Abstract
Mercury (Hg) is an environmental pollutant that impacts human and ecosystem health. In our previous works, we reported alterations in the properties of Mytilus galloprovincialis protamine-like (PL) proteins after 24 h of exposure to subtoxic doses of toxic metals such as copper and cadmium. The present work aims to assess the effects of 24 h of exposure to 1, 10, and 100 pM HgCl2 on spermatozoa and PL proteins of Mytilus galloprovincialis. Inductively coupled plasma–mass spectrometry indicated accumulation of this metal in the gonads of exposed mussels. Further, RT-qPCR analyses showed altered expression levels of spermatozoa mt10 and hsp70 genes. In Mytilus galloprovincialis, PL proteins represent the major basic component of sperm chromatin. These proteins, following exposure of mussels to HgCl2, appeared, by SDS-PAGE, partly as aggregates and showed a decreased DNA-binding capacity that rendered them unable to prevent DNA damage, in the presence of CuCl2 and H2O2. These results demonstrate that even these doses of HgCl2 exposure could affect the properties of PL proteins and result in adverse effects on the reproductive system of this organism. These analyses could be useful in developing rapid and efficient chromatin-based genotoxicity assays for pollution biomonitoring programs.
Keywords: mussel, HgCl2, sperm nuclear basic proteins, sperm chromatin, male gametes, reproduction
1. Introduction
In recent decades, large quantities of pollutants have been released into the marine environment and estuaries [1]. Of all the pollutants, toxic metals have long been considered the main contaminants in the marine environment, representing a serious danger to marine organisms [2,3,4]. In marine environments, due to their benthic and sedentary way of life, bivalves are easily exposed to environmental pollution (toxic metals, persistent organic pollutants, etc.) and bioaccumulate these toxicants [5]. For this reason, these organisms are typically used as models in the field of environmental toxicology [6,7]. In particular, the mussel Mytilus galloprovincialis has been identified by several authors as a bioindicator that responds quickly to environmental pollution, with a wide spatial distribution and economic relevance. In the life cycle of bivalves, the early developmental stages result in being most susceptible to various pollutants, such as toxic metals [8], pesticides [9], and antifouling paints [10], and it has been demonstrated that the concentrations of toxic metals able to cause lethal toxicity in embryos and larvae are much lower than those that are lethal to adults [11,12]. Among toxic metals, mercury (Hg) is one of the most toxic nonessential metals [13]. Mercury contamination in seawater is an issue to the environment and human health. In fact, concentrations of Hg species in seawater are very low, about 1 pM [14], but are sufficient to drive bio-accumulation and -magnification of this toxic metal to levels in marine organisms that can pose human and ecological health risks [15,16]. In addition, Hg has also resulted in being more toxic in comparison to other metals, such as lead (Pb) and cadmium (Cd), to M. galloprovincialis embryos [17]. The toxicity of mercury has been evaluated at high doses (micromolar), in some organisms, such as the European clam (Ruditapes decussatus), in which Hg significantly reduced sperm viability [18] and also in Ciona intestinalis on which mercury affected early developmental stages. In addition, this metal impaired the testes function of the tropical fish Gymnotus carapo (L.), producing a decrease in sperm count and the alteration in sperm morphology [19]. Moreover, an in vivo study performed by Lahnsteiner et al. (2004) [20] showed that mercury had significantly decreased the percentage of sperm motility and velocity of Clarias gariepinus and Lota lota [20]. Further, early transcriptional changes induced in vivo in the mussel gills by a combination of nanomolar concentration of Cd, Cu, and Hg were also reported [21]. Differential toxicity has also been observed among species. In particular, the embryos and larvae of Crassostrea gigas were more sensitive to copper, yet Paracentrotus lividus embryos and larvae were more sensitive to lead and mercury [22]. Finally, Beiras et al. [23] exposed different life stages of M. galloprovincialis to mercury and reported a consistent decreased sensitivity to the metal as developmental stages increased in age. To the best of our knowledge, the literature lacks information regarding the effects of mercury concentrations, similar to those present in the waters of the Mediterranean basin and the North Atlantic oceans, on M. galloprovincialis sperm nuclear basic proteins (SNBP) and DNA. To this aim, in the present work, we exposed M. galloprovincialis for 24 h to three picomolar doses (1, 10, and 100 pM) of HgCl2. After exposures, we measured the accumulation of mercury in the gonads of exposed mussels and the expression of the stress genes mt10 and hsp70 in spermatozoa. Further, we analyzed the possible changes in the electrophoretic pattern and in the DNA binding ability of protamine-like (PL) proteins. Finally, we analyzed the ability of PL proteins to protect DNA from the action of free radicals, as sperm DNA fragmentation is one of the main causes of infertility [24].
2. Results
2.1. Evaluation of Accumulation of Mercury in Male Gonads
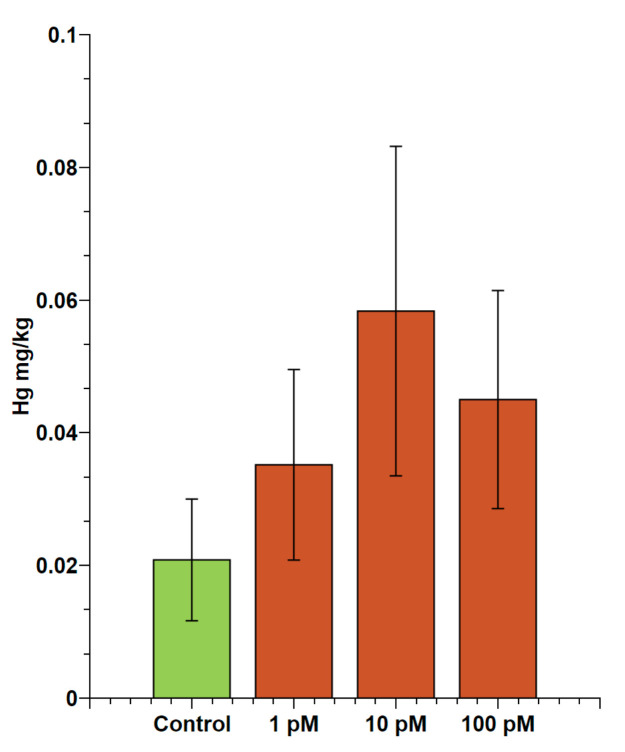
The possible accumulation of mercury in the gonads of male mussels exposed to HgCl2 was evaluated using inductively coupled plasma–mass spectrometry (ICP-MS). These analyses showed mercury accumulation in gonads after exposure for 24 h to 1, 10 and 100 pM HgCl2. In particular, the amount of mercury found in the gonads of unexposed mussels was 0.02 ± 0.01 mg/kg. As regards the exposed mussels, the values were: 0.04 ± 0.01 mg/kg for 1 pM; 0.05 ± 0.03 mg/kg for 10 pM; and 0.04 ± 0.02 mg/kg for 100 pM (Figure 1).
Figure 1.
