Abstract
Simple Summary
Hepatocellular carcinoma (HCC) is one of the deadliest cancer. Clinical guidelines for the management of HCC endorse algorithms deriving from clinical variables whose performances to prognosticate HCC is limited. Liquid biopsy is the molecular analysis of tumor by-products released into the bloodstream. It offers minimally-invasive access to circulating analytes like DNA, RNA, exosomes and cells. This technology demonstrated promising results for various applications in cancers, including prognostication. This review aimed to provide a comprehensive overview of the contribution of liquid biopsy in HCC prognostication. The results suggested that liquid biopsy may be a polyvalent and valuable tool to prognosticate HCC.
Abstract
Showing a steadily increasing cancer-related mortality, the epidemiological evolution of hepatocellular carcinoma (HCC) is concerning. Numerous strategies have attempted to prognosticate HCC but their performance is modest; this is partially due to the heterogeneous biology of this cancer. Current clinical guidelines endorse classifications and scores that use clinical variables, such as the Barcelona Clinic Liver Cancer (BCLC) classification. These algorithms are unlikely to fully recapitulate the genomic complexity of HCC. Integrating molecular readouts on a patient-basis, following a precision-medicine perspective, might be an option to refine prognostic systems. The limited access to HCC tissue samples is an important limitation to these approaches but it could be partially circumvented by using liquid biopsy. This concept consists of the molecular analysis of products derived from a solid tumor and released into biological fluids, mostly into the bloodstream. It offers an easy and minimally-invasive access to DNA, RNA, extracellular vesicles and cells that can be analyzed with next-generation sequencing (NGS) technologies. This review aims to investigate the potential contributions of liquid biopsy in HCC prognostication. The results identified prognostic values for each of the components of liquid biopsy, suggesting that this technology may help refine HCC prognostication.
Keywords: liver cancer, biomarkers, precision medicine, circulating, prognosis
1. Background
Hepatocellular carcinoma (HCC) shows a worrisome epidemiological trajectory [1]. The WHO predicts over 1 million HCC-related deaths in 2030 [2]. A particular feature of HCC is that it typically arises on a chronically damaged organ, mostly cirrhosis, for which viral hepatitis, alcohol use disorder and NAFLD are common causes. This facilitates the identification of patients at risk and has enabled successful surveillance programs for early cancer detection. However, having two potentially life-threatening diseases in the same patient (i.e., HCC and cirrhosis) complicates its clinical management and prognosis prediction. Numerous attempts have been pursued to reliably prognosticate HCC, using various strategies. The most widely used classifications, like the Barcelona Clinic Liver Cancer (BCLC) algorithm [3,4], relies on clinical variables. Other approaches investigate the contribution of tumor markers like α-fetoprotein (AFP) [5] and molecular markers derived from the tumor or adjacent non-tumor samples [6,7,8,9]. Regardless of the strategy, the prognostic performance of these algorithms needs to be improved. Following a ‘precision medicine’ perspective may be a way of improving HCC prognostication. This implies access to genomic data on a patient-basis, which requires biopsy or surgical specimens for tissue samples of the tumor. This is particularly difficult in HCC for which, unlike most solid tumors, diagnosis mostly relies on imaging and tissues samples are rarely available [10,11]. In addition, tissue biopsies are associated with potential complications and should not be sequentially repeated [12].
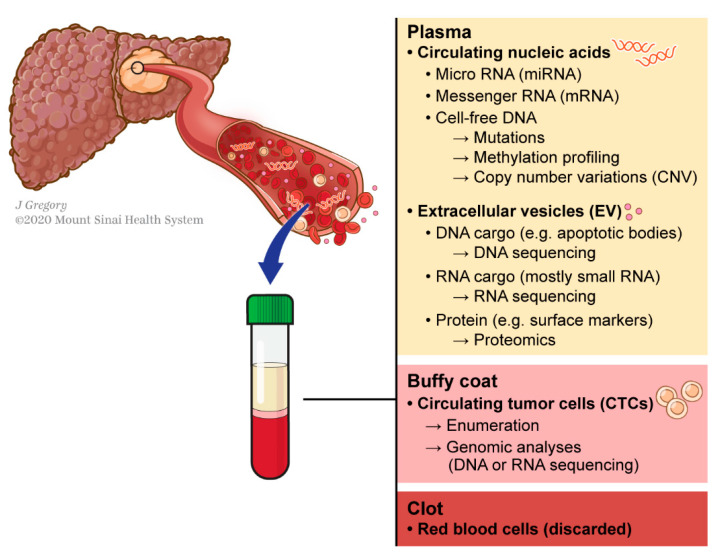
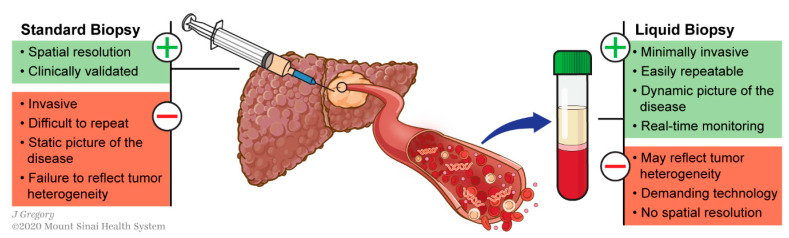
This can be circumvented using liquid biopsy, which refers to the molecular analysis of tumor components released from a solid tumor into biological fluids like blood. These analytes include circulating tumor nucleic acids (DNA and RNAs), cells (CTCs) and exosomes (Figure 1). This technology has shown promising results for various applications in the management of different types of cancers including HCC [13,14,15]: early diagnosis [16,17], detection of minimal residual disease [18], decision-making for systemic therapies [19,20] or even to decipher complex biological traits of cancers [21,22,23,24]. This technology offers a valuable alternative to standard biopsy. Tissue biopsy is indeed invasive and associated with potential risks such as pain, bleeding or even seeding of the cancer (PMID: 18669577). Conversely to standard biopsy, liquid biopsy displays the advantages of being easily repeatable and can thereby help for monitoring, providing a dynamic picture of the disease course. In addition, it may reflect different regions of the tumor and thus recapitulate eventual intra-tumoral heterogeneity (ITH) (Figure 2) [25].
Figure 1.
Concept of liquid biopsy referring to the molecular analysis of tumor-byproducts released into the bloodstream.
Figure 2.
Pros and cons of liquid biopsy versus standard biopsy.
This study aims to provide a comprehensive overview of the potential contributions of each component of liquid biopsy (i.e., DNA, RNAs and cells) to HCC prognostication.
1.1. Circulating Tumor DNA (ctDNA)
DNA fragments are released from solid tumors into the bloodstream via active and passive mechanisms. The latter seems to predominate, being partially driven by cell necrosis and apoptosis [26]. This occurs at any tumor stage and offers minimally-invasive access to key molecular information of the tumor including genomic (copy number variations (CNV) or point mutations) as well as epigenetic (DNA methylations changes) data. Numerous studies show the value of ctDNA as a polyvalent biomarker in cancer. For example, ctDNA allowed detection of minimal residual disease (MRD) in a prospective cohort of 230 patients undergoing surgical resection of stage II colon cancer. Postoperative detection of ctDNA outperformed prognostic factors such as TNM stage, for the prediction of recurrence-free survival [18].
In HCC, a pilot study demonstrated that detection of mutations in the plasma of HCC patients was feasible and recapitulated the ones detected in tumor tissue [27]. Table 1 summarizes reports investigating ctDNA in HCC prognostication.
Table 1.
Circulating tumor DNA (ctDNA).
