Abstract
(1) Background: Elizabethkingia spp. is an emerging nosocomial pathogen which causes mostly blood stream infection and nosocomial pneumonia. Among Elizabethkingia species, Elizabethkingia anophelis is the major pathogen, but misidentification as Elizabethkingia meningoseptica is a common problem. Elizabethkingia also possesses broad antibiotic resistance, resulting in high morbidity and mortality of the infection. The aim of our study was to review Elizabethkingia intra-abdominal infections and investigate resistance mechanisms against TMP/SMX in Elizabethkingia anophelis by whole genome sequencing. (2) Methods: We retrospectively searched records of patients with Elizabethkingia intra-abdominal infection between 1990 and 2019. We also conducted whole genome sequencing for a TMP/SMX-resistant Elizabethkingia anophelis to identify possible mechanisms of resistance. (3) Results: We identified a total of nine cases of Elizabethkingia intra-abdominal infection in a review of the literature, including our own case. The cases included three biliary tract infections, three CAPD-related infection, two with infected ascites, and two postoperation infections. Host factor, indwelling-catheter, and previous invasive procedure, including surgery, play important roles in Elizabethkingia infection. Removal of the catheter is crucial for successful treatment. Genomic analysis revealed accumulated mutations leading to TMP/SMX-resistance in folP. (4) Conclusions: Patients with underlying disease and indwelling catheter are more susceptible to Elizabethkingia intra-abdominal infection, and successful treatment requires removal of the catheter. The emerging resistance to TMP/SMX may be related to accumulated mutations in folP.
Keywords: Elizabethkingia anopheles, trimethoprim-sulfamethoxazole, sequence alignment, whole genome sequencing
1. Introduction
The genus Elizabethkingia was proposed by Kim in 2005 [1], and soon attracted attention due to nosocomial infection and broad antibiotic resistance. In the genus, Elizabethkingia anophelis was first discovered in 2011 from the midgut of mosquitoes in Africa [2]. With advances in identification, including 16s RNA sequencing and the availability of the MALDI-ToF system with new databases, Elizabethkingia anophelis was recognized as the dominant pathogen in Elizabethkingia spp., rather than Elizabethkingia meningoseptica [3,4].
The first report of Elizabethkingia anophelis nosocomial outbreak was in a Singapore intensive care unit in 2012. Similar outbreaks were subsequently reported in hospitals in Wisconsin, USA, during 2015–2016 [5,6]. Most infections caused by Elizabethkingia species were blood stream infections, but pneumonia, septic arthritis, infected ascites, meningitis, and eye infection were also reported [6,7,8]. Recently, reports have emerged of intra-abdominal infections, such as postoperation infection, biliary tract infection, ascites infection, and CAPD infection. Elizabethkingia anophelis exhibit broad antibiotic resistance, including to most types of penicillin, cephazolin, carbapenem, aminoglycoside, and macrolide [9,10]. Fluoroquinolones, trimethoprim-sulfamethoxazole (TMP/SMX), and piperacillin/tazobactam were given as first-line therapy for Elizabethkingia infection, but emerging resistance has made treatment more challenging recently [9,10,11]. The development of sequencing technologies and genomic analyses are improving our understanding of the genetic background of resistance to antibiotics [12,13].
In this study, we present a case of Elizabethkingia anophelis infection with bacteremia and infected ascites and review the literature on Elizabethkingia infection with intra-abdominal infection. Whole genome sequencing and genome comparison were conducted to determine the genetic factors leading to resistance against TMP/SMX in Elizabethkingia anophelis.
2. Results
2.1. Case Report
A 69-year-old male who visited our hospital for severe pitting edema and dyspnea was admitted for autoimmune disease-related protein-losing enteropathy. A high-dose steroid was administered. Recurrent infection developed during the hospital course, including pneumonia, empyema, and several episodes of bacteremia.
Elizabethkingia meningoseptica bacteremia developed with fever, chills, dyspnea, and septic shock. Blood culture yielded two sets of Elizabethkingia meningoseptica, but re-identification by whole genome sequencing detected Elizabethkingia anophelis. Levofloxacin 750mg QD with TMP/SMX was administered. A survey for fever focus found ascite infection, favoring spontaneous bacterial peritonitis, and culture from ascites also yielded Elizabethkingia anophelis.
Follow-up blood culture after three days of antibiotic treatment still found a positive result. Repeat culture found a change in susceptibility. A new culture report showed E. anophelis, which was resistant to trimethoprim/sulfamethoxazole (TMP/SMX) but sensitive to piperacillin/tazobactam. Piperacillin/tazobactam was administered and TMP/SMX with levofloxacin was discontinued. There was no improvement in fever and progression of sepsis, and the patient expired about 10 days after piperacillin/tazobactam use, due to sepsis-related profound DIC and massive GI bleeding.
2.2. Reported Elizabethkingia Intra-Abdominal Infection in the Literature
A total of eight cases of E. anophelis- or E. meningoseptica-associated intra-abdominal infection were identified with detailed information. With the addition of our case, a total of nine cases are presented in this study [6,14,15,16,17,18] (Table 1 and Table 2).
