Abstract
Simple Summary
Melanotic neuroectodermal tumor of infancy (MNTI) is a rare tumor of uncertain origin, morphologically overlapping other rare neoplasms such as pineal anlage tumor (PAT) and a subset of medulloblastomas (i.e., melanotic medulloblastoma). Despite the similarities with MNTI, their possible histogenetic relationship has been traditionally disregarded based on their aggressive behavior and dismal prognosis. The aim of this study was to further characterize the molecular features of MNTI and PAT based on DNA-methylation and copy number variation profiling analysis. We found that MNTI shares a methylation profile with group 3 high-risk medulloblastoma, and potentially with PAT, suggesting a common histogenesis. Most MNTIs in our series lacked copy number variation alterations, whereas their presence in the one PAT deserves further study in larger cohorts to better determine their impact in prognosis and biologic behavior.
Abstract
MNTI is a rare tumor of indeterminate histogenesis and molecular signature. We performed methylation and copy number variation (CNV) profiles in patients with MNTI (n = 7) and PAT (n = 1) compared to the methylation brain tumor classifier v11b4 (BT-C) and the medulloblastoma (MB) classifier group 3/4 v1.0 (MB3/4-C). The patients’ mean age was 8 months (range: 4–48). The BT-C classified five MNTIs and one PAT (relapse) as class family MB-G3/G4, subclass group 3 (score: >0.9). The remaining two MNTIs and PAT (primary) were classified as class family plexus tumor, subclass pediatric (scores: >0.45). The MB3/4-C classified all MNTIs as high-risk MB-G3, Subtype II (score: >0.45). The primary PAT was classified as subtype III (score: 0.99) and its relapse as subtype II/III. MNTI and PAT clustered close to MB-G3. CNV analysis showed multiple rearrangements in one PAT and two MNTIs. The median follow-up was 54 months (four MNTIs in remission, one PAT died). In conclusion, we demonstrated that MNTI shares a homogenous methylation profile with MB-G3, and possibly with PAT. The role of a multipotent progenitor cell (i.e., early cranial neural crest cell) in their histogenesis and the influence of the anatomical site, tumor microenvironment, and other cytogenetic events in their divergent biologic behavior deserve further investigation.
Keywords: melanotic neuroectodermal tumor, melanotic progonoma, medulloblastoma, pineal anlage tumor, DNA methylation, copy number variation
1. Introduction
Since its original description in 1918 by Krompecher et al., under the name of “congenital melanocarcinoma of the alveolar process” [1], the histogenesis of melanotic neuroectodermal tumor of infancy (MNTI) has been disputed, as evidenced by different names attributed to this entity over the past century. Halpert and Patzer proposed a histogenetic relationship of these tumors with the ciliary process and retinal nuclear layers by introducing the term “retinal anlage tumor” [2], a notion later contested by Stowens, who instead coined the term “melanotic progonoma” to emphasize the absence of an anatomical precursor (and its putative origin from misplacement of tissue due to fetal atavism) while highlighting the characteristic presence of pigmented cells [3]. Finally, the production of vanillylmandelic acid (VMA) observed in one such case led Borello and Gorlin to propose a neuroectodermal origin, coining the current terminology of MNTI [4]. Although MNTI is currently considered a neurocristic tumor [5,6], only very few studies have attempted to further address its underlying molecular profile, most of them on a case report basis [7,8,9].
Interestingly, MNTI overlaps with the rare pineal anlage tumor (PAT), which is regarded as a variant of pineoblastoma [10]. It also shows morphologic overlap with melanotic medulloblastoma, which was proposed as a possible variant of MNTI [11]. However, this theory is debatable given the usual aggressive course of melanotic medulloblastomas in contrast to the generally benign behavior of MNTI [12]. Recently, Ebel et al. reported a rapidly growing skull tumor in a male infant with a methylation profile of group 3 medulloblastoma despite showing conventional MNTI morphology, raising further questions regarding the underlying pathobiology of these neoplasms [13]. Intriguingly, we recently came across a PAT showing a similar methylation profile. On this ground, we sought to explore the DNA methylation and copy number variation profiling in a series of seven MNTIs and one PAT to better delineate their histogenesis.
2. Results
2.1. Clinical Findings
The index case was a PAT, diagnosed in a 12-month-old boy. This patient presented with a local relapse, occurring 2 months after the original diagnosis. Additionally, seven MNTIs were identified in various anatomic locations, including the maxilla (n = 4), hard palate (n = 1), neck (n = 1), and pretesticular region (n = 1). The median age at diagnosis was 8 months (range: 4–48 months) with a male-to-female ratio of 6:2. All MNTI patients with available clinical information (n = 4) were in clinical remission. In addition, the PAT patient died of disease 16 months after the original diagnosis following a multistage surgical resection approach (https://doi.org/10.1007/978-3-030-16006-7 (accessed on 5 February 2021)). The median follow-up was 54 months. Follow-up information was lost in the remaining cases.
2.2. Morphologic and Immunophenotypic Findings
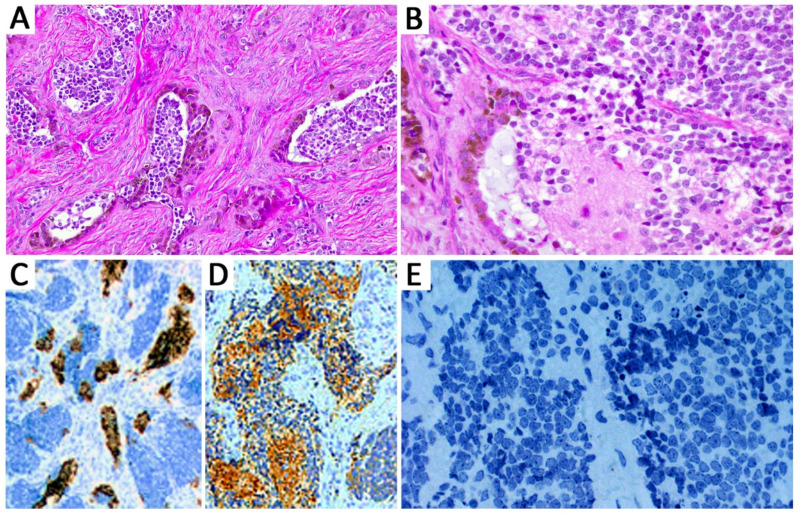
The index case, a PAT, demonstrated a variably pigmented epithelial component similar to MNTIs (Figure 1), admixed with areas featuring ectomesenchymal differentiation (fibrous tissue and multiple foci of cartilage), whereas the recurrence was predominantly composed of primitive embryonal cells, with areas of neuronal differentiation resembling neuroblastic areas of MNTIs (Figure 2). All MNTI exhibited a characteristic biphasic pattern composed of small round, neuroblast-like cells forming clusters/nests alternating with larger cells with more abundant cytoplasm and variable melanotic pigment in a dense fibrous stroma (Figure 3A,B). The mitotic activity was overall low (average: 0–2 mitoses per 10 high-power fields), and no significant atypia or tumoral necrosis was identified. All MNTI had a large cell component consistently positive for HMB-45 (Figure 3C) and cytokeratin AE1/AE3 with variable synaptophysin expression. The small, neuroblast-like cell component was strongly reactive for NSE and synaptophysin (Figure 3D), while Phox2b was constantly negative in all components (Figure 3E).
Figure 1.
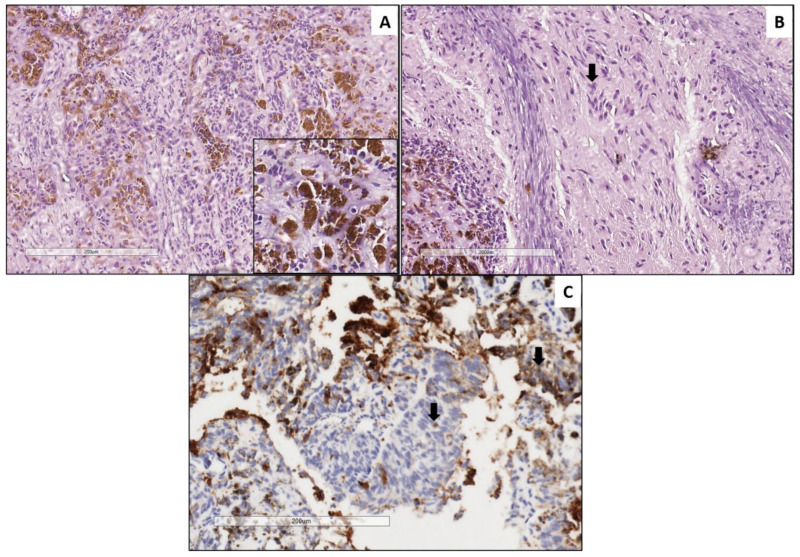
Pineal anlage tumor (PAT, primary specimen). Neuroepithelial component characterized by pineoblastoma-like nests of small round blue cells with hyperchromatic nuclei admixed with tubular structures composed of epithelioid cells containing melanin pigment (inset) (A). Ganglion cells characterized by round nuclei and abundant eosinophilic cytoplasm (arrow) (B). Positive immunoreactivity for synaptophysin in a cytoplasmic granular pattern (arrow) (C).
