Abstract
Drug-induced kidney injury (DIKI) is a major concern in both drug development and clinical practice. There is an unmet need for biomarkers of glomerular damage and more distal renal injury in the loop of Henle and the collecting duct (CD). A cross-laboratory program to identify and characterize urinary microRNA (miRNA) patterns reflecting tissue- or pathology-specific DIKI was conducted. The overall goal was to propose miRNA biomarker candidates for DIKI that could supplement information provided by protein kidney biomarkers in urine. Rats were treated with nephrotoxicants causing injury to distinct nephron segments: the glomerulus, proximal tubule, thick ascending limb (TAL) of the loop of Henle and CD. Meta-analysis identified miR-192-5p as a potential proximal tubule-specific urinary miRNA candidate. This result was supported by data obtained in laser capture microdissection nephron segments showing that miR-192-5p expression was enriched in the proximal tubule. Discriminative miRNAs including miR-221-3p and -222-3p were increased in urine from rats treated with TAL versus proximal tubule toxicants in accordance with their expression localization in the kidney. Urinary miR-210-3p increased up to 40-fold upon treatment with TAL toxicants and was also enriched in laser capture microdissection samples containing TAL and/or CD versus proximal tubule. miR-23a-3p was enriched in the glomerulus and was increased in urine from rats treated with doxorubicin, a glomerular toxicant, but not with toxicants affecting other nephron segments. Taken together these results suggest that urinary miRNA panels sourced from specific nephron regions may be useful to discriminate the pathology of toxicant-induced lesions in the kidney, thereby contributing to DIKI biomarker development needs for industry, clinical, and regulatory use.
Keywords: microRNAs, biomarkers, kidney injury, safety assessment
Drug-induced kidney injury (DIKI) is a major concern in both drug development and clinical practice. Approximately 20% of clinical cases of renal injury are due to drug-induced toxicity, with many affecting the elderly population (Khan et al., 2017; Patel and Sapra, 2020). The incidence of acute kidney injury (AKI) in hospitalized patients worldwide ranges from 17% to 31% (Susantitaphong et al., 2013) and can exceed 50% in the intensive care setting (Hoste et al., 2015). Furthermore, nephrotoxicity is an important reason for drug development failure both in preclinical and clinical stages (Cook et al., 2014). Traditionally used clinical pathology biomarkers of AKI and DIKI that are measured in biofluids suffer from several shortcomings. For example, blood urea nitrogen (BUN) and serum creatinine lack sensitivity and specificity to ensure optimal renal safety monitoring of potential DIKI in preclinical and clinical situations. There is a need for improved translational kidney biomarkers which would enable renal toxicity potential to be identified earlier during clinical development programs, thereby improving health protection of trial subjects and preventing promising drug candidates from being discontinued due to lack of appropriate safety monitoring tools.
During the last decade, progress has been made to identify and characterize novel biomarkers of DIKI that are more sensitive and specific than the traditional pathology parameters BUN and serum creatinine. Several urine-based protein biomarkers of drug-induced acute tubular injury such as kidney injury molecule-1 (Kim-1), albumin, clusterin, neutrophil gelatinase-associated lipocalin (NGAL), and osteopontin were recently qualified or received Letters of Support from health authorities for use in both preclinical (Ennulat and Adler, 2015) and clinical studies (EMA, 2016; FDA, 2016). However, for some of these biomarkers, there is a lack of specificity for kidney and a lack of biomarkers specific for each nephron segment. For example, systemic release of low molecular weight protein NGAL during inflammatory processes could lead to a false positive response through urinary excretion in the absence of kidney injury (Vlasakova et al., 2014). Clusterin has no nephron segment specificity (Harpur et al., 2011). Despite this progress there is an unmet need for biomarkers of glomerular damage and for more distal injury in the loop of Henle and collecting duct (CD).
MicroRNAs (miRNAs) are single-stranded, noncoding 17–28 nucleotide small RNAs (sRNAs), involved in posttranscriptional regulation of gene expression. miRNAs are abundant in the kidney and involved in many physiological and pathological processes (Bhatt et al., 2011). miRNAs have been shown to be differentially expressed in different regions of the kidney (Kriegel and Liang, 2013a). Since they are described to be released into urine under injury conditions and to be stable in urine (Mall et al., 2013) they have been suggested as kidney injury biomarkers (Ramachandran et al., 2013; Saikumar et al., 2012). Additionally, given the highly conserved nature of miRNA sequences across mammalian species (Friedman et al., 2009) and the overlap in kidney miRNA expression between humans and mice (Pavkovic and Vaidya, 2016), there is potential translatability of miRNAs as kidney-specific injury markers from preclinical species to humans to monitor DIKI in drug development. For clinical use, the predicted area under the curve (AUC) for AKI using a panel of microRNAs (miRNAs) demonstrated a near perfect AUC (1.0) using receiver operating characteristic (ROC) curve analysis for AKI (Aguado-Fraile et al., 2015) outperforming creatinine, BUN, NGAL, Kim-1, and other urinary biomarkers for renal injury (Brandenburger and Lorenzen, 2020). Taken together, these characteristics make miRNAs promising candidates for nephron segment-specific biomarkers which could complement novel protein kidney biomarkers in urine.
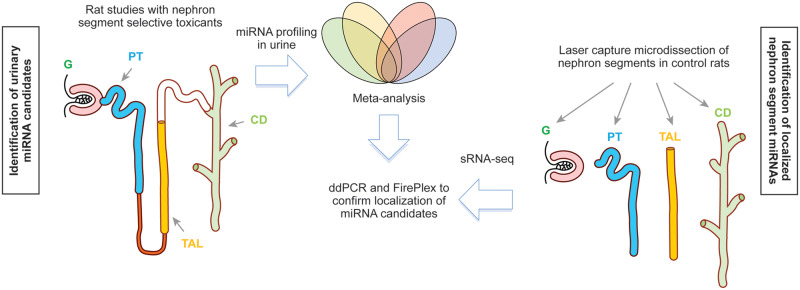
In order to address the ability of urinary miRNA to inform underlying kidney pathology and site-specific injury of the renal nephron as a “liquid biopsy,” a coordinated effort was required. To meet this need, a cross-laboratory study was formed by the Health and Environmental Sciences Institute (HESI) Biomarkers of Nephrotoxicity Technical Committee, whose mission is to develop a systematic approach for the evaluation of biomarkers that bridge from the preclinical to clinical stages of drug development and to provide a scientific forum for building consensus regarding how to apply biomarkers of toxicity in risk assessment. The goal of the committee’s project was the identification and characterization in urine of tissue- or pathology-specific miRNA patterns (ie, changes in individual miRNAs and/or a panel of differentially expressed miRNAs) for DIKI in rats. The overall strategy to identify nephron segment-specific urinary miRNAs is depicted in Figure 1. In 11 studies, rats were dosed with nephrotoxicants known to result in injury to distinct nephron segments, ie, the glomerulus, proximal tubule, thick ascending limb (TAL) of the loop of Henle, and CD. A meta-analysis of the miRNA data from these 11 studies was performed to identify significantly modulated urinary miRNAs which were specific to a histopathology finding in 1 nephronal segment or common to more than 1. In parallel, sRNA-sequencing (sRNA-seq) experiments were conducted to identify enriched expression of miRNAs in nephron segments of the kidney isolated by laser capture microdissection (LCM). Confirmatory experiments were conducted using digital droplet (dd)PCR and FirePlex platforms on LCM enriched nephron segments to further characterize the localization of miRNAs candidates in the kidney. It was envisaged that appearance of such nephron segment-enriched miRNAs in urine upon nephrotoxicant administration would indicate their potential value as biomarkers of segment-specific DIKI in rats.
Figure 1.
Strategy to identify and characterize the localization of nephron segment specific miRNAs. Abbreviations: G, glomerulus; PT, proximal tubule; TAL, thick ascending limb of the loop of Henle; CD, collecting duct.
