Abstract
Food allergy (FA) and, in particular, IgE-mediated cow’s milk allergy is associated with compositional and functional changes of gut microbiota. In this study, we compared the gut microbiota of cow’s milk allergic (CMA) infants with that of cow’s milk sensitized (CMS) infants and Healthy controls. The effect of the intake of a mixture of Bifidobacterium longum subsp. longum BB536, Bifidobacterium breve M-16V and Bifidobacterium longum subsp. infantis M-63 on gut microbiota modulation of CMA infants and probiotic persistence was also investigated. Gut microbiota of CMA infants resulted to be characterized by a dysbiotic status with a prevalence of some bacteria as Haemophilus, Klebsiella, Prevotella, Actinobacillus and Streptococcus. Among the three strains administered, B. longum subsp. infantis colonized the gastrointestinal tract and persisted in the gut microbiota of infants with CMA for 60 days. This colonization was associated with perturbations of the gut microbiota, specifically with the increase of Akkermansia and Ruminococcus. Multi-strain probiotic formulations can be studied for their persistence in the intestine by monitoring specific bacterial probes persistence and exploiting microbiota profiling modulation before the evaluation of their therapeutic effects.
Keywords: cow’s milk allergic (CMA), cow’s milk sensitized (CMS), probiotics, quantitative real-time PCR assays, gut microbiota profiling
1. Introduction
Food allergy (FA) is a common chronic condition characterized by hypersensitivity reactions to a certain food, such as cow’s milk proteins [1].
The prevalence of IgE-mediated cow’s milk allergy (CMA) in infancy and early childhood has been estimated at 2–3% worldwide [2]. In Europe, a large cohort study confirmed challenge-proven cow’s milk allergy in <1% of children up to 2 years old [3]. Children with CMA in the first year of life carry an increased risk of being affected by subsequent other atopic diseases [4,5]. Therefore, understanding CMA etiology is needed for the management and prevention of the disease and its later impact on patients.
Several epidemiologic factors suggest that CMA may be linked to microbiota alterations. Among them, its association with C-section, low maternal intake of fermented foods during pregnancy, maternal dysbiosis, high maternal socioeconomic status, intrapartum antibiotic prophylaxis and the use of antibiotics in the nursery [6].
These observations generated the hypothesis that a lack of microbial exposure during infancy may be one of the responsible factors of cow’s milk allergy. According to the extended hygiene hypothesis, also known as the “microflora hypothesis of allergic disease” [7,8], the gut microbiota seems to be a key player in the early development of the host immune system [9]. Early microbial exposure moves the Th1/Th2 immune balance towards a Th1 phenotype [10,11] while a failure of the normal intestinal bacterial colonization is associated with a shift to a Th2 response, thus sustaining allergic manifestations [12,13]. Additionally, the establishment of oral tolerance to dietary antigens is, at least in part, dependent on the presence of commensal microbes [14,15]. Thus, it is not surprising that FA and particularly CMA have been associated with compositional and functional changes of gut microbiota [16,17].
Among gut microbiota phyla, Bacteroidetes, Proteobacteria, and Actinobacteria resulted significantly reduced in FA children, while Firmicutes was highly enriched. In particular, Clostridium and Anaerobacter have been identified as bacterial markers in these patients [16].
Furthermore, the gut microbiota dysbiosis, characterized by the enrichment of Bacteroides and Alistipes, has been described in non-IgE-mediated CMA [17].
In children with food sensitization, the increase of Enterobacteriaceae/Bacteroidaceae ratio and a decrease of Ruminococcaceae abundance have been described, suggesting that early gut dysbiosis contributes to subsequent development of FA [7].
To the best of our knowledge, only one study evaluated gut microbiota differences in food allergic versus food sensitized and healthy controls. Particularly, Haemophilus, Dialister, Dorea and Clostridium were decreased in subjects with food sensitization, while Citrobacter, Oscillospira, Lactococcus and Dorea were less abundant in subjects with FA [18].
On this premise, probiotics have gained widespread attention in recent years as a preventive and therapeutic option for FA. They may act as immune modulators that stimulate Th1-mediated responses. Their positive effects on FA symptoms are largely described in the literature [19].
The most common bacteria used as probiotics belong to Lactobacillus and Bifidobacterium species. Several studies found that Bifidobacterium spp. have anti-allergic properties in both animal models and clinical trials [20,21,22]. However, the timing and duration of probiotic treatment, the optimal probiotic strains and factors that may alter their action on the gut microbiota still need further research. Before proposing specific probiotic strains for clinical use it would be necessary to explore their ability to reach the intestinal lumen and to modify gut host microbiota [9].
This is one of the reasons why univocal recommendations about probiotics have not been yet drawn up for FA. A multitude of administration approaches has been proposed for CMA, investigating the effect of different doses and intake timing [22].
In the present study, we aimed to describe the gut microbiota ecology and composition of CMA infants, cow’s milk sensitized (CMS) infants and healthy controls. In this context, we also evaluated the persistence of a probiotic mix in the gastrointestinal tract of CMA infants and its impact on gut microbiota modulation.
2. Results
2.1. Demographic and Clinical Profiling of Subjects
During the study period of 3-years, 124 patients aged 10–15 months were evaluated for suspected CMA at our hospital units. Ninety–seven patients were excluded: 14 were treated with proton-pump inhibitors (PPI), 22 were still partially breastfed, 38 had taken a course of antibiotic therapy in the previous 6 months, 52 had probiotics and 55 had functional foods in the last 2 months; 67 infants had more than one reason for exclusion. The main reasons for exclusion were the use of probiotics and feeding an extensively hydrolysed cow’s milk formula with added functional foods (probiotics or prebiotics), prescribed by the paediatrician to replace breast milk (52 cases).
The study was proposed to 27 children. As one of them was diagnosed with celiac disease soon after the double-blind placebo-controlled food challenge (DBPCFC) he was excluded, hence 26 children were enrolled (median age 12.8 months).
The CMA group included 14 infants, the CMS group 12 DBPCFC-negative infants, and the Healthy group 14 age- and sex-matched healthy infants (Table 1). There was no significant difference between groups in anthropometric characteristics including sex, age, mode of delivery, duration of breastfeeding, and duration of cow’s milk-free diet (Table 1).
Table 1.
Demographic and clinical characteristics of subjects. N/A, not applicable.
| Features | CMA | CMS | Healthy | p-Value |
|---|---|---|---|---|
| Mean age ± SD (months) | 12.9 ± 1.7 | 13.4 ± 1.0 | 12.6 ± 1.5 | 0.08 |
| Male/Female | 8/6 | 7/5 | 6/8 | 0.70 |
| Cesarean section (%) | 21% | 25% | 43% | 0.60 |
| Mean breastfeeding duration (months) | 7.2 | 7.5 | 8 | 0.44 |
| Mean cow’s milk free diet duration ± SD (months) | 6.3 ± 4.1 | 8.1 ± 4.6 | N/A | 0.51 |
| Mean diameter Prick by Prick with Cow’s Milk (mm) | 5.0 | 1.3 | 0.0 | 0.09 |
| Mean diameter Skin Prick Test with Alpha-Lactalbumin (mm) | 4.4 | 1.0 | 0.0 | 0.04 |
| Mean diameter Skin Prick Test with Beta-Lactoglobulin (mm) | 3.8 | 0.9 | 0.0 | 0.20 |
| Mean diameter Skin Prick Test with Casein (mm) | 6.4 | 1.0 | 0.0 | 0.04 |
| Total IgE (kU/L) | 228.2 | 51.2 | N/A | <0.01 |
| Specific IgE for Cow’s milk (kUA/L) | 23.8 | 0.8 | N/A | 0.05 |
| Specific IgE for α-Lactalbumin (kUA/L) | 3.5 | 0.1 | N/A | 0.14 |
| Specific IgE for β-Lactoglobulin (kUA/L) | 9.0 | 0.5 | N/A | 0.26 |
| Specific IgE for Casein (kUA/L) | 9.4 | 0.4 | N/A | 0.18 |
Three out of 14 (21.4%) in the CMA group, 3/12 (25%) in the CMS group and 6/14 (43%) in the Healthy group were delivered by caesarean section (p = 0.6). Thirty-eight infants had been breastfed for more than two months, exclusively or complementarily (Table 1).
