Abstract
Deciphering the genetic basis of plant secondary metabolism will provide useful insights for genetic improvement and enhance our fundamental understanding of plant biological processes. Although citrus plants are among the most important fruit crops worldwide, the genetic basis of secondary metabolism in these plants is largely unknown. Here, we use a high-density linkage map to dissect large-scale flavonoid metabolic traits measured in different tissues (young leaf, old leaf, mature pericarp, and mature pulp) of an F1 pseudo-testcross citrus population. We detected 80 flavonoids in this population and identified 138 quantitative trait loci (QTLs) for 57 flavonoids in these four tissues. Based on transcriptional profiling and functional annotation, twenty-one candidate genes were identified, and one gene encoding flavanone 3-hydroxylase (F3H) was functionally verified to result in naturally occurring variation in dihydrokaempferol content through genetic variations in its promoter and coding regions. The abundant data resources collected for diverse citrus germplasms here lay the foundation for complete characterization of the citrus flavonoid biosynthetic pathway and will thereby promote efficient utilization of metabolites in citrus quality improvement.
Subject terms: Secondary metabolism, Agricultural genetics
Introduction
Citrus plants, belonging to the genus Citrus L. of the family Rutaceae, are widely grown in tropical and subtropical areas worldwide and are among the most important fruit crops in the world1,2. The global citrus production was 152.4 million metric tons in 2018 (http://www.fao.org/faostat/en/#data/QC), ranking first among all fruit crops. Citrus fruits and juices provide us with rich natural products containing high amounts of vitamin C, carotenoids, and flavonoids, thereby conferring significant nutritional and pharmacological benefits3,4.
As the largest class of polyphenols, estimated to comprise over 8000 metabolites, flavonoids are widely distributed in the plant kingdom5. They usually exist in multitudinous decorated forms, and modification of their common phenol moiety is catalyzed by glycosyltransferase, acyltransferase, hydroxylase, and methyltransferase6. These diverse forms enhance the solubility and stability of flavonoids in plants and have a variety of functions. Flavonoids play important roles in several plant biological and physiological processes, such as ultraviolet-B (UV-B, 280–315 nm) protection, which is becoming increasingly critical for obtaining stable yields in deteriorating environments7,8. In addition, their pharmaceutical effects on human health as a dietary source of antioxidants have attracted increasing attention9,10.
Citrus plants are among the most important dietary sources of flavonoids, which influence their flavor and medicinal value11,12. Flavonoids exhibit tissue and species specificity in citrus plants13. The major flavonoids in citrus fruits are flavanone-O-glycosides, flavone-O/C-glycosides, and their derivatives14. Rich citrus resources provide favorable opportunities for the analysis of natural variation in flavonoids. Studies on specific flavonoids among citrus varieties have been carried out to screen outstanding citrus germplasms and breed citrus plants with optimal levels of these target compounds15,16. However, a global view and a systematic understanding of flavonoid diversity in citrus plants and the underlying genetic mechanisms are still lacking.
Reverse genetic approaches have allowed direct determination of the effects of the deficiency, or surplus, of a given protein on the biochemistry of plants17. In parallel, top-down approaches, which rely on the screening of a broad range of natural genetic resources for metabolic diversity, have also been taken18. In recent years, forward genetic approaches have been successfully applied to dissect plant metabolic traits, and a considerable number of metabolic quantitative trait loci (mQTLs) have been identified in model plants alongside several horticultural plants19,20. For example, four QTLs responsible for α-farnesene production in ripe fruit were identified in a segregating ‘Royal Gala’ × ‘Granny Smith’ population, with one major QTL on LG10 colocalizing with the MdAFS1 (α-farnesene synthase-1) gene in apple21. Concomitantly, two high-density maps based on two F2 populations were developed, and two novel loci, namely, qgf5.1 and qgf3.1, which regulate green-flesh formation resulting from the accumulation of chlorophyll in cucumber, were characterized22. In citrus plants, several fruit aroma QTLs and candidate genes in the terpenoid biosynthetic pathway were found using a mandarin linkage population23. Recently, based on a high-density integrated genetic map containing 3817 specific lengths amplified fragment (SLAF) sequencing-based molecular markers constructed from a Citrus reticulata × Poncirus trifoliata F1 pseudo-testcross population (used in this study), a total of 17 significant QTLs responsible for carotenoid content in citrus pulp were detected24.
Here, we performed ultra-performance liquid chromatography coupled with electrospray ionization and mass spectrometry (UPLC-ESI-MS/MS)-based metabolite profiling in multiple tissues (young leaf, old leaf, mature pericarp, and mature pulp) of 94 lines from the abovementioned Citrus. reticulata × Poncitrus. trifoliata F1 pseudo-testcross population25. The same metabolite profiling was also conducted on 14 varieties covering six citrus taxa. QTLs associated with the flavonoid content in these four tissues were detected based on the abovementioned high-density integrated genetic map. By combining transcriptomic analysis, gene functional annotation, and correlation analysis between metabolites and gene expression levels, twenty-one genes were identified, and an important structural gene, CitF3H, in the flavonoid pathway was characterized. The function of CitF3H in flavonoid biosynthesis has been elucidated in several other plant species; however, its functional verification and naturally occurring functional genetic variation in citrus plants have not yet been reported26. This study validates the role of CitF3H, responsible for the biosynthesis of dihydrokaempferol; a naturally occurring allelic variation of this gene exerted its effects at the enzyme activity and transcriptional levels in citrus plants. Taken together, the findings of this study lay a foundation for elucidating the citrus flavonoid pathway and provide useful resources to support breeding and engineering efforts for citrus quality improvement and the biosynthesis of beneficial natural products.
Materials and methods
Plant materials and growth conditions
The C. reticulata (C. reticulata Blanco, female parent) × P. trifoliata (P. trifoliata, male parent) F1 population, including 94 siblings, was established in 200325 and used for linkage analysis in this study. In addition, 14 citrus varieties, including C. grandis ‘Gaoban’ (GBY), C. grandis ‘Siam acidless’ (TAI), C. aurantium ‘Daidai’ (DD), C. sinensis ‘Washington’ (HSD), C. sinensis ‘Valencia’ (XC), C. reticulata ‘Mangshanyeju’ (MS), C. reticulata ‘Daoxianyeju’ (DX), C. chuana ‘Bendizao’ (BDZ), C. unshiu ‘Wenzhou’ (WZ), C. medica ‘Foshou’ (FS), C. limon ‘Eureka’ (YLK), F. hindsii ‘QK’ (QK) and F. crassifolia (PTJG), and the hybrid citrus variety C. grandis × C. sp. ‘Sugan’ (SG) were used for gene–metabolome association analysis. The citrus germplasms were collected from the National Citrus Breeding Center of Huazhong Agricultural University, Wuhan, China. Young and old leaves were collected in May and September 2018, respectively (Fig. S1). All fruits were collected at full maturity (at 210 days after flowering; 210 DAF) in 2018 (Fig. S2) and subsequently dissected to obtain two tissues, namely, pericarp (OF) and pulp (OJ), using sterilized scalpels. For each genotype of the F1 population, three biological replicates were used for metabolite profiling. Two biological replicates were sampled for metabolite profiling, and one pooled sample was used for transcriptomic analysis in each tissue of the 14 citrus varieties.
Reagents and standards
HPLC-grade acetonitrile, ethyl acetate, and methanol were purchased from Merck (Darmstadt, Germany). Water was purified using a Milli-Q ULTRA purification system (Millipore, Vimodrone, Italy). Authentic standards of eriocitrin, linarin, hesperidin, naringin, naringenin, methyl hesperidin, didymin, neodiosmin, dihydrokaempferol, and poncirin were purchased from Yuanye S&T Co. (Shanghai, China) (Data S1). Stock solutions of authentic standards were prepared as follows: the compounds were first dissolved in dimethyl sulfoxide and then in methanol.
