Abstract
We present an analysis and comparison study of genetic variants and mutations of about 1200 genomes of SARS-CoV-2 virus sampled across the first seven months of 2020. The study includes 12 sets of about 100 genomes each collected between January and September. We analyzed the mutations, mutation frequency and count trends over time, and genomes trends over time from January through September. We show that certain mutations in the SARS-CoV-2 genome are not occurring randomly as it has been commonly believed. This finding is in agreement with other recently published research in this domain. Therefore, this validates other findings in this direction. This study includes approximately 1000 genomes and was able to identify over 35 different mutations most of which are common to almost all genomes groups. Some mutations' ratios (frequency percentage) fluctuate over time to adapt the virus to various environmental factors, climate, and populations. One of the interesting findings in this paper is that the coding region, at the nucleotide level for NSP13 protein is relatively conserved compared with other protein regions in the ORF1ab gene which makes this protein a good candidate for developing drug targets and treatment for the COVID-19 disease. Although this outcome was already reported by other researchers, we corroborated their result with our work in a different approach and another experimental setting with almost one thousand complete genome sequences. We presented and discussed all these results and findings with tables of results and illustrating figures.
Keywords: SARS-CoV-2, Novel coronavirus, 2019 H-CoV 2, Genetic variants, SARS-CoV-2 genetic variants, SARS-CoV-2 genome
1. Introduction
Mutagens in the environment such as UV radiation, metals, and even endogenous substances produced by organisms create genetic variation in the genetic make-up of the SARS-CoV-2 genome. Over time, these random, naturally occurring mutations fluctuate in frequency in the genome pool, which contains implications about evolutionary mechanisms favoring certain mutations due to their adaptive values. The variations in the genetic sequence at the nucleotide level of an important virus like SARS-CoV-2 can help us in understanding and unfolding important facts and knowledge about its virulence. Such information and facts shall assist in drug and treatment design and discovering and determining new chemicals and therapies for the COVID-19 disease caused by this virus. Knowing that this virus is an RNA single -stranded virus helps researchers understand more details on the mutation mechanisms followed by this virus, SARS-CoV-2. Viruses in general exhibit one of the highest mutation rate variations among all organisms. In their comprehensive study about mutation mechanisms of viruses, Sanjuan and Domingo-Calap reported that RNA viruses tend to mutate faster than DNA viruses (Sanjuan and Domingo-Calap, 2016). Moreover, they found that single-stranded viruses, in general, show higher rates of mutations compared with double-stranded (Sanjuan and Domingo-Calap, 2016; Koyama et al., 2020). Besides, viruses with smaller genome found to have higher mutation rates (Sanjuan and Domingo-Calap, 2016; Koyama et al., 2020). Among the many factors that affect mutation rates, they found the largest difference being DNA and RNA viruses.
SARS-CoV-2 is a highly contagious and quickly transmissible virus among various populations and with different (and perhaps difficult) conditions. This means it is highly adaptable to many situations and many conditions, which is apparent from the fast and broad spread of the disease all over the world. There were about 200,000 new cases of COVID-19 every day in the world during the months of June – September (The Center for Systems Science and Engineering (CSSE) at JHU and the Worldmeter, n.d.). All these indicate that the virus has an exceptional ability to changing itself and adapting to survive and keep transmitting itself. Typically, genetic changes, including sequence variations, help the virus become more transmissible and more vicious (Center for Disease Control and Prevention (CDC), n.d.). Most importantly, SARS-CoV-2, like most other viruses and pathogens, mutates to produce variants that are not known to the host immune system (Biological Sciences, National Institutes of Health (US), 2007; Saha et al., 2020). The availability of a large number of (complete) genomes (for SARS-CoV-2) freely online makes it easier for researchers to study and analyze the mutation and sequence variation mechanisms of this virus. In this paper, we present a study and analysis of the genetic variations and mutations of the SARS-CoV-2 genomic sequences at the nucleotide level of the complete genome. For that, the study includes about 1000 genomes grouped according to the month of collection from January through September. The SARS-CoV-2 virus causes the COVID-19 disease which started back in December 2019 in the Wuhan region in China and transferred to the rest of the world quickly during the first three months of 2020.