Skin prick test with fresh cow‘s milk returned positive in 11/14, 6/12, 0/14 in CMA, CMS and Healthy groups, respectively. Specific IgE for cow’s milk was positive in 13/14 and 9/12, in CMA and CMS groups respectively.
After DBPCFC, all CMA infants followed a cow’s milk protein’s free diet and regularly took the probiotic mixture twice daily for 30 days without any relevant adverse events.
2.2. RT-PCR Analysis
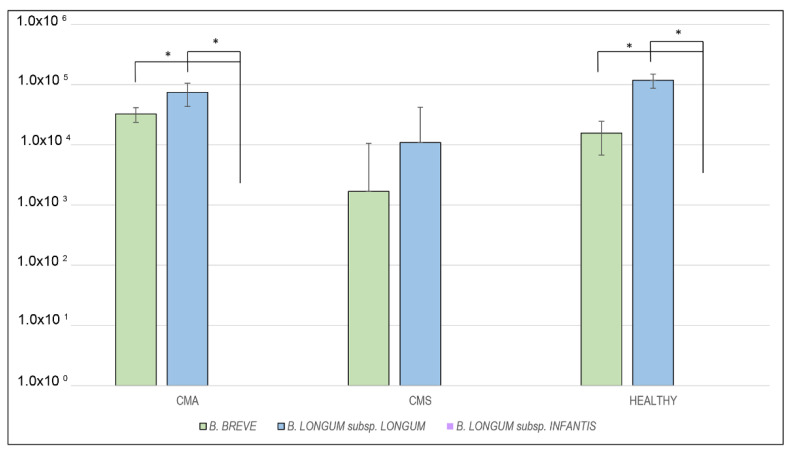
At baseline, significant differences between B. breve and B. infantis and between B. longum and B. infantis were found for CMA and Healthy groups, respectively (Figure 1, Supplementary Table S2).
Figure 1.
Median differences between B. breve, B. longum subsp. longum and B. longum subsp. infantis levels expressed as molecules/ul at T0 for the three analyzed groups (* p < 0.05).
B. breve and B. longum were present in the gut microbiota of the three groups in the order of 103–105 molecules/ul (Figure 1), without significant differences (Supplementary Table S3). No trace of B. longum subsp. infantis was detected in the three groups.
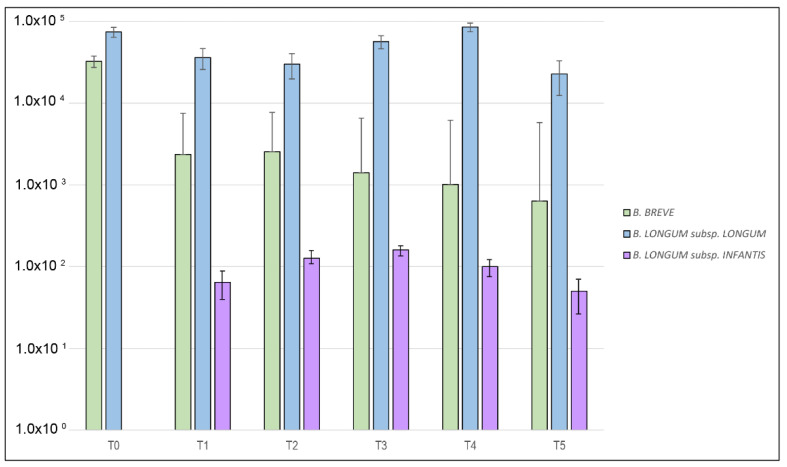
During the probiotic intervention, no significant variations of B. breve and B. longum subsp. longum amounts were detected in the CMA group, while the amount of B. longum subsp. infantis, significantly increased from T1 to T3 and decreased since point T4 (Figure 2, Supplementary Table S4).
Figure 2.
Median differences between B. breve, B. longum subsp. longum and B. longum subsp. infantis levels expressed as molecules/ul at each point of the time-course for the cow’s milk allergic (CMA) group.
2.3. Gut Microbiota Profiling
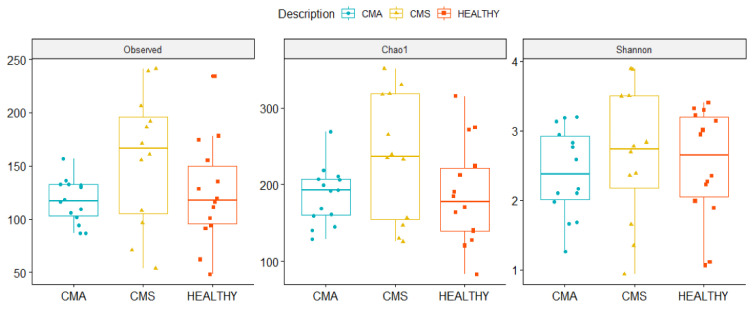
Alpha diversity was calculated using Observed, Chao1 and Shannon indices in order to evaluate Operational Taxonomic Units (OTUs) evenness, rarity and richness for CMA, CMS, and Healthy groups, at baseline. No statistically significant differences were found in the three indices comparisons. However, CMS showed the highest number of total and rare OTUs (i.e., Observed and Chao I index, respectively), while CMA was characterized by the lowest OTUs richness (i.e., Shannon index) (Figure 3).
Figure 3.
Boxplots representing alpha-diversity computed by Observed, Chao1 and Shannon indexes in the three CMA, cow’s milk sensitized (CMS), and HEALTHY groups. Boxes represent the median, 25th and 75th percentiles calculated for CMA (blue), CMS (yellow), and HEALTHY (red) groups.
Beta diversity was computed to estimate the phylogenetic relatedness (i.e., unweighted UniFrac algorithm) of CMA, CMS and Healthy groups. CMA group was clearly separated from the other groups, as verified by the PERMANOVA test (p = 0.019) (Supplementary Figure S1).
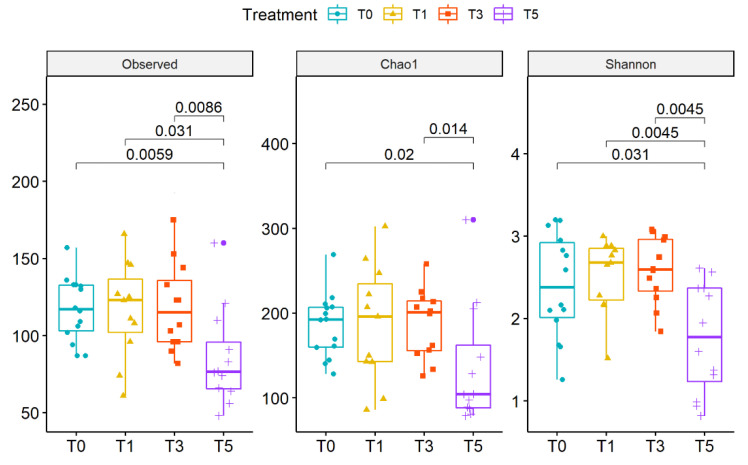
After probiotics intake, no statistically significant changes of alpha diversity were observed in the CMA group. At T5 a statistically significant decrease was observed for all alpha diversity indices (Figure 4).