MATERIALS AND METHODS
Animal Studies
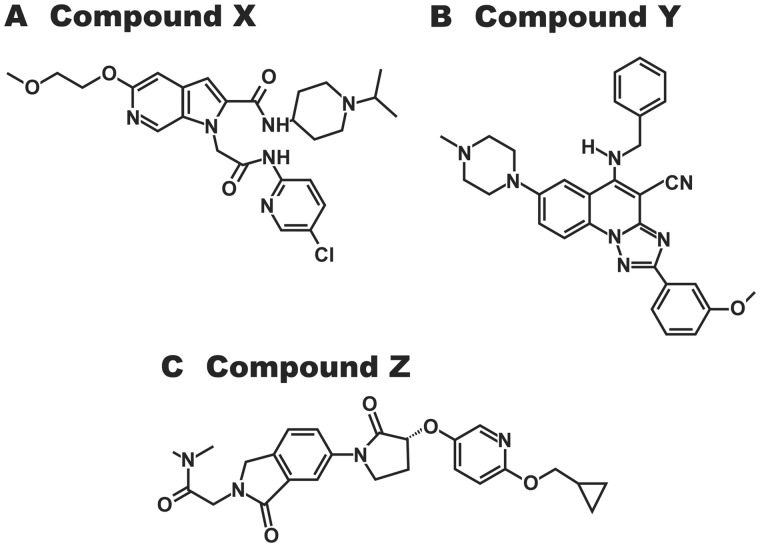
Participating scientists on the HESI Biomarkers of Nephrotoxicity Committee acted as principal investigator for one or more of the individual studies. All animal studies were conducted by the participating laboratories according to the national guidelines for care and use of laboratory animals and were approved by the Institutional Animal Care and Use Committee where the study was performed. Each study was designed taking account of the particular requirements of the compound under study. The studies described here used a total of 454 male Sprague Dawley (SD) or Han-Wistar or Wistar Kyoto rats. The first 2 strains are routinely used in nonclinical safety studies to support clinical development and acceptable to regulatory authorities worldwide. The Wistar Kyoto strain was used in only 1 study (Pavkovic et al., 2015) for comparison of strain differences. SD rats (Crl: CD[SD]) at 6–8 weeks of age and weighing between 225 and 270 g at dose of initiation were treated with the glomerular toxicants puromycin, doxorubicin, sheep anti-Fx1A serum (passive Heymann nephritis [HN] rat model of immune-mediated glomerulonephritis), sheep anti-rat glomerular basement membrane serum (nephrotoxic serum model of immune-mediated glomerulonephritis), and with the proximal tubular toxicant gentamicin. For these studies, euthanization was performed by CO2 inhalation (Church et al., 2014) or isoflurane gas (Nassirpour et al., 2014, 2015; Pavkovic et al., 2014) and exsanguination via vena cava blood collection (see Table 1 for more details on the study design). The doses of nephrotoxicants and the duration of the studies were chosen in such a way as to produce kidney lesions of similar intensity (minimal to moderate degeneration/necrosis in the expected tubular segments of the nephron) to allow comparison of the biomarker results in the meta-analysis. Han Wistar rats, 8 weeks old, were treated with the proximal tubular toxicant cisplatin as described previously (Pavkovic et al., 2014). Wistar Kyoto rats, 8 weeks old, were treated with sheep antirat glomerular basement membrane serum (Pavkovic et al., 2015). Additional studies (not previously published) were conducted in SD rats, 7 weeks old, with the proximal tubular toxicant potassium dichromate and the CD toxicant N-phenylanthranilic acid (NPAA). Three proprietary compounds, Sanofi compounds X, Y, and Z (small molecules differing in their chemical structures and pharmacological targets—see Figure 2) which in preclinical rodent studies produced lesions in the tubular epithelial cells of the loop of Henle (compounds X and Y), and the CD (compound Z) were also tested in 7 weeks old SD rats (see Table 1 for doses, mode of administration and duration used in this study). As for the other compounds included in this program of work, the dosage regimens were based on the information available from the studies in which the lesions were first observed. The vehicle was 0.9% sodium chloride for potassium dichromate, sterile water for compound X, and an aqueous solution of 0.6% (w/w) methylcellulose/0.5% (w/w) polysorbate 80 for NPAA, compounds Y and Z. The volumes of administration were 4 ml/kg for potassium dichromate given subcutaneously, and 10 ml/kg for NPAA, compounds X, Y, and Z given by gavage. Rats were anesthetized with isoflurane and euthanized by exsanguination from the abdominal aorta in the additional studies with potassium dichromate, NPAA, compounds X, Y, and Z.
Table 1.
Main Kidney Histopathological Findings and Urinary Protein Biomarker Changes in Rats Treated With Segment Selective Nephrotoxicants
| Nephron Site Injury | Compounds | Study Allocation | Rat Strain | Study Design (Dose, Route of Administration, Day of Necropsy, and Number of Animals Per Group) | Key Histopathological Findings in Kidney | Main Urinary Protein Biomarker Changesa | References |
|---|---|---|---|---|---|---|---|
| Glomerulus | Puromycin | Pfizer | SD | 0 and 150 mg/kg ip—days 2, 3, and 6 (n = 10) | Glomerular alterations, grades 1–5 | Microalbumin | Nassirpour et al. (2015) |
| HN | Pfizer | SD | 0 and 1 ml/200 g body weight iv—days 3, 6, 9, and 16 (n = 10) | Glomerular alterations, grades 2 and 3 | Microalbumina | Nassirpour et al. (2015) | |
| Doxorubicin | Janssen | SD | 0 and 5 mg/kg iv—days 8 and 15 (n = 6) | Glomerular alterations, grade 2 | Microalbumin, Kim-1, and NGAL | Church et al. (2014) | |
| Tubular dilatation, cortico-medullary junction, grades 2 and 3 | |||||||
| Nephrotoxic serum | Bayer | SD | 0 and 5 ml/kg iv—days 8 and 14 (n = 5 or 6) | Glomerular alterations, grades 1–3 | Microalbumin, Kim-1, NGAL, and clusterin | Pavkovic et al. (2015) | |
| Tubular regeneration, cortex, grades 1–4 | |||||||
| Wistar Kyoto | 0, 1, 2.5, and 5 ml/kg iv—day 14 (n = 3 or 5) | Glomerular alterations, grades 1–3 | Microalbumin, Kim-1, NGAL, and clusterin | Pavkovic et al. (2015) | |||
| Tubular regeneration, cortex, grades 1–3 | |||||||
| Proximal tubule | Potassium dichromate | Sanofi | SD | 0, 5, and 15 mg/kg subcutaneously—day 2 (n = 10) | Tubular deg/necrosis, procalcitonin (s1-2), grades 1–3 | Microalbumin and NAGa | NA |
| Cisplatin | Bayer | Han Wistar | 0, 1, 3 mg/kg ip—days 3, 5, 8, and 26 (n = 6) | Tubular deg/necrosis, Thick descending tubule (s3), grades 1–5 | Microalbumin, Kim-1, NGAL, and clusterin | Pavkovic et al. (2014) | |
| Gentamicin | Pfizer | SD | 0 and 50 mg/kg/day subcutaneously—day 7 (n = 10) | Tubular deg/necrosis, procalcitonin (s1-2), grades 1–3 | Microalbumin and Kim-1 | Nassirpour et al. (2014) | |
| TAL of the loop of Henle | Proprietary compound X | Sanofi | SD | 0, 50, and 200 mg/kg/day orally—days 4 and 11 (n = 4–10) | Tubular deg/necrosis, Thick ascending tubule, grades 1–3 | Kim-1, microalbumin, osteopontin, and clusterin | NA |
| Proprietary compound Y | Sanofi | SD | 0, 100, and 400 mg/kg/day orally—days 4 and 7 (n = 10) | Tubular deg/necrosis, Thick ascending tubule, grades 1 and 2 | Microalbumina | NA | |
| CD | NPAA | Sanofi | SD | 0 and 400 mg/kg/day orally—days 4, 8, and 15 (n = 10) | CD necrosis with loss of papilla tip, grades 1 and 2 | RPA-1 and clusterin | NA |
| Proprietary compound Z | Sanofi | SD | 0 and 200 mg/kg/day orally—days 4, 8, and 15 (n = 10) | Necrosis of renal papilla, grades 1 and 2 | RPA-1 and clusterin | NA |
Abbreviation: NA, non applicable.
Kim-1, clusterin, and osteopontin were not measured in the studies with potassium dichromate, HN model, and compound Y.
Figure 2.
Compound structures for Sanofi compounds (A) X, (B) Y, and (C) Z. The pharmacological targets of compounds X, Y, and Z are Factor Xa, adenosine A3 receptor, and GPR119, respectively.
Biochemical and Histopathological Analyses
The methods used to measure the traditional biomarkers in blood (serum creatinine and BUN) and in urine (total protein, albumin, and N-acetyl-β-d-glucosaminidase [NAG]), as well as the novel urinary protein biomarkers Kim-1, NGAL, osteopontin, clusterin, and renal papillary antigen-1 (RPA-1) in the studies with puromycin, doxorubicin, HN model, nephrotoxic serum, cisplatin, and gentamicin have been previously described (Church et al., 2014; Nassirpour et al., 2014, 2015; Pavkovic et al., 2014, 2015). The methods used to measure the above protein urinary biomarkers in the studies with potassium dichromate, NPAA, compounds X, Y, and Z were previously published (Gautier et al., 2014).
Animals were necropsied and tissues, including kidneys, fixed in 10% neutral buffered formalin for subsequent microscopic examination. Sections were cut from these blocks, stained by hematoxylin and eosin and examined by a qualified veterinary pathologist from the respective study sites. Observations were recorded using the histopathology lexicon of the Predictive Safety Testing Consortium (Sistare et al., 2010) in order to compare the histopathological findings across studies. The microscopic lesion severities were defined and graded as follows; minimal—grade 1, mild (slight)—grade 2, moderate—grade 3, marked—grade 4, or severe—grade 5. Slides and histopathology data from all studies used in this HESI-based project were reviewed by a resident pathologist at Sanofi, Pfizer, Bayer, and Janssen, depending on where the study was generated, to assess morphological diagnoses and consistency in grading and to identify key treatment-related findings for each toxicant.
Urinary miRNA Profiling Using TaqMan Low-Density Arrays
Using previously described methods (Pavkovic et al., 2014), total RNA including sRNA species (eg, miRNAs) was isolated from 200 µl urine using the miRNeasy Mini Kit (Qiagen, Hilden, Germany), which also included 2.5 fmol synthetic ath-miR-159a as a spike-in within each urine sample. Urinary miRNA profiles were subsequently analyzed using 3 µl of the RNA eluate with TaqMan low-density array (TLDA) cards A (all studies) and B (all studies, except the nephrotoxic serum study performed at Bayer), representing 381 miRNA TaqMan assays per card, including 225 and 131 rat valid miRNAs and 5–6 (depending on card version) sRNAs (Thermo Fisher Scientific, Darmstadt, Germany).
Statistical analysis workflow
Urine samples corresponding to marked or severe grades of histopathological kidney lesions in studies with puromycin on day 6 and cisplatin on days 8 and 26 were excluded from the analysis to reinforce the probability to detect normalizers. A consistent analysis workflow was used for each study using Array Studio software (Omicsoft, USA). Given that the TLDA profiling of miRNA extracts includes a preamplification step of 12 cycles, the probability to introduce spurious amplification of miRNAs not present in a sample must be taken into account; consequently, the background level in terms of Ct value was evaluated in preliminary validation trials with No Template Control samples in which RNA extract was replaced by water. As a result, a Ct cutoff value of 32 was chosen as higher Ct values were considered unreliable. Normalization with least variant miRNAs was possible in all studies (see Supplementary File 1). Briefly, a normalization was performed after selection of the least variant miRNA after ranking of the Ct max – Ct min value of miRNAs of interest. The fold changes were computed using classical ΔΔCt method and inferential statistics were performed with 1- or 2-way ANOVA depending on the study design. A fold change > 2 and false discovery rate adjusted p values below .05 were used for selection of modulated miRNAs. For the NPAA study, 2 independent analyses were conducted: a first analysis with all animals at day 15, and a more focused analysis with animals presenting renal papilla necrosis grade 2 (animal numbers 21, 24, 27, 29) and control rats (animal numbers 1, 4, 6, 9). Then miRNAs apparently distinct for 1 kidney lesion or overlapping between 2 or more kidney lesions were determined using Venn diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/; last accessed December, 21 2020).
