Abstract
Objectives
As the world transitions into a new era of the COVID‐19 pandemic in which vaccines become available, there is an increasing demand for rapid reliable serological testing to identify individuals with levels of immunity considered protective by infection or vaccination.
Methods
We used 34 SARS‐CoV‐2 samples to perform a rapid surrogate virus neutralisation test (sVNT), applicable to many laboratories as it circumvents the need for biosafety level‐3 containment. We correlated results from the sVNT with five additional commonly used SARS‐CoV‐2 serology techniques: the microneutralisation test (MNT), in‐house ELISAs, commercial Euroimmun‐ and Wantai‐based ELISAs (RBD, spike and nucleoprotein; IgG, IgA and IgM), antigen‐binding avidity, and high‐throughput multiplex analyses to profile isotype, subclass and Fc effector binding potential. We correlated antibody levels with antibody‐secreting cell (ASC) and circulatory T follicular helper (cTfh) cell numbers.
Results
Antibody data obtained with commercial ELISAs closely reflected results using in‐house ELISAs against RBD and spike. A correlation matrix across ten measured ELISA parameters revealed positive correlations for all factors. The frequency of inhibition by rapid sVNT strongly correlated with spike‐specific IgG and IgA titres detected by both commercial and in‐house ELISAs, and MNT titres. Multiplex analyses revealed strongest correlations between IgG, IgG1, FcR and C1q specific to spike and RBD. Acute cTfh‐type 1 cell numbers correlated with spike and RBD‐specific IgG antibodies measured by ELISAs and sVNT.
Conclusion
Our comprehensive analyses provide important insights into SARS‐CoV‐2 humoral immunity across distinct serology assays and their applicability for specific research and/or diagnostic questions to assess SARS‐CoV‐2‐specific humoral responses.
Keywords: antibody‐secreting cells, ELISA, neutralisation assay, SARS‐CoV‐2 antibodies, T follicular helper cells
We describe robust correlations across six SARS‐CoV‐2 serology assays detecting distinct antibody features. Our comprehensive analyses provide important insights into SARS‐CoV‐2 humoral immunity across distinct serology assays and their applicability for specific research and/or diagnostic questions to assess SARS‐CoV‐2‐specific humoral responses.

Introduction
The coronavirus disease 2019 (COVID‐19) pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) has infected > 100 million individuals, caused over 2 million deaths 1 and greatly disrupted economies worldwide. While awaiting global vaccination programmes, countries have focused on rapid testing, isolating and contact tracing of SARS‐CoV‐2‐infected individuals using the nucleic acid amplification test (NAAT; RT‐PCR), 2 , 3 while implementing social distancing and lockdown measures to reduce virus transmission and prevent healthcare systems from being over‐whelmed with COVID‐19 patients. 4 , 5 Concurrently, scientists have endeavoured to unravel the correlates of protective immunity 2 , 6 , 7 and there has been an unprecedented effort to develop and produce vaccines. 8 The world is now transitioning into a new stage of the pandemic, in which vaccines should become available globally. It is likely that identifying individuals with protective immunity induced by infection or vaccination, and whether previously induced immune responses are protective against potential new variants, will assume increasing importance. 9 Furthermore, from a scientific standpoint, one of the key knowledge gaps is related to the duration of protective immunity in the population after infection and/or vaccination. This will be closely monitored in the years to come. Thus, from both a diagnostic and scientific standpoint, there will be an increasing demand for reliable rapid serological tests.
In general, virus neutralising antibodies measured by a plaque‐reduction neutralisation test (PRNT) 10 or a virus microneutralisation test (MNT) 11 , 12 are considered the gold standard correlate of protection, 13 although it is currently unclear what titre is required for absolute protection from SARS‐CoV‐2 infection. These assays detect antibodies that can inhibit viral entry and thus can prevent reinfection with the same virus. 13 However, SARS‐CoV‐2‐specific PRNT and MNT can only be performed in biosafety level (BSL) 3 containment laboratories by specially trained personnel and take up to 5 days to perform. Furthermore, as they are biological assays, samples must be run in the same assay for accurate comparisons between samples. These assays are also limited by their inability to discriminate different antibody isotypes, and to detect antibodies directed against other non‐neutralising epitopes known to contribute to protection against severe disease. 14
Alternative serology assays are required to gain a rapid and comprehensive profile of humoral responses against SARS‐CoV‐2. Recent advances include the use of (1) the surrogate virus neutralisation test (sVNT), a competition enzyme‐linked immunosorbent assay (ELISA) which uses purified receptor binding domain (RBD) of the spike (S) protein and host cell receptor angiotensin‐converting enzyme 2 (ACE2) to mimic the virus–host interaction 15 , 16 and allows detection of antibodies that block RBD binding to ACE2 without the need for infectious virus or cells and therefore only requiring BSL 2 containment, (2) ELISAs which detect SARS‐CoV‐2‐specific antibodies of different isotypes directed towards viral proteins such as the full S protein, RBD and nucleocapsid (N) protein, 11 (3) multiplex‐based serology allowing detection of different antibody isotypes against a large set of viral antigens and provides information on Fc‐receptor specificity, while requiring small sera or plasma volumes, 14 , 17 (4) chaotropic‐based dissociation assays to measure antigen‐binding avidity, and (5) commercial semi‐quantitative ELISA systems, including assays developed by Euroimmun (Lübeck, Germany) and Wantai (Beijing, China) to detect the level of SARS‐CoV‐2‐specific antibodies in patient samples. 18 , 19 , 20 , 21 These assays are rapid, circumvent the use of infectious virus and cells, and detect responses against a broader range of viral epitopes, thus revealing and identifying different antibody features beyond virus neutralisation that contribute to the prevention of infection and/or severe disease.
Although some studies have compared the specificity and sensitivity of different diagnostic tests, 22 , 23 , 24 , 25 , 26 relatively limited data are available on correlations between distinct serology assays and understanding how they relate to the classical MNT readout. Such comparisons are needed to understand the applicability of SARS‐CoV‐2 serology assays for specific research questions. In our study, we used a cohort of 34 serum/plasma samples from 15 COVID‐19 patients to perform a rapid sVNT which is highly applicable to many laboratories as it circumvents the need for BSL 3 containment. We further correlated the antibody data obtained from the sVNT to five additional commonly used SARS‐CoV‐2 serology techniques as described above for the same COVID‐19 patient samples. These included a MNT, in‐house and commercial Euroimmun‐ and Wantai‐based ELISAs (RBD, S and N; IgG, IgA and IgM), antigen‐binding avidity assays and serology multiplex analysis. 6 , 11 , 14 Furthermore, we correlated the antibody levels obtained from these assays with cellular immune responses important for antibody production, antibody‐secreting cells (ASC) and circulatory T follicular helper (cTfh) cells. Our comprehensive analyses provide important insights into SARS‐CoV‐2 humoral immunity across distinct serology assays as well as their applicability for specific research questions to assess humoral immunity in COVID‐19 clinical samples.
Results
COVID‐19 patient IgM, IgG and IgA titres correlate between commercial and in‐house ELISAs
Our study included 34 SARS‐CoV‐2 samples from 15 PCR‐confirmed COVID‐19 cases. Seven patients were recruited in hospital during acute infection (with three being on supplemental oxygen), and eight patients from the community during convalescence. The median age of the COVID‐19 patients was 57 (range 25–74 years) and 67% were females. Patient blood was longitudinally sampled up to three times, between days 2 and 188 post‐symptom onset (Supplementary table 1). Acute samples were defined as those obtained within 14 days of symptom onset (range 2–14 days), while convalescent samples were obtained more than 5 weeks post‐symptom onset (range 38–188 days). We also recruited 27 pre‐pandemic healthy non‐exposed controls, with a median age of 52 (range 24–75 years) and 67% were female.