Table 1.
Clinical features of intra-abdominal infection with Elizabethkingia spp.
| No. | Location | Age | Sex | Underlining Disease | Clinical Presentation | Culture- Positive Specimens | Elizabethkingia Speices | Method for Identification | References |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Taichung, Taiwan | 69 | M | Autoimmune protein losing enteropathy, with ascites, pleural effusion | Fever, abdominal distention, Spontaneous bacterial peritonitis | Blood and ascites | E. anophelis | Whole genome sequencing | NA |
| 2 | Wisconsin, USA | 84 | M | Chronic HCV infection, cirrhosis, Type 2 DM, alcohol abuse | Abdominal distention, fever, Suspected spontaneous bacterial peritonitis | blood and ascites | E. anophelis | Verigene system /MALDI-ToF MS | [6] |
| 3 | Saudi Arabia | 55 | F | Type 2 DM, liver cirrhosis, s/p liver transplantation. with post-transplant anastomotic biliary stricture and bile leakage, s/p PTBD | RUQ abdominal pain, no fever. Intra-abdominal infection with subphrenic fluid accumulation | Bile * | E. meningoseptica | NA | [14] |
| 4 | Hongkong, China | 52 | M | biliary pancreatitis, recurrent pyogenic cholangitis, cirrhosis with biliary stent | Acute cholangitis | Blood | E. meningoseptica | 16S rRNA gene sequence analysis | [18] |
| 5 | Hongkong, China | 89 | F | Hypertension, Atrial fibrillation, painless obstructive jaundice on palliative stenting | Biliary tract infection with sepsis | Blood | E. meningoseptica | 16S rRNA gene sequence analysis | [18] |
| 6 | Tamil Nadu, India | 72 | M | Type 2 DM, hypertension, ESRD under CAPD for 22 months | Abdominal pain, and cloudy CAPD fluid. CAPD peritonitis |
CAPD fluid | E. meningoseptica | Vitek-2 compact | [15] |
| 7 | Taipei, Taiwan | 54 | F | ESRD under CAPD for eight years | Turbid CAPD fluid, abdominal pain and fever. CAPD peritonitis with Tenckhoff tube infection | CAPD fluid | E. meningoseptica (previously Chryseobacterium meningoseptica) | API and Vitek test system | [16] |
| 8 | New Delhi, India | 8 | F | ESRD since 18 m/o, under CAPD for six years | CAPD peritonitis | CAPD fluid | Elizabethkingia meningoseptica | Vitek system and Vitek AST-N090 card | [17] |
| 9 | New Delhi, India | 23 | F | s/p Medical Termination of Pregnancy by suction and evacuation for suspected blighted ovum or missed abortion | Postposture fever and bleeding. Peritonitis, secondary to uterine perforation | blood culture * 2 set | Elizabethkingia meningoseptica | Vitek system and Vitek AST-N090 card | [17] |
Under immunosuppressant with oral tacrolimus 1mg, oral mycophenolate mofetil 500 mg, and oral prednisolone 5 mg; * from PTBD; NA, nonavailable; HCV, Hepatitis C virus; type 2 DM, type 2 diabetes mellitus; PTBD, percutaneous biliary drainage; ESRD, End-stage renal disease; CAPD, Continuous Ambulatory Peritoneal Dialysis.
Table 2.
Treatment for Intra-abdominal infection with Elizabethkingia spp.
| No. | Elizabethkingia spp. | Clinical Presentation | Antibiotic Susceptibility (Susceptible Drugs) | Antibiotic Use | Removal of Catheter | Survival |
|---|---|---|---|---|---|---|
| 1 | E. anophelis | Spontaneous bacterial peritonitis | Pipercacillin/tazobactam, Cefepime, Cefoperazone/sulbactam; TMP/SMX * | TMP/SMX+ Levofloxacin, then Piperacillin/tazobactam | No catheter | Expired |
| 2 | E. anophelis | Suspected spontaneous bacterial peritonitis | Ciprofloxacin, Piperacillin/tazobactam, TMP/SMX, Cefepime | Ciprofloxacin and Piperacillin/tazobactam | No catheter | Survived |
| 3 | E. meningoseptica | Intra-abdominal infection with subphrenic fluid accumulation | Ciprofloxacin, Minocycline, Tigecycline, TMP/SMX | Ciprofloxacin and metronidazole | NA | Survived |
| 4 | E. meningoseptica | Acute cholangitis | NA | Levofloxacin and metronidazole | NA | Survived |
| 5 | E. meningoseptica | Biliary tract infection with sepsis | NA | Levofloxacin | NA | Survived |
| 6 | E. meningoseptica | CAPD peritonitis | Cefoperazone/sulbactam, Ciprofloxacin, Levofloxacin, Minocycline, TMP/SMX | PO TMP/SMX + IV Cefoperazone/sulbactam, then shift to PO Minocycline + IV Cefoperazone/sulbactam | Removal of CAPD tube. | Survived |
| 7 | E. meningoseptica (previously Chryseobacterium meningoseptica) | CAPD peritonitis with Tenckhoff tube infection | Gentamicin, Ciprofloxacin, Piperacillin-tazobactam, Levofloxacin | Piperacillin-tazobactam, then shift to Levofloxacin | Removal of CAPD tube. | Survived |
| 8 | E. meningoseptica | CAPD peritonitis | Cefoperazone-sulbactam and nalidixic acid | Cefoperazone-sulbactam | NA | Survived |
| 9 | E. meningoseptica | Peritonitis, secondary to uterine perforation | TMP/SMX | TMP/SMX, Piperacillin- tazobactam, amikacin, teicoplanin and metronidazole | No catheter | Survived |
* TMP/SMX-susceptible at first but became resistant in repeated blood culture and ascites culture; NA, nonavailable; CAPD, Continuous Ambulatory Peritoneal Dialysis; TMP/SMX, Trimethoprim/sulfamethoxazole.