Meta-analysis of urinary miRNA results
All of the studies in this project were conducted with a common purpose, namely, to identify urinary miRNA changes that occurred in response to nephrotoxin-induced injury to specific nephron segments. The choice of doses, frequency and duration of administration of the nephrotoxins was selected in each study based on prior experience with the test compound with the goal of producing lesions of minimal to moderate intensity. Irrespective of study dosing regimen, all of the miRNA results were anchored to the histopathology findings in different segments of the nephron. The morphological diagnoses and severity gradings were harmonized by discussion within a working group of the pathologists who read the studies. These steps ensured that the meta-analysis was agnostic with regard to the nature of the chemical and the dosing regimen used.
LCM: Serial Section Staining
Kidneys from 3 naïve adult male SD rats, 6 months old, were collected at necropsy into optimum cutting temperature embedding medium, frozen on dry ice and isopentane, and stored at ≤ −70°C until use. Frozen blocks containing kidney tissue were sectioned at 8 µm and adhered to glass slides. Serial sections were stained with rabbit α-glutathione-s-transferase, sheep anti-Tamm-Horsfall protein, or rabbit anti-aquaporin-2 (all from EMD Millipore) to identify proximal tubules, the TAL of the loop of Henle, and CD, respectively (Bauchet et al., 2011) using the HistoGene LCM immunofluorescence staining kit (Thermo Fisher Scientific) and Alexa Fluor 488-conjugated donkey anti-rabbit or donkey anti-sheep secondary antibodies (Thermo Fisher Scientific) as appropriate. Additional serial sections were stained with a histochemical stain (HistoGene LCM frozen section staining kit; Thermo Fisher Scientific) to identify glomeruli. Nephron segments of interest were identified under fluorescence or light microscopy as appropriate and collected from the stained slides using an Arcturus XT LCM instrument onto CapSure LCM Macro caps (Thermo Fisher Scientific). For samples used for sRNA-seq and ddPCR, RNA was isolated using the miRNeasy Micro kit (Qiagen). For samples used in the FirePlex analysis, RNA isolation was not performed.
Small RNA-Sequencing
RNA was isolated using the RNeasy Micro kit (Qiagen). Samples were eluted in 20 µl RNase-free water, and RNA concentrations were determined using a NanoDrop 8000 at 260 nm. RNA quality was assessed using an Agilent Bioanalyzer and an Agilent sRNA Analysis Kit. Samples were stored at −80°C prior to sRNA-seq conducted at the Laboratory Corporation, Inc (Seattle, Washington; acquired by Covance).
Library construction was performed according to Illumina’s TruSeq sRNA sample prep protocol (Illumina, San Diego, California). Each of the twelve samples was barcoded. Library quality was assessed using the Agilent Bioanalyzer DNA chip and quantified using Ribo Green dye and standard methods. Sample libraries were pooled and divided across 2 lanes and 4 flow cells to reach the targeted read count. Illumina HiSeq2000 analysis was performed using 50 bp single-end reads, with a targeted read depth per sample of 40 million reads. FASTQ files are available for download at the Gene Expression Omnibus (GSE156590). Subsequent analysis was performed using Partek Flow (Partek Inc., St. Louis, Missouri). Briefly, raw sequence was filtered for rRNA, tRNA, and mitochondrial DNA sequence using Bowtie 2 (v 2.2.5). Adapters were then removed from the remaining sequence and subsequently aligned to rat genome (rn6) with Bowtie (v 1.0.0) using a seed mismatch limit of 1 and total seed length of 10. Samples that had <500 000 aligned counts were removed from further analysis (rat 2, glomerulus). miRNA counts were generated by quantifying to miRbase (v 22, rn6) using a minimum of 10 reads and 100% minimum read overlap with features. To identify those miRNA features with higher expression, counts were filtered out if the total raw counts for all samples were less than 100. Raw counts were then normalized by “trimmed mean of M-values” (TMM). For principal component analysis (PCA), raw counts were normalized by TMM, adding 1.0 to all counts, then log2-transforming the data.
Digital Droplet PCR
ddPCR for specific miRNAs present in RNA isolated from LCM sections was performed according to the “Protocol for miRNA PCR profiling using miRNA LNA PCR primer sets with the QX200 Droplet Digital PCR System (Qiagn (Exiqon) protocol 923996—v1.0—06/2015).” Annealing temperatures (51.5–57°C) and amount of input RNA (0.5–20 ng per 20 µl RT-reaction, 0.5–2.5 ng cDNA per PCR setup) and of primer volumes (0.5–1 μl) were first optimized with total rat kidney RNA for each specific miRNA. Furthermore, FAM-labeled Ppia and Hex-labeled B2m primer sets were used in a separate one-step RT-ddPCR reaction (Bio-Rad, Hercules, CA; catalog number 1864021) as endogenous reference mRNAs, for comparison with potential endogenous reference miRNAs. This analysis and comparison with sRNA-seq results suggested miR-30d-5p as a potential endogenous reference because it was expressed with no significant differences in the 4 nephron segments isolated by LCM, at a medium level concerning the number of positive droplets and with low coefficient of variation. Considering the overall very low amounts of RNA present in microdissected tissue regions, the absolute copy number of each miRNA as detected with ddPCR, was expressed as a relative ratio compared with miR-30d-5p to account for potential RNA quantification errors.
Abcam FirePlex miRNA Assays
The FirePlex miRNA Assay (Abcam, Cambridge, MA; catalog number ab218342) was performed on a custom panel consisting of 64 rat miRNAs and 6 off-species and spike in controls. LCM samples were stored on CapSure Macro LCM Caps at −80°C until use. 250 µl Digest Mix was added to a fresh 0.5 ml tube and the LCM Cap securely affixed. The sample was inverted and shaken for 1 h at 60°C. Lysate was then filtered and 1.2 × 106 copies of cel-miR-39 synthetic spike in added per 25 μl of lysate. The FirePlex assay was from this point performed following the manufacturer’s protocol to profile miRNAs directly from the lysate without RNA extraction. In brief, 25 µl lysate was hybridized to the custom miRNA panel for 60 min with shaking at 37°C. After 2 rinses Labeling Buffer was added to each well and incubated at room temperature for 60 min with shaking, resulting in the ligation of universal adapters to the ends of the hybridized miRNAs. After 3 additional rinses the adapter-ligated miRNAs were then eluted from the particles using boiling water and were amplified by PCR with biotinylated primers that hybridize to the universal adapters. Following PCR amplification, miRNAs were rehybridized to the probes with shaking at 37°C for 30 min. After additional rinses Reporter Mix was used to fluorescently label the biotinylated probes now hybridized to the particles. After final rinses the samples were then scanned on an EMD Millipore Guava 6HT flow cytometer.
RESULTS
Histopathological and Urinary Protein Biomarker Changes in Rats Exposed to Segment Selective Nephrotoxicants
Rats were treated for up to 26 days with several nephrotoxicants affecting different tubular regions of the nephron. The key histopathological findings in kidney and the main urinary protein biomarker changes are summarized in Table 1. Incidence tables for histopathology and urinary biomarker values are shown in Supplementary Files 2 and 3, respectively. Overall, the nephrotoxicants used in the study produced minimal to moderate degeneration/necrosis in the expected tubular segments of the nephron.
Whereas BUN and serum creatinine were not or only slightly changed, novel urinary protein biomarker profiles confirmed the nephron-segment selectivity of the damages. Indeed, the glomerular toxicants puromycin, doxorubicin, nephrotoxic serum and HN model induced the expected morphological alterations in glomeruli together with increases in urinary microalbumin and total protein. Of note, doxorubicin and nephrotoxic serum also lead to tubular alterations secondary to glomerular lesions which was corroborated by increases in urinary Kim-1 and NGAL. The proximal tubule toxicants potassium dichromate, cisplatin, and gentamicin similarly caused the expected proximal tubular lesions with concomitant increases in urinary Kim-1. In agreement with prior results (unpublished) the 2 proprietary compounds X and Y produced tubular degeneration/regeneration in the TAL, together with elevations in urinary microalbumin, Kim-1, osteopontin, and clusterin. Immunohistochemistry (IHC) experiments with anti-glutathion s-transferase-alpha and antiaquaporin-1 antibodies confirmed that the proximal tubules were not morphologically affected by compounds X and Y (data not shown). IHC experiments with anti-Kim-1 antibody showed that both compounds X and Y lead to increased expression of Kim-1 protein in some tubules at the junction between outer and inner stripes of the outer medulla (data not shown), suggesting effects on the S3 segment/transition to thin descending limb of Henle’s loop. Kim-1 staining may indicate, in this case, early (reflux) lesions in the straight portion of the proximal tubule. The CD toxicants NPAA and proprietary compound Y induced lesions in the CD and the corresponding expected increases in urinary RPA-1 and clusterin.