To determine isotype‐specific antibody responses against a range of SARS‐CoV‐2 antigens in COVID‐19 patients, we used commercial ELISAs detecting IgM against RBD (Wantai), IgG and IgA against S (Euroimmun) and IgG against N (Euroimmun). Antibody levels against these antigens were significantly higher in acute (median index value IgM (RBD); 13.791, IgG (S); 3.883, IgA (S), IgG (N); 2.633) and convalescent COVID‐19 samples (median index value IgM (RBD); 12.986, IgG (S); 4.369, IgA (S); 3.791, IgG (N); 2.378), when compared to healthy non‐exposed individuals (median index value IgM (RBD); 0.184, IgG (S); 0.132, IgA (S); 0.260, IgG (N); 0.188) (Figure 1a). Across all isotypes and specificities, the antibody levels were maintained between acute and convalescent samples, although IgM (RBD) and IgA (S) started to decrease at about day 40, while IgG (S and N) were maintained for slightly longer decreasing at about day 80 (Figure 1b). Antibody data obtained with the commercial ELISAs closely reflected the antibody profiles detected with the in‐house performed ELISAs against RBD and S (Figure 1c), 6 in which all COVID‐19 patients seroconverted by convalescence as measured by the in‐house IgG (RBD) ELISA. Longitudinally, titres for both RBD and S specificities trended towards decreased levels with time post‐symptom onset (Figure 1d), which was similar to our observation using the commercial ELISAs.
Figure 1.

Antibody signatures in COVID‐19 patients determined by commercial and in‐house ELISAs. (a) Antibody levels against SARS‐CoV‐2 proteins for IgM (RBD), IgG (S and N) and IgA (S) in acute (n = 7) and convalescent (n = 14) COVID‐19 donors and non‐exposed healthy individuals (n = 25) were measured by commercial ELISA. Grey dotted lines indicate borderline antibody levels as determined by the manufacturer. (b) Kinetics of antibody levels determined by commercial ELISA from days post‐symptom onset for IgM, IgG and IgA. (c) In‐house ELISA end‐point titres of SARS‐CoV‐2 RBD and S antibodies where the dotted line indicates the seroconversion cut‐off. (d) Kinetics of antibody levels determined by in‐house ELISA from days post‐symptom onset for IgM, IgG and IgA. (e) Correlation between commercial ELISA antibody isotopes and specificities (n = 59 samples per isotype). (f) Correlation between IgG and IgM/IgA for RBD (n = 58 samples) and S (n = 49 samples) specificities as measured by in‐house ELISA. (g) Correlation between matched isotype and specificities for commercial and in‐house ELISAs (n = 34 samples). (h) Correlation matrix of commercial and in‐house ELISA antibody levels in all acute and convalescent COVID‐19 samples. Spearman's ranked correlation coefficients (r s) are depicted for each paired correlation, with positive correlations shown in shades of blue and negative correlations in shades of red. (a, c) Median is shown, and statistical significance was determined by a Kruskal–Wallis multiple comparisons test. (b, d) LOESS regression lines with 95% confidence intervals shaded in grey are shown. (e–g) Spearman's correlation coefficients and P‐values shown.
IgG (S) antibody levels measured by the commercial ELISA correlated strongly across isotypes (IgM, IgA) and specificities (RBD, N) (P < 0.0001) (Figure 1e). Similarly, IgG (RBD, S) titres measured by the in‐house ELISA positivity correlated with IgM and IgA with matched antigen specificities (P < 0.0001) (Figure 1f). To confirm that the antibody levels determined via the two different ELISA methodologies were comparable, we analysed paired antigen specificities and found strong positive correlations across all three isotypes (r s = 0.7525–0.9636, P < 0.0001) (Figure 1g). We also generated a correlation matrix across the 10 parameters derived from the commercial and in‐house ELISAs, revealing positive correlations between all factors. The weakest correlates (r s < 0.7) were between the in‐house IgM‐RBD versus the commercial IgA‐S, IgG‐N, IgG‐S and in‐house IgA‐S, IgG‐S and IgA‐S, whereas the strongest correlates were between commercial IgG‐S versus in‐house IgG‐S (r s = 0.96) and in‐house IgG‐RBD (r s = 0.95) (Figure 1h).
ACE2‐RBD binding inhibition correlates with IgG titres
The rapid surrogate neutralising assay measures neutralising activity of SARS‐CoV‐2‐specific antibodies in COVID‐19 patients at a lower biosafety level (BSL 2). The sVNT used in this study was based on antibody‐mediated blockade of the interaction between the ACE2 receptor protein and RBD, 15 and unlike the conventional MNT, does not require infectious virus, cells or a BSL 3 laboratory. The sVNT detected neutralising antibodies in all COVID‐19‐infected individuals except one by convalescence (despite a positive PCR test, this individual was also negative by sVNT and MNT). Both acute and convalescent COVID‐19 groups displayed significantly higher ACE2‐RBD inhibition than healthy non‐exposed individuals (P = 0.0007 and P < 0.0001, respectively) (Figure 2a). Although we observed a decrease in neutralising antibody titres over time, neutralising activity was maintained into convalescence, with detectable neutralising antibodies found in 3 out of 5 donors at ~180 days post‐symptom onset, with the remaining two donors displaying borderline inhibition (21% and 22.9%).
Figure 2.
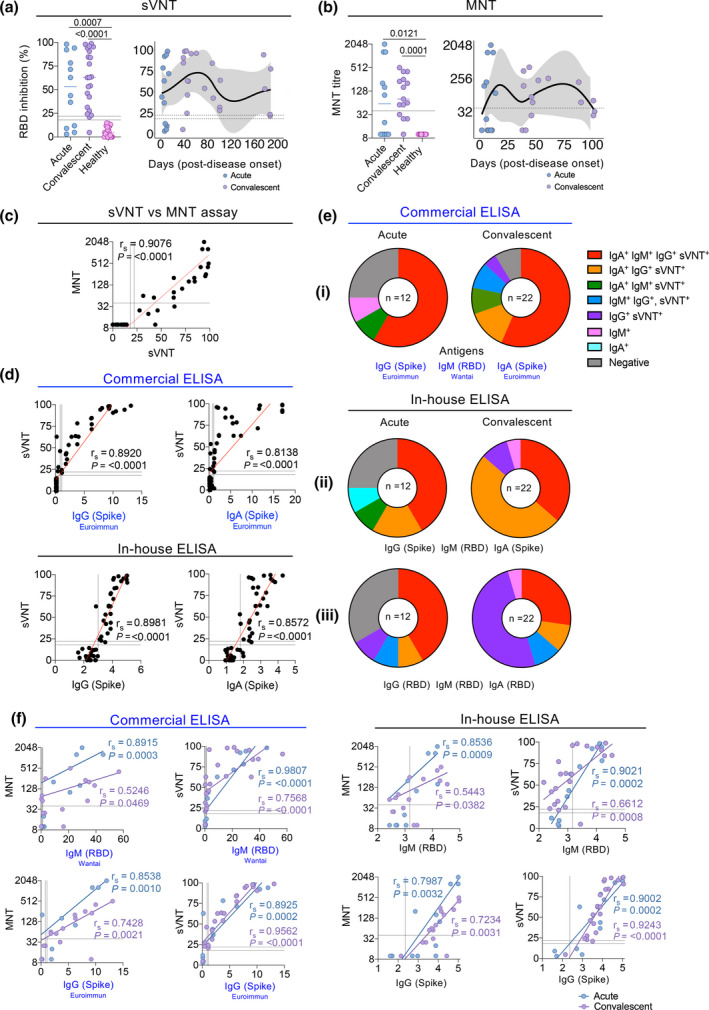
ACE2‐RBD binding inhibition correlates with MNT, IgG and IgA titres. (a) Percentage inhibition of ACE2 and RBD binding by neutralising antibodies in acute (n = 12) and convalescent (n = 22) COVID‐19 samples and non‐exposed healthy individuals (n = 25). (b) MNT titres are shown in a subset of donors (12 acute, 15 convalescent and 12 non‐exposed healthy serum samples) as previously described 6 . (a, b) Medians are shown, and statistical significance was determined by a Kruskal–Wallis multiple comparisons test (left panels). Kinetics of neutralising antibodies from days post‐symptom onset (right panels). LOESS regression lines with 95% confidence intervals shaded in grey are shown. (c, d) Correlation between sVNT percentage inhibition and (c) MNTs (titre log2), and (d, top) commercial (n = 59 samples per isotype) and (d, bottom) in‐house (n = 49 samples per isotype) ELISA for S‐specific IgG and IgA antibody levels. Spearman's correlation coefficients and P‐values shown. (e) Isotype and neutralisation profiles for RBD and Spike‐specific IgM, IgG and IgA at acute and convalescent timepoints, measured by sVNT and (i) commercial and (ii, iii) in‐house ELISAs. (f) Correlation between commercial and in‐house IgM (RBD) and IgG (S) ELISAs with sVNT and MNT divided into acute and convalescent phases. Spearman's correlation coefficients and P‐values for each phase are shown.
