Abstract
Schistosomiasis remains the fourth most prevalent parasitic disease affecting over 200 million people worldwide. Control efforts have focussed on the disruption of the life cycle targeting the parasite, vector and human host. Parasite burdens are highly skewed, and the majority of eggs are shed into the environment by a minority of the infected population. Most morbidity results from hepatic fibrosis leading to portal hypertension and is not well-correlated with worm burden. Genetics as well as environmental factors may play a role in these skewed distributions and understanding the genetic risk factors for intensity of infection and morbidity may help improve control measures. In this review, we focus on how genetic factors may influence parasite load, hepatic fibrosis and portal hypertension. We found 28 studies on the genetics of human infection and 20 studies on the genetics of pathology in humans. S. mansoni and S. haematobium infection intensity have been showed to be controlled by a major quantitative trait locus SM1, on chromosome 5q31-q33 containing several genes involved in the Th2 immune response, and three other loci of smaller effect on chromosomes 1, 6, and 7. The most common pathology associated with schistosomiasis is hepatic and portal vein fibroses and the SM2 quantitative trait locus on chromosome six has been linked to intensity of fibrosis. Although there has been an emphasis on Th2 cytokines in candidate gene studies, we found that four of the five QTL regions contain Th17 pathway genes that have been included in schistosomiasis studies: IL17B and IL12B in SM1, IL17A and IL17F in 6p21-q2, IL6R in 1p21-q23 and IL22RA2 in SM2. The Th17 pathway is known to be involved in response to schistosome infection and hepatic fibrosis but variants in this pathway have not been tested for any effect on the regulation of these phenotypes. These should be priorities for future studies.
Keywords: schistosomiasis, fibrosis, Th17, intensity of infection, QTL, linkage
Introduction
Schistosomiasis is caused by digenic trematodes of the genus Schistosoma with Schistosoma mansoni and Schistosoma haematobium causing the majority of human infections. Adult parasites live in the veins around the gut and bladder and eggs are excreted in feces or urine and infect snails in fresh water. Parasite numbers are amplified in the snail intermediate host and human infective stages then emerge that can penetrate human skin when people enter the water. Schistosomiasis induces acute, severe, and chronic morbidity among those who are infected and can cause liver and bladder fibrosis and eventually bladder or colorectal cancer (1). Although exposure to water infested with the infective schistosome cercariae is the main risk factor for schistosomiasis there is considerable variation in infection intensity between people with similar exposures and schistosomiasis cases aggregate in families, some of this variation has been attributed to the genetics of the human immune response (2–4).
A review of the genetics of human susceptibility to schistosomiasis related fibrosis has been published recently (5), but there has been no review of the genetics of schistosome infection since 2008 (2).
A fundamental understanding of the genetics of schistosomiasis susceptibility and high worm load may contribute to rational design of interventions, including vaccines (6). For example, it has recently been shown that a set of 32 SNPs in 10 genes can predict susceptibility to severe hepatic disease among Brazilians with 63% sensitivity and 90% specificity (5). In the present review of the genetics of human susceptibility to schistosomiasis we focus on loci associated with egg/worm burden and hepatic fibrosis.
We therefore present an updated review of the genes and variants that have been found associated with schistosomiasis infection intensity and liver disease, together with a review of genes within QTL that could be prioritized for future analyses. We have excluded the HLA region since we have only identified one study of genes in this region since they were was last reviewed (7, 8).
Epidemiology and Treatment
The disease affects almost 240 million people, and 700 million are at risk of infection in 74 countries, the majority being in Africa, Asia and South America (9, 10). Between 3 and 56 million disability-adjusted life years (DALYs) are lost per annum and 280,000 deaths per annum have been attributed to effects of schistosomiasis (11–13). Approximately 85% of infections occur in sub-Saharan Africa and at least 90% of people requiring treatment for schistosomiasis live in Africa (14). Schistosomiasis can also be associated with chronic anemia, childhood growth stunting, protein calorie malnutrition, cognitive disability, and poor school performance (15–20).
Control of schistosomiasis has continued to rely mainly on mass drug administration (MDA) of school-aged children using the anti-schistosomal drug praziquantel (PZQ) (21, 22). Although this strategy has reduced morbidity, the impact on prevalence has been more limited, because praziquantel does not kill immature schistosomes (23, 24), coverage remains restricted and only school age children are routinely treated. Vaccines are in development but phase three trials have not been successful (25).
Immune Responses During Schistosome Infections
Immune responses to penetrating and migrating Schistosoma larvae (schistosomula) and maturing adults are predominantly Th1 (26). Excretory/secretory Schistosoma antigens damage host barrier cells which release alarmins, activate innate cells and induce proinflammatory cytokines (IL1B, IL6, IL17, TNF, and IFNG) (27). About 6 weeks after infection, Schistosoma eggs are deposited in tissues (the liver and the intestine or the bladder) and trigger an expansion of Th2 cells (28). Schistosoma egg antigens also directly bind receptors on antigen presenting cells inhibiting IL12 production and consequently Th1 responses (29). Th2 responses can also be induced independently of egg deposition as infection with single sex schistosomes induce pre-patent IL4 production by CD4 T cells (30). Schistosome specific Th2 responses are downmodulated in long-standing infections (31) and this is associated with a development of regulatory cells producing IL10 and transforming growth factor beta (TGFB). This not only allows the parasite to survive in the host and minimize host tissue damage but also modulates host immune responses to unrelated antigens including allergens, self-antigens, and vaccines.
Schistosome egg secretions are highly antigenic (31) and typically induce polarized granulomatous Th2 responses (32). Granulomas form around eggs lodged in tissues to protect tissue cells (33) but persistent host CD4 Th2 cell mediated responses to parasite eggs cause fibrosis (34). The pro-fibrotic Th2 cytokine IL13 is associated with periportal fibrosis in humans (35). Beyond Th2 cytokine responses, intensified hepatic granulomatous inflammation in S. mansoni infected mice is associated with high levels of IL17 and controlled by IFNG (36). In human schistosomiasis, IL17 producing CD4 T helper cells are associated with ultrasound textural abnormalities while T regulatory cells are associated with reductions in this pathology (37).
Phenotypes
Schistosomal Fecal Egg Count and Worm Burden
Studies of the human genetics of susceptibility to schistosomiasis have focussed on two classes of phenotype; infection associated phenotypes and pathology related phenotypes. Infection associated phenotypes are usually egg counts or worm burden estimates but sometimes total IgE as a marker of the immune response (38). Eggs counts are obtained by the Kato Katz (KK) method for S. mansoni and by urine filtration for S. haematobium and worm burdens are estimated by measuring the circulating cathodic antigen (CCA) in urine or circulating anodic antigen (CAA) in plasma (39) that are produced by adult worms.
Approximately 80% of the environmental egg burden from helminths including schistosomes, derives from ~20% of the cases (40). For example 22 out of 119 Kenyan school children had developed high S. mansoni egg burden (>100 eggs per gram of feces) 12 months after treatment, whilst 70 children still had low (<30 epg) egg burdens (41) and this effect was not correlated with the amount of water contact. This tendency for some people to develop high infections even after treatment has been attributed to variation in genetic risk (42).
Schistosome egg burdens are also highly skewed by age with egg burdens increasing up to the age of puberty and declining thereafter (43–46). A study in Brazil found that children under 19 had egg burdens that were over ten times higher than older adults (42). The higher intensity and frequency of infections in children may be due to the slow development of immunity to schistosomes. Possibly, the antigens that are exposed by dying worms cross react with larval antigens and stimulate a protective anti-larval IgE response. The long life span of adult worms (5–15 years) means that it takes many years for children to be exposed to sufficient dying worms to develop an IgE response to the larvae (47). High anti-parasite IgE levels have been associated with resistance and high specific IgG4 has been associated with susceptibility and it has been proposed that the ratio of these two immunoglobulins controls resistance to schistosomiasis (48–51).
Schistosomiasis Associated Hepatic and Periportal Fibrosis and Portal Vein Hypertension
Although schistosomes cause a wide range of symptoms and fibrotic lesions can form around egg granulomas in many tissues, the main indicator of S. mansoni and S. japonicum pathology is hepatic fibrosis (HF) and periportal fibrosis (PPF) (52). WHO guidelines provide scoring scales for HF and PPF (53) and both scales are used as phenotypes in genetic research (54). HF and PPF is caused by extracellular matrix forming around schistosome eggs. In the hepatic portal vein this can lead to portal hypertension (PH) (55, 56), ultimately, some patients with PH die of internal bleeding, superinfections, or heart or kidney failure. S. haematobium is associated with bladder cancer and S. mansoni may be associated with hepatocellular carcinoma (57) but genetic studies of the pathology of schistosomiasis have focussed almost exclusively on hepatic and periportal fibrosis and in this review all references to pathology are to these closely related conditions unless otherwise indicated. Fibrosis can be measured by ultra-sound scan, although there are concerns about the accuracy and reproducibility of ultrasound (58); additional markers and protocols for grading pathology are being developed but are not well-validated (59–61).
Genome Wide Linkage Studies Discover Schistosomiasis Susceptibility Loci
The reviews of linkage and candidate gene studies of schistosomiasis are broken down into two sections by phenotype: (1) studies of infection status, which is usually determined by egg count in urine or feces and (2) studies of pathology which is mainly periportal fibrosis determined by ultra-sound. Relevant publications were identified by searching PubMed using the terms in Supplementary Table 1.
Heritability of Schistosomiasis Infection Risk
Heritability, the proportion of risk attributable to genetic variation, must be substantial to be detectable. A summary of heritability estimates for schistosomiasis are shown in Table 1. Studies in Brazil (62–64); Kenya (46); and China (65, 66) have each estimated similar proportions of the variance of S. mansoni egg count that are attributable to genetic variation, with additive heritability (h2) estimates of 23–31%. However, there were striking differences in the two estimates for heritability of infection with S. japonicum in China using variance components (VC) (0 & 58%) (Table 1), which have been discussed by others (2).
Table 1.