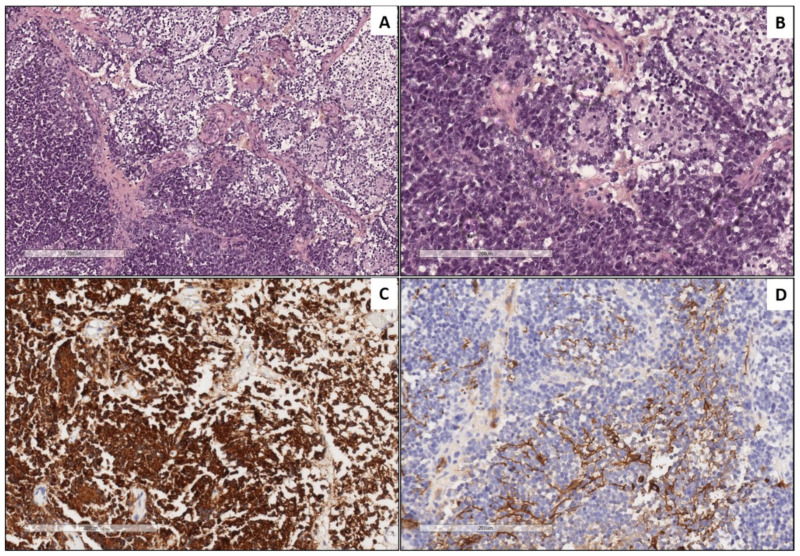
Figure 2.
Pineal anlage tumor (relapse specimen). Neuroepithelial component characterized by solid areas of small round blue cells with hyperchromatic nuclei and high N:C ratio (left side) associated to Homer-Wright rosettes (right side) ((A) and (B), respectively). Synaptophysin staining neuroepithelial cells (C). Scattered neoplastic glial cells positive for glial fibrillary acidic protein (D).
Figure 3.
Melanotic neuroectodermal tumor of infancy (MNTI). Tumors showed a biphasic pattern composed of small round, neuroblast-like cells forming clusters/nests alternating with larger cells with more abundant cytoplasm and variable melanotic pigment in a dense fibrous stroma (A,B). All MNTIs had a large cell component consistently positive for HMB-45 (C). The small, neuroblast-like cell component was strongly reactive for synaptophysin (D), while Phox2b was constantly negative in all components (E).
2.3. Methylation Profiling
The methylation data of the PAT were first categorized using the brain tumor classifier v11b4 (https://www.molecularneuropathology.org/mnp/classifier/2 (accessed on 5 February 2021)) [14], which also generated a copy number variation (CNV) plot. The primary tumor clustered in the class family plexus tumor, subclass pediatric (PLEX, PED B), with suboptimal raw and calibrated scores (>0.45), while its relapse was classified as group 3 medulloblastoma (MB G3) with optimal scores (Table 1).
Table 1.
Clinicopathological and molecular features of seven MNTIs and one PAT.
| Case No. | Sex | Age (mo) | Size (cm) | Location | Morphological Diagnosis | Idat | Brain Tumor Classifier Result | Calibrated Scores | MB G3/G4 Classifier | Calibrated Scores |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 12 | N/A | Pineal | PAT | 200992280039_R07 | PLEX PED B | 0.63 | Subtype III | 0.99 |
| 2 | M | N/A | Pineal | PAT (relapse) | 200992280039_R08 | MB, G3 | 0.98 | Subtype II /III | 0.45/0.4 | |
| 3 | M | 48 | 2 | Neck | MNTI | 203939360062_R07 | MB, G3 | 0.99 | Subtype II | 0.99 |
| 4 | F | 3 | 4 | Left maxilla | MNTI | 203724130138_R05 | MB, G3 | 0.98 | Subtype II | 0.99 |
| 5 | M | 7 | 3.5 | Left maxilla | MNTI | 203724130138_R06 | PLEX PED B/MB, G3 | 0.38/0.27 | Subtype II/III | 0.48/0.31 |
| 6 | M | 4 | 4.3 | Left maxilla | MNTI | 203724130138_R07 | MB, G3 | 0.95 | Subtype II | 0.85 |
| 7 | M | 9 | 2.5 | Right paratesticular | MNTI | 203939360072_R02 | PLEX PED B | 0.29 | Subtype II | 0.64 |
| 8 | M | 4 | 4.5 | Right maxilla | MNTI | 203939360072_R03 | MB, G3 | 0.97 | Subtype II | 0.72 |
| 9 | F | 8 | 3 | Left hard palate | MNTI | 203939360072_R04 | MB, G3 | 0.93 | Subtype II | 0.72 |
Legend: PLEX PED B = Methylation class plexus tumor, subclass pediatric B. MB, G3 = Methylation class medulloblastoma, subclass group 3. PAT = Pineal anlage tumor. MNTI = Melanotic neuroectodermal tumor of infancy.
However, it has been reported that tumors that cannot be correctly classified (with a score > 0.95) are often classified as PLEX, PED B while there is no clear connection to this tumor group. Classifier developers suggest that a low score for PLEX PED B in an unexpected setting should therefore be considered with caution [15].
No significant methylation scores were reached in the sarcoma classifier v12.2 (https://www.molecularneuropathology.org/mnp/classifier/9 (accessed on 5 February 2021)). When exploiting the “Medulloblastoma classifier group 3/4 v1.0”, which takes into account the new consensus on the second-generation molecular subgrouping of medulloblastoma [16], the PAT was classified as subtype III (calibrated score 0.99) and its relapse as subtype II/III (Table 1).
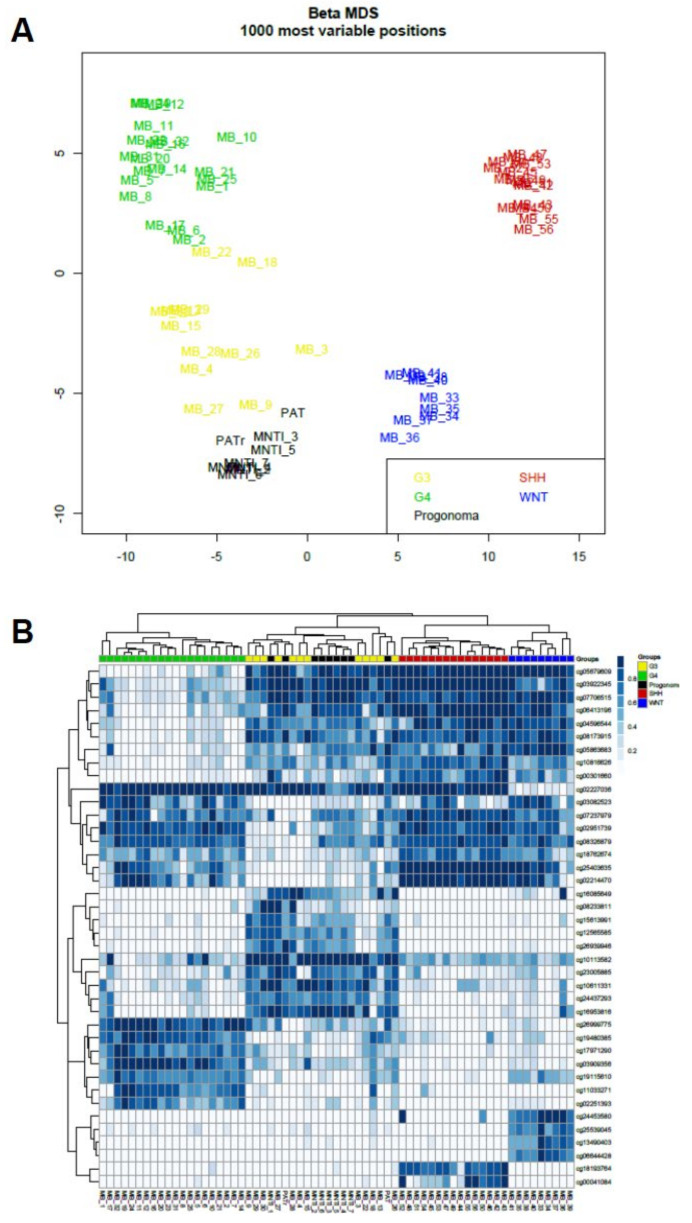
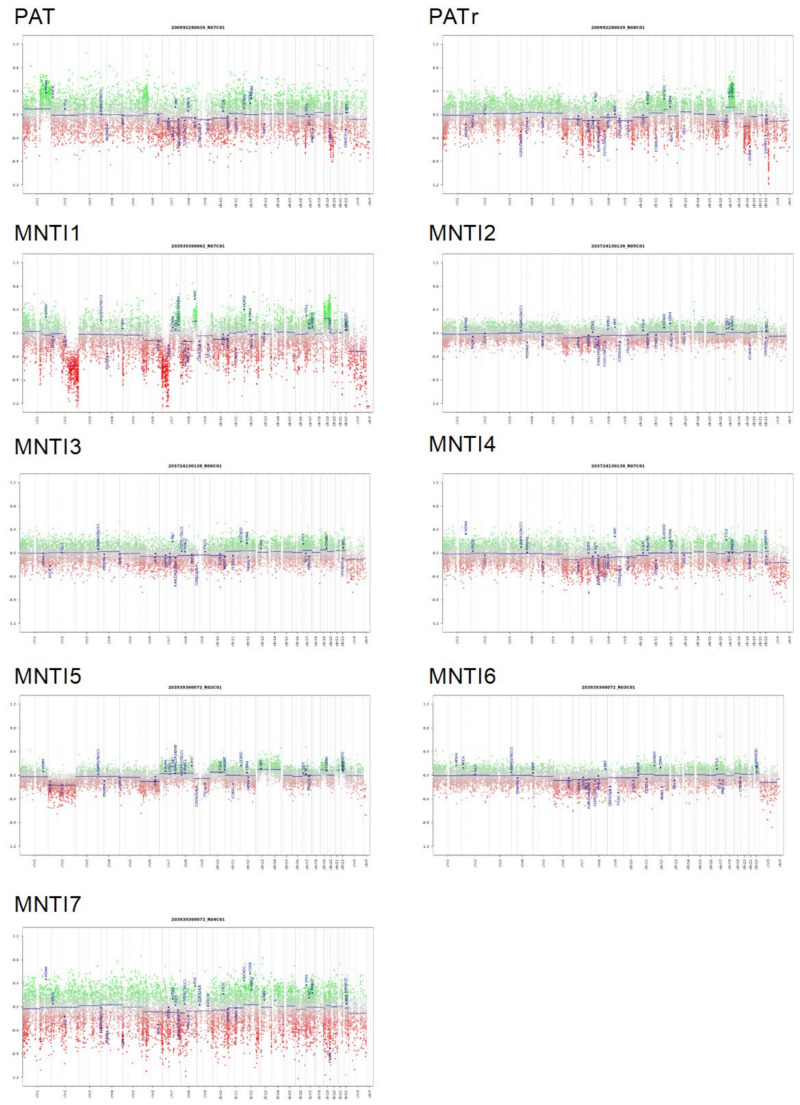
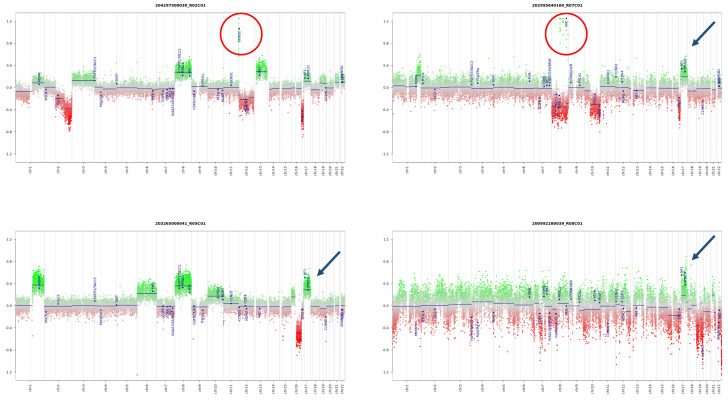
Subsequently, the global methylation profile of MNTI (n = 7) was analyzed. Five tumors clustered in the class MB G3 with optimal scores (Figure S1 and Supplementary information). The two remaining MNTIs clustered in the PLEX, PED B methylation class, with suboptimal calibrated scores (>0.45), while the maximum raw classification scores for both samples were reached in the MB G3 methylation class (Figure S1 and Supplementary information). In the “Medulloblastoma classifier group 3/4 v1.0”, all MNTI were classified as high-risk medulloblastoma G3 tumors, subtype II with a score > 0.45 (Table 1 and Supplementary information). In light of these results, we further analyzed and compared MNTI and PAT methylation data with those derived from a cohort of 36 medulloblastoma patients, diagnosed and treated at the Ospedale Pediatrico Bambino Gesù (OPBG Rome) [17]. All MNTI samples and PATs displayed a global methylation profile close to those of MB G3, as evidenced by both multidimensional scaling analysis performed on the 1000 most variable islands in the cohort (Figure 4A) and hierarchical clustering analysis (Figure 4B and Figure S2) performed on the reduced 48 CpG islands signature used to characterize the medulloblastoma subgroups [18]. Copy number variation (CNV) analysis showed an almost flat profile in five MNTIs (cases 2–6), multiple whole chromosomal gains and losses (including MYC amplification) in one MNTI (case 1) and PAT (both primary and recurrent specimen), and a highly variable profile in MNTI7 (Figure 5), similar to those observed in MB G3 in the control cohort (Figure 6).
