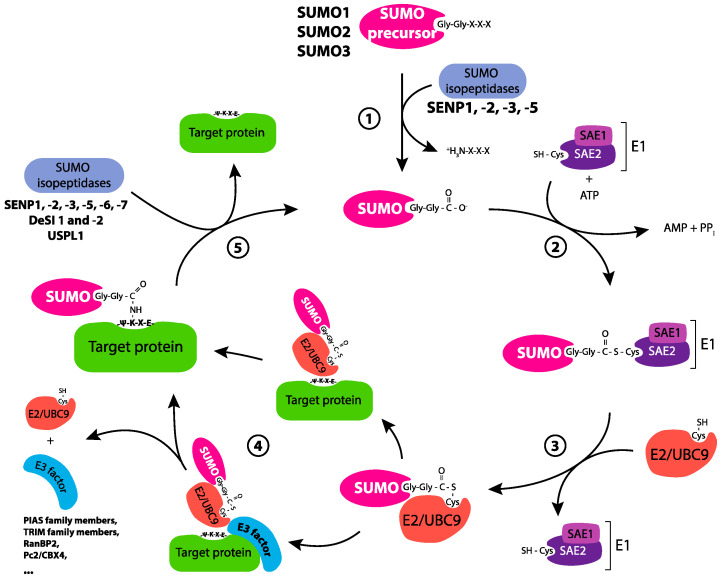
Figure 1.
The SUMOylation cycle. The C-terminal extensions of SUMO-1, -2 and -3 precursors are removed by specific peptidases from the SENP family (SENP1, -2, -3 and -5, which also harbor isopeptidasic activities) to expose a C-terminal Gly-Gly motif essential for SUMO activation and, then, conjugation to protein substrates (1). SUMO is activated in an ATP-dependent manner via transfer to a reactive cysteine of the SAE2 subunit of the heterodimeric E1 SUMO-activating enzyme (2). This occurs in two steps. First, the SUMO GG motif is adenylated by the E1 enzyme. Then, full activation is obtained by transfer of the adenylated SUMO intermediate to the SAE2 reactive cysteine. Activated SUMO is then transferred to the reactive cysteine of the E2 SUMO-conjugating enzyme also called Ubc9 (3) and covalently coupled to the ε-NH2 group of target lysines of protein substrates owing to an isopeptide bond. This can be achieved directly or with the help of SUMO E3 factors assisting the SUMO E2 enzyme (4) and often, but not always, occurs at a ψKxE consensus motif (where ψ corresponds to a bulky hydrophobic amino acid and x to any amino acid). Some SUMO E3 factors are indicated but others probably remain to be discovered. Certain enzymes display an E4 elongase activity facilitating the formation of polySUMO chains (not represented). SUMO can be removed from its substrates and SUMO chains can be depolymerized by proteases showing SUMO isopeptidasic activity (5). These include the members of the SENP family as well as a few other enzymes such as DeSI-1 and -2 and USPL1.