Figure 4.
Boxplots representing alpha-diversity by Observed, Chao1 and Shannon indexes during the time-course for the CMA group. Boxes represent the median, 25th and 75th percentiles calculated for CMA group at T0 (blue), T1 (yellow), T3 (red) and T5 (purple). p-values are reported.
No clearly defined patient clusters along the entire time-course was detected (PERMANOVA = 0.829), (Supplementary Figure S2).
To detect differences in OTU composition, we compared the three groups, at phylum and genus levels.
At the phylum level, Verrucomicrobia was highest in Healthy and gradually decreased from CMS to CMA group (p < 0.05) (Supplementary Table S5). On the contrary, Firmicutes resulted more abundant in the CMS group and lowest in the healthy subjects (Supplementary Figure S3).
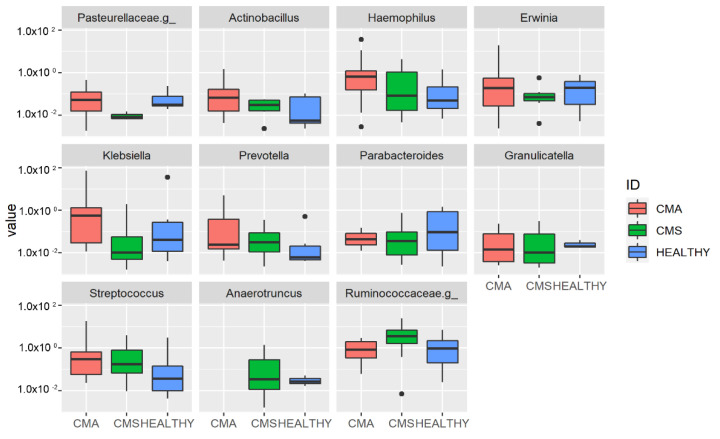
At the genus level, Haemophilus, Actinobacillus, Prevotella, Klebsiella and Streptococcus were associated with the CMA group (p < 0.05, Supplementary Table S6) while Parabacteroides and Granulicatella were more abundant in the Healthy group (Figure 5).
Figure 5.
Operational Taxonomic Unit (OUT) distribution of the gut microbiota for the three groups’ comparison assessed by DeSeq2 test at genus level. The interquartile range is represented by the box and the line in the box is the median. The whiskers indicate the largest and the lowest data points, respectively, while the dots symbolize outliers. Only statistically significant OTUs (p < 0.05) are plotted.
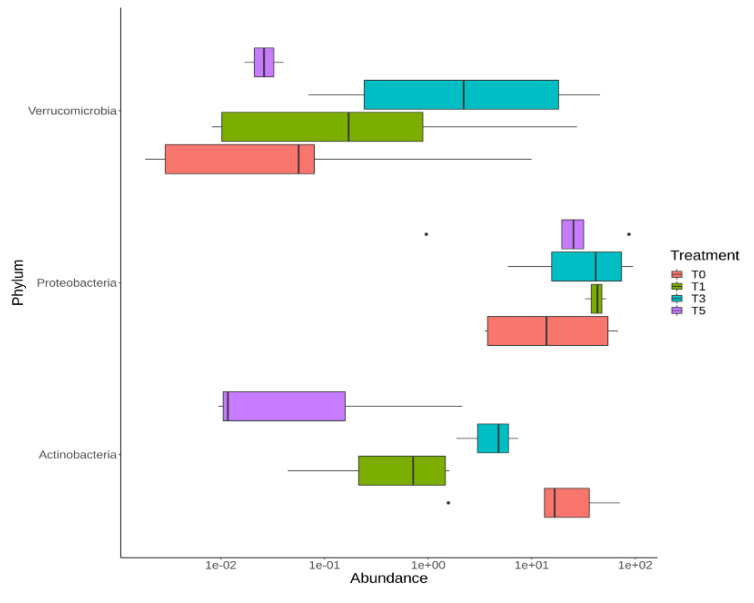
To evaluate the influence of probiotic intake on gut microbiota modulation, we tested the OTU distribution during the T0–T5 time-course in the CMA group. At the phylum level, there was an increase of Verrucomicrobia during probiotic intake with a peak of abundance at T3 and a rapid decrease at T5. Proteobacteria showed a gradual increase during probiotic intake and this increment was maintained also at T5. On the contrary, Actinobacteria decreased during the time-course, even if an increase was registered from point T1 to point T3 (Figure 6, Supplementary Table S7).
Figure 6.
OTU distribution of the gut microbiota during the time-course of the CMA group computed by DeSeq2 test at phylum level. The interquartile range is represented by the box and the line in the box is the median. The whiskers indicate the largest and the lowest data points, respectively, while the dots symbolize outliers. Only statistically significant OTUs (p < 0.05) are plotted.
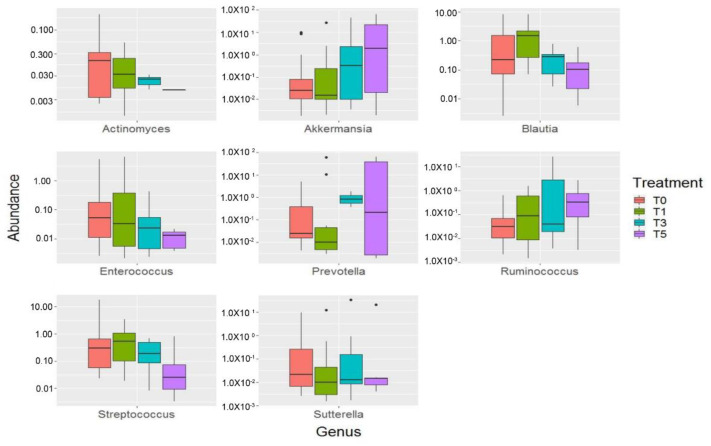
At the genus level, probiotic intake determined an increase of Akkermansia, Prevotella and Ruminococcus that was maintained also at T5. Blautia increased during the entire period of probiotic intake, until T3 point, while at T5 it started to decrease showing a level of abundance lower than that observed at T0. On the opposite way, Actinomyces, Enterococcus, Streptococcus and Sutterella resulted diminished after probiotic intervention (Figure 7, Supplementary Table S8).
Figure 7.
OTU distribution of the gut microbiota during the time-course of the CMA group computed by DeSeq2 test at the genus level. The interquartile range is represented by the box and the line in the box is the median. The whiskers indicate the largest and the lowest data points, respectively, while the dots symbolize outliers. Only statistically significant OTUs (p < 0.05) are plotted.
3. Discussion
Several comparative studies have been conducted on the intestinal microbiota composition of healthy versus allergic children, but their results are controversial [7,8,23,24,25]. To the best of our knowledge only one study evaluated gut microbiota differences in food-allergic versus food sensitized and healthy controls [18].
We observed a lower microbial diversity in CMA infants, both in terms of richness and evenness indicating that bacterial OTUs are not equally abundant in the gut ecosystem, hence generating a disequilibrium or compositional dysbiosis. Indeed, previous studies found low intestinal diversity in CMA patients [26], supporting the theory of “microbial deprivation syndromes of affluence” [27]. According to this theory, the reduced microbial stimulation could lead to abnormal immune development in early life, a possible contributory cause of allergic disease [28]. This is in line with other previous results obtained for atopy and food allergy [29,30,31].