Figure 4.
Multidimensional scaling (MS) analysis performed on the 1000 most variable probes of the whole-genome DNA methylation data shows a close similarity between (MNTI/PAT) and MBG3. Color legend of the MDS plot as follows: Progonoma (MNTI/PAT, black); WNT, Wingless MB (blue); SHH Sonic Hedgehog (red); G3, Group 3 MB (yellow); G4, Group 4 MB (green) (A). Hierarchical clustering and heatmap of beta values relative to the 39 high-quality CpG islands better discriminating MB subgroup in the Hovestadt set (Hovestadt et al.). The heatmap shows normalized methylation levels in MNTI/PAT samples and MB samples. Clusters were obtained by means of Ward’s minimum variance method using the Euclidean distance. Color legend: Progonoma (MNTI/PAT, black); WNT, Wingless MB (blue); SHH-A, Sonic Hedgehog MB (red); G3, Group 3 MB (yellow); G4, Group 4 MB (green) (B).
Figure 5.
Copy number variation (CNV) profile analysis of the indicated PAT/MNTI. Depiction of structural rearrangements involving autosomes and X/Y chromosome. Gains/amplifications represent positive (green) and losses represent negative (red) deviations from the baseline. Twenty-nine tumor relevant genomic regions are highlighted.
Figure 6.
CNV of four different MBG3, subtype II analyzed at Ospedale Pediatrico Bambino Gesù. The CNV analysis shows recurrent gene and chromosomal alterations (e.g., MYC amplification [red circles] and Chr 17q gain (blue arrows)).
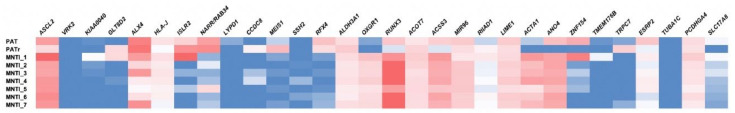
To gain insight into the functional significance of the methylation data, we then analyzed the differentially methylated regions (DMRs) between MNTI/PAT and Group 3 medulloblastoma (Figure 7, Tables S1–S3). Functional enrichment analysis showed few statistically significant pathways (Table S4) (p values < 0.001, none with False Discovery Rate (FDR) < 0.01), supporting the hypothesis of common histogenesis and developmental programs.
Figure 7.
Heatmap depicting the top 30 differentially regulated genes among MNTI/PAT samples vs. MBG3. Color scale: Hypermethylated genes are reported in red, while hypomethylated genes are in blue.
3. Discussion
MNTI is a rare neoplasm of putative neural crest origin, preferentially arising in craniofacial sites. The histogenesis of MNTI remains elusive, with divergent theories (odontogenic, primordial germ cell, and a neural crest origin) being hypothesized over the years, leading to inconsistent—and confusing—nomenclature in the literature [6,19]. Although MNTIs are currently considered neoplasms of neurocristic origin [5,6], only very few studies have attempted to further address their underlying molecular and epigenetic profile [7,8,9]. This has been partly due to their rarity, with only about 500 cases reported until now [19,20,21]. As such, our understanding of their biology and prognosis remains elusive. Although most appear indolent despite their primitive “embryonal” morphology, there are reports of rare metastasizing cases with poor outcomes [6,22,23].
The histologic diagnosis of MNTI is relatively straightforward and rarely represents a diagnostic challenge, provided it arises from the usual anatomic sites and adequate tissue material is available for examination. In our series, most MNTI originated from the head and neck region except in case 7, which presented as a right paratesticular mass. These findings are consistent with other reported cases as most MNTI involve the head and neck [20], with a subset occurring in the paratesticular region [24,25,26] and other unusual locations [19]. Histologically, all MNTI had biphasic morphology characterized by small primitive neuroblast-like cells and larger peripheral epithelioid cells containing melanin granules confirmed by immunohistochemistry. Remarkably, the possibility of metastatic neuroblastoma was excluded in all cases by negative imaging studies and negative expression of Phox2b, which is considered a highly sensitive and specific immunohistochemical marker for tumors of neuroblastic origin, whereas it was reported negative in MNTI [27]. This immunostain can be used as a diagnostic aid in patients presenting with increased urinary levels of VMA, particularly in cases with limited biopsy material containing a predominantly neuroblastic-like morphology and expression of neuronal markers (i.e., synaptophysin, chromogranin A, PGP9.5, CD56).
