various scores. In this regard, both deep learning and machine learning techniques are implemented for automatic disease prediction in the current resource-con-strained environment. In Ref. [7], a frame-based classification architecture is introduced based on the spatial transformer network [18] to classify the disease severity into four scores ranging from 0 to 3. Apart from the classification, video-level grading and pathological artifact segmentation by stacking three models are performed there. On top of that, they published a subset of the dataset they utilized in the study, comprising a total of 60 videos, among which 58 videos are fully labeled at the frame level. In Ref. [17], an SVM-based classification model is proposed following the automatic localization of the pleural line by the hidden Markov Model and the Viterbi algorithm. Here, the authors utilized the dataset released by Ref. [7] and to deal with the limitation of available data, they limited the study on hospital-specific cases and deployed a machine learning model. Therefore, an efficient deep learning-based architecture for automatic disease severity prediction with satisfactory performance under the resource-constrained and hospital-independent environment to assist clinicians in the COVID-19 diagnosis process is still in great demand.
In this study, a deep CNN is proposed to perform frame-based classification of the LUS images into four severity scores, followed by a recurrent neural network, Long Short-Term Memory (LSTM) that effectively handles the temporal features of the LUS videos. Apart from the spatial features between the frames in an LUS video, temporal features are also present, which can be effectively handled by the proposed implementation of the LSTM network. The proposed CNN architecture is introduced by an autoencoder block and separable convolutional branches that adjoin the various parallel convolutional paths along with the DenseNet-201 network at different points to ensure noise-free, edge-dominant features for the classification task. The integration of LSTM layers after the proposed CNN block achieves a propitious increase in the 4-score disease severity prediction performance. Expansive analysis along with the five-fold cross-validation at each of the stages is performed to prove the potency of this study. All the codes and architectures of this study are publicly available at: https://github.com/ankangd/HybridCovidLUS.
2. Materials and methods
2.1. Dataset
In this paper, the Italian COVID-19 Lung Ultrasound DataBase (ICLUS-DB) [19] is utilized, which currently holds a total of 60 lung ultrasound (LUS) videos from 29 patients, accessible after the manual approval of the account request. Data came from different clinical centers of Italy: BresciaMed, Brescia (BS); Fondazione Policlinico Universitario A. Gemelli IRCCS, Rome (RM); Valle del Serchio General Hospital, Lucca (LU); Tione General Hospital, Tione (TN); and Fondazione Policlinico Universitario San Matteo IRCCS, Pavia. Both linear and convex probes were used to acquire data following the acquisition protocol. Out of the 60 videos, 39 were acquired with convex probes, and the other 21 were acquired with a linear probe. Scores of the individual frames have been taken from Ref. [7], where all the frames of 58 videos (38 with a convex probe, 20 with a linear probe, comprises of a total of 14,311 frames) are scored based on a 4-level scoring system, ranging from 0 to 3, which can efficiently predict the condition of the patient in a rapid manner. Here 0 is the most healthy case, and in a declining manner 3 is the worst-case [14], which is proved to be an effective tool for adult respiratory distress syndrome (ARDS) affected people, and a potential substitute to the existing methods of COVID-19 severity assessment [20].
The correctness of scoring is ensured by a 4-level process involving four master students with background knowledge on ultrasound, a Ph.D. student with LUS expertise, a biomedical engineer, and clinicians with 10+ years of experience [7]. Throughout the study, training-testing is separately done on the images acquired from convex and linear probes, because images from these two sources are of different patterns and dimensions. Details of the dataset are provided in Table 1 . As evident from the table, the proposed network has to deal with an imbalanced type of data on various categories, with a greater amount of data having score 0 and score 2 in linear and convex cases, respectively.
Table 1.
Dataset used for this study.
| Type | No. of Videos | No. of Frames |
|||
|---|---|---|---|---|---|
| Score 0 | Score 1 | Score 2 | Score 3 | ||
| Linear | 20 | 1987 | 305 | 1365 | 958 |
| Convex | 38 | 1953 | 1704 | 4481 | 1558 |
2.2. Preprocessing
In this study, frame-based disease severity prediction is performed, for which the first task is to extract the frames from the videos. The frames contain unwanted non-ultrasound white regions, which remain stationary during the continuous motion of the ultrasound video. The desired region can easily be extracted by eliminating the unwanted portions, and subsequently passed to the classification network.
2.3. Proposed convolutional neural network
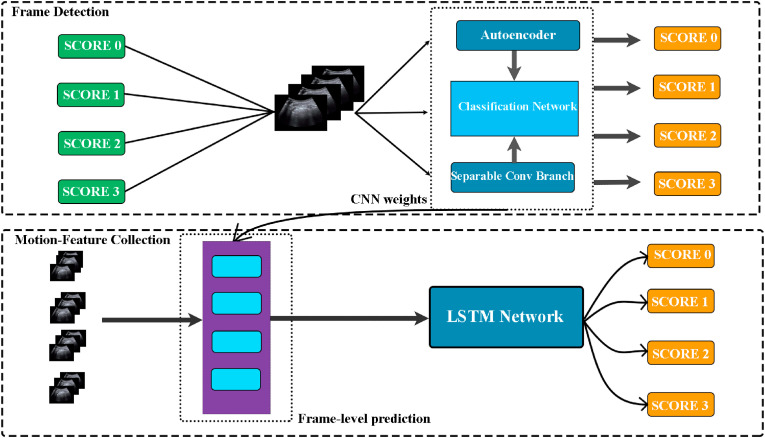
In this paper, a novel classification network is introduced on the basis of a modified CNN adjoined to the second stage of LSTM to classify the given LUS image in one of the four severity scores between 0 and 3. The simplified pipeline of the proposed scheme is shown in Fig. 1 . Here the top portion exhibits the proposed CNN stage, which consists of an autoencoder block passing vigorous features to the different convolutional branches originating from the given LUS images and a block of separable convolutional branch to extract the edge-dominant features accordingly. Later, the images are passed to the LSTM block sequentially for each LUS video, as shown in the bottom region of Fig. 1, to perform the final predictions based on the joint weight vectors generated from the proposed CNN and LSTM blocks.