In our study harmful bacteria as Haemophilus and Klebsiella, Prevotella were associated with CMA.
Prior studies investigating the association of gut microbiota and allergic disease during infancy demonstrated inverse associations of Haemophilus with eczema [32] and food sensitization [18]. In vitro studies demonstrated that the pro-inflammatory activity of this bacteria leads to activation of inflammatory Th2 pathways suggesting a possible role in allergy manifestation [33].
Additionally, Klebsiella spp. have been associated with pro-inflammatory responses [34,35]. Moreover, a higher abundance of this genus has been correlated with atopy in infants [23]. So, our results confirm its possible association to later development of pediatric allergy [36,37].
Interestingly, Haemophilus, Prevotella, Actinobacillus and Streptococcus gradually decreased in the gut microbiota from CMA to CMS to Healthy infants, leading to suppose a correlation of these bacteria to allergic status.
Probiotics have been proposed as a strategy for the management of CMA allergy. However, not all the studies based on probiotic intake demonstrate the actual ability of probiotics to colonize the GT. This makes it difficult to decide which strain and at what doses to administer it.
For this reason, we investigated the persistence of TRIBIF® probiotic strains in the gut microbiota at defined time-points. The mixture we studied is reported to get a significant improvement of symptoms and quality of life in allergic rhinitis and intermittent asthma [38], but no data have yet been provided about the real presence of probiotics in patients’ microbiota.
From our study, B. breve and B. longum subsp. longum are present in the gut microbiota without significant differences in CMA, CMS and Healthy groups. B. breve and B. longum subsp. longum, commonly present in the gut of breastfed infants, are able to metabolize certain human milk oligosaccharides (HMOs) and carbohydrates released by other bifidobacteria. Moreover, B. longum subsp. longum persist in the gut microbiota also after weaning, due to its ability to utilize such diet-derived carbohydrates [39]. Our data confirm that they persist after weaning, and the probiotic supplementation has not produced any evaluable effect on the concentration of these species, which were already abundant in the infant gut microbiota. By contrast, B. longum subsp. infantis was totally absent at baseline in all groups. During probiotic intake, its amount increased significantly during the first 30 days, while it decreased after probiotic discontinuation. This indicates the survival of the bacteria during their transit in the gastrointestinal tract and their ability to colonize and to persist at least for some time in the gut microbiota of children affected by IgE-mediated CMA.
B. longum subsp. infantis is unique among gut microbes for its capacity to transport into its cytoplasm and to consume the full range of HMOs [40]. Then, it is presumable that B. infantis is a very early colonizer of the infant gut during breastfeeding, whereas after weaning this species is replaced by others more adaptable to the new environment. This may be the cause of the absence of B. infantis in all the groups at baseline.
It is known that the prolonged colonization of this species is a rare phenomenon [41], while both probiotic inoculation and continued provision of HMOs are likely to foster long-term colonization [42]. Our results are in agreement with these studies, indicating that between 10 and 15 months the colonization of B. infantis persists only if there is a continuous intake.
A beneficial impact of probiotic intake on gut microbiota modulation could be associated with the significant increase of Akkermansia that was also maintained after probiotic suspension. A. muciniphila is involved in the maintenance of intestinal barrier functions and in the containment of intestinal inflammation, playing a pivotal role in the host’s overall health status [43].
Additionally, the persisting reduction of Streptococcus after the probiotic supplementation [44] can be considered a positive effect of the intervention, as Streptococcus is a potentially pathogenic microorganism that could be directly involved in atopy [45] especially considering that it resulted in a biomarker of CMA in the present study.
Blautia distribution increased during all the period of probiotic intake, until T3 point, while at T5 it started to decrease, suggesting a possible synergistic mechanism with the probiotic species for gut microbiota colonization.
Additionally, Ruminoccocus increased during the probiotic intake and after probiotic suspension. Interestingly, low relative abundance of Ruminococcaceae were registered at one week of age in infants that developed IgE-associated eczema, and low abundance of the genus Ruminococcus at that age was associated with exaggerated Toll-Like Receptor (TLR)-2-induced interleukin (IL)-6 and Tumor necrosis factor (TNF)-α responses at 6 months [46]. The increase of Ruminococcus during and after probiotic intake may be beneficial also considering its ability to produce ruminococcins, such as ruminococcin A, a bacteriocin that can inhibit the development of Clostridium species [47].
Our results indicate that B. infantis could have the biological plausibility for therapeutic actions for CMA infants, while the two other strains administered probably do not. As multi-strain probiotic formulations are largely marketed, we suggest that, according to the 5th Bradford Hill criterion [48], an evaluation of the colonization and persistence of their specific probiotics in the intestine should precede clinical studies.
This study has few limitations. It is not an efficacy study: we chose don’t include any clinical evaluation of the trend of milk allergy symptoms, because we had no indication of the kinetics of the probiotic. Moreover, this probiotic formulation has not been described in efficacy studies in allergic children.
Another limitation is the small number of patients involved. However, we performed this study on a sample of children with homogeneous feeding, in a strict range of age which guarantees the uniformity stability of the gut microbiota composition.
4. Materials and Methods
4.1. Patients Enrolment
This was a prospective multicentric study. Infants aged 10–15 months, evaluated for suspected IgE-mediated CMA, were consecutively enrolled at the Allergy Division of Bambino Gesù Children’s Hospital in Rome between January 2016 and December 2018. Infants who were still breastfed, those with gastrointestinal disease in the last 30 days, metabolic or congenital disorders were not included. We excluded infants who had been administered with antibiotics in the last 6 months, or probiotics, proton-pump inhibitors [49], fermented milk and other functional foods in the 4 weeks prior to the onset of the study.
For each patient, informed consent was obtained from their parents, and anamnestic data were recorded. The sensitivity to cow’s milk was documented by a positive skin prick test (cut-off 3 mm wheel diameter) and/or allergen-specific IgE (ImmunoCAP® Thermo Fisher Scientific, Uppsala, Sweden) with 0.35 kU/L as a lower limit of eligibility, as described elsewhere [50].
In positive patients, DBPCFC was then performed.
4.2. Food Challenges
Cow’s milk was administered in seven incremental doses of 0.1, 0.3, 1, 3, 10, 30, and 100 mL at 30 min intervals for three and a half hours. Neocate® was used as a placebo and the milk without lactose was blinded according to the standardized AAAAI/Europrevall protocol [51,52]. Oral food challenge was discontinued at the first onset of objective symptoms [53]. Patients were observed up to 6 h after starting the test.
4.3. Study Design and Faecal Sample Collection
The included patients were divided into two groups:
CMA: patients sensitized to cow’s milk proteins with DBPCFC-confirmed IgE-mediated cow’s milk allergy;
CMS: sensitized but tolerant to cow’s milk proteins patients (negative DBPCFC).
A group of age- and sex-matched children without suspect of CMA, regularly formula-fed and subjected to the same exclusion criteria, was recruited at our immunization clinic (Healthy group).
For each subject (CMA, CMS, Healthy groups), a stool sample was collected at enrolment and stored at −80°C at the Human Microbiome Unit of Bambino Gesù Children’s Hospital in Rome until further processing.
Patients belonging to the CMA group were treated with a probiotic mixture (Bifidobacterium breve M-16V, Bifidobacterium longum subsp. longum BB536 and Bifidobacterium longum subsp. infantis M-63; TRIBIF®, Valeas SpA, Milan, Italy). A dose of 3.5 × 109 colony forming units (CFU)/dose of each species was administered twice per day for 30 consecutive days. Fecal samples were collected during clinical visits at time T0 (no probiotic intake), T1 (7 days of probiotic intake), T2 (15 days of probiotic intake), T3 (30 days of probiotic intake), T4 (after 60 days from probiotic discontinuation), T5 (after 90 days after probiotic discontinuation). All samples were stored at −80 °C at the Human Microbiome Unit of Bambino Gesù Children’s Hospital in Rome until further processing.
