Abstract
Analysis of pooled genomic short read sequence data revealed the presence of nudivirus-derived sequences from U.S. populations of both southern corn rootworm (SCR, Diabrotica undecimpunctata howardi Barber) and western corn rootworm (WCR, Diabrotica virgifera virgifera LeConte). A near complete nudivirus genome sequence was assembled from sequence data for an SCR population with relatively high viral titers. A total of 147,179 bp was assembled from five contigs that collectively encode 109 putative open reading frames (ORFs) including 20 nudivirus core genes. In contrast, genome sequence recovery was incomplete for a second nudivirus from WCR, although sequences derived from this virus were present in three geographically dispersed populations. Only 48,989 bp were assembled with 48 putative ORFs including 13 core genes, representing about 20% of a typical nudivirus genome. Phylogenetic analysis indicated that both corn rootworm nudiviruses grouped with the third known nudivirus of beetles, Oryctes rhinoceros nudivirus in the genus Alphanudivirus. On the basis of phylogenetic and additional analyses, we propose further taxonomic separation of nudiviruses within Alphanudivirus and Betanudivirus into two subfamilies and five genera. Identification of nudivirus-derived sequences from two species of corn rootworm highlights the diversity of viruses associated with these agricultural insect pests.
Keywords: Nudiviridae, Alphanudivirus, beetle, corn rootworm, Diabrotica spp.
1. Introduction
Corn rootworms are major insect pests of corn in the Americas and Europe [1]. The four North American corn rootworm species and subspecies are the western corn rootworm (WCR, Diabrotica virgifera virgifera LeConte), Mexican corn rootworm (MCR, Diabrotica virgifera zeae Krysan & Smith), and northern corn rootworm (NCR, Diabrotica barberi Smith & Lawrence) in the virgifera species group of Diabrotica beetles; and the southern corn rootworm (SCR, Diabrotica undecimpunctata howardi Barber) in the fucata species group [2]. Of these, WCR, MCR, and NCR can cause devastating damage to corn, and are serious threats to agricultural productivity in the U.S. Together, these pests cause losses estimated at nearly $2 billion annually in revenue in the U.S. (75% in yield loss, 25% in treatment costs) [3]. Traditionally, WCR, MCR, and NCR have been managed through crop rotation, extensive application of insecticides to the soil to kill the underground larvae, and, in some regions, late-season aerial applications of insecticides targeting egg-laying adults. However, biotypes have evolved that are adapted to crop rotation, and in areas of high insecticide use, insecticide resistance has become a problem [4]. More recently, transgenic corn varieties expressing WCR-active pesticidal proteins derived from the bacterium Bacillus thuringiensis (Bt) have been widely adopted, but resistant populations have evolved in Iowa and Nebraska within the U.S. [5,6,7]. New transgenic lines have been developed that use a combination of gut-active Bt-derived proteins and silencing RNAs to silence essential rootworm genes through RNA interference (RNAi) [8,9,10]. While promising, there are indications that WCR will also readily develop resistance and cross-resistance to RNAi corn varieties, and care must be taken to preserve their efficacy after deployment [11,12]. Given the significant losses resulting from rootworm damage, and limitations associated with current management techniques, new tools to suppress rootworm populations are urgently needed. Pathogenic viruses could play a role in rootworm management as demonstrated for another coleopteran pest, Asiatic rhinoceros beetle, Oryctes rhinoceros (L.) [13,14].
Little is known about the DNA viruses that infect Diabrotica spp. Virus particles have been observed in the spermatheca of WCR [15], and baculovirus-like and filamentous virus-like particles have been described from hemocytes and from the midgut, respectively, of SCR [16]. As no occlusion bodies typical of nucleopolyhedroviruses or granuloviruses were observed, the baculovirus-like particles observed in SCR were likely derived from a nudivirus. No pathologic symptoms were observed in beetles infected by these viruses [17,18].
Nudiviridae, formerly known as “non-occluded” baculoviruses, is a family of large, rod-shaped viruses with genomes composed of circular, double-stranded DNA (dsDNA). Nudivirus genomes range from 97 to 232 kbp and encode 93–154 putative proteins [19]. Nudiviruses are classified into two genera, Alphanudivirus and Betanudivirus. Members of Alphanudivirus have been isolated from insects, while the betanudiviruses infect both insects and marine arthropods (Amphipods and Decapods). Nudivirus-like viral particles have been widely observed, including from 10 insect orders (Coleoptera, Lepidoptera, Orthoptera, Diptera, Siphonaptera, Hymenoptera, Thysanura, Trichoptera, Neuroptera, and Hemiptera), and three additional arthropod orders (Acarina, Araneina, and Crustacea) [20]. A total of 13 nudiviruses have been sequenced (Table 1). The Oryctes rhinoceros nudivirus (OrNV), isolated from O. rhinoceros, is the only fully sequenced and annotated nudivirus from a coleopteran host. Short sequences derived from a second putative beetle-infecting nudivirus were isolated from the Japanese rhinoceros beetle, Allomyrina dichotoma (Table 1).
Table 1.
Characteristics of exogenous nudivirus genome sequences (current as of December 2020). NA, not applicable.
| Virus | Abbr. | Length (bp) | Accession | No. ORFs | %G + C | Host | Isolates | Ref |
|---|---|---|---|---|---|---|---|---|
| Alphanudivirus | ||||||||
| Gryllus bimaculatus nudivirus | GrBNV | 96,944 | NC_009240.1 | 98 | 27.99 | Gryllus bimaculatus | Starnberg, Germany | [28,29] |
| Kallithea virus | KNV | 152,388 | NC_033829.1 | 95 | 32.88 | Drosophila melanogaster | Kharkiv, Ukraine | [30] |
| Drosophila innubila nudivirus | DiNV | 155,555 | NC_040699.1 | 106 | 30.29 | Drosophila innubila | Arizona, USA | [31] |
| Oryctes rhinoceros nudivirus | OrNV | 127,615 | EU747721.1 | 139 | 41.63 | Oryctes rhinoceros | Malaysia | [32,33] |
| Esparto virus | EV | 183,261 | NC_040536.1 | 87 | 29.45 | Drosophila melanogaster | Esparto, CA, USA | NA |
| Mauternbach virus | MNV | 154,465 | MG969167.1 | 93 | 30.67 | Drosophila melanogaster | DrosEU50 Mauternbach 2015, Austria | NA |
| Tomelloso virus | TNV | 112,307 | NC_040789.1 | 93 | 39.63 | Drosophila melanogaster | DrosEU28 Tomelloso 2015, Spain | NA |
| Betanudivirus | ||||||||
| Heliothis zea nudivirus 1 | HzNV1 | 228,089 | AF451898.1 | 154 | 41.81 | Heliothis zea | Taiwan, PRC | [34,35] |
| Helicoverpa zea nudivirus 2 | HzNV2 | 231,621 | NC_004156.2 | 113 | 41.88 | Helicoverpa zea | Massachusetts, USA | [36,37] |
| Homarus gammarus nudivirus | HgNV | 107,063 | MK439999.1 | 97 | 35.03 | Homarus gammarus | 52S104HLG2, hepatopancreas, UK | [38] |
| Penaeus monodon nudivirus | PmNV | 119,638 | NC_024692.1 | 115 | 34.53 | Penaeus monodon | Indonesia | [39] |
| Tipula oleracea nudivirus | ToNV | 145,704 | NC_026242.1 | 131 | 25.53 | Tipula oleracea | England, UK | [40] |
| Dikerogammarus haemobaphes nudivirus | DhNV | 119,754 | MT488302 | 106 | Dikerogammarus haemobaphes | UK | [41] | |
| Unclassified | ||||||||
| Allomyrina virus | NA | AIY68660 | NA | NA | Allomyrina dichotoma | Youngdong-1, Korea | NA |
In contrast to the exogenous nudiviruses described above, nudiviral genome sequence fragments are also frequently integrated into the genome of the host insect [21]. Such an association gave rise to the Bracovirus genus of Polydnaviridae with sequences integrated into the genomes of parasitic wasps playing a critical role in the survival of wasp larvae within the parasitized host [22,23,24,25]. Such endogenous nudiviruses have also been documented in several hemipteran insects [26,27].
Our goal was to characterize the virome of two corn rootworm species, drawing on genomic and transcriptomic sequence data [42] toward the identification of viruses with potential for use in rootworm management. From this work, sequences derived from three novel small RNA viruses were identified from the WCR transcriptome [43,44,45], and two from the SCR transcriptome [46,47]. Here, we report on sequences derived from two novel nudiviruses of SCR and WCR assembled from corresponding short genomic sequencing read data. Both viruses appear to be closely related to the third known coleopteran nudivirus, OrNV in the genus Alphanudivirus. Based on phylogenetic analysis and assessment of protein homology among nudiviruses, a revised taxonomic classification is proposed. This work expands upon the described diversity of viruses infecting corn rootworm species, with potential for use in management of these devastating pests.
2. Materials and Methods
2.1. Insect Collection, DNA Isolation, Library Preparation and Illumina Sequencing
Adult SCR were collected live from a field near Ames, IA, USA in late July 2012, transported to the laboratory where all samples were flash frozen in liquid nitrogen and stored at −80 °C. A total of 35 males and 36 females (n = 71) were pooled and ground to a powder in liquid nitrogen. DNA was extracted from 3.0 mg of the ground SCR sample using Qiagen DNeasy Blood and Tissue Extraction kits (Germantown, MD, USA), with modifications to avoid DNA shearing as described previously [48]. Purified DNA was submitted to the Iowa State University DNA Facility (Ames, IA, USA), where genomic DNA was size selected and used to generate ~500-bp insert libraries using the Illumina TruSeq v2 Library Construction Kit (Illumina, San Diego, CA, USA). Single-end 100-bp Illumina HiSeq2500 reads were generated. Data were received in raw fastq format and submitted to the National Center for Biotechnology Information (NCBI) Short Read Archive (BioProject PRJNA689866; SRA Accession SRR13364002) [48].