Sample preparation and metabolite profiling based on LC-MS
Samples were homogenized to fine powders using liquid nitrogen and then stored at −80 °C until use. One hundred milligrams of freeze-dried powder were weighed and extracted overnight at 4 °C using 1.0 mL of 70% methanol.
The extract was first analyzed by liquid chromatography coupled with Q Exactive Plus mass spectrometry (LC-MS; Thermo Fisher Scientific, California, USA). The mass spectrometer was operated in full-scan data-dependent MS/MS acquisition mode. Characteristic peaks from the liquid chromatography coupled with high-resolution mass spectrometry (LC-HRMS) runs were processed using the commercial software Compound Discoverer (Thermo Fisher Scientific, California, USA). Putative flavonoids were annotated by the mzCloud and mzVault libraries integrated in Compound Discoverer. After identification, the precursor ions and product ions as well as retention times of all the target compounds were exported for subsequent analysis. Selective reaction monitoring (SRM) was performed using triple-quadrupole MS (TSQ Altis; Thermo Fisher Scientific, California, USA). Briefly, high-resolution mass spectra of precursor and product ions as well as the retention times acquired from previous nontargeted metabolite profiling were selected and transferred to construct a transition list for the SRM assay. Then, metabolites of all samples were detected in the SRM scanning mode. The LC conditions were set as follows: solvent system: 0.1% formic acid in water (solution A) and ACN (solution B); gradient program: 0–0.5 min, 98% A; 0.5–15 min, 70% A; 15–28 min, 2% A; 28–28.1 min, 98% A; 28.1–30 min, 98% A; flow rate: 0.4 mL/min.
The relative intensity of each sample was normalized by determining the average of the two adjacent quality controls after deducting the intensity of the blank sample. A data matrix containing the intensity of 80 flavonoids in 108 genotypes from 1041 runs (94 siblings × three samples × four tissue sets of F1 population and 14 citrus varieties × two samples × two-three tissue sets) was generated.
QTL mapping
A SNP-based high-density genetic map of the C. reticulata × P. trifoliata F1 population was previously constructed24. Metabolite and expression quantitative trait locus (mQTL and eQTL) detection was carried out using MapQTL software (version 6.0) with the interval mapping (IM) algorithm. The final QTL confidence intervals were assigned as the 1-LOD drop of the peak27. In this study, a QTL designated with a LOD value higher than 5 was counted and further analyzed.
Gene expression profiling using RNAseq and qRT-PCR
Total RNA was extracted using the MAGEN RNA Extraction Kit (Magen Biotechnology, Guangzhou, China) following the manufacturer’s instructions. The library consisted of RNA samples from three different tissues (young leaf, old leaf, and mature pericarp) of 14 citrus varieties. Stranded mRNA-Seq libraries with an insert fragment size of 300 bp were constructed using the KAPA Stranded mRNA-seq Kit (Kapa Biosystems, Cape Town, SA) following the manufacturer’s recommendations and sequenced using the Illumina HiSeq2000 system (paired-end 150-bp reads). Poor-quality reads, adapters, and contaminated reads among the raw reads were trimmed by Trimmomatic software (Version 0.36)28 under the default parameters. The clean reads were mapped to the C. clementina genome using HISAT (with the -G parameter). The uniquely mapped reads were extracted to estimate the expression levels of protein-coding genes using StringTie. The Ballgown procedure was followed to identify differentially expressed genes (FDR < 0.05)29.
Synthesis of the first-strand cDNA and qRT-PCR were conducted as described previously30. The gene-specific primers used for qRT-PCR are listed in Table S1.
Construction of expression plasmids and incubation in yeast
The pESC vector series (Agilent, California, US) was used for the expression of plant genes, and S. cerevisiae strain INVSc-1 (Invitrogen) was used as the expression host. Seed cultures were grown for 16–22 h at 30 °C and 210 rpm (New Brunswick C25KC shaker) in 30 mL of selection medium (minus URA) with 2% glucose. After 24 h, 20 μL the old medium was added into 30 mL of fresh medium with 2% glucose, and 24 h later, 1 mL of all substrates (10 μM solutions) was supplied per culture vessel at a final concentration of 0.2 μM along with 2% galactose. Culture broth with cells was harvested after 2 days.
Extraction of yeast fermentation products and LC-MS detection
Aliquots (500 μL) of culture broth with cells were removed from the culture vessels. When necessary, cells were separated from the culture medium by centrifugation at 14,000 rpm for 1 min. The supernatants were then transferred to another plastic tube, while the cell pellet was suspended in 500 μL of water. All samples were acidified using 25 μL of 6 N HCl and extracted using 550 μL of ethyl acetate. Combined organic fractions were dried in a centrifugal vacuum concentrator. The residues were dissolved in 200 μL of 50% aqueous methanol with 0.1% formic acid and then filtered for LC-MS detection31. LC conditions were set as follows: solvent system: 0.05% acetic acid in water (solution A) and 0.05% acetic acid in ACN (solution B); gradient program: 0–1 min, 90% A; 1.1–12 min, 5% A; 12.1–14 min, 90% A; flow rate: 0.3 mL/min.
Transient overexpression in N. benthamiana
N. benthamiana was used for transformation. The coding sequence of the CitF3H cDNA was amplified and cloned into the PH7WG2D vector. The constructs and vector-only controls were transformed by heat shock into Agrobacterium tumefaciens strain GV3101. The monoclonal bacteria were selected in LB containing 100 mg/L spectinomycin and incubated until the OD600 was 0.6–0.8. Then, the cells were resuspended in buffer solution (2 mg/mL MES and 2 mg/mL MgCl2·6H2O), supplemented with 100 mg/L AS and 700 μL of 1 mg/mL naringenin. Tobacco leaves were injected after incubation with buffer solution for 1 h at 28 °C. After 3 days, the leaves were extracted with 70% methanol-water for LC-MS analysis.
GUS staining
Two types of promoters from P. trifoliata and the allele from C. reticulata were cloned into the Gateway vector PKGWFS7 and then transferred into Agrobacterium strain GV3101. Monoclonal bacteria were selected in LB, incubated until the OD600 was 0.6–0.8, and then injected into tobacco leaves. Two days later, the leaves were cut into squares around the injection hole and placed in GUS dye solution. The leaves were kept in the dark at 37 °C and shaken for 24 h, and then, 75% ethanol was used to decolorize the leaves. When all the chlorophyll faded completely, blue areas in the leaves were clearly observed.
Recombinant protein analysis and enzyme assay
CitF3H of C. reticulata and the P. trifoliata alleles were cloned into the prokaryotic expression vector pET-32a and transformed into E. coli strain BL21 (DE3). A single colony was picked and added to 10 mL of LB containing 50 mg/L ampicillin and grown at 37 °C for 5 h. Then, the entire 10 mL LB culture was added into fresh LB (1 L) containing 50 mg/L ampicillin and grown until the OD600 reached 0.6–0.8. Isopropyl β-d-1-thiogalactopyranoside (IPTG) was added at 0.5 mM, and the strain was grown at 20 °C and 160 rpm for 16 h. The bacteria were harvested by centrifugation at 4500 rpm for 15 min and washed with lysis buffer (300 mM NaCl, 50 mM sodium phosphate buffer, 10 mM imidazole; pH 8.0). The bacterial pellet was suspended in 10 mL of lysis buffer, and the cells were sonicated 100 times for 4 s with an interval of 4 s. The supernatant was collected by centrifugation at 10,000 rpm for 1 h at 4 °C. The recombinant protein was further purified by Ni-NTA Sefinose resin. The purified protein was washed extensively with elution buffer (300 mM NaCl, 50 mM sodium phosphate buffer, 250 mM imidazole; pH 8.0) and used for enzyme assays and kinetics determination.