Among the nine cases, there were two infected ascites, three CAPD peritonitis, three hepato-biliary infections, and one peritonitis after gynecologic procedure. Patients ages ranged from 8 to 89 years, and the female to male was 6:3. All patients had previous underlying disease, except for one with peritonitis after gynecologic procedure. The underlying diseases included type II diabetes mellitus, liver cirrhosis, end-stage renal disease, hypertension, and HCV infection. In two of nine (22%) cases of Elizabethkingia-related abdominal infection, they had previous procedure: liver transplantation and medical termination of pregnancy. Six of nine cases (66.7%) had indwelling catheter or stent inserted before the infection, with one PTBD, two biliary stent, and three CAPD tube. Most patients presented with fever and abdominal pain, but fever may be absent under immunosuppressant use.
Among the nine cases, three had Elizabethkingia anophelis infection and six had Elizabethkingia meningoseptica infection. Five positive cultures were yielded from blood culture, and bacteria growth from PTBD drainage in one case and from ascites or CAPD fluid in five cases was also identified. The pathogen was identified by Vitek 2 system in four cases, two by 16s rRNA sequencing, and one by MOLDI-ToF. Our case was re-identified by whole genome sequencing. The identification method was unavailable in the case with postliver transplantation infection from India.
Most cases with Elizabethkingia infection had broad antibiotic resistance but were susceptible to ciprofloxacin, levofloxacin, Cefoperazone/sulbactam, minocycline, TMP/SMX, piperacillin, piperacillin/tazobactam, and tigecycline. Most cases survived after antibiotic treatment, but for those with catheter—especially CAPD tube—infection were controlled only after removal of the CAPD tube.
Genomics Revealed Key Mutations Leading to TMP/SMX and Quinolone Resistance
The genome of Elizabethkingia anophelis SUE was sequenced by Nanopore and Illumina sequencing plateforms (Supplementary Table S1). The sequencing reads were assembled into a circular genome of 4.2Mbp (NCBI accession number CP034247). Gene annotation revealed 3869 genes in the genome, including 3744 protein-coding genes, 73 rRNAs/tRNAs/ncRNAs, and 52 pseudogenes. We compared the resistance determinants of SUE with six other public Elizabethkingia anophelis genomes with antibiotic-resistant profiles provided (Table 3, Supplementary Table S2). The Minimum Inhibitory Concentration (MIC) indicated three of them were resistant to TMP/SMX and to ciprofloxacin (Table 3, Supplementary Table S3), while the other four were sensitive to these agents.
Table 3.
Comparison of seven Elizabethkingia anophelis genomes and antibiotic resistance.
| Strains | Genome Size | Genes | Sequencing Technology | TMP/SMX | Quinolone | Status | Assembly Number |
|---|---|---|---|---|---|---|---|
| SUE | 4,201,198 bp | 3869 | Nanopore; Illumina MiSeq |
160 R | ≥4 R | Circ. | GCA_014702245.1 |
| 12012 | 4,023,312 bp | 3700 | Illumina MiSeq | >2/38 R | >2 R | Linear | GCA_001482795.1 |
| EM361-97 | 4,077,699 bp | 3752 | Illumina HiSeq | >4/76 R | >2 R | Linear | GCA_001703835.1 |
| NUH1 | 4,334,661 bp | 4031 | Illumina MiSeq | S | S | Linear | GCA_000495995.1 |
| NUHP1 | 4,369,828 bp | 4034 | Illumina | S | S | Linear | GCA_000495935.2 |
| Po0527107 | 4,032,057 bp | 3717 | Illumina HiSeq-2000 | S | S | Linear | GCA_000689515.1 |
| V0378064 | 4,036,754 bp | 3804 | Illumina HiSeq-2001 | S | I | Linear | GCA_000689455.1 |
S: susceptible; I: intermediate; R: resistance.