ICP-MS analyses of mercury bioaccumulation in gonads of M. galloprovincialis. Mercury concentration was evaluated after 24 h of exposure of mussels to 1, 10, and 100 pM HgCl2 in laboratory tanks. The values are expressed based on the wet weight basis. All values represent the mean ± S.D. obtained from 6 gonads of mussels from the same tank.
2.2. Spermatozoa Gene Expression
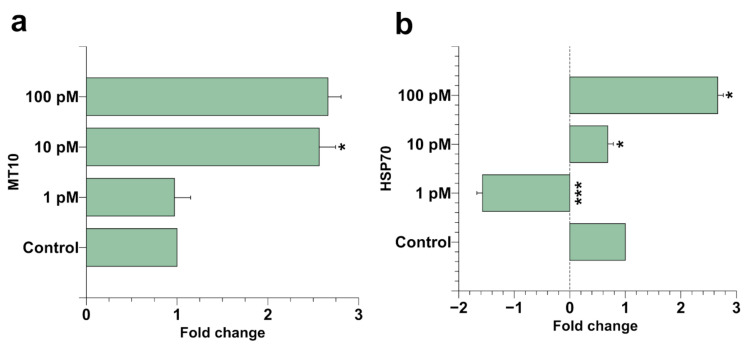
After mussel exposure, a possible stress at the M. galloprovincialis spermatozoa level was assessed by quantitative reverse transcription polymerase chain reaction (RT-qPCR). For this aim, we measured the level of the stress genes hsp70 and mt10 in spermatozoa of mussels exposed to the three different concentrations of HgCl2 (1, 10, and 100 pM). It was found that mt10 was on the order of about threefold over the control after 10 and 100 pM HgCl2 exposure. hsp70 was hyper-expressed at the same extent after 100 pM HgCl2 exposure, while after 10 and 1 pM HgCl2 exposure, this gene resulted in being hypo-expressed, particularly at 1 pM HgCl2. In this latter condition, the hypo-expression of this gene was about 1.5 times compared with the control condition. These results showed that the exposure of mussels to these HgCl2 doses produced a stress condition for M. galloprovincialis spermatozoa (Figure 2).
Figure 2.
RT-qPCR expression analysis of M. galloprovincialis spermatozoa mt10 and hsp70. In the figure, the fold change of the expression of the transcripts of mt10 (a) and hsp70 (b) is reported in the three HgCl2 exposure conditions with respect to the control condition (unexposed mussels), after determining their relative expression in comparison to the reference housekeeping 18S gene. Mussels were unexposed and exposed to 1, 10, and 100 pM HgCl2. Values are presented as mean ± S.D. (n = 6); asterisks indicate a statistically significant difference compared to unexposed mussels: * = p < 0.05; *** = p < 0.001.
2.3. Acid-Urea (AU)-PAGE of M. galloprovincialis PL-Proteins
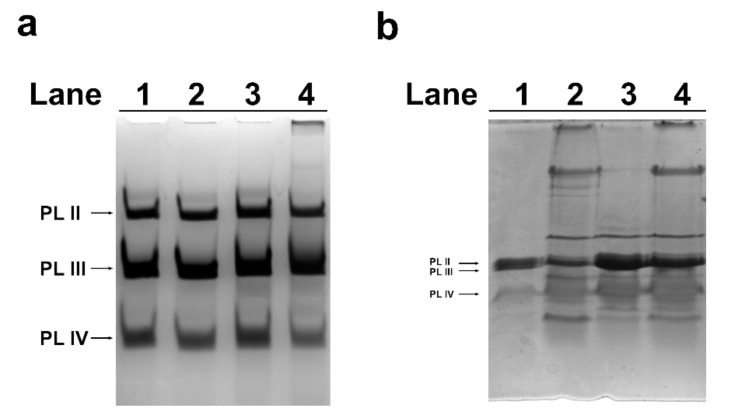
The alteration of hsp70 and mt10 genes expression in spermatozoa was indicative of a stress condition after mussels exposure to HgCl2. For this reason, the electrophoretic pattern of PL proteins extracted from mussels exposed to the three different concentrations of HgCl2 was determined by acetic acid-urea polyacrylamide gel electrophoresis (AU-PAGE). No significant differences were found in the electrophoretic pattern of PL proteins extracted from mussels exposed to the three different conditions compared with unexposed mussels (Figure 3a). By contrast, differences were found by analyzing the same proteins by SDS-PAGE (Figure 3b). As shown in this figure, the samples of PL proteins extracted from exposed mussels (lanes 2–4) presented, especially at 1 and 100 pM (Figure 3b, lane 2 and 4), additional protein bands with reduced mobility compared to control PL, likely indicative of the formation of several protein aggregates.
Figure 3.
Electrophoretic analyses by acid-urea (AU)-PAGE (a) and SDS PAGE (b) of M. galloprovincialis protamine-like (PL) proteins extracted from unexposed (lane 1) and exposed mussels (lanes 2, 3, and 4): To 1, 10, and 100 pM HgCl2, respectively.
2.4. Electrophoretic Mobility Shift Assay (EMSA) Assay
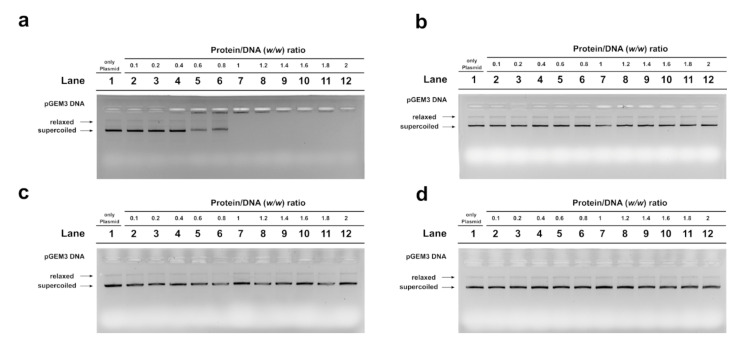
Because several differences were found in the SDS-PAGE analysis of PL proteins extracted from mussels exposed to the three different concentrations of HgCl2, the DNA binding affinity of these PL proteins was investigated. For this aim, electrophoretic mobility shift assay (EMSA) was performed, using pGEM3 plasmid DNA as a probe, under the same conditions described in Vassalli et al. (2015) [25]. It was referred to as “DNA saturation” when all the plasmid DNA was close to the well. In the control condition (unexposed mussels), the DNA saturation was achieved at a PL proteins/DNA ratio of 1.0 (Figure 4a, lane 7), while at 1, 10, and 100 pM HgCl2 conditions, DNA saturation was not observed until a PL proteins/DNA ratio of 2 (Figure 4b–d, lanes 12). These results indicated a lower DNA binding ability of the PL proteins extracted from exposed mussels with respect to those obtained from control mussels.
Figure 4.
Electrophoretic mobility shift assay (EMSA) analyses of DNA binding ability of PL proteins from unexposed (a) and 1, 10, and 100 pM HgCl2-exposed mussels (b–d), using plasmid DNA. Numbers on the wells indicate the PL protein/DNA (w/w) ratios used; only plasmid denotes pGEM3 plasmid DNA with no proteins added.