Three sets of WCR genomic sequencing data were downloaded from the NCBI SRA database (BioProject PRJNA222656). Each adult WCR sequence dataset represented one of three locations, and each sample from a location comprised 5 pooled individuals (Table 2). Sequences were determined by Flagel et al. by Illumina HiSeq 2000 (200-nt sequence reads) [49].
Table 2.
Summary of southern corn rootworm (SCR) and western corn rootworm (WCR) sequence assembly and identification of nudivirus-derived sequences.
| Species | Sequencing Dataset | U.S. Collection Site |
Total Reads (M) | Trimmed Reads (M) | Average Quality Score (q) | N50 (bp) | No. of Contigs (≥200) | No. of Contigs (≥1000) |
|---|---|---|---|---|---|---|---|---|
| D. undecimpunctata | SRR13364002 | Ames, IA | 9.7 | 9.7 | 37 | 420 | 33,539 | NA |
| D. virgifera virgifera | SRR1107649 | Adams County, IN | 195 | 184 | 37 | 403 | 4.4 M | 84,419 |
| D. virgifera virgifera | SRR1107695 | Seneca, KS | 194 | 186 | 38 | 414 | 4.1 M | 71,902 |
| D. virgifera virgifera | SRR1107698 | Colfax, NE | 41 | 40 | 37 | 422 | 3.8 M | 97,060 |
2.2. Sequence Assembly
The quality of the Illumina sequencing reads from SCR was assessed by FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and raw reads were trimmed to remove adapter and low-quality nucleotides, as described previously [48]. The low-quality nucleotides (q < 30) were trimmed from raw WCR sequence reads using Trimmomatic (v0.36) [50]. Trinity (v2.6.6) [51] was used to separately assemble trimmed reads de novo from SCR and WCR libraries. Redundancy was reduced among the resulting contigs using the CAP3 assembler [52]. Nudiviral-associated contigs were manually analyzed and further joined based on sequence overlap. To estimate read coverage of nudiviral sequences, trimmed sequence reads were mapped to the assembled nudiviral fragments with a Perl mapping script [26].
2.3. Identification of Contigs and Viral Sequence Discovery
Methods for annotation of the assembled DNA contigs for identification of virus-derived DNA sequences were as described previously [26]. Briefly, to identify potential viral sequences, the assembled contigs were queried against a local viral sequence database (built using the viral protein sequences extracted from the NCBI nr database) and filtered for BLASTx alignments with minimum E-value ≤ 0.001). Contigs with “hits” to viral proteins were extracted and used as BLASTx queries against the NCBI nr database. Results from BLASTx were analyzed to identify sequences derived from potential viruses. Based on the protein query results, nudiviral-derived sequences were either reassembled with the CAP3 DNA assembly program [52], or manually assembled by sequence alignment with BioEdit v7.2 [53] (https://bioedit.software.informer.com/). The nudiviral DNA sequences were further used to query the NCBI non-redundant nucleotide (nr/nt) database using the BLASTn algorithm, and alignments filter for E-values ≤ 0.001.
2.4. Viral Sequence Analysis and Annotation
Structural annotation of contigs derived from nudivirus sequences was performed via putative nudivirus open reading frame (ORF) translation using SnapGene Viewer (v4.1.9) (https://www.snapgene.com/). ORFs ≥ 50 aa with limited overlap were considered to be nudivirus-encoded genes as applied previously [33]. The selected ORFs were annotated using multiple BLAST algorithms including BLASTp, PSI-BLAST (Position-Specific Iterated BLAST), PHI-BLAST (Pattern Hit Initiated BLAST) or DELTA-BLAST (Domain Enhanced Lookup Time Accelerated BLAST) against the NCBI nr database. Viral genes encoding hypothetical proteins with unknown function were also analyzed by searching for conserved protein domains using the Conserved Domain Architecture Retrieval Tool (CDART) [54]. To assess interspecific similarities using BioEdit [53], translated proteins from SCR and WCR nudiviruses were used as BLASTp queries against a local BLAST database of nudivirus (proteins downloaded from NCBI and results filtered for E-values < 0.0001. Baculoviral core genes and nudivirus essential genes were used to construct a supermatrix phylogenetic tree as described previously [55]. The tree builder programs of the ETE Toolkit v3.0 [56] were used with default parameters employing the JTT (Jones–Taylor–Thorton) or WAG (Whelan and Goldman) models for maximizing the tree’s likelihood, and the resulting phylogeny viewed using FigTree (v1.4.4) (http://tree.bio.ed.ac.uk/software/figtree/).
3. Results
3.1. Assembly of Genomic Sequence Data for Nudiviral Sequence Identification
The genome sequence data for SCR and the three DNA sequence datasets for WCR were assembled separately, resulting in 33,593 contigs for SCR, and 3.8 to 4.3 million contigs for WCR (≥200 bp; Table 2). Contigs of at least 200 bp for SCR and 1000 bp for WCR were selected and annotated by BLASTx to search for sequences potentially derived from DNA viruses. BLASTx results showed that multiple contigs from all four samples (ranging from 20 to 26) had “hits” to known nudivirus proteins genes (Supplementary Tables S1 and S2). The 20 contigs from SCR putatively encoding proteins homologous to those from known nudiviruses were further joined into five nudiviral fragments (F1 to F5) totaling 147,179 bp, which was close to a size typical of known nudiviral genomes (Table 1). The sequence read coverage across the five SCR nudiviral genome fragments ranged from 16.1- to 19.5-fold (mean ~18.5-fold) (Table 3).
Table 3.
Sequence read coverage of five nudivirus genomic fragments (F1 to F5) from D. undecimpunctata.
| Fragment | Length (bp) | G + C (%) | No. Putative CDS | Total Reads Mapped | Total Bases Mapped | Read Coverage |
|---|---|---|---|---|---|---|
| F1 | 6515 | 28.49 | 5 | 1210 | 121,000 | 18.6 |
| F2 | 52,843 | 27.93 | 40 | 10,280 | 1,028,000 | 19.5 |
| F3 | 51,592 | 27.88 | 40 | 9858 | 985,800 | 19.1 |
| F4 | 31,968 | 27.56 | 23 | 6202 | 620,200 | 19.4 |
| F5 | 4261 | 29.46 | 2 | 687 | 68,700 | 16.1 |
| Total | 147,179 | 109 | 28,237 | 2,823,700 | ||
| Average | 28.264 | 18.5 |
BLASTx “hits” were obtained to known nudivirus protein coding regions on contigs independently assembled from WCR data in three accessions SRR1107649 (n = 26), SRR1107695 (n = 25), and SRR1107698 (n = 24). There was a total of 75 putative nudivirus-derived contigs across all assemblies. Comparison of nudivirus-derived contigs from these three assemblies indicated that ~70% of the viral sequences were shared among all three. This result indicates that there is a single nudivirus species shared by all three WCR population samples. Further assembly and depletion of duplicated sequences resulted in a final set of 22 unique fragments, a 3.4-fold reduction from the initial 75 fragments. These unique fragments totaled 48,989 bp, or about one-fifth of the average nudivirus genome size (Table 1). Most of the nudiviral sequences from WCR libraries were assembled into short contigs, suggesting that the samples contained limited nudiviral DNA. Sequence homology from BLASTx query results indicated that both the putative SCR and WCR nudivirus sequences are novel nudiviruses. We tentatively name the viruses Diabrotica undecimpunctata howardi nudivirus (DuhNV) and Diabrotica virgifera virgifera nudivirus (DvvNV).