Since the sVNT has been developed relatively recently, we next asked how the ACE2‐RBD inhibition detected by sVNT compared to classical MNT titres, as detected on a subset of donors described previously. 6 Indeed, very similar patterns of neutralisation were observed with the MNT compared to the sVNT, with MNT antibodies detected at significantly higher levels in acute (P = 0.0121) and convalescent (P = 0.0001) COVID‐19 donors when compared to healthy non‐exposed individuals (Figure 2b). Antibody neutralisation activity strongly correlated between the two tests (r s = 0.9076; P < 0.0001) (Figure 2c). Importantly, results obtained using the RBD in‐house ELISA also correlated with the classical MNT, as shown previously. 6
Having shown that the frequency of ACE2‐RBD inhibition measured by the sVNT is comparable to the neutralisation activity determined by MNT, we tested whether the ACE2‐RBD inhibition correlated with the ELISA antibody levels. We observed the frequency of ACE2‐RBD inhibition strongly correlated with the S‐specific IgG and IgA titres measured by both commercial (IgG (S) r s = 0.8920 P < 0.0001, IgA (S) r s = 0.8138 P < 0.0001) and in‐house ELISAs (IgG (S) r s = 0.8981 P < 0.0001, IgA (S) r s = 0.8572 P < 0.0001) (Figure 2d). Analysis of antibody titres and ACE2‐RBD inhibition revealed that the proportion of COVID‐19 samples with undetectable IgM‐RBD, IgG‐S and IgA‐S antibodies (measured in the commercial ELISA) and low neutralising activity (below 18%) decreased from 25% at the acute phase of infection to 4.5% during convalescence (Figure 2ei). Using the in‐house ELISA, this trend in antibody responses (against the same antigens used in the commercial ELISAs) and neutralisation activity was also observed, decreasing from 25% at the acute phase to 0% at convalescence as all the recovered individuals had at least one positive isotype response (Figure 2eii). This suggests that a combined detection of IgM, IgG and IgA S or RBD‐specific antibody isotypes via the in‐house ELISA might be more sensitive and/or has a lower threshold of detection than the commercial ELISA. Interestingly, when antibody levels were measured by commercial ELISA, more than half (59%) of the convalescent donors were positive for all antibody isotypes (IgG‐S, IgM‐RBD and IgA‐S) and ACE2‐RBD inhibition (Figure 2ei). In contrast, when titres were measured by in‐house ELISA, a majority of the participants were positive for neutralisation activity featuring either all isotypes (IgG‐S, IgM‐RBD and IgA‐S) (36%) or only IgA‐S and IgG‐S (50%) (Figure 2eii), indicating potentially a lower sensitivity for IgM in the in‐house assay. However, when only RBD‐specificity was considered, unresponsive frequencies were higher at the acute phase (33%). Moreover, half (50%) of convalescent donors exhibiting neutralising activity had only antibodies with IgG isotype (Figure 2eiii), indicating that a proportion of antibody responses observed in the commercial (Figure 2ei) and in‐house ELISA (Figure 2eii) against the S protein are directed to non‐RBD regions of the protein.
Given that IgM can mediate a significant proportion of the early neutralisation activity, 27 we correlated commercial and in‐house ELISA IgM and IgG antibody levels with neutralisation measured by sVNT and MNT. We observed that the IgM levels at acute timepoints correlated more strongly with neutralisation activity (commercial IgM with MNT r s = 0.8915 P = 0.0003, sVNT r s = 0.9807 P < 0.0001; in‐house IgM with MNT r s = 0.8536 P = 0.0009, sVNT r s = 0.9021 P = 0.0002) than IgM levels at convalescence (commercial IgM with MNT r s = 0.5246 P = 0.0469, sVNT r s = 0.7568 P < 0.0001; in‐house IgM with MNT r s = 0.5443 P = 0.0382, sVNT r s = 0.6612 P = 0.0008) (Figure 2f). In comparison, the strong correlations between IgG levels and neutralisation activity were maintained across both acute and convalescent phases (acute r s = 0.7987–0.9002; convalescence r s = 0.7234–0.9562). These correlations are thus in line with the view that both IgM and IgG contribute to protection during the acute phase; however, IgG responses may have a greater contribution to the overall neutralisation activity over time.
Avidity of IgG antibodies increases at convalescence
Data obtained using the sVNT and the commercial and in‐house ELISA assays, as well as our previous MNT data, 6 clearly demonstrated that the majority of COVID‐19 patients had robust antibody titres and neutralising activity, but provided little information on how these parameters were related to the qualitative features of humoral immunity. Since antibody neutralisation quality and potency is often correlated with antibody avidity, a urea dissociation assay (Figure 3a and d, representative donors) was used to determine the proportion of antigen‐bound IgM and IgG antibody remaining after 6 m urea treatment (expressed as an avidity index). 6 , 14 The avidity of IgM was greater than 50% at all timepoints for all COVID‐19 donors tested (Figure 3b). In paired COVID‐19 plasma samples (n = 12) obtained 4 to 141 days apart (median 40.5 days between first and last visits), we found a significant increase (P = 0.023) in IgM avidity on the second visit (Figure 3c), with avidity from day 40 post‐symptom onset apparently maintained to at least day ~188 (Figure 3b). IgM avidity also correlated with the IgM titres from in‐house ELISAs (r s = 0.8158, P < 0.0001) (Figure 3c), an indication of the presence of IgM at high levels after initial pathogen exposure and its pentameric conformation providing antigen binding strength.
Figure 3.

IgG antibody avidity increases at convalescence. (a) Representative avidity analysis for IgM RBD‐specific antibodies. Plasma was diluted across antigen‐coated wells before treatment in the presence (red) or absence (blue) of 6 m Urea. Shown is the percentage of antibody bound at each dilution relative to the amount detected at the lowest dilution (1:31.6) in the absence of urea (100%). (b) Longitudinal antibody avidity levels of acute (n = 7) and convalescent (n = 13) COVID‐19 patient samples for RBD‐specific IgM. The avidity index reflects the percentage of antibody remaining by comparing the AUC of each antibody titration curve with or without urea treatment. (c) Avidity analysis for IgM RBD‐specific antibodies in paired samples (left panel). Correlation between in‐house IgM (RBD) ELISA titres and IgM avidity scores (right panel). (d) Representative avidity analysis for IgG RBD‐specific antibodies. (e) Longitudinal antibody avidity levels of acute (n = 7) and convalescent (n = 13) COVID‐19 patient samples for RBD‐specific IgG. (f) Avidity analysis for IgG RBD‐specific antibodies in paired samples (left panel). Correlation between IgG (RBD) ELISA titres and IgG avidity scores (right panel). (c, f, left panels) First and last samples were collected between 4 and 141 days apart. Statistical significance was assessed by the Wilcoxon matched‐pairs signed rank test, n = 12. (c, f, right panels) Spearman's correlation coefficients and P‐values are shown.
In contrast, RBD‐specific IgG antibodies in COVID‐19 donors (n = 12) exhibited lower avidity than IgM at the first visit during the acute phase of disease, which then increased during convalescence (Figure 3e). Paired analysis of IgG avidity during first and last visits, however, showed a more significant increase by the second visit (P = 0.009) compared to the IgM (Figure 3f). The fact that IgG avidity did not correlate as strongly with the IgG titre (r s = 0.6085, P = 0.0004) indicates that despite high initial titres, IgG responses require time to undergo somatic hypermutation to acquire stronger avidity, or alternatively that the IgG titre and avidity are less interconnected than their IgM counterparts.