After the oral challenge, all the positive children were prescribed a milk-free diet with an extensively hydrolysed whey formula not containing prebiotics or probiotics (Aptamil SP, Danone-Nutricia, Utrecht NL, The Netherlands). Healthy controls continued to take their regular formula, while CMS children were allowed a free diet with the substitution of a formula not enriched with functional foods (Mio 2, Nestlé Vevey, Switzerland).
This study, designed according to the Declaration of Helsinki, was registered on Clinicaltrials.gov (ClinicalTrials.gov Identifier: NCT03639337; 21 August 2018), and was approved by Bambino Gesù Children’s Hospital IRCCS Ethics Committee (protocol number 787_OPBG_2014).
4.4. Isolation of B. breve M-16V, B. longum subsp. longum BB536 and B. longum subsp. Infantis M-63, Bacterial DNA Extraction and ITS Locus Amplification
B. breve M-16V, B. longum subsp. longum BB536 and B. longum subsp. infantis M-63 were cultured, isolated and confirmed for identification as already described [54]. DNA was automatically extracted from bacterial colonies using biorobot EZ1 following the manufacturer’s instructions (Qiagen, Hilden, Germany). The entire ITS (internal transcription spacer) locus (1430 bp) was amplified using universal primers (FW: 5′-GGT GTG AAA GTC CAT CGC T-3′; RV: 5′-GTC TGC CAA GGC ATC CAC CA-3′). PCR was performed in a reaction mix containing 5 µL 10× Buffer, 2 µL 2.5 mM MgCl2, 1 µL each primer (10 µmol/L), 2 µL dNTPs (10 mmol/L), 1 µL Taq DNA polymerase (5 U/µL) (KAPA Taq PCR kit, KAPA Biosystems, Boston MA, USA), 5 µL DNA template (10 ng/µL) and molecular-grade H2O to a final reaction volume of 50 µL. The amplification protocol consisted of one cycle of initial denaturation at 94 °C for 5 min, 30 cycles of denaturation at 94 °C for 1 min, annealing at 56.5 °C for 1 min, and extension at 72 °C for 1 min followed by a final extension at 72 °C for 10 min. Amplicons were purified using centrifugal filter units (Amicon Ultra-0.5 mL Centrifugal filters 30 K, Sigma-Aldrich, St. Louis, MO, USA) and quantified using the NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
4.5. Cloning and RT-PCR Assay Design
The pGEM®-T Easy Vector System kit (Promega, Milan, Italy) was used to clone the purified PCR fragment into the PGEM vector with the use of Escherichia coli competent cells. The extraction of recombinant plasmids pGEM-BB (pGEM+B. breve), pGEM-BL (pGEM+B. longum subsp. longum) and pGEM-BI (pGEM+B. longum subsp. infantis) was then performed (Plasmid Miniprep Kit, Promega, Italy). Obtained DNA was quantified using the NanoDrop ND-1000 spectrophotometer and serial dilutions of plasmids were prepared in the range from 106 to 101 vector copy numbers and used to create a standard curve for the quantitative RT-PCR (qRT-PCR) assays. ITS rDNA cloned fragments were amplified and sequenced (Genetic Analyser 3500, Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. FASTA sequences were aligned with CLUSTAL-W software (http://www.ebi.ac.uk/clustalw/) (accessed on 1 February 2021) and species-specific primers and TaqMan probes (Roche Diagnostics) design was carried out (Supplementary Table S1).
4.6. Stool Samples DNA Extraction
DNA extraction was executed on 200 mg of stool sample by the use of QIAmp Fast DNA Stool mini kit (Qiagen). The obtained DNA was quantified by NanoDrop ND-1000 and roughly diluted to 16 ng/µL for a total amount of 80 ng per assay.
4.7. qRT-PCR Assay
The amounts of B. breve, B. longum subsp. longum and B. longum subsp. infantis in faecal samples were measured by qRT-PCR in the Light Cycler 480 instrument (Roche Diagnostics, Mannheim, Germany) and following the procedure already described in detail [54].
4.8. 16S rRNA Targeted-Metagenomics
The 16S rRNA targeted-metagenomics was performed on T0 samples of CMA, CMS, and Healthy groups and on the follow-up samples of the CMA group (T1, T3 and T5 time-points).
The variable region V3-V4 of the 16S rRNA gene (∼460 bp) was amplified by using the primer pairs 16S_F 5′ -(TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG CCT ACG GGN GGC WGC AG)-3′ and 16S_R 5′ -(GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GGA CTA CHV GGG TAT CTA ATC C)-3′ reported in the 16S Metagenomic Sequencing Library Preparation protocol (Illumina, San Diego, CA, USA). The first PCR reaction was set up using Fast Start Hifi Taq (Roche Diagnostics) with the following conditions: initial denaturation at 95 °C for 3 min, 32 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 30 s, and a final extension step at 72 °C for 5 min.
Twenty ul of KAPA Pure Beads (Roche Diagnostics) were employed to purify DNA amplicons. To obtain a unique combination of bar-code sequences, a second amplification step was prepared using Nextera technology (Illumina). The final library was cleaned-up using 50 μL of KAPA Pure Beads, quantified using Quant-iT™ PicoGreen® dsDNA Assay Kit (Thermo Fisher Scientific) and diluted in equimolar concentrations (4 nM). The following steps consisted of samples’ pooling, denaturation and dilution to 7 pM. Samples were then sequenced on an Illumina MiSeqTM platform (Illumina) following the manufacturer’s specifications to generate paired-end reads of 250 base-length.
4.9. Biocomputational and Statistical Analyses
Illumina Miseq reads were first analyzed for their quality, length and chimera presence using the Qiime v1.8 pipeline. Then, sequences were organized into Operational Taxonomic Units (OTUs) with a 97% of clustering threshold of pairwise identity. PyNAST v.0.1 program was used to carry out a multiple sequence alignment against Greengenes 13_08 database with a 97% similarity for bacterial sequences [55]. The OTU multiple sequence alignment was used as well to build a phylogenetic tree [56]. In order to filter rarely distributed OTUs, and avoid noise (i.e., artifact taxa), taxa with a threshold of prevalence greater than 0.05% within the whole dataset were selected. Alpha and beta diversity were performed by phyloseq of R package [57] and PERMANOVA test was applied on beta diversity metrics with 9999 permutations to compare samples at different time points. With the aim to identify microbial biomarkers, differences in the relative abundances of taxa between groups and during the time-course of the CMA group was assessed by DESeq2 test at phylum and genera taxonomic level. In particular, for the genus level, only taxa with a relative abundance higher than 0.01 were considered for the computational analysis [58]. SPSS statistical software (version 21) was used for graphical analysis of probiotic species persistence during the time-course and to perform Wilcoxon signed rank test for paired comparison between time-points (Supplementary Tables S2 and S3). Student’s t-test was applied for comparison of clinical variables of the population.
4.10. Metagenomic Data Open Access Repository
All raw sequencing reads are available at NCBI BioProject database (PRJNA644788) (https://www.ncbi.nlm.nih.gov/bioproject/) (accessed on 1 February 2021).