3.2. Sequence Analysis of Diabrotica undecimpunctata Howardi Nudivirus (DuhNV)
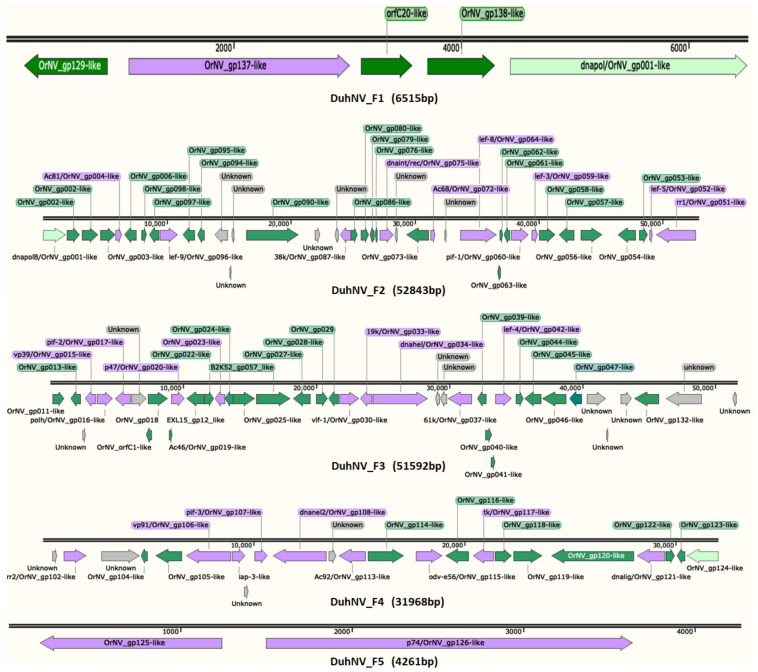
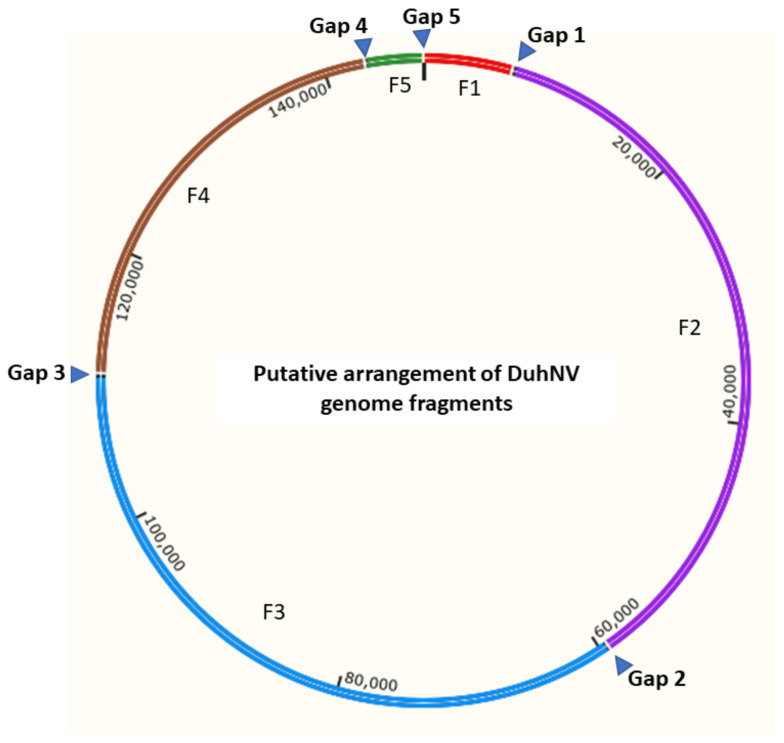
The 147,179 bp of the DuhNV genome sequence were assembled from five fragments, with fragment size ranging from 4261 to 52,843 bp (Table 3). The numbers of putative genes encoded by each fragment is also shown in Table 3. The nucleotide composition of DuhNV is about 28.3% G + C, which is similar to the percentage of G + C in the GrBNV genome (28.0%), but much less than that of OrNV (41.6% G + C) (Table 1). The arrangement of the five DuhNV sequence fragments has yet to be confirmed, but available evidence was used to putatively order and orient fragments. Specifically, given that the 5′ end of dnapol was located at the 3′ end of DuhNV F1 and the 3′ end of dnapol was located at the 5′ end of DuhNV F2, DuhNV F2 was determined to follow DuhNV F1 (Figure 1). The ORFs identified in DuhNV F4 and DuhNV F5 are homologous to the same cluster of genes in OrNV, thus indicating that DuhNV F4 and DuhNV F5 may be adjacent. The two ORFs of DuhNV F5 are homologous to genes OrNV_gp124 and OrNV_gp125, while the 5′ end of DuhNV F1 encodes a gene homologous to OrNV_ gp129 that is positioned downstream of OrNV_gp124 and OrNV_gp125 in the OrNV genome. In the event that the genomic arrangement is conserved between coleopteran nudiviruses, the circular DuhNV genome fragment order was predicted to be F1–F2–F3–F4–F5 (Figure 2).
Figure 1.
Arrangement of putative genes located on five Diabrotica undecimpunctata howardi nudivirus (DuhNV) genomic fragments (F1–F5). All ORFs > 50 aa were predicted by BLASTp query against the National Center for Biotechnology Information (NCBI) nr protein database. Arrows indicate ORF direction and orientation. Green, ORFs with BLASTx “hits” to nudiviruses (E-values ≤ 0.01); purple, core genes; grey, ORFs for hypothetical genes with unknown protein function; light green, partial ORFs (lacking 5′-start or 3′-stop codons). The maps were generated with SnapGene Viewer v5.2.1.
Figure 2.
Putative arrangement of fragments F1–F2–F3–F4–F5 in the 147,179 bp assembled DuhNV genome sequence. Triangles represent sequence gaps. The putative genome schematic was generated by SnapGene Viewer v5.2.1.
A total of 109 putative ORFs of > 50 aa were predicted on the five DuhNV genome fragments. The ORFs were named based on the linear order within each fragment (Table 4), and putative structural BLASTp annotations are shown for encoded protein sequences from DuhNV genes and OrNV homologs (Table 4; Supplementary Table S1). Fifteen hypothetical genes of unknown function were predicted in the DuhNV genome (Table 4; Figure 1). The alignment of in silico translated protein sequences encoded by DuhNV genes with nudiviral orthologs indicated that the majority of the DuhNV genes had high sequence identities with the homologous genes of OrNV or nudiviruses isolated from Drosophila. The protein sequence identity of 74 predicted DuhNV proteins (~81%) showed >40% sequence identity with those encoded by other nudiviruses (Supplementary Table S1). The protein annotation revealed that the majority of the DuhNV genes are similar to those of known nudiviruses, with 56 of the top BLAST “hits” to homologs from OrNV (Table 4; E-values ≤ 6.0 × 10−8). The other top “hits” were from nudiviruses isolated from Drosophila (Supplementary Table S1), with the exception of DuhNV_F3_ORF35, which showed highest similarity to an ATP-binding cassette (ABC) transporter encoded by Gammaproteobacteria (Supplementary Table S1). Taken together, these results suggest that DuhNV is a novel species within the genus Alphanudivirus.
Table 4.
Putative genes of Diabrotica undecimpunctata howardi nudivirus (DuhNV).
| ORF | Gene/Domain | Length (aa) | OrNV Homologs |
|---|---|---|---|
| Replication, Repair and Recombination of DNA | |||
| DuhNV_F1_ORF5 * | Dnapol | 695 | OrNV_gp001 |
| DuhNV_F2_ORF1 * | Dnapol | 633 | OrNV_gp001 |
| DuhNV_F3_ORF23 | Dnahel | 1390 | OrNV_gp034 |
| DuhNV_F2_ORF23 | dnaint/rec | 389 | OrNV_gp075 |
| DuhNV_F2_ORF33 | lef-3 | 177 | OrNV_gp059 |
| DuhNV_F4_ORF10 | dnahel2 | 839 | OrNV_gp108 |
| DuhNV_F4_ORF20 | Dnalig | 423 | OrNV_gp121 |
| Nucleotide Metabolism | |||
| DuhNV_F2_ORF40 | rr1 | 1045 | OrNV_gp051 |
| DuhNV_F4_ORF2 | rr2 | 358 | OrNV_gp102 |
| DuhNV_F1_ORF2 | tk/GrBNV_gp17-like | 648 | OrNV_gp137 |
| DuhNV_F3_ORF31 | tk/GrBNV-gp74_like | 197 | OrNV_gp044 |
| DuhNV_F4_ORF16 | tk(guanosine monophosphate kinase) | 321 | OrNV_gp117 |
| DuhNV_F5_ORF1 | tk/GrBNV_gp044_like | 352 | OrNV_gp125 |
| Transcription | |||
| DuhNV_F3_ORF11 | p47 | 324 | OrNV_gp020 |
| DuhNV_F3_ORF30 | lef-4 | 417 | OrNV_gp042 |
| DuhNV_F2_ORF39 | lef-5 | 82 | OrNV_gp052 |
| DuhNV_F2_ORF28 | lef-8 | 988 | OrNV_gp064 |
| DuhNV_F2_ORF9 | lef-9 | 505 | OrNV_gp096 |
| DuhNV_F3_ORF21 | vlf-1 | 504 | OrNV_gp030 |
| Oral Infectivity | |||
| DuhNV_F5_ORF2 | p74 | 712 | OrNV_gp126 |
| DuhNV_F2_ORF32 | pif-1 | 489 | OrNV_gp060 |
| DuhNV_F3_ORF6 | pif2 | 376 | OrNV_gp017 |
| DuhNV_F4_ORF9 | pif-3 | 202 | OrNV_gp107 |
| Packaging, Assembly, and Morphogenesis | |||
| DuhNV_F3_ORF5 | polh/gran | 404 | OrNV_gp016 |
| DuhNV_F3_ORF22 | 19k/pif-5 | 281 | OrNV_gp033 |
| DuhNV_F2_ORF26 | Ac68 | 136 | OrNV_gp072 |
| DuhNV_F2_ORF18 | 38k | 276 | OrNV_gp087 |
| Inhibition of Apoptosis | |||
| DuhNV_F4_ORF7 | iap-3 | 202 | OrNV_gp134 |
| Unknown Function | |||
| DuhNV_F2_ORF5 | Ac81 | 173 | OrNV_gp004 |
| DuhNV_F3_ORF4 | vp39 | 250 | OrNV_gp015 |
| DuhNV_F4_ORF6 | vp91 | 695 | OrNV_gp106 |
| DuhNV_F4_ORF12 | Ac92 | 410 | OrNV_gp113 |
| DuhNV_F4_ORF14 | odv-e56 | 420 | OrNV_gp115 |
| Other | |||
| DuhNV_F1_ORF1 | ND | 240 | OrNV_gp129/gp136 |
| DuhNV_F1_ORF3 | ND | 149 | OrNV_orfC20 |
| DuhNV_F1_ORF4 | ND | 594 | OrNV_gp138 |
| DuhNV_F2_ORF2 | trypsin-like serine protease-like protein | 339 | OrNV_gp002 |
| DuhNV_F2_ORF3 | trypsin-like serine protease-like protein | 427 | OrNV_gp002 |
| DuhNV_F2_ORF4 | ND | 395 | OrNV_gp003 |
| DuhNV_F2_ORF6 | thymidylate synthase/pyrimidine hydroxymethylase-like | 296 | OrNV_gp006 |
| DuhNV_F2_ORF7 | ND | 143 | OrNV_gp098 |
| DuhNV_F2_ORF8 | mRNA decapping enzyme 2-like | 245 | OrNV_gp097 |
| DuhNV_F2_ORF10 | ND | 314 | OrNV_gp095 |