2.5. DNA Protection Analysis
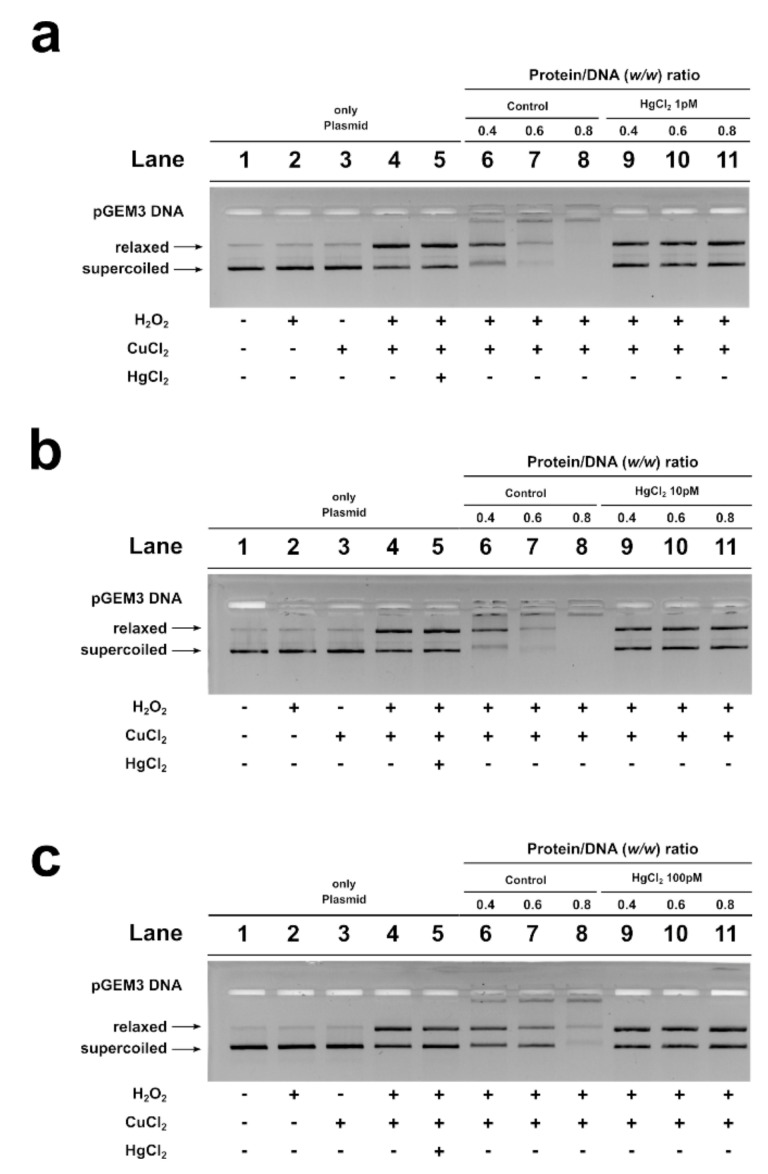
An assay was also performed to determine the potential of M. galloprovincialis PL proteins to protect DNA from oxidative damage. A condition was created in which plasmid DNA damage occurred. In this condition, the plasmid DNA was placed in the presence of 10 µM H2O2 and 5 µM CuCl2 in order to cause the Fenton reaction and produce DNA breakage. The result obtained under this condition is shown in lane 4 of all agarose gels in Figure 5, and it can be seen that more than 50% of the plasmid DNA was in the relaxed form. The addition of PL proteins from unexposed and exposed mussels to this mixture, in protein/DNA ratios of 0.4, 0.6, and 0.8, produced different effects. In particular, when PL proteins from unexposed mussels were added, already at a protein/DNA ratio of 0.4, the entity of DNA breakage was lower with respect to the damage condition (compare lanes 6 and 4 in Figure 5). At 0.6 and 0.8 protein/DNA ratios, DNA damage was not observed, suggesting that these PL proteins produced complexes capable of protecting DNA. In fact, in this latter case, at increasing PL proteins/DNA ratios, the plasmid DNA bands corresponding to supercoiled and relaxed forms became less intense as DNA saturation took place, detectable by the appearance of a high-molecular-weight DNA band close to the well. The same assays performed by using the PL proteins from exposed mussels produced different results. In fact, the PL proteins obtained from mussels exposed to all HgCl2 doses were unable to produce complexes that protected DNA from oxidative damage (lanes 9–11 of panels a (1 pM), b (10 pM), and c (100 pM)).
Figure 5.
DNA protection analysis on 1% agarose gel of pGEM3 plasmid DNA in the presence of increasing (0.4, 0.6, and 0.8) PL/DNA ratios. Control (lanes 6, 7, and 8 of all the panels); 1 pM HgCl2 (lanes 9, 10, and 11 of panel (a)); 10 pM HgCl2 (lanes 9, 10, and 11 of panel (b)); 100 pM HgCl2 (lanes 9, 10, and 11 of panel (c)).
3. Discussion
Some metals are important to maintain several biochemical and physiological functions in living organisms when they are in very low concentrations; however, they become toxic when they exceed certain threshold concentrations and represent a problem of increasing relevance for ecological, evolutionary, nutritional, and environmental reasons [26,27,28,29,30].
Human industrial activities have caused an increase in levels of Hg in the air, soil, and fresh and sea waters, and bioaccumulation along the food chain. Given the widespread exposure of organisms and the well-known toxicity of this metal, there is growing concern that exposure to mercury also at low levels may have many different adverse effects on different biological functions, including reproduction. In our previous works, we have reported alterations in the properties of M. galloprovincialis PL proteins after 24 h of exposure to subtoxic doses of toxic metals such as copper and cadmium [31,32]. Thus, in this work, we have evaluated the effects on the reproductive health of male M. galloprovincialis after exposure to mercury doses similar to those present in the waters of the Mediterranean basin and the North Atlantic oceans, sites where metal pollution has previously been reported [14,33,34].
For this aim, we exposed mussels for 24 h in laboratory tanks to 1, 10, and 100 pM HgCl2 and measured the mercury accumulation and the mt10 and hsp70 expression, in the gonads and spermatozoa of exposed mussels, respectively. This is because, in our previous studies, it has been demonstrated that mussel gonad has the same accumulation capacity as gill, at least for copper. In addition, this latter metal induces alterations in the expression level of mt10 in spermatozoa [31]. As reported by several authors [35,36], Mytilus species are able to synthesize the metal binding protein, metallothionein, for their detoxification.
In general, mt10 genes appear to be highly expressed at baseline and can respond to both essential (Cu, Zn) and nonessential (Cd, Hg) toxic metals [37,38].