Heritability estimates for genetic component of risk of schistosomiasis in different populations.
| Parasite | Phenotype | Country and district | Heritability estimate and method eg h2 | References |
|---|---|---|---|---|
| S. mansoni | Egg count | Brazil, Minas Gerais | 23% (h2) | (62) |
| S. mansoni | Egg count | Brazil, Minas Gerais | 27% (VC) | (63) |
| S. mansoni | Egg count | Brazil, Bahia | 31% (VC) | (64) |
| S. mansoni | IgE | Brazil, Bahia | 59% (VC) | (64) |
| S. haematobium | Egg count | Kenya, Coast | 9% (h2) | (46) |
| S. haematobium | Bladder morbidity | Kenya, Coast | 14% (h2) | (46) |
| S. japonicum | Infection | China, Jiangxi | 58% (VC) | (65) |
| S. japonicum | Infection | China Sichuan | 0% (VC) | (66) |
h2 additive heritability, VC variance components.
Linkage Studies for Schistosoma Egg Count
The initial study of the genetics of human schistosomiasis used segregation analysis, which determines whether the distribution of the disease on family pedigrees is consistent with the presence of a major gene (3). This study demonstrated the presence of a major gene which was subsequently named SM1 and located on chromosome five by parametric linkage analysis (4). A major gene has alleles that cause a difference in phenotype between family members that is large enough to be able to categorize individuals as carriers or non-carriers on the basis of phenotype alone (67) and parametric linkage analysis requires estimates of the disease allele frequency and penetrance of the phenotype for the three possible genotypes.
The SM1 quantitative trait locus (QTL) on chromosome 5 5q31-q33 for S. mansoni fecal egg count was the first QTL to be mapped in humans for any infectious disease (Table 2, Figure 1) (4). The great success of this study was partially attributable to the very large effect size of the SM1 locus (66% of the variance after accounting for water contact, age and sex). This is in striking contrast to the very modest proportions of the variance explained by most loci identified by GWAS in which loci rarely explain more than 10% of the variance of the trait that is not attributable to covariates (96). Three further loci (1p22.2, 7q36 and 21q22–22-qter) had evidence of association and contained genes known to be involved in the response to schistosomes but did not achieve genome wide significance (68). A reanalysis of the same data controlling for SM1 genotype identified additional genome wide significant loci at 1p21-q23 and 6p21-q21 (Table 2, Figure 1) (69), since the effect of these loci was small in comparison to the effect of SM1, they were only identifiable when using SM1 genotype as a parameter of the model.
Table 2.
Loci associated with S. mansoni infection discovered by linkage studies.
| Phenotypes | Locus name | 5' Marker (Position) | 3'Marker (Position) | References | Candidate genes, Tested (Untested) |
|---|---|---|---|---|---|
| Egg count | SM1 5q31-q33 | D5S642 (128Mb) | D5S412 (158Mb) | (4, 68–70) | IL4, IL5, IL9, IL13, (IL3, CXCL14, CD14, 1IL17B, IL12B) |
| Egg count | 1p21-q23 | D1S236 (95Mb) | D1S196 (168Mb) | (69) | (IL6R, CRP) |
| Egg count | 6p21-q21 | D6S271 (43Mb) | D6S283 (67Mb) | (69) | (VEGFA, IL17A, IL17F) |
| Hepatic fibrosis | SM2 6q22-q23 | D6S1009 (137Mb) | D6S310 (142Mb) | (71) | CTGF, IFNGR1, IL22RA2 |
| Egg count | 7q35-q36 | D7S483 (152Mb) | D7S550 (156Mb) | (69) | (TRB, NOS3, SHH) |
Marker positions are shown in GRCh37 co-ordinates. Candidate genes that have been found associated with the phenotype are shown in bold (Tables 3, 4). Other plausible candidates (See Figure 2) in the regions are in brackets. Genes associated with the Th17 response are underlined. These loci are shown graphically in Figures 1, 2.
Figure 1.
Genes and quantitative trait loci associated with schistosomiasis plotted on a human karyotype. Blue lines indicate QTL, with reported −log p-value for association shown on the y axis. Genes containing SNP associated with schistosomiasis infection (Table 3) are shown in blue, genes associated with pathology (Table 4) are shown in red and genes associated with both pathology and infection are shown in black. Genes are arranged vertically on the plot for clarity and their position on the y axis is arbitrary.
A further study on a Senegalese population, by the same group that conducted the original study in Brazil, confirmed an association at the SM1 locus. However, the effect was not as strong and the association could only be demonstrated by non-parametric pedigree tests for an effect at the SM1 locus (70). Non-parametric analysis requires no prior knowledge of the disease allele frequency or the disease penetrance of the different genotypes.
Linkage Studies Identify QTL for Pathology That Are Independent of QTL for Parasite Burden
A study in Sudan found that 12% of the study population had moderate or advanced fibrosis and that half of these also had portal hypertension (97). A linkage study of four candidate gene regions in the same population identified a locus (SM2) on chromosome six near the interferon-gamma receptor (IFNGR1) associated with hepatic fibrosis (Table 2, Figure 1) (98). Fifty percentage of people with risk alleles at SM2 had some continuous thickening of periportal vein branches within 19 years of coming to live within the study area. IFNG is strongly anti-fibrogenic and polymorphisms in IFNGR1 could plausibly regulate fibrosis. No linkage was found with the SM1 locus suggesting that control of infection and pathology were independent. The SM2 locus at 6q22-q23 did not overlap either the HLA region on chromosome six or the 6p21-q21 region that was associated with S. mansoni worm burden in Brazil (69) (Table 2, Figure 1).
The SM2 locus was replicated in a linkage study of 11 candidate gene regions in Egypt where 32.7% of individuals 11 years and older had significant fibrosis and rs1327475 in IFNGR1 was significantly associated with severe PPF. In contrast to the earlier study in Sudan, this study found a weak association with the Th2 cytokine cluster (IL4, IL13) in SM1 (54), suggesting that worm burden does contribute to risk of pathology. There is evidence for a potentially protective role of a high IFNG response to schistosome infection, consistent with the anti-fibrogenic properties of IFNG [reviewed by Abath et al. (99)] and a SNP in IFNG has been associated with time to reinfection (Table 3) (72). Consequently, it appears that the variation in the IFNG system is involved in both the outcome of infection and pathology. Variation in IFNGR1 has only been shown to be involved in the development of pathology but it has yet to be tested in a candidate gene study for effect on infection response.
Table 3.
SNP which have been found to be associated with schistosomiasis infection phenotypes in candidate gene studies.
| SNP | Gene | Phenotype | Parasite | Neg refs | Pos refs |
|---|---|---|---|---|---|
| rs2430561 | IFNG | T2R | Sm | none | (72) |
| rs3024495 | IL10 | FEC | Sm | None | (38)* |
| rs1800896 | IL10 | IgE | Sm | None | (38) |
| rs1800871 | IL10 | IgE | Sm | None | (38)* |
| rs1800872 | IL10 | IgE | Sm | None | (38)* |
| IL10(-1082/-819/-592) | IL10 | UEC | Sh | (73) | (74)* |
| rs20541 | IL13 | FEC | Sm | (75, 76) | (77) |
| rs2066960 | IL13 | FEC | Sm | (78) | (77) |
| rs7719175 | IL13 | UEC | Sh | (73) | (79)* |
| rs2069743 | IL13 | UEC | Sh | (78) | (80) |
| rs1800925 | IL13 | UEC, FEC, T2R | Sh, Sm | None | (72, 77–79)* |
| rs2243250 | IL4 | UEC, T2R | Sh | (80) | (73), (72)* |
| rs2079103 | IL5 | IF | Sj | None | (75)* |
| rs2706399 | IL5 | IF | Sj | None | (75)* |
| rs3024974 | STAT6 | UEC | Sh | None | (73)* |
| rs324013 | STAT6 | UEC | Sh | None | (78)* |
| rs733618 | CTLA4 | UEC | Sh | None | (81) |
| rs11571316 | CTLA4 | UEC | Sh | None | (81) |
| rs231775 | CTLA4 | UEC | Sh | None | (81) |
| rs3124952 | FCN2 | UEC | Sh | None | (82) |
| rs17514136 | FCN2 | UEC | Sh | None | (82) |
| rs7567833 | COLECC11 | UEC | Sh | None | (83) |
| COLEC11*TCCA | COLECC11 | UEC | Sh | None | (83) |
| Blood group O | ABO | FEC,UEC | Sh,Sm | None | (84) |
| rs746822072 | RNASE3 | FEC | Sm | None | (85) |
Loci that have been found associated with schistosomiasis in more than one study are shown in bold. * Loci that are not significant after Bonferroni correction. IL10 (−1082/−819/−592) = a haplotype of rs1800870, rs1800871 and rs1800872. COLEC11*TCCA is a haplotype of rs1864480 (−676T/C), rs4849953 (−472T/C), rs6714770 (−469C>G), and (−276C>T). Blood Group O is most commonly defined by genotypes at three SNP rs8176719, rs8176746(CC), rs8176747(GG). IF, Infection Frequency; T2R Time to Reinfection; UEC, Urine Egg Count; FEC, Fecal Egg Count: Sh, S. haematobium; Sm, S. mansoni; Sj, S. japonicum. “Pos refs” column contains citations for the studies that showed a significant association with phenotype and “Neg refs” column contains citations of studies that failed to show a significant association at that locus.
Annotation of Genes in QTL Regions
Very few genes were known in the 5 QTL regions for schistosomiasis (Table 2) at the time that the QTL were discovered, and we are not aware of any attempts to identify the genes that are responsible for the QTL effect. Hundreds of genes are now known in these loci, each of which could potentially regulate the phenotype and we prepared a short list of the most likely candidates in each region. In order to discover which genes in each schistosomiasis QTL region might be involved in the response to schistosomiasis we used a custom Perl script to search PubMed with terms for schistosomiasis and each gene name and its aliases and obtained a count of the number of publications returned as detailed in Supplementary Table 4.
We assumed that the genes that are most likely to be the QTL genes will already have been shown to be involved in the response to schistosome infection. In order to identify these genes, we systematically searched PubMed for papers that included terms for schistosomiasis and each of the gene names in the 5 QTL regions in Table 2 or their synonyms (Supplementary Table 4). The genes that have been mentioned most frequently in the abstract of a paper that also mentions schistosomiasis and that are in one of the QTL are shown in Figure 2, Table 2. A complete list of all genes that are in the QTL and that have been studied in the context of schistosomiasis is in Supplementary Table 4. The number of papers shown in Figure 2 is an indicator of the genes most commonly associated with schistosomiasis in these regions. The genes with the largest literature were the Th2 cytokine genes originally identified by Marquet (4) in SM1 (IL3, IL4, IL5, IL9, and IL13), that each had between 17 and 511 publications associated with schistosomiasis. Only CSF1 and TRB (beta T cell receptor) were identified as candidate genes by the original authors in the 1p21-q23 and 7q35-q36 egg burden loci (69) and IFNGR1 was the candidate gene that was used for the linkage study at SM2 (98). The large literature on Th2 cytokines and schistosomiasis is expected given the important role of this pathway in response to egg antigens and the development of pathology. The Th2 cytokines in SM1 are therefore credible candidate genes at this locus.