| DuhNV_F2_ORF11 | ND | 162 | OrNV_gp094 |
| DuhNV_F2_ORF12 | cl17238, RING-Ubox | 329 | no hit |
| DuhNV_F2_ORF13 | None | 62 | no hit |
| DuhNV_F2_ORF14 | None | 63 | no hit |
| DuhNV_F2_ORF15 1 | cl25745, pfam05764 | 1412 | OrNV_gp090 |
| DuhNV_F2_ORF16 | None | 148 | no hit |
| DuhNV_F2_ORF17 | None | 78 | no hit |
| DuhNV_F2_ORF19 | ND | 189 | OrNV_gp086 |
| DuhNV_F2_ORF20 | ND | 227 | OrNV_gp080 |
| DuhNV_F2_ORF21 | ND | 89 | OrNV_gp079 |
| DuhNV_F2_ORF22 | ND | 52 | OrNV_gp076 |
| DuhNV_F2_ORF24 | None | 61 | no hit |
| DuhNV_F2_ORF25 | ND | 593 | OrNV_gp073 |
| DuhNV_F2_ORF27 | None | 56 | no hit |
| DuhNV_F2_ORF29 | ND | 74 | OrNV_gp063 |
| DuhNV_F2_ORF30 | ND | 88 | OrNV_gp062 |
| DuhNV_F2_ORF31 | ND | 140 | OrNV_gp061 |
| DuhNV_F2_ORF34 | HZV_115-like protein | 425 | OrNV_gp058 |
| DuhNV_F2_ORF35 | patatin-like phospholipase-like protein | 397 | OrNV_gp057 |
| DuhNV_F2_ORF36 | ND | 576 | OrNV_gp056 |
| DuhNV_F2_ORF37 | ND | 458 | OrNV_gp054 |
| DuhNV_F2_ORF38 | ND | 234 | OrNV_gp053 |
| DuhNV_F3_ORF1 | mitochondrial carrier protein-like protein | 293 | OrNV_gp011 |
| DuhNV_F3_ORF2 | ND | 231 | OrNV_gp013 |
| DuhNV_F3_ORF3 | None | 76 | no hit |
| DuhNV_F3_ORF7 | None | 388 | no hit |
| DuhNV_F3_ORF8 | ND | 131 | OrNV_orfC1 |
| DuhNV_F3_ORF9 | ND | 509 | OrNV_gp018 |
| DuhNV_F3_ORF10 | Ac46 | 89 | OrNV_gp019 |
| DuhNV_F3_ORF12 2 | EXL15_gp12_like/DUF4679 (pfam15728) | 432 | no hit |
| DuhNV_F3_ORF13 | ND | 241 | OrNV_gp022 |
| DuhNV_F3_ORF14 | guanylate kinase-like protein | 245 | OrNV_gp023 |
| DuhNV_F3_ORF15 | ND | 196 | OrNV_gp024 |
| DuhNV_F3_ORF16 | ND | 550 | OrNV_gp025 |
| DuhNV_F3_ORF17 3 | B2K52_gp057/Smc cl341474 | 858 | no hit |
| DuhNV_F3_ORF18 | ND | 422 | OrNV_gp027 |
| DuhNV_F3_ORF19 4 | PPK13561/cl32896, EAL domain containing protein | 297 | OrNV_gp028 |
| DuhNV_F3_ORF20 | ND | 232 | OrNV_gp029 |
| DuhNV_F3_ORF24 | None | 96 | no hit |
| DuhNV_F3_ORF25 | None | 157 | no hit |
| DuhNV_F3_ORF26 | 61k AcORF9 | 575 | OrNV_gp037 |
| DuhNV_F3_ORF27 | ND | 199 | OrNV_gp039 |
| DuhNV_F3_ORF28 | ND | 166 | OrNV_gp040 |
| DuhNV_F3_ORF29 | ND | 110 | OrNV_gp041 |
| DuhNV_F3_ORF32 | ND | 403 | OrNV_gp045 |
| DuhNV_F3_ORF33 | ND | 591 | OrNV_gp046 |
| DuhNV_F3_ORF34 | ND | 293 | OrNV_gp047 |
| DuhNV_F3_ORF35 5 | cl34121, energy-coupling factor ABC transporter | 490 | no hit |
| DuhNV_F3_ORF36 | None | 66 | no hit |
| DuhNV_F3_ORF37 | None | 278 | no hit |
| DuhNV_F3_ORF38 | ND | 592 | OrNV_gp132 |
| DuhNV_F3_ORF39 | None | 868 | no hit |
| DuhNV_F3_ORF40 | None | 85 | no hit |
| DuhNV_F4_ORF1 | None | 76 | no hit |
| DuhNV_F4_ORF3 | None | 616 | no hit |
| DuhNV_F4_ORF4 | ND | 91 | OrNV_gp104 |
| DuhNV_F4_ORF5 | ND | 408 | OrNV_gp105 |
| DuhNV_F4_ORF8 | None | 68 | no hit |
| DuhNV_F4_ORF11 | None | 120 | no hit |
| DuhNV_F4_ORF13 | ND | 566 | OrNV_gp114 |
| DuhNV_F4_ORF15 | ND | 345 | OrNV_gp116 |
| DuhNV_F4_ORF17 | ND | 266 | OrNV_gp118 |
| DuhNV_F4_ORF18 | ND | 454 | OrNV_gp119 |
| DuhNV_F4_ORF19 | ND | 1285 | OrNV_gp120 |
| DuhNV_F4_ORF21 | ND | 143 | OrNV_gp122 |
| DuhNV_F4_ORF22 | ND | 124 | OrNV_gp123 |
| DuhNV_F4_ORF23 | ND | 476 * | OrNV_gp124 |
Bold, baculovirus core genes; italic, common nudivirus genes. *, separate fragments of the same gene; ND, domain search not done for genes with Oryctes rhinoceros nudivirus (OrNV) homologs; none, no pfam domains identified. 1 YL1 nuclear protein; 2 cl29686, domain of unknown function (DUF4679); 3 chromosome segregation ATPase (cell cycle control, cell division, chromosome partitioning); 4 putative diguanylate cyclase (provisional); 5 ABC transporter protein.
Of the 109 proteins encoded by putative DuhNV ORFs, about 83% had a top BLASTp “hit” to a nudiviral protein represented in the NCBI nr database. These genes included the ~20 core genes of baculoviruses and other essential genes identified in nudiviruses (Table 4). A majority of the DuhNV ORFs show top BLAST “hits” to hypothetical proteins of unknown function previously characterized in other nudivirus genomes. To assess the possible functions, we searched for potential functional domains in the protein sequences encoded by these hypothetical DuhNV genes. CDART results identified known protein domains encoded by 6 of the 24 queried hypothetical DuhNV genes: DvNV_F2_ORF12, _ORF15, DvNV_F3_ORF12, _ORF17, _ORF19, and _ORF35. Most of the unknown DuhNV genes that showed no similarity to previously described nudivirus genes were relatively short ORFs, but some encode relatively large proteins, e.g., DuhNV_F2_ORF12 (329 aa), DuhNV_F3_ORF7 (388 aa), DuhNV_F3_ORF37 (278 aa), DuhNV_F3_ORF39 (868 aa), and DuhNV_F4_ORF3 (616 aa). CDART did not predict any known protein domains encoded by these genes, with the exception of DuhNV_F2_ORF12 that encodes a RING_Ubox (cl7238) zinc-binding domain. RING_Ubox-containing proteins serve various functions, including involvement in virus replication [57], which may reflect the function of this nudivirus ORF.
3.3. Analysis of Diabrotica virgifera virgifera Nudivirus Sequences
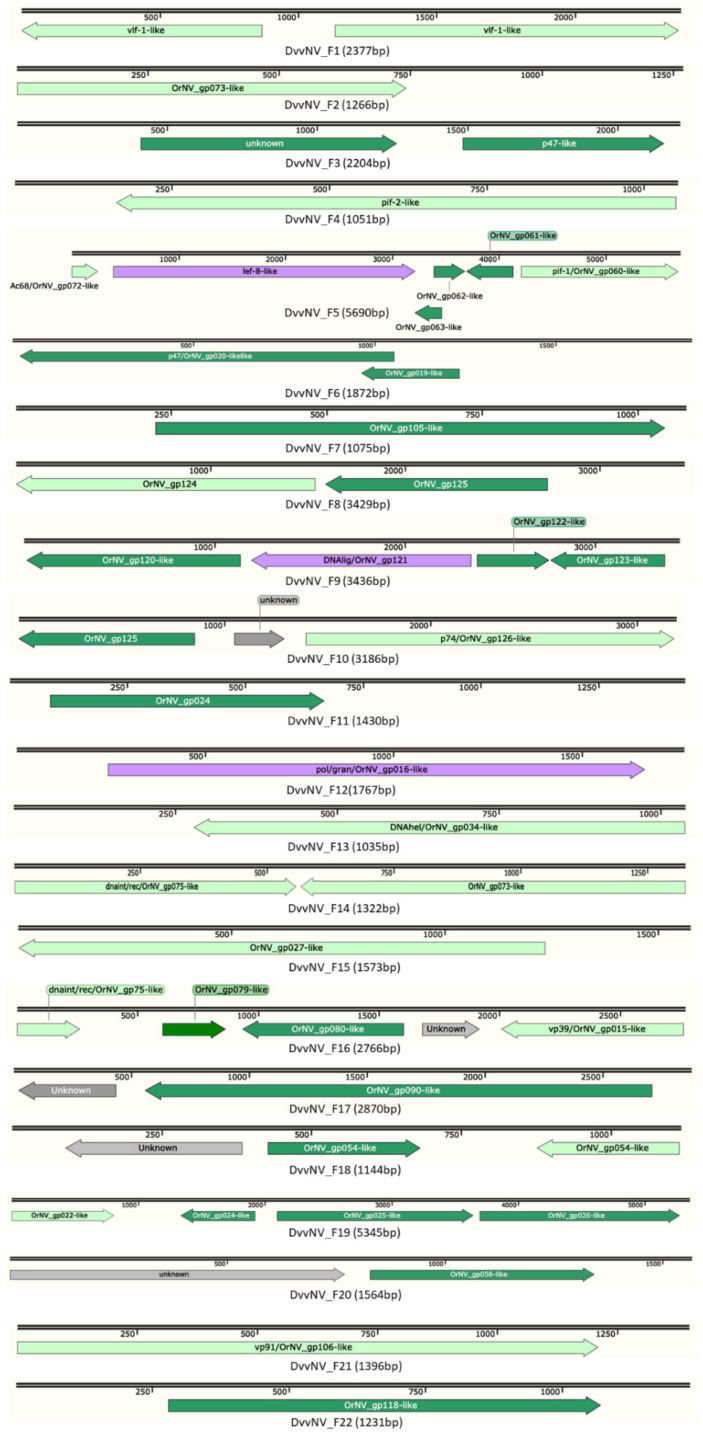
The sequence assembly for the DvvNV genome was not highly contiguous using the available short read data, and resulted in 22 short sequence fragments spanning a total of 53,293 bp (median contig length (N50) = 1867; range 1035 to 5690). Two fragments were >5000 bp in length, and 12 were ≤1872 bp. Despite their relatively short lengths, 48 ORFs were predicted on these 22 DvvNV genome fragments (range 1 to 6 ORFs per contig; Figure 3). Thirteen of the ORFs were considered “partial” sequences due to BLAST alignment coverages of <80%; Supplementary Table S2). Among the 48 predicted DvvNV ORFs, 43 showed similarity to previously described genes from nudiviruses (Table 5), with most top BLAST “hits” to OrNV or nudiviruses isolated from Drosophila (E-values ≤ 0.009; Supplementary Table S2). Six of the predicted DvvNV ORFs showed no similarity to orthologs from other nudiviruses, and were designated as “unknown” genes. Only 13 core genes were found in the 48 predicted DvvNV ORFs. Pairwise comparisons by BLASTp among the homologous proteins of encoded proteins in the three beetle-infecting nudivirus genomes, DvvNV, DuhNV and OrNV, indicated that most share high (>35%) amino acid sequence identities.