| Number of Patients | Treatment | Biomarkers | Technique | Main Finding | [Ref.] |
|---|---|---|---|---|---|
| CNV | |||||
| 34 HCC | Surgery | ctDNA (harboring SNV or CNV) | Targeted-sequencing and low coverage whole-genome sequencing | ctDNA can detect minimal residual disease (MRD) and predict survival | [28] |
| 151 HCC; 14 healthy controls | Sorafenib | VEGFA amplification | Whole-genome sequencing | High concentration of cell-free DNA (cfDNA) was associated with poor outcomes but VEGFA ratio was not a prognostic factor. | [29] |
| Mutations | |||||
| 46 HCC | Surgery Transplant |
ctDNA | Targeted-sequencing and exome-sequencing | Detection of ctDNA was associated with increased recurrence | [30] |
| 41 HCC; 10 controls | Surgery | TERT, TP53 and CTNNB1 | Targeted-sequencing | Detection of ctDNA predicted shorter recurrence-free survival | [31] |
| 10 HCC | Surgery TACE RFA |
Methylation of GSTP1 and RASSF1A or TP53 mutation | Methylation-specific PCR and sanger sequencing | Detecting ctDNA in urine was feasible and predicted recurrence | [32] |
| 218 HCC; 81 cirrhotic | NA | TERT promoter mutation (C228T and C250T) | Droplet digital PCR (ddPCR) and sanger sequencing | TERT promoter mutation can be used as an early biomarker of HCC and is associated with survival | [33] |
| 34 HCC | Surgery | ctDNA (harboring SNV or CNV) | Targeted-sequencing and low coverage whole-genome sequencing | ctDNA can detect minimal residual disease (MRD) and predict survival | [28] |
| 95 HCC; 45 cirrhotic | Surgery | TERT promoter mutation (C228T) | Droplet digital PCR (ddPCR) | Detection of mutated TERT promoter was associated with lower survival | [34] |
| 59 HCC | Surgery TACE RFA Systemic chemotherapy BSC |
Single nucleotide variant (SNV) in a panel of 69 genes | Targeted-sequencing | Mutated MLH1 in plasma was associated with lower survival | [35] |
| 130 HCC | TACE Systemic chemotherapy |
TERT promoter mutation | Droplet digital PCR (ddPCR) | Detection of mutated TERT promoter was associated with lower survival | [36] |
| 895 HCC | Surgery/NA | TP53 mutation (R249S) | Droplet digital PCR (ddPCR) | Detection of mutated TP53 was associated with lower survival | [37] |
| 22 HCC | TKI (tyrosine kinase inhibitors) | Genes of the PI3K/MTOR pathway | Targeted-sequencing and ddPCR | Mutations of genes in the PI3K/MTOR pathway are associated with lower survival in patients treated with TKI | [38] |
| Methylation Changes | |||||
| 72 HCC; 37 benign liver diseases; 41 healthy controls | - | APC, GSTP1, RASSF1A, and SFRP1 | Methylation-specific PCR | Methylation of RASSF1A was associated with poor survival | [39] |
| 1098 HCC; 835 controls | NA | 8-marker panel | Targeted bisulfite sequencing | Methylation-based classifier predicted survival | [40] |
| 10 HCC | TACE RFA Surgery |
Methylation of GSTP1 and RASSF1A or TP53 mutation | Methylation-specific PCR and sanger sequencing | Detecting ctDNA in urine was feasible and predicted recurrence | [32] |
| 203 HCC; 104 chronic viral hepatitis B or C; 50 healthy controls | NA | APC, COX2, RASSF1A (+miR-203) | Methylation-specific PCR | Classifier predicted survival | [41] |
| 172 HCC | NA | LINE-1 | Methylation-specific PCR | Hypomethylation of LINE-1 was associated with lower survival | [42] |
| 155 HCC; 60 chronic HBV; 20 healthy controls | Surgery | IGFBP7 | Methylation-specific PCR | Methylation of IGFBP7 was associated with lower survival | [43] |
| 43 HCC (+347 HCC from TCGA Atlas); 5 cirrhotic; 6 benign liver lesions | - | CTCFL | Methylation-specific PCR | Hypomethylation of CTCFL was associated with higher recurrence and lower survival | [44] |
1.1.1. Copy Number Variations (CNVs)
CNVs are potential drivers of hepatocarcinogenesis; they predominantly affect chromosomes (chr) 8 and 11 [45]. A pioneer study took advantage of this knowledge and of a mathematical model to infer CNVs using ctDNA in HCC and non-HCC patients [26]. First, it determined the typical size of DNA fragments, approximating 160 bp, thereby suggesting that DNA fragments are foremost released by apoptotic cancer cells. Second, the model performed well in distinguishing patients with chronic HBV with and without HCC with an area under the curve (AUC) of 0.93.
In plasma, a recent study targeted both CNVs and mutations using ctDNA in 34 HCC patients undergoing surgery. CtDNA was reported as a prognostic factor for survival, and it was also able to detect MRD [28]. Another study targeted VEGFA amplification in circulating-free DNA (cfDNA), assuming that it would predict response to sorafenib. Although a high concentration of cfDNA was associated with lower survival, the VEGFA ratio was not a predictor of response to therapy [29].
1.1.2. Mutations
The genomic complexity of HCC–characterized by a wide spectrum of different potential driver mutations [46,47]–and the low amount of tumor DNA among the pool of cell free DNA are two major challenges when conducting mutation calling from the plasma of HCC patients. Various techniques can be applied to detect plasma mutations: droplet digital PCR (ddPCR) is better suited to targeting a small number of genes, whereas targeted-sequencing allows investigation of a larger panel of candidates. Although whole-genome sequencing is feasible, it is associated with relatively low coverage, making the interpretation of mutations calling cumbersome.
Several studies focused on TERT promoter, TP53 and CTNNB1, as they are commonly mutated genes in HCC. In a cohort of 41 HCC patients and 10 controls, detection of these mutations was associated with shorter recurrence-free survival after liver resection [31]. This was confirmed by other studies identifying TERT prom and TP53 mutations as prognostic factors of poor survival [33,34,36,37]. Targeted-sequencing of various panels of genes further confirmed the prognostic impact of ctDNA detection, associated with worse survival or higher recurrence [30]. Providing more detailed analyses than just the simple presence/absence of ctDNA, Kim et al. showed that MLH1 mutation was specifically associated with lower survival [35], whereas von Felden et al. recently demonstrated that mutations of genes from the PI3K/mTOR pathway were predictors of non-response to tyrosine kinase inhibitors (TKI) in patients with advanced HCC [38].
Although most reports used blood samples, liquid biopsy is applicable to other types of biological fluids like urine or saliva. This was illustrated by a study demonstrating the feasibility of detecting mutations in urine samples of HCC patients. Moreover, detection of mutation preceded tumor recurrence as detected by magnetic resonance imaging (MRI) [32].
1.1.3. DNA Methylation Changes
The carcinogenic role of epigenetic events like DNA methylation is well known in HCC [48,49]. Changes in DNA methylation can also be detected in cfDNA. Similar to mutations, focus can be either on specific candidate or interrogate multiple CpG sites. Several studies identified methylation changes of specific genes associated with HCC outcomes: hypomethylation of LINE-1 [42] and methylation of IGFBP7 [43] were associated with lower survival, whereas hypomethylation of CTCFL predicted higher tumor recurrence and lower survival [44]. A study including 1095 HCC and 835 controls generated a classifier of 8 markers for diagnosis and prognosis (SH3PXD2A, C11orf9, PPFIA1, Chr 17:78, SERPINB5, NOTCH3, GRHL2 and TMEM8B). In addition to providing a high diagnostic accuracy, the score was also associated with survival [40]. Two other studies used comparable approaches and provided similar findings [39,41].
1.2. Circulating Free RNAs (cfRNAs)
RNAs include a large family of members: micro, long non-coding, messenger or exosomal RNAs. Herein, the present review will focus on the most commonly investigated circulating RNAs in liquid biopsies (Table 2).
Table 2.
Circulating free RNAs (cfRNAs) and exosomes.
| Number of Patients | Treatment | Biomarkers | Technique | Main Finding | [Ref.] |
|---|---|---|---|---|---|
| miRNA | |||||
| 195 HCC 54 cirrhotic |
Surgery Transplant TACE RFA sorafenib |
miR-1 and miR-12 | qRT-PCR | Low level of miR-1 was associated with lower survival | [50] |
| 122 HCC | Surgery | miR-122 | qRT-PCR | Low level of miR-122 was associated with lower survival | [51] |
| 120 HCC | Surgery RFA |
MiR-21, miR-26a, and miR-29a | qRT-PCR | Low levels of miR-26a and miR-29a were associated with lower survival | [52] |
| 30 HCC; 30 controls | Surgery | miR-155, miR-96 and miR-99a | qRT-PCR | High levels of miR-155 and miR-96 were associated with lower survival | [53] |
| 116 HCC | NA | Circulating miR | Whole miRNome profling | Low levels of miR-424-5p, miR-101-3p or high levels of miR-128, miR-139-5p, miR-382-5p and miR410 were associated with lower survival | [54] |
| 41 HCC; 20 controls | Surgery transplant |
miR193a-5p | qRT-PCR | High level of miR193a-5p was associated with lower survival | [55] |
| 70 HBV-related HCC 70 HBV 50 healthy controls |
Surgery | miRNA-223-3p | qRT-PCR | Low level of miRNA-223-3p was associated with lower survival | [56] |
| mRNA | |||||
| 50 HCC; 50 controls | Surgery | VEGF-165 | qRT-PCR | Detection of circulating VEGF mRNA (isoform 165) was associated with higher recurrence and recurrence-related mortality | [57] |
| 38 HCC | Surgery | AFP | qRT-PCR | Detection of AFP mRNA was associated with extrahepatic recurrence and shorter disease-free survival | [58] |
| 343 HCC | Surgery TACE RFA Systemic chemotherapy Radiotherapy BSC |
AFP and hTERT | qRT-PCR | Detection of AFP mRNA or hTERT mRNA was not associated with survival | [59] |
| Exosomes | |||||
| 59 HCC | Transplant | miR-718 | qRT-PCR | Recurrence was associated with higher level of exosomal miR-718 | [60] |
| 30 HCC | Surgery | miR-665 | qRT-PCR | High level of exosomal miR-665 was associated with lower survival | [61] |
| 79 HCC | Surgery Transplant TACE RFA Sorafenib BSC |
miR-21 and lncRNA-ATB | qRT-PCR | High levels of exosomal miR-21 and lncRNA-ATB were associated with lower survival | [62] |
| 126 HCC; 21 healthy controls | Surgery | miR-638 | qRT-PCR | Low level of exosomal miR-638 was associated with lower survival | [63] |
| 31 HCC; 3 CLD; 11 healthy controls | NA | RN7SL1 S fragment | qRT-PCR | High expression of RN7SL1 S fragment was associated with lower survival | [64] |
| 124 HCC; 100 healthy controls | Surgery | AKT3 | qRT-PCR | High level of exosomal circulating AKT3 was associated with higher recurrence and lower survival rates | [65] |
| 104 HCC; 55 CLD; 50 healthy controls | Surgery | miR-320a | qRT-PCR | Low serum exosomal miR-320a was associated with lower survival | [66] |
1.2.1. Micro-RNAs (miRNAs)
MiRNAs have gained increased interest as cancer biomarkers, as they have key properties, in particular their molecular stability. Lin et al. established a classifier based on seven miRNAs, which was able to detect preclinical HCC [67]. This could be a valuable tool for HCC surveillance, which could outperform the recommended bi-annual ultrasound (US) and AFP measurement.