The SARS-CoV-2 virus is highly contagious and easily transmissible and is the main cause of the coronavirus disease. The coronavirus disease, COVID-19, began at the end of 2019 in China and quickly spread to the rest of the world. By March 2020, the rapid spread had caused many countries around the world to administer a lockdown and apply stay-at-home and work from home policies. By the end of March 2020, there were one million COVID-19 cases and 45,000 deaths globally. By end of June 2020, the number of cases exceeded 10 million while deaths exceeded half a million worldwide. By the last week of August 2020, there were 850,000 deaths, 26 million cases, and the number of daily new cases in the world exceeded 250,000 new cases every day in the world (The Center for Systems Science and Engineering (CSSE) at JHU and the Worldmeter, n.d.).
2. Background and related work
2.1. Background
Started in Wuhan city in China around December 2019, the coronavirus disease is caused by a SARS family virus named SARS-CoV-2 by the World Health Organization (WHO). This novel disease spread throughout the world starting in January 2020 wherein Italy and Spain were among the first few countries to experience initial outbreaks outside of China (The Center for Systems Science and Engineering (CSSE) at JHU and the Worldmeter, n.d.; Center for Disease Control and Prevention (CDC), n.d.; Biological Sciences, National Institutes of Health (US), 2007; Saha et al., 2020). Around mid-March, COVID-19 became the most difficult and biggest problem, impacting every aspect of life around the globe.
The SARS-CoV-2 virus is not lab manipulated; it is naturally produced as a result of many years of changes, as most significant studies showed, but it is not proved as the origin of this virus (Andersen et al., 2020).
The disease was named COVID-19 by the World Health Organization WHO, and the virus called the novel coronavirus (novel because it is a new member of the coronaviruses family). Initially, they named it n-Cov-19 or 2019-nCoV then changed the name to SARS-CoV-2. Thus, the virus is now named SARS-CoV-2, and the disease is COVID-19 or the ‘Novel Coronavirus Disease 2019’. From the family of human coronaviruses (H-CoV's), three highly pathogenic H-CoV's have been identified so far (Center for Disease Control and Prevention (CDC), n.d.), including: (1) Middle East respiratory syndrome coronavirus (MERS-CoV); (2) severe acute respiratory syndrome (SARS) coronavirus (SARS-CoV), and (3) the 2019 novel coronavirus (or SARS-CoV-2), previously called 2019-nCoV (Center for Disease Control and Prevention (CDC), n.d.). Among these three, the MERS-CoV (Li et al., 2020) was responsible for 2494 cases and 858 deaths in 27 countries during the 2012 MERS outbreak; whereas the SARS-CoV pathogen was responsible for >8000 cases and 774 deaths in 37 countries during the 2002 to 2003 SARS outbreak; and SARS-CoV-2 is causing close to one million deaths so far (The Center for Systems Science and Engineering (CSSE) at JHU and the Worldmeter, n.d.; Li et al., 2020).
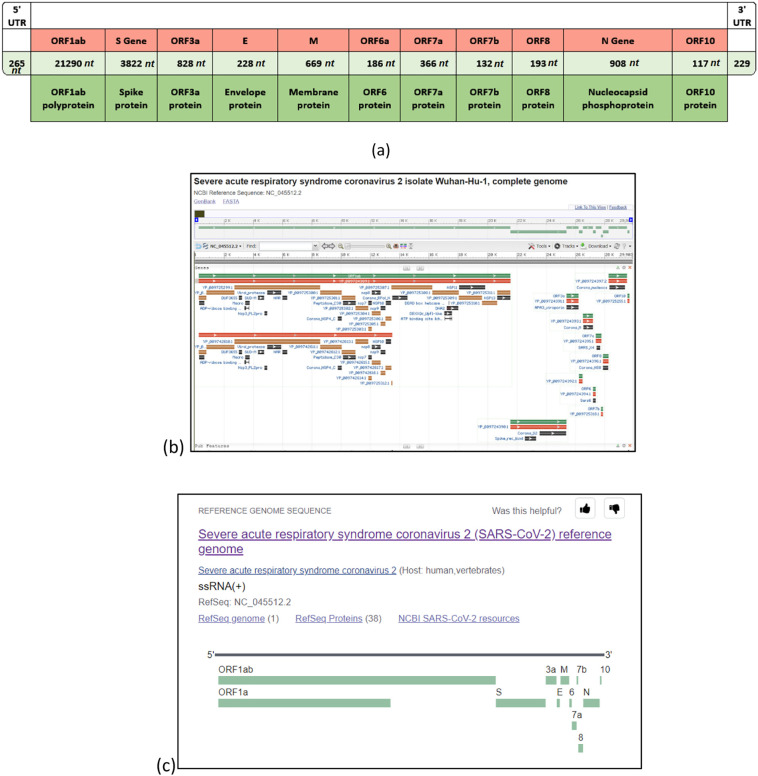
The complete genome (reference) of the virus is comprised of 11 (protein-coding) genes. The map of virus SARS-CoV-2 is shown in Fig. 1 while Table 1 contains all the genes and proteins of this virus. There are 16 nonstructural proteins (NSP): NSP1 to NSP11 by gene ORF1a and NSP12 to NSP16 by ORF1ab. As shown in the gene map in Fig. 1, the first gene ORF1ab occupies more than two-thirds of the virus, and ORF2 (S gene; Spike protein) is the second largest (8319 nt); Fig. 1 and Table 1. Moreover, Fig. 2 illustrates the position of the Spike protein (S gene) within the reference genome (Andersen et al., 2020). Table 2 presents other details about coded proteins (and genes) in the virus as laid in the NCBI database.