Indeed, our data are in accordance with the literature; the mussels showed higher expression levels of mt10 after mercury contamination (Figure 2); in particular, the treatment of 100 pM showed an expression level increased about three times compared to the control condition (Figure 1). Expression levels of hsp70 in mussel spermatozoa were also analyzed. HSP acts to prevent protein aggregation and to maintain functional conformations. Our RT-qPCR analysis showed significant and different responses in hsp70 expression levels in mussel spermatozoa after the three HgCl2 tested exposure doses. In particular, the extremely low expression of hsp70 was found at the lowest concentration used (Figure 2b), suggesting an impairment of the biochemical mechanism underlying the heat shock response. Other studies have suggested that the downregulation of this gene is due to some factors that influence the stability and translation of mRNA [39].
Furthermore, even at the concentration of 10 pM of HgCl2, a slight downregulation of the hsp70 gene was observed in the spermatozoa of mussels. To our knowledge, this is the first study demonstrating the downregulation of the hsp70 gene in spermatozoa under metallic stress at low concentrations and within short acute exposure, which suggests a high sensitivity of spermatozoa in the response to mercury.
Interestingly, the increased HgCl2 concentration (100 pM) upregulated the transcription of the hsp70 gene in mussel spermatozoa, differently to low concentrations (1–10 pM). This transcriptional regulation of hsp70 could probably be needed to improve mercury tolerance, as high levels of HSP have been shown to protect against the negative impact of metals on protein integrity [40,41].
Considering the high efficiency of spermatozoa in the response to mercury, possible alterations in the properties of PL proteins, which represent the main component of the basic nuclear proteins that organize sperm DNA, were also evaluated. In particular, the electrophoretic pattern and DNA binding affinity of these proteins were analyzed after exposure of mussels to HgCl2. No significant differences in the electrophoretic pattern of PL proteins were found by AU-PAGE between unexposed and exposed mussels (Figure 3a). However, by SDS-PAGE, several protein bands with reduced mobility were found in the samples of PL proteins extracted from spermatozoa of mussels exposed to all the HgCl2 doses (Figure 3b). These protein bands could represent aggregates of PL proteins. Considering these alterations in PL proteins, their ability to bind DNA was tested. As a matter of fact, the decreased DNA binding capacity of PL proteins from exposed mussels was found for the pGEM3 DNA plasmid (Figure 4); in fact, DNA saturation was not achieved even at a proteins/DNA ratio of 2. Consistent with our previous work, it was confirmed by EMSA that the extracts of PL proteins (containing PL-II, PL-III, and PL-IV) from unexposed M. galloprovincialis interacted with DNA in “all or nothing” mode [4], as sperm H1 histones and C. variopedatus PL protein [42,43,44,45,46] and the DNA saturation were achieved at a proteins/DNA ratio of 1. The PL proteins obtained after all HgCl2 exposures maintained the same DNA binding mode of control PL proteins, differently from those obtained after copper mussels exposure [31]. In that case, we observed an increase in DNA binding affinity and also a change in DNA binding mode from “all or nothing” to “intermediate mode,” the typical DNA binding mode of somatic H1 histone [45]. These alterations could result in a change in the ability of PL proteins to bind and condense DNA and, in turn, abrogate their canonical role of DNA protection. In fact, in our previous studies, it has been reported that, in some cases, sperm nuclear basic proteins are involved in DNA oxidative damage [47].
Indeed, it was observed that all PL proteins extracted from spermatozoa of exposed mussels to mercury were unable to protect DNA from the action of copper and hydrogen peroxide (Figure 5). Taken together, our findings provide additional information that offers new insights into the mechanisms of mercury toxicity on the reproductive system of M. galloprovincialis. Additionally, PL protein studies could be useful for developing rapid and effective chromatin-based genotoxicity tests for biomonitoring programs for heavy metal impact assessment and species management. Therefore, further investigations are needed to understand the specific mechanisms of the action of mercury, leading to a more complete understanding of the mussel’s response to stress of this toxic metal.
4. Materials and Methods
4.1. Ethics Statement
This research was performed on the marine invertebrate M. galloprovincialis (Lamarck, 1819), which is not protected by any environmental agency in Italy. This study was conducted in strict accordance with European (Directive 2010/63) and Italian (Legislative Decree n. 116/1992) legislation on the care and use of animals for scientific purposes.
4.2. Evaluation of Accumulation of Mercury in Male Mussel Gonads
Mercury quantification was performed as follows: Samples consisting of single gonads of male mussels were digested by using 1 mL of ultrapure nitric acid (HNO3 ≥ 69%, v/v, Sigma Aldrich) in a microwave system equipped with an autosampler (CEM DISCOVER SP-D, CEM Srl, Cologno al Serio, Bergamo, Italy), according to the digestion procedure UNI EN 13805:2014 [48]. Samples were diluted to 10 mL with a solution of HNO3, 2%, v/v. For each digestion batch, a blank digestion of ultrapure nitric acid was performed to check for metal contamination, and a quality control shellfish-based sample (QMAS, Sample 741, LGC Standard) was analyzed to control the effectiveness of digestion.
The elemental analysis was conducted by inductively coupled plasma–mass spectrometry (ICP-MS, Aurora M90 Bruker, Billerica, MA, USA). Element concentrations were determined from a calibration curve calculated on the basis of five concentrations for the analyzed elements obtained from certified standard solutions. All the materials and reagents used were checked for metal contamination through repeat analysis of method blanks. Standard samples were processed at the beginning and every 10 samples in the analytical batch to verify the instrument calibration.
The accuracy of the method was evaluated by analyzing the CRM MT-742 of the fish-based sample from interlaboratory comparison (QMAS, Round 295, LGC Standard), obtaining a recovery value of 93%.
The analyses were performed on 6 samples of each type (control, exposed to 1 pM, exposed to 10 pM, and exposed to 100 pM HgCl2).
4.3. Mussels Sampling and HgCl2 Exposure
To investigate the specific effects of HgCl2, mixed-sex and medium-shell-size specimens of M. galloprovincialis were used (length 4.95 ± 0.17 cm), kindly provided by Eurofish Napoli S.R.L. Bacoli. The mussels were exposed to three concentrations of HgCl2 (1, 10, and 100 pM) as previously described for other toxic metals in Piscopo et al., 2016 [5], in laboratory plastic tanks (36 × 22 × 22 cm), each containing 6 L of 33‰ artificial sea water (ASW) with the following composition for 1 L: NaCl 29.2 g; KCl 0.60 g; MgCl2 1.2 g; NaHCO3 0.20 g; and CaCl2 1.08 g. In particular, 13 mussels were placed in any tank for 24 h at 18 ± 1 °C. Water and metal salts were changed every 12 h during treatment. For each system, dissolved oxygen and temperature were recorded at predetermined time intervals. The experiments were performed in the winter period, January–February 2020. Tanks containing only ASW were used as a control for unexposed mussels. Two tanks were used for each condition for a total of eight tanks as already described in Lettieri et al., 2019 [49].