Coronavirus multiplex array reveals a range of key antibody features beyond neutralisation
Apart from neutralisation activity exerted by a variety of antibody isotypes, antibodies also have the capacity to engage Fc Receptors (FcRs) or complement as a mechanism to mediate Fc effector functions; they are not limited to targeting the RBD. The coronavirus multiplex array that has previously been applied to systems serology studies, 14 allows detection of a broad range of antibody levels directed against a range of antigen specificities (including trimeric S, spike 1 (S1) and 2 (S2), RBD, N and antigens from seasonal coronaviruses to test cross‐reactivity), 14 isotypes (IgG, IgA, IgM) and subclasses (IgG1, IgG2, IgG3, IgG4, IgA1, IgA2), along with functions like C1q [a predictor of classical Antibody‐Dependent Complement Activation (ADCA)] and soluble dimer engagement via FcRs (FcγRIIa, FcγRIIb and FcγRIIIa), which have previously been shown to correlate with a range of effector functions. 28
A subset of donor samples was examined using the multiplex approach (COVID‐19 donors n = 9, non‐exposed healthy individuals n = 22) across five SARS‐CoV‐2 antigens (trimeric S, S1, S2, RBD and N) and 14 detectors (IgM, IgG, IgG1, IgG2, IgG3, IgG4, IgA1, IgA2, FcγRIIaH, FcγRIIaR, FcγRIIIaV, FcγRIIIaF, FcγRIIb and C1q), as previously described. 14 Significantly higher levels of S‐specific IgM (P = 0.0056), IgG (P = 0.0037), IgG1 (P = 0.013), IgG2 (P = 0.017), IgG3 (P = 0.002), IgA1 (P = 0.0026) and IgA2 P = 0.002) antibodies were detected in convalescent COVID‐19 donors compared to healthy controls, whereas only IgM (P < 0.0001), IgG2 (P = 0.0076), IgG3 (P = 0.01), IgA1 (P = 0.015) and IgA2 (P = 0.0317) showed significantly higher levels in acute donors (Figure 4a and Supplementary figure 1a). Significant differences between convalescent COVID‐19 donors and healthy individuals were maintained when only the RBD‐specific antibody response was considered for all antibody isotypes apart from IgM and IgG2, with significantly higher antibodies in acute donors observed only for IgG (P = 0.0225) and IgA1 (P = 0.0292) (Figure 4b). In comparison, significant differences in N‐specific antibodies were limited to convalescent IgG (P = 0.0212), IgG1 (P = 0.0109) and IgG3 (convalescent P = 0.0162 and acute P = 0.0266) (Figure 4c). When FcRs and C1q were examined across antigen specificities, similar to the antibody isotypes, the acute and convalescent COVID‐19 donors had significantly higher levels than non‐exposed individuals for S, S1 and RBD, but less so for N and S2 (Figure 4a–c and Supplementary figure 1b). The increased binding to high avidity FcγRIIaH131 and FcγRIIIaV158 soluble dimers for the S and RBD‐specific antibodies may suggest that, in addition to the potential neutralising activity, antibody‐mediated activity such as antibody‐dependent cellular cytotoxicity (ADCC) and antibody‐dependent cellular phagocytosis (ADCP) could potentially contribute to viral clearance. 14 Furthermore, we observed a trend towards increasing FcR and C1q engagement over time, suggesting that the Fc function is being regulated or modulated in COVID‐19 donors during and possibly even after infection. Significant binding to C1q in acute (S P = 0.0112, S1 P = 0.0013, S2 P = 0.0002, RBD P < 0.0001, N P = 0.0008) and convalescent (S P < 0.0001, S1 P < 0.0001, S2 P = 0.0124, RBD P = 0.0033) COVID‐19 donors compared to non‐exposed healthy individuals also indicates that viral infection induces antibodies that could clear the virus infected cells via ADCA. Overall, the multiplex data positively correlated with each other, with the strongest correlations observed for IgG, IgG1, FcR and C1q detectors against S, S1 and RBD antigens (Figure 4d). In context with the neutralising data, the high avidity FcR and C1q detectors against S and RBD were found to positively correlate with neutralising activity determined by both sVNT and MNT, with the S‐specific antibodies generally displaying a stronger correlation than the RBD antibodies (Figure 4e), indicating a potential contribution of neutralising antibody directed against epitopes outside the RBD.
Figure 4.
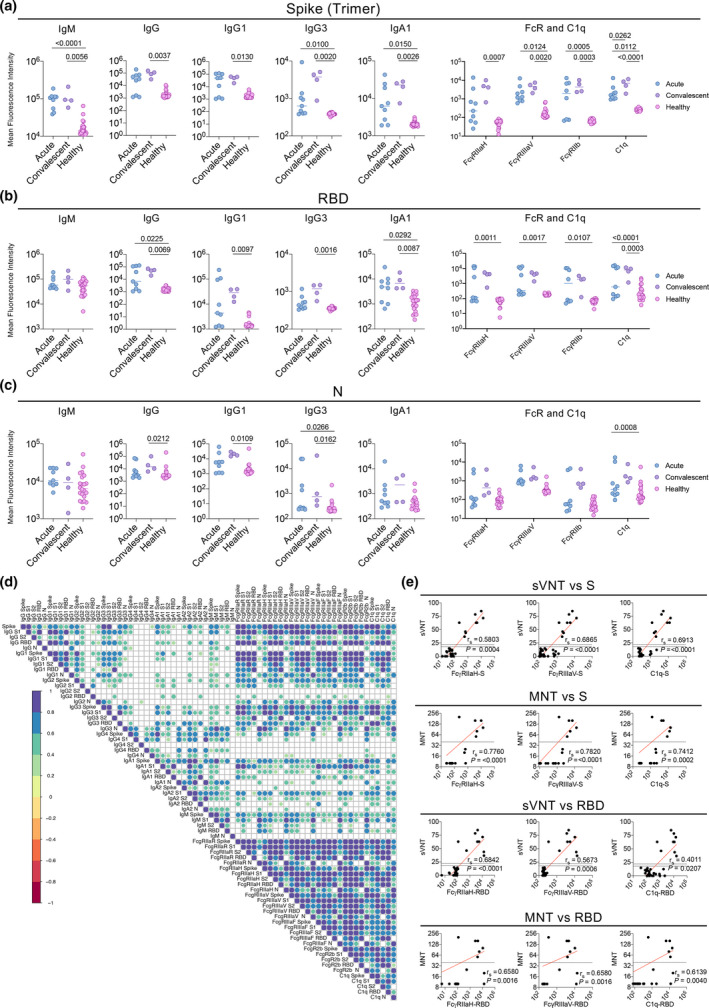
Multiplex analysis of COVID‐19 patients. Median fluorescence intensity of selected Ig isotypes, FcR and C1q against (a) S trimer, (b) RBD and (c) N protein within acute (n = 6), convalescent (n = 4) and non‐exposed healthy (n = 22) plasma samples. (d) Correlation heatmap of MFI's measured across all 14 detectors (IgM, IgG, IgG1, IgG2, IgG3, IgG4, IgA1, IgA2, FcγRIIaH, FcγRIIaR, FcγRIIIaV, FcγRIIIaF, FcγRIIb and C1q) against the 5 SARS‐CoV‐2 antigens (trimeric S, S1, S2, RBD and N). Only significant correlations (FDR corrected P < 0.05) are shown. (e) Correlations of sVNT percentage inhibition (n = 33 COVID‐19 positive and healthy samples) or MNT (n = 20 samples) between S‐ or RBD‐specific FcγRIIaH, FcγRIIIaV, or C1q MFI are shown. Spearman's correlation coefficients and P‐values are shown.
Acute cTfh‐type 1 numbers are indicative of antibody‐mediated immunity
We and others have recently shown that SARS‐CoV‐2 antibody responses correlate with the rise in ASCs and cTfh‐type 1 (cTfh1) responses in COVID‐19 patients. 2 , 6 , 12 , 29 In our previous study, 6 CD38hiCD27hi ASCs peaked rapidly and transiently post infection, with the number of ASCs in a subset of acute COVID‐19 donors included in this comparative study (n = 15) also being significantly higher than convalescent COVID‐19 donors (P = 0.001) and non‐exposed healthy individuals P = 0.0084) (Figure 5a). In contrast, total activated PD1+ICOS+ CXCR3+CXCR5+CD4+ cTfh1 cells peaked later post‐symptom onset, 6 with significantly higher levels in acute COVID‐19 donors compared to healthy individuals (P = 0.0083) included in this study (Figure 5b). The early and temporary increase in ASC did not correlate with antibody levels measured by commercial or in‐house ELISA (IgG‐S/RBD) or sVNT. However, the number of cTfh1 cells correlated positively with levels of S and RBD‐specific IgG antibodies measured by both the commercial and in‐house ELISA, as well as ACE2‐RBD inhibition by sVNT.
Figure 5.