Fig. 1.
Pipeline of the proposed method. The ROI of the respective frames are passed into the proposed CNN block, which is then integrated with the LSTM blocks to make the final predictions into one of the four severity scores.
In this study, the images are at first passed to an autoencoder block to reduce noise and therefore, pull out the robust features which identify the best discriminatory characteristics between the scores. The idea behind introducing an autoencoder section is to take input and map them into a latent representation , where the mapping occurs through the function . This hidden representation is then mapped back into a reconstruction of the same shape as input x through . Here s is a non-linear function e. g sigmoid function. The first part is known as the encoder and the latter one is the decoder. The parameters of this model are optimized in such a way that the average reconstruction error is minimized. In this paper, the input images are refined with the assistance of a denoising autoencoder [21], which drives the hidden layers to discover the significant features from any input image. The input image of size is encoded into size of feature maps which are the latent space representations of the input. This layer contains robust features of the image. These features are decoded serially with upsampling, and finally the reconstructed output of size is achieved which is the same size as the input. One of the major problems of using traditional autoencoder blocks is its nature of erasing the dominant features. In order to solve this problem, connections between encoded and decoded features are added through concatenation to preserve the ruling features and keep the resemblance between input and output in the same way as used in supervised segmentation networks, such as Fully Connected Network (FCN). The decoded part implemented in the study can be presented as:
| (1) |
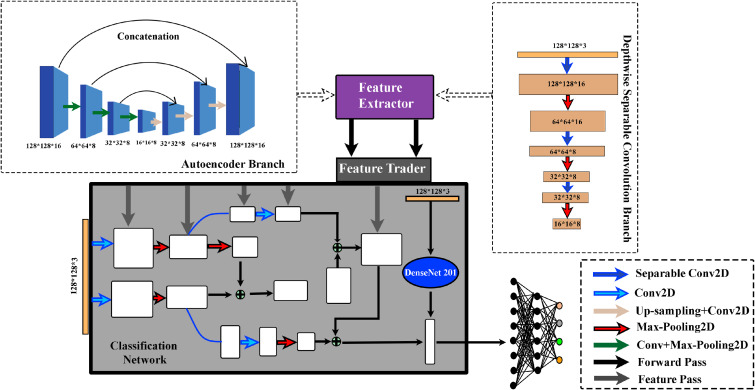
Here, the symbol is used to indicate concatenation. For a better understanding of the implementation details, referring to the autoencoder branch shown in Fig. 2 [top left corner], each step is explained hereafter. Prior to entering the autoencoder branch, a given input image of size is convolved into a matrix of size , and the resulting image of size is fed to the autoencoder branch.
Fig. 2.
Proposed deep CNN architecture.
In the autoencoder branch shown in Fig. 2, the first three encoding operations generate the encoded feature matrix of size (located at the middle of the figure), and then the next three decoding operations are performed. In order to get the first decoded output , is used to generate that is then concatenated with . It is to be noted that for the encoding part, convolution and max-pooling operations are carried out; and for the decoding part, convolution and upsampling operations are performed (in Fig. 2, corresponding arrowheads are denoted). Following the convolution and upsampling operations, is converted to a matrix of size and then concatenated to the previously encoded branch , which is also of the same size (). This process of convolution, upsampling, and concatenation is then repeated until a feature matrix of size is obtained, as shown in Fig. 2. At the beginning stage of encoding, the filter size is changed to 16 instead of 8, which makes the matrix size . The reconstructed output of size is obtained following the deconvolution.
A loss function is required to minimize the reconstruction error. In this paper, the categorical cross-entropy loss function [22] is incorporated, which uses sigmoid/softmax as the activation function. The cross-entropy function is defined as:
| (2) |
where N is the total number of classes, t is the respective label, and is the softmax function, defined as:
| (3) |
For the autoencoder, the entropy function reduces into:
| (4) |
Apart from using the traditional autoencoder, a special type of convolution branch is introduced for classification purpose. In this CNN-based block, in place of using conventional CNN, depthwise separable convolution is utilized [23]. In the depthwise separable convolution, instead of using a single kernel of size , three separate kernels are used. Each kernel has a size of . Each kernel convolves with 1 channel of the input layer. For an input of size , each of such convolution provides a map of size . Stacking these maps together, a image is obtained. In the second step of the depthwise separable convolution, the convolution is applied with kernel size to extend the depth. Convolving the input image with each kernel provides a map of size . Thus, after applying K number of convolutions, a layer with size is obtained. As the separable convolution splits the convolution operation into depthwise convolution and pointwise convolution, the whole convolution operation takes lesser time than the time taken by the traditional convolution. This branch helps to gain some edge dominant features that can later be used for classification. As a result, some low-level features are extracted from the input image with separable convolution, and the Sobel kernel is used there. While the number of parameter is increasing due to several branches, it leads to overfitting. Hence, the depthwise separable convolution is necessary to adjust these parameters.
Features from different stages of both the autoencoder branch and the separable convolution branch are extracted and passed to the main classification architecture. The input image is separated into several paths where features from the previous two branches are concatenated. The input image is again passed into the DenseNet-201 [24] which is the backbone of the proposed CNN. The output from this network is flattened and features are added from the previous path. Finally, fully connected layers of size 128, 64, and 4 are assimilated to classify the images into 4 scores. A proper optimization is needed to minimize the loss function. Here categorical cross-entropy is utilized as a loss function, and Adam optimizer is used to minimize the loss. The proposed CNN is depicted in Fig. 2.