A number of circulating miRNAs have shown prognostic value in HCC. Low levels of miR-1, miR-122, miR-26a, miR-29a and miR-223-3p were associated with lower survival [50,51,52,56]. Patients with high levels of miR-155, miR-96 and miR-193-5p had lower survival rates [53,55]. A recent study reported whole miRNome profiling in 116 HCC patients [54]. Using three different cohorts, the study reported on miRNAs differentially expressed in HCC vs. non-HCC patients. This effort identified miRNAs with specific clinical utilities; certain biomarkers detected cirrhosis, while others detected HCC. Furthermore, six miRNAs were identified as prognostic factors. Down-regulation of miR-424-5p or miR-101-3p and up-regulated miR-128, miR-139-5p, miR-382-5p and miR410 were associated with lower survival.
1.2.2. Messenger RNAs (mRNAs)
Unlike miRNAs, circulating mRNAs are highly unstable and are thus rarely explored as liquid biopsy analytes. Studies attempting to measure mRNAs in blood samples analyzed a limited number of candidates.
A comparison of 50 HCC patients and 50 controls detected an association between VEGF expression level (isoform 165) and the risk of tumor recurrence [57]. The concentration of circulating mRNAs coding for AFP was investigated in two other studies. The first one included 38 HCC patients undergoing partial resection and showed that detection of AFP mRNA was associated with extrahepatic recurrence and shorter disease-free survival [58]. The second one tested both levels of AFP and hTERT mRNAs, but failed to identify any prognostic impact [59].
1.3. Extracellular Vesicles (EVs): Exosomes
Exosomes are a type of EV, nanoparticles encapsulating a variety of cargo including DNA and RNA fragments in a lipidic double-layer, which protects them from enzymatic degradation. With these unique features, circulating exosomes allow RNAs to circulate without being degraded plasma. Their nature and roles remain largely unknown but exosomes may not only be passively released from apoptotic cells into the bloodstream. Data suggested they may be actively secreted, acting as messengers in the cell-to-cell communication network, conferring them priceless values like accuracy and tissue-specificity [68,69,70,71].
In HCC, the data exploring the contribution of exosomes remain limited, particularly for prognosis. However, these analytes have demonstrated promising and polyvalent performance in other cancer types both for diagnosis and prognosis [72,73].
Several projects analyzed exosomal miRNAs. In a cohort of 59 HCC patients, authors found a correlation between tumor recurrence after liver transplantation and a higher level of miR-718 [60]. Similar signals were detected after liver resection and other exosomal miRNAs: high levels of miR-665 or low levels of miR-638 and miR-320a were identified as predictors of poor survival [61,63,66]. In a cohort of 79 HCC patients of different stages receiving various treatments, Lee et al. focused on two candidates: a miRNA (miR-21) and a long non-coding RNA (lncRNA) (lncRNA-ATB). On multivariable analysis, both markers were independently associated with disease progression [62]. A recent study profiled 57 plasma cell-free RNA transcriptomes and 20 exosomal RNA transcriptomes to test their diagnostic and prognostic performance. RN7SL1 and its S fragment were promising, showing a high diagnostic accuracy (AUC = 0.87). Furthermore, a higher concentration of RN7SL1 S fragment was an independent factor of worse survival [64]. The analyses of blood samples from 124 HCC patients treated with surgical resection and 100 healthy controls identified an exosomal circular RNA (circAKT3) as a prognostic factor; a high level of circAKT3 predicted both higher recurrence and lower survival [65].
1.4. Circulating Tumor Cells (CTCs)
CTCs play a pivotal role during the hematogenous dissemination of cancers. Most technologies to analyze CTCs include two steps: enrichment (isolation) and detection (identification). The development of sensitive and specific technologies is challenging. Estimated to be released by cancers of intermediate and advanced stages, CTCs are probably more useful for prognostication than for early cancer detection. In this context, studies have demonstrated the prognostic value of CTC enumeration in different cancers including HCC [74,75]. More sophisticated technologies, like single-cell RNA sequencing, have allowed further characterization of CTC subtypes [76]. Studies exploring the value of CTCs for HCC prognostication are summarized in Table 3.
Table 3.
Circulating tumor cells (CTCs).
| Number of Patients | Treatment | Technique of Detection | Main Finding | [Ref.] |
|---|---|---|---|---|
| 44 HCC 30 HCV 39 cirrhosis 38 healthy controls |
Surgery NA |
Isolation by size of epithelial tumor cells (ISET) | Presence and number of detected CTCs were associated with shorter survival | [77] |
| 85 HCC 37 benign liver diseases 20 healthy volunteers 14 miscellaneous advanced cancers other than HCC |
Surgery NA |
Antibody-coated magnetic beads | Presence and number of detected CTCs correlated with tumor size, portal vein tumor thrombus, differentiation status, TNM stage and Milan criteria | [78] |
| 82 HCC | Surgery | Multicolor flow cytometry | Circulating cancer stem cells (CSC) are associated with higher rates of intra- and extra-hepatic recurrence, decreased recurrence-free survival (RFS) and overall survival (OS) rates | [79] |
| 96 HCC 31 healthy controls 21 viral hepatitis 8 cirrhosis |
Surgery | Magnetic cell sorting (Lin28B) | Detection of CTCs expressing Lin28B was associated with early recurrence | [80] |
| 60 HCC | Surgery NA |
Flow cytometry (ICAM-1) | Detection of CTCs expressing ICAM-1 was associated with shorter disease-free survival | [81] |
| 123 HCC | Surgery | EpCAM antibody-coated magnetic beads (CellSearch) | Detection of CTCs (EpCAM+) was associated with higher recurrence | [75] |
| 59 HCC 19 controls |
NA | EpCAM antibody-coated magnetic beads (CellSearch) | Detection of CTCs was associated with lower overall survival | [82] |
| 122 HCC 120 controls |
Surgery TACE Radiotherapy |
EpCAM antibody-coated magnetic beads (CellSearch) | Peri-treatment decrease of detected CTC reflected treatment response | [83] |
| 109 HCC | Surgery TACE RFA sorafenib |
Flow cytometry (ASGPR and CPS1) | pERK+/pAkt-CTCs correlated with progression-free survival and predicted response to systemic therapy (sorafenib) | [84] |
| 72 HCC | Surgery | EpCAM antibody-coated magnetic nanoparticals (MagVigen, Nvigen) | Detection of CTCs expressing AFP was associated with metastatic disease | [85] |
| 69 HCC 31 controls |
Surgery Transplant TACE RFA Sorafenib BSC |
Imaging flow cytometry (EpCAM, AFP, glypican-3 and DNA-PK together with analysis of size, morphology and DNA content) (ImageStream) | Detection of CTCs was associated with lower survival | [86] |
| 57 HCC | Surgery | EpCAM antibody-coated magnetic beads (CellSearch) | CTCs detection was associated with higher recurrence and lower recurrence-free survival after liver resection | [87] |
| 14 HCC 16 CCA 4 GBC |
Surgery | SE-iFISH | Detection of small CTCs with CNV (chromosome 8) was associated with lower survival | [88] |
| 199 HCC | Surgery | Fluorescence-activated cell sorting (FACS) | Anterior approach was associated with a decreased dissemination of CTCs compared to conventional approach, resulting in poorer outcomes. | [89] |