The in vitro enzyme activity assay for CitF3H was performed in a total volume of 100 μL containing 200 μM naringenin, 10 mM sodium ascorbate, 0.25 mM ferrous sulfate heptahydrate, 10 mM 2-oxo-glutarate, and 35 μg of purified protein in Tris-HCl buffer (80 mM, pH 8.5), and the mixture was incubated at 35 °C. After incubating for 10 min, the reaction product was extracted by ethyl acetate. The extraction solution was dried by an evaporator, and the residue was resuspended in 200 μL of methanol and then analyzed by LC-MS.
Kinetic characterization of variants of the CitF3H enzyme
To compare the kinetic constants of recombinant CitF3H proteins with C. reticulata and P. trifoliata alleles for naringenin acceptors, their activities were determined using a 0–1000 μM naringenin gradient with a fixed amount of purified protein (35 μg). All kinetic parameters were calculated using the Michaelis-Menten model (GraphPad Prism 8.0.1). All reactions were performed in three replicates.
Results
Variation in flavonoids in multiple tissues of the C. reticulata x P. trifoliata F1 pseudo-testcross population
Based on liquid chromatography-high-resolution mass spectrometry (LC-HRMS) and liquid chromatography-triple-quadrupole mass spectrometry (LC-TQMS), a total of 80 putative flavonoids were quantified in four citrus tissues spanning the young leaf, old leaf, mature pericarp (210 DAF), and mature pulp (210 DAF) (Fig. 1). Fifty-nine flavonoids were detected in the young leaves, and more than 30 flavonoids were detected in each of the other three tissues (Table S2). These metabolites include flavones, flavonols, flavanones, isoflavones, and anthocyanins, 14 of which were detected in all four tissue types, while 28 were detected in at least three tissue types.
Fig. 1. Different tissues of C. reticulata and P. trifoliata.
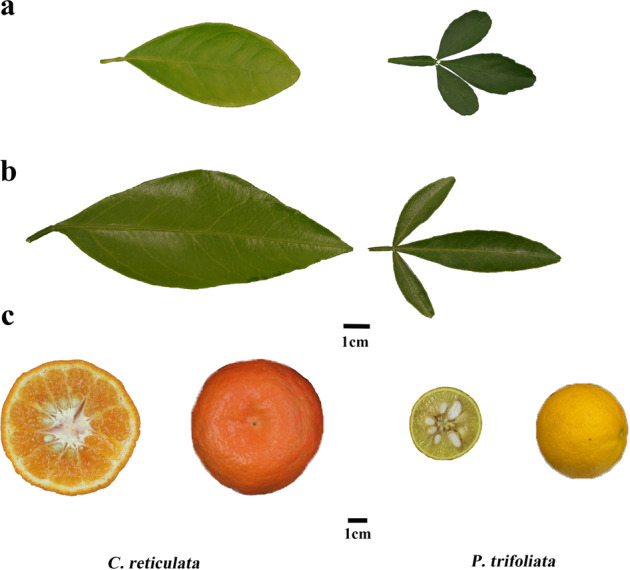
a, b and c show the young leaf, old leaf, and mature fruit of C. reticulata (left) and P. trifoliata (right), respectively
The number of flavonoids with significantly different accumulation between the two types of leaf and mature pulp was greater than that between the two types of leaf and mature pericarp (Table S3). The F1 population exhibited great diversity of flavonoids across all four tissue types, especially in the mature pulp. The mean fold changes in flavonoid content among lines were 2.27, 1.56, 14.17, and 71.54 in young leaf, old leaf, mature pericarp, and mature pulp, respectively (Table 1, Table S4). Then, correlation analysis between flavonoids in each tissue was conducted. Flavonoid accumulation patterns varied greatly between leaves and fruits in the F1 population. A similar accumulation pattern was observed in mature pericarp and mature pulp, and some of the flavanones showed high correlation, such as cmp550 (hesperidin), cmp1628 (hesperidin isomer(I)), cmp1632 (methyl hesperidin), cmp1239 (naringin isomer(I)) and cmp851 (prunin or isomer(I)). Moreover, a similar metabolic accumulation pattern was also observed between young and old leaves. For instance, the correlation coefficient between cmp829 (naringenin) and cmp856 (prunin or isomer(II)) in young and old leaves reached 0.99 (Fig. S3).
Table 1.
Range and mean fold changes of metabolic traits measured in the C. reticulata x P. trifoliata F1 Population
| Tissuea | No. of | Fold change | Fold change |
|---|---|---|---|
| metabolitesb | (Mean)c | (Range) | |
| YL | 59 | 2.27 | 1.52–4.89 |
| OL | 33 | 1.56 | 1.16–5.63 |
| OF | 35 | 14.17 | 1.69–43.88 |
| OJ | 32 | 71.54 | 1.49–221.16 |
aYL represents young leaf; OL represents old leaf; OF represents mature pericarp; OJ represents mature pulp
bNumber of flavonoids detected in this study
cAverage fold change of all flavonoids in each tissue
QTL mapping of the flavonoid content
QTLs associated with variations in the flavonoid content in the four aforementioned tissues of the F1 population were mapped (Fig. 2). A total of 138 QTLs (LOD > 5) affecting the levels of 57 flavonoids were identified. Nineteen QTLs were colocalized in at least three tissues, and two QTLs were present in all four tissues (Table S5).
Fig. 2. Chromosomal distribution of metabolic QTLs identified in this study.
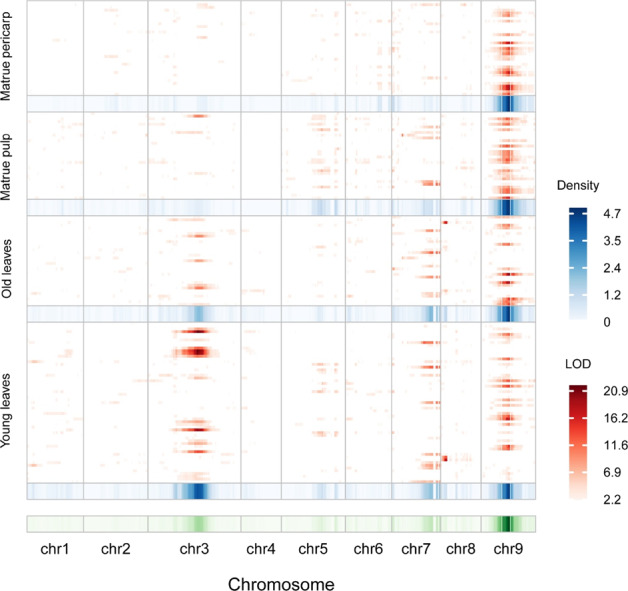
QTL regions (represented by the confidence interval) across the C. clementina genome are responsible for metabolite levels from mature pericarp, mature pulp, old leaves and young leaves. The x axis indicates genetic positions across the C. clementina genome in cM. The heat map under the x axis illustrates the density of metabolic QTLs across the genome. The window size is 10 cM. The y axis indicates every flavonoid trait detected in the four tissues. The heat map shown in red on the right illustrates LOD values for each trait
In the young leaves, 50 QTLs were mapped for 45 metabolic traits. The percentage of phenotypic variation that each QTL could explain ranged from 22.3 to 65.3%, with a mean of 35.0%. In the old leaf, 28 QTLs were mapped for 24 traits. In this case, the phenotypic variation that each QTL could explain ranged from 22.1 to 63.3%, with a mean of 32.3%. Twenty-five QTLs were mapped for 22 traits in mature pericarp, and the phenotypic variation that each QTL could explain ranged from 30.2 to 71.8%, with a mean of 43.4%. By contrast, 35 QTLs were mapped for 26 traits in the mature pulp, with the phenotypic variation explained by each QTL ranging from 26.7 to 63.0%, with a mean of 37.2% (Table 2, Table S6 and Table S7).