Figure 3.
Map of the 22 DvvNV genomic fragments. Arrows indicate ORFs and ORF orientation. Green, ORFs that hit nudiviruses; purple, core genes; grey, unknown ORFs; light green, partial ORFs.
Table 5.
Sequence analysis of Diabrotica virgifera virgifera nudivirus (DvvNV). Baculovirus core genes are shown in bold. Genes common to nudiviruses are shown in italics.
| ORF | Length (Some Partial) |
Gene | Similar OrNV ORF | Similar DuhNV ORF |
|---|---|---|---|---|
| DvvNV_F1_ORF1 | 414 | vlf-1 | OrNV_gp030 | DuhNV_F3_ORF21 |
| DvvNV_F1_ORF2 | 289 | vlf-1 | OrNV_gp030 | DuhNV_F3_ORF21 |
| DvvNV_F2_ORF1 | 246 | OrNV_gp073-like | OrNV_gp073 | DuhNV_F2_ORF25 |
| DvvNV_F3_ORF1 | 223 | p47 | OrNV_gp020 | DuhNV_F3_ORF11 |
| DvvNV_F3_ORF2 | 284 | no hit | DuhNV_F3_ORF9 | |
| DvvNV_F4_ORF1 | 293 | pif-2 | OrNV_gp017 | DuhNV_F3_ORF6 |
| DvvNV_F5_ORF1 | 82 | Ac68-like | OrNV_gp072 | no hit |
| DvvNV_F5_ORF2 | 943 | lef-8 | OrNV_gp064 | DuhNV_F2_ORF28 |
| DvvNV_F5_ORF3 | 81 | OrNV_gp063-like | OrNV_gp063 | no hit |
| DvvNV_F5_ORF4 | 98 | OrNV_gp062-like | OrNV_gp062 | DuhNV_F2_ORF30 |
| DvvNV_F5_ORF5 | 144 | GrBNV_gp51-like | OrNV_gp061 | no hit |
| DvvNV_F5_ORF6 | 493 | pif-1 | OrNV_gp060 | DuhNV_F2_ORF32 |
| DvvNV_F6_ORF1 | 343 | p47 | OrNV_gp020 | DuhNV_F3_ORF11 |
| DvvNV_F6_ORF2 | 89 | Ac146-like | OrNV_gp019 | DuhNV_F3_ORF10 |
| DvvNV_F7_ORF1 | 272 | GrBNV gp43-like | OrNV_gp105 | DuhNV_F4_ORF5 |
| DvvNV_F8_ORF1 | 379 | GrBNV_gp44-like | OrNV_gp125 | DuhNV_F5_ORF1 |
| DvvNV_F8_ORF2 | 512 | OrNV_gp124-like | OrNV_gp124 | DuhNV_F4_ORF23 |
| DvvNV_F9_ORF1 | 373 | GrBNVgp37-like | OrNV_gp120 | DuhNV_F4_ORF19 |
| DvvNV_F9_ORF2 | 384 | DNAlig | OrNV_gp121 | DuhNV_F4_ORF20 |
| DvvNV_F9_ORF3 | 125 | GrBNV_gp39-like | OrNV_gp122 | no hit |
| DvvNV_F9_ORF4 | 199 | GrBNVgp41-like | OrNV_gp123 | DuhNV_F4_ORF22 |
| DvvNV_F10_ORF1 | 284 | GrBNV_gp44-like | OrNV_gp125 | DuhNV_F5_ORF1 |
| DvvNV_F10_ORF2 | 80 | unknown | no hit | |
| DvvNV_F10_ORF3 | 596 | p74 | OrNV_gp126 | DuhNV_F5_ORF2 |
| DvvNV_F11_ORF1 | 193 | GrBNVgp75-like | OrNV_gp024 | DuhNV_F3_ORF15 |
| DvvNV_F12_ORF1 | 474 | pol/gran | OrNV_gp016 | DuhNV_F3_ORF5 |
| DvvNV_F13_ORF1 | 251 | DNAhel | _OrNV_gp034 | DuhNV_F3_ORF23 |
| DvvNV_F14_ORF1 | 184 | dnaint/rec | OrNV_gp075 | DuhNV_F2_ORF23 |
| DvvNV_F14_ORF2 | 251 | putative gene | OrNV_gp073 | DuhNV_F2_ORF25 |
| DvvNV_F15_ORF1 | 411 | GrBNV_gp78-like | OrNV_gp027 | DuhNV_F3_ORF18 |
| DvvNV_F16_ORF1 | 86 | dnaint/rec | OrNV_gp075 | DuhNV_F2_ORF23 |
| DvvNV_F16_ORF2 | 222 | GrBNVgp60-like | OrNV_gp080 | no hit |
| DvvNV_F16_ORF3 | 86 | GrBNVgp59-like | OrNV_gp079 | DuhNV_F2_ORF21 |
| DvvNV_F16_ORF4 | 79 | unknown | ||
| DvvNV_F16_ORF5 | 250 | vp39 | OrNV_gp015 | DuhNV_F3_ORF4 |
| DvvNV_F17_ORF1 | 715 | GrBNVgp28-like | OrNV_gp090 | DuhNV_F2_ORF15 |
| DvvNV_F17_ORF2 | 136 | unknown | ||
| DvvNV_F18_ORF1 | 84 | GrBNV gp83-like | OrNV_gp054 | DuhNV_F2_ORF37 |
| DvvNV_F18_ORF2 | 97 | unknown | DuhNV_F2_ORF37 | |
| DvvNV_F18_ORF3 | 78 | GrBNV gp83-like | OrNV_gp054 | no hit |
| DvvNV_F19_ORF1 | 270 | GrBNV_gp72-like | OrNV_gp022 | DuhNV_F3_ORF13 |
| DvvNV_F19_ORF2 | 516 | GrBNV_gp76-like | OrNV_gp025 | DuhNV_F3_ORF16 |
| DvvNV_F19_ORF3 | 526 | unknown | OrNV_gp026 | DuhNV_F3_ORF17 |
| DvvNV_F19_ORF4 | 193 | GrBNVgp75-like | OrNV_gp024 | DuhNV_F3_ORF15 |
| DvvNV_F20_ORF1 | 171 | OrNV_gp056 | OrNV_gp056 | DuhNV_F2_ORF36 |
| DvvNV_F20_ORF2 | 256 | unknown | ||
| DvvNV_F21_ORF1 | 402 | vp91 | OrNV_gp106 | DuhNV_F4_ORF6 |
| DvvNV_F22_ORF1 | 263 | GrBNV_gp35-like | OrNV_gp118 | DuhNV_F4_ORF17 |
3.4. Phylogenetic Analysis of WCR Nudiviruses and Other Nudiviruses
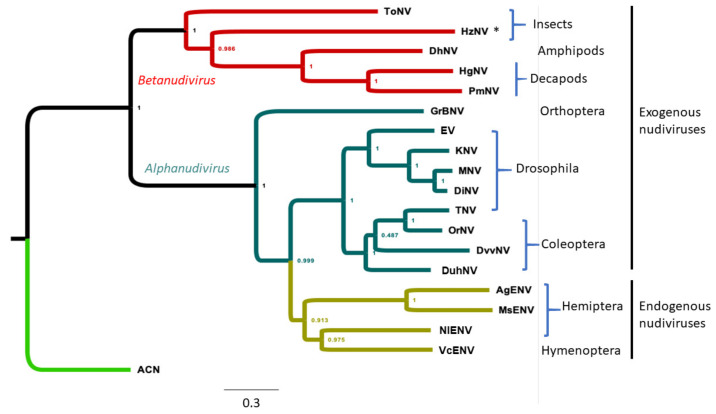
The phylogenetic relationships among DuhNV, DvvNV and other nudiviruses were reconstructed as a supermatrix phylogenetic tree based on aligned protein sequences of core nudiviral genes (Figure 4; Supplementary Table S3). The tree shows two major divisions representing alpha- and beta-nudiviruses. The six betanudiviruses (HzNV represents HzNV1 and HzNV2) were closely related and grouped into a clade distinct from the alphanudiviruses. In the alphanudivirus clade, GrBNV formed an individual branch and was more closely related to ancestral nudiviruses than to other alphanudiviruses. The remaining alphanudiviruses were clustered into two major sub-clades. One sub-clade included a sub-branch of four of the five nudiviruses isolated from Drosophila. The exception was TNV, which was more closely related to OrNV and grouped into the sub-branch with the three beetle-infecting nudiviruses. The four endogenous nudiviruses analyzed previously [26] form the second major sub-clade in the alphanudiviral clade.