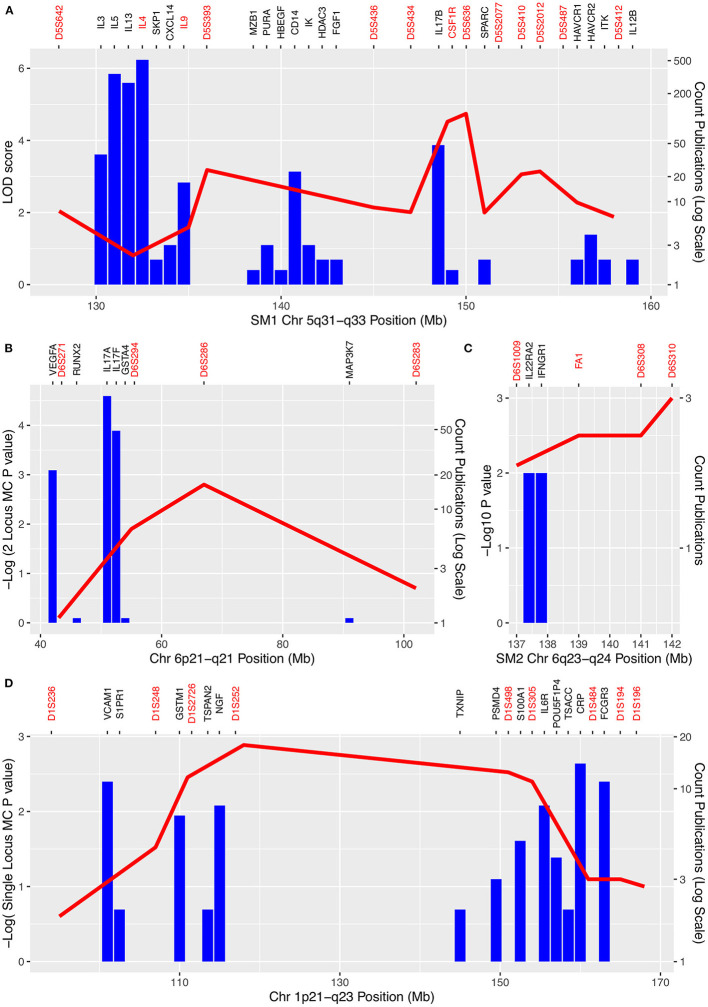
Figure 2.
Schistosomiasis QTL and numbers of publications mentioning each gene in each QTL regions. (A) SM1 region 5q31-q33; (B) 6p21-q21; (C) SM2 6q23-q24; (D) 1p21-q23. Negative log p-values for associations between markers and schistosomiasis are shown on the left-hand axis. Counts of publications which mention both schistosomiasis and genes in the QTL are shown by the blue columns and on a log scale on the right-hand y axis. Markers used in mapping are shown in red on the top axis (including genes used as markers). Genes for which there was only one publication are omitted for clarity and positions of genes and markers have been adjusted by up to 1Mb for clarity. Note the cluster of Th2 cytokine genes (IL3, IL4, IL5, IL9, IL13) in SM1 with large numbers of publications between 131 and 135 Mb but low LOD scores. However, in a reanalysis of the same data using weighted pairwise correlations the peak of the QTL shifted toward this cytokine cluster (69).
Our annotation of these QTL also revealed the presence of Th17 related genes in four of the five QTL: IL17B and IL12B in SM1, IL22RA2 in SM2, IL17A and IL17F in 6p21-q21 and IL6R in1p21-q23. Although IL12B (IL12p40) in SM1 is primarily known as a Th1 cytokine it is also a component of the hetero-dimeric IL23 cytokine which is important for Th17 maintenance and expansion (100) and IL6 is important in Th17 T helper cell differentiation (101). IL17 cytokines are involved in regulation of worm and egg burdens as well as the development of fibrosis and granuloma in response to eggs (102). The presence of Th17 related cytokines in four of the five QTL suggests that variation in this system may also contribute to variation in outcome of infection in addition to that caused by variation in the Th2 system.
Candidate Gene Studies
Infection Status and Intensity
We have identified 28 candidate gene studies of Schistosoma infection or worm burden that reported associations between 24 loci in eleven candidate genes and seven different phenotypes (Table 3, Supplementary Tables 2, 3). The genes with associations were IFNG, IL10, IL13, IL4, IL5, STAT6, CTLA4, FCN2, COLECC11, ABO, and RNASE3. These genes were all chosen because their protein products were known to be involved in the response to infection. One study of MASP2 (103) and one on LTA (104) only reported negative results and are not included in Table 3. We have not attempted any formal meta-analysis since few loci were replicated and there were important differences in study design and data reporting in the studies of loci that were replicated, making any meta-analysis hard to interpret.
Schistosome eggs induce granulomatous Th2 responses (IL4, IL5, IL9, IL10, and IL13) (32) and antigen-specific IgG1, IgG4, and IgE (105). The SM1 region on chromosome five identified by Marquet et al. (4) (Figure 1) included the prototypical Th2 cytokines IL4, IL5, IL9, and IL13, these are strong candidates for the QTL gene(s) and SNP and all except IL9 have been found associated with schistosomiasis in candidate gene studies (Table 3). IL13 and IL4 regulate STAT6 expression which in turn regulates IgE class switching (106) and STAT6 variants are also associated with schistosomiasis (Table 3). IL13, IL4, IL5, and STAT6 are also involved in regulation of the Th2 response to schistosomiasis (72, 75, 77–80).
Th1 cytokines and IFNG in particular are involved in the resistance to the immature worms. Studies of mice and ex vivo human PBMC have shown that IFNG levels increase in response to schistosome antigens and are correlated with resistance or tolerance to infection (27, 107, 108) and a candidate gene study found an association between the IFNG SNP rs2430561 and time to reinfection (72). IL10 and CTLA4 downregulate immune responses in long standing infections (31). COLECC11 and FCN2 are involved in the innate immune response, they both bind to specific pathogen-associated molecular patterns (PAMPs) on the pathogen surface and stimulate the complement lectin cascade, thereby clearing the pathogens by opsonization (82, 83). ABO regulates blood group and a meta-analysis found evidence for a protective effect for blood group O (84). RNASE3 also known as eosinophil cationic protein (ECP) is a secretory protein of eosinophil granulocytes that efficiently kills the larval stage of S. mansoni (85).
The Ensembl Variant Effect Predictor was used to provide functional annotations for these variants (Supplementary Table 5). In the functional annotation only rs231775 in CTLA4 and rs20541 in IL13 were predicted to have an effect on function. Both of which were non-synonymous variants and were classified as risk factors by ClinVar (109), although SIFT (110), and Polyphen (111) predicted the effect of these SNP would be benign. Other SNP had no predicted effects on function, possibly because they are not functional but are linked to functional SNP nearby. However, the functional annotation cannot detect all functional variants and experimental work has shown that IL13 expression is regulated by rs1800925 (112). Further detailed studies will be required to determine which of the SNP are truly functional and which are not functional but still potentially useful markers for risk of schistosomiasis.
Pathology: Hepatic and Periportal Fibrosis
It has been noted since at least 1974 that the development of severe fibrosis is clustered in families and is not well-correlated with intensity of infection suggesting that the mechanisms regulating infection intensity and pathology are not closely coupled (5). We found 20 studies which identified associations with schistosomiasis related pathology at 46 candidate SNP or haplotypes in 21 genes outside the HLA complex (Table 4, Supplementary Table 2). Few of the studies applied any multiple testing corrections and 15 out of the 43 associations would not be significant after a Bonferroni correction (Table 4).
Table 4.
SNP which have been found to be associated with schistosomiasis pathology related phenotypes in candidate gene studies.