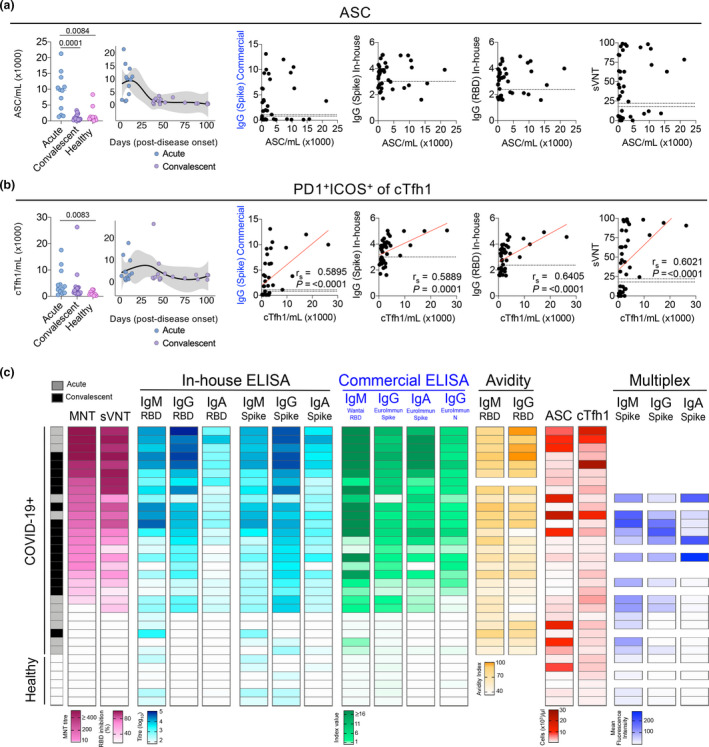
Contribution of cellular factors to antibody‐mediated immunity. (a, b) Absolute numbers and longitudinal kinetics of (a) ASCs and (b) cTfh1 cells have previously been described. 6 Correlations of cellular subsets between commercial and in‐house IgG‐S, in‐house IgG‐RBD and sVNT % inhibition are shown in the right panels. (c) Heatmap of neutralisation (MNT, sVNT), antibody levels (in‐house and commercial ELISA), antibody avidity, ASC and cTfh1 cells and multiplex analyses of acute (n = up to 11), convalescent (n = up to 16) and non‐exposed healthy (n = up to 6). Each row represents a different sample with their matched measurements (where available) in each column.
To provide insights into SARS‐CoV‐2 humoral immunity across the distinct serological assays, we collated a heatmap of neutralisation (MNT, sVNT), antibody levels (in‐house and commercial ELISA), antibody avidity, ASC and cTfh1 cells and multiplex analyses from a subset of patients (acute n = 11, convalescent n = 15) and non‐exposed healthy individuals (n = 6). When we ranked the COVID‐19 patients according to their MNT neutralising titres, COVID‐19 patients with the highest neutralising titres also had a high frequency of ACE2‐RBD inhibition detected by sVNT. Interestingly, patients with high neutralising activity also had high levels of all antibody isotypes, specificities, avidity, as well as cTfh1 cells (Figure 5c), suggesting a broad range of antibodies likely leads to improved virus neutralisation. This was particularly true for donors with high IgG antibodies levels against S and RBD (as RBD is captured within the S‐specific response) also having high sVNT and MNT, suggesting a pivotal role for IgG in neutralisation, although higher levels of neutralising IgA could be expected in the mucosa. 17 While the multiplex assay lacked donors with very strong MNT and sVNT scores, S‐specific antibody levels based on MFI decreased with lower neutralisation activity, particularly for IgG and IgA. Finally, an extended correlation heatmap of ASC, cTfh1 and antibody responses across the six different serology assays evaluated in this study displayed mostly positive correlations, with 2277 positive correlations from a total of 2279 significant correlations (Supplementary figure 2). The exception was IgG avidity with IgG1‐S and IgG2‐S MFI, as determined by multiplex analyses, which may be due to limited convalescent sampling in this assay (n = 4), rather than biological effects as we showed that IgG avidity increases at the later timepoints. Although positive correlations were observed earlier between the FcR and C1q MFIs (S and RBD) with the sVNT and MNT (Figure 4e), these correlations were not significant for MNT after correcting for multiple comparisons in the extended heatmap (Supplementary figure 2). However, clusters of very strong positive correlations were observed as expected between (1) the ELISAs, both in‐house and commercial, (2) the ELISAs to the multiplex FcR detectors, (3) multiplex IgG with FcR and (4) within the different the FcR detectors. Therefore, our comparative analysis showed that antibody levels obtained from the above six assays were highly correlated with each other, and with the cTfh1 response.
Discussion
The COVID‐19 pandemic is expected to gradually transition to a new phase as vaccines become available that will lead to a rapid increase in protective immune responses within the population. Such increased immunity at the population level can exert immune pressure on the virus and induce escape mutations, which could coincide with lower viral fitness. It is likely that the virus will shift from being a pandemic strain to a seasonally circulating strain, which like other seasonal coronaviruses, will cause repetitive and (hopefully) mild infections, 30 as observed for pandemic influenza virus strains, including the deadly 1918 influenza pandemic strain. 31 Introduction of viral variants, which could potentially evade protective immune responses is also a possibility, as happens frequently with influenza viruses. 32 , 33 Thus, there is a need and an increasing demand for rapid reliable serological testing to identify individuals with protective immunity induced by infection or vaccination. Furthermore, from a scientific standpoint, these assays will be useful to monitor the longevity of humoral immunity, especially in high‐risk groups, in order to re‐vaccinate before protective immune responses wane. 34 Serological testing can also be used to assess whether previously induced SARS‐CoV‐2 specific antibodies recognise viral variants that have mutated within known neutralising antibody binding sites. Virus neutralisation assays, including the MNT, are considered the gold standard to detect protective antibody responses. However, this assay may not be suitable for every situation and/or laboratory. The MNT needs to be performed under BSL 3 containment, requires highly qualified personnel, needs a relatively large volume of serum, is labour intensive and does not provide information on antibody isotypes or potential protective features beyond virus neutralisation. Another technical limitation is that the current MNT assay is a biological assay, so all samples need to be performed in the same assay for direct comparison. To circumvent these limitations, several alternative assays have been developed (Table 1). However, knowledge on how distinct serology assays correlate with each other and understanding how they relate to the classical MNT readout is limited. In addition, it will be of increasing importance to compare data generated by different laboratories using different serological assays. Our comprehensive analyses provided important insights into SARS‐CoV‐2 humoral immunity across distinct serology assays, which will enable scientists to select the best assay to answer their research questions.
Table 1.
Antibody assay comparison
| MNT | sVNT Genscript | ELISA Euroimmun (IgG/IgA; Spike & IgG; N) | ELISA Wantai (IgM; RBD) | In‐house ELISA (IgM/IgA/IgG; RBD/Spike) | Chaotropic‐based dissociation assays | Multiplex | |
|---|---|---|---|---|---|---|---|
| Test time | 5 days | 65 min | 120 min (S); 105 min (N) | 75 min | 48 h | 48 h | 24 h per detector (antibody isotype or FcR) |
| Hands‐on user time | 7 h | 30 min | 30 min | 30 min | ~2.5 h (6 plates) | ~3 h (6 plates) | ~4 h per detector |
| Capacity; samples/run | 96 samples | 96 samples | 96 samples | 96 samples | 10 samples per plate | 5 samples per plate | 384 samples per detector |
| Volume of sample per test | 28 µL | 10 µL | 10 µL | 10 µL | 7.3 µL | 7.3 µL | 1 µL per detector |
| Plasma vs serum | Either a | Either | Either | Either | Either | Either | Either |
| Samples run in duplicate or singularly | Duplicate in independent assays | Singularly (duplicate for samples within the equivocal zone) | Singularly (duplicate for samples within the equivocal zone) | Singularly (duplicate for samples within the equivocal zone) | Singularly | Singularly | Duplicate in independent assays |
| Cost per sample (AUD) | – | ~$14 | ~$8 | ~$8 | ~$1.60–$2.20 | ~$1.80–$2.40 | ~$16 |
| TGA approval 42 | – | Emergency access scheme | Emergency access scheme | Emergency access scheme | – | – | – |
| FDA approval 43 | – | Emergency use authorisation | Emergency use authorisation (IgG) | – | – | – | – |
| Biosafety level | BSL 3 | BSL 2 | BSL 2 | BSL 2 | BSL 2 | BSL 2 | BSL 2 |
| Antigen specificity | Whole virus | RBD | S or N | RBD | S, RBD | S, RBD | S, S1, S2, RBD, N |
| Class switching | No | No | Yes | Yes | Yes | No | Yes |
| Antibody avidity | No | No | No | No | No | Yes | Yes |
| Level of expertise | High | Moderate | Moderate | Moderate | Moderate | Moderate | High |
| Standardisation | Run specific | Yes | Yes | Yes | Yes | Yes | Bead set specific |
| Result readout | End‐point titre | % inhibition | Index value | Index value | End‐point titre | Avidity Index | MFI |
Serum preferred.