Table 2.
Summary of QTL mapping for metabolic traits measured in the C. reticulata x P. trifoliata F1 population
| Tissuea | No. of | No. of QTLs | No. of QTLs |
|---|---|---|---|
| metabolic traitsb | (Mean)c | (Range) | |
| YL | 45 (59) | 1.11 | 1–2 |
| OL | 24 (33) | 1.17 | 1–2 |
| OF | 22 (35) | 1.14 | 1–2 |
| OJ | 26 (32) | 1.35 | 1–3 |
aYL represents young leaf; OL represents old leaf; OF represents mature pericarp; OJ represents mature pulp
bNumber of flavonoids that have at least one QTL identified in this study; the number in parentheses represents all the metabolic traits detected in this tissue
cAverage number of QTLs identified for each flavonoid
Of the identified QTLs, 13.8% were located on Chr3, 26.1% were located on Chr7, and 46.4% were mapped on Chr9. The remaining 13.7% were distributed randomly across the other chromosomes. Three QTL hotspots (on chromosomes 3, 7, and 9) were identified with an interval size of 8.05 Mb (Ciclev19777128 bp_27830405bp), 3.10 Mb (Ciclev852254bp_3948461bp) and 3.68 Mb (Ciclev17388036 bp_21073008 bp), including 463, 391 and 98 genes, respectively (with reference to the C. clementina genome; Table S5, S8 and Fig. 232).
Candidate gene selection
We further screened candidate genes within the abovementioned hotspots. First, correlations between the expression levels of the 952 genes encoded within the hotspots (Table S9) and the relative metabolite levels (Table S10) were calculated using the transcriptomic and metabolic data collected for the 14 citrus varieties. There were 184, 228, and 278 highly correlated gene-metabolite pairs in the young leaf, old leaf, and mature pericarp, respectively (|R | ≥ 0.8, p < 0.01) (Table S11). Among these metabolite-gene correlations, 43 were identified in at least two tissues, and three were found in all three tissues (Table S12). In addition, based on functional annotation, 21 genes putatively associated with the flavonoid pathway were selected. Detailed information, including the corresponding metabolic trait, functional annotation, physical position, tissue type, and approach used for cross-validation, for these 21 candidate genes is provided in Table 3.
Table 3.
Summary of candidate genes and related information
| Gene | Metabolic trait | Functional annotation | Chr | Physical position | Tis | Cross-validation | ||
|---|---|---|---|---|---|---|---|---|
| Ciclev10021069m.g | 1532 | Eriocitrin isomer(I) | 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | Chr_3 | 21579109 | 21583976 | YL | RG-M, eQTL |
| 1232 | Kaempferitrin isomer(I) | YL | RG-M, eQTL | |||||
| 1251 | Neodiosmin | YL,OL | RG-M, eQTL | |||||
| 1249 | Linarin | YL | mQTL, RG-M, eQTL | |||||
| Ciclev10023815m.g | 1532 | Eriocitrin isomer(I) | 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | Chr_3 | 21651769 | 21652245 | YL,OL | RG-M |
| 1232 | Kaempferitrin isomer(I) | YL,OL | RG-M | |||||
| 1251 | Neodiosmin | YL,OL | RG-M | |||||
| 1249 | Linarin | YL,OL | RG-M | |||||
| Ciclev10024496m.g | 1632 | Methyl hesperidin | S-adenosyl-L-methionine-dependent methyltransferases superfamily protein | Chr_3 | 26666978 | 26668075 | YL | RG-M |
| 178 | 3,4-Dihydroxy-6,7,3′,4′-tetramethoxyflavonol | YL | RG-M | |||||
| Ciclev10026967m.g | 1027 | Hesperetin or isomer(I) | basic helix-loop-helix (bHLH) DNA-binding superfamily protein | Chr_7 | 3255156 | 3255959 | YL | RG-M |
| 856 | Prunin or isomer(II) | OL | mQTL, RG-M | |||||
| 642 | Artabotryside A or isomer(I) | OL | RG-M | |||||
| Ciclev10024361m.g | 734 | Luteolin-3′,7-Diglucoside or isomer(I) | S-adenosyl-L-methionine-dependent methyltransferases superfamily protein | Chr_3 | 25649848 | 25653317 | YL | RG-M |
| Ciclev10021734m.g | 1232 | Kaempferitrin isomer(I) | S-adenosyl-L-methionine-dependent methyltransferases superfamily protein | Chr_3 | 26976583 | 26978658 | YL,OL | RG-M |
| 1249 | Linarin | OL | mQTL, RG-M | |||||
| 1532 | Eriocitrin isomer(I) | OL | RG-M | |||||
| Ciclev10026557m.g | 1232 | Kaempferitrin isomer(I) | myb-like transcription factor family protein | Chr_7 | 2881144 | 2882746 | OL | RG-M |
| 1251 | Neodiosmin | OL | RG-M | |||||
| Ciclev10027167m.g | 877 | Keracyanin or isomer(I) | cytochrome P450, family 722, subfamily A, polypeptide 1 | Chr_7 | 1758740 | 1762044 | OL | RG-M |
| Ciclev10027097m.g | 1632 | Methyl hesperidin | cytochrome P450, family 722, subfamily A, polypeptide 1 | Chr_7 | 936705 | 940639 | OL | RG-M |
| 178 | 3,4-Dihydroxy-6,7,3′,4′-tetramethoxyflavonol | OL | RG-M | |||||
| Ciclev10023900m.g | 879 | Keracyanin or isomer(II) | S-adenosyl-L-methionine-dependent methyltransferases superfamily protein | Chr_3 | 26724892 | 26726825 | OL | RG-M |
| Ciclev10006413m.g | 1251 | Neodiosmin | basic helix-loop-helix (bHLH) DNA-binding superfamily protein | Chr_9 | 20145029 | 20146263 | OL | RG-M |
| Ciclev10027234m.g | 829 | Naringenin | cytochrome P450, family 81, subfamily D, polypeptide 2 | Chr_7 | 2678933 | 2679557 | OL | RG-M |
| 1620 | Naringin | OL | RG-M | |||||
| 856 | Prunin or isomer(II) | OL | mQTL, RG-M | |||||
| Ciclev10021728m.g | 1232 | Kaempferitrin isomer(I) | S-adenosyl-L-methionine-dependent methyltransferases superfamily protein | Chr_3 | 26847877 | 26850055 | OL | RG-M |
| 1249 | Linarin | OL | mQTL, RG-M | |||||
| 1532 | Eriocitrin isomer(I) | OL | RG-M | |||||
| Ciclev10025490m.g | 660 | Methyl hesperidin isomer(I) | UDP-Glycosyltransferase superfamily protein | Chr_7 | 2356952 | 2358892 | OF | RG-M |
| Ciclev10025311m.g | 1249 | Linarin | cytochrome P450, family 711, subfamily A, polypeptide 1 | Chr_7 | 1492717 | 1495961 | OF | RG-M |
| 1251 | Neodiosmin | OF | RG-M | |||||
| 1232 | Kaempferitrin isomer(I) | OF | RG-M | |||||
| 1532 | Eriocitrin isomer(I) | OF | RG-M | |||||
| Ciclev10027189m.g | 1249 | Linarin | O-methyltransferase family protein | Chr_7 | 1085045 | 1086475 | OF | RG-M |
| 1251 | Neodiosmin | OF | RG-M | |||||
| 1232 | Kaempferitrin isomer(I) | OF | RG-M | |||||