Urinary miRNA Profiles in Individual Rat Studies
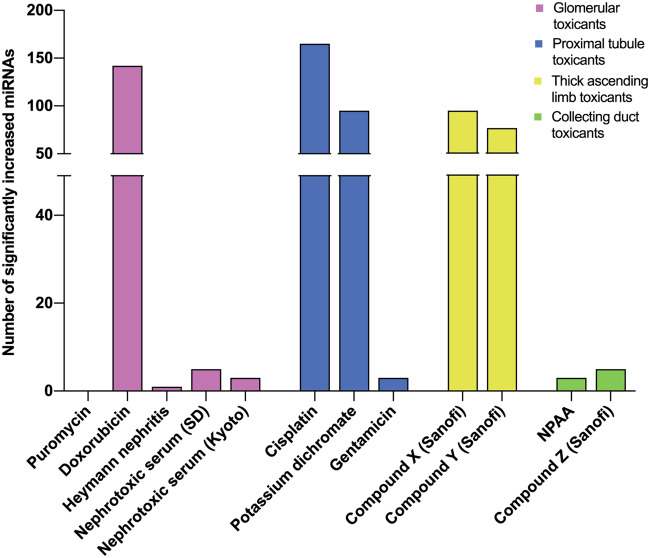
Applying the statistical analysis workflow, the highest number of significantly modulated urinary miRNAs using TLDA cards were found in studies conducted with doxorubicin, cisplatin, potassium dichromate and the 2 loop of Henle toxicants (see Figure 3 for overall numbers of increased miRNAs and Supplementary File 4 for full list of modulated urinary miRNAs and Ct values in all rat studies). Among the urinary miRNAs modulated at the end of treatment several miRNAs were also modulated at earlier interim time-points (ie, on both days 3 and 5 with cisplatin; on days 10 and 14 with doxorubicin). Several urinary miRNAs were increased only at interim time-points (eg, in the study with NPAA where miR-27a and -208b were significantly increased on day 8 but not on day 15, and miR-193 was significantly increased on day 4 only). In a similar manner to the urinary protein biomarkers, several urinary miRNAs were increased with doses of compounds resulting in minimal to moderate degeneration/necrosis in the expected tubular segments with no or slight changes in BUN and serum creatinine (eg, miR-672-5p and miR-210-3p which were modulated in urine from rats treated with the TAL toxicant compound Y at 400 mg/kg/day on day 7). Only few urinary miRNAs were found altered with the glomerular toxicants puromycin, nephrotoxic serum, HN model, with the proximal tubular toxicant gentamicin, and with the 2 CD toxicants NPAA and compound Z.
Figure 3.
Urinary microRNAs measured in rat studies using TaqMan low-density array A cards.
Identification of Potential Nephron Segment-Specific Urinary miRNAs Biomarker Candidates
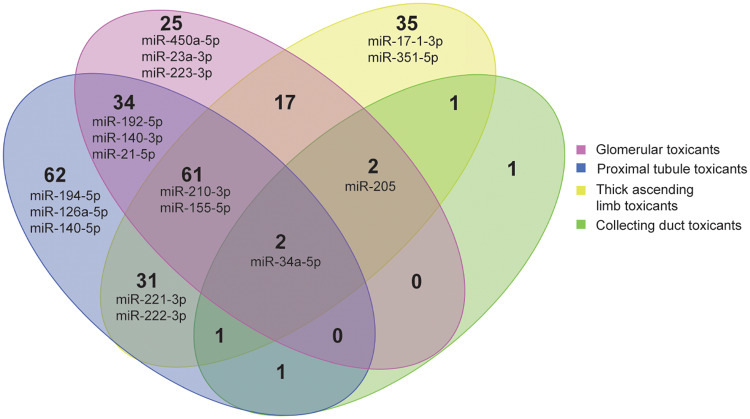
A Venn analysis was carried out to identify significantly increased urinary miRNAs which were common and/or specific between studies with toxicants causing injury to specific nephron segments. Results from this Venn diagram analysis showed that several urinary miRNAs were exclusively increased after treatments of rats with either glomerular toxicants (eg, miR-450a-5p, -223-3p, -23a-3p), proximal tubular toxicants (eg, miR-194-5p, -140-5p, -126a-5p) or loop of Henle toxicants (eg, miR-17-1-3p, -351-5p; see Figure 4 and Supplementary File 5 for full list of significantly increased urinary miRNAs from Venn analysis). Sixteen top miRNA urinary candidates were selected based on the following criteria: (1) significant increases detected with 2 different nephrotoxicants affecting the same nephron segment, and/or (2) highest fold changes (Table 2). For example, miR-140-5p was increased with the proximal tubular toxicants cisplatin and potassium dichromate, whereas miR-17-1-3p was increased the 2 TAL toxicants compounds X and Y. Distinct miRNAs were increased to a greater extent in urine from rats treated with the TAL toxicants versus the proximal tubular toxicants (eg, miR-221-3p and -222-3p; Table 2). miR-210-3p was increased in urine from rats treated with doxorubicin and cisplatin, but it was one of the most strongly increased miRNAs (40-fold) with the TAL toxicants. miR-192-5p was preferentially selected instead of miR-194-5p, even if it was apparently less specific of the proximal tubule because it was increased in the urine about 10-fold more after treatment with the proximal tubular toxicant cisplatin. miR-223-3p was selected as it was highly increased with both the glomerular toxicants doxorubicin in the current analysis and with puromycin in previously published results (Nassirpour et al., 2015). miR-34c-5p was selected as glomerular candidate despite observed increase with cisplatin given that it was highly increased with the glomerular toxicant doxorubicin, thus confirming previously published results (Church et al., 2014). miR-208b was specifically increased in urine from rats treated with the CD toxicant NPAA at day 8. This miRNA was not selected for further characterization because it was not increased with the other CD toxicant tested. Of note, another miRNA miR-34a-5p was increased in urine from rats treated with all types of nephrotoxicants tested regardless the site of lesion in the nephron.
Figure 4.
Venn diagram of urinary microRNAs (miRNAs) in rats treated with segment selective nephrotoxicants. miRNAs highlighted in the diagram correspond to miRNAs candidates further characterized in nephron segments isolated by laser capture microdissection.
Table 2.
Urinary Changes (Fold-Changes vs Respective Controls) of miRNA Candidates in Rats Treated With Site Selective Nephrotoxicants
| Glomerulus Toxicants |
Proximal Tubule Toxicants |
TAL Toxicants |
CD Toxicants |
|||
|---|---|---|---|---|---|---|
| Doxorubicin | Cisplatin | Potassium Dichromate | Sanofi X | Sanofi Y | NPAA | |
| miR-421-5p | — | 47.9 | 3.3 | — | — | — |
| miR-872-5p | — | 17.4 | 2.4 | — | — | — |
| miR-140-5p | — | 3.1 | 3.5 | — | — | — |
| miR-1306 | — | — | — | — | 29.6 | — |
| miR-17-1-3p | — | — | — | 18.0 | 4.7 | — |
| miR-351-5p | — | — | — | 8.2 | 3.7 | — |
| miR-347 | — | — | — | 5.8 | 10.6 | — |
| miR-222-3p | — | 23.5 | — | 38.8 | 6.3 | — |
| miR-221-3p | — | 15.5 | — | 42.8 | - | — |
| miR-210-3p | 28.7 | 650.0 | — | 39.6 | 12.8 | — |
| miR-192-5p | 5.7 | 68.3 | — | — | — | — |
| miR-184 | 76.1 | — | — | — | — | — |
| miR-223-3p a | 41.4 | — | — | — | — | — |
| miR-369-3p | 17.4 | — | — | — | — | — |
| mir-9a-3p | 10.0 | — | — | — | — | — |
| miR-34c-3p | 12.1 | 11.3 | — | — | — | 6.4 |
miRNA candidates selected for further characterization are shown in bold.
miR-223-3p was also previously reported to be increased in the study with the glomerular toxicant puromycin (Nassirpour et al., 2015).
miRNA Expression in Different Nephron Segments Quantified Using ddPCR
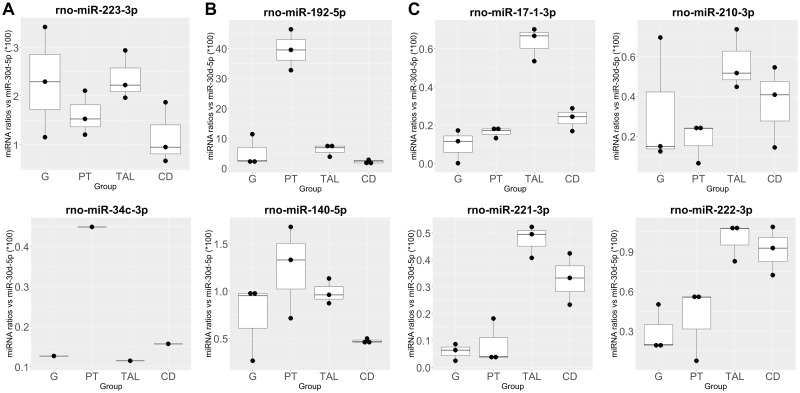
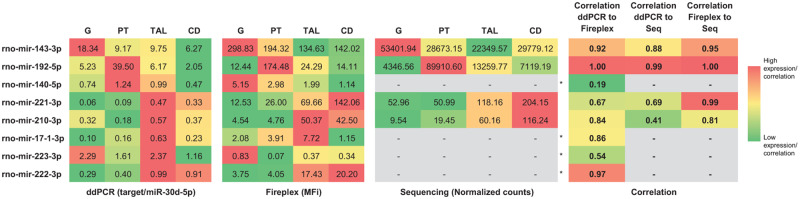
A selected set of 8 miRNA candidates that were highly increased in urine and that presented some degree of nephron segment-specificity based on the current Venn analysis and/or previously published data were selected for further characterization of their localization in nephron segments isolated by LCM using ddPCR (Figure 5 and Supplementary File 6): miR-223-3p and -34c-3p as potential biomarkers of glomerular injury, miR-192-5p and -140-5p as potential biomarkers of proximal tubular injury, miR-17-1-3p, -221-3p, -222-3p, and -210-3p as potential biomarkers of TAL injury. The level of overall expression varied from candidate to candidate; however each of these candidates demonstrated nephron segment enrichment. The highest expressed candidate, miR-192-5p was enriched in the proximal tubule, by at least 6-fold compared with the next highest segment. Other candidates were expressed 2 magnitudes lower or less than miR-192-5p; however, some still showed regional enrichment. For example, miRs-221-3p, -222-3p, -210-3p, and -17-1-3p were 2-fold or greater enriched in the TAL and/or the CD fractions isolated by LCM compared with other segments.
Figure 5.