| SNP | Gene | Phenotype | Species | Not associated | Associated | References |
|---|---|---|---|---|---|---|
| rs1327475 | IFNGR1 | HF | Sm | NA | Egypt | (54) |
| rs2243250§ | IL4 | HF | Sm | NA | Egypt | (54) |
| rs1056854 | TGFB1 | HF | Sm | NA | Egypt* | (54) |
| rs373880612 | IFNG | PPF, HF, PH | Sm | NA | Sudan* | (55) |
| rs1861494 | IFNG | PPF, HF, PH | Sm | NA | Sudan* | (55) |
| IFN-gR1 6q22-q23 | IFNGR1 | HF, PH | Sm | NA | Sudan | (71) |
| rs3037970 | CCN2(CTGF) | HF, PH | Sj Sm |
China Sudan Brazil |
China | (71) |
| rs1257705 | CCN2(CTGF) | HF, PH | Sj Sm |
Sudan Brazil China |
China* | (86) |
| rs2151532 | CCN2(CTGF) | HF, PH | Sj Sm |
China Egypt |
China* | (54, 86) |
| rs9402373 | CCN2(CTGF) | HF, PH | Sj Sm |
NA | 2 China* Sudan Brazil* |
(86) |
| rs9399005 | CCN2(CTGF) | HF, PH | Sj | China | China* | (86) |
| rs6918698 | CCN2(CTGF) | HF, PH | Sj Sm |
China Sudan |
China* Brazil* |
(86) |
| rs1931002 | CCN2(CTGF) | HF, PH | Sj | China | China | (86) |
| rs12526196 | CCN2(CTGF) | HF, PH | Sj Sm |
Sudan Brazil |
2 China* | (86) |
| rs1800925C-rs20541A§ | IL13 | HF, PH | Sj | NA | China | (87) |
| rs1800925§ | IL13 | HF PH | Sj | NA | China* | (87) |
| rs12712135 | ST2 (IL1RL1) | IL1RL1 | Sj Sm |
NA | China Brazil |
(88) |
| rs1420101 | ST2 (IL1RL1) | IL1RL1 | Sj Sm |
NA | China Brazil |
(88) |
| rs6543119 | ST2 (IL1RL1) | IL1RL1 | Sj Sm |
NA | China Brazil |
(88) |
| rs7412, rs429358 APOE3 | APOE | TC, LDL | Sm | Brazil | NA | (89) |
| rs6570136 | IL22RA2 | HF, PH | Sj Sm |
NA | China Brazil* Sudan* |
(90) |
| rs7774663 | IL22RA2 | HF, PH | Sj Sm |
China | Sudan Brazil* |
(90) |
| rs2064501 | IL22RA2 | HF, PH, IL22 | Sj Sm |
NA | China* Brazil* Sudan* |
(90) |
| rs11154915 | IL22RA2 | HF, PH, IL22 | Sj Sm |
China Brazil |
Sudan* | (90) |
| rs7749054 | IL22RA2 | HF, PH, IL22 level | Sj Sm |
China Sudan |
Brazil* | (90) |
| rs1800870 | IL10 | PPF | Sm | NA | Brazil | (91) |
| IL10 (-1082/-819/-592)§ | IL10 | PPF | Sm | NA | Brazil | (91) |
| rs1800629 | TNFA | PPF, TNFA | Sm | NA | Brazil | (92) |
| rs10118570 | MAPKAP1 | HF, infection | Sj | NA | 2 China | (93) |
| rs391957 | HSPA5 | HF, infection | Sj | NA | China | (93) |
| rs17025963 rs185882198 rs71093915 |
TGFBR2 | HF | Sm | NA | Brazil | (5) |
| rs1690215 rs56368010 |
ACVRL1(ALK1) | HF | Sm | NA | Brazil | (5) |
| rs114046193 rs12345675 |
TGFbR1 (ALK5) | HF | Sm | NA | Brazil | (5) |
| rs138079455 rs10555873 rs77414361 |
SMAD9 | HF | Sm | NA | Brazil | (5) |
| rs12913547 rs12914140 rs12439500 |
SMAD3 | HF | Sm | NA | Brazil | (5) |
| rs746822072 | RNASE3 (ECP) | HF | Sm | NA | Uganda | (85) |
| rs1800629 | TNFA | PPF regression after treatment | Sm | NA | Brazil | (94) |
| rs2295080 | mTOR | HF, infection | Sj | NA | China | (95) |
| rs7254617 | AKT2 | HF, infection | Sj | NA | China* | (95) |
Loci that have been found associated with schistosomiasis in more than one population are shown in bold. *Loci that are not significant after Bonferroni correction. §SNP which are also associated with infection (Table 3). Studies of classical HLA loci have been excluded since they have been fully reviewed elsewhere (7, 8). Sh, S. haematobium; Sm, S. mansoni; Sj, S. japonicum. The “Associated” column shows the locations of studies where a significant association has been found and the “Not Associated” column shows the location of studies that failed to show a significant association. PPF Periportal Fibrosis; PH Portal Hypertension; PT Portal thickness, including portal vein diameter and portal vein branch point thickness, HF Hepatic Fibrosis; AHSD Advanced HepatoSplenic Disease, TC Total Cholesterol, HDL High Density Lipoprotein, TNFA Median levels TNFA higher in patients with PPF, IL22 level of IL22 in plasma, IL1RL1 level of soluble IL1RL1 in plasma. IL-10 (−1082/−819/−592) represents a haplotype of rs1800870, rs1800871, and rs1800872. NA, Not applicable.
There are sixteen genes for which an effect has only been reported for fibrosis and not for infection: APOE, CCN2, HSPA5, IFNGR1, IL22RA2, MAPKAP1, IL1RL1, TNFA, mTOR, AKT2, TGFB1, TGFBR1, TGFBR2, ACVRL1, SMAD9, and SMAD3 (Tables 3, 4). Five genes have been associated with both fibrosis and intensity of infection (RNASE3, IL4, IL10, IFNGI, and IL13). Four genes were associated with fibrosis in more than one population: IFNGR1, IL22RA2, CCN2, and MAPKAP1 (Table 4), the two former genes are also in the SM2 QTL (55, 71, 86, 90, 93). Although these sixteen genes have not been tested for associations with infection status or intensity it is plausible that some of them are only associated with pathology. All of these genes except APOE and IL1RL1 have been associated with the regulation of fibrosis (Table 4) and variation in these may regulate risk of pathology irrespective of intensity of infection.
The Ensembl Variant Effect Predictor was used to provide functional annotations for the SNP in Table 4 (Supplementary Table 5). Two non-synonymous SNP in APOE (rs7412 and rs429358) were predicted by ClinVar to be a risk factor, pathogenic and involved in drug response (109), the rs7412 SNP was also predicted to be deleterious or damaging by SIFT (110) and Polyphen (111). A non-coding SNP in LTA (rs1800629) was predicted to be involved in drug response by ClinVar (109) and a non-coding SNP (rs1800872) in IL10 was predicted to be risk factor by ClinVar. Other SNP in Table 4 did not have functional annotations, and many may be marker SNP that are linked to functional variants rather than functional themselves.
A recent study found that just 32 SNPs could predict who gets severe hepatic fibrosis in Brazil with 63% sensitivity and 90% specificity (5). This review emphasized the importance of TGFB signaling pathway and IL22. TGFB is also involved in the differentiation of Th17 cells (113), and together with SMAD regulates Th17 in response to another worm infection Echinococus multilocularis (114) providing further justification for systematic investigation of the role of variants in the Th17 pathway in differences in response to infection. IL22 and IL17 are co-expressed by Th17 CD4+ T cells and polymorphisms have been associated with hepatic fibrosis in the IL22 receptor IL22RA2 (5, 90). IL22 also has protective effects on the intestinal epithelium against toxic bacterial products (5).
Associations With the HLA Region
The HLA region is associated with response to many communicable and non-communicable diseases. The importance of CD4+ T helper cells in the response to schistosomiasis, and the role of HLA class II alleles in recruiting these, suggests that variation in the HLA region may play an important role in control of schistosome infections. However, associations were not found in this region in the whole genome linkage scans either for worm burden or pathology (4, 68, 71).
Two reviews reported 17 and 18 studies, respectively, of associations of HLA markers with schistosomiasis induced PPF (7, 8), but surprisingly we could not find any studies of HLA genes and worm burden (Table 3, Supplementary Table 3). We have found only one study of genes in the HLA region and schistosomiasis that has been published in the 9 years since those reviews. A SNP in Major histocompatibility complex class I chain-related A (MICA) was associated with liver fibrosis in a Han Chinese population (76).
It has been emphasized that the problems of extensive linkage disequilibrium within the HLA region, the small sizes of the studies reviewed, the allelic diversity and large variations in allele frequencies between populations mean that these studies may not replicate in different populations and need further confirmation from larger studies (7). However, some alleles of HLA class II loci DQA1, DQB1 and DRB1 and HLA class I HLA-A and HLA-B were associated with PPF in a meta-analysis that combined evidence from 2 to 3 studies for each allele evaluated (8) and these associations may be robust.
Discussion
Replication of Candidate Gene Studies for Infection
Few of the candidate gene studies shown in Tables 3, 4 applied correction for testing multiple SNPs and the 32 associations that would not remain significant after such a correction are indicated by asterisks. The lack of Bonferroni corrections suggests that some of these studies will not replicate and, for some of these loci, there are studies that have not replicated the association (Tables 3, 4), although this is often when using a different phenotype. Notably, half of these studies that did not replicate an association were at loci that were significant after a Bonferroni correction.
There are many instances of failures to replicate candidate gene studies. One review found that only 6 out of 166 associations replicated in more than 75% of studies, although 97 of the 166 associations (58%) were reproduced in at least one study (115). Failure to replicate can be due to the initial observation being due to random variation in allele frequencies between test and control samples (a type one error). However, genuine associations can fail to replicate because of linkage between the marker SNP and the functional SNP varying between populations, small study sizes, variable penetrance, population variation at modifier loci, (occult) population stratification within study populations or differences in allele frequencies between populations leading to type two errors. In addition, it is also possible for different SNP in the same gene to be most important in regulating a response in different populations or individuals (116). Therefore, most of these observations should be considered provisional until adequately powered meta-analyses can be conducted.
Associations with infection were replicated at two SNP (IL13 rs1800925; IL4 rs2243250), and both of these SNP and also the IL10 (−1082/−819/−592) haplotype were also found to be associated with pathology (Tables 3, 4), despite the studies using different phenotypes and in one case different parasite species. The IL13 rs1800925 SNP was associated with schistosomiasis in four infection related studies and one pathology study and all studies that included this SNP found an association with it, despite the association not being significant after a Bonferroni correction in any of these studies. The high level of replication at this SNP suggests that these associations may be robust despite the lack of significance in individual studies. Functional data also supports a role for rs1800925, which is in the promoter of IL13, and is associated with increased expression of IL13 from stimulated cells in vitro (112). Since IL13 regulates IgE levels via STAT6 (106), there is a plausible mechanism for a role for this SNP in response to infection, increasing the confidence that it is a genuine association. Although IL4 expression is also associated with IgE levels, the rs2243250 SNP is not (117), so its impact on intensity of infection must be via some other mechanism.
The Th17 Pathway Has Been Neglected in Schistosomiasis Association Studies
Since the Th2 pathway is the dominant response to helminth infections and is the main pathway for response to egg antigens (28, 30), genes in this pathway have been well-represented in association studies (Tables 3, 4) and these have confirmed the importance of variation in this pathway for outcome of infection. However, the Th1 and Th17 pathways are also important, particularly in the early stages of the infection (26, 29, 36, 37). Variants in IL17F and IL17RA have been found associated with cerebral malaria (118) and similar variation may contribute to the outcome of schistosome infection. Our annotation of QTL, with the genes that have published associations with schistosomiasis, revealed an excess of Th17 pathway genes in these QTL (Figures 1, 2). There have been no association studies to test candidate gene hypothesis for three out of the five QTL (Table 2) and there are Th17 genes in four of the five QTL (underlined in Table 2), which could be priorities for future association studies.
It is possible that variation in other Th17 pathway genes outside of the QTL also contribute to variation in response to infection. A KEGG pathway diagram of Th17 cell differentiation and a list of 108 genes in this pathway is shown in Supplementary Table 6. We have obtained a list of the 1,742,019 SNP in these genes from dbSNP and kept the 1,052 SNP that were predicted to be “pathogenic” by ClinVar, irrespective of minor allele frequency (Supplementary Table 7). We also kept SNP with minor allele frequency > 5% and that had any of the following functional classifications: splice acceptor variant, stop gained, initiator codon variant, stop lost, splice donor variant, missense variant, terminator codon variant, frameshift variant. This left a list of 2,701 SNP in the Th17 pathway which are most likely to have an effect on function and that could be priorities for further testing (Supplementary Table 7).