Figure 4.
Phylogenetic tree based on nudivirus core sequences. Protein sequences encoded by 31 core genes were downloaded from the NCBI database for 13 sequenced nudiviruses, four endogenous nudiviruses and a baculovirus, Autographa californica nucleopolyhedrovirus (ACN, outgroup; Supplementary Table S3). Abbreviations for exogenous nudiviruses from Table 1. AgENV, Aphis glycines endogenous nudivirus; MsENV, Melanaphis sacchari endogenous nudivirus; NlENV, Nilaparvata lugens endogenous nudivirus; VcENV, Venturia canescens endogenous nudivirus. Core sequences of AgENV and MsENV were previously reported [26]. The phylogenetic tree was built using supermatrix methods with ETE Toolkit v3. The tree was edited and viewed using FigTree. Taxonomic categorization of host species for most nudiviruses is indicated. * HzNV represents both HzNV1 and HzNV2. Numerals at branch points indicate bootstrap support.
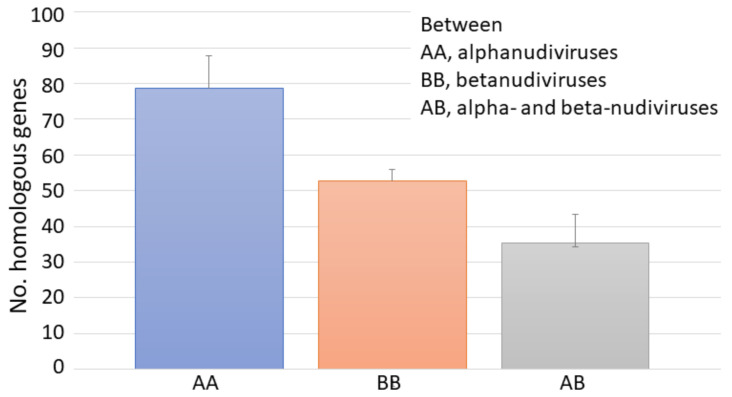
We further analyzed gene similarities within and between the alpha- and beta-nudivirus groups (Figure 5, Supplementary Table S4). Comparison of homologous genes indicated that, on average, ~80 (out of an average nudivirus gene number of ~100) are homologous among Alphanudivirus, with ~50 of these Betanudivirus genes having high similarities (E-values < 0.0001) (Figure 5). Only ~35 genes between alpha- and beta-nudiviruses show high levels of similarity.
Figure 5.
Numbers of homologous proteins among nudiviruses as determined by BLASTp. The cutoff E-value for determination of homology was set at 0.0001. Bars represent SD of the mean.
4. Discussion
Insect-derived DNA and RNA sequence data provide useful resources for metagenomic analyses, including the identification of sequences derived from viruses [42,58]. For this investigation, we report assembled DNA sequences derived from two novel nudiviruses embedded in SCR and WCR short shotgun genomic read data. The 147,179 bp DuhNV genome sequence assembly was considered near complete based on the high representation of genes homologous with those from OrNV, including a core set of protein coding genes. Although near complete, five gaps remain in the DuhNV assembly. Attempts to fill the gaps in the DuhNV genome sequence by PCR were unsuccessful (data not shown). The gaps could contain tandem sequence repeats that make amplification difficult. Third-generation sequencing technologies [59], which provide longer reads, may provide a means for completion of both the DuhNV and DvvNV genome sequences. Putative nudiviral particles were reported from SCR field populations more than three decades ago [16]; however, it remains unknown if those particles were from the same DuhNV species described herein. Given the assembly of the majority of the DuhNV genome (Figure 1; Figure 2) at read coverage depths >18-fold (Table 2), the viral titer was likely high in the Iowa SCR population sample used for this work. Despite the previous observation of nudivirus-like particles [16] and discovery of DuhNV sequences described herein, it is not possible to comment on nudivirus distribution in the U.S. as these insects move very long distances: SCR migrate to the northern states in late spring or early summer from their overwintering range along the Gulf Coast [60,61], in contrast to WCR, which are present in the northern states year-round [62].
DNA viruses have not previously been documented from WCR. The discovery of DvvNV-derived sequences within short shotgun genomic read data from WCR suggests that a nudivirus infects WCR. The viral sequences were observed in WCR populations from three Midwestern states, suggesting the virus has a wide geographic distribution. However, due to relatively low read coverage, the DvvNV genome sequence assembly was highly fragmented, preventing detailed characterization. The low level of viral sequences in these WCR genomic DNA samples may be due to a low rate of DvvNV infection. The sample size of five used for WCR DNA isolation and sequencing [42] compared to 71 SCR adults pooled from the Iowa sample suggests that sampling bias may have impacted our results. Low recovery also may be due to lower viral titers in each of the WCR individuals that were sampled. A third possible explanation for the presence of limited number of DvvNV sequences is that the DvvNV-derived reads are from an endogenous nudivirus. The insertion of nudivirus genomic sequence into the host genome can involve loss or disruption of most of the sequence. To investigate this possibility, we used the same methods described herein to examine DNA sequence data and assembled sequences from more than five datasets for WCR collected from disparate geographical locations in the U.S. (Arizona, Pennsylvania, Iowa and South Dakota), and from Hungary. No nudivirus sequences were identified in these additional datasets when queried with homologous nudiviruses sequences using the BLASTn. Hence, DvvNV is unlikely to be an endogenous nudivirus as DvvNV sequence would otherwise have been present in the genomes of other WCR. In addition, phylogenetic analysis of nudiviruses grouped DvvNV into the exogenous nudivirus clade, rather than the clade composed of endogenous nudiviruses. Hence, we conclude that DvvNV is likely an exogenous alphanudivirus (Figure 4).
The supermatrix phylogenetic tree based on 31 core nudiviral genes showed an interesting evolutionary relationship among 18 nudiviruses, including DuhNV and DvvNV described here. This phylogeny suggests that alpha- and beta-nudiviruses evolved independently from ancestral dsDNA viruses. The bifurcation of marsh crane fly-infecting nudivirus ToNV and HzNV, from the moth pest Helicoverpa (formerly Heliothis) zea, is the closest to the base of the phylogeny rooted by the ACN outgroup. Analysis of the similarity of homologous genes between alpha- and betanudiviruses (Figure 5) suggests that the genes of the betanudiviruses, for which we have sequences, are more diverse than those of alphanudiviruses. This suggests a greater evolutionary time over which mutations could have accumulated, and is consistent with the phylogenetic analysis based on nudivirus core genes (Figure 4). Based on the nudivirus sequences available to date, however, we can only conclude that the alpha- and betanudiviruses are derived from a common ancestor and that the two groups are, therefore, of the same evolutionary age.
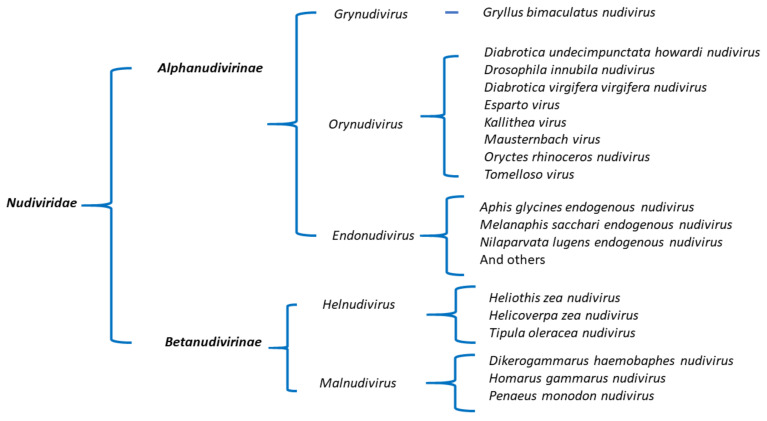
The phylogenetic analysis clearly divided nudiviruses into two clusters, namely alpha- and beta- nudiviruses. However, the phylogenetic tree also suggests that the current taxonomic classification of Nudiviridae can be further expanded. Based on our analysis, we propose that alpha- and beta- nudiviruses could be assigned to two subfamilies, Alphanudivirinae and Betanudivirinae, under Nudiviridae (Figure 6). In Alphanudivirinae, GrBNV is in a separate clade, distant from other alphanudiviral species (Figure 4). GrBNV shared only 63–72 homologous genes with other alphanudiviruses (Supplementary Table S3), while the other nudiviruses in this subfamily share similar numbers (75–90) of homologous genes and were isolated from Drosophila or beetles. We propose that GrBNV represents a genus, Grynudivirus (Gry from Gryllus), and OrNV represents the type species of the other genus, named Orynudivirus. Notably, endogenous nudiviruses, e.g., Aphis glycines endogenous nudivirus, are in the branch of the Alaphanudivirinae, and form a unique clade. We propose the name of Endonudivirus for the genus of endogenous nudiviruses that are integrated in host genomes. Hence Alphanudivirinae contains three new genera. Similarly, two novel genera could be established in the Betanudivirinae based on the class of host species: Helnudivirus and Malnudivirus. The name Helnudivirus is from HzNV, which includes three nudiviral species isolated from the class Insecta. The three nudiviruses isolated from the class Malacostraca could be assigned a new genus, Malnudivirus. The proposed new nudiviral classification, based on our phylogenetic tree (Figure 4), and evidence of homologous gene similarities among the nudiviruses (Figure 5), is illustrated in Figure 6.
Figure 6.
Proposed new taxonomic classification of Nudiviridae. Two subfamilies and five genera are proposed.