The MNTI morphology overlaps with other more aggressive entities, including PAT [1,28,29] and medulloblastoma with melanotic differentiation [11,30]. Both lesions reveal a primitive neuroblastic-like component and epithelioid cells containing melanin pigment and were considered in the past as intracranial variants of MNTI [31,32]. As a matter of fact, PATs were initially named based on their similarities with MNTI (historically designated as retinal anlage tumor). However, their highly aggressive biologic behavior (in contrast to MNTI) has led to a critical reappraisal of these tumors, which are now considered variants of medulloblastoma and pineoblastoma, respectively [33]. Interestingly, PAT is characterized by a distinctive morphologic spectrum that includes a combination of neuroectodermal and heterologous ectomesenchymal components without endodermal elements [34]. This histological heterogeneity has raised the question of whether they truly represent a variant of classic pineoblastoma or a separate entity [33]. Interestingly, a recent study revealed different molecular subgroups among pineoblastomas, exhibiting substantial molecular heterogeneity based on DNA-methylation profiling. In particular, two cases diagnosed as PAT were classified by DNA-methylation profiling in the “pineoblastoma, MYC-FOXR2-activated group” [35]. In addition, Uro-Coste et al. reported a posterior fossa tumor (morphologically reminiscent of PAT) composed of undifferentiated monotonous cells, brisk mitotic activity, and foci of chondroid and rhabdomyoblastic differentiation exhibiting molecular features of an embryonal tumor with multilayered rosettes (ETMR)-like with DICER1-mutation in an infant [36].
Medulloblastomas are currently divided into four histologically distinct entities (classic, desmoplastic/nodular, extensive nodularity, and large cell/anaplastic) and four genetically defined subgroups (WNT-activated, SHH- activated TP53-wildtype, SHH-activated TP53-mutant, and non-WNT/non-SHH) [31]. Non-WNT/non-SHH medulloblastomas are consistently separated into group 3 (MB G3) and 4 by clustering large methylation array datasets. MB G3 are mainly known for their aggressive clinical behavior, with the worst and shortest survival among infants [37]. Notably, medulloblastoma with melanotic differentiation is an unusual tumor whose cell of origin is still debated (despite being considered a variant of medulloblastoma). Interestingly, a neural crest origin has been proposed based on the ultrastructure and proteomic features of its pigment resembling oculocutaneous melanin, including premelanosomes (rather than neuromelanin) [38]. Recently, Rajeshwari et al. reported a WNT-activated medulloblastoma with melanotic differentiation and speculated about the potential contribution of neural crest cell progenitors into their pathobiology [39].
This study found a compelling relationship between MNTI, PAT, and MB G3 based on their overlapping methylation profiles. Our results are in line with a recent report by Ebel et al., who described a 4-month-old boy with an MNTI arising from the skull and clustering in the class family of MB G3 by methylation studies [13]. In our series, 5/7 MNTI clustered in the class family MB G3 with an optimal score (>0.9), supporting the findings reported by Ebel et al. [13], as well as our index case (PAT, score 0.99). The remaining two MNTI cases clustered in the class family PLEX, PED B, with suboptimal scores (>0.45). However, when analyzed with the “Medulloblastoma classifier group 3/4 v1.0”, all seven MNTIs classified as high-risk MB G3 tumors, subtype II, with a score >0.45. Furthermore, compared with methylation data from a cohort of 36 medulloblastoma patients [17], all MNTIs and PATs displayed a global methylation profile very close to those of MB G3.
Interestingly, among the top 30 differentially methylated genes shown in Figure 3, 10 are reported to be implicated in development, morphogenesis or neural patterning (VRK2, ALK4, ISLR2, NARR/RAB34, LYPD1, MEIS1, SSH2, RFX4, MIR96, TUBA1C) (Table 2). Of note, three of these (ALK4, MEIS1, and MIR96), together with another six (ASCL2, CCDC8, RUNX3, MIR96, ACTA1, ESRP2), have been associated with WNT/β-catenin signaling as direct target genes or indirect regulators. It is known that WNT/β-catenin signaling has a widespread influence in cellular differentiation, tumor initiation, and progression [40,41]. This suggests there is a closer relationship between WNT/β-catenin signaling and non-WNT medulloblastomas (i.e., group 3 and group 4 medulloblastoma) [42].
Table 2.
Differentially methylated genes and their relationship with MNTI and PAT.
| Gene Name | Gene Function Description | Status MNTI-PAT vs. MBG3 | References |
|---|---|---|---|
| ACOT7 | Lipid metabolism | Hypermethylated | [49] |
| ACSS3 | Lipid metabolism | Hypermethylated | [50] |
| ANO4 | Ca2+ activated Cl- channels | Hypomethylated in PAT Hypermethylated in MNTI | [51] |
| GLT8D2 | GLT8D2 is a glycosyltransferase of apoB100 that regulates apoB100 levels ER | Hypomethylated | [52] |
| HLA-J | Major histocompatibility complex, class I, J (pseudogene) | Hypermethylated | [53] |
| KIAA0040 | HLA-DR11-restricted T-cell epitope encoded by KIAA0040, alcohol dependency | Hypomethylated | [54] |
| LIME1 | Adaptor protein involved in CD4 and CD8 coreceptor signaling | Hypermethylated | [55] |
| OXGR1 | Receptor for alpha-ketoglutarate, expressed in adrenal glands. Frequently hypermethylated in hepatocellular carcinoma | Hypermethylated in PAT, Hypomethylated in MNTI | [56] |
| PCDHGA4 | Neural cadherin-like cell adhesion genes, hypermethylated in neuroblastoma | Hypomethylated | [57,58] |
| RIIAD1 | Regulatory subunit of type II PKA R-subunit domain containing 1 | Hypermethylated | [59] |
| SLC17A8 | GLUT3 (Slc17a8) is expressed in neurons classically defined by their use of another transmitter, such as acetylcholine and serotonin | Hypomethylated | [60] |
| TMEM176B | Innate immune checkpoint | Hypomethylated | [61] |
| TRPC7 | Ca2+ signaling pathway | Hypomethylated | [62] |
| ZNF154 | Candidate Tumor Suppressor ZNF154 suppresses invasion and Metastasis in NPC by inhibiting the EMT via Wnt/β-catenin signaling | hypomethylated in PAT Hypermethylated in MNTI | [63] |
| ACTA1† | α-skeletal actin, WNT target | Hypomethylated in PAT Hypermethylated in MNTI | [64] |
| ALDH3A1† | Target of WNT pathway | Hypermethylated | [65] |
| ASCL2† |
SMYD3 controls a Wnt-responsive epigenetic switch for ASCL2 activation and cancer stem cell maintenance Achaete-scute like 2 (ASCL2) is a target of Wnt signaling and is upregulated in intestinal neoplasia |
Hypermethylated | [64,66,67,68] |
| CCDC8† | Wnt inhibition of CCDC8 phosphorylation or patient-derived mutations | Hypomethylated | [69] |
| ESRP2† | Wnt pathway genes are enriched in genes downregulated by ESRP2 | Hypomethylated | [70] |
| RUNX3† | RUNX3 inhibits glioma survival and invasion via suppression of the β-catenin/TCF-4 signaling pathway | Hypermethylated | [71] |
| ISLR2 * | Essential role of Linx/Islr2 in the development of the forebrain anterior commissure The LRR receptor Islr2 is required for retinal axon routing at the vertebrate optic chiasm |
Hypermethylated in PAT, Hypomethylated in MNTI | [72,73] |
| LYPD1 * | Synaptic signaling | Hypomethylated | [74] |
| NARR/RAB34 * | Genomic characterization of Gli-activator targets in Sonic Hedgehog-mediated neural patterning | Hypermethylated in PAT, Hypomethylated in MNTI | [75] |
| RFX4 * | Zebrafish Rfx4 controls dorsal and ventral midline formation in the neural tube | Hypermethylated in PAT, Hypomethylated in MNTI | [76] |
| SSH2 * | Regulation of actin filaments; neural development and function | Hypomethylated | [77] |
| TUBA1C * | Down-regulation of TUBA1C significantly reduces proliferation and migration in HCC cells Transcriptome analysis of adherens junction pathway-related genes after peripheral nerve injury |
Hypomethylated | [78,79] |
| VRK2 * | VRK2A is an A-type lamin-dependent nuclear envelope kinase that phosphorylates BAF. Associated with schizophrenia and epilepsy | Hypomethylated | [80,81] |
| ALX4 *† |
ALX4, an epigenetically down regulated tumor suppressor, inhibits breast cancer progression by interfering Wnt/β-catenin pathway ALX4 relays sequential FGF signaling to induce lacrimal gland morphogenesis ALX4 and MSX2 play phenotypically similar and additive roles in skull vault differentiation ALX4, a transcriptional activator whose expression is restricted to sites of epithelial-mesenchymal interactions |
Hypermethylated | [82] |
| MEIS1 *† | Cardiac regeneration, stem cells and cancer Meis1 coordinates cerebellar granule cell development by regulating Pax6 transcription, BMP signaling, and Atoh1 degradation. Meis1 is specifically expressed by Sox2(+) stem cells, which give rise to all dental epithelial cell lineages Differential transcriptional regulation of meis1 by Gfi1b and its cofactors LSD1 and CoREST. Potential RA downstream targets play a crucial role in normal development; Combined overexpression of Hoxa9 and Meis1 in hematopoietic stem and progenitor cells (HSPCs) in mice leads to AML and an associated increase in the level of unphosphorylated (“activated”) β-catenin; WNT/β-catenin pathway activation in Myc immortalized cerebellar progenitor cells inhibits neuronal differentiation and generates tumors resembling medulloblastoma (whereas ptf1a, Meis1 and Lhx2 were used as markers of cells in the cerebellar VZ; Meis1 expression was seen in both WNT and Shh groups) |
Hypomethylated | [40] |
| MIR96 *† | Regulates GFI1 MIR-96
is required for normal development of the auditory hindbrain. MIR-96 was identified to indirectly regulate the Wnt/β-catenin pathway by suppressing HMG-box transcription factor protein 1 (HBP-1) |
Hypermethylated | [83] |
* Genes involved in neural function/development. † Genes involved in WNT pathway.