Fig. 1.
Structure of the SARS-CoV-2 genome in three different views: (a) basic illustration of the complete genome structure (b) illustration of coronavirus 2 isolate Wuhan-Hu-1, NC_045512 (complete genome 29,903 bp) as presented in the GenBank/NCBI https://www.ncbi.nlm.nih.gov/nuccore/NC_045512.2?report=graph (c) another view of the NC_045512.2 reference genome from NCBI showing all location of all genes. {Note: (b) and (c) are both courtesy of The US National Center for Biotechnology Information NCBI www.ncbi.nlm.nih.org.}
Table 1.
Description and details of all genes and proteins of the SARS-CoV-2.
(a) The description and division of regions of a complete SARS-CoV-2 genome (reference genome nc-045512 from the GenBank).
(b) This table shows the location and number of amino acids for each gene/protein {the first gene ORF1ab expresses a polyprotein comprised of 16 nonstructural proteins (NSP's) shown in (c)}.
(c) The 16 NSP's {nonstructural proteins} from the SARS-CoV-2 polyprotein.
| (a) | |||||
|---|---|---|---|---|---|
|
NC_045512.2 29903 nt | |||||
| Start | Stop | Gene symbol | Strand | NCBI gene ID | |
| 1 | 266 | 21555 | ORF1ab | Plus | 43740578 |
| 2 | 21563 | 25384 | S | Plus | 43740568 |
| 3 | 25393 | 26220 | ORF3a | Plus | 43740569 |
| 4 | 26245 | 26472 | E | Plus | 43740570 |
| 5 | 26523 | 27191 | M | Plus | 43740571 |
| 6 | 27202 | 27387 | ORF6 | Plus | 43740572 |
| 7 | 27394 | 27759 | ORF7a | Plus | 43740573 |
| 8 | 27756 | 27887 | ORF7b | Plus | 43740574 |
| 9 | 27894 | 28259 | ORF8 | Plus | 43740577 |
| 10 | 28274 | 29533 | N | Plus | 43740575 |
| 11 | 29558 | 29674 | ORF10 | Plus | 43740576 |
| (b) | |||||
|---|---|---|---|---|---|
| Gene | Number (#) | Gene | Gene ID | Location | Protein |
| 1 | 1 (7096) | ORF1ab | 43740578 | 266–21555 | ORF1ab polyprotein |
| 1 | 1 (4405) | ORF1a | 43740578 | 266–13483 | ORF1a polyprotein |
| 2 | 2 (1273) | ORF2 (S) | 43740568 | 21563–25384 | Spike protein (S protein) |
| 3 | 3 (275) | ORF3a | 43740569 | 25393–26220 | ORF3a protein |
| 4 | 4 (75) | ORF4 (E) | 43740570 | 26245–26472 | Envelope protein (E protein) |
| 5 | 5 (222) | ORF5 (M) | 43740571 | 26523–27191 | Membrane protein (M protein) |
| 6 | 6 (61) | ORF6 | 43740572 | 27202–27387 | ORF6 protein |
| 7 | 7 (121) | ORF7a | 43740573 | 27394–27759 | ORF7a protein |
| 8 | 8 (43) | ORF7b | 43740574 | 27756–27887 | ORF7b protein |
| 9 | 9 (121) | ORF8 | 43740577 | 27894–28259 | ORF8 protein |
| 10 | 10 (419) | ORF9 (N) | 43740575 | 28274–29533 | Nucleocapsid phosphoprotein (N protein) |
| 11 | 11 (38) | ORF10 | 43740576 | 29558–29674 | ORF10 protein |
| (c) | ||||
|---|---|---|---|---|
| # | Name | Accession | Amino acids | Proposed function |
| (i) | NSP1 | YP_009725297.1 | 180 amino acids | Induce host mRNA (leader protein) cleavage |
| (ii) | NSP2 | YP_009725298.1 | 638 amino acids | Binds to PHBs 1, 2 |
| (iii) | NSP3a | YP_009725299.1 | 1945 amino acids | Release NSPs 1, 2, 3 (Papain like proteinase) |
| (iv) | NSP4 | YP_009725300.1 | 500 amino acids | Membrane rearrangement |
| (v) | NSP5a | YP_009725301.1 | 306 amino acids | Cleaves at 11 sites of (3C-like proteinase) NSP polyprotein |
| (vi) | NSP6 | YP_009725302.1 | 290 amino acids | Generates autophagosomes |
| (vii) | NSP7 | YP_009725303.1 | 83 amino acids | Dimerizes with NSP8 |