We demonstrated that all evaluated serological assays strongly correlated with the classical MNT and with each other. Furthermore, all assays had the capability to detect the longevity of antibody responses over time. The decision to choose one of the six assays studied here will therefore greatly depend on alternative factors such as biosafety level, labour (hands‐on and total duration of the assay), costs, approval by Therapeutic Goods Administration (TGA; Australia) or Food and Drug Administration (FDA; USA) for diagnostic use, the type of readout and level of expertise (Table 1), and the research question at hand. Access to BSL 3 containment for the MNT, or the high level of expertise and relatively high costs of the multiplex assay means that these assays may not be suitable for most laboratories, and that the other antibody assays such as the in‐house and/or commercial ELISAs are likely to be sufficient if the research aim is to detect protective antibody responses.
However, to understand whether SARS‐CoV‐2 vaccines can induce similar or even stronger/broader protective immune responses than natural infection, additional serological features beyond virus neutralisation need to be studied. These include class‐switching, antibody maturation, recognition of (non‐)neutralising epitopes and/or their ability to engage FcRs or complement, both contributing to protection from severe viral infections via ADCC, ADCP and ADCA. 35 , 36 , 37 , 38 , 39 When it comes to class‐switching, either one of the commercial or in‐house ELISAs evaluated herein can be used, as the isotype data of all three assays strongly correlated with each other. However, compared to the commercial ELISAs, the in‐house ELISA might be slightly more sensitive for the detection of IgG and IgA, but slightly less sensitive for IgM. Furthermore, the Euroimmun ELISAs have the additional advantage of not only detecting spike or RBD‐specific antibodies but also those directed against the nucleocapsid protein. Detection of nucleocapsid‐specific antibodies could potentially help differentiate vaccinated individuals (S‐based vaccines such as those manufactured by Pfizer, Moderna or AstraZeneca) from naturally infected individuals (who would theoretically have both S and N‐specific antibodies). Affinity maturation, for which antibody avidity could be used as a surrogate measure, could best be studied using the chaotropic‐based dissociation assay for both IgG and IgM. However, given that IgA RBD titres were often lower than IgM and IgG, this affected the detection range of the assay, limiting its ability to measure accurate differences in antibody binding following urea treatment. Interestingly, when we correlated IgG titres with the IgG avidity data, we showed that, during the course of the immune response, the initial increase in IgG titres was followed by enhanced avidity, which indicates that despite initial high titres, time is needed to acquire strong avidity.
Although multiplex analysis allows one to study isotype switching, the strength lies in its ability to simultaneously study antibody responses at the subclass level to a broad range of viral epitopes, and additionally provide an in‐depth understanding of their ability to engage FcRs or complement. Here, we enlisted 14 detectors against 5 antigens, which totalled to measuring 70 different antibody features in a single assay but could easily be further expanded, as described previously. 14 This makes the multiplex analysis of great interest for an in‐depth systems serology comparison of antibody responses induced by natural infection or vaccination. However, this assay is technically challenging, and requires specialised equipment and bioinformatic expertise to grasp the complexity of the large data sets, which makes the assay less suitable for most laboratories.
Germinal centre derived ASCs and Tfh1 cells play an important role in the induction and maintenance of antibody responses. However, since germinal centres are largely inaccessible for most immune and vaccination studies, we and others 2 , 6 , 12 , 29 , 40 correlated our serological findings with previously acquired circulating cellular immune responses. We observed an increase of ASCs early during the acute phase of infection, potentially driving the initial increase in SARS‐CoV‐2 specific antibodies; however, long‐lived plasma cells rather than ASCs likely support the production of long‐term serological memory. This response was followed by an increase in cTfh1 numbers in the late acute phase. Interestingly, only acute cTfh1 cell numbers, but not ASCs, correlated with spike and RBD‐specific IgG antibodies measured by ELISAs and ACE2‐RBD inhibition. These results are in agreement with our recent findings that acute cTfh1 cells correlate with and predict antibody titres, especially towards spike, in convalescence as measured by in‐house ELISA. 6 These data may suggest a role for cTfh1 cells in the establishment, class‐switching and longevity of humoral immunity in SARS‐CoV‐2 infection. The caveat there is that only ASC and cTfh1 cells measured in the acute phase can be used as an indicator for humoral immunity, since this is the only timepoint where they can be accurately detected in peripheral blood.
Overall, the scientific community is well equipped with a broad range of serological assays to answer a variety of biologically relevant questions on SARS‐CoV‐2‐specific antibody responses that will arise over the coming months and years. Our study establishes that strong correlations exist between different assays despite the fact that the assays are designed to detect distinct antibody features, thus allowing laboratories with different technologies to compare their findings to a certain extent.
Methods
Study participants and ethics statement
Our study enrolled 15 SARS‐CoV‐2 PCR‐positive participants, seven of whom were hospitalised acute patients and eight of whom were sampled at the convalescent phase 38–188 days post‐symptom onset (Supplementary table 1). We included 27 pre‐pandemic individuals as healthy non‐exposed controls. Experiments conformed to the Declaration of Helsinki Principles and the Australian National Health and Medical Research Council Code of Practice. Written informed consents were obtained from all blood donors prior to the study. The study was approved by the University of Melbourne (#2056761, #1955465, #1442952, #1443389), the Alfred Hospital (#280/14) and James Cook University (H7886) Human Research Ethics Committees.
Commercial enzyme‐linked immunosorbent assays (ELISAs)
ELISA testing was performed according to manufacturers' ‘Instructions For Use’ (IFU), with results reported semi‐quantitatively as either a signal/cut‐off ratio (for Euroimmun and Wantai assays) or percentage inhibition (for surrogate virus neutralisation assay).
Euroimmun anti‐SARS‐CoV‐2 S IgG, S IgA and N IgG (Euroimmun Medizinische Labordiagnostika, Lubeck, Germany); indirect ELISAs for detection of IgG or IgA against SARS‐CoV‐2 antigens. Microtitre plate wells were coated with either (1) S1‐domain or (2) modified N protein. SARS‐CoV‐2‐binding antibodies were detected using enzyme‐labelled anti‐human‐IgG or anti‐human‐IgA conjugates and a colourimetric substrate. The results were read spectrophotometrically.
Wantai IgM (Beijing Wantai Biological Pharmacy Enterprise, Beijing China); Wantai SARS‐CoV‐2 IgM is a capture ELISA for detection of IgM‐class antibodies to SARS‐CoV‐2 virus. Anti‐µ chain antibodies on the microtitre plate captured any patient IgM‐class antibodies, and detection was achieved by recombinant SARS‐CoV‐2‐antigen‐horseradish peroxidase (HRP) conjugate followed by a colourimetric substrate. The results were read spectrophotometrically.
SARS‐CoV‐2 surrogate virus neutralisation test (sVNT)
Manufactured by GenScript (NJ, USA), the sVNT is a blocking ELISA which mimics the virus neutralisation process, detecting circulating neutralising SARS‐CoV‐2 antibodies that block the interaction between RBD and ACE2 on the cell surface receptor of the host. The test is species and isotype independent. HRP‐conjugated recombinant SARS‐CoV‐2 RBD fragment bound to any circulating neutralising antibodies to RBD preventing capture by the human ACE2 protein in the well, which was subsequently removed in the following wash step. Substrate reaction incubation time was determined by temperature. The IFU reported that the ideal reaction temperature and time were 25°C for 15 min but that for temperatures lower than 25°C, the time could be extended. Since our control values did not meet the assay validity criteria at 15 min, but fell within the acceptable ranges at 20 min, we used, a 20‐min substrate incubation time and results were read spectrophotometrically. Colour intensity was inversely dependent on the titre of anti‐SARS‐CoV‐2 neutralising antibodies.