| 73 HCC | Surgery | EpCAM antibody-coated magnetic beads (CellSearch) | Analyzes of blood samples collected in different vessels revealed a spatial heterogeneity of CTCs distribution whose biology was associated with recurrence pattern. | [90] |
| 130 HCC | Surgery TACE |
qRT-PCR test platform | CTCs detection was associated with recurrence after liver resection | [91] |
| 112 HCC | Surgery | CanPatrolTM system (filtration by size) and Tri-color RNA-ISH assay | The presence of CTCs and the proportion of mesenchymal-CTC (M-CTCs) were associated with recurrence | [92] |
| 61 HCC 19 non-HCC |
TACE TARE RFA Systemic therapy |
Antibody-based platform | Vimentin (VIM)-positive CTCs predicted OS and faster recurrence after curative-intent surgical or locoregional therapy in potentially curable early-stage HCC | [93] |
| 139 HCC 23 controls |
Surgery | EpCAM antibody-coated magnetic beads (CellSearch) | Surgical resection induces a release of CTCs | [94] |
| 105 HCC | Surgery | ISET | ΔCTCs is an independent predictor of lower survival and higher recurrence in patients | [95] |
| 85 HCC 27 non-HCC |
Surgery | Flow cytometry (GPC3) | GPC3 positive-CTCs detection was associated with lower survival | [96] |
| 50 HCC | Transplant | Negative enrichment and immunofluorescence in situ hybridization (imFISH) | CTCs detection was associated with early recurrence after liver transplant | [97] |
| 137 HCC | Surgery | ISET | CTCs detection was associated with early recurrence after liver resection | [98] |
| 87 HCC 7 cirrhosis 8 healthy controls |
Transplant Surgery TACE TARE RFA Systemic therapy |
Antibody-based platform | Detection of CTCs expressing PD-L1 were associated with shorter OS and predicted response to immunotherapy | [99] |
| 128 HCC | Surgery ± TACE | EpCAM antibody-coated magnetic beads (CellSearch) | Adjuvant TACE provided survival and recurrence benefits in patients with positive preoperative CTCs | [100] |
| 193 HCC | Transplant | Antibody-based platform (ChimeraX®-i120) | CTCs detection was associated with recurrence after liver transplant | [101] |
Most studies investigating CTCs included HCC patients undergoing surgical resection. In 2004, Vona G et al. isolated and enumerated CTCs based on their size and morphology, showing that the presence and number of detected CTCs were associated with shorter survival [77]. CellSearch® is an isolation system that targets EpCAM positive cells. It was approved by the Food and Drug Administration (FDA) and became the most commonly used technique for CTC enumeration. Its use remains debated in HCC as only around 30% of HCC cells express EpCAM [102]. Nonetheless, several studies utilized this approach in HCC, demonstrating that the detection of EpCAM positive CTCs was associated with higher tumor recurrence [75] or lower survival [82,87]. Thereafter, more sophisticated technologies for CTC isolation have been reported, like ImageStream flow cytometry. A study has provided the proof-of-concept of this technology, demonstrating its capacity to detect CTCs using a panel of markers. This technology also generates high-resolution images of isolated CTCs [103]. Its value in detecting CTCs was confirmed and the CTC count was further confirmed as an independent prognostic factor [86]. Other reports have aimed at exploring the impact of subgroups of CTCs, clustered based on cell surface markers, RNA expression or genomic aberrations. Studies using surface markers to detect CTCs with cancer stem cell-like [79,80,81] or mesenchymal [92] features, revealed their clinical value to predict tumor recurrence. CTCs expressing AFP were also associated with an increased risk of metastasis [85], whereas CTCs harboring CNV (chr 8) predicted worse survival [88]. Ha et al. used a simple isolation technique but introduced the concept of ΔCTC, referring to the perioperative fluctuation of detected CTCs, which appeared as an independent factor of lower survival and higher recurrence rates after partial hepatectomy [95]. Besides their intrinsic biological traits, CTC dissemination seemed to be impacted by treatment. Data suggested that surgery-induced manipulation of the liver is associated with a release of CTCs [94]. A comparison between anterior and conventional surgical approaches suggested that the latter was associated with a higher release of CTCs as well as poorer outcomes [89]. Toso et al. described five steps during orthotopic liver transplant (OLT) in HCC patients to minimize CTC dissemination and thereby the risk of recurrence [104].
Recent studies also underscored the relevance of CTC analysis in patients undergoing OLT for HCC, highlighting an association between CTC detection and recurrence [97,101].
Guiding decision-making would be another application of CTCs. For example, selection of patients who would benefit from adjuvant transarterial chemoembolization (TACE) after surgery or to predict response to systemic therapies like tyrosine kinase inhibitors or immunotherapy [84,99,100]. An interesting study analyzed CTCs using samples collected from different vessels. By doing so, they were able to demonstrate spatial heterogeneity in the distribution of CTCs, with a predominance of epithelial status at release, which gradually switched to EMT-activated phenotype during hematogenous transit [90]. Overall, data consistently identified that the number of CTCs was a surrogate of poor prognosis, predicting higher recurrence and/or lower survival. A recent meta-analysis and data from experimental models corroborated these findings [105,106].
2. Challenges and Future Perspectives
The field of liquid biopsy has numerous challenges. Besides those related to cost and technology, there is a limited understanding of the fundamental mechanisms responsible for the release of tumor molecular components to the bloodstream. A better understanding of these mechanisms would provide new tools and targets to improve the diagnostic and prognostic performance of liquid biopsy analytes. Gasparello et al. performed one of the few studies of liquid biopsies in animal models, identifying potential gateways regulating the detection of ctDNA [107]. Experimental models will be instrumental to better understand these mechanisms. ITH has emerged as a major drawback for single-biopsy biomarker development. The clinical impact of ITH is progressively recognized, even at early tumor stages [108]. Liquid biopsy can help address the clinical issues posed by ITH as it likely includes a molecular composite of tumor components released by any potential tumor area. Thus, it is not restricted by the specific tumor section sampled by a needle-biopsy. There are few data of integrative analysis of different analytes within the liquid biopsy space (e.g., simultaneous evaluation of ctDNA and CTCs). Finally, it is key to have prospective data to determine the exact role of liquid biopsy as a prognostic biomarker in HCC, and which is the clinical niche that will be better suited for this transformative technology.
3. Conclusions
While data on liquid biopsy in HCC remain scanter than for other malignancies, there has been numerous recent publications demonstrating its prognostic value in HCC patients. Potential contributions in HCC prognostication were detected for each of the tumor by-products (e.g., DNA, RNA, exosomes and cells). The next step will be to determine the optimal way of integrating liquid biopsy in the clinical management of HCC patients and to modify current clinical practice guidelines accordingly.
Acknowledgments
The authors would like to thank Jill Gregory for the design of the figures.