The discovery of sequences derived from two novel nudiviruses from corn rootworms increases our understanding of diversity within the rootworm virome, and sheds light on the evolution of this remarkable virus family. Analysis of virus-derived sequences from a limited number of SCR, WCR and NCR (unpublished data) DNA sequence datasets indicated that DuhNV sequences were observed only from SCR, and that DvvNV sequences were found only in WCR. A large-scale sequence investigation, in addition to experimentation, would be required to determine the host range of each of these viruses. Further work is needed to complete the genome sequences of DuhNV and DvvNV, to examine the distribution and host range of these viruses, and to address their impact on corn rootworm populations. Furthermore, our analyses suggest that revision of the current taxonomic classification of Nudiviridae may be warranted.
Acknowledgments
The mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply a recommendation or endorsement by USDA. USDA ARS is an equal opportunity provider and employer.
Supplementary Materials
The following Supplementary Materials are available online at https://www.mdpi.com/1999-4915/13/2/269/s1. Table S1, Analysis of DuhNV ORFs; Table S2, Analysis of DvvNV ORFs; Table S3, List of core genes used for phylogenetic analysis; Table S4, Number of homologous genes among nudiviruses. Virus sequence text files: DuhNV_sequence, DvvNV_gb_format.
Author Contributions
Conceptualization, B.C.B. and S.L.; methodology, S.L. and B.S.C.; formal analysis, S.L.; investigation, S.L., T.W.S., B.S.C. and B.C.B.; resources, T.W.S. and B.S.C.; writing—original draft preparation, S.L. and B.C.B.; writing—review and editing, T.W.S., B.S.C. and B.C.B.; visualization, S.L. and B.C.B.; project administration, B.C.B.; funding acquisition, B.C.B. and T.W.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Monsanto Company’s Corn Rootworm Knowledge Research Program, USDA National Institute of Food and Agriculture, Hatch project IOW05450, USDA Agricultural Research Service project 5030-22000-019-00D, by State of Iowa funds, and by the University of Florida Institute of Food and Agricultural Sciences.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Raw sequence data used for this analysis are available as indicated in Section 2.1 of the manuscript. The sequences of DuhNV and DvvNV are provided as text files in Supplementary Materials. GenBank accession numbers DuhNV_F1 to 5, MW503925 to 9 respectively.
Conflicts of Interest
The authors declare no conflict of interest.
Author Notes
A 2020 ICTV proposal that places HgNV and PmNV into a new genus Gammanudivirus, and ToNV into a new genus Deltanudivirus, has come to our attention: https://talk.ictvonline.org/files/proposals/animal_dna_viruses_and_retroviruses/m/animal_dna_ec_approved/10386 These changes will be added to the 2020 release of official virus taxonomy (https://talk.ictvonline.org/taxonomy/) after ratification by the ICTV in 2021. While the supermatrix tree presented in the current manuscript (Figure 4) does not support creation of additional nudivirus genera, the reader is referred to the ICTV proposal to review additional analyses that provide the basis for the revised classification.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cabrera Walsh G., Ávila C.J., Cabrera N., Nava D.E., de Sene Pinto A., Weber D.C. Biology and management of pest Diabrotica species in South America. Insects. 2020;11:421. doi: 10.3390/insects11070421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krysan J.L., Smith R.F. Systematics of the virgifera species group of Diabrotica (Coleoptera: Chrysomelidae: Galerucinae) Entomography. 1987;5:375–484. [Google Scholar]
- 3.Wechsler S., Smith D. Has resistance taken root in U.S. corn fields? Demand for insect control. Am. J. Agric. Econ. 2018;100:1136–1150. doi: 10.1093/ajae/aay016. [DOI] [Google Scholar]
- 4.Souza D., Peterson J.A., Wright R.J., Meinke L.J. Field efficacy of soil insecticides on pyrethroid-resistant western corn rootworms (Diabrotica virgifera virgifera LeConte) Pest Manag. Sci. 2020;76:827–833. doi: 10.1002/ps.5586. [DOI] [PubMed] [Google Scholar]
- 5.Gassmann A.J., Petzold-Maxwell J.L., Keweshan R.S., Dunbar M.W. Field-evolved resistance to Bt maize by western corn rootworm. PLoS ONE. 2011;6:e22629. doi: 10.1371/journal.pone.0022629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gassmann A.J. Resistance to Bt maize by western corn rootworm: Insights from the laboratory and the field. Curr. Opin. Insect Sci. 2016;15:111–115. doi: 10.1016/j.cois.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Gassmann A.J., Shrestha R.B., Kropf A.L., St Clair C.R., Brenizer B.D. Field-evolved resistance by western corn rootworm to Cry34/35Ab1 and other Bacillus thuringiensis traits in transgenic maize. Pest Manag. Sci. 2020;76:268–276. doi: 10.1002/ps.5510. [DOI] [PubMed] [Google Scholar]
- 8.Baum J.A., Bogaert T., Clinton W., Heck G.R., Feldmann P., Ilagan O., Johnson S., Plaetinck G., Munyikwa T., Pleau M., et al. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 2007;25:1322–1326. doi: 10.1038/nbt1359. [DOI] [PubMed] [Google Scholar]
- 9.Price D.R., Gatehouse J.A. RNAi-mediated crop protection against insects. Trends Biotechnol. 2008;26:393–400. doi: 10.1016/j.tibtech.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 10.Fishilevich E., Vélez A.M., Storer N.P., Li H., Bowling A.J., Rangasamy M., Worden S.E., Narva K.E., Siegfried B.D. RNAi as a management tool for the western corn rootworm, Diabrotica virgifera virgifera. Pest Manag. Sci. 2016;72:1652–1663. doi: 10.1002/ps.4324. [DOI] [PubMed] [Google Scholar]
- 11.Khajuria C., Ivashuta S., Wiggins E., Flagel L., Moar W., Pleau M., Miller K., Zhang Y., Ramaseshadri P., Jiang C., et al. Development and characterization of the first dsRNA-resistant insect population from western corn rootworm, Diabrotica virgifera virgifera LeConte. PLoS ONE. 2018;13:e0197059. doi: 10.1371/journal.pone.0197059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Velez A.M., Fishilevich E., Rangasamy M., Khajuria C., McCaskill D.G., Pereira A.E., Gandra P., Frey M.L., Worden S.E., Whitlock S.L., et al. Control of western corn rootworm via RNAi traits in maize: Lethal and sublethal effects of Sec23 dsRNA. Pest Manag. Sci. 2020;76:1500–1512. doi: 10.1002/ps.5666. [DOI] [PubMed] [Google Scholar]
- 13.Bedford G.O. Biology and management of palm dynastid beetles: Recent advances. Annu. Rev. Entomol. 2013;58:353–372. doi: 10.1146/annurev-ento-120710-100547. [DOI] [PubMed] [Google Scholar]
- 14.Jackson T.A., Crawford A.M., Glare T.R. Oryctes virus--time for a new look at a useful biocontrol agent. J. Invertebr. Pathol. 2005;89:91–94. doi: 10.1016/j.jip.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 15.Degrugillier M.E., Degrugillier S.S., Jackson J.J. Nonoccluded, Cytoplasmic Virus-Particles and Rickettsia-Like Organisms in Testes and Spermathecae of Diabrotica-virgifera. J. Invertebr. Pathol. 1991;57:50–58. doi: 10.1016/0022-2011(91)90040-W. [DOI] [Google Scholar]
- 16.Kim K.S., Kitajima E.W. Nonoccluded Baculovirus-Like and Filamentous Virus-Like Particles in the Spotted Cucumber Beetle, Diabrotica-Undecimpunctata (Coleoptera, Chrysomelid) J. Invertebr. Pathol. 1984;43:234–241. doi: 10.1016/0022-2011(84)90142-3. [DOI] [Google Scholar]
- 17.Kim K.S. Cytopathology of spotted cucumber beetle hemocytes containing virus-like particles. J. Invertebr. Pathol. 1980;36:292–301. doi: 10.1016/0022-2011(80)90038-5. [DOI] [Google Scholar]
- 18.Kim K.S., Scott H.A., Fulton J.P. Viruses discovered in beetle pests. Ark. Farm Res. 1981;30 [Google Scholar]
- 19.Harrison R.L., Herniou E.A., Bézier A., Jehle J.A., Burand J.P., Theilmann D.A., Krell P.J., van Oers M.M., Nakai M. and ICTV Report ConsortiumICTV Virus Taxonomy Profile: Nudiviridae. J. Gen. Virol. 2020;101:3–4. doi: 10.1099/jgv.0.001381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huger A.M., Krieg A. Baculoviridae. Nonoccluded Baculoviruses. In: Adams J.R., Bonami J.R., editors. Atlas of Invertebrate Viruses. CRC Press, Inc.; Boca Raton, FL, USA: 1991. pp. 287–319. [Google Scholar]
- 21.Cheng R.L., Li X.F., Zhang C.X. Nudivirus Remnants in the Genomes of Arthropods. Genome Biol. Evol. 2020;12:578–588. doi: 10.1093/gbe/evaa074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drezen J.M., Leobold M., Bézier A., Huguet E., Volkoff A.N., Herniou E.A. Endogenous viruses of parasitic wasps: Variations on a common theme. Curr. Opin. Virol. 2017;25:41–48. doi: 10.1016/j.coviro.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 23.Leobold M., Bézier A., Pichon A., Herniou E.A., Volkoff A.N., Drezen J.M. The Domestication of a Large DNA Virus by the Wasp Venturia canescens Involves Targeted Genome Reduction through Pseudogenization. Genome Biol. Evol. 2018;10:1745–1764. doi: 10.1093/gbe/evy127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pichon A., Bézier A., Urbach S., Aury J.M., Jouan V., Ravallec M., Guy J., Cousserans F., Thézé J., Gauthier J. Recurrent DNA virus domestication leading to different parasite virulence strategies. Sci. Adv. 2015;1:e1501150. doi: 10.1126/sciadv.1501150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bézier A., Annaheim M., Herbinière J., Wetterwald C., Gyapay G., Bernard-Samain S., Wincker P., Roditi I., Heller M., Belghazi M., et al. Polydnaviruses of braconid wasps derive from an ancestral nudivirus. Science. 2009;323:926–930. doi: 10.1126/science.1166788. [DOI] [PubMed] [Google Scholar]