| 1532 | Eriocitrin isomer(I) | OF | RG-M | |||||
| Ciclev10025931m.g | 1532 | Eriocitrin isomer(I) | flavanone 3-hydroxylase | Chr_7 | 1207141 | 1210820 | OF | eQTL, RG-M |
| 1249 | Linarin | OF | eQTL, RG-M | |||||
| Ciclev10004785m.g | 1532 | Eriocitrin isomer(I) | cytochrome P450, family 71, subfamily B, polypeptide 34 | Chr_9 | 20764326 | 20767162 | OF | RG-M |
| Ciclev10023355m.g | 1249 | Linarin | O-methyltransferase family protein | Chr_3 | 19971303 | 19972959 | OF | RG-M |
| Ciclev10026344m.g | 178 | 3,4-Dihydroxy-6,7,3′,4′-tetramethoxyflavonol | S-adenosyl-L-methionine-dependent methyltransferases superfamily protein | Chr_7 | 3362860 | 3364363 | OF | RG-M |
| Ciclev10024440m.g | 1620 | Naringin | myb domain protein 14 | Chr_3 | 23417440 | 23417807 | OF | RG-M |
| 856 | Prunin or isomer(II) | OF | RG-M | |||||
| 1027 | Hesperetin or isomer(I) | OF | RG-M | |||||
| 879 | Keracyanin or isomer(II) | OF | RG-M | |||||
| 829 | Naringenin | OF | RG-M | |||||
aYL represents young leaf; OL represents old leaf; OF represents mature pericarp
bRG-M represents correlation between gene expression and metabolic level; mQTL represents QTL mapping of flavonoid contents; eQTL represents QTL mapping of relative expression level quantified by qRT-PCR
Nine of the candidate genes are located on chromosome 3, including genes encoding an O-methyltransferase, a MYB14 domain protein, two 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily proteins and five S-adenosyl-L-methionine-dependent methyltransferase superfamily proteins. Ten of the candidate genes are located on chromosome 7, including genes encoding a flavanone 3-hydroxylase (F3H), an S-adenosyl-L-methionine-dependent methyltransferase superfamily protein, a UDP-glycosyltransferase superfamily protein, an O-methyltransferase family protein, two transcription factors (a MYB-like transcription factor and a basic helix-loop-helix DNA-binding superfamily protein), and four cytochrome P450s. Another two candidate genes located on chromosome 9 are genes encoding a cytochrome P450 (family 71, subfamily B) and a basic helix-loop-helix (bHLH) DNA-binding superfamily protein (Table 3). Among these candidate genes, Ciclev10025931m.g encodes a flavanone 3-hydroxylase (F3H), which is the key enzyme that converts flavanone (naringenin) to flavonol (dihydrokaempferol) (Fig. 3). Notably, based on the relative expression level profiling conducted using qRT-PCR, eQTLs were identified for 16 candidate genes (Table S13), and the eQTL of Ciclev10021069m.g and mQTL of the associated metabolite linarin were also colocalized (Tables S7, S8, and S14). Some candidate genes selected here are likely to function as gene clusters. For example, both Ciclev10021069m.g and Ciclev10023815m.g, annotated as 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenases, are adjacent to one another.
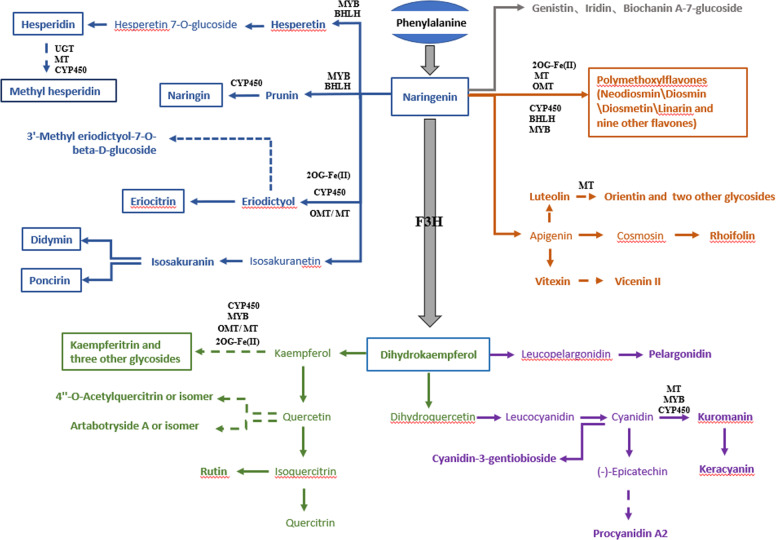
Fig. 3. A proposed citrus flavonoid metabolic network.
Candidate genes identified in this study are shown in their respective pathways. Metabolites and genes identified in this study are shown in bold. Flavonoids in blue, green, gray, orange and purple are flavanones, flavonols, isoflavones, flavones, and anthocyanins, respectively. The flavonoids in the box were verified by standards. UGT, UDP-glycosyltransferase; MT, S-adenosyl-L-methionine-dependent methyltransferase; OMT, O-methyltransferase family protein; CYP450, cytochrome P450; bHLH, basic helix-loop-helix (bHLH) DNA-binding superfamily protein; MYB, myb-like transcription factor family protein; 2OG-Fe(II), 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase; F3H, flavanone 3-hydroxylase
A proposed citrus flavonoid metabolic network involving key genes and metabolites
Based on the highlighted candidate genes and metabolites found in this study, a citrus flavonoid metabolic network is proposed (Fig. 3). The network contains five modules of flavonoid synthesis, with three modules starting from naringenin, including flavanones, flavones, and isoflavones and two modules consisting of flavonols and anthocyanins derived from dihydrokaempferol. Within this network, most genes may possess functions in metabolite modification, while most detected flavonoids are downstream products of pathways modified via glycosylation, methylation, oxygen methylation or hydroxylation. Notably, three putative isoflavones (cmp843, cmp1166 and cmp1208) are included within this network, and eight pathway branches (represented by the dotted arrow in Fig. 3) leading to methyl hesperidin and seven other flavonoids, annotated as 3′-methyl eriodictyol-7-O-beta-D-glucoside, kaempferitrin, 4”-O-acetylquercitrin or an isomer thereof, artabotryside A or an isomer thereof, orientin, vicenin II and procyanidin A2, are reported for the first time here. According to the correlation between gene expression and the level of metabolite accumulation, candidate genes were mapped onto the corresponding pathway branches. For instance, Ciclev10024496m.g, which is annotated as an S-adenosyl-L-methionine-dependent methyltransferase and correlated with the levels of cmp178 (3,4-dihydroxy-6,7,3′,4′-tetramethoxyflavonol) and cmp1632 (methyl hesperidin), is very likely associated with the branch of methyl hesperidin.
Notably, some flavonoids in the network have medicinal value and are enriched in P. trifoliata, which is utilized as traditional Chinese medicine. For example, here, the linarin content in P. trifoliata, which was shown to possess antipyretic, analgesic, and anti-inflammatory activities33, was 1672 times higher than that in C. reticulata. Moreover, the levels of cmp1234 (rhoifolin) and naringin in P. trifoliata, both of which have been regarded as indicators of the efficacy of ECG (Exocarpium Citri Grandis, a precious Chinese medicine with a history spanning hundreds of years)34, are over 700 times more abundant than those in C. reticulata (Table S15).