Which Are the Optimal Study Designs to Discover Susceptibility Loci?
Approaches for discovering susceptibility loci for parasitic infections have been reviewed previously (119). The major approaches are association studies in unrelated individuals and linkage studies within families, the merits of which have been evaluated by Abel and Dessein (120). We noted in this review that family based designs were the first to discover QTL loci in schistosomiasis (4), and these were followed up with considerable success by candidate gene studies in these QTL. Schistosomiasis affected communities are frequently geographical clusters of related individuals where case control studies can be confounded by cryptic relatedness. In contrast family-based association studies exploit this relatedness by estimating disequilibrium in transmission of alleles within families.
Schistosomiasis is also an excellent setting for family-based linkage studies of infection intensity because children are the most heavily infected; therefore, parents are often available for genotyping to create complete families, unlike adult onset diseases. However, it is more difficult to collect full families for complications of chronic schistosomiasis such as fibrosis that affect adults. Whole genome linkage studies have only been undertaken on two populations, one in Brazil and one in Senegal, further studies to identify loci regulating intensity of infection in additional populations should be undertaken and could enlarge our understanding of the mechanisms of response to infection. It has already been shown that 32 SNP can be used to identify those at highest risk of developing pathology after S. mansoni infection (5). If the 20% of the people that shed 80% of the eggs could also be identified they could be targeted for regular treatment which could dramatically reduce the number of eggs in the environment and the pressure of infection on the whole community.
Conclusion
Despite the remarkable success of the early linkage studies that identified major QTL loci, no further whole genome scans for association have been conducted and the QTL genes underlying these loci have not been definitively identified. All subsequent studies have been candidate gene linkage and association studies focussing on genes within the QTL regions SM1, SM2, and the Th2 pathway that are hypothesized to play a role in schistosomiasis progression. No candidate gene studies have attempted to identify QTL genes in three of the QTL for S. mansoni egg count (Table 2). This review has presented evidence that the Th17 pathway has been overlooked in studies of the genetics of schistosomiasis and should be prioritized in future investigations of susceptibility genes.
The rapid development of genotyping technologies makes large scale genomic studies easier than ever, provided that well-characterized samples can be obtained. The studies of Dessein et al. (86) on populations from China, Sudan and Brazil have demonstrated the value of replicating analyses in multiple populations and similar replication is needed for other candidate SNP and genes.
Author Contributions
GS and EMa conceived the review. EMe and ON wrote the draft and collected all the relevant literature. HN significantly reviewed the draft and analyzed QTL data. ME contributed to the draft and revised the manuscript. All authors read and approved the final manuscript.
The TrypanoGEN+ Research Group of the H3Africa Consortium
Membership of the TrypanoGEN+ research group is available at http://www.trypanogen.net/.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The handling editor declared a past co-authorship with the authors HN, EMa, and GS.
Acknowledgments
The authors are grateful to Alison Elliott of the London School of Hygiene and Tropical Medicine for helpful discussions and reviewing the manuscript.
Footnotes
Funding. This study was funded by the African Academy of Sciences grant to the TrypanoGEN+ H3Africa program, Grant no. H3A/18/004. ME received funds from HIC-Vac, a GCRF Network in Vaccines Research and Development which was co-funded by the MRC and BBSRC and is part of the EDCTP2 programme supported by the European Union. Additional support was received through Wellcome Trust grants 215993/Z/19/Z and 218454/Z/19/Z, and through the Makerere University-Uganda Virus Research Institute Centre of Excellence for Infection and Immunity Research and Training (MUII) which was funded by the DELTAS Africa Initiative (Grant no. 107743).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.613468/full#supplementary-material
Search strategy: Summary of PubMed search strategy.
All included references: Papers included in the review.
Details of Refs Populations, study design, phenotypes, and effects from studies.
Genes in QTL. List of genes in QTL regions with lists of publications associated with each gene and schistosomiasis.
VEP SNP Annotations. Functional annotation of SNP from candidate genes studies with the Ensembl Variant Effect predictor.
Th17 genes. List of Genes in Th17 pathway with KEGG pathway diagram.
Th17 SNP. List of potentially function SNP in genes in the Th17 pathway.
References
- 1.Ismail HAHA, Hong ST, Babiker ATEB, Hassan RMAE, Sulaiman MAZ, Jeong HG, et al. Prevalence, risk factors, and clinical manifestations of schistosomiasis among school children in the White Nile River basin, Sudan. Parasites Vectors. (2014) 7:478. 10.1186/s13071-014-0478-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bethony JM, Quinnell RJ. Genetic epidemiology of human schistosomiasis in Brazil. Acta Trop. (2008) 108:166–174. 10.1016/j.actatropica.2007.11.008 [DOI] [PubMed] [Google Scholar]
- 3.Abel L, Demenais F, Prata A, Souza AE, Dessein A. Evidence for the segregation of a major gene in human susceptibility/resistance to infection by Schistosoma mansoni. Am J Hum Genet. (1991) 48:959–70. [PMC free article] [PubMed] [Google Scholar]
- 4.Marquet S, Abel L, Hillaire D, Dessein H, Kalil J, Feingold J, et al. Genetic localization of a locus controlling the intensity of infection by Schistosoma mansoni on chromosome 5q31-q33. Nat Genet. (1996) 14:181–4. 10.1038/ng1096-181 [DOI] [PubMed] [Google Scholar]
- 5.Dessein H, Duflot N, Romano A, Opio C, Pereira V, Mola C, et al. Genetic algorithms identify individuals with high risk of severe liver disease caused by schistosomes. Hum Genet. (2020) 139:821–31. 10.1007/s00439-020-02160-4 [DOI] [PubMed] [Google Scholar]
- 6.Alsallaq RA, Gurarie D, Ndeffo Mbah M, Galvani A, King C. Quantitative assessment of the impact of partially protective anti-schistosomiasis vaccines. PLoS Negl Trop Dis. (2017) 11:e0005544. 10.1371/journal.pntd.0005544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blackwell JM, Jamieson SE, Burgner D. HLA and infectious diseases. CMR. (2009) 22:370–85. 10.1128/CMR.00048-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huy NT, Hamada M, Kikuchi M, Lan NTP, Yasunami M, Zamora J, et al. Association of HLA and post-schistosomal hepatic disorder: a systematic review and meta-analysis. Parasitol Int. (2011) 60:347–56. 10.1016/j.parint.2011.05.008 [DOI] [PubMed] [Google Scholar]
- 9.Colley DG, Bustinduy AL, Secor WE, King CH. Human schistosomiasis. Lancet. (2014) 383:2253–64. 10.1016/S0140-6736(13)61949-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hotez PJ, Kamath A. Neglected tropical diseases in sub-saharan Africa: review of their prevalence, distribution, and disease burden. PLoS Negl Trop Dis. (2009) 3:e412. 10.1371/journal.pntd.0000412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steinmann P, Keiser J, Bos R, Tanner M, Utzinger J. Schistosomiasis and water resources development: systematic review, meta-analysis, and estimates of people at risk. Lancet Infect Dis. (2006) 6:411–25. 10.1016/S1473-3099(06)70521-7 [DOI] [PubMed] [Google Scholar]
- 12.Murray CJL, Vos T, Lozano R, Naghavi M, Flaxman AD, Michaud C, et al. Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990-2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. (2012) 380:2197–223. 10.1016/S0140-6736(12)61690-0 [DOI] [PubMed] [Google Scholar]
- 13.van der Werf MJ, de Vlas SJ, Brooker S, Looman CWN, Nagelkerke NJD, Habbema JDF, et al. Quantification of clinical morbidity associated with schistosome infection in sub-Saharan Africa. Acta Trop. (2003) 86:125–39. 10.1016/S0001-706X(03)00029-9 [DOI] [PubMed] [Google Scholar]
- 14.Gbalégba NGC, Silué KD, Ba O, Ba H, Tian-Bi NTY, Yapi GY, et al. Prevalence and seasonal transmission of Schistosoma haematobium infection among school-aged children in Kaedi town, southern Mauritania. Parasites Vectors. (2017) 10:353. 10.1186/s13071-017-2284-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ezeamama AE, Friedman JF, Acosta LP, Bellinger DC, Langdon GC, Manalo DL, et al. Helminth infection and cognitive impairment among filipino children. Am J Trop Med Hyg. (2005) 72:540–8. 10.4269/ajtmh.2005.72.540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ezeamama AE, Bustinduy AL, Nkwata AK, Martinez L, Pabalan N, Boivin MJ, et al. Cognitive deficits and educational loss in children with schistosome infection—a systematic review and meta-analysis. PLoS Negl Trop Dis. (2018) 12:e0005524. 10.1371/journal.pntd.0005524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman JF, Kanzaria HK, McGarvey ST. Human schistosomiasis and anemia: the relationship and potential mechanisms. Trends Parasitol. (2005) 21:386–92. 10.1016/j.pt.2005.06.006 [DOI] [PubMed] [Google Scholar]