RBD and Spike protein in‐house enzyme‐linked immunosorbent assays (ELISAs)
RBD‐ and S‐specific ELISAs for detection of IgM, IgG and IgA antibodies were performed as described, 6 , 11 , 41 with some modifications: Nunc MaxiSorp flat bottom 96‐well plates (Thermo Fisher Scientific, Carlsbad, CA, USA) were used for antigen coating, blocking with PBS (containing w/v 1% BSA) and serial dilutions performed in PBS (containing v/v 0.05% Tween and w/v 0.5% BSA). For detection of IgG and IgA, peroxidase‐conjugated goat anti‐human IgG (Fcγ fragment specific; Jackson ImmunoResearch, West Grove, PA, USA) or alkaline phosphate‐conjugated rat anti‐human IgA (mAb MT20; MabTech, Stockholm, Sweden) was used and developed with TMB (Sigma‐Aldrich, St Louis, MO, USA) substrate for IgG or pNPP (Sigma‐Aldrich) for IgA. For IgM, biotinylated mAb MT22 and peroxidase‐conjugated streptavidin (Pierce; Thermo Fisher Scientific) was used in conjunction with TMB. Peroxidase reactions were stopped using 1 m H3PO4 and plates read on a Multiskan plate reader (Labsystems, Vantaa, Finland). Inter‐ and intra‐experimental measurements were normalised using a positive control plasma from a COVID‐19 patient (#1‐073) run on each plate. End‐point titres were determined by interpolation from a sigmodial curve fit (all R‐squared values > 0.95; GraphPad Prism 8, San Diego, CA, USA) as the reciprocal dilution of plasma that produced ≥ 15% (for IgA and IgG) or ≥ 30% (for IgM) absorbance of the positive control at a 1:31.6 (IgG and IgM) or 1:10 dilution (IgA). Seroconversion was defined when titres were above the mean titres plus 2 standard deviations of healthy non‐COVID‐19 donors (24 for RBD and 15 for Spike).
Antibody avidity assay
The avidity of RBD‐specific IgG and IgM antibodies in plasma samples was measured using urea as the chaotropic agent. Plasma was titrated in half‐log dilutions (1:31.6 to 1:1000) onto antigen‐coated wells and incubated for 2 h. Wells were washed and 6 m urea added and incubated for a further 15 min at room temperature. Bound antibodies were then detected using the respective secondary detection reagents as described above in the in‐house ELISAs. The amount (in percentage) of antibody remaining was determined by comparing the total area of the antibody titration curve (across 4 dilutions) in the presence and absence of urea treatment and is expressed as the avidity index.
Microneutralisation assay (MNTs)
Microneutralisation activity of serum samples was previously described. 6 Briefly, SARS‐CoV‐2 isolate CoV/Australia/VIC01/2020 3 was propagated in Vero cells and stored at −80°C. Sera were heat‐inactivated at 56°C for 30 min. Samples were serially diluted in two‐fold dilutions starting at 1:20 and 100 TCID50 of SARS‐CoV‐2 in MEM/0.5% BSA were added and incubated at room temperature for 1 h. Residual virus infectivity in the serum/virus mixtures was assessed in quadruplicate wells of Vero cells incubated in serum‐free media containing 1 μg mL−1 of TPCK trypsin at 37°C and 5% CO2; viral cytopathic effect was read on day 5. The neutralising antibody titre was calculated using the Reed–Muench method, as previously described. 12
Coronavirus multiplex
A coronavirus multiplex array was previously described. 14 Briefly, SARS‐CoV‐2 antigens were covalently coupled to magnetic carboxylated beads using a two‐step carbodiimide reaction. Antigen‐coupled beads were pooled and combined with diluted plasma overnight before washing and staining with detectors (PE‐conjugated anti‐human IgM, IgG, IgA (and their respective subclasses) antibodies or soluble dimeric FcR or C1q protein followed by streptavidin PE conjugate). Plates were washed and read by the FlexMap 3D, with the binding of the PE detectors measured to calculate the median fluorescence intensity (MFI).
ASCs and cTfh1 analysis
Absolute numbers of ASCs and activated cTfh1 cells have been previously described. 6 Briefly, whole blood was freshly stained with antibody cocktails, RBC lysed then fixed with 1% PFA before acquiring on a LSRII Fortessa (BD Biosciences, San Jose, CA, USA) and analysed using FlowJo v10 software (FlowJo, Ashland, OR, USA).
Conflict of interest
The authors declare no conflict of interest.
Author contributions
Louise Rowntree: Conceptualization; Data curation; Formal analysis; Investigation; Methodology; Writing‐original draft. Brendon Chua: Conceptualization; Data curation; Formal analysis; Investigation; Methodology; Writing‐original draft. Suellen Nicholson: Formal analysis; Methodology; Supervision; Validation; Writing‐review & editing. Marios Koutsakos: Formal analysis; Writing‐review & editing. Luca Hensen: Formal analysis; Investigation; Writing‐review & editing. Celia Douros: Formal analysis; Writing‐review & editing. Kevin Selva: Formal analysis; Methodology; Writing‐review & editing. Francesca Mordant: Formal analysis; Writing‐review & editing. Chinn Wong: Formal analysis; Writing‐review & editing. Jennifer Habel: Investigation; Writing‐review & editing. Wuji Zhang: Investigation; Writing‐review & editing. Xiaoxiao Jia: Investigation; Writing‐review & editing. Lily Allen: Investigation; Writing‐review & editing. Denise Doolan: Investigation; Writing‐review & editing. David Jackson: Supervision; Writing‐review & editing. Adam Wheatley: Formal analysis; Methodology; Writing‐review & editing. Stephen Kent: Supervision; Writing‐review & editing. Fatima Amanat: Methodology; Resources; Writing‐review & editing. Florian Krammer: Methodology; Resources; Supervision; Writing‐review & editing. Kanta Subbarao: Methodology; Supervision; Writing‐review & editing. Allen Cheng: Conceptualization; Resources; Writing‐review & editing. Amy Chung: Methodology; Supervision; Writing‐review & editing. Mike Catton: Methodology; Resources; Supervision; Writing‐review & editing. Oanh Nguyen: Conceptualization; Investigation; Methodology; Project administration; Supervision; Writing‐original draft. Carolien van de Sandt: Conceptualization; Investigation; Methodology; Formal analysis; Supervision; Writing‐original draft. Katherine Kedzierska: Conceptualization; Resources; Supervision; Writing‐original draft; Writing‐review & editing.
Supporting information
Supplementary figure 1
Supplementary figure 2
Supplementary table 1
Acknowledgments
We thank all the participants involved in the study; Bernie McCudden and Jenny Mitchell for support with the cohort; and Jill Garlick, Janine Roney, Anne Paterson and the research nurses at the Alfred Hospital. This work was supported by the Australian National Health and Medical Research Council (NHMRC) Leadership Investigator Grant to KK (#1173871), NHMRC Program Grant to DLD (#1132975), Research Grants Council of the Hong Kong Special Administrative Region, China (#T11‐712/19‐Ncdf) to KK, the Jack Ma Foundation to KK, KS and AWC, the Medical Research Future Fund (#2005544) to KK, SJK, AKW and AWC, the a2 Milk Foundation to KS, MRFF Award (#1202445) to KK, NHMRC Program Grant (#1071916) to KK. KK was supported by NHMRC Senior Research Fellowship (1102792), DLD by a NHMRC Principal Research Fellowship (#1137285) and KS by an NHMRC Investigator grant (#1177174). THON is supported by NHMRC EL1 Fellowship (#1194036). LH is supported by the Melbourne International Research Scholarship (MIRS) and the Melbourne International Fee Remission Scholarship from The University of Melbourne. JRH and WZ are supported by the Melbourne Research Scholarship from The University of Melbourne. XJ is supported by China Scholarship Council‐University of Melbourne joint Scholarship. CES has received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska‐Curie grant agreement (#792532). CES and KS received funding from the Doherty Collaborative Seed Grant. KK and AWC were supported by the University of Melbourne Dame Kate Campbell Fellowship. The Melbourne WHO Collaborating Centre for Reference and Research on Influenza is supported by the Australian Government Department of Health.