Overexpression of CitF3H leads to the generation of dihydrokaempferol in yeast and tobacco
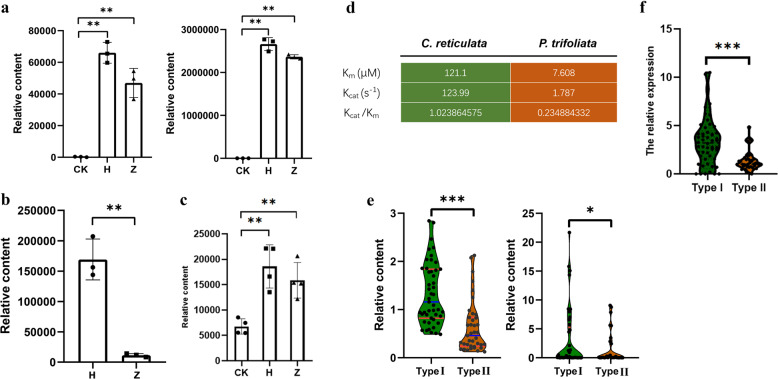
Flavanone 3-hydroxylase (F3H) is a much-studied upstream structural gene of the flavonoid biosynthetic pathway (Fig. 3 and Fig. S4a). To characterize the function of CitF3H (Ciclev10025931m.g), yeast lines overexpressing C. reticulata and P. trifoliata alleles were constructed. Following the provision of the putative substrate naringenin, the supernatant and precipitate of the yeast culture were extracted for metabolite measurement. Dihydrokaempferol was enriched mainly in the supernatant, and the intensity of dihydrokaempferol increased dramatically in the yeast overexpression lines expressing both the C. reticulata and P. trifoliata alleles. Moreover, the intensity of dihydrokaempferol in the line expressing the C. reticulata allele was significantly higher than that with the P. trifoliata allele (Fig. 4a and Fig. S4d–i), which is consistent with the content of dihydrokaempferol detected in the young leaves of C. reticulata and P. trifoliata, respectively (Fig. 4b).
Fig. 4. The level of dihydrokaempferol is associated with the genetic variants of CitF3H in the coding and promoter regions.
a Validation of the candidate gene CitF3H in yeast. The left panel shows the yeast precipitate, and the right panel shows the supernatant. CK represents the control line, H represents the overexpression line with the C. reticulata allele, and Z represents the overexpression line with the P. trifoliata allele. b The relative content of dihydrokaempferol in the young leaf. c The relative content of dihydrokaempferol in the leaves of two transient overexpression lines of N. benthamiana. d The kinetic parameters of recombinant CitF3H proteins with the C. reticulata and P. trifoliata alleles, and the data are presented for three replicates in one assay. e The relative content of dihydrokaempferol in the type I and type II genotypes. The left panel shows the young leaf, and the right panel shows mature pulp. f The relative expression level of CitF3H in type I and type II genotypes. All the results are based on the t-test of independent samples (*p < 0.05, **p < 0.01 and ***p < 0.001)
To assess the role of the proteins in vivo, transient overexpression of CitF3H in N. benthamiana was performed. The content of dihydrokaempferol in lines overexpressing either the C. reticulata or P. trifoliata allele was significantly higher than that in the negative control. Consistent with the results in yeast, the dihydrokaempferol content in the line expressing the C. reticulata allele was higher than that in the line expressing the P. trifoliata allele (Fig. 4c). Similar results were observed when the substrate naringenin was coinjected with Agrobacterium into tobacco leaves (Fig. S5).
Identification of functional genetic variants of CitF3H
By comparing the coding regions of CitF3H of C. reticulata and P. trifoliata, we found 13 SNPs resulting in five amino acid differences and one indel in P. trifoliata (Fig. S6). Recombinant CitF3H proteins of C. reticulata and P. trifoliata were then expressed and purified for enzymatic assays. The Michaelis-Menten equations of the recombinant CitF3H proteins with the C. reticulata and P. trifoliata alleles were fitted (Table S16 and Fig. S7). The kinetic parameters of C. reticulata were quantified, yielding a Km (μM) of 121.1 and Kcat (s −1) of 123.99, while for P. trifoliata, the Km (μM) and Kcat (s −1) were 7.608 and 1.787, respectively. The recombinant CitF3H protein of the C. reticulata allele therefore clearly shows superior catalytic efficiency compared with that of the P. trifoliata allele (Fig. 4d).
To further dissect the relationship between the genetic variations in the promoters of CitF3H and the content of dihydrokaempferol, the activity of three different types (one is the allele of C. reticulata, which is homologous at this locus; the other two are alleles of P. trifoliata, which is heterozygous at this locus; Data S2) of promoter sequences was compared through GUS staining. The staining area of the two P. trifoliata promoter types was larger than that of C. reticulata, and the staining area of the P. trifoliata promoter allele A (short deletion) was larger than that of allele B (long deletion; Fig. S8). The promoter activity was consistent with the expression level of CitF3H in C. reticulata and P. trifoliata. In both the young leaf and mature pulp, the expression level of CitF3H in P. trifoliata was higher than that in C. reticulata (Fig. S9).
Furthermore, a molecular marker designed for discriminating the different promoter sequences was validated in the F1 population. The progeny can be grouped into two types based on their genotypes: type I (allele of C. reticulata and allele A of P. trifoliata) and type II (allele of C. reticulata and allele B of P. trifoliata) (Fig. S10). The relative intensity of dihydrokaempferol in the type I lines was significantly higher than that in the type II lines (p = 0.05, t-test; Fig. 4e), which is consistent with the relative expression between the type I and type II lines (p = 0.05, t-test; Fig. 4f). Hence, genetic variations in both the coding and promoter regions of CitF3H are responsible for the variation in dihydrokaempferol content in this population.
Discussion
Forward genetic studies integrating metabolic profiling with quantitative genetics have been conducted in many plant species, including mainly model plants and crops such as maize, rice, and wheat35,36. These studies successfully elucidated the genetic basis underlying metabolic variation. Owing to the long growth cycle and breeding process, it is far more difficult to conduct forward genetic studies in perennial woody plants. However, some important traits, such as fruit flavor and quality, have still been dissected using QTL analysis37–39, such as identification of QTLs controlling aroma volatile production in a Citrus reticulata population23 and mapping of carotenoid content in citrus fruits using a high-density genetic map24. As important fruit crops worldwide, citrus plants are rich in specialized metabolites, including flavonoids and carotenoids. Detection and comparison of flavonoids at different developmental stages in different citrus species and tissues and mining of candidate genes related to flavonoid metabolism have been previously performed40–42. However, studies based on QTL mapping and identification of candidate genes underlying the flavonoid content in citrus plants are lacking.
By adopting widely targeted metabolomic and transcriptomic analyses and combining results from the segregating population and diverse citrus varieties, this study provided valuable insights allowing the dissection of flavonoid biosynthesis in citrus plants. A number of QTLs overlapped in at least three tissues, indicating that the content of at least some flavonoids may be under common genetic control across different tissue types. However, most QTLs were tissue specific, suggesting distinct genetic and biochemical regulatory bases between tissues, which is consistent with the reported tissue specificity of flavonoids in citrus plants13,42. Moreover, the metabolic diversity among different tissues might result from spatially or temporally distinct expression patterns of genes, such as tissue-specific transcript expression and alternative splicing43.
Flavanone 3-hydroxylase (F3H) is a key enzyme channeling carbon flow toward the production of 3-hydroxylated flavonoids, including flavonols, proanthocyanidins, and anthocyanidins, and it has been well characterized in a wide range of plant species44–46. Petunia hybrida tends to accumulate flavonol glycosides and anthocyanins; however, Citrus paradisi is known for its accumulation of flavanone diglycosides47. Different species display considerably different flavonoid accumulation patterns, which possibly results from the function of F3H. Recently, it was reported that in a sorghum variety lacking endogenous F3H activity, heterologous F3H overexpression led to enhanced accumulation of flavonols and flavan-3-ols as well as considerable changes in the flavonoid profile. However, no such changes in the flavonoid profile occurred in tobacco with endogenous F3H activity, further demonstrating the functional complexity of F3H48.