Digital droplet PCR profiles of selected urinary microRNA top candidates in nephron segments isolated by laser capture microdissection (LCM) in Sprague Dawley rats. Candidates for (A) glomerulus (G), proximal tube (PT), and thick ascending limb (TAL) of the loop of Henle and collecting duct (CD) were measured in their respective nephron segments. Ratios calculated by using miR-30d-5p as normalizer per region and rat, multiplied by 100 to obtain more easily comparable numbers. miR-34c-3p was measured in LCM samples from only 1 rat.
Identification of Nephron Segment-Enriched miRNAs by sRNA-Seq and FirePlex in Nephron Segments Isolated by LCM
A limitation of the current dataset and meta-analysis is that few nephrotoxicants produce highly discrete histopathological lesions, there is often some degree of secondary lesions at other sites, and elevated urinary protein biomarkers can be associated with injury to multiple nephron sites. To increase the chance of finding miRNA candidates that may be associated with specific nephron segment expression but not necessarily with urine enrichment due to secondary effects of chemical exposure, miRNAome-wide measurements were performed with sRNA-seq using independent LCM-isolated SD rat nephron in control samples and validated with medium-scaled candidate measurements using the Abcam FirePlex platform.
sRNA-seq results
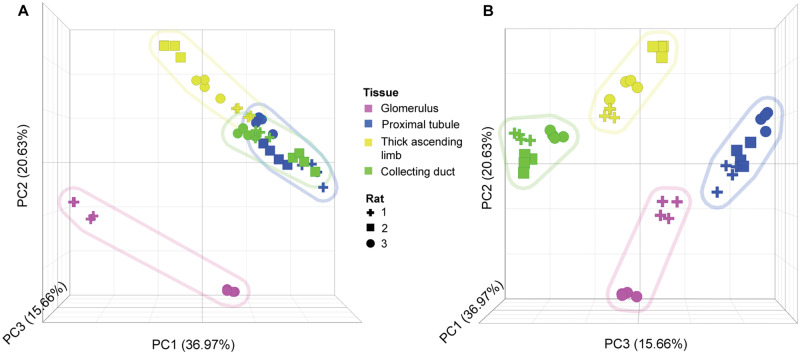
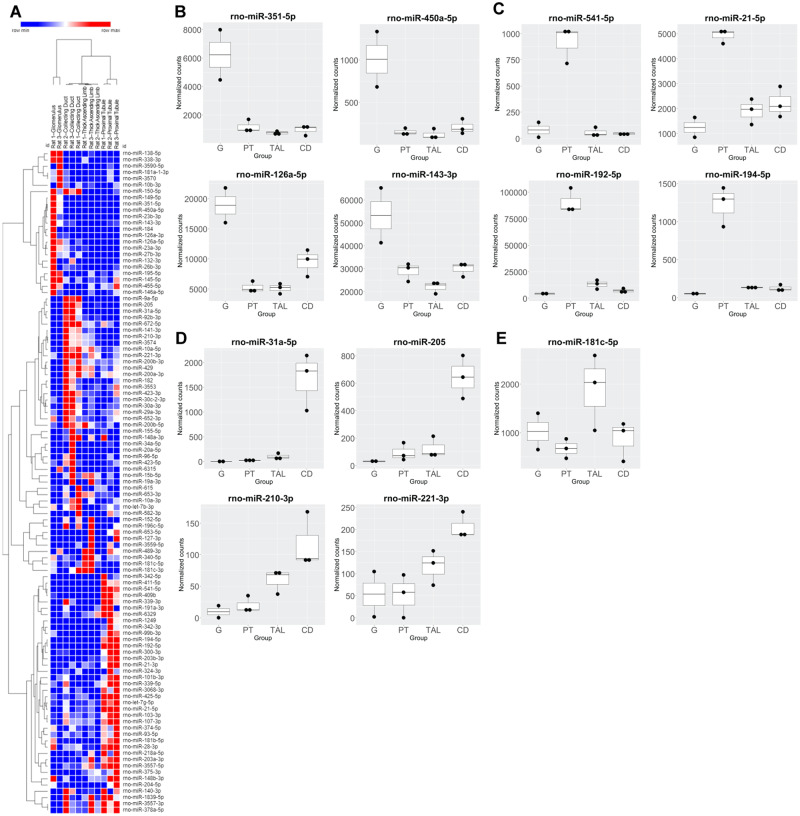
PCA analysis of normalized miRNA count data demonstrated distinct patterns according to different nephron segments (Figure 6). Principal component 2 (PC2) readily distinguished glomerular-derived miRNAs from the other nephron segments, whereas the other segments were distinguished by PC3, indicating more similarity of these segments compared with the glomerulus. To identify segmental concentrated miRNAs, normalized counts were summed for all samples and compared. Segments that commanded 80% or greater of the total reads for a particular miRNA was considered segment specific, whereas segments that demonstrated between 40-80% were considered enriched for that miRNA (Supplementary File 7). This analysis identified 2 and 35 collecting ductal, 2 and 18 glomerular, and 4 and 37 proximal tubular specific and enriched miRNAs, respectively. There were no TAL-specific miRNAs identified, but 10 were found to be enriched in this segment. Not surprisingly, 3 of these were also enriched in the neighboring CD region. This was further indicated by hierarchical clustering of these candidates (Figure 7A). A selection of top candidates, based on specificity/enrichment scores and level of expression, is shown in Figures 7B–E. Taken individually, miR-351-5p, -450a-5p, -126a-5p, 126a-3p, -23a-3p, 27b-3p, and -143-3p were considered enriched and expressed at medium to high levels in the glomerulus. It should be noted that miR-143-3p was also measured by ddPCR in LCM samples and was found to be consistently expressed at a higher level in the glomerulus as compared with the other nephron segments (Supplementary File 6). In the proximal tubule, miR-541-5p was found to be specific for that segment and expressed at moderate counts. In addition, miRs-192-5p, -194-5p, and -21-5p were regarded as enriched in the proximal tubule and expressed at medium to high level, with miR-192-5p was overall highly expressed. In the CD, miR-31a-5p was considered specific and was moderately expressed. A series of miRNAs including miRs-205, -141-3p, -92b-3p, -182, -672-5p, -3553, and -423-3p were found enriched in the CD and were moderately to highly expressed (miRs-182 and -10a-5p were the highest expressed enriched miRNA in the CD region). miR-208b, which was selectively increased in urine from rats treated with the CD toxicant NPAA, was not detected by sRNA-seq experiments in LCM samples. Only one TAL-enriched miRNA demonstrated appreciable expression, rno-miR-181c-5p (Figure 7E).
Figure 6.
Principal component analysis of sRNA-seq normalized microRNA counts in nephron segments isolated by laser capture microdissection (LCM) in Sprague Dawley rats. A, Principal component 1 (PC1; x-axis) and PC2 (y-axis) view. B, PC3 (z-axis) view. First 3 PCs shown, representing 73% of the total variance. Each data point represents different sequencing lanes. Shapes designate individual rat samples. Colors represent different nephron regions (see key). Colored shaded regions represent groupings of each nephron segment sampled via LCM.
Figure 7.
Small RNA-sequencing (sRNA-seq) profiles of selected miRNA top candidates in nephron segments isolated by laser capture microdissection in Sprague Dawley rats. A, Hierarchical clustering of region-specific or -enriched miRNAs based on sRNA-seq. Top (B) glomerulus (G), (C) proximal tube (PT), (D) collecting duct (CD), and (E) thick ascending limb (TAL) candidates of medium to high expression based on normalized counts (>500 in target regions) are shown with the exception of miR-210-3p and -221-3p, which had lower expression but which were identified as potential candidates in the urinary miRNA analysis.
FirePlex assay results and correlation with ddPCR and sRNA-seq data
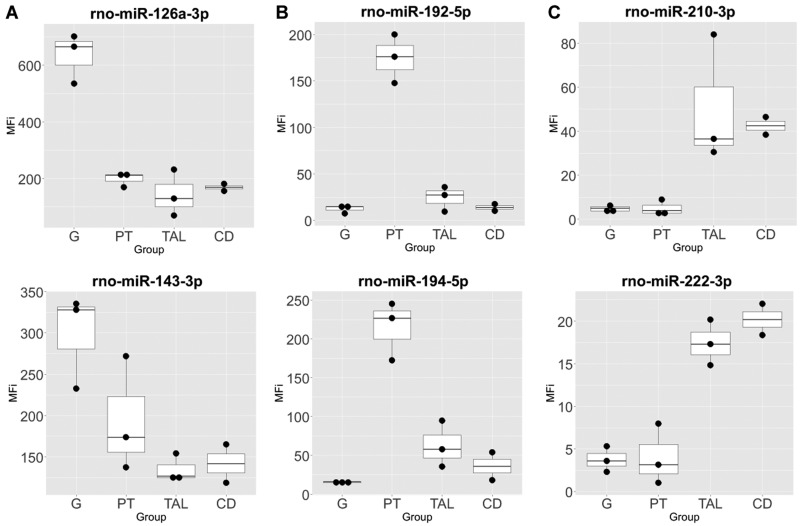
Sixty-four miRNA candidates that demonstrated some level of segmental specificity/enrichment in sRNA-seq and/or were identified as urinary injury miRNA candidates were further characterized using a targeted, multiplexed method (FirePlex) with independently isolated LCM samples (Figure 8 and Supplementary File 8). Approximately one-third of the measured miRNA candidates measured (22 of the 64) demonstrated a strong correlation coefficient to sRNA-seq of 0.8 or greater based on average segmental measurement values (Supplementary File 9). In addition, we measured 8 miRNAs using ddPCR. Four candidates demonstrated a correlation coefficient of 0.8 or greater across all measurements, whereas the other 4 candidates were below the expression cutoff in sRNA-seq (Figure 9). Specifically, miRs-143-3p, -192-5p, -221-3p, and -210-3p demonstrated expression over the cutoff threshold in sRNA-seq. The glomerulus and proximal tube injury candidates, miR-143-3p and -192-5p, respectively, both demonstrated high and well-correlated patterns of expression using all 3 methods. TAL injury candidates, selected on the basis of the urinary miRNA data and Venn analysis (miRs-221-3p, -210-3p, -222-3p, and -17-1-3p), demonstrated low to medium expression and, depending on the platform, displayed a distal expression pattern (TAL and/or CD). Correlation of ddPCR and FirePlex measurements for miR-221-3p was modest (0.67) because of disagreement of a more dominant expression in either distal segments, CD or TAL. The glomerular injury candidate miR-223-3p also demonstrated modest correlation (0.54) driven by dominant TAL expression with ddPCR measurement that was not observed in FirePlex data. Similarly, the proximal tubular injury candidate, miR-140-5p, displayed poor correlation between the 2 measurement methods, only agreeing on suppressed comparative expression in the CD.