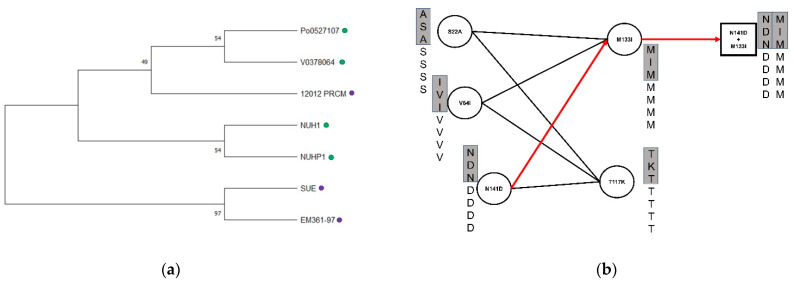
Genomic analysis revealed nine mutations in folp possibly related to TMP/SMX resistance (Figure 1), which were completely nonoverlapped with seventeen known folp mutations in the literature (Supplementary Table S4). Phylogeny reconstruction of seven folp amino acid sequences revealed their similarity and clusters (Figure 2a). Although two resistant strains, SUE and EM361-97, were clustered in the same clade, the resistant strain 12012 was clustered with other sensitive strains. This implies that the entire folp sequence is not sufficient for distinguishing resistant from susceptible strains, probably owing to many neutral mutations unrelated to the resistance.
Figure 1.
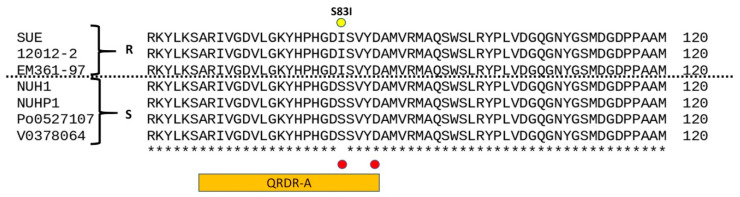
Sequence alignment of seven E anopheles DHPS orthologs. Residues marked in red dots are loci of known sulfa resistance mutations, and blue/yellow dots indicate those residues mutated in the current study. Loop1, loop2, loop5, and helix αLoop7 are boxed in bars with different colors.
Figure 2.
(a) Phylogeny of folp (DHPS) of seven E. anopheles strains. Green and purple circles indicate sensitive and resistance strains, respectively; (b) Identification of epi-static mutation via combinatorial enumeration. Each circle indicates a mutation point of seven E. anopheles, including three of them boxed in a gray bar, which were resistant to TMP/SMX. A line drawn between two circles represents a feasible mutation combination. An example of a combination (D141N+M133I) is shown on the right.
We then assessed the statistical significance of each mutation associated with TMP/SMX resistance. One mutation, T184A, was significantly associated (p = 0.0286, Fisher’s exact test), while the others only showed weak or no correlation. In order to identify weakly associated variants yet with synergistic effects (i.e., epistatic interactions), we investigated their combinations associated with resistance (Figure 2b). All possible combinations of nine mutations were enumerated and validated for the ability to distinguish resistant strains from sensitive strains. Eleven combinations were found to be associated with TMP/SMX resistance. Among them, the M133I+D141N mutational combination was located on loop 5 (Figure 3), in which known mutations have been reported. Although the T184A mutation exhibited statistical significance, the locus was not within the conserved loop.
Figure 3.
Sequence alignment of seven E anopheles DNA gyrase subunit A orthologs. Residues marked in red dots are sites of common quinolone drug resistance mutations. The yellow dot indicates a specific residue mutated in the current study, which was Isoleucine in the three quinolone-resistant strains and Serine in the four quinolone-sensitive strains QRDR-A are boxed in an orange bar.
Similarly, a comparison of the DNA gyrase subunit A (GyrA) orthologous in the seven E anopheles genomes was revealed on mutation S83I located within the quinolone-resistance determining region (QRDR) (Figure 3). The amino acid changes were the same as those reported for fluoroquinolone resistance in Proteus mirabilis and Proteus stuartii, as confirmed experimentally by site-directed mutagenesis.
3. Discussion
Elizabethkingia species, previously named Chryseobacterium, are gram-negative, aerobic, nonmotile rod-shaped bacteria, and were re-identified by Kim, 2005 [1]. Elizabethkingia anophelis was first discovered in 2011 from the midgut of mosquitoes in Africa, and the first clinical infection, a case with neonatal meningitis, was reported in the same year in the Central Africa Republic [2,19].
The first nosocomial outbreak of E. anophelis infection occurred in a Singapore intensive care unit in 2012, and another outbreak occurred in hospitals in Wisconsin, USA, in 2015–2016 [5,6]. Most cases presented as blood stream infection and nosocomial pneumonia. Other infections including neonatal meningitis, biliary tract infection, septic arthritis, infective ascites, and eye infection were also reported [6,7,8]. The majority of patients with E. anophelis infection had previous underlying disease, including malignancy, diabetes mellitus, and recent major operation. [10] Most cases were nosocomial infection, and community-acquired disease accounted for a small proportion (<20% in HongKong), although the Wisconsin outbreak had a community infection rate of 84.6%. [6,10,18] Mortality and morbidity rates were relatively high among patients infected with E. anophelis, due to its broad antibiotic resistance. A cases series in Hong Kong, 2016, found a mortality rate of around 24–30%, and during the outbreak in Wisconsin, USA, the mortality rate in 30 days was 18.2% [6,18].