Author Contributions
Study concept and design: I.L., A.V., O.D., N.D., E.M.; Acquisition of data: I.L.; Analysis and interpretation of data: I.L., A.V., E.M.; Drafting of the manuscript: I.L.; Critical revision of the manuscript for important intellectual content: I.L., A.V., O.D., N.D., E.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
AV has received consulting fees from Guidepoint, Fujifilm, Boehringer Ingelheim, FirstWord, and MHLife Sciences; advisory board fees from Exact Sciences, Nucleix, Gilead and NGM Pharmaceuticals; and research support from Eisai.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Villanueva A. Hepatocellular Carcinoma. N. Engl. J. Med. 2019;380:1450–1462. doi: 10.1056/NEJMra1713263. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization Projections of Mortality and Causes of Death, 2016 to 2060. [(accessed on 14 October 2018)]; Available online: http://www.who.int/healthinfo/global_burden_disease/projections/en/
- 3.Llovet J.M., Bru C., Bruix J. Prognosis of hepatocellular carcinoma: The BCLC staging classification. Semin. Liver. Dis. 1999;19:329–338. doi: 10.1055/s-2007-1007122. [DOI] [PubMed] [Google Scholar]
- 4.Bruix J., Sherman M., American Association for the Study of Liver Diseases Management of hepatocellular carcinoma: An update. Hepatology. 2011;53:1020–1022. doi: 10.1002/hep.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He C., Peng W., Liu X., Li C., Li X., Wen T.F. Post-treatment alpha-fetoprotein response predicts prognosis of patients with hepatocellular carcinoma: A meta-analysis. Medicine (Baltimore) 2019;98:e16557. doi: 10.1097/MD.0000000000016557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Labgaa I., Torrecilla S., Martinez-Quetglas I., Sia D. Genetics of Hepatocellular Carcinoma: Risk Stratification, Clinical Outcome, and Implications for Therapy. Digest. Disease Interv. 2017;01:055–065. doi: 10.1055/s-0037-1603577. [DOI] [Google Scholar]
- 7.Goossens N., Labgaa I., Villanueva A. Nontumor Prognostic Factors in Hepatocellular Carcinoma. In: Carr B.I., editor. Hepatocellular Carcinoma. Current Clinical Oncology. Springer; Cham, Switzerland: 2016. [DOI] [Google Scholar]
- 8.Hoshida Y., Villanueva A., Kobayashi M., Peix J., Chiang D.Y., Camargo A., Gupta S., Moore J., Wrobel M.J., Lerner J., et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N. Engl. J. Med. 2008;359:1995–2004. doi: 10.1056/NEJMoa0804525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villanueva A., Hoshida Y., Battiston C., Tovar V., Sia D., Alsinet C., Cornella H., Liberzon A., Kobayashi M., Kumada H., et al. Combining clinical, pathology, and gene expression data to predict recurrence of hepatocellular carcinoma. Gastroenterology. 2011;140:1501–1512. doi: 10.1053/j.gastro.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.European Association for the Study of the Liver EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2018;69:182–236. doi: 10.1016/j.jhep.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 11.Heimbach J.K., Kulik L.M., Finn R.S., Sirlin C.B., Abecassis M.M., Roberts L.R., Zhu A.X., Murad M.H., Marrero J.A. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology. 2018;67:358–380. doi: 10.1002/hep.29086. [DOI] [PubMed] [Google Scholar]
- 12.Silva M.A., Hegab B., Hyde C., Guo B., Buckels J.A., Mirza D.F. Needle track seeding following biopsy of liver lesions in the diagnosis of hepatocellular cancer: A systematic review and meta-analysis. Gut. 2008;57:1592–1596. doi: 10.1136/gut.2008.149062. [DOI] [PubMed] [Google Scholar]
- 13.Labgaa I., Villanueva A. Liquid biopsy in liver cancer. Discov. Med. 2015;19:263–273. [PubMed] [Google Scholar]
- 14.von Felden J., Garcia-Lezana T., Schulze K., Losic B., Villanueva A. Liquid biopsy in the clinical management of hepatocellular carcinoma. Gut. 2020;69:2025–2034. doi: 10.1136/gutjnl-2019-320282. [DOI] [PubMed] [Google Scholar]
- 15.Labgaa I., Craig A.J., Villanueva A. Diagnostic and Prognostic Performance of Liquid Biopsy in Hepatocellular Carcinoma. In: Giordano A., Rolfo C., Russo A., editors. Liquid Biopsy in Cancer Patients. Current Clinical Pathology. Humana Press; Cham, Switzerland: 2017. [DOI] [Google Scholar]
- 16.Cohen J.D., Li L., Wang Y., Thoburn C., Afsari B., Danilova L., Douville C., Javed A.A., Wong F., Mattox A., et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bettegowda C., Sausen M., Leary R.J., Kinde I., Wang Y., Agrawal N., Bartlett B.R., Wang H., Luber B., Alani R.M., et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014;6:224. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tie J., Wang Y., Tomasetti C., Li L., Springer S., Kinde I., Silliman N., Tacey M., Wong H.L., Christie M., et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl. Med. 2016;8:346. doi: 10.1126/scitranslmed.aaf6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tie J., Cohen J.D., Wang Y., Christie M., Simons K., Lee M., Wong R., Kosmider S., Ananda S., McKendrick J., et al. Circulating Tumor DNA Analyses as Markers of Recurrence Risk and Benefit of Adjuvant Therapy for Stage III Colon Cancer. JAMA Oncol. 2019 doi: 10.1001/jamaoncol.2019.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee B., Lipton L., Cohen J., Tie J., Javed A.A., Li L., Goldstein D., Burge M., Cooray P., Nagrial A., et al. Circulating tumor DNA as a potential marker of adjuvant chemotherapy benefit following surgery for localized pancreatic cancer. Ann. Oncol. 2019;30:1472–1478. doi: 10.1093/annonc/mdz200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parikh A.R., Leshchiner I., Elagina L., Goyal L., Levovitz C., Siravegna G., Livitz D., Rhrissorrakrai K., Martin E.E., Van Seventer E.E., et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat. Med. 2019;25:1415–1421. doi: 10.1038/s41591-019-0561-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Russo M., Misale S., Wei G., Siravegna G., Crisafulli G., Lazzari L., Corti G., Rospo G., Novara L., Mussolin B., et al. Acquired Resistance to the TRK Inhibitor Entrectinib in Colorectal Cancer. Cancer Discov. 2016;6:36–44. doi: 10.1158/2159-8290.CD-15-0940. [DOI] [PubMed] [Google Scholar]
- 23.Russo M., Siravegna G., Blaszkowsky L.S., Corti G., Crisafulli G., Ahronian L.G., Mussolin B., Kwak E.L., Buscarino M., Lazzari L., et al. Tumor Heterogeneity and Lesion-Specific Response to Targeted Therapy in Colorectal Cancer. Cancer Discov. 2016;6:147–153. doi: 10.1158/2159-8290.CD-15-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siravegna G., Mussolin B., Buscarino M., Corti G., Cassingena A., Crisafulli G., Ponzetti A., Cremolini C., Amatu A., Lauricella C., et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat. Med. 2015;21:795–801. doi: 10.1038/nm.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Rubis G., Rajeev Krishnan S., Bebawy M. Liquid Biopsies in Cancer Diagnosis, Monitoring, and Prognosis. Trends Pharmacol. Sci. 2019;40:172–186. doi: 10.1016/j.tips.2019.01.006. [DOI] [PubMed] [Google Scholar]
- 26.Jiang P., Chan C.W., Chan K.C., Cheng S.H., Wong J., Wong V.W., Wong G.L., Chan S.L., Mok T.S., Chan H.L., et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc. Natl. Acad. Sci. USA. 2015;112:E1317–E1325. doi: 10.1073/pnas.1500076112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Labgaa I., Villacorta-Martin C., D’Avola D., Craig A.J., von Felden J., Martins-Filho S.N., Sia D., Stueck A., Ward S.C., Fiel M.I., et al. A pilot study of ultra-deep targeted sequencing of plasma DNA identifies driver mutations in hepatocellular carcinoma. Oncogene. 2018;37:3740–3752. doi: 10.1038/s41388-018-0206-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai Z., Chen G., Zeng Y., Dong X., Li Z., Huang Y., Xin F., Qiu L., Xu H., Zhang W., et al. Comprehensive Liquid Profiling of Circulating Tumor DNA and Protein Biomarkers in Long-Term Follow-Up Patients with Hepatocellular Carcinoma. Clin. Cancer Res. 2019;25:5284–5294. doi: 10.1158/1078-0432.CCR-18-3477. [DOI] [PubMed] [Google Scholar]