Figure 8.
FirePlex profiles of selected microRNA top candidates in nephron segments isolated by laser capture microdissection in Sprague Dawley rats. (A) glomerulus (G), (B) proximal tubule (PT), (C) thick ascending limb (TAL) of the loop of Henle and collecting duct (CD).
Figure 9.
Correlation coefficient comparison between digital droplet PCR, FirePlex, and small RNA-sequencing measurements of nephrotoxicity microRNA (miRNA) candidates across nephron segments isolated by laser capture microdissection in Sprague Dawley rats. Color gradient was adjusted per miRNA per method across 4 nephron segments. Correlation gradient was based on all comparisons. *, raw sequencing count for these miRNA were below cutoff (<100 total counts for all samples).
DISCUSSION
Rat studies were conducted with region-selective nephrotoxicants to identify urinary miRNAs which could report lesion at sites in the nephron for which biomarkers have not previously been characterized. As confirmed by histopathological findings, aided in some cases by urinary protein biomarker profiles, relatively pure site-selective lesions were observed as follows: (1) glomerular lesions with puromycin and with the HN model; (2) proximal tubule lesions with potassium dichromate, cisplatin and gentamicin; (3) TAL of the loop of Henle lesions with the 2 proprietary compounds X and Y; and (4) CD lesions with NPAA and the proprietary compound Z. However, doxorubicin and nephrotoxic serum lead to a mixture of primary glomerular lesions and secondary tubular lesions in rats which complicated the search for nephron segment-specific urinary miRNAs in those studies. In case of compounds X and Y which produced lesions in the TAL but not proximal tubules, the observed increase in urinary microalbumin and Kim-1 was suggestive of functional alterations in the proximal tubules in the absence of histologically detectable lesions.
Several miRNAs were increased in rat urine following exposure to nephrotoxic compounds with histologic evidence characterized by minimal to moderate degeneration/necrosis in the expected nephron segments despite a lack of or only slight changes in serum creatinine and BUN levels. In this cross study analysis, using the same data, there was a lower number of miRNAs judged to have been increased compared with the previously published individual rat studies (Nassirpour et al., 2014, 2015; Pavkovic et al., 2015). The comparative numbers are; with puromycin on day 3 (0 vs 11 modulated miRNAs), with HN model on day 16 (1 vs 11 modulated miRNAs), with nephrotoxic serum in SD rats on day 14 (4 vs 28 modulated miRNAs), and with gentamicin on day 7 (3 vs 20 modulated miRNAs). This can be explained at least in part by the fact that the analysis workflow used in this work was more stringent with respect to a fold change cutoff of a miRNA to be selected as modulated, and/or due to the fact that in the current analysis samples from animals with severe lesions, eg, with puromycin were excluded. Indeed, the fold-change cutoff in the prior published studies with gentamicin, puromycin, and the HN model was 1.5 instead of 2.0 in this analysis. In addition, a more relaxed adjusted p value cutoff to < .15 instead of .05 was used in the previously published studies with puromycin and the HN model. Another reason to explain the differences in the number of modulated urinary miRNAs between this work versus previously published results may be related to variation in the methods of normalization used. In any case, more stringent criteria were used in the present workflow to decrease the risk of false positives in the subsequent meta-analysis.
Six miRNAs were found significantly increased in urine from rats treated with the CD toxicant NPAA in this study. On the other hand, Glineur et al. (2018) reported that the expression profiles of 36 different urinary miRNAs were significantly altered in male Wistar rats treated with NPAA at 500 mg/kg/day for up to 28 days, with miR-210-3p displaying the most robust changes. These discrepancies may be explained at least in part by a higher susceptibility of Wistar rats to the kidney lesions induced by NPAA (Betton et al., 2012) as compared with SD rats which were used in this study. In addition, a lower dose of NPAA was used in this work (400 mg/kg/day for 14 days).
A meta-analysis was carried out across rat studies to identify significantly modulated urinary miRNAs which were common and/or specific across the rat studies. In this approach, the regional enrichment of urinary miRNA leakage markers is linked to renal histopathology findings and elevated urinary protein biomarkers that are associated with injury to multiple nephron sites. Consequently, assessment of site specificity of urinary miRNAs was complicated by observation of “collateral damage” (although minimal) to surrounding nephron segments. To further characterize the nephron segment localization of potential urinary miRNA injury marker candidates, additional experiments were conducted in nephron segments isolated by LCM. Results of the meta-analysis across rat studies suggest that a subset of miRNAs is associated with some degree of kidney segment specificity for tubular injury. Indeed, urinary miR-192-5p was preferentially increased upon treatment with proximal tubule toxicants, whereas urinary miR-17-1-3p was selectively increased upon treatment with TAL toxicants. This was supported by data from LCM samples showing that (1) miR-192-5p was clearly enriched in proximal tubules in accordance with previously published data (Kriegel et al., 2013b) and (2) ddPCR data from LCM samples showed a trend for enrichment of miR-17-1-3p in the TAL fraction. Additional miRNAs were increased to a greater extent in urine from rats treated with TAL versus proximal tubule toxicants (eg, miRs-221-3p and -222-3p), which is in agreement with data from LCM experiments showing that these 2 latter miRNAs were overall preferentially enriched in the TAL and/or CD fractions versus the proximal tubules. Interestingly, miR-210-3p was clearly increased in urine from rats treated with TAL toxicants and was also previously reported to be increased in urine from rats treated with the CD toxicant NPAA (Glineur et al., 2018). This is in accordance with results from our localization experiments with LCM samples showing that miR-210-3p was enriched in the TAL and/or CD fractions. These results were also in accordance with the observation that miRs-221-3p, 222-3p, and -210-3p were more abundant in the kidney medulla versus cortex in a previous rat miRNA body atlas study (Smith et al., 2016; Aaron Smith, personal communication). On the other hand, miRs-223-3p and -34c-3p which were increased in urine from rats treated with the glomerular toxicant doxorubicin were not found enriched in glomeruli isolated by LCM from naive rats. It is hypothesized that miRs-223-3p and -34c-3p are expressed at very low level in basal conditions and that their observed increase in urine after treatment with doxorubicin may result from induction of their intracellular miRNA abundance at the gene expression level; however, future studies would need to explore this further.
Several miRNAs not identified following meta-analysis of urine from the rat studies using TLDA cards were found using sRNA-seq and FirePlex-based measurements to be specific or enriched in nephron segments isolated by LCM. This has allowed identification of novel kidney miRNAs which can potentially be additional candidates for kidney-region-specific urinary miRNAs. Specifically, miR-126a and -143-3p were consistently enriched in the glomerulus using sRNA-seq and FirePlex platforms—and also ddPCR for miR-143-3p—in agreement with previously published results showing that miR-126 is a glomerular-endothelial-specific miRNA and that miR-143-3p is a podocyte specific miRNA (Muller-Deile et al., 2019). We also found that miR-23a-3p was more abundant in the glomerulus as compared with the other nephron segments. We retrospectively examined whether these latter 3 miRNAs were increased in urine from animals exposed to glomerular toxicants and found that miR-23a-3p was increased in urine from rats treated with the glomerular toxicant doxorubicin but not with compounds affecting other segments of the nephron. For the 2 other miRNAs, there was no clear link between their enrichment in the glomerulus and a release in urine in relation with nephron segment site selectivity of the nephrotoxicants used in the study. Indeed, miR-126a-5p was increased in urine from rats treated with proximal tubule toxicants exclusively. Although miR-143-3p was not measured in the urine from rats treated with nephrotoxic serum given that TLDA card B was not analyzed in this study, miR-143-3p was not found to be significantly increased in urine from rats treated with any of the other nephrotoxicants tested in the rat HESI studies. On the other hand, miR-126a-5p and -143-3p were both found increased in total urine and exosome-enriched urine fractions of ZSF1 obese vs ZSF1 lean rats (H. Ellinger-Ziegelbauer, personal communication). Obese ZSF1 rats are associated with diabetic nephropathy characterized by both tubular and glomerular injury (Su et al., 2016), and thus increased levels of miR-126a and -143-3p in urine of the ZSF1 obese rats together with the LCM results reported in this manuscript suggest that these miRNAs can appear in urine and are derived at least partly from the glomerular region. The data also suggest that exosome enrichment may increase the sensitivity to detect at least certain miRNAs. As another example, miR-423-3p was found consistently enriched in CD using sRNA-seq and FirePlex platforms. However, miR-423-3p was not increased in urine from rats treated with the 2 nephrotoxicants which induced lesions in the CD (NPAA and compound Z). On the other hand, miR-205 was found to be enriched in the CD by sRNA-seq, and was significantly increased in urine from rat treated with compound Z. More experiments, including in situ detection of miRNA in the tissue samples to visualize the nephron segment specific distribution, are needed to further explore those novel urinary miRNAs which were concomitantly enriched within a given nephron segment and increased in urine from rats treated with the corresponding nephron segment selective toxicant.