2.4. Integrating the LSTM units
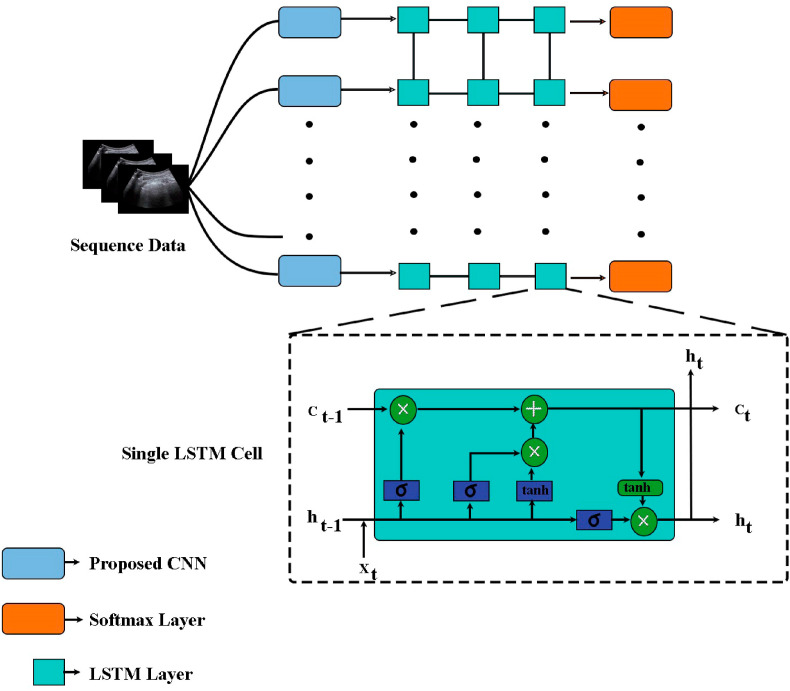
Since an LUS video comprises a sequential representation of the lung images, it bears temporal features e. g motion information like other video recognition scenarios. A naive approach is to input stacks of images depending on their respective classes, and make predictions through performing the CNN alone. But this kind of approach tends to lose motion information. In this paper, the units of recurrent neural network architecture LSTM [25] are introduced, which utilize memory cells to store, modify, and access the internal state, and therefore discover the temporal information. Traditional CNN is sequence invariant whereas the recurrent neural network considers a sequence of frames by encapsulating the real-time temporal information that enhances the overall performance of the network. A standard recurrent neural network computes the hidden vector sequence and the output vector sequence from an input sequence .
| (5) |
| (6) |
Where W denotes the weight matrices, b denotes the bias vectors, and σ is the hidden layer activation function. The LSTM architecture uses memory cells to store and output information, allowing it to better discover long-range temporal relationships which is highly reliable for long time dependencies. Each LSTM unit consists of three gates, namely input gate, forget gate, and output gate. A single LSTM unit is shown in the inset of Fig. 3 . Here the cell captures the data over a certain time interval and the information flow is regulated by the other gates. The input gate adds information to the cell, the forget gate removes unnecessary information from the cell, and the output gate selectively chooses necessary information from the current cell [26]. The sigmoid (σ) and hyperbolic tangent () functions are used as the activation functions inside the unit cell, as shown in Fig. 2. Optimizing these three gates, the LSTM layers try to calculate one weight parameter which adds temporal information. In this paper, the LSTM takes input from the output of the proposed CNN layer at each consecutive video frame. The output from one LSTM layer is the input for the next layer. The output from the proposed CNN is refined forward through time and upwards through three layers of stacked LSTMs. A softmax layer is used for the normalization of the probability vector for the four classes.
Fig. 3.
The block of LSTMs next to the proposed CNN block. The LSTM block consists of three types of gates, namely forget gate, input gate, and output gate. Collectively, they decide which information is relevant from the input data and updates accordingly. A single cell LSTM takes cell state , hidden state , and input data at each timestamp t to perform its operations. The forget gate decides which previous information is not required at the moment, the input gate selects relevant information from the input data , and the output gate produces the hidden state for time t.
If a single LUS frame is given, the proposed CNN weight will be dominant to predict the score. However, if a video is provided, the efficacy of the integrated CNN-LSTM weight will be able to predict the frames based on both sequential or temporal and spatial features with higher accuracy, as examined in the results section of this paper.