This is the first literature review for Elizabethkingia intra-abdominal infection. In our case series of Elizabethkingia intra-abdominal infection, infected ascites, biliary tract infection, postoperative peritonitis, and intra-abdominal abscess formation were found. However, in our review of the literature, no enterocolitis was found and no patients presented with diarrhea. For Elizabethkingia intra-abdominal infections, host factor was an important factor. Overall, 88.9% patients had underlying disease, indicating that Elizabethkingia species does not tend to infect healthy, immunocompetent individuals, which is in line with previous studies [9,10]. Diseases in infected patients included type II diabetes mellitus, hypertension, liver cirrhosis, end-stage kidney disease, and postliver transplantation, which are known to increase susceptibility to nosocomial infection [18,20].
In Elizabethkingia intra-abdominal infection, indwelling catheter, stenting, and recent operation were also important risk factors, and removal of the prosthesis plays an important role in successful treatment. In our literature review, two-thirds (66.7%) of our patients had indwelling catheter or stent, including one PTBD, two biliary stent, and three CAPD tube. Previous studies on blood stream infection also found catheter use was a significant factor for Elizabethkingia blood-stream infection [18]. Among the cases with CAPD peritonitis, infection was finally brought under control in two out of three cases after removal of the CAPD tube, suggesting that the removal of the catheter may be an important factor in successful treatment [15,16,17]. In addition to indwelling catheter or stent, recent operation or invasive procedure was also a significant risk factor for Elizabethkingia intra-abdominal infection. In our literature review, one patient had recent liver transplantation from a living donor within one month and the other patients had peritonitis just after medication termination of pregnancy [14,17]. The first outbreak in Singapore was noted in a surgical intensive care unit and in a cardiothoracic intensive care unit; three out of five patients were found to have E. anophelis infection after surgery [5]. Therefore, Elizabethkingia infection may be an important pathogen in postoperation infection and nosocomial infection.
Elizabethkingia anophelis caused the majority of Elizabethkingia infection, but true prevalence was underestimated because of misidentification. Most Elizabethkingia anophelis was misidentified as E. meningoseptica, by Vitek 2 and MOLDI-ToF [3,4,21]. E. anophelis, which was identified by 16s rRNA, accounted for 96.2% of Elizabethkingia infections in Singapore [3]. In another study in Korea, E. anopheles, re-identified by 16s rRNA sequencing, caused 59.3% of clinical Elizabethkingia infections [4]. In the study in Singapore during the period of 2009–2017, 76/79 of (96.2%) E. anophelis infections were misidentified as E. meningoseptica infections by MOLDI-ToF (bioMérieux) [3]. In a study by Lin. et al. in Taiwan, MALDI-TOF with knowledge base v 2.0/3.0 and Vitek 2 compact could only identify 26.5% Elizabethkingia species correctly when using 16s rRNA sequencing as the gold standard, and misidentified most Elizabethkingia species as Elizabethkingia meningoseptica [21]. In our case series, most cases with E. meningoseptica intra-abdominal infection were identified by the Vitek 2 compact, and misidentification was shown to be a problem. Misidentification can be improved by using 16s rRNA as the current standard and using the MOLDI-ToF system with change in databases and inclusion of mass spectra from seven E. anophelis isolates or SARAMIS database [4,18].
Elizabethkingia anophelis has exhibited broad antibiotic resistance; fluoroquinolones, TMP/SMX and piperacillin/tazobactam have been used as the first-line therapy [9,11]. However, there is large variability in susceptibility to fluoroquinolones (9.8%~70%), including ciprofloxacin and levofloxacin [11,22,23]. Elizabethkingia infection with resistance to fluoroquinolone can cause increased mortality [24]. Most resistance to fluoroquinolone in E. anophelis was caused by mutations in quinolone-resistance determining regions (QRDR), i.e., a single amino acid alteration in DNR gyrase or DNA topoisomerase IV [25,26]. Most common QRDR were noted in GyrA, including Ser83Ile, Ser83Arg [11,25,26]. Lin also reported other nonsynonymous alteration sites in the QRDR: two in GyrA (positions 95 and 102), and three in GyrB (positions 425, 452, and 470) [11]. Asp87Asn in GyrA was also reported in E. miricola [25]. In a study by Ming-Jr Jian, a 12.7-fold increase in the fluoroquinolone-related efflux pump AcrB was noted in fluoroquinolone-resistant Elizabethkingia anophelis strains, which may play a role in fluoroquinolone resistance [26]. Currently, no mutations in ParC or ParE have been reported in Elizabethkingia species. Further studies on other possible mechanisms for resistance are required to gain a more detailed understanding of quinolone resistance in Elizabethkingia species, such as efflux pump, drug-modifying enzyme, or plasmid mediated quinolone resistance [27].