Urinary abundance of several miRNAs was found to be similarly altered in rats exposed to toxicants affecting different nephron segments, thus suggesting a lack of region-specificity of these miRNAs. For example, miR-34a-5p, a known p53-target (Navarro and Lieberman, 2015), was increased in urine from rats exposed to toxicants affecting all 4 nephron segments under investigation. miR-34a-5p thus may have value only as a general kidney injury marker. miR-155-5p, which is involved in the regulation of the injury response in kidneys (Pellegrini et al., 2014), was increased in urine from rats exposed to nephrotoxicants that induce lesions in the glomeruli, the proximal tubules, and the TAL in this study. Conversely, miRs-34a-5p and -155-5p were detected at very low levels in nephron segments isolated by LCM from control rats. This suggests that the increase of miRs-34a-5p and -155-5p in urine upon treatment with nephrotoxicants affecting different nephron segments may be related to their biological function and thus induction at the gene expression level in the kidney, regardless of the localization of the kidney lesions. Interestingly, another miRNA, miR-21-5p which has an important role in kidney fibrosis (Chau et al., 2012), was increased in the urine from rats exposed to cisplatin and doxorubicin but not with the TAL toxicants, and was found enriched in the proximal tubules in sRNA-seq experiments. This suggests that in the case of miR-21-5p there is some degree of proximal nephron region specificity at both constitutive and gene expression level.
Another challenge when evaluating urinary miRNAs as biomarkers of DIKI is the potential lack of kidney organ specificity. Indeed, although kidney enriched miRNAs have been identified, none seems to be solely specific to the kidney (Nassirpour et al., 2016; Smith et al., 2016). As an example, miR-192-5p which is enriched in the kidney is also expressed in other organs including liver, duodenum, jejunum, and ileum. miR-126 is an endothelial-specific miRNA which modulates angiogenesis in vivo (Wang et al., 2008). miR-210-3p has been found significantly overexpressed in different human malignancies, including breast and lung cancers (Pasculli et al., 2019). In addition, whereas urine is directly associated with the kidney and thus was assumed to be the most proximal noninvasive biofluid to find kidney specific miRNAs, miRNAs have also been reportedly increased in urine following injury to organs other than the kidney, eg, the liver (Wolenski et al., 2017; Yang et al., 2012) or the heart (Cheng et al., 2012; Zhou et al., 2013). These findings were not corroborated; however, with similar studies where increases of plasma miRNAs following liver or heart damage did not result in corresponding miRNA increases in urine (Pavkovic et al., 2014; Thompson et al., 2016). Importantly, these discrepancies highlight that further development of miRNAs as site-specific kidney biomarkers will require confirmatory studies assessing organ specificity, as well as investigation of renal clearance of miRNAs present in plasma.
To determine nephron segment specificity, we utilized 3 different methods of detection: ddPCR, the Abcam FirePlex assay, and sRNA-seq for analysis of LCM samples. These technologies represent targeted, panel, and global approaches for miRNA measurements and we highlighted those miRNA measurements that agreed among the data obtained by these methods. Indeed, when determining nephron specificity or enrichment of miRNAs, there was general consensus—33 of the 56 (59%) exhibited moderate to strong correlation (>0.50) when comparing average expression between nephron segments for a miRNA measured on the FirePlex and sRNA-seq platforms. In addition, a strong average correlation coefficient (0.84) for segment miRNA enrichment was observed among the 4 miRNAs detected by all 3 methods (Figure 9). However, discrepancies were noted, especially when comparing rank-ordered expression of miRNAs in individual segments for miRNAs commonly measured by FirePlex and sRNA-seq (average correlation coefficient of 0.10). There are many potential factors that could contribute to these discrepancies, including natural variation due to different samples used, differences in lengths of storage and transit before use, differences between direct lysate measurements (FirePlex) versus RNA isolated before sample measurement (sRNA-seq) and associated stabilities and recovery of certain miRNAs (Brown et al., 2018), and utilization of different normalization and analysis approaches associated with platform-specific data, among other reasons. These factors could have certainly influenced our final measurements in the LCM samples, as well as the urinary miRNAs assessed, thereby limiting the number of biomarker candidates that may be appropriate for further development to identify nephron-specific toxicity. It was, however, beyond the scope of this study to comprehensively compare and contrast the relative advantages and disadvantages of these techniques. We simply took the approach to prioritize potential miRNA candidates that exhibited nephron enrichment when consistent with multiple measurements; but it should be noted that some candidates may have been overlooked due to differences in limits of detection or variation. Until more is known about relative miRNA stabilities and influences of the method utilized to detect for candidate miRNAs, multiple methods should be employed to lend higher confidence in miRNA measurements when determining relative expression levels.
CONCLUSION
Several urinary miRNAs were increased following administration of site-selective nephrotoxicants resulting in minimal to moderate degeneration/necrosis in the expected segments of the nephron. Meta-analysis of the rat studies identified miR-192-5p as a potential nephron segment specific urinary miRNA candidate for the proximal tubule. This result was aligned with miRNA measurements obtained in nephron segments isolated by LCM showing that miR-192-5p was enriched in the proximal tubule. Discriminative miRNAs such as miR-221-3p and -222-3p were increased to a higher extent in urine from rats treated with at least 1 of the 2 the TAL toxicants tested versus proximal tubule toxicants in accordance with data from LCM experiments showing that these miRNAs were preferentially enriched in the TAL and/or CD fractions versus the proximal tubules. Urinary miR-210-3p, which was highly increased upon treatment with TAL-specific toxicants, was also enriched in the TAL and/or CD fractions versus proximal tubule. miR-23a-3p which was enriched in the glomerulus, was increased in urine collected from rats treated with doxorubicin, a glomerular toxicant, but not with nephrotoxicants affecting other segments of the nephron. Taken together, these results suggest that a weighted panel of urinary miRNAs with preferential or at least partial localization in the glomerulus (eg, miR-23a-3p), proximal tubule (eg, miR-192-5p), TAL (eg, miRs-221-3p, -22-3p, and -210-3p), and CD (eg, miR-210-3p) may be useful to discriminate the localization of drug-induced kidney lesions. Other urinary miRNA candidates that demonstrated regional nephron toxicity and enrichment in sRNA-seq/FirePlex measurements (such as miR-450a-5p in glomerulus, miR-194-5p in the proximal tubule, and miR-17-1-3p in TAL), are also worth future consideration as part of these panels. Evaluation using a panel approach could reduce the risk associated with a potential lack of kidney organ specificity of individual urinary miRNA candidates. Additional studies are warranted to assess utility of kidney nephron region enriched miRNAs and associated fold-changes cutoff as urine-based biomarkers of DIKI in rats and additional preclinical species like dog and Cynomolgus monkey, and to evaluate if these miRNAs candidates are translatable to kidney injury detection in humans.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
Supplementary Material
ACKNOWLEDGMENTS
The authors gratefully acknowledge Carolina Morell-Perez of the Health and Environmental Sciences Institute for her enduring support of the committee work and resulting publications. We would like to thank Claire Mariet, Lydie Pilatre et Martine Courcol from Sanofi for technical assistance. We also would like to thank Deidre Dalmas, Gregory Travlos, and Julie Foley for critical reading of the article. In addition, we appreciate support from B. Alex Merrick and Julie Foley of NIEHS and Dr Matthew Toma of Covance Genomics Laboratory for technical support of sRNA-seq. Members of the Health and Environmental Sciences Institute (HESI) Biomarkers of Nephrotoxicity Technical Committee. HESI is a nonprofit organization whose mission is to collaboratively create science-based solutions to various challenges affecting human health and the environment. HESI’s activities are organized around scientific committees comprised of scientists from academia, NGOs, government and industry.
FUNDING
This work was provided for in-kind by the HESI Biomarkers of Nephrotoxicity Technical Committee, which is supported by sponsorships from member companies. HESI’s scientific initiatives are primarily supported by the in-kind contributions (from public and private sector participants) of time, expertise, and experimental effort. Small RNA-sequencing was supported by NIEHS Contract (HHSN273201500005I). These contributions are supplemented by direct funding (that primarily supports program infrastructure and management) provided primarily by HESI’s corporate sponsors. The content of this article is the sole responsibility of the authors and does not necessarily represent the official views or policy of the U.S. Environmental Protection Agency, the National Institutes of Health, or the U.S. Government.
DECLARATION OF CONFLICTING INTERESTS
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
REFERENCES
- Aguado-Fraile E., Ramos E., Conde E., Rodríguez M., Martín-Gómez L., Lietor A., Candela Á., Ponte B., Liaño F., García-Bermejo M. L. (2015). A pilot study identifying a set of microRNAs as precise diagnostic biomarkers of acute kidney injury. PLoS One 10, e0127175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauchet A. L., Masson R., Guffroy M., Slaoui M. (2011). Immunohistochemical identification of kidney nephron segments in the dog, rat, mouse, and cynomolgus monkey. Toxicol. Pathol. 39, 1115–1128. [DOI] [PubMed] [Google Scholar]
- Betton G. R., Ennulat D., Hoffman D., Gautier J. C., Harpur E., Pettit S. (2012). Biomarkers of collecting duct injury in Han-Wistar and Sprague-Dawley rats treated with N-phenylanthranilic acid. Toxicol. Pathol. 40, 682–694. [DOI] [PubMed] [Google Scholar]
- Bhatt K., Mi Q. S., Dong Z. (2011). microRNAs in kidneys: Biogenesis, regulation, and pathophysiological roles. Am. J. Physiol. Renal Physiol. 300, F602–F610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenburger T., Lorenzen J. M. (2020). Diagnostic and therapeutic potential of microRNAs in acute kidney injury. Front. Pharmacol. 11, 657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R. A. M., Epis M. R., Horsham J. L., Kabir T. D., Richardson K. L., Leedman P. J. (2018). Total RNA extraction from tissues for microRNA and target gene expression analysis: Not all kits are created equal. BMC Biotechnol. 18, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau B. N., Xin C., Hartner J., Ren S., Castano A. P., Linn G., Li J., Tran P. T., Kaimal V., Huang X., et al. (2012). MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Sci. Transl. Med. 4, 121ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Wang X., Yang J., Duan X., Yao Y., Shi X., Chen Z., Fan Z., Liu X., Qin S., et al. (2012). A translational study of urine miRNAs in acute myocardial infarction. J. Mol. Cell Cardiol. 53, 668–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church R. J., McDuffie J. E., Sonee M., Otieno M., Ma J. Y., Liu X., Watkins P. B., Harrill A. H. (2014). MicroRNA-34c-3p is an early predictive biomarker for doxorubicin-induced glomerular injury progression in male Sprague-Dawley rats. Toxicol. Res. 3, 384–394. [Google Scholar]
- Cook D., Brown D., Alexander R., March R., Morgan P., Satterthwaite G., Pangalos M. N. (2014). Lessons learned from the fate of AstraZeneca’s drug pipeline: A five-dimensional framework. Nat. Rev. Drug Discov. 13, 419–431. [DOI] [PubMed] [Google Scholar]
- EMA. (2016). EMA Letter of Support for Drug-Induced Renal Tubular Injury Biomarker(s). Available at: http://www.ema.europa.eu/docs/en_GB/document_library/Other/2017/01/WC500219666.pdf. Accessed December, 21 2020.