3. Experimental results
3.1. Training-testing strategy
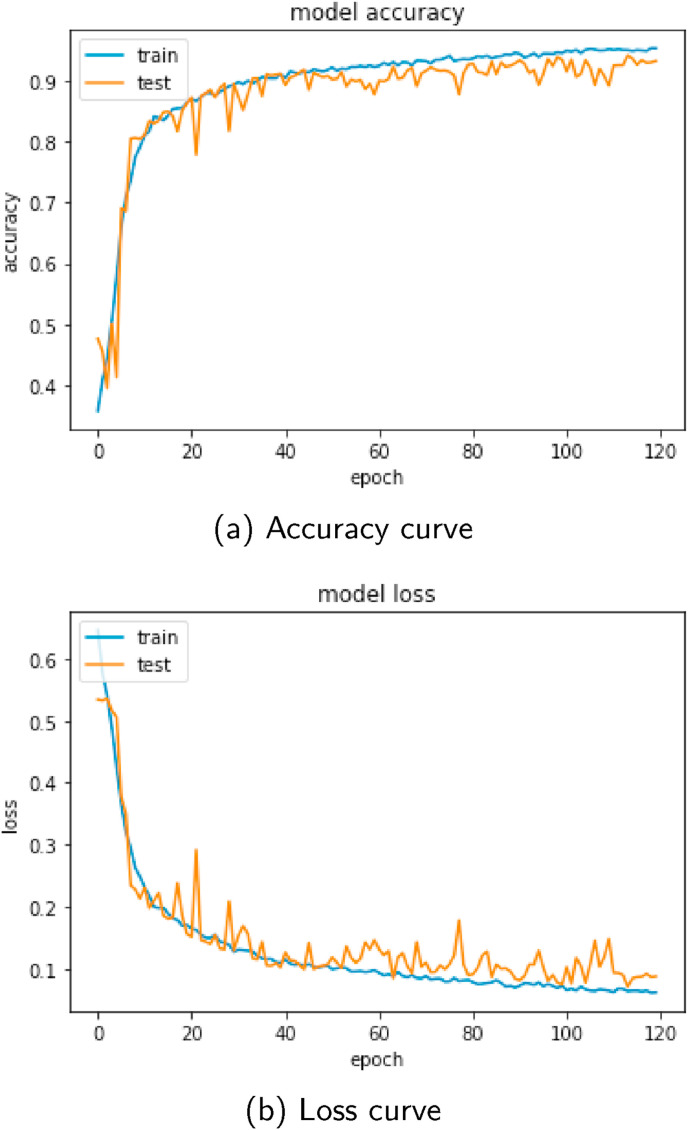
The dataset is split into train and test set separately for both convex and linear probes. On the given dataset, the 5-fold cross-validation technique is adapted to obtain the train and test data. For linear probe data, frames from 4 videos (20%) are kept in the test set, whereas the training set consists of the other 16 (80%) videos for each of the cross-validation stages. Similarly for convex probe data, at each stage, 8 videos (20%) are kept for testing, and the remaining 30 videos (80%) are used for training. The Adam optimizer [27] is used in each of the stages, with a learning rate of 1e-3, batch size 64, and 120 epochs, for both CNN and LSTM. Several dropout layers are used after the convolutional layers to prevent overfitting [28]. The model accuracy and loss curve for one of the cross-validation stages is shown in Fig. 4 . The minimal gap between training and validation curves in the figure depicts a good fit at the training stage. It is understandable from Table 1 that class imbalance is present in the dataset, which is a common scenario for medical imaging. To deal with the problem, data augmentation is performed on the training data including rotation (0° 360°), horizontal and vertical shift (), scaling (), horizontal and vertical flips at the training stage, so that a balance over the frame quantity is achieved in the respective four classes.
Fig. 4.
Accuracy and loss curve at one of the training phases.
3.2. Classification results
In this paper, the performance of the proposed model is demonstrated on the basis of three baselines: (1) the DenseNet-201 architecture alone, (2) proposed CNN architecture without LSTM block, and (3) proposed CNN architecture integrated with the LSTM block. Accuracy, sensitivity, specificity, and score are considered as the evaluation parameters for these three baselines. A comprehensive analysis followed by the detailed results of 5-fold cross-validation is also presented in the following part.
At first, the DenseNet-201 model with the pre-trained weight of ImageNet [29] is applied to the dataset to classify the LUS images into the 4 categories, representing the four severity scores. The DenseNet is fine-tuned to extract the best result from it, and the process has undergone the five-cross validation stages by training with and testing on the remaining unseen test data. The overall accuracy, in this case, is for linear-probe data, and for convex probe data, which is quite unsatisfactory.
Next, the proposed CNN architecture is implemented on the dataset instead of traditional DenseNet-201. All the evaluating parameters improve noticeably after the implementation of the proposed CNN. Later, the proposed combined network consisting of the CNN and LSTM blocks is employed which achieves the best result. The gradual development in results by the implementation of the proposed network is summarized in Table 2 where the margin of error is shown for a confidence level. The enhancement in overall performance by adjoining the proposed CNN along with the LSTM block is quite promising, with an increase of accuracy from the traditional DenseNet-201 architecture, and an increase of from the proposed CNN, which itself increases the accuracy by from the DenseNet-201 alone at the first stage. Other parameters also improve significantly, with minimal deviation from the average values.
Table 2.
Gradual development by imposing the proposed network, examination of CNN and CNN + LSTM has shown separately. The margin of error is also specified at confidence level.
| Type |
Model |
Evaluating Parameter |
|||
|---|---|---|---|---|---|
| Accuracy | Sensitivity | Specificity | F1 Score | ||
| Linear | DenseNet-201 | 0.575 0.107 | 0.575 0.107 | 0.878 0.048 | 0.57 0.130 |
| Proposed CNN | 0.700 0.091 | 0.700 0.091 | 0.908 0.053 | 0.702 0.123 | |
| Proposed CNN + LSTM | 0.791 0.058 | 0.791 0.058 | 0.901 0.034 | 0.786 0.057 | |
| Convex | DenseNet-201 | 0.535 0.039 | 0.535 0.039 | 0.662 0.082 | 0.515 0.076 |
| Proposed CNN | 0.610 0.040 | 0.610 0.040 | 0.756 0.097 | 0.586 0.035 | |
| Proposed CNN + LSTM | 0.677 0.032 | 0.677 0.032 | 0.768 0.140 | 0.666 0.034 | |
It is to be noted that the five-fold cross validation technique is performed to appraise the proposed model. Detailed results including each of the cross-validations stages are shown in Table 3 for the proposed CNN + LSTM network. The average values are shown with a margin of error at confidence level. Consistent performance is manifested at each of the validation stages with an average accuracy of for linear probe data, and for convex probe data. The overall deviation from the average value is insignificant as evident from the table. Analyzing the results, it is perceptible that the overall performance is better in the images acquired from the linear probe than the convex ones. In consideration of image quality, linear transducers are preferred [30], which is a probable answer to the better-predicting performance with the linear-probe images than that is obtained with the convex-probe images. Although lung consolidations are visible in every sort of probes, linear probes are considered to be more efficient in magnifying smaller consolidations [31].