Our series proposes that MNTI, PAT, and MB G3 could be part of a spectrum of neoplasms primarily determined by the influence of neural crest-derived stem cells, an appealing hypothesis deserving further investigation. Our findings support the longstanding notion of a histogenetic relationship between these three entities based on their molecular profiles and highlights a risk for potential diagnostic pitfalls. Nonetheless, the divergent benign behavior of MNTIs in contrast to PATs and MB G3 deserves further investigation. Possibly, additional cytogenetic events and site-related growth factors could play a role in these tumors’ biologic behavior. In fact, several subtypes (high risk vs. low risk) of MB G3 have recently emerged based on methylation profiles and frequency of MYC amplification, among others [43]. Interestingly, most MNTIs in our series (5/7; 71%) showed no CNVs in clear contrast to our PAT and MB G3 in the control cohort (Figure S4). We speculate that the absence of CNV may account, at least in part, for the indolent clinical behavior typically seen in most MNTIs. This discrepancy could also be related to other factors, including differences in oncogene activation, anatomical location, and tumor microenvironment. The divergence in biological behavior and treatment response based upon location has been suggested previously in other tumors [44]. Furthermore, most MNTIs arise from extracranial sites, potentially allowing for earlier identification and complete surgical resection [45] in comparison to PATs and medulloblastomas. Besides, most cancers coexist with other cells and cytokines, collectively known as the tumor microenvironment [46]. Some of these cellular and chemical mediators appear overexpressed in medulloblastomas, including molecules such as IL-8 and TGF-beta, as well as tumor-infiltrating T lymphocytes [47], that may differ from PATs and medulloblastomas. Finally, the role of mutations in epigenetic regulators is a known phenomenon in medulloblastomas [48]. In contrast, these events are still poorly characterized in PATs or MNTIs [8,9]. The contribution of these mediators to the divergent pathobiology between these three entities remains elusive, and further studies are needed to support these hypotheses.
4. Materials and Methods
4.1. Patient Population
Based on the methylation profile of a PAT (index case) showing morphologic overlap with a MNTI (from Sapienza University of Rome), institutional and consultation files from 3 large pediatric institutions (The Children’s Hospital of Philadelphia, Philadelphia, PA, United States; Ospedale Bambino Gesù, Roma, Italy; Università di Padova, Padua, Italy) were queried. Formalin-fixed, paraffin-embedded (FFPE) tumor samples with a morphologic diagnosis of MNTI (age 3 to 48 months) were retrieved following institutional review board approval. The pertinent clinical information was obtained from electronic medical records and available consultation material. A cohort of 36 previously published medulloblastomas diagnosed at Ospedale Bambino Gesù, Rome, Italy [17], was used as control group. This study was reviewed and approved by Bambino Gesù Children’s Hospital Ethical Committee (Protocol n° 1556-OPBG, 26th October 2018, final approval 15th January 2019).
4.2. Histology and Immunohistochemistry
Hematoxylin-eosin-stained sections and available immunohistochemical material were retrieved. The slides were reviewed by at least 3 pediatric pathologists. The mitotic count was determined by identification of the number of mitotic figures (cell divisions) in 10 high-power (400× magnification) fields. Additional immunohistochemical and molecular studies were conducted as needed. Immunohistochemistry for Phox2b (H-20, Santa Cruz Biotechnology, Dallas, TX, USA) was conducted in all cases.
4.3. Methylation Studies
DNA methylation profiling was performed as previously described [17,84], following protocols approved by the institutional review board. Tumor areas with highest tumor cell content (≥ 70%) were selected for analysis. DNA was extracted according to MagPurix FFPE DNA Extraction Kit (Resnova, Rome, Italy) for automatic extraction of genomic DNA. Samples were analyzed using Illumina Infinium Human Methylation EPIC BeadChip (EPIC) arrays (Illumina, San Diego, CA, USA), according to the manufacturer’s instructions, on the Illumina iScan Platform (Illumina, San Diego, CA, USA). In detail, 250 ng DNA was used as input material. Generated methylation data were compared to brain tumor classifier v11b4 [14], sarcoma classifier v12.2, and medulloblastoma classifier group 3/4 classifier v1.0, developed by Heidelberg University and DKFZ (https://www.molecularneuropatholog.org/mnp/classifier/all (accessed on 5 February 2021)) to assign a subgroup score for the tumors in the recognized methylation classes. EPIC BeadChip data were analyzed as previously reported [17,85] by means of R (V.3.6.1), using ChAMP package (V.2.16.1) for quality checks and filters to calculate methylation levels and functionally annotate probes at the gene level. Multidimensional scaling (MDS) on the cohort samples was performed using the cmdscale function with the Euclidean distance. A heatmap depicting normalized beta values was created by means of the pheatmap function, using the complete linkage method and Euclidean distance to cluster samples and probes. Low-quality CpG islands among the 48 islands previously identified [18] were removed from this clustering analysis. Differentially methylated regions (DMR) were identified by means of the Bumphunter tool (as implemented in ChAMP pipeline), setting the adjusted p-value threshold at 0.001 [86]. Functional enrichment analysis of DMRs was carried out by means of the WebGestalt tool [87], using overrepresentation analysis (ORA) on the Reactome and wikipathway cancer databases.
5. Conclusions
In summary, our study provides evidence that MNTI consistently shares a homogenous methylation profile with MB G3, and possibly with PAT. The morphologic and molecular overlap of these entities may represent a diagnostic pitfall in the routine diagnostic workup of biopsy material. Thus, further correlation with imaging and clinical features may prevent a potential misdiagnosis. A comprehensive methylation profiling supports our results and, for the first time, confirms the notion of a histogenetic relationship between these three tumors. However, our study also has several limitations, including a cohort with a relatively small number of cases (primarily due to these tumors’ rare nature) and its retrospective format (facilitating selection bias and incomplete or lost follow-up information). Besides, we did not conduct gene expression analysis or identification of single nucleotide variants in epigenetic modifiers in our cohort. The possible role of a multipotent progenitor cell (derived from early migratory cranial neural crest cells) in the histogenesis of these tumors and the influence of the anatomical site, tumor microenvironment, and additional cytogenetic events in their divergent biologic behavior deserve further investigation.
Acknowledgments
The authors would like to thank Claudia Nardini for her valuable technical support.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/13/4/706/s1, Figure S1: Box-and-whisker plots depicting the maximum raw classification scores (highlighted in red) of the indicated tumor samples (red dot) in the first methylation class of the brain tumor classifier assigned to each sample. Grey dots represent the reference cases in the methylation class, Figure S2: Cluster dendrogram with bootstrap analysis (1000 iterations) values on the 39 high-quality CpG islands in the Hovestadt set (Hovestadt et al., 2013) for the samples in the cohort. The bootstrap probability (BP) values are depicted in green, the approximately unbiased (AU) probability values (p-values) are reported in red. p-value of a cluster is a value between 0 and 1, which indicates how strongly the cluster is supported by data. Values are expressed in percentage. Edge numbers are in grey. Clusters with AU larger than 95% are highlighted by red boxes. Progonoma (MNTI/PA); Medulloblastoma (MB), Figure S3: Copy number variation (CNV) profile analysis of the indicated PAT/MNTI. Depiction of structural rearrangements involving autosomes and X/Y chromosome. Gains/amplifications represent positive (green) and losses represent negative (red) deviations from the baseline. Twenty-nine tumor relevant genomic regions are highlighted, Table S1: Annotated probes located in differentially methylated regions (DMRs, p-value < 0.001) between MNTI/PAT and Group 3, Table S2: Differentially methylated regions (DMRs, p-value < 0.001) between MNTI/PAT and Group 3. Top 30 DMRs used for functional enrichment analyses are highlighted in blue, Table S3: Methylation level differences in probes from significant DMRs in MNTI/PAT (blue) vs. MBG3 (black), Table S4: Webgestalt analysis, from total DMR genes Supplementary information (zip file)
Author Contributions
Conceptualization: R.A., E.M. Methodology: R.A., E.M., L.F.S., A.C., L.P., S.R. Molecular interpretation: E.M., L.F.S., M.T. Patients management and samples source: A.M., V.D., F.G. (Felice Giangaspero), G.C., F.G. (Francesca Gianno), L.F.S. Data curation: O.L.-N. Writing—original draft: O.L.-N., R.A. Writing—review & editing: O.L.-N., R.A., E.M., I.J., F.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Italian Ministry of Health Ricerca Corrente 5 × 1000 funds 2020 to EM (202005_ONCO_MIELE), Ministry of Health, Ricerca Corrente 5 × 1000 funds 2020 to R.A (202005_ONCO_ALAGGIO), the Department of Pathology Research Funds, University of Pittsburgh Medical Center, and The Department of Pathology, The Children’s Hospital of Philadelphia.