- 29.Oh C.R., Kong S.Y., Im H.S., Kim H.J., Kim M.K., Yoon K.A., Cho E.H., Jang J.H., Lee J., Kang J., et al. Genome-wide copy number alteration and VEGFA amplification of circulating cell-free DNA as a biomarker in advanced hepatocellular carcinoma patients treated with Sorafenib. BMC Cancer. 2019;19:292. doi: 10.1186/s12885-019-5483-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ono A., Fujimoto A., Yamamoto Y., Akamatsu S., Hiraga N., Imamura M., Kawaoka T., Tsuge M., Abe H., Hayes C.N., et al. Circulating Tumor DNA Analysis for Liver Cancers and Its Usefulness as a Liquid Biopsy. Cell Mol. Gastroenterol. Hepatol. 2015;1:516–534. doi: 10.1016/j.jcmgh.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao W., Yang H., Xu H., Wang Y., Ge P., Ren J., Xu W., Lu X., Sang X., Zhong S., et al. Noninvasive detection of tumor-associated mutations from circulating cell-free DNA in hepatocellular carcinoma patients by targeted deep sequencing. Oncotarget. 2016;7:40481–40490. doi: 10.18632/oncotarget.9629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hann H.W., Jain S., Park G., Steffen J.D., Song W., Su Y.H. Detection of urine DNA markers for monitoring recurrent hepatocellular carcinoma. Hepatoma Res. 2017;3:105–111. doi: 10.20517/2394-5079.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiao J., Watt G.P., Stevenson H.L., Calderone T.L., Fisher-Hoch S.P., Ye Y., Wu X., Vierling J.M., Beretta L. Telomerase reverse transcriptase mutations in plasma DNA in patients with hepatocellular carcinoma or cirrhosis: Prevalence and risk factors. Hepatol. Commun. 2018;2:718–731. doi: 10.1002/hep4.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oversoe S.K., Clement M.S., Pedersen M.H., Weber B., Aagaard N.K., Villadsen G.E., Gronbaek H., Hamilton-Dutoit S.J., Sorensen B.S., Kelsen J. TERT promoter mutated circulating tumor DNA as a biomarker for prognosis in hepatocellular carcinoma. Scand. J. Gastroenterol. 2020;55:1433–1440. doi: 10.1080/00365521.2020.1837928. [DOI] [PubMed] [Google Scholar]
- 35.Kim S.S., Eun J.W., Choi J.H., Woo H.G., Cho H.J., Ahn H.R., Suh C.W., Baek G.O., Cho S.W., Cheong J.Y. MLH1 single-nucleotide variant in circulating tumor DNA predicts overall survival of patients with hepatocellular carcinoma. Sci. Rep. 2020;10:17862. doi: 10.1038/s41598-020-74494-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirai M., Kinugasa H., Nouso K., Yamamoto S., Terasawa H., Onishi Y., Oyama A., Adachi T., Wada N., Sakata M., et al. Prediction of the prognosis of advanced hepatocellular carcinoma by TERT promoter mutations in circulating tumor DNA. J. Gastroenterol. Hepatol. 2020 doi: 10.1111/jgh.15227. [DOI] [PubMed] [Google Scholar]
- 37.Shen T., Li S.F., Wang J.L., Zhang T., Zhang S., Chen H.T., Xiao Q.Y., Ren W.H., Liu C., Peng B., et al. TP53 R249S mutation detected in circulating tumour DNA is associated with Prognosis of hepatocellular carcinoma patients with or without hepatectomy. Liver Int. 2020;40:2834–2847. doi: 10.1111/liv.14581. [DOI] [PubMed] [Google Scholar]
- 38.von Felden J., Craig A.J., Garcia-Lezana T., Labgaa I., Haber P.K., D’Avola D., Asgharpour A., Dieterich D., Bonaccorso A., Torres-Martin M., et al. Mutations in circulating tumor DNA predict primary resistance to systemic therapies in advanced hepatocellular carcinoma. Oncogene. 2020 doi: 10.1038/s41388-020-01519-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang Z.H., Hu Y., Hua D., Wu Y.Y., Song M.X., Cheng Z.H. Quantitative analysis of multiple methylated genes in plasma for the diagnosis and prognosis of hepatocellular carcinoma. Exp. Mol. Pathol. 2011;91:702–707. doi: 10.1016/j.yexmp.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 40.Xu R.H., Wei W., Krawczyk M., Wang W., Luo H., Flagg K., Yi S., Shi W., Quan Q., Li K., et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat. Mater. 2017;16:1155–1161. doi: 10.1038/nmat4997. [DOI] [PubMed] [Google Scholar]
- 41.Lu C.Y., Chen S.Y., Peng H.L., Kan P.Y., Chang W.C., Yen C.J. Cell-free methylation markers with diagnostic and prognostic potential in hepatocellular carcinoma. Oncotarget. 2017;8:6406–6418. doi: 10.18632/oncotarget.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeh C.C., Goyal A., Shen J., Wu H.C., Strauss J.A., Wang Q., Gurvich I., Safyan R.A., Manji G.A., Gamble M.V., et al. Global Level of Plasma DNA Methylation is Associated with Overall Survival in Patients with Hepatocellular Carcinoma. Ann. Surg. Oncol. 2017;24:3788–3795. doi: 10.1245/s10434-017-5913-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li F., Qiao C.Y., Gao S., Fan Y.C., Chen L.Y., Wang K. Circulating cell-free DNA of methylated insulin-like growth factor-binding protein 7 predicts a poor prognosis in hepatitis B virus-associated hepatocellular carcinoma after hepatectomy. Free Radic. Res. 2018;52:455–464. doi: 10.1080/10715762.2018.1443448. [DOI] [PubMed] [Google Scholar]
- 44.Chen M.M., Zhao R.C., Chen K.F., Huang Y., Liu Z.J., Wei Y.G., Jian Y., Sun A.M., Qin L., Li B., et al. Hypomethylation of CTCFL promoters as a noninvasive biomarker in plasma from patients with hepatocellular carcinoma. Neoplasma. 2020;67:909–915. doi: 10.4149/neo_2020_190819N789. [DOI] [PubMed] [Google Scholar]
- 45.Chiang D.Y., Villanueva A., Hoshida Y., Peix J., Newell P., Minguez B., LeBlanc A.C., Donovan D.J., Thung S.N., Sole M., et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008;68:6779–6788. doi: 10.1158/0008-5472.CAN-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schulze K., Imbeaud S., Letouze E., Alexandrov L.B., Calderaro J., Rebouissou S., Couchy G., Meiller C., Shinde J., Soysouvanh F., et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat. Genet. 2015;47:505–511. doi: 10.1038/ng.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Totoki Y., Tatsuno K., Covington K.R., Ueda H., Creighton C.J., Kato M., Tsuji S., Donehower L.A., Slagle B.L., Nakamura H., et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat. Genet. 2014;46:1267–1273. doi: 10.1038/ng.3126. [DOI] [PubMed] [Google Scholar]
- 48.Hernandez-Meza G., von Felden J., Gonzalez-Kozlova E.E., Garcia-Lezana T., Peix J., Portela A., Craig A.J., Sayols S., Schwartz M., Losic B., et al. DNA methylation profiling of human hepatocarcinogenesis. Hepatology. 2020 doi: 10.1002/hep.31659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Villanueva A., Portela A., Sayols S., Battiston C., Hoshida Y., Mendez-Gonzalez J., Imbeaud S., Letouze E., Hernandez-Gea V., Cornella H., et al. DNA methylation-based prognosis and epidrivers in hepatocellular carcinoma. Hepatology. 2015;61:1945–1956. doi: 10.1002/hep.27732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koberle V., Kronenberger B., Pleli T., Trojan J., Imelmann E., Peveling-Oberhag J., Welker M.W., Elhendawy M., Zeuzem S., Piiper A., et al. Serum microRNA-1 and microRNA-122 are prognostic markers in patients with hepatocellular carcinoma. Eur. J. Cancer. 2013;49:3442–3449. doi: 10.1016/j.ejca.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 51.Xu Y., Bu X., Dai C., Shang C. High serum microRNA-122 level is independently associated with higher overall survival rate in hepatocellular carcinoma patients. Tumour. Biol. 2015;36:4773–4776. doi: 10.1007/s13277-015-3128-5. [DOI] [PubMed] [Google Scholar]
- 52.Cho H.J., Kim S.S., Nam J.S., Kim J.K., Lee J.H., Kim B., Wang H.J., Kim B.W., Lee J.D., Kang D.Y., et al. Low levels of circulating microRNA-26a/29a as poor prognostic markers in patients with hepatocellular carcinoma who underwent curative treatment. Clin. Res. Hepatol. Gastroenterol. 2017;41:181–189. doi: 10.1016/j.clinre.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 53.Ning S., Liu H., Gao B., Wei W., Yang A., Li J., Zhang L. miR-155, miR-96 and miR-99a as potential diagnostic and prognostic tools for the clinical management of hepatocellular carcinoma. Oncol. Lett. 2019;18:3381–3387. doi: 10.3892/ol.2019.10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jin Y., Wong Y.S., Goh B.K.P., Chan C.Y., Cheow P.C., Chow P.K.H., Lim T.K.H., Goh G.B.B., Krishnamoorthy T.L., Kumar R., et al. Circulating microRNAs as Potential Diagnostic and Prognostic Biomarkers in Hepatocellular Carcinoma. Sci. Rep. 2019;9:10464. doi: 10.1038/s41598-019-46872-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Loosen S.H., Wirtz T.H., Roy S., Vucur M., Castoldi M., Schneider A.T., Koppe C., Ulmer T.F., Roeth A.A., Bednarsch J., et al. Circulating levels of microRNA193a-5p predict outcome in early stage hepatocellular carcinoma. PLoS ONE. 2020;15:e0239386. doi: 10.1371/journal.pone.0239386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pratedrat P., Chuaypen N., Nimsamer P., Payungporn S., Pinjaroen N., Sirichindakul B., Tangkijvanich P. Diagnostic and prognostic roles of circulating miRNA-223-3p in hepatitis B virus-related hepatocellular carcinoma. PLoS ONE. 2020;15:e0232211. doi: 10.1371/journal.pone.0232211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jeng K.S., Sheen I.S., Wang Y.C., Gu S.L., Chu C.M., Shih S.C., Wang P.C., Chang W.H., Wang H.Y. Prognostic significance of preoperative circulating vascular endothelial growth factor messenger RNA expression in resectable hepatocellular carcinoma: A prospective study. World J. Gastroenterol. 2004;10:643–648. doi: 10.3748/wjg.v10.i5.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morimoto O., Nagano H., Miyamoto A., Fujiwara Y., Kondo M., Yamamoto T., Ota H., Nakamura M., Wada H., Damdinsuren B., et al. Association between recurrence of hepatocellular carcinoma and alpha-fetoprotein messenger RNA levels in peripheral blood. Surg. Today. 2005;35:1033–1041. doi: 10.1007/s00595-005-3077-5. [DOI] [PubMed] [Google Scholar]