There is limited information on resistance to Trimethoprim/sulfamethoxazole in Elizabethkingia species. Positive dfrA12, sul I and sul II gene in Elizabethkingia with resistance to TMP/SMX was found by PCR in a previous study [28]. The Sul I gene may be associated with integron, but no type I nor type II integron was noted in their study. However, there were still several strains with resistance to TMP/SMX that showed a negative finding in a previous study, suggesting other possible mechanisms than sul I, II, and dfr A1-12. We reported mutations in folP, including T184A, M133I and D141N, which may be associated with TMP/SMX resistance. However, as in previous studies, these mutations were not present in all resistant strains. There may be multiple other mutations and mechanisms involved in TMP/SMX resistance in Elizabethkingia, and thus, further investigation is needed.
4. Materials and Methods
4.1. Literature Review
We searched the English-language medical literature using PubMed/MEDLINE and Google Scholar from 1990 to 2019, using the following keywords: Elizabethkingia meningoseptica, Elizabethkingia anophelis, intra-abdominal infection, ascites infection, biliary infection. The references of articles found using this search were also reviewed to identify other potential cases that were not located using the search terms.
4.2. Whole Genome Sequencing and Bioinformatics Analysis
The SUE genome was deeply sequenced using Nanopore long-read sequencing and Illumina short-read sequencing (Supplementary Table S2). Adaptor sequences left in long reads were trimmed using Porechop. The remaining reads were hybrid assembled by Unicycler (v0.4.7) into a 4.2 Mbp circular genome. Protein-coding genes, coding and noncoding RNAs in the chromosomes, and plasmids were annotated by NCBI PGAP pipeline. Antibiotic-resistant genes were predicted by aligning protein-coding genes against the Comprehensive Antibiotic Resistance Database (CARD) using Diamond. Only ARGs with alignment coverage greater than 90% were retained. Efflux pumps were excluded from ARG analysis.
Multiple sequence alignment of DHPS (folP) and GyrA of the seven E anopheles genomes were carried out by MEGA X in order to identity mutation loci. The multiple sequence alignment of DHPS was also used to generate a phylogeny tree by MEGA X. The nine mutations within DHPS were tested for association with sulfa resistance via Fisher’s exact test. All possible combinations of these nine mutations (29) were tested for correlation with TMP resistance by solving a combinatorial problem known as the minimum test collection; that is, only the combinations able to distinguish resistant strains from sensitive strains are retained as solutions.
5. Conclusions
Elizabethkingia intra-abdominal infection is a relatively uncommon condition to which patients with underlying disease are more susceptible. In our review of intra-abdominal infections, infective ascites, biliary tract infection, CAPD peritonitis, and postprocedural peritonitis were reported. Indwelling catheter and recent invasive procedure may be important risk factors, and removal of catheter was shown to be key to successful treatment. Elizabethkingia anophelis infection rates may be underestimated due to misidentification. We also conducted a genomic analysis to investigate antibiotic resistance genes in Elizabethkingia anophelis. Fol p was found to be associated with SMX/TMP resistance, and Gyr A was related to fluroquinolone resistance. However, these mutations were not present in all resistant strains, and multiple mutations may be associated with resistance. Further studies on the mechanism of resistance to SMX/TMP in Elizabethkingia infection are needed.
Supplementary Materials
The following are available online at https://www.mdpi.com/2079-6382/10/2/173/s1, Table S1: Summary of sequencing and assembly statistics, Table S2: Genome features of strains analysed in the study, Table S3: Antibiotic susceptibility profile of strains analyzed in the study, Table S4: Identified folp mutations in SUE, Table S5: folP mutation combinations in SUE.
Author Contributions
Conceptualization, Y.-T.H. and P.-Y.L.; methodology, L.-C.T., J.-M.W., H.-Y.L. and Y.-C.M.; software, H.-Y.L., Y.-C.M. and K.-L.L.; validation, H.-Y.L., Y.-C.M., K.-L.L. and C.-H.T.; formal analysis, H.-Y.L., Y.-T.H. and P.-Y.L.; investigation, H.-Y.L., Y.-C.M., K.-L.L. and C.-H.T.; resources, Y.-C.M., K.-L.L., Y.-T.H. and P.-Y.L.; data curation, H.-Y.L., Y.-C.M. and K.-L.L.; writing—original draft preparation, L.-C.T., J.-M.W., Y.-T.H. and P.-Y.L.; writing—review and editing, L.-C.T., J.-M.W., Y.-T.H. and P.-Y.L.; visualization, L.-C.T., J.-M.W., Y.-T.H. and P.-Y.L.; supervision, J.-M.W., Y.-T.H. and P.-Y.L.; project administration, J.-M.W., Y.-T.H. and P.-Y.L.; funding acquisition, Y.-T.H. and P.-Y.L. All authors have read and agreed to the published version of the manuscript.