- Ennulat D., Adler S. (2015). Recent successes in the identification, development, and qualification of translational biomarkers: The next generation of kidney injury biomarkers. Toxicol Pathol 43, 62–69. [DOI] [PubMed] [Google Scholar]
- FDA. (2016). FDA Letter of support for drug-induced (DIKI) renal tubular injury biomarker(s). Available at: https://www.fda.gov/media/102623/download. Accessed December, 21 2020.
- Friedman R. C., Farh K. K., Burge C. B., Bartel D. P. (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier J. C., Gury T., Guffroy M., Khan-Malek R., Hoffman D., Pettit S., Harpur E. (2014). Normal ranges and variability of novel urinary renal biomarkers in Sprague-Dawley Rats: Comparison of constitutive values between males and females and across assay platforms. Toxicol. Pathol. 42, 1092–1104. [DOI] [PubMed] [Google Scholar]
- Glineur S. F., Hanon E., Dremier S., Snelling S., Berteau C., De Ron P., Nogueira da Costa A. (2018). Assessment of a urinary kidney microRNA panel as potential nephron segment-specific biomarkers of subacute renal toxicity in preclinical rat models. Toxicol. Sci. 166, 409–419. [DOI] [PubMed] [Google Scholar]
- Harpur E., Ennulat D., Hoffman D., Betton G., Gautier J. C., Riefke B., Bounous D., Schuster K., Beushausen S., Guffroy M., et al. ; on behalf of the HESI Committee on Biomarkers of Nephrotoxicity. (2011). Biological qualification of biomarkers of chemical-induced renal toxicity in two strains of male rat. Toxicol. Sci. 122, 235–252. [DOI] [PubMed] [Google Scholar]
- Hoste E. A., Bagshaw S. M., Bellomo R., Cely C. M., Colman R., Cruz D. N., Edipidis K., Forni L. G., Gomersall C. D., Govil D., et al. (2015). Epidemiology of acute kidney injury in critically ill patients: The multinational AKI-EPI study. Intensive Care Med. 41, 1411–1423. [DOI] [PubMed] [Google Scholar]
- Khan S., Loi V., Rosner M. H. (2017). Drug-induced kidney injury in the elderly. Drugs Aging 34, 729–741. [DOI] [PubMed] [Google Scholar]
- Kriegel A. J., Liang M. (2013a). MicroRNA in situ hybridization for formalin fixed kidney tissues. J. Vis. Exp. (81), 50785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegel A. J., Liu Y., Liu P., Baker M. A., Hodges M. R., Hua X., Liang M. (2013b). Characteristics of microRNAs enriched in specific cell types and primary tissue types in solid organs. Physiol. Genomics 45, 1144–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mall C., Rocke D. M., Durbin-Johnson B., Weiss R. H. (2013). Stability of miRNA in human urine supports its biomarker potential. Biomark. Med. 7, 623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Deile J., Dannenberg J., Liu P., Lorenzen J., Nystrom J., Thum T., Schiffer M. (2019). Identification of cell and disease specific microRNAs in glomerular pathologies. J. Cell Mol. Med. 23, 3927–3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassirpour R., Homer B. L., Mathur S., Li Y., Li Z., Brown T., Carraher D., Warneke J., Bailey S., Percival K., et al. (2015). Identification of promising urinary microRNA biomarkers in two rat models of glomerular injury. Toxicol. Sci. 148, 35–47. [DOI] [PubMed] [Google Scholar]
- Nassirpour R., Mathur S., Gosink M. M., Li Y., Shoieb A. M., Wood J., O’Neil S. P., Homer B. L., Whiteley L. O. (2014). Identification of tubular injury microRNA biomarkers in urine: Comparison of next-generation sequencing and qPCR-based profiling platforms. BMC Genomics 15, 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassirpour R., Ramaiah S. K., Whiteley L. O. (2016). Nephron segment specific microRNA biomarkers of pre-clinical drug-induced renal toxicity: Opportunities and challenges. Toxicol. Appl. Pharmacol. 312, 34–41. [DOI] [PubMed] [Google Scholar]
- Navarro F., Lieberman J. (2015). miR-34 and p53: New insights into a complex functional relationship. PLoS One 10, e0132767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasculli B., Barbano R., Rendina M., Fontana A., Copetti M., Mazza T., Valori V. M., Morritti M., Maiello E., Graziano P., et al. (2019). Hsa-miR-210-3p expression in breast cancer and its putative association with worse outcome in patients treated with Docetaxel. Sci. Rep. 9, 14913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel J. B., Sapra A. (2020). Nephrotoxic medications. In: StatPearls. StatPearls, Treasure Island (FL: ). [Google Scholar]
- Pavkovic M., Vaidya V. S. (2016). MicroRNAs and drug-induced kidney injury. Pharmacol. Ther. 163, 48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavkovic M., Riefke B., Ellinger-Ziegelbauer H. (2014). Urinary microRNA profiling for identification of biomarkers after cisplatin-induced kidney injury. Toxicology 324, 147–157. [DOI] [PubMed] [Google Scholar]
- Pavkovic M., Riefke B., Frisk A. L., Groticke I., Ellinger-Ziegelbauer H. (2015). Glomerulonephritis-induced changes in urinary and kidney microRNA profiles in rats. Toxicol. Sci. 145, 348–359. [DOI] [PubMed] [Google Scholar]
- Pellegrini K. L., Han T., Bijol V., Saikumar J., Craciun F. L., Chen W. W., Fuscoe J. C., Vaidya V. S. (2014). MicroRNA-155 deficient mice experience heightened kidney toxicity when dosed with cisplatin. Toxicol. Sci. 141, 484–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran K., Saikumar J., Bijol V., Koyner J. L., Qian J., Betensky R. A., Waikar S. S., Vaidya V. S. (2013). Human miRNome profiling identifies microRNAs differentially present in the urine after kidney injury. Clin. Chem. 59, 1742–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikumar J., Hoffmann D., Kim T. M., Gonzalez V. R., Zhang Q., Goering P. L., Brown R. P., Bijol V., Park P. J., Waikar S. S., et al. (2012). Expression, circulation, and excretion profile of microRNA-21, -155, and -18a following acute kidney injury. Toxicol. Sci. 129, 256–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sistare F. D., Dieterle F., Troth S., Holder D. J., Gerhold D., Andrews-Cleavenger D., Baer W., Betton G., Bounous D., Carl K., et al. (2010). Towards consensus practices to qualify safety biomarkers for use in early drug development. Nat. Biotechnol. 28, 446–454. [DOI] [PubMed] [Google Scholar]
- Smith A., Calley J., Mathur S., Qian H. R., Wu H., Farmen M., Caiment F., Bushel P. R., Li J., Fisher C., et al. (2016). The Rat microRNA body atlas; Evaluation of the microRNA content of rat organs through deep sequencing and characterization of pancreas enriched miRNAs as biomarkers of pancreatic toxicity in the rat and dog. BMC Genomics 17, 694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z., Widomski D., Ma J., Namovic M., Nikkel A., Leys L., Olson L., Salte K., Donnelly-Roberts D., Esbenshade T., et al. (2016). Longitudinal changes in measured glomerular filtration rate, renal fibrosis and biomarkers in a rat model of type 2 diabetic nephropathy. Am. J. Nephrol. 44, 339–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susantitaphong P., Cruz D. N., Cerda J., Abulfaraj M., Alqahtani F., Koulouridis I., Jaber B. L.; Acute Kidney Injury Advisory Group of the American Society of Nephrology. (2013). World incidence of AKI: A meta-analysis. Clin. J. Am. Soc. Nephrol. 8, 1482–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson K. L., Boitier E., Chen T., Couttet P., Ellinger-Ziegelbauer H., Goetschy M., Guillemain G., Kanki M., Kelsall J., Mariet C., et al. (2016). Absolute measurement of cardiac injury-induced microRNAs in biofluids across multiple test sites. Toxicol. Sci. 154, 115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasakova K., Erdos Z., Troth S. P., McNulty K., Chapeau-Campredon V., Mokrzycki N., Muniappa N., Gu Y. Z., Holder D., Bailey W. J., et al. (2014). Evaluation of the relative performance of 12 urinary biomarkers for renal safety across 22 rat sensitivity and specificity studies. Toxicol. Sci. 138, 3–20. [DOI] [PubMed] [Google Scholar]
- Wang S., Aurora A. B., Johnson B. A., Qi X., McAnally J., Hill J. A., Richardson J. A., Bassel-Duby R., Olson E. N. (2008). The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell 15, 261–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolenski F. S., Shah P., Sano T., Shinozawa T., Bernard H., Gallacher M. J., Wyllie S. D., Varrone G., Cicia L. A., Carsillo M. E., et al. (2017). Identification of microRNA biomarker candidates in urine and plasma from rats with kidney or liver damage. J. Appl. Toxicol. 37, 278–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Greenhaw J., Shi Q., Su Z., Qian F., Davis K., Mendrick D. L., Salminen W. F. (2012). Identification of urinary microRNA profiles in rats that may diagnose hepatotoxicity. Toxicol. Sci. 125, 335–344. [DOI] [PubMed] [Google Scholar]
- Zhou X., Mao A., Wang X., Duan X., Yao Y., Zhang C. (2013). Urine and serum microRNA-1 as novel biomarkers for myocardial injury in open-heart surgeries with cardiopulmonary bypass. PLoS One 8, e62245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.