5. Conclusions
In conclusion, our study revealed that the gut microbiota of children with CMA is characterized by the prevalence of harmful bacteria such as Haemophilus, Klebsiella and Prevotella which can be considered as possible biomarkers associated with the disease.
Moreover, we can confirm the ability of B. longum subsp. infantis to pass the gastrointestinal tract and to persist in the gut microbiota of infants with CMA.
Potentially beneficial modulation of the gut microbiota during the probiotic intake was also observed and was maintained for some genera as Akkermansia and Ruminococcus.
Considering the protective and anti-inflammatory properties of B. infantis [40], it would be interesting to investigate, in a clinical trial, its clinical effect on food allergy symptoms.
Moreover, it would be interesting in a future study to evaluate the gut microbiota profile in children with a history of allergy with a negative oral challenge and sensitized at the enrollment, and compare that data with our results on sensitized children in order to investigate common fingerprints in terms of gut microbiota composition.
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/22/4/1649/s1.
Author Contributions
Conceptualization, A.G.F.; and L.P.; methodology, S.R. and M.M.; formal analysis, S.G. and A.Q.; investigation, S.R., M.M., F.D.C., R.L.V., V.F., T.N., C.R. and P.V.; resources, L.P.; data curation, S.R. and M.M.; writing—original draft preparation, S.R. and M.M.; writing—review and editing, S.R., M.M., A.G.F., L.P. and F.D.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Ministry of Health (Ricerca Corrente 201587X003556): “Persistence of probiotics in the intestinal microbiota of children: integrated systems biology study”. The study was partially supported by Valeas S.p.A.
Institutional Review Board Statement
This study, designed in according to the Declaration of Helsinki, was registered on Clinicaltrials.gov (ClinicalTrials.gov Identifier: NCT03639337; 21 August 2018), and was approved by Bambino Gesù Children’s Hospital IRCCS Ethics Committee (protocol number 787_OPBG_2014).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
All raw sequencing reads are available at NCBI BioProject database (PRJNA644788) (https://www.ncbi.nlm.nih.gov/bioproject/) (accessed on 1 February 2021).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fiocchi A., Schünemann H.J., Brozek J., Restani P., Beyer K., Troncone R., Martelli A., Terracciano L., Bahna S.L., Rancé F., et al. Diagnosis and Rationale for Action Against Cow’s Milk Allergy (DRACMA): A Summary Report. J. Allergy Clin. Immunol. 2010;126:1119–1128.e12. doi: 10.1016/j.jaci.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 2.Hossny E., Ebisawa M., El-Gamal Y., Arasi S., Dahdah L., El-Owaidy R., Galvan C.A., Lee B.W., Levin M., Martinez S., et al. Challenges of Managing Food Allergy in the Developing World. World Allergy Organ. J. 2019;12:100089. doi: 10.1016/j.waojou.2019.100089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schoemaker A.A., Sprikkelman A.B., Grimshaw K.E., Roberts G., Grabenhenrich L., Rosenfeld L., Siegert S., Dubakiene R., Rudzeviciene O., Reche M., et al. Incidence and Natural History of Challenge-Proven Cow’s Milk Allergy in European Children—EuroPrevall Birth Cohort. Allergy. 2015;70:963–972. doi: 10.1111/all.12630. [DOI] [PubMed] [Google Scholar]
- 4.D’Auria E., Salvatore S., Pozzi E., Mantegazza C., Sartorio M.U.A., Pensabene L., Baldassarre M.E., Agosti M., Vandenplas Y., Zuccotti G. Cow’s Milk Allergy: Immunomodulation by Dietary Intervention. Nutrients. 2019;11:1399. doi: 10.3390/nu11061399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fox A., Bird J.A., Fiocchi A., Knol J., Meyer R., Salminen S., Sitang G., Szajewska H., Papadopoulos N. The Potential for Pre-, pro-and Synbiotics in the Management of Infants at Risk of Cow’s Milk Allergy or with Cow’s Milk Allergy: An Exploration of the Rationale, Available Evidence and Remaining Questions. World Allergy Organ. J. 2019;12:100034. doi: 10.1016/j.waojou.2019.100034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levin M.E., Botha M., Basera W., Facey-Thomas H.E., Gaunt B., Gray C.L., Kiragu W., Ramjith J., Watkins A., Genuneit J. Environmental Factors Associated with Allergy in Urban and Rural Children from the South African Food Allergy (SAFFA) Cohort. J. Allergy Clin. Immunol. 2020;145:415–426. doi: 10.1016/j.jaci.2019.07.048. [DOI] [PubMed] [Google Scholar]
- 7.Azad M.B., Konya T., Guttman D.S., Field C.J., Sears M.R., HayGlass K.T., Mandhane P.J., Turvey S.E., Subbarao P., Becker A.B., et al. Infant Gut Microbiota and Food Sensitization: Associations in the First Year of Life. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2015;45:632–643. doi: 10.1111/cea.12487. [DOI] [PubMed] [Google Scholar]
- 8.Bisgaard H., Li N., Bonnelykke K., Chawes B.L.K., Skov T., Paludan-Müller G., Stokholm J., Smith B., Krogfelt K.A. Reduced Diversity of the Intestinal Microbiota during Infancy Is Associated with Increased Risk of Allergic Disease at School Age. J. Allergy Clin. Immunol. 2011;128:646–652.e5. doi: 10.1016/j.jaci.2011.04.060. [DOI] [PubMed] [Google Scholar]
- 9.Fiocchi A., Burks W., Bahna S.L., Bielory L., Boyle R.J., Cocco R., Dreborg S., Goodman R., Kuitunen M., Haahtela T., et al. Clinical Use of Probiotics in Pediatric Allergy (CUPPA): A World Allergy Organization Position Paper. World Allergy Organ. J. 2012;5:148–167. doi: 10.1097/WOX.0b013e3182784ee0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Round J.L., Mazmanian S.K. Inducible Foxp3+ Regulatory T-Cell Development by a Commensal Bacterium of the Intestinal Microbiota. Proc. Natl. Acad. Sci. USA. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vernocchi P., Del Chierico F., Fiocchi A.G., El Hachem M., Dallapiccola B., Rossi P., Putignani L. Understanding Probiotics’ Role in Allergic Children: The Clue of Gut Microbiota Profiling. Curr. Opin. Allergy Clin. Immunol. 2015;15:495–503. doi: 10.1097/ACI.0000000000000203. [DOI] [PubMed] [Google Scholar]
- 12.Cosmi L., Liotta F., Maggi E., Romagnani S., Annunziato F. Th17 and Non-Classic Th1 Cells in Chronic Inflammatory Disorders: Two Sides of the Same Coin. Int. Arch. Allergy Immunol. 2014;164:171–177. doi: 10.1159/000363502. [DOI] [PubMed] [Google Scholar]
- 13.Elazab N., Mendy A., Gasana J., Vieira E.R., Quizon A., Forno E. Probiotic Administration in Early Life, Atopy, and Asthma: A Meta-Analysis of Clinical Trials. Pediatrics. 2013;132:e666–e676. doi: 10.1542/peds.2013-0246. [DOI] [PubMed] [Google Scholar]