Funding
YTH was supported in part by the Ministry of Science and Technology (109-2221-E-194 -038 -MY3). PYL was supported in part by the by the Ministry of Science and Technology (109-2314-B-075A-009) and Taichung Veterans General Hospital (TCVGH-1103901C).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board of Taichung Veterans General Hospital.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
All the sequencing data have been deposited in GenBank under BioProject ID no. PRJNA507867.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kim K.K., Kim M.K., Lim J.H., Park H.Y., Lee S.-T. Transfer of Chryseobacterium meningosepticum and Chryseobacterium miricola to Elizabethkingia gen. nov. as Elizabethkingia meningoseptica comb. nov. and Elizabethkingia miricola comb. nov. Int. J. Syst. Evol. Microbiol. 2005;55:1287–1293. doi: 10.1099/ijs.0.63541-0. [DOI] [PubMed] [Google Scholar]
- 2.Kämpfer P., Matthews H., Glaeser S.P., Martin K., Lodders N., Faye I. Elizabethkingia anophelis sp. nov., isolated from the midgut of the mosquito Anopheles gambiae. Int. J. Syst. Evol. Microbiol. 2011;61:2670–2675. doi: 10.1099/ijs.0.026393-0. [DOI] [PubMed] [Google Scholar]
- 3.Chew K.L., Cheng B., Lin R.T.P., Teo J.W.P. Elizabethkingia anophelis is the dominant Elizabethkingia species found in blood cultures in singapore. J. Clin. Microbiol. 2017;56:e01445-17. doi: 10.1128/JCM.01445-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han M.-S., Kim H., Lee Y., Kim M., Ku N.S., Choi J.Y., Yong D., Jeong S.H., Lee K., Chong Y. Relative prevalence and antimicrobial susceptibility of clinical isolates of Elizabethkingia species based on 16S rRNA gene sequencing. J. Clin. Microbiol. 2017;55:274–280. doi: 10.1128/JCM.01637-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teo J., Tan S.Y.-Y., Tay M., Ding Y., Kjelleberg S., Givskov M., Lin R.T., Yang L. First case of E anophelis outbreak in an intensive-care unit. Lancet. 2013;382:855–856. doi: 10.1016/S0140-6736(13)61858-9. [DOI] [PubMed] [Google Scholar]
- 6.Figueroa Castro C.E., Johnson C., Williams M., VanDerSlik A., Graham M.B., Letzer D., Ledeboer N., Buchan B.W., Block T., Borlaug G., et al. Elizabethkingia anophelis: Clinical experience of an academic health system in southeastern wisconsin. Open Forum Infect. Dis. 2017;4:ofx251. doi: 10.1093/ofid/ofx251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin J.-N., Lai C.-H., Yang C.-H., Huang Y.-H. Elizabethkingia infections in humans: From genomics to clinics. Microorganisms. 2019;7:295. doi: 10.3390/microorganisms7090295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bulagonda E.P., Manivannan B., Mahalingam N., Lama M., Chanakya P.P., Khamari B., Jadhao S., Vasudevan M., Nagaraja V. Comparative genomic analysis of a naturally competent Elizabethkingia anophelis isolated from an eye infection. Sci. Rep. 2018;8:1–10. doi: 10.1038/s41598-018-26874-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang M., Gao H., Lin N., Zhang Y., Huang N., Walker E.D., Ming D., Chen S., Hu S. The antibiotic resistance and pathogenicity of a multidrug-resistant Elizabethkingia anophelis isolate. Microbiologyopen. 2019;8:e804. doi: 10.1002/mbo3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janda J.M., Lopez D.L. Mini review: New pathogen profiles: Elizabethkingia anophelis. Diagn. Microbiol. Infect. Dis. 2017;88:201–205. doi: 10.1016/j.diagmicrobio.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 11.Lin J.-N., Lai C.-H., Yang C.-H., Huang Y.-H. Comparison of clinical manifestations, antimicrobial susceptibility patterns, and mutations of fluoroquinolone target genes between Elizabethkingia meningoseptica and Elizabethkingia anophelis isolated in Taiwan. J. Clin. Med. 2018;7:538. doi: 10.3390/jcm7120538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burnard D., Gore L., Henderson A., Ranasinghe A., Bergh H., Cottrell K., Sarovich D.S., Price E.P., Paterson D.L., Harris P. Comparative genomics and antimicrobial resistance profiling of Elizabethkingia isolates reveal nosocomial transmission and in vitro susceptibility to fluoroquinolones, tetracyclines, and trimethoprim-sulfamethoxazole. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00730-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang C.-Y., Yang C.-H., Lai C.-H., Huang Y.-H., Lin J.-N. Comparative genomics of 86 whole-genome sequences in the six species of the Elizabethkingia genus reveals intraspecific and interspecific divergence. Sci. Rep. 2019;9:1–11. doi: 10.1038/s41598-019-55795-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Musalem H.M., Honjol Y.N., Tuleimat L.M., Al Abbad S.I., Alsohaibani F.I. Elizabethkingia Meningoseptica in a case of biliary tract infection following liver transplantation. Am. J. Case Rep. 2017;18:1014–1019. doi: 10.12659/AJCR.905247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ranjan S., Veerappan I., Patil S., Sethuraman R. Elizabethkingia meningoseptica peritonitis in continuous ambulatory peritoneal dialysis patient: A rare case report with diagnostic challenges. Indian J. Pathol. Microbiol. 2017;60:626. doi: 10.4103/IJPM.IJPM_28_17. [DOI] [PubMed] [Google Scholar]