- 18.King CH, Dangerfield-Cha M. The unacknowledged impact of chronic schistosomiasis. Chronic Illn. (2008) 4:65–79. 10.1177/1742395307084407 [DOI] [PubMed] [Google Scholar]
- 19.King CH, Dickman K, Tisch DJ. Reassessment of the cost of chronic helmintic infection: a meta-analysis of disability-related outcomes in endemic schistosomiasis. Lancet. (2005) 365:1561–9. 10.1016/S0140-6736(05)66457-4 [DOI] [PubMed] [Google Scholar]
- 20.Koukounari A, Gabrielli AF, Toure S, Bosque-Oliva E, Zhang Y, Sellin B, et al. Schistosoma haematobium infection and morbidity before and after large-scale administration of praziquantel in Burkina Faso. J Infect Dis. (2007) 196:659–69. 10.1086/520515 [DOI] [PubMed] [Google Scholar]
- 21.Utzinger J, Raso G, Brooker S, De Savigny D, Tanner M, Ornbjerg N, et al. Schistosomiasis and neglected tropical diseases: towards integrated and sustainable control and a word of caution. Parasitology. (2009) 136:1859–74. 10.1017/S0031182009991600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evan Secor W. Water-based interventions for schistosomiasis control. Pathogens Global Health. (2014) 108:246–54. 10.1179/2047773214Y.0000000149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ross AGP, Olveda RM, Li Y. An audacious goal: the elimination of schistosomiasis in our lifetime through mass drug administration. Lancet. (2015) 385:2220–1. 10.1016/S0140-6736(14)61417-3 [DOI] [PubMed] [Google Scholar]
- 24.Gray DJ, McManus DP, Li Y, Williams GM, Bergquist R, Ross AG. Schistosomiasis elimination: lessons from the past guide the future. Lancet Infect Dis. (2010) 10:733–6. 10.1016/S1473-3099(10)70099-2 [DOI] [PubMed] [Google Scholar]
- 25.Riveau G, Schacht AM, Dompnier JP, Deplanque D, Seck M, Waucquier N, et al. Safety and efficacy of the rSh28GST urinary schistosomiasis vaccine: a phase 3 randomized, controlled trial in Senegalese children. PLoS Negl Trop Dis. (2018) 12:e0006968. 10.1371/journal.pntd.0006968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Egesa M, Lubyayi L, Tukahebwa EM, Bagaya BS, Chalmers IW, Wilson S, et al. Schistosoma mansoni schistosomula antigens induce Th1/Pro-inflammatory cytokine responses. Parasite Immunol. (2018) 40:e12592. 10.1111/pim.12592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perrigoue JG, Marshall FA, Artis D. On the hunt for helminths: innate immune cells in the recognition and response to helminth parasites. Cellular Microbiology. (2008) 10:1757–1764. 10.1111/j.1462-5822.2008.01174.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grzych JM, Pearce E, Cheever A, Caulada ZA, Caspar P, Heiny S, et al. Egg deposition is the major stimulus for the production of Th2 cytokines in murine schistosomiasis mansoni. J Immunol. (1991) 146:1322–7. [PubMed] [Google Scholar]
- 29.Pearce EJ, M Kane C, Sun JJ, Taylor J, McKee AS, Cervi L. Th2 response polarization during infection with the helminth parasite Schistosoma mansoni. Immunol Rev. (2004) 201:117–26. 10.1111/j.0105-2896.2004.00187.x [DOI] [PubMed] [Google Scholar]
- 30.de Oliveira Fraga LA, Torrero MN, Tocheva AS, Mitre E, Davies SJ. Induction of type 2 responses by schistosome worms during prepatent infection. J Infect Dis. (2010) 201:464–72. 10.1086/649841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joseph S, Jones FM, Kimani G, Mwatha JK, Kamau T, Kazibwe F, et al. Cytokine production in whole blood cultures from a fishing community in an area of high endemicity for Schistosoma mansoni in Uganda: the differential effect of parasite worm and egg antigens. IAI. (2004) 72:728–34. 10.1128/IAI.72.2.728-734.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Everts B, Perona-Wright G, Smits HH, Hokke CH, van der Ham AJ, Fitzsimmons CM, et al. Omega-1, a glycoprotein secreted by Schistosoma mansoni eggs, drives Th2 responses. J Exp Med. (2009) 206:1673–80. 10.1084/jem.20082460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaplan MH, Whitfield JR, Boros DL. Th2 Cells are required for the schistosoma mansoni egg-induced granulomatous response. J Immunol. (1998) 160:1850–6. [PubMed] [Google Scholar]
- 34.Dunne DW, Lucas S, Bickle Q, Pearson S, Madgwick L, Bain J, et al. Identification and partial purification of an antigen (ω1 from Schistosoma mansoni eggs which is putatively hepatotoxic in T-cell deprived mice. Trans R Soc Trop Med Hyg. (1981) 75:54–71. 10.1016/0035-9203(81)90013-4 [DOI] [PubMed] [Google Scholar]
- 35.Alves Oliveira LF, Moreno EC, Gazzinelli G, Martins-Filho OA, Silveira AMS, Gazzinelli A, et al. Cytokine production associated with periportal fibrosis during chronic schistosomiasis mansoni in humans. Infect Immun. (2006) 74:1215–21. 10.1128/IAI.74.2.1215-1221.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rutitzky LI, Stadecker MJ. Exacerbated egg-induced immunopathology in murine Schistosoma mansoni infection is primarily mediated by IL-17 and restrained by IFN-γ. Eur J Immunol. (2011) 41:2677–87. 10.1002/eji.201041327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mbow M, Larkin BM, Meurs L, Wammes LJ, de Jong SE, Labuda LA, et al. T-Helper 17 cells are associated with pathology in human schistosomiasis. J Infect Dis. (2013) 207:186–95. 10.1093/infdis/jis654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grant AV, Araujo MI, Ponte EV, Oliveira RR, Cruz AA, Barnes KC, et al. Polymorphisms in IL10 are associated with total Immunoglobulin E levels and Schistosoma mansoni infection intensity in a Brazilian population. Genes Immun. (2011) 12:46–50. 10.1038/gene.2010.50 [DOI] [PubMed] [Google Scholar]
- 39.Corstjens PLAM, De Dood CJ, Kornelis D, Tjon Kon Fat EM, Wilson RA, Kariuki TM, et al. Tools for diagnosis, monitoring and screening of Schistosoma infections utilizing lateral-flow based assays and upconverting phosphor labels. Parasitology. (2014) 141:1841–55. 10.1017/S0031182014000626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woolhouse MEJ, Dye C, Etard JF, Smith T, Charlwood JD, Garnett GP, et al. Heterogeneities in the transmission of infectious agents: implications for the design of control programs. PNAS. (1997) 94:338–42. 10.1073/pnas.94.1.338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Butterworth AE, Capron M, Cordingley JS, Dalton PR, Dunne DW, Kariuki HC, et al. Immunity after treatment of human Schistosomiasis mansoni. II. Identification of resistant individuals, and analysis of their immune responses. Trans R Soc Trop Med Hyg. (1985) 79:393–408. 10.1016/0035-9203(85)90391-8 [DOI] [PubMed] [Google Scholar]
- 42.Abel L, Demenais F, Prata A, Souza AE, Dessein A. Evidence for the segregation of a major gene in human susceptibility/resistance to infection by Schistosoma mansoni. Am J Hum Genet. (1991) 48:959–70. [PMC free article] [PubMed] [Google Scholar]
- 43.Wilkins HA, Goll PH, Marshall TF, Moore PJ. Dynamics of Schistosoma haematobium infection in a Gambian community. III. Acquisition and loss of infection. Trans R Soc Trop Med Hyg. (1984) 78:227–32. 10.1016/0035-9203(84)90283-9 [DOI] [PubMed] [Google Scholar]
- 44.Bradley DJ, McCullough FS. Egg output stability and the epidemiology of Schistosoma haematobium. II. An analysis of the epidemiology of endemic S. haematobium. Trans R Soc Trop Med Hyg. (1973) 67:491–500. 10.1016/0035-9203(73)90080-1 [DOI] [PubMed] [Google Scholar]
- 45.Warren KS. Regulation of the prevalence and intensity of schistosomiasis in man: immunology or ecology? J Infect Dis. (1973) 127:595–609. 10.1093/infdis/127.5.595 [DOI] [PubMed] [Google Scholar]
- 46.King CH, Kariuki HC, Kadzo H, Koech DK, Magak P, Ouma JH, et al. Low heritable component of risk for infection intensity and infection-associated disease in urinary schistosomiasis among Wadigo village populations in Coast province, Kenya. Am J Trop Med Hyg. (2004) 70:57–62. 10.4269/ajtmh.2004.70.57 [DOI] [PubMed] [Google Scholar]
- 47.Fitzsimmons CM, Jones FM, Pinot de Moira A, Protasio AV, Khalife J, Dickinson HA, et al. Progressive cross-reactivity in IgE responses: an explanation for the slow development of human immunity to schistosomiasis? Infect Immun. (2012) 80:4264–70. 10.1128/IAI.00641-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Demeure CE, Rihet P, Abel L, Ouattara M, Bourgois A, Dessein AJ. Resistance to Schistosoma mansoni in humans: influence of the IgE/IgG4 Balance and IgG2 in immunity to reinfection after chemotherapy. J Infect Dis. (1993) 168:1000–8. 10.1093/infdis/168.4.1000 [DOI] [PubMed] [Google Scholar]
- 49.Figueiredo JP, Oliveira RR, Cardoso LS, Barnes KC, Grant AV, Carvalho EM, et al. Adult worm-specific IgE/IgG4 balance is associated with low infection levels of Schistosoma mansoni in an endemic area. Parasite Immunol. (2012) 34:604–10. 10.1111/pim.12001 [DOI] [PubMed] [Google Scholar]
- 50.Hagan P, Dunn D, Simpson G, Wilkins HA. Human lgE, lgG4 and resistance to reinfection with Schistosoma haematobium. (1991) 349:3. 10.1038/349243a0 [DOI] [PubMed] [Google Scholar]
- 51.Ndombi EM, Abudho B, Kittur N, Carter JM, Korir H, Riner DK, et al. Effect of four rounds of annual school-wide mass praziquantel treatment for Schistosoma mansoni control on schistosome-specific immune responses. Parasite Immunol. (2018) 40:e12530. 10.1111/pim.12530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McManus DP, Dunne DW, Sacko M, Utzinger J, Vennervald BJ, Zhou XN. Schistosomiasis. Nat Rev Dis Primers. (2018) 4:13. 10.1038/s41572-018-0013-8 [DOI] [PubMed] [Google Scholar]