Institutional Review Board Statement
This study was approved by the Ethics Committee of Bambino Gesù Children’s Hospital (protocol code 1556_OPBG_2018; date of approval 15 January 2019).
Informed Consent Statement
A written informed consent was obtained from the patients’ legal guardians.
Data Availability Statement
The data presented in this study are available in the provided Supplementary Materials.
Conflicts of Interest
The authors declare that they have no conflict of interest to disclose for the current study.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dooling E.C., Chi J.G., Gilles F.H. Melanotic neuroectodermal tumor of infancy: Its histological similarities to fetal pineal gland. Cancer. 1977;39:1535–1541. doi: 10.1002/1097-0142(197704)39:4<1535::AID-CNCR2820390426>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 2.Halpert B., Patzer R. Maxillary tumor of retinal anlage. Oral Surg. Oral Med. Oral Pathol. 1948;1:568–572. doi: 10.1016/0030-4220(48)90248-5. [DOI] [PubMed] [Google Scholar]
- 3.Stowens D. A pigmented tumour of infancy: The melanotic progonoma. J. Pathol. Bacteriol. 1957;73:43–51. doi: 10.1002/path.1700730107. [DOI] [Google Scholar]
- 4.Borello E.D., Gorlin R.J. Melanotic neuroectodermal tumor of infancy—A neoplasm of neural crese origin. Report of a case associated with high urinary excretion of vanilmandelic acid. Cancer. 1966;19:196–206. doi: 10.1002/1097-0142(196602)19:2<196::AID-CNCR2820190210>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 5.Nitta T., Endo T., Tsunoda A., Kadota Y., Matsumoto T., Sato K. Melanotic neuroectodermal tumor of infancy: A molecular approach to diagnosis. Case report. J. Neurosurg. 1995;83:145–148. doi: 10.3171/jns.1995.83.1.0145. [DOI] [PubMed] [Google Scholar]
- 6.Dehner L.P., Sibley R.K., Sauk J.J., Jr., Vickers R.A., Nesbit M.E., Leonard A.S., Waite D.E., Neeley J.E., Ophoven J. Malignant melanotic neuroectodermal tumor of infancy. A clinical, pathologic, ultrastructural and tissue culture study. Cancer. 1979;43:1389–1410. doi: 10.1002/1097-0142(197904)43:4<1389::AID-CNCR2820430429>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 7.Khoddami M., Squire J., Zielenska M., Thorner P. Melanotic neuroectodermal tumor of infancy: A molecular genetic study. Pediatr. Dev. Pathol. 1998;1:295–299. doi: 10.1007/s100249900042. [DOI] [PubMed] [Google Scholar]
- 8.Barnes D.J., Hookway E., Athanasou N., Kashima T., Oppermann U., Hughes S., Swan D., Lueerssen D., Anson J., Hassan A.B. A germline mutation of CDKN2A and a novel RPLP1-C19MC fusion detected in a rare melanotic neuroectodermal tumor of infancy: A case report. BMC Cancer. 2016;16:629. doi: 10.1186/s12885-016-2669-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomes C.C., Diniz M.G., de Menezes G.H., Castro W.H., Gomez R.S. BRAFV600E Mutation in Melanotic Neuroectodermal Tumor of Infancy: Toward Personalized Medicine? Pediatrics. 2015;136:e267-9. doi: 10.1542/peds.2014-3331. [DOI] [PubMed] [Google Scholar]
- 10.Ramdasi R., Kothari K., Goel N., Mahore A. An unusual variant of anlage tumor of pineal region in an infant. Brain Tumor Res. Treat. 2015;3:52–55. doi: 10.14791/btrt.2015.3.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borcek A.O., Durdag E., Emmez H., Kurt G., Baykaner M.K. Myogenic and melanotic differentiated medulloblastoma: Case report. Turk. Neurosurg. 2011;21:438–442. doi: 10.5137/1019-5149.JTN.2964-10.2. [DOI] [PubMed] [Google Scholar]
- 12.Li W.Y., Ma L., Liu J., Yang H., Mao Q. Melanotic medulloblastoma in children. Childs Nerv. Syst. 2011;27:517–521. doi: 10.1007/s00381-011-1389-x. [DOI] [PubMed] [Google Scholar]
- 13.Ebel F., Thieringer F.M., Kunz C., Klein-Franke A., Scheinemann K., Guzman R., Soleman J. Melanotic neuroectodermal tumor of infancy to the skull: Case-based review. Childs Nerv. Syst. 2020;36:679–688. doi: 10.1007/s00381-020-04509-6. [DOI] [PubMed] [Google Scholar]
- 14.Capper D., Jones D.T.W., Sill M., Hovestadt V., Schrimpf D., Sturm D., Koelsche C., Sahm F., Chavez L., Reuss D.E., et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555:469–474. doi: 10.1038/nature26000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Capper D., Stichel D., Sahm F., Jones D.T.W., Schrimpf D., Sill M., Schmid S., Hovestadt V., Reuss D.E., Koelsche C., et al. Practical implementation of DNA methylation and copy-number-based CNS tumor diagnostics: The Heidelberg experience. Acta Neuropathol. 2018;136:181–210. doi: 10.1007/s00401-018-1879-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma T., Schwalbe E.C., Williamson D., Sill M., Hovestadt V., Mynarek M., Rutkowski S., Robinson G.W., Gajjar A., Cavalli F., et al. Second-generation molecular subgrouping of medulloblastoma: An international meta-analysis of Group 3 and Group 4 subtypes. Acta Neuropathol. 2019;138:309–326. doi: 10.1007/s00401-019-02020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ballabio C., Anderle M., Gianesello M., Lago C., Miele E., Cardano M., Aiello G., Piazza S., Caron D., Gianno F., et al. Modeling medulloblastoma in vivo and with human cerebellar organoids. Nat. Commun. 2020;11:583. doi: 10.1038/s41467-019-13989-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hovestadt V., Remke M., Kool M., Pietsch T., Northcott P.A., Fischer R., Cavalli F.M., Ramaswamy V., Zapatka M., Reifenberger G., et al. Robust molecular subgrouping and copy-number profiling of medulloblastoma from small amounts of archival tumour material using high-density DNA methylation arrays. Acta Neuropathol. 2013;125:913–916. doi: 10.1007/s00401-013-1126-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soles B.S., Wilson A., Lucas D.R., Heider A. Melanotic Neuroectodermal Tumor of Infancy. Arch. Pathol. Lab. Med. 2018;142:1358–1363. doi: 10.5858/arpa.2018-0241-RA. [DOI] [PubMed] [Google Scholar]
- 20.Rachidi S., Sood A.J., Patel K.G., Nguyen S.A., Hamilton H., Neville B.W., Day T.A. Melanotic Neuroectodermal Tumor of Infancy: A Systematic Review. J. Oral Maxillofac. Surg. 2015;73:1946–1956. doi: 10.1016/j.joms.2015.03.061. [DOI] [PubMed] [Google Scholar]
- 21.Kruse-Losler B., Gaertner C., Burger H., Seper L., Joos U., Kleinheinz J. Melanotic neuroectodermal tumor of infancy: Systematic review of the literature and presentation of a case. Oral. Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2006;102:204–216. doi: 10.1016/j.tripleo.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 22.Block J.C., Waite D.E., Dehner L.P., Leonard A.S., Ogle R.G., Gatto D.J. Pigmented neuroectodermal tumor of infancy. An example of rarely expressed malignant behavior. Oral Surg. Oral Med. Oral Pathol. 1980;49:279–285. doi: 10.1016/0030-4220(80)90133-4. [DOI] [PubMed] [Google Scholar]
- 23.Batsakis J.G. Pathology consultation. Melanotic neuroectodermal tumor of infancy. Ann. Otol. Rhinol. Laryngol. 1987;96:128–129. doi: 10.1177/000348948709600132. [DOI] [PubMed] [Google Scholar]
- 24.Denadai E.R., Zerati Filho M., Verona C.B., Martucci R.C., Zerati S., Suzigan S. Tumor of the testicle: A case of melanotic neuroectodermal tumor of infancy. J. Urol. 1986;136:117–118. doi: 10.1016/S0022-5347(17)44752-5. [DOI] [PubMed] [Google Scholar]
- 25.Ulbright T.M., Young R.H. Testicular and paratesticular tumors and tumor-like lesions in the first 2 decades. Semin. Diagn. Pathol. 2014;31:323–381. doi: 10.1053/j.semdp.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 26.Al-Obaidy K.I., Alruwaii F.I., Ulbright T.M., Idrees M.T. Appendageal tumors and tumor-like lesions of the testis and paratestis: A 32-year experience at a single institution. Hum. Pathol. 2020;103:25–33. doi: 10.1016/j.humpath.2020.06.006. [DOI] [PubMed] [Google Scholar]
- 27.Warren M., Matsuno R., Tran H., Shimada H. Utility of Phox2b immunohistochemical stain in neural crest tumours and non-neural crest tumours in paediatric patients. Histopathology. 2018;72:685–696. doi: 10.1111/his.13412. [DOI] [PubMed] [Google Scholar]