- 14.Rolinck-Werninghaus C., Staden U., Mehl A., Hamelmann E., Beyer K., Niggemann B. Specific Oral Tolerance Induction with Food in Children: Transient or Persistent Effect on Food Allergy? Allergy. 2005;60:1320–1322. doi: 10.1111/j.1398-9995.2005.00882.x. [DOI] [PubMed] [Google Scholar]
- 15.Fazlollahi M., Chun Y., Grishin A., Wood R.A., Burks A.W., Dawson P., Jones S.M., Leung D.Y.M., Sampson H.A., Sicherer S.H., et al. Early-Life Gut Microbiome and Egg Allergy. Allergy. 2018;73:1515–1524. doi: 10.1111/all.13389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ling Z., Li Z., Liu X., Cheng Y., Luo Y., Tong X., Yuan L., Wang Y., Sun J., Li L., et al. Altered Fecal Microbiota Composition Associated with Food Allergy in Infants. Appl. Environ. Microbiol. 2014;80:2546–2554. doi: 10.1128/AEM.00003-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berni Canani R., De Filippis F., Nocerino R., Paparo L., Di Scala C., Cosenza L., Della Gatta G., Calignano A., De Caro C., Laiola M., et al. Gut Microbiota Composition and Butyrate Production in Children Affected by Non-IgE-Mediated Cow’s Milk Allergy. Sci. Rep. 2018;8:12500. doi: 10.1038/s41598-018-30428-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savage J.H., Lee-Sarwar K.A., Sordillo J., Bunyavanich S., Zhou Y., O’Connor G., Sandel M., Bacharier L.B., Zeiger R., Sodergren E., et al. A Prospective Microbiome-Wide Association Study of Food Sensitization and Food Allergy in Early Childhood. Allergy. 2018;73:145–152. doi: 10.1111/all.13232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan-Lim C.S.C., Esteban-Ipac N.A.R. Probiotics as Treatment for Food Allergies among Pediatric Patients: A Meta-Analysis. World Allergy Organ. J. 2018;11:25. doi: 10.1186/s40413-018-0204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Kivit S., Kostadinova A.I., Kerperien J., Morgan M.E., Muruzabal V.A., Hofman G.A., Knippels L.M.J., Kraneveld A.D., Garssen J., Willemsen L.E.M. Dietary, Nondigestible Oligosaccharides and Bifidobacterium Breve M-16V Suppress Allergic Inflammation in Intestine via Targeting Dendritic Cell Maturation. J. Leukoc. Biol. 2017;102:105–115. doi: 10.1189/jlb.3A0516-236R. [DOI] [PubMed] [Google Scholar]
- 21.Liu M.-Y., Yang Z.-Y., Dai W.-K., Huang J.-Q., Li Y.-H., Zhang J., Qiu C.-Z., Wei C., Zhou Q., Sun X., et al. Protective Effect of Bifidobacterium Infantis CGMCC313-2 on Ovalbumin-Induced Airway Asthma and β-Lactoglobulin-Induced Intestinal Food Allergy Mouse Models. World J. Gastroenterol. 2017;23:2149–2158. doi: 10.3748/wjg.v23.i12.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qamer S., Deshmukh M., Patole S. Probiotics for Cow’s Milk Protein Allergy: A Systematic Review of Randomized Controlled Trials. Eur. J. Pediatr. 2019;178:1139–1149. doi: 10.1007/s00431-019-03397-6. [DOI] [PubMed] [Google Scholar]
- 23.Candela M., Rampelli S., Turroni S., Severgnini M., Consolandi C., De Bellis G., Masetti R., Ricci G., Pession A., Brigidi P. Unbalance of Intestinal Microbiota in Atopic Children. BMC Microbiol. 2012;12:95. doi: 10.1186/1471-2180-12-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nylund L., Satokari R., Nikkilä J., Rajilić-Stojanović M., Kalliomäki M., Isolauri E., Salminen S., de Vos W.M. Microarray Analysis Reveals Marked Intestinal Microbiota Aberrancy in Infants Having Eczema Compared to Healthy Children in At-Risk for Atopic Disease. BMC Microbiol. 2013;13:12. doi: 10.1186/1471-2180-13-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mennini M., Fierro V., Di Nardo G., Pecora V., Fiocchi A. Microbiota in Non-IgE-Mediated Food Allergy. Curr. Opin. Allergy Clin. Immunol. 2020;20:323–328. doi: 10.1097/ACI.0000000000000644. [DOI] [PubMed] [Google Scholar]
- 26.Berni Canani R., Sangwan N., Stefka A.T., Nocerino R., Paparo L., Aitoro R., Calignano A., Khan A.A., Gilbert J.A., Nagler C.R. Lactobacillus Rhamnosus GG-Supplemented Formula Expands Butyrate-Producing Bacterial Strains in Food Allergic Infants. ISME J. 2016;10:742–750. doi: 10.1038/ismej.2015.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.West C.E., Jenmalm M.C., Prescott S.L. The Gut Microbiota and Its Role in the Development of Allergic Disease: A Wider Perspective. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2015;45:43–53. doi: 10.1111/cea.12332. [DOI] [PubMed] [Google Scholar]
- 28.Rodríguez J.M., Murphy K., Stanton C., Ross R.P., Kober O.I., Juge N., Avershina E., Rudi K., Narbad A., Jenmalm M.C., et al. The Composition of the Gut Microbiota throughout Life, with an Emphasis on Early Life. Microb. Ecol. Health Dis. 2015;26 doi: 10.3402/mehd.v26.26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abrahamsson T.R., Jakobsson H.E., Andersson A.F., Björkstén B., Engstrand L., Jenmalm M.C. Low Diversity of the Gut Microbiota in Infants with Atopic Eczema. J. Allergy Clin. Immunol. 2012;129:434–440.e2. doi: 10.1016/j.jaci.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 30.Marrs T., Flohr C. How Do Microbiota Influence the Development and Natural History of Eczema and Food Allergy? Pediatr. Infect. Dis. J. 2016;35:1258–1261. doi: 10.1097/INF.0000000000001314. [DOI] [PubMed] [Google Scholar]
- 31.Nylund L., Nermes M., Isolauri E., Salminen S., de Vos W.M., Satokari R. Severity of Atopic Disease Inversely Correlates with Intestinal Microbiota Diversity and Butyrate-Producing Bacteria. Allergy. 2015;70:241–244. doi: 10.1111/all.12549. [DOI] [PubMed] [Google Scholar]
- 32.Zheng H., Liang H., Wang Y., Miao M., Shi T., Yang F., Liu E., Yuan W., Ji Z.-S., Li D.-K. Altered Gut Microbiota Composition Associated with Eczema in Infants. PLoS ONE. 2016;11:e0166026. doi: 10.1371/journal.pone.0166026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goleva E., Jackson L.P., Harris J.K., Robertson C.E., Sutherland E.R., Hall C.F., Good J.T., Gelfand E.W., Martin R.J., Leung D.Y.M. The Effects of Airway Microbiome on Corticosteroid Responsiveness in Asthma. Am. J. Respir. Crit. Care Med. 2013;188:1193–1201. doi: 10.1164/rccm.201304-0775OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garrett W.S., Gallini C.A., Yatsunenko T., Michaud M., DuBois A., Delaney M.L., Punit S., Karlsson M., Bry L., Glickman J.N., et al. Enterobacteriaceae Act in Concert with the Gut Microbiota to Induce Spontaneous and Maternally Transmitted Colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Weerth C., Fuentes S., Puylaert P., de Vos W.M. Intestinal Microbiota of Infants with Colic: Development and Specific Signatures. Pediatrics. 2013;131:e550–e558. doi: 10.1542/peds.2012-1449. [DOI] [PubMed] [Google Scholar]