Table 3.
Detailed result of 5-fold CV for the proposed CNN + LSTM network. The margin of error for the average result is specified at confidence level.
| Type |
Parameter |
Cross Validation |
Average |
||||
|---|---|---|---|---|---|---|---|
| K = 1 | K = 2 | K = 3 | K = 4 | K = 5 | |||
| Linear | Accuracy | 0.870 | 0.814 | 0.687 | 0.791 | 0.791 | 0.791 0.058 |
| Sensitivity | 0.870 | 0.814 | 0.687 | 0.791 | 0.791 | 0.791 0.058 | |
| Specificity | 0.947 | 0.902 | 0.844 | 0.923 | 0.923 | 0.901 0.034 | |
| F1 Score | 0.860 | 0.829 | 0.691 | 0.770 | 0.780 | 0.786 0.057 | |
| Convex | Accuracy | 0.719 | 0.668 | 0.622 | 0.676 | 0.700 | 0.677 0.032 |
| Sensitivity | 0.719 | 0.668 | 0.622 | 0.676 | 0.700 | 0.677 0.032 | |
| Specificity | 0.500 | 0.910 | 0.827 | 0.848 | 0.759 | 0.768 0.140 | |
| F1 Score | 0.660 | 0.728 | 0.622 | 0.655 | 0.665 | 0.666 0.034 | |
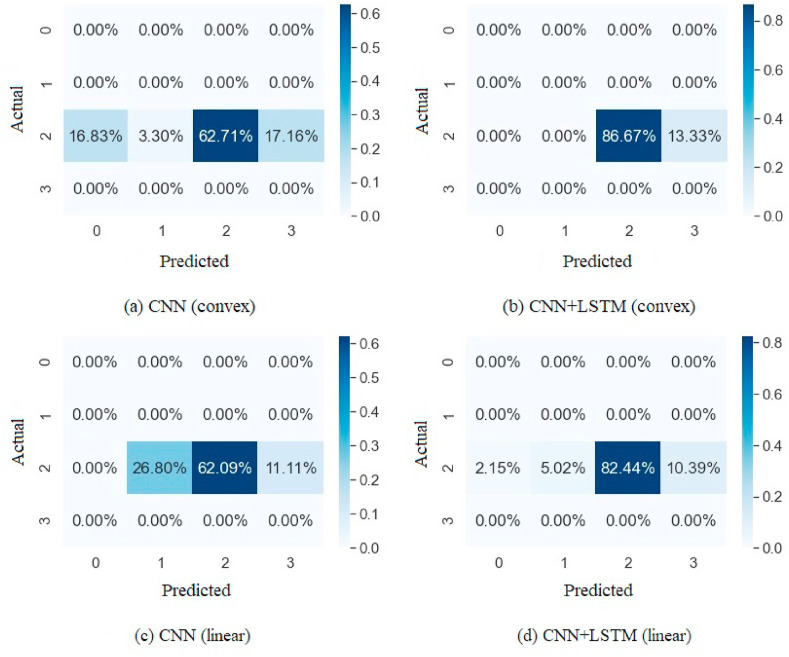
In Fig. 5 , a visualization of the gradual improvement by implementing the proposed network is shown on two LUS videos (one from the convex probe and the other is from the linear probe) collected from two different patients, from two different hospitals. The matrices indicate a significant improvement in the results of the proposed integration method. In the case of linear probe data in Fig. 5(a) and (b), the proposed CNN + LSTM network is capable of identifying most of the score 2 frames accurately, which are mispredicted as score 0 and 1 after the implementation of the CNN model alone. The number of false negatives in score 3 has also been reduced in the CNN + LSTM network. Similarly, in Fig. 5(c) and (d), the number of false negatives has reduced significantly after imposing the proposed CNN + LSTM network. The demonstration of the real LUS images is shown in Fig. 7, where the left and right images are predicted as score 1 and 3, respectively, although both are score 2 images. In the healthy lung condition, ultrasound imaging generates horizontal lines parallel to the pleural line known as A-lines. On the contrary, B-lines are vertical comet-tail shaped artifacts reflecting various pathological conditions of the lung [32]. Pleural lines are affected by these vertical artifacts, which are completely continuous in a healthy lung but become more and more obscure due to these artifacts. However, scores 2 and 3 are related and differ slightly depending on the magnification of consolidations [14]. The implementation of CNN along with the LSTM blocks can identify the subtle differences between the images and is capable of predicting the severity score accurately.
Fig. 5.
Confusion matrices for the proposed CNN (left) and CNN + LSTM (right) networks. Figure (a), (b) are generated for the frames of a video acquired by the convex probe, and figure (c), (d) are for the frames of a video acquired by the linear probe.
Fig. 7.
LUS frames acquired from convex (left) and linear (right) probe. Both of them are annotated as score 2. The proposed CNN model predicts them as score 1 and 3, respectively. The proposed CNN + LSTM model predicts them correctly.
In order to verify the performance of the proposed scheme, hospital-based data are considered. For example, the model is applied to the videos collected from BresciaMed, Brescia (BS) with the convex probe, which holds a total of 21 videos from 12 patients. The proposed model is trained with 16 videos from 8 patients and tested on the unseen 6 videos () from 4 patients. The proposed model achieves the best result with tremendous improvement in all the parameters than the previous case, as shown in Table 4 , where the model is trained regardless of its source. Frame-based prediction results achieved an average of accuracy for convex-probe videos in the hospital-independent case, whereas the same proposed network achieves an accuracy of for the hospital-dependent case. Sensitivity, specificity, and score also meet significant improvement than the previous case.
Table 4.