References
- 1. Dong E, Du H, Gardner L. An interactive web‐based dashboard to track COVID‐19 in real time. Lancet Infect Dis 2020; 20: 533–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Thevarajan I, Nguyen THO, Koutsakos M et al. Breadth of concomitant immune responses prior to patient recovery: a case report of non‐severe COVID‐19. Nat Med 2020; 26: 453–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Caly L, Druce J, Roberts J et al. Isolation and rapid sharing of the 2019 novel coronavirus (SARS‐CoV‐2) from the first patient diagnosed with COVID‐19 in Australia. Med J Aust 2020; 212: 459–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kucharski AJ, Klepac P, Conlan AJK et al. Effectiveness of isolation, testing, contact tracing, and physical distancing on reducing transmission of SARS‐CoV‐2 in different settings: a mathematical modelling study. Lancet Infect Dis 2020; 20: 1151–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Prem K, Liu Y, Russell TW et al. The effect of control strategies to reduce social mixing on outcomes of the COVID‐19 epidemic in Wuhan, China: a modelling study. Lancet Public Health 2020; 5: e261–e270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Koutsakos M, Rowntree LC, Hensen L et al. Integrated immune dynamics define correlates of COVID‐19 severity and antibody responses. Cell Rep Med 2021; 10.1016/j.xcrm.2021.100208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mathew D, Giles JR, Baxter AE et al. Deep immune profiling of COVID‐19 patients reveals distinct immunotypes with therapeutic implications. Science 2020; 369: eabc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Krammer F. SARS‐CoV‐2 vaccines in development. Nature 2020; 586: 516–527. [DOI] [PubMed] [Google Scholar]
- 9. Public Health England . Investigation of novel SARS‐CoV‐2 variant; Variant of Concern 202012/01. 2020.
- 10. Okba NMA, Müller MA, Li W et al. Severe acute respiratory syndrome coronavirus 2‐specific antibody responses in coronavirus disease patients. Emerg Infect Dis 2020; 26: 1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Amanat F, Stadlbauer D, Strohmeier S et al. A serological assay to detect SARS‐CoV‐2 seroconversion in humans. Nat Med 2020; 26: 1033–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Juno JA, Tan HX, Lee WS et al. Humoral and circulating follicular helper T cell responses in recovered patients with COVID‐19. Nat Med 2020; 26: 1428–1434. [DOI] [PubMed] [Google Scholar]
- 13. Berry JD, Hay K, Rini JM et al. Neutralizing epitopes of the SARS‐CoV S‐protein cluster independent of repertoire, antigen structure or mAb technology. MAbs 2010; 2: 53–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Selva KJ, van de Sandt CE, Lemke MM et al. Distinct systems serology features in children, elderly and COVID patients. medRxiv 2020; 2020.2005.2011.20098459. [Google Scholar]
- 15. Tan CW, Chia WN, Qin X et al. A SARS‐CoV‐2 surrogate virus neutralization test based on antibody‐mediated blockage of ACE2–spike protein–protein interaction. Nat Biotechnol 2020; 38: 1073–1078. [DOI] [PubMed] [Google Scholar]
- 16. Rodda LB, Netland J, Shehata L et al. Functional SARS‐CoV‐2‐specific immune memory persists after mild COVID‐19. Cell 2021; 184: 169–183.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tosif S, Neeland MR, Sutton P et al. Immune responses to SARS‐CoV‐2 in three children of parents with symptomatic COVID‐19. Nat Commun 2020; 11: 5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Beavis KG, Matushek SM, Abeleda APF et al. Evaluation of the EUROIMMUN Anti‐SARS‐CoV‐2 ELISA assay for detection of IgA and IgG antibodies. J Clin Virol 2020; 129: 104468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Manalac J, Yee J, Calayag K et al. Evaluation of Abbott anti‐SARS‐CoV‐2 CMIA IgG and Euroimmun ELISA IgG/IgA assays in a clinical lab. Clin Chim Acta 2020; 510: 687–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nilsson AC, Holm DK, Justesen US et al. Comparison of six commercially available SARS‐CoV‐2 antibody assays‐choice of assay depends on intended use. Int J Infect Dis 2020; 103: 381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bond K, Nicholson S, Lim SM et al. Evaluation of serological tests for SARS‐CoV‐2: implications for serology testing in a low‐prevalence setting. J infect Dis 2020; 222: 1280–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. GeurtsvanKessel CH, Okba NMA, Igloi Z et al. An evaluation of COVID‐19 serological assays informs future diagnostics and exposure assessment. Nat Commun 2020; 11: 3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Coste AT, Jaton K, Papadimitriou‐Olivgeris M, Greub G, Croxatto A. Comparison of SARS‐CoV‐2 serological tests with different antigen targets. J Clin Virol 2021; 134: 104690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Meschi S, Colavita F, Bordi L et al. Performance evaluation of Abbott ARCHITECT SARS‐CoV‐2 IgG immunoassay in comparison with indirect immunofluorescence and virus microneutralization test. J Clin Virol 2020; 129: 104539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Naaber P, Hunt K, Pesukova J et al. Evaluation of SARS‐CoV‐2 IgG antibody response in PCR positive patients: comparison of nine tests in relation to clinical data. PLoS One 2020; 15: e0237548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kohmer N, Westhaus S, Rühl C, Ciesek S, Rabenau HF. Brief clinical evaluation of six high‐throughput SARS‐CoV‐2 IgG antibody assays. J Clin Virol 2020; 129: 104480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gasser R, Cloutier M, Prévost J et al. Major role of IgM in the neutralizing activity of convalescent plasma against SARS‐CoV‐2. bioRxiv 2020; 2020.2010.2009.333278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wines BD, Billings H, McLean MR, Kent SJ, Hogarth PM. Antibody functional assays as measures of Fc receptor‐mediated immunity to HIV – new technologies and their impact on the HIV vaccine field. Curr HIV Res 2017; 15: 202–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gong F, Dai Y, Zheng T et al. Peripheral CD4+ T cell subsets and antibody response in COVID‐19 convalescent individuals. J Clin Invest 2020; 130: 6588–6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Edridge AWD, Kaczorowska J, Hoste ACR et al. Seasonal coronavirus protective immunity is short‐lasting. Nat Med 2020; 26: 1691–1693. [DOI] [PubMed] [Google Scholar]
- 31. Short KR, Kedzierska K, van de Sandt CE. Back to the future: lessons learned from the 1918 influenza pandemic. Front Cell Infect Microbiol 2018; 8: 343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van de Sandt CE, Kreijtz JH, Rimmelzwaan GF. Evasion of influenza A viruses from innate and adaptive immune responses. Viruses 2012; 4: 1438–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Quiñones‐Parra S, Loh L, Brown LE, Kedzierska K, Valkenburg SA. Universal immunity to influenza must outwit immune evasion. Front Microbiol 2014; 5: 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hodgson SH, Mansatta K, Mallett G, Harris V, Emary KRW, Pollard AJ. What defines an efficacious COVID‐19 vaccine? A review of the challenges assessing the clinical efficacy of vaccines against SARS‐CoV‐2. Lancet Infect Dis 2021; 21: e26–e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Winkler ES, Gilchuk P, Yu J et al. Human neutralizing antibodies against SARS‐CoV‐2 require intact Fc effector functions and monocytes for optimal therapeutic protection. bioRxiv 2020; 2020.2012.2028.424554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee WS, Selva KJ, Davis SK et al. Decay of Fc‐dependent antibody functions after mild to moderate COVID‐19. medRxiv 2020; 2020.2012.2013.20248143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ilinykh PA, Huang K, Santos RI et al. Non‐neutralizing antibodies from a Marburg infection survivor mediate protection by Fc‐effector functions and by enhancing efficacy of other antibodies. Cell Host Microbe 2020; 27: 976–991.e911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Henry Dunand Carole J, Leon Paul E, Huang M et al. Both neutralizing and non‐neutralizing human H7N9 influenza vaccine‐induced monoclonal antibodies confer protection. Cell Host Microbe 2016; 19: 800–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schäfer A, Muecksch F, Lorenzi JCC et al. Antibody potency, effector function and combinations in protection from SARS‐CoV‐2 infection in vivo . bioRxiv 2020; 2020.2009.2015.298067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Crotty S. T follicular helper cell biology: a decade of discovery and diseases. Immunity 2019; 50: 1132–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stadlbauer D, Amanat F, Chromikova V et al. SARS‐CoV‐2 seroconversion in humans: a detailed protocol for a serological assay, antigen production, and test setup. Curr Protoc Microbiol 2020; 57: e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Therapeutic Goods Australia . COVID‐19 test kits included in the ARTG for legal supply in Australia. [updated 24/12/2020]. Available from: https://www.tga.gov.au/covid‐19‐test‐kits‐included‐artg‐legal‐supply‐australia
- 43. U.S. Food & Drug Administration . EUA Authorized Serology Test Performance [updated 12/07/20]. Available from: https://www.fda.gov/medical‐devices/coronavirus‐disease‐2019‐covid‐19‐emergency‐use‐authorizations‐medical‐devices/eua‐authorized‐serology‐test‐performance
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1
Supplementary figure 2
Supplementary table 1