- 53.Richter J, Hatz C, Campagne G, Bequist N, Jenkins J. Ultrasound in Schistosomiasis. A Practical Guide to the Standardized Use of Ultrasonography for the Assessment of Schistosomiasis-Related Morbidity. Niamey: WHO. (2000). [Google Scholar]
- 54.Blanton RE, Salam EA, Ehsan A, King CH, Goddard KA. Schistosomal hepatic fibrosis and the interferon gamma receptor: a linkage analysis using single-nucleotide polymorphic markers. Eur J Hum Genet. (2005) 13:660–8. 10.1038/sj.ejhg.5201388 [DOI] [PubMed] [Google Scholar]
- 55.Chevillard C, Moukoko CE, Elwali N-EMA, Bream JH, Kouriba B, Argiro L, et al. IFN-gamma polymorphisms (IFN-gamma +2109 and IFN-gamma +3810) are associated with severe hepatic fibrosis in human hepatic schistosomiasis (Schistosoma mansoni). J Immunol. (2003) 171:5596–601. 10.4049/jimmunol.171.10.5596 [DOI] [PubMed] [Google Scholar]
- 56.Hirayama K, Chen H, Kikuchi M, Yin T, Gu X, Liu J, et al. HLA-DR-DQ alleles and HLA-DP alleles are independently associated with susceptibility to different stages of post-schistosomal hepatic fibrosis in the Chinese population. Tissue Antigens. (1999) 53:269–74. 10.1034/j.1399-0039.1999.530307.x [DOI] [PubMed] [Google Scholar]
- 57.Palumbo E. Association between schistosomiasis and cancer: a review. Infect Dis Clin Pract. (2007) 15:145–8. 10.1097/01.idc.0000269904.90155.ce [DOI] [Google Scholar]
- 58.Asztely MS, Eriksson B, Gabone RM, Nilsson L-Å. Is ultrasonography useful for population studies on schistosomiasis mansoni? An evaluation based on a survey on a population from Kome Island, Tanzania. Acta Radiol Open. (2016) 5:2058460116686392. 10.1177/2058460116686392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barreto AVMS, Alecrim VM, Medeiros TB, de Domingues ALC, Lopes EP, Martins JRM, et al. New index for the diagnosis of liver fibrosis in Schistosomiasis mansoni. Arq Gastroenterol. (2017) 54:51–6. 10.1590/s0004-2803.2017v54n1-10 [DOI] [PubMed] [Google Scholar]
- 60.Olveda DU, Inobaya M, Olveda RM, Vinluan ML, Ng S-K, Weerakoon K, et al. Diagnosing schistosomiasis-induced liver morbidity: implications for global control. Int J Infect Dis. (2017) 54:138–44. 10.1016/j.ijid.2016.10.024 [DOI] [PubMed] [Google Scholar]
- 61.Pereira TA, Syn WK, Pereira FEL, Lambertucci JR, Secor WE, Diehl AM. Serum osteopontin is a biomarker of severe fibrosis and portal hypertension in human and murine Schistosomiasis mansoni. Int J Parasitol. (2016) 46:829–32. 10.1016/j.ijpara.2016.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pullan RL, Bethony JM, Geiger SM, Correa-Oliveira R, Brooker S, Quinnell RJ. Human helminth co-infection: no evidence of common genetic control of hookworm and Schistosoma mansoni infection intensity in a Brazilian community. Int J Parasitol. (2010) 40:299–306. 10.1016/j.ijpara.2009.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gazzinelli A, Coelho L, Alves-Fraga L, Soares-Filho B, Williams-Blangero S, Bethony J, et al. Additive host genetic factors influence fecal egg excretion rates during Schistosoma mansoni infection in a rural area in Brazil. Am J Med Hyg. (2002) 67:336–43. 10.4269/ajtmh.2002.67.336 [DOI] [PubMed] [Google Scholar]
- 64.Grant A, Araujo MI, Ponte EV, Oliveira RR, Cruz AA, Barnes KC, et al. High heritability but uncertain mode of inheritance for total serum IgE level and Schistosoma mansoni infection intensity in a Schistosomiasis-Endemic Brazilian Population. J Infect Dis. (2008) 198:1227–36. 10.1086/591946 [DOI] [PubMed] [Google Scholar]
- 65.Ellis MK, McManus DP. Familial aggregation of human helminth infection in the Poyang lake area of China with a focus on genetic susceptibility to schistosomiasis japonica and associated markers of disease. Parasitology. (2009) 136:699–712. 10.1017/S003118200900612X [DOI] [PubMed] [Google Scholar]
- 66.Spear RC, Zhong B, Kouch J, Seto EYW, Hubbard A. Genetic and household risk factors for Schistosoma japonicum infection in the presence of larger scale environmental differences in the mountainous transmission areas of China. Am J Trop Med Hyg. (2005) 73:1145–50. 10.4269/ajtmh.2005.73.1145 [DOI] [PubMed] [Google Scholar]
- 67.King R.C, Stransfield W.D. Dictionary of Genetics. Oxford: Oxford University Press. (1998). [Google Scholar]
- 68.Marquet S, Abel L, Hillaire D, Dessein A. Full results of the genome-wide scan which localises a locus controlling the intensity of infection by Schistosoma mansoni on chromosome 5q31-q33. Eur J Hum Genet. (1999) 7:88–97. 10.1038/sj.ejhg.5200243 [DOI] [PubMed] [Google Scholar]
- 69.Zinn-Justin A, Marquet S, Hillaire D, Dessein A, Abel L. Genome search for additional human loci controlling infection levels by Schistosoma mansoni. Am J Trop Med Hyg. (2001) 65:754–8. 10.4269/ajtmh.2001.65.754 [DOI] [PubMed] [Google Scholar]
- 70.Müller-Myhsok B, Stelma FF, Guissé-Sow F, Muntau B, Thye T, Burchard GD, et al. Further evidence suggesting the presence of a locus, on human chromosome 5q31-q33, influencing the intensity of infection with Schistosoma mansoni. Am J Hum Genet. (1997) 61:452–4. 10.1086/524851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dessein AJ, Hillaire D, Elwali NE, Marquet S, Mohamed-Ali Q, Mirghani A, et al. Severe Hepatic Fibrosis in Schistosoma mansoni Infection Is Controlled by a Major Locus That Is Closely Linked to the Interferon-Gamma Receptor Gene. Am J Hum Genet. (1999) 65:709–21. 10.1086/302526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gatlin MR, Black CL, Mwinzi PN, Secor WE, Karanja DM, Colley DG. Association of the gene polymorphisms IFN-gamma +874, IL-13−1055 and IL-4−590 with patterns of reinfection with Schistosoma mansoni. PLoS Negl Trop Dis. (2009) 3:e375. 10.1371/journal.pntd.0000375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Adedokun SA, Seamans BN, Cox NT, Liou G, Akindele AA, Li Y, et al. Interleukin-4 and STAT6 promoter polymorphisms but not interleukin-10 or 13 are essential for schistosomiasis and associated disease burden among Nigerian children. Infect Genetics Evol. (2018) 65:28–34. 10.1016/j.meegid.2018.07.012 [DOI] [PubMed] [Google Scholar]
- 74.Adedoja A, Hoan NX, Tong H, van Adukpo S, Tijani DB, Akanbi AA, et al. Differential contribution of interleukin-10 promoter variants in malaria and schistosomiasis mono- and co-infections among Nigerian children. Trop Med Int Health. (2018) 23:45–52. 10.1111/tmi.13007 [DOI] [PubMed] [Google Scholar]
- 75.Ellis MK, Zhao ZZ, Chen HG, Montgomery GW, Li YS, McManus DP. Analysis of the 5q31 33 locus shows an association between single nucleotide polymorphism variants in the IL-5 gene and symptomatic infection with the human blood fluke, Schistosoma japonicum. J Immunol. (2007) 179:8366–71. 10.4049/jimmunol.179.12.8366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gong Z, Luo Q-Z, Lin L, Su Y-P, Peng H-B, Du K, et al. Association of MICA gene polymorphisms with liver fibrosis in schistosomiasis patients in the Dongting Lake region. Braz J Med Biol Res. (2012) 45:222–9. 10.1590/S0100-879X2012007500024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grant AV, Araujo MI, Ponte EV, Oliveira RR, Gao P, Cruz AA, et al. Functional polymorphisms in IL13 are protective against high Schistosoma mansoni infection intensity in a Brazilian population. PLoS ONE. (2012) 7:e35863. 10.1371/journal.pone.0035863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.He H, Isnard A, Kouriba B, Cabantous S, Dessein A, Doumbo O, et al. A STAT6 gene polymorphism is associated with high infection levels in urinary schistosomiasis. Genes Immun. (2008) 9:195–206. 10.1038/gene.2008.2 [DOI] [PubMed] [Google Scholar]
- 79.Isnard A, Kouriba B, Doumbo O, Chevillard C. Association of rs7719175, located in the IL13 gene promoter, with Schistosoma haematobium infection levels and identification of a susceptibility haplotype. Genes Immun. (2011) 12:31–9. 10.1038/gene.2010.43 [DOI] [PubMed] [Google Scholar]
- 80.Kouriba B, Chevillard C, Bream JH, Argiro L, Dessein H, Arnaud V, et al. Analysis of the 5q31-q33 Locus Shows an Association between IL13-1055C/T IL-13-591A/G Polymorphisms and Schistosoma haematobium Infections. J Immunol. (2005) 174:6274–81. 10.4049/jimmunol.174.10.6274 [DOI] [PubMed] [Google Scholar]
- 81.Idris ZM, Yazdanbakhsh M, Adegnika AA, Lell B, Issifou S, Noordin R. A pilot study on Cytotoxic T Lymphocyte-4 gene polymorphisms in urinary schistosomiasis. Genetic Testing Mol Biomarkers. (2012) 16:488–92. 10.1089/gtmb.2011.0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ouf EA, Ojurongbe O, Akindele AA, Sina-Agbaje OR, Van Tong H, Adeyeba AO. Ficolin-2 Levels and FCN2 genetic polymorphisms as a susceptibility factor in schistosomiasis. J InfectDis. (2012) 206:562–70. 10.1093/infdis/jis396 [DOI] [PubMed] [Google Scholar]
- 83.Antony JS, Ojurongbe O, Kremsner PG, Velavan TP. Lectin complement protein collectin 11 (CL-K1) and susceptibility to urinary schistosomiasis. PLoS Negl Trop Dis. (2015) 9:e0003647. 10.1371/journal.pntd.0003647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tiongco RE, Paragas NA, Dominguez MJ, Lasta SL, Pandac JK, Pineda-Cortel MR. ABO blood group antigens may be associated with increased susceptibility to schistosomiasis: a systematic review and meta-analysis. J Helminthol. (2018) 94:e21. 10.1017/S0022149X18001116 [DOI] [PubMed] [Google Scholar]