- 26.Liu S., Coates B.S., Bonning B.C. Endogenous viral elements integrated into the genome of the soybean aphid, Aphis glycines. Insect Biochem. Mol. Biol. 2020;123:103405. doi: 10.1016/j.ibmb.2020.103405. [DOI] [PubMed] [Google Scholar]
- 27.Cheng R.L., Xi Y., Lou Y.H., Wang Z., Xu J.Y., Xu H.J., Zhang C.X. Brown Planthopper Nudivirus DNA Integrated in Its Host Genome. J. Virol. 2014;88:5310–5318. doi: 10.1128/JVI.03166-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huger A.M. A new virus-disease of crickets (Orthoptera, Gryllidae) causing macronucleosis of fat-body. J. Invertebr. Pathol. 1985;45:108–111. doi: 10.1016/0022-2011(85)90055-2. [DOI] [Google Scholar]
- 29.Wang Y., Kleespies R.G., Huger A.M., Jehle J.A. The genome of Gryllus bimaculatus nudivirus indicates an ancient diversification of baculovirus-related nonoccluded nudiviruses of insects. J. Virol. 2007;81:5395–5406. doi: 10.1128/JVI.02781-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palmer W.H., Medd N.C., Beard P.M., Obbard D.J. Isolation of a natural DNA virus of Drosophila melanogaster, and characterisation of host resistance and immune responses. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Unckless R.L. A DNA Virus of Drosophila. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0026564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huger A.M. A virus disease of Indian rhinoceros beetle Oryctes rhinoceros (Linnaeus) caused by a new type of insect virus Rhabdionvirus oryctes Gen N sp. N. J. Invertebr. Pathol. 1966;8:38–51. doi: 10.1016/0022-2011(66)90101-7. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y., Bininda-Emonds O.R., van Oers M.M., Vlak J.M., Jehle J.A. The genome of Oryctes rhinoceros nudivirus provides novel insight into the evolution of nuclear arthropod-specific large circular double-stranded DNA viruses. Virus Genes. 2011;42:444–456. doi: 10.1007/s11262-011-0589-5. [DOI] [PubMed] [Google Scholar]
- 34.Granados R.R. Early events in the infection of Heliothis zea midgut cells by a baculovirus. Virology. 1978;90:170–174. doi: 10.1016/0042-6822(78)90347-1. [DOI] [PubMed] [Google Scholar]
- 35.Cheng C.H., Liu S.M., Chow T.Y., Hsiao Y.Y., Wang D.P., Huang J.J., Chen H.H. Analysis of the complete genome sequence of the Hz-1 virus suggests that it is related to members of the Baculoviridae. J. Virol. 2002;76:9024–9034. doi: 10.1128/JVI.76.18.9024-9034.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raina A.K., Adams J.R. Gonad-specific virus of corn-earworm. Nature. 1995;374:770. doi: 10.1038/374770a0. [DOI] [PubMed] [Google Scholar]
- 37.Burand J.P., Kim W., Afonso C.L., Tulman E.R., Kutish G.F., Lu Z., Rock D.L. Analysis of the genome of the sexually transmitted insect virus Helicoverpa zea nudivirus 2. Viruses. 2012;4:28–61. doi: 10.3390/v4010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holt C.C., Stone M., Bass D., Bateman K.S., van Aerle R., Daniels C.L., van der Giezen M., Ross S.H., Hooper C., Stentiford G.D. The first clawed lobster virus Homarus gammarus nudivirus (HgNV n. sp.) expands the diversity of the Nudiviridae. Sci. Rep. 2019;9:10086. doi: 10.1038/s41598-019-46008-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Y.T., Lee D.Y., Wang Y., Hu J.M., Li W.H., Leu J.H., Chang G.D., Ke H.M., Kang S.T., Lin S.S., et al. The genome and occlusion bodies of marine Penaeus monodon nudivirus (PmNV, also known as MBV and PemoNPV) suggest that it should be assigned to a new nudivirus genus that is distinct from the terrestrial nudiviruses. BMC Genomics. 2014;15:628. doi: 10.1186/1471-2164-15-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bézier A., Thézé J., Gavory F., Gaillard J., Poulain J., Drezen J.M., Herniou E.A. The genome of the nucleopolyhedrosis-causing virus from Tipula oleracea sheds new light on the Nudiviridae family. J. Virol. 2015;89:3008–3025. doi: 10.1128/JVI.02884-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allain T.W., Stentiford G.D., Bass D., Behringer D.C., Bojko J. A novel nudivirus infecting the invasive demon shrimp Dikerogammarus haemobaphes (Amphipoda) Sci. Rep. 2020;10:14816. doi: 10.1038/s41598-020-71776-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonning B.C. The Insect Virome: Opportunities and Challenges. Curr. Issues Mol. Biol. 2020;34:1–12. doi: 10.21775/cimb.034.001. [DOI] [PubMed] [Google Scholar]
- 43.Liu S., Chen Y., Sappington T.W., Bonning B.C. Genome Sequence of a Novel Positive-Sense, Single-Stranded RNA Virus Isolated from Western Corn Rootworm, Diabrotica virgifera virgifera LeConte. Genome Announc. 2017;5:e00366-17. doi: 10.1128/genomeA.00366-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu S., Chen Y., Sappington T.W., Bonning B.C. Genome Sequence of Diabrotica virgifera virgifera virus2, a Novel Small RNA Virus of the Western Corn Rootworm, Diabrotica virgifera virgifera LeConte. Genome Announc. 2017;5:e00365-17. doi: 10.1128/genomeA.00365-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu S., Chen Y., Sappington T.W., Bonning B.C. Genome Sequence of the First Coleopteran Iflavirus Isolated from Western Corn Rootworm, Diabrotica virgifera virgifera LeConte. Genome Announc. 2017;5:e01530-16. doi: 10.1128/genomeA.00366-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu S., Valencia-Jiménez A., Darlington M., Vélez A.M., Bonning B.C. Diabrotica undecimpunctata virus 2, a Novel Small RNA Virus Discovered from Southern Corn Rootworm, Diabrotica undecimpunctata howardi Barber (Coleoptera: Chrysomelidae) Microbiol. Resour Announc. 2020;9:e00380-20. doi: 10.1128/MRA.00380-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu S., Valencia-Jiménez A., Darlington M., Vélez A.M., Bonning B.C. Genome Sequence of a Small RNA Virus of the Southern Corn Rootworm, Diabrotica undecimpunctata howardi Barber (Coleoptera: Chrysomelidae) Microbiol. Resour Announc. 2020;9:e00379-20. doi: 10.1128/MRA.00379-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coates B.S. Assembly and annotation of full mitochondrial genomes for the corn rootworm species, Diabrotica virgifera virgifera and Diabroticabarberi (Insecta: Coleoptera: Chrysomelidae), using Next Generation Sequence data. Gene. 2014;542:190–197. doi: 10.1016/j.gene.2014.03.035. [DOI] [PubMed] [Google Scholar]
- 49.Flagel L.E., Bansal R., Kerstetter R.A., Chen M., Carroll M., Flannagan R., Clark T., Goldman B.S., Michel A.P. Western corn rootworm (Diabrotica virgifera virgifera) transcriptome assembly and genomic analysis of population structure. BMC Genomics. 2014;15:195. doi: 10.1186/1471-2164-15-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang X., Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hall T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program. for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 54.Geer L.Y., Domrachev M., Lipman D.J., Bryant S.H. CDART: Protein homology by domain architecture. Genome Res. 2002;12:1619–1623. doi: 10.1101/gr.278202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Y.J., Jehle J.A. Nudiviruses and other large, double-stranded circular DNA viruses of invertebrates: New insights on an old topic. J. Invertebr. Pathol. 2009;101:187–193. doi: 10.1016/j.jip.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 56.Huerta-Cepas J., Serra F., Bork P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Molec. Biol. Evol. 2016;33:1635–1638. doi: 10.1093/molbev/msw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu S., Wang J., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R., Gwadz M., Hurwitz D.I., Marchler G.H., Song J.S., et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020;48:D265–D268. doi: 10.1093/nar/gkz991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Webster C.L., Waldron F.M., Robertson S., Crowson D., Ferrari G., Quintana J.F., Brouqui J.M., Bayne E.H., Longdon B., Buck A.H., et al. The Discovery, Distribution, and Evolution of Viruses Associated with Drosophila melanogaster. PLoS Biol. 2015;13:e1002210. doi: 10.1371/journal.pbio.1002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Dijk E.L., Jaszczyszyn Y., Naquin D., Thermes C. The Third Revolution in Sequencing Technology. Trends Genet. 2018;34:666–681. doi: 10.1016/j.tig.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 60.Drees B.M. Southern corn rootworm (spotted cucumber beetle) In: Steffey K., Rice M., All J., Andow D., Gray M., Van Duyn J.., editors. Handbook of Corn Insects. Entomological Society of America; Lanham, MD, USA: 1999. p. 63. [Google Scholar]
- 61.Sappington T.W., Hesler L.S., Allen K.C., Luttrell R.G., Papiernik S.K. Prevalence of sporadic insect pests of seedling corn, and factors affecting risk of infestation. J. Integr. Pest Manag. 2018;9:1–27. doi: 10.1093/jipm/pmx020. [DOI] [Google Scholar]
- 62.Gray M.E., Sappington T.W., Miller N.J., Moeser J., Bohn M.O. Adaptation and invasiveness of western corn rootworm: Intensifying research on a worsening pest. Annu. Rev. Entomol. 2009;54:303–321. doi: 10.1146/annurev.ento.54.110807.090434. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence data used for this analysis are available as indicated in Section 2.1 of the manuscript. The sequences of DuhNV and DvvNV are provided as text files in Supplementary Materials. GenBank accession numbers DuhNV_F1 to 5, MW503925 to 9 respectively.