Results for hospital-specific case and hospital-independent case for CNN and CNN + LSTM. For the hospital-specific case, the convex-probe data acquired from BresciaMed, Brescia are considered.
| Parameter | Hospital-dependent |
Hospital-independent |
||
|---|---|---|---|---|
| (Brescia) | (Proposed) | |||
| CNN | CNN + LSTM | CNN | CNN + LSTM | |
| Accuracy | 0.785 | 0.792 | 0.610 | 0.677 |
| Sensitivity | 0.785 | 0.792 | 0.610 | 0.677 |
| Specificity | 0.857 | 0.872 | 0.756 | 0.768 |
| F1 score | 0.784 | 0.790 | 0.586 | 0.666 |
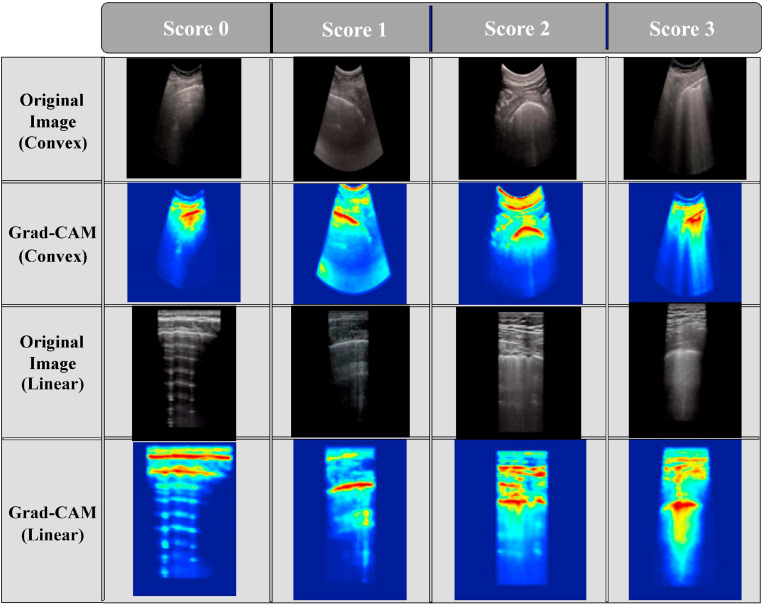
Finally, in order to extract a better visual understanding of the targeted regions in each image, the heatmaps are generated by the gradient-based class activation mapping (Grad-CAM) algorithm [33]. Following the implementation of Grad-CAM, information preserved through the various layers of the proposed architecture can be exploited and the heatmap shows which part of the input images activated the final prediction result. In Fig. 6 , some LUS frames with localization by the Grad-CAM are shown. For the score-0 frames in both convex and linear probe data, the model is activated in a broader region around the pleural line as the expected findings for a COVID-19 affected lung are not present here. The Grad-CAM does not exhibit the most germane areas on an image always and sometimes focuses on additional areas apart from the most relevant ones [7,34]. Hence, in this case, the focus of the Grad-CAM algorithm has been spread over a comparatively larger region. As the score rises up to 3, the model activates in a more indicative manner as patchy areas of B-lines, irregular pleural lines, and consolidations in the subpleural region, i.e., symptoms for an unhealthy lung [35] are prevalent there.
Fig. 6.
The heatmaps indicate which portions instigated the classification decision in the frames acquired from both convex and linear probes.
It is to be noted that the proposed algorithm can predict the severity scores of the given LUS frames with a minimal amount of time. For an ultrasound video with a 100-ms frame interval, it took on an average less than 6 ms to perform the test with a reasonable machine configuration. Therefore, it can be integrated with point-of-care devices like ultrasound machines to predict the condition of a patient in a real-time scenario. The model is also applicable to predict the overall condition of the patient if a whole video is fed to the algorithm. If a video is used as input, whether in real-time or as a recorded version, the temporal features will be considered along with the spatial features and the proposed hybrid model can predict the severity scores with higher accuracy. However, the proposed CNN model can predict the score for a certain frame as well.
3.3. Comparison with other studies
Studies devoted to LUS on COVID-related subjects are rarely attempted comparing with the works on CT scan or X-Ray imaging. Among them, in Ref. [7], the authors utilized a total of 277 LUS videos to classify the severity scores, and claimed to publish them; but until now, only 60 videos are publicly accessible. Hence, a direct comparison with their work is not possible in this case. In Ref. [17], the classification results are hospital-specific. Data from the same source holds similarity as the LUS image quality varies depending on the type of apparatus used to examine by the clinician, which enables the machine learning or deep learning models to predict with greater accuracy. One example is presented in Table 4 to check the consistency of the proposed model on similar types of hospital-specific data, where the accuracy for the BresciaMed convex cases is increased by , than the average hospital-independent cases. In most of the cases, hospital-specific data are significantly limited in quantity to train DL models on them. In a broader sense, a model trained on data regardless of its source like the one proposed in this study is desired considering its global application, which can efficiently predict hospital-specific cases as well. In Ref. [7], the frame-based prediction task was hospital-independent as well.
4. Conclusion
In this work, an integrated model of the convolutional and recurrent neural network is proposed for frame-based disease severity prediction to classify the LUS frames into four severity levels with scores ranging from 0 to 3. The proposed CNN block introduces embranchments along with the DenseNet-201 model with an initial autoencoder branch to enhance the performance of the classification network ensuring noise-free, robust features resulting in a boost in the classification accuracy with respect to the DenseNet-201 model. The CNN block is followed by a block of LSTM layers to consider the real-time sequence of the LUS frames within a particular video which at the end offers an improvement of the prediction accuracy by an average of than the proposed CNN alone. The sensitivity, specificity, and F1 score also improve by , , and , respectively, compared with the proposed CNN; which itself increases the sensitivity and specificity by and than the DenseNet-201. The proposed hybrid network can predict regardless of the source of data with a drastic improvement in hospital-specific cases, where it shows an increase in the prediction accuracy than the hospital-independent cases. Consistent performance is perceived in each of the five cross-validation stages with a minimal deviation of from the average value. The proposed model is capable of predicting severity scores at a minimal amount of time and therefore can be implemented in real-time scenarios. Meticulous evaluation and comparison with relative studies show that the proposed integrated network achieves an auspicious performance at predicting the disease severity scores, thereby diagnosing the immediate condition of the patient, which might be a great assistance for the clinicians in the present condition. The performance of the proposed model is relatively limited for the convex-probe cases. One possible way to improve the performance would be to utilize more training data, if available. Moreover, various processing schemes can be tested on the frames before entering the classification network. Finally, one possible future work of the proposed work would be to design deep learning-based segmentation architecture to carry out pathological artifact segmentation.