- 28.Raisanen J., Vogel H., Horoupian D.S. Primitive pineal tumor with retinoblastomatous and retinal/ciliary epithelial differentiation: An immunohistochemical study. J. Neurooncol. 1990;9:165–170. doi: 10.1007/BF02427838. [DOI] [PubMed] [Google Scholar]
- 29.Schmidbauer M., Budka H., Pilz P. Neuroepithelial and ectomesenchymal differentiation in a primitive pineal tumor (“pineal anlage tumor”) Clin. Neuropathol. 1989;8:7–10. [PubMed] [Google Scholar]
- 30.Best P.V. A medulloblastoma-like tumour with melanin formation. J. Pathol. 1973;110:109–111. doi: 10.1002/path.1711100114. [DOI] [PubMed] [Google Scholar]
- 31.Parízek J., Nĕmecek S., Cernoch Z., Heger L., Nozicka Z., Spacek J. Melanotic neuroectodermal neurocranial tumor of infancy of extra-intra-and subdural right temporal location: CT examination, surgical treatment, literature review. Neuropediatrics. 1986;17:115–123. doi: 10.1055/s-2008-1052511. [DOI] [PubMed] [Google Scholar]
- 32.Yu J.S., Moore M.R., Kupsky W.J., Scott R.M. Intracranial melanotic neuroectodermal tumor of infancy: Two case reports. Surg. Neurol. 1992;37:123–129. doi: 10.1016/0090-3019(92)90188-S. [DOI] [PubMed] [Google Scholar]
- 33.Louis D.N., Cancer IAfRo. Wiestler O.D., Ohgaki H. WHO Classification of Tumours of the Central Nervous System. International Agency for Research on Cancer; Lyon, France: 2016. [Google Scholar]
- 34.Ahuja A., Sharma M.C., Suri V., Sarkar C., Sharma B.S., Garg A. Pineal anlage tumour—A rare entity with divergent histology. J. Clin. Neurosci. 2011;18:811–813. doi: 10.1016/j.jocn.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 35.Li B.K., Vasiljevic A., Dufour C., Yao F., Ho B.L.B., Lu M., Hwang E.I., Gururangan S., Hansford J.R., Fouladi M., et al. Pineoblastoma segregates into molecular sub-groups with distinct clinico-pathologic features: A Rare Brain Tumor Consortium registry study. Acta Neuropathol. 2020;139:223–241. doi: 10.1007/s00401-019-02111-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uro-Coste E., Masliah-Planchon J., Siegfried A., Blanluet M., Lambo S., Kool M., Roujeau T., Boetto S., Palenzuela G., Bertozzi A.I., et al. ETMR-like infantile cerebellar embryonal tumors in the extended morphologic spectrum of DICER1-related tumors. Acta Neuropathol. 2019;137:175–177. doi: 10.1007/s00401-018-1935-7. [DOI] [PubMed] [Google Scholar]
- 37.Northcott P.A., Robinson G.W., Kratz C.P., Mabbott D.J., Pomeroy S.L., Clifford S.C., Rutkowski S., Ellison D.W., Malkin D., Taylor M.D., et al. Medulloblastoma. Nat. Rev. Dis. Primers. 2019;5:11. doi: 10.1038/s41572-019-0063-6. [DOI] [PubMed] [Google Scholar]
- 38.Dolman C.L. Melanotic medulloblastoma. A case report with immunohistochemical and ultrastructural examination. Acta Neuropathol. 1988;76:528–531. doi: 10.1007/BF00686394. [DOI] [PubMed] [Google Scholar]
- 39.Rajeshwari M., Kakkar A., Nalwa A., Suri V., Sarkar C., Satyarthee G.D., Garg A., Sharma M.C. WNT-activated medulloblastoma with melanotic and myogenic differentiation: Report of a rare case. Neuropathology. 2016;36:372–375. doi: 10.1111/neup.12281. [DOI] [PubMed] [Google Scholar]
- 40.Rogers H.A., Sousa S., Salto C., Arenas E., Coyle B., Grundy R.G. WNT/β-catenin pathway activation in Myc immortalised cerebellar progenitor cells inhibits neuronal differentiation and generates tumours resembling medulloblastoma. Br. J. Cancer. 2012;107:1144–1152. doi: 10.1038/bjc.2012.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morris S.-A.L., Huang S. Crosstalk of the Wnt/β-catenin pathway with other pathways in cancer cells. Genes Dis. 2016;3:41–47. doi: 10.1016/j.gendis.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manoranjan B., Venugopal C., Bakhshinyan D., Adile A.A., Richards L., Kameda-Smith M.M., Whitley O., Dvorkin-Gheva A., Subapanditha M., Savage N., et al. Wnt activation as a therapeutic strategy in medulloblastoma. Nat. Commun. 2020;11:4323. doi: 10.1038/s41467-020-17953-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Juraschka K., Taylor M.D. Medulloblastoma in the age of molecular subgroups: A review. J. Neurosurg. Pediatr. 2019;24:353–363. doi: 10.3171/2019.5.PEDS18381. [DOI] [PubMed] [Google Scholar]
- 44.Pio L., Milanaccio C., Mascelli S., Raso A., Nozza P., Sementa A.R., Cama A., Buffa P., Avanzini S., Vannati M., et al. Congenital multifocal rhabdoid tumor: A case with peculiar biological behavior and different response to treatment according to location (central nervous system and kidney) Cancer Genet. 2014;207:441–444. doi: 10.1016/j.cancergen.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 45.Ren Q., Chen H., Wang Y., Xu J. Melanotic Neuroectodermal Tumor of Infancy Arising in the Skull and Brain: A Systematic Review. World Neurosurg. 2019;130:170–178. doi: 10.1016/j.wneu.2019.07.017. [DOI] [PubMed] [Google Scholar]
- 46.Quail D.F., Joyce J.A. The Microenvironmental Landscape of Brain Tumors. Cancer Cell. 2017;31:326–341. doi: 10.1016/j.ccell.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Byrd T., Grossman R.G., Ahmed N. Medulloblastoma-biology and microenvironment: A review. Pediatr. Hematol. Oncol. 2012;29:495–506. doi: 10.3109/08880018.2012.698372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roussel M.F., Stripay J.L. Epigenetic Drivers in Pediatric Medulloblastoma. Cerebellum. 2018;17:28–36. doi: 10.1007/s12311-017-0899-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamada J., Kurata A., Hirata M., Taniguchi T., Takama H., Furihata T., Shiratori K., Iida N., Takagi-Sakuma M., Watanabe T., et al. Purification, molecular cloning, and genomic organization of human brain long-chain acyl-CoA hydrolase. J. Biochem. 1999;126:1013–1019. doi: 10.1093/oxfordjournals.jbchem.a022544. [DOI] [PubMed] [Google Scholar]
- 50.Yoshimura Y., Araki A., Maruta H., Takahashi Y., Yamashita H. Molecular cloning of rat acss3 and characterization of mammalian propionyl-CoA synthetase in the liver mitochondrial matrix. J. Biochem. 2017;161:279–289. doi: 10.1093/jb/mvw067. [DOI] [PubMed] [Google Scholar]
- 51.Tian Y., Schreiber R., Kunzelmann K. Anoctamins are a family of Ca2+-activated Cl- channels. J. Cell Sci. 2012;125:4991–4998. doi: 10.1242/jcs.109553. [DOI] [PubMed] [Google Scholar]
- 52.Wei H.S., Wei H.L., Zhao F., Zhong L.P., Zhan Y.T. Glycosyltransferase GLT8D2 positively regulates ApoB100 protein expression in hepatocytes. Int. J. Mol. Sci. 2013;14:21435–21446. doi: 10.3390/ijms141121435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Messer G., Zemmour J., Orr H.T., Parham P., Weiss E.H., Girdlestone J. HLA-J, a second inactivated class I HLA gene related to HLA-G and HLA-A. Implications for the evolution of the HLA-A-related genes. J. Immunol. 1992;148:4043–4053. [PubMed] [Google Scholar]
- 54.Hill S.Y., Jones B.L., Zezza N., Stiffler S. Family-based association analysis of alcohol dependence implicates KIAA0040 on Chromosome 1q in multiplex alcohol dependence families. Open J. Genet. 2013;3:243–252. doi: 10.4236/ojgen.2013.34027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brdicková N., Brdicka T., Angelisová P., Horváth O., Spicka J., Hilgert I., Paces J., Simeoni L., Kliche S., Merten C., et al. LIME: A new membrane Raft-associated adaptor protein involved in CD4 and CD8 coreceptor signaling. J. Exp. Med. 2003;198:1453–1462. doi: 10.1084/jem.20031484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu C.Y., Hsieh S.Y., Lu Y.J., Wu C.S., Chen L.C., Lo S.J., Wu C.T., Chou M.Y., Huang T.H., Chang Y.S. Aberrant DNA methylation profile and frequent methylation of KLK10 and OXGR1 genes in hepatocellular carcinoma. Genes Chromosomes Cancer. 2009;48:1057–1068. doi: 10.1002/gcc.20708. [DOI] [PubMed] [Google Scholar]
- 57.Yagi T., Takeichi M. Cadherin superfamily genes: Functions, genomic organization, and neurologic diversity. Genes Dev. 2000;14:1169–1180. [PubMed] [Google Scholar]