4.4. Spermatozoa Sampling and Processing
After 24 h of exposure to 1, 10, and 100 pM HgCl2, collection of spermatozoa from male mussels was performed. Mussels were then opened using a knife, taking care not to cut the soft tissues. Subsequently, after stimulating male gonads with a glass Pasteur pipette and the help of seawater, gametes were obtained and subjected to microscopic examination to identify the sex of the mussels and the sexual maturity on the basis of a morphological and seminal analysis as previously described in Piscopo et al., 2018 [3]. Spermatozoa were collected as reported in Vassalli et al., 2015 [25], with a glass Pasteur pipette. In brief, the semen collected from all the male mussels contained in the tanks corresponding to a specific µM HgCl2 condition were pooled and centrifuged at 1000× g for 2 min at 4 °C in order to remove the debris. To collect the spermatozoa, the supernatant obtained was centrifuged at 9000× g for 10 min at 4 °C. Pellets containing spermatozoa of about 200 mg were recovered and stored at −80 °C for the further investigations.
4.5. RNA Extraction and RT-qPCR
Total RNA was purified from spermatozoa of mussels unexposed (control) and exposed to the three different concentrations of HgCl2 by using the Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The quantification and quality of RNA samples were controlled with a UV-Vis spectrophotometer (NanoDropH ND-1000, Waltham, MA, USA) and by 1% agarose gel electrophoretic analysis under denaturing conditions [50]. Equal amounts of RNA obtained from the spermatozoa of mussels unexposed and exposed to HgCl2 were used in qPCR analyses. We used the procedure described in Basile et al., 2017 [26] with few modifications. First, we purified the RNA samples from genomic DNA with an Ambion (Austin, TX, USA) DNA-free kit and then performed the retrotranscription with M-MLV reverse transcriptase (ImpProm II kit, Promega, Madison, WIS, USA). Then, 1 μg of RNA from each condition was used to perform cDNA syntheses using random hexamers (0.5 µg/µg RNA). For the determination of genes expression by real-time PCR, 100 ng of the cDNA was used and 10 μM of each forward and reverse primers was used in a final volume of 50 μL using SYBR Green PCR Master Mix Kit (Applied Biosystems, Foster City, CA, USA) with the 7500 Real Time PCR System (Applied Biosystems, Foster City, CA, USA). The conditions of qPCR were as follows: All PCR reactions were performed for 40 cycles with the following specifications: Denaturation at 95 °C for 15 min; annealing and elongation at 60 °C for 1 min. The primer set used was designed using the open-source software Primer3, starting from the sequences indicated with the accession numbers reported in the last column of the table shown in Table 1. Before real-time PCR, it was verified by PCR that each primer pair produced a single primary amplicon. qPCR product dissociation curves for all transcripts gave single peaks.
Table 1.
List of forward and reverse primers used for amplification of each gene analyzed and for the reference housekeeping 18S gene.
The results were analyzed using the ViiA™-7 Software (Foster City, CA, USA) and exported into Microsoft Excel (Redmond, WA, USA, ver. 2009—build 13231.20262). To determine relative gene expression values, the ΔΔCt method was used [51]. All samples were processed with technical triplicates. Data for each gene were normalized against 18S ribosomal RNA. The expression levels of this gene were essentially stable as also reported by other authors [38]. The change in expression of mt10 and hsp70 transcripts related to reference 18S rRNA in the spermatozoa samples from mussels exposed to HgCl2 compared with control mussels was measured.
4.6. PL Proteins from M. galloprovincialis Spermatozoa Extraction and Analyses
Extraction of protamine-like proteins from spermatozoa was performed using 5% perchloric acid (PCA) as previously described in [52]. For this work, we used n = 10 spermatozoa pellets deriving from each tank corresponding to a specific µM HgCl2 condition. Spermatozoa pellets were homogenized in a potter with 15 mL of distilled water, and then PCA was added. Acid extraction was performed as described by Vassalli and coworkers [25], and at the end of procedure, we extensively dialyzed the samples containing PCA-soluble PL-proteins against distilled water, in order to guarantee all PCA was removed. Finally, the extracted proteins were lyophilized and stored at −80 °C.
PL proteins were analyzed by AU-PAGE as previously described in Piscopo et al., 2018 [53]. Gels were then acquired using a Gel-Doc system (BioRad, Hercules, CA, USA) via Quantity One v.4.4.0 (BioRad, Hercules, CA, USA) software. The software ImageJ ver 1.50d (https://imagej.nih.gov/ij/), supported by the National Institute of Health (Wayne Rasband, National Institute of Mental Health, Bethesda, MD, USA), was used for the densitometric analysis of the protein bands on gel.
SDS-PAGE of PL proteins was performed as previously described in De Guglielmo et al., 2019 [32] with a few modifications. In particular, the stacking gel was constituted by 5.0% (w/v) acrylamide (acrylamide/bis-acrylamide 30:0.15) and the separating gel was at 18.0% (w/v) acrylamide (acrylamide/bis-acrylamide 30:0.15). At the end of the run, the gels were stained with Coomassie Brilliant Blue and acquired using a GelDoc system via Quantity One v.4.4.0 software (BioRad, Hercules, CA, USA). The densitometric analysis of gel bands was carried out using the software ImageJ.
4.7. Plasmid DNA Preparation and Analysis
pGEM3 plasmid DNA was prepared according to Carbone and coworkers [54]. The quantification and quality of plasmid DNA were conducted with a UV-Vis spectrophotometer (NanoDropH ND-1000, Waltham, MA, USA) and the integrity of DNA was analyzed by gel electrophoresis on 1% agarose gels in 89 mM Tris-HCl pH 8.0, 2 mM EDTA, and 89 mM boric acid (TBE).
4.8. Analysis of the Effect of M. galloprovincialis PL Proteins on DNA Electrophoretic Mobility
The DNA binding affinity of PL proteins was evaluated by performing Electrophoretic Mobility Shift Assay (EMSA), as described in Fioretti et al., 2012 [42]. For this analysis, we used 150 ng of plasmid pGEM3 DNA in the circular form and increasing amounts of proteins, expressed as protein/DNA w/w ratios in a final volume of 30 µL. The protein/DNA w/w ratios used ranged from 0.1 to 2. The results obtained were analyzed on 1% agarose gels. On the wells of the gels shown in the results, the protein/DNA w/w ratios in the different conditions tested are reported. DNA migration was visualized by staining gels with ethidium bromide (2 µg/mL) after electrophoresis.
4.9. DNA Protection Analysis
The PL ability to protect DNA from oxidative damage in the presence of 10 µM H2O2 and 5 µM CuCl2 was performed by using 150 ng of plasmid DNA (pGEM3) and PL extracted from mussels unexposed and exposed to 1, 10, and 100 pM HgCl2. The conditions of DNA oxidative damage were redefined by using a fixed amount of H2O2 and increasing CuCl2 concentrations, in order to obtain more than 50% of the plasmid in the relaxed form, as shown in Figure S1. These conditions were obtained starting from those reported in Lettieri et al., 2020 [24]. The samples were prepared using the EMSA protocol described in paragraph 4.8. Plasmid DNA and proteins/DNA w/w ratios of 0.4, 0.6, and 0.8 were used. The interaction between DNA and PL proteins was for 5 min at room temperature, and then H2O2 and CuCl2 were added and the samples were incubated in the dark for 30 min at 37 °C. Just before electrophoresis analysis, in order to avoid the EDTA coordination of eventual metals, a sample buffer at 1X final concentration was added to the samples. The samples obtained were then analyzed on 1% agarose gel at 100 V for 30 min in TEB 1X, staining the gels with ethidium bromide (2 µg/mL) at the end. The images of the gels were acquired at the GelDoc Biorad (Hercules, CA, USA). All experiments were performed at least three times.