Here, we verified the function of CitF3H in citrus plants, demonstrating that it can convert naringenin into dihydrokaempferol. We further revealed that genetic variations in the coding region of CitF3H between the two parental lines resulted in differences in its enzyme activity. Moreover, an indel variation in the promoter region of CitF3H can affect the content of dihydrokaempferol by varying the transcriptional level of CitF3H. Considering the key position of F3H in the flavonoid pathway, these findings regarding natural variations in CitF3H could aid in the genetic improvement and synthesis of beneficial flavonoids in citrus plants. It has been indicated that enzymes that enable the formation of flavonoid scaffold structures probably first appeared by recruiting enzymes from primary metabolic pathways. Subsequently, enzymes that belong to superfamilies, such as 2-oxoglutarate-dependent dioxygenase, cytochrome P450, and short-chain dehydrogenase/reductase, modified these structures49. Interestingly, most of the 21 candidate genes highlighted here were annotated as downstream modification enzymes, including CYP450s, 2OG-Fe(II)s, methyltransferases, oxymethyltransferases and glycosyltransferases. Enzymes responsible for the final steps of metabolite synthesis generally make greater contributions to the natural variation in metabolite abundance than those earlier in the pathway5. Further functional and biochemical investigation of these remaining candidate genes will undoubtably provide insights into the expansion and supplementation of the flavonoid pathway in citrus plants.
Genes in our proposed flavonoid metabolic network were mapped based on the correlation of metabolite accumulation and gene expression level, in which one gene may be associated with multiple metabolites or, conversely, multiple genes may be associated with the same flavonoid. The network contained genes that were adjacent to each other and had the same functional annotation, namely, Ciclev10023815m.g and Ciclev10021069m.g, both of which encode a 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase. Whether these gene clusters are variable among different citrus species warrants further study. On the other hand, the similarity in the chemical structure of flavonoids presents a massive challenge in the identification of these compounds. In this study, authentic standards and high-resolution m/z and MS/MS fragmentation were used for the identification and annotation of flavonoids. That said, further accurate structural determination of additional metabolites would be helpful for elucidation of a more complete network.
Although they exhibit an astringent taste, the Poncirus trifoliata [L.] Raf. the fruit has been widely used as a traditional medicine in East Asia owing to the proposed anticancer and anti-inflammatory activities of the metabolites they contain50. Some specific flavonoids with important pharmacological value, such as linarin, cmp1234 (annotated as rhoifolin) and naringin, were more enriched in P. trifoliata than in C. reticulata. Therefore, P. trifoliata and the F1 population represent useful resources for the improvement of the popular fruit crop C. reticulata. The enhancement of compounds with high medicinal value and lacking undesirable flavors, such as bitterness and astringency, could be a direction for future fruit breeding.
In summary, the multiomics analysis based on a citrus genetic population and diverse varieties generated rich data resources and identified a number of QTLs for flavonoid content in multiple tissues, laying a foundation for elucidation of the flavonoid biosynthetic pathway and presents a blueprint that can be followed for unveiling further metabolic pathways in citrus.
Supplementary information
Author contributions
W.W.W. and X.X.D. conceived and supervised the research. J.L.M., Z.H.Z., X.Z., H.J.Q., Z.Q.F., Q.H.Z., and L.Y. performed the experiments. J.L.Y., J.L.M., and H.J.Q. conducted data analysis. W.W.W. and J.L.M. wrote the manuscript. A.R.F., Y.J.C., and X.X.D. edited the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Xiuxin Deng, Email: xxdeng@mail.hzau.edu.cn.
Weiwei Wen, Email: wwwen@mail.hzau.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41438-021-00472-8.
References
- 1.Zou Z, Xi W, Hu Y, Nie C, Zhou Z. Antioxidant activity of Citrus fruits. Food Chem. 2016;196:885–896. doi: 10.1016/j.foodchem.2015.09.072. [DOI] [PubMed] [Google Scholar]
- 2.Mehl F, et al. Differentiation of lemon essential oil based on volatile and non-volatile fractions with various analytical techniques: a metabolomic approach. Food Chem. 2014;143:325–335. doi: 10.1016/j.foodchem.2013.07.125. [DOI] [PubMed] [Google Scholar]
- 3.Yoo KM, Moon B. Comparative carotenoid compositions during maturation and their antioxidative capacities of three citrus varieties. Food Chem. 2016;196:544–549. doi: 10.1016/j.foodchem.2015.09.079. [DOI] [PubMed] [Google Scholar]
- 4.Teixeira, F. et al. Addition of orange peel in orange jam: evaluation of sensory, physicochemical, and nutritional characteristics. Molecules10.3390/molecules25071670 (2020). [DOI] [PMC free article] [PubMed]
- 5.Wen W, Alseekh S, Fernie AR. Conservation and diversification of flavonoid metabolism in the plant kingdom. Curr. Opin. Plant Biol. 2020;55:100–108. doi: 10.1016/j.pbi.2020.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Yonekura-Sakakibara K, Hanada K. An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J.: Cell Mol. Biol. 2011;66:182–193. doi: 10.1111/j.1365-313X.2011.04493.x. [DOI] [PubMed] [Google Scholar]
- 7.Peng M, et al. Differentially evolved glucosyltransferases determine natural variation of rice flavone accumulation and UV-tolerance. Nat. Commun. 2017;8:1975. doi: 10.1038/s41467-017-02168-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tohge T, et al. Characterization of a recently evolved flavonol-phenylacyltransferase gene provides signatures of natural light selection in Brassicaceae. Nat. Commun. 2016;7:12399. doi: 10.1038/ncomms12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bojic M, Males Z, Antolic A, Babic I, Tomicic M. Antithrombotic activity of flavonoids and polyphenols rich plant species. Acta Pharm. 2019;69:483–495. doi: 10.2478/acph-2019-0050. [DOI] [PubMed] [Google Scholar]
- 10.Grigalius I, Petrikaite V. Relationship between antioxidant and anticancer activity of trihydroxyflavones. Molecules. 2017;22:2169. doi: 10.3390/molecules22122169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, et al. Cit1,2RhaT and two novel CitdGlcTs participate in flavor-related flavonoid metabolism during citrus fruit development. J. Exp. Bot. 2019;70:2759–2771. doi: 10.1093/jxb/erz081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teixeira F, et al. Addition of orange peel in orange jam: evaluation of sensory, physicochemical, and nutritional characteristics. Molecules. 2020;25:1670. doi: 10.3390/molecules25071670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durand-Hulak M, et al. Mapping the genetic and tissular diversity of 64 phenolic compounds in Citrus species using a UPLC–MS approach. Ann. Bot. 2015;115:861–877. doi: 10.1093/aob/mcv012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abad‑García B, et al. Polyphenolic contents in Citrus fruit juices: authenticity assessment. Eur. Food Res. Technol. 2014;238:803–818. doi: 10.1007/s00217-014-2160-9. [DOI] [Google Scholar]
- 15.Huang R, et al. Antioxidant and pancreatic lipase inhibitory effects of flavonoids from different citrus peel extracts: an in vitro study. Food Chem. 2020;326:126785. doi: 10.1016/j.foodchem.2020.126785. [DOI] [PubMed] [Google Scholar]