Declaration of competing interest
The authors hereby declare that they have no relevant financial or non-financial interests, or competing interests to disclose.
Acknowledgments
The authors would like to acknowledge the Department of Electrical and Electronic Engineering, Bangladesh University of Engineering and Technology for conducting this study.
Footnotes
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
References
- 1.Xie Xingzhi, Zheng Zhong, Zhao Wei, Zheng Chao, Wang Fei, Liu Jun. Chest CT for typical coronavirus disease 2019 (COVID-19) pneumonia: relationship to negative RT-PCR testing. Radiology. 2020;296(2):E41–E45. doi: 10.1148/radiol.2020200343. PMID: 32049601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tao Ai, Yang Zhenlu, Hou Hongyan, Zhan Chenao, Chen Chong, Lv Wenzhi, Qian Tao, Sun Ziyong, Xia Liming. Radiology; 2020. Correlation of Chest CT and RT-PCR Testing in Coronavirus Disease 2019 (COVID-19) in China: a Report of 1014 Cases; p. 200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Chen, Horby Peter W., Hayden Frederick G., Gao George F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmadi Mohsen, Sharifi Abbas, Dorosti Shadi, Ghoushchi Saeid Jafarzadeh, Ghanbari Negar. Investigation of effective climatology parameters on COVID-19 outbreak in Iran. Sci. Total Environ. 2020;729:138705. doi: 10.1016/j.scitotenv.2020.138705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hassantabar Shayan, Stefano Novati, Ghanakota Vishweshwar, Ferrari Alessandra, Nicola Gregory N., Bruno Raffaele, Marino Ignazio R., Hamidouche Kenza, Jha Niraj K. 2020. CovidDeep: SARS-CoV-2/COVID-19 Test Based on Wearable Medical Sensors and Efficient Neural Networks. [Google Scholar]
- 6.Chung Michael, Adam Bernheim, Mei Xueyan, Zhang Ning, Huang Mingqian, Zeng Xianjun, Cui Jiufa, Xu Wenjian, Yang Yang, Fayad Zahi A., et al. CT imaging features of 2019 Novel Coronavirus (2019-nCoV) Radiology. 2020;295(1):202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roy S., Menapace W., Oei S., Luijten B., Fini E., Saltori C., Huijben I., Chennakeshava N., Mento F., Sentelli A., Peschiera E., Trevisan R., Maschietto G., Torri E., Inchingolo R., Smargiassi A., Soldati G., Rota P., Passerini A., van Sloun R.J.G., Ricci E., Demi L. Deep learning for classification and localization of COVID-19 markers in point-of-care lung ultrasound. IEEE Trans. Med. Imag. 2020;39(8):2676–2687. doi: 10.1109/TMI.2020.2994459. [DOI] [PubMed] [Google Scholar]
- 8.Mahmud Tanvir, Rahman Md Awsafur, Fattah Shaikh Anowarul. CovXNet: a multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization. Comput. Biol. Med. 2020;122:103869. doi: 10.1016/j.compbiomed.2020.103869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hassantabar Shayan, Ahmadi Mohsen, Sharifi Abbas. Diagnosis and detection of infected tissue of COVID-19 patients based on lung X-ray image using convolutional neural network approaches. Chaos, Solitons & Fractals. 2020;140:110170. doi: 10.1016/j.chaos.2020.110170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan Li, Li Dong, Xue Huadan, Zhang Longjiang, Liu Zaiyi, Zhang Bing, Zhang Lina, Yang Wenjie, Xie Baojun, Duan Xiaoyi, et al. Progress andprospect on imaging diagnosis of COVID-19. Chinese Journal of Academic Radiology. 2020:1–10. doi: 10.1007/s42058-020-00031-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horry M.J., Chakraborty S., Paul M., Ulhaq A., Pradhan B., Saha M., Shukla N. COVID-19 detection through transfer learning using multimodal imaging data. IEEE Access. 2020;8:149808–149824. doi: 10.1109/ACCESS.2020.3016780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amatya Yogendra, Rupp Jordan, Russell Frances M., Saunders Jason, Bales Brian, House Darlene R. Diagnostic use of lung ultrasound compared to chest radiograph for suspected pneumonia in a resource-limited setting. Int. J. Emerg. Med. 2018;11(1) doi: 10.1186/s12245-018-0170-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soldati Gino, Smargiassi Andrea, Inchingolo Riccardo, Buonsenso Danilo, Perrone Tiziano, Briganti Domenica Federica, Perlini Stefano, Torri Elena, Mariani Alberto, Mossolani Elisa Eleonora, Tursi Francesco, Mento Federico, Demi Libertario. Is there a role for lung ultrasound during the COVID-19 pandemic? J. Ultrasound Med. 2020;39(7):1459–1462. doi: 10.1002/jum.15284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soldati Gino, Smargiassi Andrea, Inchingolo Riccardo, Buonsenso Danilo, Perrone Tiziano, Briganti Domenica Federica, Perlini Stefano, Torri Elena, Mariani Alberto, Eleonora Mossolani Elisa, Tursi Francesco, Mento Federico, Demi Libertario. Proposal for international standardization of the use of lung ultrasound for patients with COVID-19. J. Ultrasound Med. 2020;39(7):1413–1419. doi: 10.1002/jum.15285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andrea Smargiassi, Gino Soldati, Elena Torri, Federico Mento, Domenico Milardi, Paola Del Giacomo, Giuseppe De Matteis, Maria Livia Burzo, Anna Rita Larici, Maurizio Pompili, Libertario Demi, and Riccardo Inchingolo, Lung ultrasound for COVID-19 patchy pneumonia,” J. Ultrasound Med.. [DOI] [PubMed]