- 59.Kong S.Y., Park J.W., Kim J.O., Lee N.O., Lee J.A., Park K.W., Hong E.K., Kim C.M. Alpha-fetoprotein and human telomerase reverse transcriptase mRNA levels in peripheral blood of patients with hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 2009;135:1091–1098. doi: 10.1007/s00432-009-0549-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sugimachi K., Matsumura T., Hirata H., Uchi R., Ueda M., Ueo H., Shinden Y., Iguchi T., Eguchi H., Shirabe K., et al. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br. J. Cancer. 2015;112:532–538. doi: 10.1038/bjc.2014.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qu Z., Wu J., Wu J., Ji A., Qiang G., Jiang Y., Jiang C., Ding Y. Exosomal miR-665 as a novel minimally invasive biomarker for hepatocellular carcinoma diagnosis and prognosis. Oncotarget. 2017;8:80666–80678. doi: 10.18632/oncotarget.20881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee Y.R., Kim G., Tak W.Y., Jang S.Y., Kweon Y.O., Park J.G., Lee H.W., Han Y.S., Chun J.M., Park S.Y., et al. Circulating exosomal noncoding RNAs as prognostic biomarkers in human hepatocellular carcinoma. Int. J. Cancer. 2019;144:1444–1452. doi: 10.1002/ijc.31931. [DOI] [PubMed] [Google Scholar]
- 63.Shi M., Jiang Y., Yang L., Yan S., Wang Y.G., Lu X.J. Decreased levels of serum exosomal miR-638 predict poor prognosis in hepatocellular carcinoma. J. Cell Biochem. 2018;119:4711–4716. doi: 10.1002/jcb.26650. [DOI] [PubMed] [Google Scholar]
- 64.Tan C., Cao J., Chen L., Xi X., Wang S., Zhu Y., Yang L., Ma L., Wang D., Yin J., et al. Noncoding RNAs Serve as Diagnosis and Prognosis Biomarkers for Hepatocellular Carcinoma. Clin. Chem. 2019;65:905–915. doi: 10.1373/clinchem.2018.301150. [DOI] [PubMed] [Google Scholar]
- 65.Luo Y., Liu F., Gui R. High expression of circulating exosomal circAKT3 is associated with higher recurrence in HCC patients undergoing surgical treatment. Surg. Oncol. 2020;33:276–281. doi: 10.1016/j.suronc.2020.04.021. [DOI] [PubMed] [Google Scholar]
- 66.Hao X., Xin R., Dong W. Decreased serum exosomal miR-320a expression is an unfavorable prognostic factor in patients with hepatocellular carcinoma. J. Int. Med. Res. 2020;48:300060519896144. doi: 10.1177/0300060519896144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin X.J., Chong Y., Guo Z.W., Xie C., Yang X.J., Zhang Q., Li S.P., Xiong Y., Yuan Y., Min J., et al. A serum microRNA classifier for early detection of hepatocellular carcinoma: A multicentre, retrospective, longitudinal biomarker identification study with a nested case-control study. Lancet Oncol. 2015;16:804–815. doi: 10.1016/S1470-2045(15)00048-0. [DOI] [PubMed] [Google Scholar]
- 68.Maisano D., Mimmi S., Russo R., Fioravanti A., Fiume G., Vecchio E., Nistico N., Quinto I., Iaccino E. Uncovering the Exosomes Diversity: A Window of Opportunity for Tumor Progression Monitoring. Pharmaceuticals. 2020;13:180. doi: 10.3390/ph13080180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hoshino A., Costa-Silva B., Shen T.L., Rodrigues G., Hashimoto A., Tesic Mark M., Molina H., Kohsaka S., Di Giannatale A., Ceder S., et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hoshino A., Kim H.S., Bojmar L., Gyan K.E., Cioffi M., Hernandez J., Zambirinis C.P., Rodrigues G., Molina H., Heissel S., et al. Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell. 2020;182:1044–1061.e1018. doi: 10.1016/j.cell.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Costa-Silva B., Aiello N.M., Ocean A.J., Singh S., Zhang H., Thakur B.K., Becker A., Hoshino A., Mark M.T., Molina H., et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Iaccino E., Mimmi S., Dattilo V., Marino F., Candeloro P., Di Loria A., Marimpietri D., Pisano A., Albano F., Vecchio E., et al. Monitoring multiple myeloma by idiotype-specific peptide binders of tumor-derived exosomes. Mol. Cancer. 2017;16:159. doi: 10.1186/s12943-017-0730-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nistico N., Maisano D., Iaccino E., Vecchio E., Fiume G., Rotundo S., Quinto I., Mimmi S. Role of Chronic Lymphocytic Leukemia (CLL)-Derived Exosomes in Tumor Progression and Survival. Pharmaceuticals. 2020;13:244. doi: 10.3390/ph13090244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cristofanilli M., Budd G.T., Ellis M.J., Stopeck A., Matera J., Miller M.C., Reuben J.M., Doyle G.V., Allard W.J., Terstappen L.W., et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 75.Sun Y.F., Xu Y., Yang X.R., Guo W., Zhang X., Qiu S.J., Shi R.Y., Hu B., Zhou J., Fan J. Circulating stem cell-like epithelial cell adhesion molecule-positive tumor cells indicate poor prognosis of hepatocellular carcinoma after curative resection. Hepatology. 2013;57:1458–1468. doi: 10.1002/hep.26151. [DOI] [PubMed] [Google Scholar]
- 76.D’Avola D., Villacorta-Martin C., Martins-Filho S.N., Craig A., Labgaa I., von Felden J., Kimaada A., Bonaccorso A., Tabrizian P., Hartmann B.M., et al. High-density single cell mRNA sequencing to characterize circulating tumor cells in hepatocellular carcinoma. Sci. Rep. 2018;8:11570. doi: 10.1038/s41598-018-30047-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vona G., Estepa L., Beroud C., Damotte D., Capron F., Nalpas B., Mineur A., Franco D., Lacour B., Pol S., et al. Impact of cytomorphological detection of circulating tumor cells in patients with liver cancer. Hepatology. 2004;39:792–797. doi: 10.1002/hep.20091. [DOI] [PubMed] [Google Scholar]
- 78.Xu W., Cao L., Chen L., Li J., Zhang X.F., Qian H.H., Kang X.Y., Zhang Y., Liao J., Shi L.H., et al. Isolation of circulating tumor cells in patients with hepatocellular carcinoma using a novel cell separation strategy. Clin. Cancer Res. 2011;17:3783–3793. doi: 10.1158/1078-0432.CCR-10-0498. [DOI] [PubMed] [Google Scholar]
- 79.Fan S.T., Yang Z.F., Ho D.W., Ng M.N., Yu W.C., Wong J. Prediction of posthepatectomy recurrence of hepatocellular carcinoma by circulating cancer stem cells: A prospective study. Ann. Surg. 2011;254:569–576. doi: 10.1097/SLA.0b013e3182300a1d. [DOI] [PubMed] [Google Scholar]
- 80.Cheng S.W., Tsai H.W., Lin Y.J., Cheng P.N., Chang Y.C., Yen C.J., Huang H.P., Chuang Y.P., Chang T.T., Lee C.T., et al. Lin28B is an oncofetal circulating cancer stem cell-like marker associated with recurrence of hepatocellular carcinoma. PLoS ONE. 2013;8:e80053. doi: 10.1371/journal.pone.0080053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu S., Li N., Yu X., Xiao X., Cheng K., Hu J., Wang J., Zhang D., Cheng S., Liu S. Expression of intercellular adhesion molecule 1 by hepatocellular carcinoma stem cells and circulating tumor cells. Gastroenterology. 2013;144:1031–1041.e1010. doi: 10.1053/j.gastro.2013.01.046. [DOI] [PubMed] [Google Scholar]
- 82.Schulze K., Gasch C., Staufer K., Nashan B., Lohse A.W., Pantel K., Riethdorf S., Wege H. Presence of EpCAM-positive circulating tumor cells as biomarker for systemic disease strongly correlates to survival in patients with hepatocellular carcinoma. Int. J. Cancer. 2013;133:2165–2171. doi: 10.1002/ijc.28230. [DOI] [PubMed] [Google Scholar]
- 83.Guo W., Yang X.R., Sun Y.F., Shen M.N., Ma X.L., Wu J., Zhang C.Y., Zhou Y., Xu Y., Hu B., et al. Clinical significance of EpCAM mRNA-positive circulating tumor cells in hepatocellular carcinoma by an optimized negative enrichment and qRT-PCR-based platform. Clin. Cancer Res. 2014;20:4794–4805. doi: 10.1158/1078-0432.CCR-14-0251. [DOI] [PubMed] [Google Scholar]