- 16.Chen, Q. et al. Profiling of flavonoid and antioxidant activity of fruit tissues from 27 Chinese local citrus cultivars. Plants10.3390/plants9020196 (2020). [DOI] [PMC free article] [PubMed]
- 17.Stitt M, Sonnewald U. Regulation of metabolism in transgenetic plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1995;46:341–368. doi: 10.1146/annurev.pp.46.060195.002013. [DOI] [Google Scholar]
- 18.Fernie AR, Tohge T. The genetics of plant metabolism. Annu. Rev. Genet. 2017;51:287–310. doi: 10.1146/annurev-genet-120116-024640. [DOI] [PubMed] [Google Scholar]
- 19.Bauchet G, et al. Identification of major loci and genomic regions controlling acid and volatile content in tomato fruit: implications for flavor improvement. N. Phytologist. 2017;215:624–641. doi: 10.1111/nph.14615. [DOI] [PubMed] [Google Scholar]
- 20.Jia D, et al. Apple fruit acidity is genetically diversified by natural variations in three hierarchical epistatic genes: MdSAUR37, MdPP2CH and MdALMTII. Plant J.: Cell Mol. Biol. 2018;95:427–443. doi: 10.1111/tpj.13957. [DOI] [PubMed] [Google Scholar]
- 21.Souleyre EJF, et al. Genetic control of alpha-farnesene production in apple fruit and its role in fungal pathogenesis. Plant J.: Cell Mol. Biol. 2019;100:1148–1162. doi: 10.1111/tpj.14504. [DOI] [PubMed] [Google Scholar]
- 22.Bo K, et al. QTL mapping and genome-wide association study reveal two novel loci associated with green flesh color in cucumber. BMC Plant Biol. 2019;19:243. doi: 10.1186/s12870-019-1835-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu Y, et al. Identification of QTLs controlling aroma volatiles using a ‘Fortune’ x ‘Murcott’ (Citrus reticulata) population. BMC Genomics. 2017;18:646. doi: 10.1186/s12864-017-4043-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng X, et al. SLAF-based construction of a high-density genetic map and its application in QTL mapping of carotenoids content in citrus fruit. J. Agric. Food Chem. 2018;67:994–1002. doi: 10.1021/acs.jafc.8b05176. [DOI] [PubMed] [Google Scholar]
- 25.Tan M, Song J, Deng X. Production of two mandarin × trifoliate orange hybrid populations via embryo rescue with verification by SSR analysis. Euphytica. 2007;157:155–160. doi: 10.1007/s10681-007-9407-5. [DOI] [Google Scholar]
- 26.Han Y, et al. Functional analysis of two flavanone-3-hydroxylase genes from Camellia sinensis: a critical role in flavonoid accumulation. Genes. 2017;8:300. doi: 10.3390/genes8110300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graham IA, et al. The genetic map of Artemisia annua L. identifies loci affecting yield of the antimalarial drug artemisinin. Science. 2010;327:328–331. doi: 10.1126/science.1182612. [DOI] [PubMed] [Google Scholar]
- 28.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu S, et al. The citrus transcription factor CsMADS6 modulates carotenoid metabolism by directly regulating carotenogenic genes. Plant Physiol. 2018;176:2657–2676. doi: 10.1104/pp.17.01830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berim A, Gang DR. Production of methoxylated flavonoids in yeast using ring A hydroxylases and flavonoid O-methyltransferases from sweet basil. Appl. Microbiol. Biotechnol. 2018;102:5585–5598. doi: 10.1007/s00253-018-9043-0. [DOI] [PubMed] [Google Scholar]
- 32.Wu GA, et al. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat. Biotechnol. 2014;32:656–662. doi: 10.1038/nbt.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quintin J, Lewin G. Semisynthesis of linarin, acacetin, and 6-iodoapigenin derivatives from diosmin. J. Nat. Prod. 2004;67:1624–1627. doi: 10.1021/np040079b. [DOI] [PubMed] [Google Scholar]
- 34.Kong F, et al. Optimization of extraction flavonoids from Exocarpium Citri Grandis and evaluation its hypoglycemic and hypolipidemic activities. J. Ethnopharmacol. 2020;262:113178. doi: 10.1016/j.jep.2020.113178. [DOI] [PubMed] [Google Scholar]
- 35.Gong L, et al. Genetic analysis of the metabolome exemplified using a rice population. Proc. Natl Acad. Sci. USA. 2013;110:20320–20325. doi: 10.1073/pnas.1319681110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riedelsheimer C, et al. Genome-wide association mapping of leaf metabolic profiles foe dissecting complex traits in maize. Proc. Natl Acad. Sci. USA. 2012;109:8872–8877. doi: 10.1073/pnas.1120813109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goto, S. et al. QTL mapping of male sterility and transmission pattern in progeny of Satsuma mandarin. PLoS ONE13, e0200844 (2018). 10.1371/journal.pone.0200844. [DOI] [PMC free article] [PubMed]
- 38.Huang M, et al. Construction of high-density genetic maps and detection of QTLs associated with Huanglongbing tolerance in citrus. Front. Plant Sci. 2018;9:1694. doi: 10.3389/fpls.2018.01694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lima RPM, Curtolo M, Merfa MV, Cristofani-Yaly M, Machado MA. QTLs and eQTLs mapping related to citrandarins’ resistance to citrus gummosis disease. BMC Genomics. 2018;19:516. doi: 10.1186/s12864-018-4888-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ito T, Fujimoto S, Suito F, Shimosaka M, Taguchi G. C-Glycosyltransferases catalyzing the formation of di-C-glucosyl flavonoids in citrus plants. Plant J.: Cell Mol. Biol. 2017;91:187–198. doi: 10.1111/tpj.13555. [DOI] [PubMed] [Google Scholar]
- 41.Frydman A, et al. Citrus fruit bitter flavors: isolation and functional characterization of the gene Cm1,2RhaT encoding a 1,2 rhamnosyltransferase, a key enzyme in the biosynthesis of the bitter flavonoids of citrus. Plant J.: Cell Mol. Biol. 2004;40:88–100. doi: 10.1111/j.1365-313X.2004.02193.x. [DOI] [PubMed] [Google Scholar]
- 42.Wang S, et al. Characterization and metabolic diversity of flavonoids in citrus species. Sci. Rep. 2017;7:10549. doi: 10.1038/s41598-017-10970-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thatcher SR, et al. Genome-wide analysis of alternative splicing in Zea mays: landscape and genetic regulation. Plant Cell. 2014;26:3472–3487. doi: 10.1105/tpc.114.130773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Owens DK, Crosby KC, Runac J, Howard BA, Winkel BS. Biochemical and genetic characterization of Arabidopsis flavanone 3beta-hydroxylase. Plant Physiol. Biochem. 2008;46:833–843. doi: 10.1016/j.plaphy.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 45.Xiong S, et al. Molecular cloning and characterization of a flavanone 3-hydroxylase gene from Artemisia annua L. Plant Physiol. Biochem. 2016;105:29–36. doi: 10.1016/j.plaphy.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 46.Kim JH, Lee YJ, Kim BG, Lim Y, Ahn J-H. Flavanone 3β-hydroxylases from rice: key enzymes for favonol and anthocyanin biosynthesis. Mol. Cells. 2007;25:312–316. [PubMed] [Google Scholar]
- 47.Durren RL, McIntosh CA. Flavanone-7-O-glucosyltransferase activity from Petunia hybrida. Phytochemistry. 1999;52:793–798. doi: 10.1016/S0031-9422(99)00307-6. [DOI] [PubMed] [Google Scholar]
- 48.Wang, L. et al. Transgenic expression of flavanone 3-hydroxylase redirects flavonoid biosynthesis and alleviates anthracnose susceptibility in sorghum. Plant Biotechnol. J. 10.1111/pbi.13397 (2020). [DOI] [PMC free article] [PubMed]
- 49.Yonekura-Sakakibara K, Higashi Y, Nakabayashi R. The origin and evolution of plant flavonoid metabolism. Front. Plant Sci. 2019;10:943. doi: 10.3389/fpls.2019.00943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jang Y, Kim EK, Shim WS. Phytotherapeutic effects of the fruits of Poncirus trifoliata (L.) Raf. on cancer, inflammation, and digestive dysfunction. Phytother. Res. 2018;32:616–624. doi: 10.1002/ptr.6008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.