- 58.Olsson M., Beck S., Kogner P., Martinsson T., Carén H. Genome-wide methylation profiling identifies novel methylated genes in neuroblastoma tumors. Epigenetics. 2016;11:74–84. doi: 10.1080/15592294.2016.1138195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Erlbruch A., Hung C.W., Seidler J., Borrmann K., Gesellchen F., König N., Kübler D., Herberg F.W., Lehmann W.D., Bossemeyer D. Uncoupling of bait-protein expression from the prey protein environment adds versatility for cell and tissue interaction proteomics and reveals a complex of CARP-1 and the PKA Cbeta1 subunit. Proteomics. 2010;10:2890–2900. doi: 10.1002/pmic.200900593. [DOI] [PubMed] [Google Scholar]
- 60.Seal R.P., Edwards R.H. The diverse roles of vesicular glutamate transporter 3. Handb. Exp. Pharmacol. 2006:137–150. doi: 10.1007/3-540-29784-7_7. [DOI] [PubMed] [Google Scholar]
- 61.Segovia M., Russo S., Jeldres M., Mahmoud Y.D., Perez V., Duhalde M., Charnet P., Rousset M., Victoria S., Veigas F., et al. Targeting TMEM176B Enhances Antitumor Immunity and Augments the Efficacy of Immune Checkpoint Blockers by Unleashing Inflammasome Activation. Cancer Cell. 2019;35:767–781.e6. doi: 10.1016/j.ccell.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Numaga T., Wakamori M., Mori Y. TRPC7. Handb. Exp. Pharmacol. 2007:143–151. doi: 10.1007/978-3-540-34891-7_8. [DOI] [PubMed] [Google Scholar]
- 63.Hu Y., Qi M.F., Xu Q.L., Kong X.Y., Cai R., Chen Q.Q., Tang H.Y., Jiang W. Candidate tumor suppressor ZNF154 suppresses invasion and metastasis in NPC by inhibiting the EMT via Wnt/β-catenin signalling. Oncotarget. 2017;8:85749–85758. doi: 10.18632/oncotarget.20479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hirata A., Utikal J., Yamashita S., Aoki H., Watanabe A., Yamamoto T., Okano H., Bardeesy N., Kunisada T., Ushijima T., et al. Dose-dependent roles for canonical Wnt signalling in de novo crypt formation and cell cycle properties of the colonic epithelium. Development. 2013;140:66–75. doi: 10.1242/dev.084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Suwala A.K., Koch K., Rios D.H., Aretz P., Uhlmann C., Ogorek I., Felsberg J., Reifenberger G., Köhrer K., Deenen R., et al. Inhibition of Wnt/beta-catenin signaling downregulates expression of aldehyde dehydrogenase isoform 3A1 (ALDH3A1) to reduce resistance against temozolomide in glioblastoma in vitro. Oncotarget. 2018;9:22703–22716. doi: 10.18632/oncotarget.25210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guillemot F., Nagy A., Auerbach A., Rossant J., Joyner A.L. Essential role of Mash-2 in extraembryonic development. Nature. 1994;371:333–336. doi: 10.1038/371333a0. [DOI] [PubMed] [Google Scholar]
- 67.Liu X., Chen X., Zhong B., Wang A., Wang X., Chu F., Nurieva R.I., Yan X., Chen P., van der Flier L.G., et al. Transcription factor achaete-scute homologue 2 initiates follicular T-helper-cell development. Nature. 2014;507:513–518. doi: 10.1038/nature12910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van der Flier L.G., Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu. Rev. Physiol. 2009;71:241–260. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- 69.Wang P., Yan F., Li Z., Yu Y., Parnell S.E., Xiong Y. Impaired plasma membrane localization of ubiquitin ligase complex underlies 3-M syndrome development. J. Clin. Investig. 2019;129:4393–4407. doi: 10.1172/JCI129107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mizutani A., Koinuma D., Seimiya H., Miyazono K. The Arkadia-ESRP2 axis suppresses tumor progression: Analyses in clear-cell renal cell carcinoma. Oncogene. 2016;35:3514–3523. doi: 10.1038/onc.2015.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun J., Li B., Jia Z., Zhang A., Wang G., Chen Z., Shang Z., Zhang C., Cui J., Yang W. RUNX3 inhibits glioma survival and invasion via suppression of the β-catenin/TCF-4 signaling pathway. J. Neurooncol. 2018;140:15–26. doi: 10.1007/s11060-018-2927-0. [DOI] [PubMed] [Google Scholar]
- 72.Abudureyimu S., Asai N., Enomoto A., Weng L., Kobayashi H., Wang X., Chen C., Mii S., Takahashi M. Essential Role of Linx/Islr2 in the Development of the Forebrain Anterior Commissure. Sci. Rep. 2018;8:7292. doi: 10.1038/s41598-018-24064-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Panza P., Sitko A.A., Maischein H.M., Koch I., Flötenmeyer M., Wright G.J., Mandai K., Mason C.A., Söllner C. The LRR receptor Islr2 is required for retinal axon routing at the vertebrate optic chiasm. Neural Dev. 2015;10:23. doi: 10.1186/s13064-015-0050-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Petitpré C., Wu H., Sharma A., Tokarska A., Fontanet P., Wang Y., Helmbacher F., Yackle K., Silberberg G., Hadjab S., et al. Neuronal heterogeneity and stereotyped connectivity in the auditory afferent system. Nat. Commun. 2018;9:3691. doi: 10.1038/s41467-018-06033-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vokes S.A., Ji H., McCuine S., Tenzen T., Giles S., Zhong S., Longabaugh W.J., Davidson E.H., Wong W.H., McMahon A.P. Genomic characterization of Gli-activator targets in sonic hedgehog-mediated neural patterning. Development. 2007;134:1977–1989. doi: 10.1242/dev.001966. [DOI] [PubMed] [Google Scholar]
- 76.Sedykh I., Keller A.N., Yoon B., Roberson L., Moskvin O.V., Grinblat Y. Zebrafish Rfx4 controls dorsal and ventral midline formation in the neural tube. Dev. Dyn. 2018;247:650–659. doi: 10.1002/dvdy.24613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ohashi K. Roles of cofilin in development and its mechanisms of regulation. Dev. Growth Differ. 2015;57:275–290. doi: 10.1111/dgd.12213. [DOI] [PubMed] [Google Scholar]
- 78.Wang J., Chen W., Wei W., Lou J. Oncogene TUBA1C promotes migration and proliferation in hepatocellular carcinoma and predicts a poor prognosis. Oncotarget. 2017;8:96215–96224. doi: 10.18632/oncotarget.21894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yi S., Wang X.H., Xing L.Y. Transcriptome analysis of adherens junction pathway-related genes after peripheral nerve injury. Neural Regen Res. 2018;13:1804–1810. doi: 10.4103/1673-5374.237127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Birendra K., May D.G., Benson B.V., Kim D.I., Shivega W.G., Ali M.H., Faustino R.S., Campos A.R., Roux K.J. VRK2A is an A-type lamin-dependent nuclear envelope kinase that phosphorylates BAF. Mol. Biol. Cell. 2017;28:2241–2250. doi: 10.1091/mbc.e17-03-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li M., Yue W. VRK2, a Candidate Gene for Psychiatric and Neurological Disorders. Mol. Neuropsychiatry. 2018;4:119–133. doi: 10.1159/000493941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang J., Han F., Liu W., Chen H., Hao X., Jiang X., Yin L., Huang Y., Cao J., Zhang H., et al. ALX4, an epigenetically down regulated tumor suppressor, inhibits breast cancer progression by interfering Wnt/β-catenin pathway. J. Exp. Clin. Cancer Res. 2017;36:170. doi: 10.1186/s13046-017-0643-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Song J.L., Nigam P., Tektas S.S., Selva E. microRNA regulation of Wnt signaling pathways in development and disease. Cell Signal. 2015;27:1380–1391. doi: 10.1016/j.cellsig.2015.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Petruzzellis G., Alessi I., Colafati G.S., Diomedi-Camassei F., Ciolfi A., Pedace L., Cacchione A., Carai A., Tartaglia M., Mastronuzzi A., et al. Role of DNA Methylation Profile in Diagnosing Astroblastoma: A Case Report and Literature Review. Front. Genet. 2019;10:391. doi: 10.3389/fgene.2019.00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miele E., De Vito R., Ciolfi A., Pedace L., Russo I., De Pasquale M.D., Di Giannatale A., Crocoli A., Angelis B., Tartaglia M., et al. DNA Methylation Profiling for Diagnosing Undifferentiated Sarcoma with Capicua Transcriptional Receptor (CIC) Alterations. Int. J. Mol. Sci. 2020;21:1818. doi: 10.3390/ijms21051818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jaffe A.E., Murakami P., Lee H., Leek J.T., Fallin M.D., Feinberg A.P., Irizarry R.A. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int. J. Epidemiol. 2012;41:200–209. doi: 10.1093/ije/dyr238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liao Y., Wang J., Jaehnig E.J., Shi Z., Zhang B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47:W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available in the provided Supplementary Materials.