- 16.Buonsenso Danilo, Pata Davide, Chiaretti Antonio. COVID-19 outbreak: less stethoscope, more ultrasound. The Lancet Respiratory Medicine. 2020;8(5):e27. doi: 10.1016/S2213-2600(20)30120-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carrer L., Donini E., Marinelli D., Zanetti M., Mento F., Torri E., Smargiassi A., Inchingolo R., Soldati G., Demi L., Bovolo F., Bruzzone L. Automatic pleural line extraction and COVID-19 scoring from lung ultrasound data. IEEE Trans. Ultrason. Ferroelectrics Freq. Contr. 2020;(1–1) doi: 10.1109/TUFFC.2020.3005512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaderberg Max, Simonyan Karen. In: Cortes C., Lawrence N., Lee D., Sugiyama M., Garnett R., editors. vol. 28. 2015. Andrew zisserman, and koray kavukcuoglu, spatial transformer networks; pp. 2017–2025. (In Advances In Neural Information Processing Systems). Curran Associates, Inc. [Google Scholar]
- 19.Italian COVID-19 Lung Ultrasound Data Base. 2020. https://iclus-web.bluetensor.ai/ [Online] Available: ICLUS-DB. [Google Scholar]
- 20.Vetrugno Luigi, Bove Tiziana, Orso Daniele, Barbariol Federico, Bassi Flavio, Boero Enrico, Ferrari Giovanni, Kong Robert. Our Italian experience using lung ultrasound for identification, grading and serial follow-up of severity of lung involvement for management of patients with COVID-19. Echocardiography. 2020;37(4):625–627. doi: 10.1111/echo.14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodfellow Ian, Bengio Yoshua, Courville Aaron. 2016. Deep Learning.http://www.deeplearningbook.org MIT Press. [Google Scholar]
- 22.Murphy Kevin P. 2012. Machine Learning: A Probabilistic Perspective. MIT press. [Google Scholar]
- 23.Chollet F. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2017. Xception: Deep Learning with Depthwise Separable Convolutions; pp. 1800–1807. [Google Scholar]
- 24.Huang Gao, Liu Zhuang, van der Maaten Laurens, Kilian Q. Proceedings Of the IEEE Conference On Computer Vision And Pattern Recognition (CVPR) 2017. Weinberger, densely connected convolutional networks; pp. 4700–4708. [Google Scholar]
- 25.Hochreiter Sepp, Schmidhuber Jurgen. Long short-term memory. Neural Comput. 1997;9(8):1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 26.Greff K., Srivastava R.K., Koutník J., Steunebrink B.R., Schmidhuber J. LSTM: a search space odyssey. IEEE Transactions on Neural Networks and Learning Systems. 2017;28(10):2222–2232. doi: 10.1109/TNNLS.2016.2582924. [DOI] [PubMed] [Google Scholar]
- 27.Diederik P. 2014. Kingma and Jimmy Ba, Adam: A Method for Stochastic Optimization. [Google Scholar]
- 28.Srivastava Nitish, Hinton Geoffrey, Krizhevsky Alex, Sutskever Ilya, Salakhutdinov Ruslan, Dropout A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 1958;15(1):1929. [Google Scholar]
- 29.Krizhevsky Alex, Sutskever Ilya, Hinton Geoffrey E. In: Pereira F., Burges C.J.C., Bottou L., Weinberger K.Q., editors. Curran Associates, Inc.; 2012. ImageNet Classification with Deep Convolutional Neural Networks,” in Advances in Neural Information Processing Systems 25; pp. 1097–1105. [Google Scholar]
- 30.Ketelaars R., Gülpinar E., Roes T., Kuut M., van Geffen G.J. Which ultrasound transducer type is best for diagnosing pneumothorax? Crit. Ultrasound J. 2018;10(1):27. doi: 10.1186/s13089-018-0109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gargani Luna, Volpicelli Giovanni. How I do it: lung ultrasound. Cardiovasc. Ultrasound. 2014;12(1) doi: 10.1186/1476-7120-12-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lichtenstein Daniel A., Mezière Gilbert A., Lagoueyte Jean-François, Biderman Philippe, Goldstein Ivan, Gepner Agnès. A-lines and B-lines: lung ultrasound as a bedside tool for predicting pulmonary artery occlusion pressure in the critically ill. Chest. 2009;136(4):1014–1020. doi: 10.1378/chest.09-0001. [DOI] [PubMed] [Google Scholar]
- 33.Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D., Grad-Cam . 2017 IEEE International Conference on Computer Vision (ICCV) 2017. Visual explanations from deep networks via gradient-based localization,”; pp. 618–626. [Google Scholar]
- 34.Rudin Cynthia. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nature Machine Intelligence. 2019;1(5):206–215. doi: 10.1038/s42256-019-0048-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cid Ximena, Wang Andrew, Heiberg Johan, Canty David, Royse Colin, Li Xiaoqiang, El-Ansary Doa, Yang Yang, Haji Kavi, Haji Darsim, et al. Point-of-care lung ultrasound in the assessment of patients with COVID-19: a tutorial. Australasian Journal of Ultrasound in Medicine. 2020;23(4):271–281. doi: 10.1002/ajum.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]