- 84.Li J., Shi L., Zhang X., Sun B., Yang Y., Ge N., Liu H., Yang X., Chen L., Qian H., et al. pERK/pAkt phenotyping in circulating tumor cells as a biomarker for sorafenib efficacy in patients with advanced hepatocellular carcinoma. Oncotarget. 2016;7:2646–2659. doi: 10.18632/oncotarget.6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jin J., Niu X., Zou L., Li L., Li S., Han J., Zhang P., Song J., Xiao F. AFP mRNA level in enriched circulating tumor cells from hepatocellular carcinoma patient blood samples is a pivotal predictive marker for metastasis. Cancer Lett. 2016;378:33–37. doi: 10.1016/j.canlet.2016.04.033. [DOI] [PubMed] [Google Scholar]
- 86.Ogle L.F., Orr J.G., Willoughby C.E., Hutton C., McPherson S., Plummer R., Boddy A.V., Curtin N.J., Jamieson D., Reeves H.L. Imagestream detection and characterisation of circulating tumour cells—A liquid biopsy for hepatocellular carcinoma? J. Hepatol. 2016;65:305–313. doi: 10.1016/j.jhep.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 87.von Felden J., Schulze K., Krech T., Ewald F., Nashan B., Pantel K., Lohse A.W., Riethdorf S., Wege H. Circulating tumor cells as liquid biomarker for high HCC recurrence risk after curative liver resection. Oncotarget. 2017;8:89978–89987. doi: 10.18632/oncotarget.21208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang L., Li Y., Xu J., Zhang A., Wang X., Tang R., Zhang X., Yin H., Liu M., Wang D.D., et al. Quantified postsurgical small cell size CTCs and EpCAM(+) circulating tumor stem cells with cytogenetic abnormalities in hepatocellular carcinoma patients determine cancer relapse. Cancer Lett. 2018;412:99–107. doi: 10.1016/j.canlet.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 89.Hao S., Chen S., Tu C., Huang T. Anterior Approach to Improve the Prognosis in HCC Patients Via Decreasing Dissemination of EpCAM(+) Circulating Tumor Cells. J. Gastrointest Surg. 2017;21:1112–1120. doi: 10.1007/s11605-017-3410-5. [DOI] [PubMed] [Google Scholar]
- 90.Sun Y.F., Guo W., Xu Y., Shi Y.H., Gong Z.J., Ji Y., Du M., Zhang X., Hu B., Huang A., et al. Circulating Tumor Cells from Different Vascular Sites Exhibit Spatial Heterogeneity in Epithelial and Mesenchymal Composition and Distinct Clinical Significance in Hepatocellular Carcinoma. Clin. Cancer Res. 2018;24:547–559. doi: 10.1158/1078-0432.CCR-17-1063. [DOI] [PubMed] [Google Scholar]
- 91.Guo W., Sun Y.F., Shen M.N., Ma X.L., Wu J., Zhang C.Y., Zhou Y., Xu Y., Hu B., Zhang M., et al. Circulating Tumor Cells with Stem-Like Phenotypes for Diagnosis, Prognosis, and Therapeutic Response Evaluation in Hepatocellular Carcinoma. Clin. Cancer Res. 2018;24:2203–2213. doi: 10.1158/1078-0432.CCR-17-1753. [DOI] [PubMed] [Google Scholar]
- 92.Qi L.N., Xiang B.D., Wu F.X., Ye J.Z., Zhong J.H., Wang Y.Y., Chen Y.Y., Chen Z.S., Ma L., Chen J., et al. Circulating Tumor Cells Undergoing EMT Provide a Metric for Diagnosis and Prognosis of Patients with Hepatocellular Carcinoma. Cancer Res. 2018;78:4731–4744. doi: 10.1158/0008-5472.CAN-17-2459. [DOI] [PubMed] [Google Scholar]
- 93.Court C.M., Hou S., Winograd P., Segel N.H., Li Q.W., Zhu Y., Sadeghi S., Finn R.S., Ganapathy E., Song M., et al. A novel multimarker assay for the phenotypic profiling of circulating tumor cells in hepatocellular carcinoma. Liver Transpl. 2018;24:946–960. doi: 10.1002/lt.25062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yu J.J., Xiao W., Dong S.L., Liang H.F., Zhang Z.W., Zhang B.X., Huang Z.Y., Chen Y.F., Zhang W.G., Luo H.P., et al. Effect of surgical liver resection on circulating tumor cells in patients with hepatocellular carcinoma. BMC Cancer. 2018;18:835. doi: 10.1186/s12885-018-4744-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ha Y., Kim T.H., Shim J.E., Yoon S., Jun M.J., Cho Y.H., Lee H.C. Circulating tumor cells are associated with poor outcomes in early-stage hepatocellular carcinoma: A prospective study. Hepatol. Int. 2019;13:726–735. doi: 10.1007/s12072-019-09994-9. [DOI] [PubMed] [Google Scholar]
- 96.Hamaoka M., Kobayashi T., Tanaka Y., Mashima H., Ohdan H. Clinical significance of glypican-3-positive circulating tumor cells of hepatocellular carcinoma patients: A prospective study. PLoS ONE. 2019;14:e0217586. doi: 10.1371/journal.pone.0217586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen Z., Lin X., Chen C., Chen Y., Zhao Q., Wu L., Wang D., Ma Y., Ju W., Chen M., et al. Analysis of preoperative circulating tumor cells for recurrence in patients with hepatocellular carcinoma after liver transplantation. Ann. Transl. Med. 2020;8:1067. doi: 10.21037/atm-20-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhou J., Zhang Z., Zhou H., Leng C., Hou B., Zhou C., Hu X., Wang J., Chen X. Preoperative circulating tumor cells to predict microvascular invasion and dynamical detection indicate the prognosis of hepatocellular carcinoma. BMC Cancer. 2020;20:1047. doi: 10.1186/s12885-020-07488-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Winograd P., Hou S., Court C.M., Lee Y.T., Chen P.J., Zhu Y., Sadeghi S., Finn R.S., Teng P.C., Wang J.J., et al. Hepatocellular Carcinoma-Circulating Tumor Cells Expressing PD-L1 Are Prognostic and Potentially Associated With Response to Checkpoint Inhibitors. Hepatol. Commun. 2020;4:1527–1540. doi: 10.1002/hep4.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang P.X., Sun Y.F., Zhou K.Q., Cheng J.W., Hu B., Guo W., Yin Y., Huang J.F., Zhou J., Fan J., et al. Circulating tumor cells are an indicator for the administration of adjuvant transarterial chemoembolization in hepatocellular carcinoma: A single-center, retrospective, propensity-matched study. Clin. Transl. Med. 2020;10:e137. doi: 10.1002/ctm2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang P.X., Xu Y., Sun Y.F., Cheng J.W., Zhou K.Q., Wu S.Y., Hu B., Zhang Z.F., Guo W., Cao Y., et al. Detection of circulating tumour cells enables early recurrence prediction in hepatocellular carcinoma patients undergoing liver transplantation. Liver Int. 2020 doi: 10.1111/liv.14734. [DOI] [PubMed] [Google Scholar]
- 102.Yamashita T., Forgues M., Wang W., Kim J.W., Ye Q., Jia H., Budhu A., Zanetti K.A., Chen Y., Qin L.X., et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008;68:1451–1461. doi: 10.1158/0008-5472.CAN-07-6013. [DOI] [PubMed] [Google Scholar]
- 103.Dent B.M., Ogle L.F., O’Donnell R.L., Hayes N., Malik U., Curtin N.J., Boddy A.V., Plummer E.R., Edmondson R.J., Reeves H.L., et al. High-resolution imaging for the detection and characterisation of circulating tumour cells from patients with oesophageal, hepatocellular, thyroid and ovarian cancers. Int. J. Cancer. 2016;138:206–216. doi: 10.1002/ijc.29680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Toso C., Mentha G., Majno P. Liver transplantation for hepatocellular carcinoma: Five steps to prevent recurrence. Am. J. Transplant. 2011;11:2031–2035. doi: 10.1111/j.1600-6143.2011.03689.x. [DOI] [PubMed] [Google Scholar]
- 105.Cui K., Ou Y., Shen Y., Li S., Sun Z. Clinical value of circulating tumor cells for the diagnosis and prognosis of hepatocellular carcinoma (HCC): A systematic review and meta-analysis. Medicine (Baltimore) 2020;99:e22242. doi: 10.1097/MD.0000000000022242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yan J., Fan Z., Wu X., Xu M., Jiang J., Tan C., Wu W., Wei X., Zhou J. Circulating tumor cells are correlated with disease progression and treatment response in an orthotopic hepatocellular carcinoma model. Cytometry A. 2015;87:1020–1028. doi: 10.1002/cyto.a.22782. [DOI] [PubMed] [Google Scholar]
- 107.Gasparello J., Allegretti M., Tremante E., Fabbri E., Amoreo C.A., Romania P., Melucci E., Messana K., Borgatti M., Giacomini P., et al. Liquid biopsy in mice bearing colorectal carcinoma xenografts: Gateways regulating the levels of circulating tumor DNA (ctDNA) and miRNA (ctmiRNA) J. Exp. Clin. Cancer Res. 2018;37:124. doi: 10.1186/s13046-018-0788-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Losic B., Craig A.J., Villacorta-Martin C., Martins-Filho S.N., Akers N., Chen X., Ahsen M.E., von Felden J., Labgaa I., D’Avola D., et al. Intratumoral heterogeneity and clonal evolution in liver cancer. Nat. Commun. 2020;11:291. doi: 10.1038/s41467-019-14050-z. [DOI] [PMC free article] [PubMed] [Google Scholar]