4.10. Statistical Analysis
Multiple group data were analyzed using one-way ANOVA and Dunnett’s test was used to compare means between the groups. Values were considered significant when p < 0.05. Statistically significant differences are defined at the 95% confidence interval. Data are shown as mean ± S.D. Analyses were made on the pool of spermatozoa collected from mussels of any different laboratory tank.
Acknowledgments
The authors express their gratitude to the EUROFISH s.r.l. fish company (Bacoli, NA) for providing the mussels used in this study.
Abbreviations
| ASW | Artificial sea water |
| AU-PAGE | Acetic acid-urea polyacrylamide gel electrophoresis |
| SDS-PAGE | Sodium dodecyl sulphate–polyacrylamide gel electrophoresis |
| EDTA | Ethylenediaminetetraacetic acid |
| EMSA | Electrophoretic mobility shift assay |
| ICP-MS | Inductively coupled plasma–mass spectrometry |
| PCA | Perchloric acid |
| PL | Protamine-like |
| qPCR | Quantitative polymerase chain reaction |
| RT-qPCR | Reverse transcript quantitative polymerase chain reaction |
| SNBP | Sperm nuclear basic protein |
Supplementary Materials
Supplementary Materials can be found at https://www.mdpi.com/1422-0067/22/4/1618/s1: Figure S1. Settings of the ideal conditions for pGEM3 plasmid DNA breakage in presence of CuCl2 and H2O2.
Author Contributions
Conceptualization, M.P.; supervision, M.P., C.M. and M.T.; investigation, G.L., R.N., A.A., A.D.B. and A.G.; formal analysis, G.L. and R.N.; visualization, M.P., G.L., R.N. and C.M.; data curation, M.P., G.L. and R.N.; writing—original draft preparation, M.P., G.L. and R.N.; project administration, M.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zebral Y.D., da Silva Fonseca J., Marques J.A., Bianchini A. Carbonic Anhydrase as a Biomarker of Global and Local Impacts: Insights from Calcifying Animals. Int. J. Mol. Sci. 2019;20:3092. doi: 10.3390/ijms20123092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Depledge M.H., Aagaard A., Györkös P. Assessment of Trace Metal Toxicity Using Molecular, Physiological and Behavioural Biomarkers. Mar. Pollut. Bull. 1995;31:19–27. doi: 10.1016/0025-326X(95)00006-9. [DOI] [Google Scholar]
- 3.Piscopo M., Notariale R., Rabbito D., Ausió J., Olanrewaju O.S., Guerriero G. Mytilus galloprovincialis (Lamarck, 1819) Spermatozoa: Hsp70 Expression and Protamine-like Protein Property Studies. Environ. Sci. Pollut. Res. Int. 2018;25:12957–12966. doi: 10.1007/s11356-018-1570-9. [DOI] [PubMed] [Google Scholar]
- 4.Piscopo M., Trifuoggi M., Notariale R., Labar S., Troisi J., Giarra A., Rabbito D., Puoti R., de Benedictis D., Brundo M.V., et al. Protamine-like Proteins’ Analysis as an Emerging Biotechnique for Cadmium Impact Assessment on Male Mollusk Mytilus galloprovincialis (Lamarck 1819) Acta Biochim. Pol. 2018;65:259–267. doi: 10.18388/abp.2017_2533. [DOI] [PubMed] [Google Scholar]
- 5.Piscopo M., Ricciardiello M., Palumbo G., Troisi J. Selectivity of Metal Bioaccumulation and Its Relationship with Glutathione S-Transferase Levels in Gonadal and Gill Tissues of Mytilus galloprovincialis Exposed to Ni (II), Cu (II) and Cd (II) Rend. Fis. Acc. Lincei. 2016;27:737–748. doi: 10.1007/s12210-016-0564-0. [DOI] [Google Scholar]
- 6.Rittschof D., McClellan-Green P. Molluscs as Multidisciplinary Models in Environment Toxicology. Mar. Pollut. Bull. 2005;50:369–373. doi: 10.1016/j.marpolbul.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 7.Piscopo M. Seasonal Dependence of Cadmium Molecular Effects on Mytilus galloprovincialis (Lamarck, 1819) Protamine-like Protein Properties. Mol. Reprod. Dev. 2019;86:1418–1429. doi: 10.1002/mrd.23240. [DOI] [PubMed] [Google Scholar]
- 8.His E., Beiras R., Seaman M.N.L. The Assessment of Marine Pollution—Bioassays with Bivalve Embryos and Larvae. In: Southward A.J., Tyler P.A., Young C.M., editors. Advances in Marine Biology. Volume 37. Academic Press; Cambridge, MA, USA: 1999. pp. 1–178. [Google Scholar]
- 9.Wessel N., Rousseau S., Caisey X., Quiniou F., Akcha F. Investigating the Relationship between Embryotoxic and Genotoxic Effects of Benzo[a]Pyrene, 17α-Ethinylestradiol and Endosulfan on Crassostrea gigas Embryos. Aquat. Toxicol. 2007;85:133–142. doi: 10.1016/j.aquatox.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Bellas J. Comparative Toxicity of Alternative Antifouling Biocides on Embryos and Larvae of Marine Invertebrates. Sci. Total Environ. 2006;367:573–585. doi: 10.1016/j.scitotenv.2006.01.028. [DOI] [PubMed] [Google Scholar]
- 11.Connor P.M. Acute Toxicity of Heavy Metals to Some Marine Larvae. Mar. Pollut. Bull. 1972;3:190–192. doi: 10.1016/0025-326X(72)90268-8. [DOI] [Google Scholar]
- 12.Beiras R., His E. Effects of Dissolved Mercury on Embryo-Genesis, Survival, Growth and Metamorphosis of Crassostrea gigas Oyster Larvae. Mar. Ecol. Prog. Ser. 1994;113:95–103. doi: 10.3354/meps113095. [DOI] [Google Scholar]