- 36.Low J.S.Y., Soh S.-E., Lee Y.K., Kwek K.Y.C., Holbrook J.D., Van der Beek E.M., Shek L.P., Goh A.E.N., Teoh O.H., Godfrey K.M., et al. Ratio of Klebsiella/Bifidobacterium in Early Life Correlates with Later Development of Paediatric Allergy. Benef. Microbes. 2017;8:681–695. doi: 10.3920/BM2017.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakayama J., Kobayashi T., Tanaka S., Korenori Y., Tateyama A., Sakamoto N., Kiyohara C., Shirakawa T., Sonomoto K. Aberrant Structures of Fecal Bacterial Community in Allergic Infants Profiled by 16S RRNA Gene Pyrosequencing. FEMS Immunol. Med. Microbiol. 2011;63:397–406. doi: 10.1111/j.1574-695X.2011.00872.x. [DOI] [PubMed] [Google Scholar]
- 38.Miraglia Del Giudice M., Indolfi C., Capasso M., Maiello N., Decimo F., Ciprandi G. Bifidobacterium Mixture (B Longum BB536, B Infantis M-63, B Breve M-16V) Treatment in Children with Seasonal Allergic Rhinitis and Intermittent Asthma. Ital. J. Pediatr. 2017;43:25. doi: 10.1186/s13052-017-0340-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Callaghan A., van Sinderen D. Bifidobacteria and Their Role as Members of the Human Gut Microbiota. Front. Microbiol. 2016;7:925. doi: 10.3389/fmicb.2016.00925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Underwood M.A., German J.B., Lebrilla C.B., Mills D.A. Bifidobacterium Longum Subspecies Infantis: Champion Colonizer of the Infant Gut. Pediatr. Res. 2015;77:229–235. doi: 10.1038/pr.2014.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maldonado-Gómez M.X., Martínez I., Bottacini F., O’Callaghan A., Ventura M., van Sinderen D., Hillmann B., Vangay P., Knights D., Hutkins R.W., et al. Stable Engraftment of Bifidobacterium Longum AH1206 in the Human Gut Depends on Individualized Features of the Resident Microbiome. Cell Host Microbe. 2016;20:515–526. doi: 10.1016/j.chom.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 42.Frese S.A., Hutton A.A., Contreras L.N., Shaw C.A., Palumbo M.C., Casaburi G., Xu G., Davis J.C.C., Lebrilla C.B., Henrick B.M., et al. Persistence of Supplemented Bifidobacterium Longum Subsp. Infantis EVC001 in Breastfed Infants. mSphere. 2017;2 doi: 10.1128/mSphere.00501-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneeberger M., Everard A., Gómez-Valadés A.G., Matamoros S., Ramírez S., Delzenne N.M., Gomis R., Claret M., Cani P.D. Akkermansia Muciniphila Inversely Correlates with the Onset of Inflammation, Altered Adipose Tissue Metabolism and Metabolic Disorders during Obesity in Mice. Sci. Rep. 2015;5:16643. doi: 10.1038/srep16643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toscano M., De Grandi R., Stronati L., De Vecchi E., Drago L. Effect of Lactobacillus Rhamnosus HN001 and Bifidobacterium Longum BB536 on the Healthy Gut Microbiota Composition at Phyla and Species Level: A Preliminary Study. World J. Gastroenterol. 2017;23:2696–2704. doi: 10.3748/wjg.v23.i15.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park Y.M., Lee S.Y., Kang M.J., Kim B.S., Lee M.J., Jung S.S., Yoon J.S., Cho H.J., Lee E., Yang S.I., et al. Imbalance of Gut Streptococcus, Clostridium, and Akkermansia Determines the Natural Course of Atopic Dermatitis in Infant. Allergy Asthma Immunol. Res. 2020;12:322–337. doi: 10.4168/aair.2020.12.2.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.West C.E., Rydén P., Lundin D., Engstrand L., Tulic M.K., Prescott S.L. Gut Microbiome and Innate Immune Response Patterns in IgE-Associated Eczema. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2015;45:1419–1429. doi: 10.1111/cea.12566. [DOI] [PubMed] [Google Scholar]
- 47.Dabard J., Bridonneau C., Phillipe C., Anglade P., Molle D., Nardi M., Ladiré M., Girardin H., Marcille F., Gomez A., et al. Ruminococcin A, a New Lantibiotic Produced by a Ruminococcus Gnavus Strain Isolated from Human Feces. Appl. Environ. Microbiol. 2001;67:4111–4118. doi: 10.1128/AEM.67.9.4111-4118.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hill A.B. The Environment and Disease: Association or Causation? J. R. Soc. Med. 2015;108:32–37. doi: 10.1177/0141076814562718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naito Y., Kashiwagi K., Takagi T., Andoh A., Inoue R. Intestinal Dysbiosis Secondary to Proton-Pump Inhibitor Use. Digestion. 2018;97:195–204. doi: 10.1159/000481813. [DOI] [PubMed] [Google Scholar]
- 50.Fiocchi A., Artesani M.C., Riccardi C., Mennini M., Pecora V., Fierro V., Calandrelli V., Dahdah L., Valluzzi R.L. Impact of Omalizumab on Food Allergy in Patients Treated for Asthma: A Real-Life Study. J. Allergy Clin. Immunol. Pract. 2019;7:1901–1909.e5. doi: 10.1016/j.jaip.2019.01.023. [DOI] [PubMed] [Google Scholar]
- 51.Nowak-Wegrzyn A., Assa’ad A.H., Bahna S.L., Bock S.A., Sicherer S.H., Teuber S.S. Adverse Reactions to Food Committee of American Academy of Allergy, Asthma & Immunology Work Group Report: Oral Food Challenge Testing. J. Allergy Clin. Immunol. 2009;123:S365–S383. doi: 10.1016/j.jaci.2009.03.042. [DOI] [PubMed] [Google Scholar]
- 52.Keil T., McBride D., Grimshaw K., Niggemann B., Xepapadaki P., Zannikos K., Sigurdardottir S.T., Clausen M., Reche M., Pascual C., et al. The Multinational Birth Cohort of EuroPrevall: Background, Aims and Methods. Allergy. 2010;65:482–490. doi: 10.1111/j.1398-9995.2009.02171.x. [DOI] [PubMed] [Google Scholar]
- 53.Grabenhenrich L.B., Reich A., McBride D., Sprikkelman A., Roberts G., Grimshaw K.E.C., Fiocchi A.G., Saxoni-Papageorgiou P., Papadopoulos N.G., Fiandor A., et al. Physician’s Appraisal vs Documented Signs and Symptoms in the Interpretation of Food Challenge Tests: The EuroPrevall Birth Cohort. Pediatr. Allergy Immunol. Off. Publ. Eur. Soc. Pediatr. Allergy Immunol. 2018;29:58–65. doi: 10.1111/pai.12811. [DOI] [PubMed] [Google Scholar]
- 54.Reddel S., Del Chierico F., Quagliariello A., Giancristoforo S., Vernocchi P., Russo A., Fiocchi A., Rossi P., Putignani L., El Hachem M. Gut Microbiota Profile in Children Affected by Atopic Dermatitis and Evaluation of Intestinal Persistence of a Probiotic Mixture. Sci. Rep. 2019;9:4996. doi: 10.1038/s41598-019-41149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I., et al. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., Huber T., Dalevi D., Hu P., Andersen G.L. Greengenes, a Chimera-Checked 16S RRNA Gene Database and Workbench Compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McMurdie P.J., Holmes S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rivera-Pinto J., Egozcue J.J., Pawlowsky-Glahn V., Paredes R., Noguera-Julian M., Calle M.L. Balances: A New Perspective for Microbiome Analysis. mSystems. 2018;3 doi: 10.1128/mSystems.00053-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw sequencing reads are available at NCBI BioProject database (PRJNA644788) (https://www.ncbi.nlm.nih.gov/bioproject/) (accessed on 1 February 2021).