- 85.Eriksson J, Reimert CM, Kabatereine NB, Kazibwe F, Ireri E, Kadzo H, et al. The 434(G>C) polymorphism within the coding sequence of Eosinophil Cationic Protein (ECP) correlates with the natural course of Schistosoma mansoni infection. Int J Parasitol. (2007) 37:1359–66. 10.1016/j.ijpara.2007.04.001 [DOI] [PubMed] [Google Scholar]
- 86.Dessein A, Chevillard C, Arnaud V, Hou X, Hamdoun AA, Dessein H, et al. Variants of CTGF are associated with hepatic fibrosis in Chinese, Sudanese, and Brazilians infected with schistosomes. J Exp Med. (2009) 206:2321–8. 10.1084/jem.20090383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Long X, Chen Q, Zhao J, Rafaels N, Mathias P, Liang H, et al. An IL-13 promoter polymorphism associated with liver fibrosis in patients with Schistosoma japonicum. PLoS ONE. (2015) 10:e0135360. 10.1371/journal.pone.0135360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Long X, Daya M, Zhao J, Rafaels N, Liang H, Potee J, et al. The role of ST2 and ST2 genetic variants in schistosomiasis. J Allergy Clin Immunol. (2017) 140:1416–22.e6. 10.1016/j.jaci.2016.12.969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Martins da Fonseca CS, Pimenta Filho AA, dos Santos BS, da Silva CA, Domingues ALC, Owen JS, et al. Human plasma lipid modulation in schistosomiasis mansoni depends on apolipoprotein E polymorphism. PLoS ONE. (2014) 9:e101964. 10.1371/journal.pone.0101964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sertorio M, Hou X, Carmo RF, Dessein H, Cabantous S, Abdelwahed M, et al. IL-22 and IL-22 binding protein (IL-22BP) regulate fibrosis and cirrhosis in hepatitis C virus and schistosome infections. Hepatology. (2015) 61:1321–31. 10.1002/hep.27629 [DOI] [PubMed] [Google Scholar]
- 91.Silva PCV, Gomes AV, de Souza TKG, Coêlho MRCD, Cahu GG, de OM Muniz MTC, et al. Association of SNP (-G1082A) IL-10 with increase in severity of periportal fibrosis in Schistosomiasis, in the Northeast of Brazil. Genetic Testing Mol Biomarkers. (2014) 18:646–52. 10.1089/gtmb.2014.0098 [DOI] [PubMed] [Google Scholar]
- 92.Silva PCV, Gomes AV, de Britto LRPB, de Lima ELS, da Silva JL, Montenegro SML, et al. Influence of a TNF-α Polymorphism on the severity of schistosomiasis periportal fibrosis in the Northeast of Brazil. Genet Test Mol Biomarkers. (2017) 21:658–62. 10.1089/gtmb.2017.0133 [DOI] [PubMed] [Google Scholar]
- 93.Zhu X, Zhang J, Fan W, Gong Y, Yan J, Yuan Z, et al. MAPKAP1 rs10118570 polymorphism is associated with anti-infection and anti-hepatic fibrogenesis in schistosomiasis japonica. PLoS ONE. (2014) 9:e105995. 10.1371/journal.pone.0105995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Oliveira JB, Silva PCV, Vasconcelos LM, Gomes AV, Coêlho MRCD, Cahu GGOM, et al. Influence of polymorphism (-G308A) TNF-α on the periportal fibrosis regression of schistosomiasis after specific treatment. Genet Test Mol Biomarkers. (2015) 19:598–603. 10.1089/gtmb.2015.0091 [DOI] [PubMed] [Google Scholar]
- 95.Xiao Q, Yu H, Zhu X. The associations of hub gene polymorphisms in PI3K/AKT/mTOR pathway and Schistosomiasis Japonica infection and hepatic fibrosis. Infect Genet Evol. (2020) 85:104423. 10.1016/j.meegid.2020.104423 [DOI] [PubMed] [Google Scholar]
- 96.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. (2009) 461:747–53. 10.1038/nature08494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mohamed-Ali Q, Elwali NE, Abdelhameed AA, Mergani A, Rahoud S, Elagib KE, et al. Susceptibility to periportal (Symmers) fibrosis in human Schistosoma mansoni infections: evidence that intensity and duration of infection, gender, and inherited factors are critical in disease progression. J Infect Dis. (1999) 180:1298–306. 10.1086/314999 [DOI] [PubMed] [Google Scholar]
- 98.Dessein AJ, Marquet S, Henri S, El Wali NE, Hillaire D, Rodrigues V, et al. Infection and disease in human Schistosoma mansoni are under distinct major gene control. Microbes Infect. (1999) 1:561–7. 10.1016/S1286-4579(99)80096-3 [DOI] [PubMed] [Google Scholar]
- 99.Abath FGC, Morais CNL, Montenegro CEL, Wynn TA, Montenegro SML. Immunopathogenic mechanisms in schistosomiasis: what can be learnt from human studies? Trends Parasitol. (2006) 22:85–91. 10.1016/j.pt.2005.12.004 [DOI] [PubMed] [Google Scholar]
- 100.Schön MP, Erpenbeck L. The Interleukin-23/Interleukin-17 axis links adaptive and innate immunity in psoriasis. Front Immunol. (2018) 9:1323. 10.3389/fimmu.2018.01323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kimura A, Kishimoto T. IL-6: Regulator of Treg/Th17 balance. Eur J Immunol. (2010) 40:1830–5. 10.1002/eji.201040391 [DOI] [PubMed] [Google Scholar]
- 102.Wen X, He L, Chi Y, Zhou S, Hoellwarth J, Zhang C, et al. Dynamics of Th17 cells and their role in Schistosoma japonicum infection in C57BL/6 Mice. PLoS Neglected Trop Dis. (2011) 5:e1399. 10.1371/journal.pntd.0001399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ojurongbe O, Antony JS, Tong HV, Meyer CG, Akindele AA, Sina-Agbaje OR. Low MBL-associated serine protease 2 (MASP-2) levels correlate with urogenital schistosomiasis in Nigerian children. Trop Med Int Health. (2015) 20:1311–9. 10.1111/tmi.12551 [DOI] [PubMed] [Google Scholar]
- 104.Elsammak MY, Al-Sharkaweey RM, Ragab MS, Amin GM, Kandil MH. In Egyptians, a mutation in the lymphotoxin-alpha gene may increase susceptibility to hepatitis C virus but not that to schistosomal infection. Ann Trop Med Parasitol. (2008) 102:709–16. 10.1179/136485908X337599 [DOI] [PubMed] [Google Scholar]
- 105.Nutman TB. Looking beyond the induction of Th2 responses to explain immunomodulation by helminths. Parasite Immunol. (2015) 37:304–13. 10.1111/pim.12194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Goenka S, Kaplan MH. Transcriptional regulation by STAT6. Immunol Res. (2011) 50:87–96. 10.1007/s12026-011-8205-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Brito CF, Caldas IR, Filho C, Correa-Oliveira R, Oliveira SC. CD4+ T cells of schistosomiasis naturally resistant individuals living in an endemic area produce interferon-γ and tumour necrosis factor-α in response to the recombinant 14kda Schistosoma mansoni fatty acid-binding protein. Scand J Immunol. (2000) 51:595–601. 10.1046/j.1365-3083.2000.00710.x [DOI] [PubMed] [Google Scholar]
- 108.Fonseca CT, Oliveira SC, Alves CC. Eliminating schistosomes through vaccination: what are the best immune weapons? Front Immunol. (2015) 6:1–8. 10.3389/fimmu.2015.00095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. (2018) 46:D1062–7. 10.1093/nar/gkx1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sim N-L, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. (2012) 40:W452–7. 10.1093/nar/gks539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. (2010) 7:248–249. 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.van der Pouw Kraan T, van Veen A, Boeije L, van Tuyl S, de Groot E, Stapel S, et al. An IL-13 promoter polymorphism associated with increased risk of allergic asthma. Genes Immun. (1999) 1:61–65. 10.1038/sj.gene.6363630 [DOI] [PubMed] [Google Scholar]
- 113.Patel DD, Kuchroo VK. Th17 cell pathway in human immunity: lessons from genetics and therapeutic interventions. Immunity. (2015) 43:1040–51. 10.1016/j.immuni.2015.12.003 [DOI] [PubMed] [Google Scholar]
- 114.Pang N, Zhang F, Ma X, Zhu Y, Zhao H, Xin Y, et al. TGF-β/Smad signaling pathway regulates Th17/Treg balance during Echinococcus multilocularis infection. Int Immunopharmacol. (2014) 20:248–57. 10.1016/j.intimp.2014.02.038 [DOI] [PubMed] [Google Scholar]
- 115.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. (2002) 4:45–61. 10.1097/00125817-200203000-00002 [DOI] [PubMed] [Google Scholar]
- 116.McClellan J, King M-C. Genetic heterogeneity in human disease. Cell. (2010) 141:210–7. 10.1016/j.cell.2010.03.032 [DOI] [PubMed] [Google Scholar]
- 117.Choi WA, Kang MJ, Kim YJ, Seo JH, Kim HY, Kwon JW, et al. Gene–gene interactions between candidate gene polymorphisms are associated with total IgE levels in Korean Children with Asthma. J Asthma. (2012) 49:243–52. 10.3109/02770903.2012.660294 [DOI] [PubMed] [Google Scholar]
- 118.Marquet S, Conte I, Poudiougou B, Argiro L, Cabantous S, Dessein H, et al. The IL17F and IL17RA genetic variants increase risk of cerebral malaria in two African Populations. Infect Immunity. (2016) 84:590–7. 10.1128/IAI.00671-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Campino S, Kwiatkowski D, Dessein A. Mendelian and complex genetics of susceptibility and resistance to parasitic infections. Semin Immunol. (2006) 18:411–22. 10.1016/j.smim.2006.07.011 [DOI] [PubMed] [Google Scholar]
- 120.Abel L, Dessein AJ. Genetic epidemiology of infectious diseases in humans: design of population-based studies. Emerging Infect Dis. (1998) 4:593–603. 10.3201/eid0404.980409 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Search strategy: Summary of PubMed search strategy.
All included references: Papers included in the review.
Details of Refs Populations, study design, phenotypes, and effects from studies.
Genes in QTL. List of genes in QTL regions with lists of publications associated with each gene and schistosomiasis.
VEP SNP Annotations. Functional annotation of SNP from candidate genes studies with the Ensembl Variant Effect predictor.
Th17 genes. List of Genes in Th17 pathway with KEGG pathway diagram.
Th17 SNP. List of potentially function SNP in genes in the Th17 pathway.