- 13.Piscopo M., Notariale R., Tortora F., Lettieri G., Palumbo G., Manna C. Novel Insights into Mercury Effects on Hemoglobin and Membrane Proteins in Human Erythrocytes. Molecules. 2020;25:3278. doi: 10.3390/molecules25143278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cinnirella S., Bruno D.E., Pirrone N., Horvat M., Živković I., Evers D.C., Johnson S., Sunderland E.M. Mercury Concentrations in Biota in the Mediterranean Sea, a Compilation of 40 Years of Surveys. Sci. Data. 2019;6:205. doi: 10.1038/s41597-019-0219-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mergler D., Anderson H.A., Chan L.H.M., Mahaffey K.R., Murray M., Sakamoto M., Stern A.H. Panel on Health Risks and Toxicological Effects of Methylmercury Methylmercury Exposure and Health Effects in Humans: A Worldwide Concern. Ambio. 2007;36:3–11. doi: 10.1579/0044-7447(2007)36[3:MEAHEI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 16.Scheuhammer A.M., Meyer M.W., Sandheinrich M.B., Murray M.W. Effects of Environmental Methylmercury on the Health of Wild Birds, Mammals, and Fish. Ambio. 2007;36:12–18. doi: 10.1579/0044-7447(2007)36[12:EOEMOT]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 17.Beiras R., Albentosa M. Inhibition of Embryo Development of the Commercial Bivalves Ruditapes decussatus and Mytilus galloprovincialis by Trace Metals; Implications for the Implementation of Seawater Quality Criteria. Aquaculture. 2004;230:205–213. doi: 10.1016/S0044-8486(03)00432-0. [DOI] [Google Scholar]
- 18.Fathallah S., Medhioub M.N., Medhioub A., Kraiem M.M. Toxicity of Hg, Cu and Zn on Early Developmental Stages of the European Clam (Ruditapes decussatus) with Potential Application in Marine Water Quality Assessment. Environ. Monit. Assess. 2010;171:661–669. doi: 10.1007/s10661-010-1311-0. [DOI] [PubMed] [Google Scholar]
- 19.Vergílio C.S., Moreira R.V., Carvalho C.E.V., Melo E.J.T. Effects of in Vitro Exposure to Mercury on Male Gonads and Sperm Structure of the Tropical Fish Tuvira Gymnotus carapo (L.) J. Fish Dis. 2014;37:543–551. doi: 10.1111/jfd.12148. [DOI] [PubMed] [Google Scholar]
- 20.Lahnsteiner F., Mansour N., Berger B. The Effect of Inorganic and Organic Pollutants on Sperm Motility of Some Freshwater Teleosts. J. Fish Biol. 2004;65:1283–1297. doi: 10.1111/j.0022-1112.2004.00528.x. [DOI] [Google Scholar]
- 21.Varotto L., Domeneghetti S., Rosani U., Manfrin C., Cajaraville M.P., Raccanelli S., Pallavicini A., Venier P. DNA Damage and Transcriptional Changes in the Gills of Mytilus galloprovincialis Exposed to Nanomolar Doses of Combined Metal Salts (Cd, Cu, Hg) PLoS ONE. 2013;8:e54602. doi: 10.1371/journal.pone.0054602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geffard O., His E., Budzinski H., Seaman M., Garrigues P. Biological quality of seawater evaluated in situ with embryo-larval test of Crassostrea gigas and Mytilus galloprovincialis. C. R. Acad. Sci. III. 2001;324:1149–1155. doi: 10.1016/s0764-4469(01)01396-8. [DOI] [PubMed] [Google Scholar]
- 23.Beiras R., His E. Effects of Dissolved Mercury on Embryogenesis, Survival and Growth of Mytilus galloprovincialis Mussel Larvae. Mar. Ecol. Prog. Ser. 1995;126:185–189. doi: 10.3354/meps126185. [DOI] [Google Scholar]
- 24.Lettieri G., Marra F., Moriello C., Prisco M., Notari T., Trifuoggi M., Giarra A., Bosco L., Montano L., Piscopo M. Molecular Alterations in Spermatozoa of a Family Case Living in the Land of Fires. A First Look at Possible Transgenerational Effects of Pollutants. Int. J. Mol. Sci. 2020;21:6710. doi: 10.3390/ijms21186710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vassalli Q.A., Caccavale F., Avagnano S., Murolo A., Guerriero G., Fucci L., Ausió J., Piscopo M. New Insights into Protamine-like Component Organization in Mytilus galloprovincialis’ Sperm Chromatin. DNA Cell Biol. 2015;34:162–169. doi: 10.1089/dna.2014.2631. [DOI] [PubMed] [Google Scholar]
- 26.Basile A., Loppi S., Piscopo M., Paoli L., Vannini A., Monaci F., Sorbo S., Lentini M., Esposito S. The Biological Response Chain to Pollution: A Case Study from the “Italian Triangle of Death” Assessed with the Liverwort Lunularia Cruciata. Environ. Sci. Pollut. Res. Int. 2017;24:26185–26193. doi: 10.1007/s11356-017-9304-y. [DOI] [PubMed] [Google Scholar]
- 27.Bonanno G., Orlando-Bonaca M. Perspectives on Using Marine Species as Bioindicators of Plastic Pollution. Mar. Pollut. Bull. 2018;137:209–221. doi: 10.1016/j.marpolbul.2018.10.018. [DOI] [PubMed] [Google Scholar]
- 28.Antioxidant Activity and Ultrastructural Alterations in the Biosensor Lemna minor L. Exposed in Bags in Sarno River (South Italy) [(accessed on 19 December 2020)]; Available online: https://www.iris.unina.it/handle/11588/694126?mode=full.367.