- 16.Wu V.-C., Tsai T.-J., Wang R., Hsueh P.-R. Peritonitis caused by Chryseobacterium meningosepticum in a patient undergoing continuous ambulatory peritoneal dialysis. J. Formos. Med. Assoc. 2003;102:270–272. [PubMed] [Google Scholar]
- 17.Khan I.D., Lall M., Sen S., Ninawe S., Chandola P. Multiresistant Elizabethkingia meningoseptica infections in tertiary care. Med. J. Armed Forces India. 2015;71:282–286. doi: 10.1016/j.mjafi.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau S.K., Chow W.-N., Foo C.-H., Curreem S.O., Lo G.C.-S., Teng J.L., Chen J.H., Ng R.H., Wu A.K., Cheung I.Y. Eliz-abethkingia anophelis bacteremia is associated with clinically significant infections and high mortality. Sci. Rep. 2016;6:1–10. doi: 10.1038/srep26045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frank T., Gody J.C., Nguyen L.B.L., Berthet N., Le Fleche-Mateos A., Bata P., Rafaï C., Kazanji M., Breurec S. First case of Elizabethkingia anophelis meningitis in the Central African Republic. Lancet. 2013;381:734–737. doi: 10.1016/S0140-6736(13)60318-9. [DOI] [PubMed] [Google Scholar]
- 20.Choi M.H., Kim M., Jeong S.J., Choi J.Y., Lee I.-Y., Yong T.-S., Yong D., Jeong S.H., Lee K. Risk Factors for Elizabethkingia acquisition and clinical characteristics of patients, South Korea. Emerg. Infect. Dis. 2019;25:42–51. doi: 10.3201/eid2501.171985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin J.-N., Lai C.-H., Yang C.-H., Huang Y.-H., Lin H.-F., Lin H.-H. Comparison of four automated microbiology systems with 16S rRNA gene sequencing for identification of Chryseobacterium and Elizabethkingia species. Sci. Rep. 2017;7:1–5. doi: 10.1038/s41598-017-14244-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perrin A., Larsonneur E., Nicholson A.C., Edwards D.J., Gundlach K.M., Whitney A.M., Gulvik C.A., Bell M.E., Rendueles O., Cury J., et al. Evolutionary dynamics and genomic features of the Elizabethkingia anophelis 2015 to 2016 Wisconsin outbreak strain. Nat. Commun. 2017;8:15483. doi: 10.1038/ncomms15483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L., Zhang X., Li D., Hu F., Wang M., Guo Q., Yang F. Molecular characteristics and antimicrobial susceptibility profiles of Elizabethkingia clinical isolates in Shanghai, China. Infect. Drug Resist. 2020;13:247–256. doi: 10.2147/IDR.S240963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang Y.-C., Lin Y.-T., Wang F.-D., Chan Y.-J., Yang T.-C., Huang Y.-W. Risk factors and outcome of levofloxacin-resistant Elizabethkingia meningoseptica bacteraemia in adult patients in Taiwan. Eur. J. Clin. Microbiol. Infect. Dis. 2017;36:1373–1380. doi: 10.1007/s10096-017-2942-7. [DOI] [PubMed] [Google Scholar]
- 25.Lin J.-N., Lai C.-H., Yang C.-H., Huang Y.-H., Lin H.-H. Clinical manifestations, molecular characteristics, antimicrobial susceptibility patterns and contributions of target gene mutation to fluoroquinolone resistance in Elizabethkingia anophelis. J. Antimicrob. Chemother. 2018;73:2497–2502. doi: 10.1093/jac/dky197. [DOI] [PubMed] [Google Scholar]
- 26.Jian M.-J., Cheng Y.-H., Chung H.-Y., Cheng Y.-H., Yang H.-Y., Hsu C.-S., Perng C.-L., Shang H.-S. Fluoroquinolone re-sistance in carbapenem-resistant Elizabethkingia anophelis: Phenotypic and genotypic characteristics of clinical isolates with topoisomerase mutations and comparative genomic analysis. J. Antimicrob. Chemother. 2019;74:1503–1510. doi: 10.1093/jac/dkz045. [DOI] [PubMed] [Google Scholar]
- 27.Correia S., Poeta P., Hébraud M., Capelo J.L., Igrejas G. Mechanisms of quinolone action and resistance: Where do we stand? J. Med. Microbiol. 2017;66:551–559. doi: 10.1099/jmm.0.000475. [DOI] [PubMed] [Google Scholar]
- 28.Jiang X., Wang D., Wang Y., Yan H., Shi L., Zhou L.-J. Occurrence of antimicrobial resistance genes sul and dfrA12 in hospital environmental isolates of Elizabethkingia meningoseptica. World J. Microbiol. Biotechnol. 2012;28:3097–3102. doi: 10.1007/s11274-012-1119-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the sequencing data have been deposited in GenBank under BioProject ID no. PRJNA507867.