- 29.Maresca V., Fusaro L., Sorbo S., Siciliano A., Loppi S., Paoli L., Monaci F., Karam E.A., Piscopo M., Guida M., et al. Functional and Structural Biomarkers to Monitor Heavy Metal Pollution of One of the Most Contaminated Freshwater Sites in Southern Europe. Ecotoxicol. Environ. Saf. 2018;163:665–673. doi: 10.1016/j.ecoenv.2018.07.122. [DOI] [PubMed] [Google Scholar]
- 30.Maresca V., Lettieri G., Sorbo S., Piscopo M., Basile A. Biological Responses to Cadmium Stress in Liverwort Conocephalum conicum (Marchantiales) Int. J. Mol. Sci. 2020;21:6485. doi: 10.3390/ijms21186485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lettieri G., Mollo V., Ambrosino A., Caccavale F., Troisi J., Febbraio F., Piscopo M. Molecular Effects of Copper on the Reproductive System of Mytilus galloprovincialis. Mol. Reprod. Dev. 2019;86:1357–1368. doi: 10.1002/mrd.23114. [DOI] [PubMed] [Google Scholar]
- 32.De Guglielmo V., Puoti R., Notariale R., Maresca V., Ausió J., Troisi J., Verrillo M., Basile A., Febbraio F., Piscopo M. Alterations in the Properties of Sperm Protamine-like II Protein after Exposure of Mytilus galloprovincialis (Lamarck 1819) to Sub-Toxic Doses of Cadmium. Ecotoxicol. Environ. Saf. 2019;169:600–606. doi: 10.1016/j.ecoenv.2018.11.069. [DOI] [PubMed] [Google Scholar]
- 33.Dellali M., Gnassia Barelli M., Romeo M., Aissa P. The Use of Acetylcholinesterase Activity in Ruditapes decussatus and Mytilus galloprovincialis in the Biomonitoring of Bizerta Lagoon. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2001;130:227–235. doi: 10.1016/S1532-0456(01)00245-9. [DOI] [PubMed] [Google Scholar]
- 34.Smaoui-Damak W., Hamza-Chaffai A., Bebianno M.J., Amiard J.C. Variation of Metallothioneins in Gills of the Clam Ruditapes decussatus from the Gulf of Gabès (Tunisia) Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2004;139:181–188. doi: 10.1016/j.cca.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 35.Köhler K., Riisgård H.U. Formation of Metallothioneins in Relation to Accumulation of Cadmium in the Common Mussel Mytilus Edulis. Mar. Biol. 1982;66:53–58. doi: 10.1007/BF00397254. [DOI] [Google Scholar]
- 36.Ceratto N., Dondero F., van de Loo J.-W., Burlando B., Viarengo A. Cloning and Sequencing of a Novel Metallothionein Gene in Mytilus galloprovincialis Lam. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2002;131:217–222. doi: 10.1016/S1532-0456(02)00008-X. [DOI] [PubMed] [Google Scholar]
- 37.Baršyt≐ D., White K.N., Lovejoy D.A. Cloning and Characterization of Metallothionein CDNAs in the Mussel Mytilus edulis L. Digestive Gland. Comp. Biochem. Physiol. Part C Pharmacol. Toxicol. Endocrinol. 1999;122:287–296. doi: 10.1016/S0742-8413(98)10126-3. [DOI] [PubMed] [Google Scholar]
- 38.Dondero F., Piacentini L., Banni M., Rebelo M., Burlando B., Viarengo A. Quantitative PCR Analysis of Two Molluscan Metallothionein Genes Unveils Differential Expression and Regulation. Gene. 2005;345:259–270. doi: 10.1016/j.gene.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 39.Benhamed S., Guardiola F.A., Martínez S., Martínez-Sánchez M.J., Pérez-Sirvent C., Mars M., Esteban M.A. Exposure of the Gilthead Seabream (Sparus Aurata) to Sediments Contaminated with Heavy Metals down-Regulates the Gene Expression of Stress Biomarkers. Toxicol. Rep. 2016;3:364–372. doi: 10.1016/j.toxrep.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haap T., Köhler H.-R. Cadmium Tolerance in Seven Daphnia Magna Clones Is Associated with Reduced Hsp70 Baseline Levels and Induction. Aquat. Toxicol. 2009;94:131–137. doi: 10.1016/j.aquatox.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 41.Singer C., Zimmermann S., Sures B. Induction of Heat Shock Proteins (Hsp70) in the Zebra Mussel (Dreissena polymorpha) Following Exposure to Platinum Group Metals (Platinum, Palladium and Rhodium): Comparison with Lead and Cadmium Exposures. Aquat. Toxicol. 2005;75:65–75. doi: 10.1016/j.aquatox.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 42.Fioretti F.M., Febbraio F., Carbone A., Branno M., Carratore V., Fucci L., Ausió J., Piscopo M. A Sperm Nuclear Basic Protein from the Sperm of the Marine Worm Chaetopterus Variopedatus with Sequence Similarity to the Arginine-Rich C-Termini of Chordate Protamine-Likes. DNA Cell Biol. 2012;31:1392–1402. doi: 10.1089/dna.2011.1547. [DOI] [PubMed] [Google Scholar]
- 43.Piscopo M., Tomei L., De Petrocellis L., Geraci G. Anion-Mediated Lysine-Arginine Interaction. Evidence in Chaetopterus Variopedatus Sperm Protamine. FEBS Lett. 1993;334:125–127. doi: 10.1016/0014-5793(93)81696-W. [DOI] [PubMed] [Google Scholar]
- 44.Piscopo M., De Petrocellis L., Conte M., Pulcrano G., Geraci G. On the Possibility That H1 Histone Interaction with DNA Occurs through Phosphates Connecting Lysine and Arginine Side Chain Groups. Acta Biochim. Pol. 2006;53:507–513. doi: 10.18388/abp.2006_3321. [DOI] [PubMed] [Google Scholar]
- 45.Piscopo M., Conte M., Di Paola F., Conforti S., Rana G., De Petrocellis L., Fucci L., Geraci G. Relevance of Arginines in the Mode of Binding of H1 Histones to DNA. DNA Cell Biol. 2010;29:339–347. doi: 10.1089/dna.2009.0993. [DOI] [PubMed] [Google Scholar]
- 46.Salvati D., Conforti S., Conte M., Matassa D.S., Fucci L., Piscopo M. Self-Association of Chaetopterus Variopedatus Sperm Histone H1-like. Relevance of Arginine Content and Possible Physiological Role. Acta Biochim. Pol. 2008;55:701–706. doi: 10.18388/abp.2008_3030. [DOI] [PubMed] [Google Scholar]
- 47.Lettieri G., D’Agostino G., Mele E., Cardito C., Esposito R., Cimmino A., Giarra A., Trifuoggi M., Raimondo S., Notari T., et al. Discovery of the Involvement in DNA Oxidative Damage of Human Sperm Nuclear Basic Proteins of Healthy Young Men Living in Polluted Areas. Int. J. Mol. Sci. 2020;21:4198. doi: 10.3390/ijms21124198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.BS EN 13805:2014 . Foodstuffs. Determination of Trace Elements. Pressure Digestion. NSAI; Dublin, Ireland: 2014. D09 A0E4. [Google Scholar]
- 49.Lettieri G., Maione M., Ranauda M.A., Mele E., Piscopo M. Molecular Effects on Spermatozoa of Mytilus galloprovincialis Exposed to Hyposaline Conditions. Mol. Reprod. Dev. 2019;86:650–660. doi: 10.1002/mrd.23141. [DOI] [PubMed] [Google Scholar]
- 50.Rave N., Crkvenjakov R., Boedtker H. Identification of Procollagen MRNAs Transferred to Diazobenzyloxymethyl Paper from Formaldehyde Agarose Gels. Nucleic Acids Res. 1979;6:3559–3567. doi: 10.1093/nar/6.11.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Livak K.J., Schmittgen T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 52.Notariale R., Basile A., Montana E., Romano N.C., Cacciapuoti M.G., Aliberti F., Gesuele R., De Ruberto F., Sorbo S., Tenore G.C., et al. Protamine-like Proteins Have Bactericidal Activity. The First Evidence in Mytilus galloprovincialis. Acta Biochim. Pol. 2018;65:585–594. doi: 10.18388/abp.2018_2638. [DOI] [PubMed] [Google Scholar]
- 53.Piscopo M., Trifuoggi M., Scarano C., Gori C., Giarra A., Febbraio F. Relevance of Arginine Residues in Cu(II)-Induced DNA Breakage and Proteinase K Resistance of H1 Histones. Sci. Rep. 2018;8:7414. doi: 10.1038/s41598-018-25784-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carbone A., Fioretti F.M., Fucci L., Ausió J., Piscopo M. High Efficiency Method to Obtain Supercoiled DNA with a Commercial Plasmid Purification Kit. Acta Biochim. Pol. 2012;59:275–278. doi: 10.18388/abp.2012_2151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.