| (viii) | NSP8 | YP_009725304.1 | 198 amino acids | Stimulates NSP12 |
| (ix) | NSP9 | YP_009725305.1 | 113 amino acids | Binds to helicase (?) |
| (x) | NSP10 | YP_009725306.1 | 139 amino acids | Stimulates NSP16 (?) |
| (xi) | NSP11 | YP_009725312.1 | 13 amino acids | Unknown |
| (xii) | NSP12a | YP_009725307.1 | 932 amino acids | Copies viral RNA (RNA polymerase) methylation (guanine) |
| (xiii) | NSP13 | YP_009725308.1 | 601 amino acids | Unwinds duplex RNA (helicase) |
| (xiv) | NSP14 | YP_009725309.1 | 527 amino acids | 5′-cap RNA (3′ to 5′ exonuclease, guanine N7-methyltransferase) |
| (xv) | NSP15a | YP_009725310.1 | 346 amino acids | Degrade RNA to (endoRNAse/endoribonuclease) evade host defense |
| (xvi) | NSP16 | YP_009725311.1 | 298 amino acids | 5′-cap RNA (2′-O-ribose-methyltransferase—potential antiviral drug target) methylation (adenine) |
Fig. 2.
This figure shows the divisions of the genome into genes/proteins and highlights the ORF1ab and Spike protein.
Table 2.
More details about then main genes (and coded proteins) in the SARS-CoV-2.
| Gene | Gene symbol | Gene ID | Gene type | Gene description |
|---|---|---|---|---|
| S surface glycoprotein | S | 43740568 | Protein coding | Surface glycoprotein (3822 nt 21563–25384) |
| M membrane glycoprotein [severe acute respiratory syndrome coronavirus 2] | M | 43740571 | Protein coding | Gene description Membrane glycoprotein (669 nt: 26523–27191) |
| ORF1ab ORF1a polyprotein;ORF1ab polyprotein [severe acute respiratory syndrome coronavirus 2] | ORF1ab | 43740578 | Protein coding | Gene description ORF1a polyprotein; ORF1ab polyprotein {sequence: NC_045512.2: 266…21555} |
| ORF3a protein [severe acute respiratory syndrome coronavirus 2] | ORF3a | 43740569 | Protein coding | Gene description ORF3a protein{sequence: NC_045512.2: 25393…26220} |
2.2. Related work
Li et al. presents a very good review study on the evolutionary history of SARS-CoV-2 and what could be the potential intermediate transferring animals (Li et al., 2020).
Yoshimoto (2020) presented the complete set of genes and proteins of SARS-CoV-2.
Saha et al. (2020) presented a study that the virus can evolve through mutations into a better version of itself to fit best in the host environment that is, the virus uses the mutation as a mechanism to acclimatize with its environment.
Li et al. (2020) presented a very good study of the evolutionary history of the virus with the potential animals and species analysis in the context of this disease.
Tai et al. (2020) study and present the complete details of the virus and the disease in the context of its transfer to humans via the Spike protein and the sequence of molecular of biological functions involved in the process (Center for Disease Control and Prevention (CDC), n.d.).
Petropoulos and Makridakis (2020) present objective forecasts for the confirmed coronavirus disease and provide a study and timeline of the potential implications of the disease for planning and decision making. Andersen et al. present a study of the notable features and origin theories of this virus (Andersen et al., 2020). A comprehensive study and list of proteins in the coronavirus SARS-CoV-2 are presented by Yoshimoto (2020).
Khailany et al. presented a study of the genetic variations and mutation comparison of the reported SARS-CoV-2 genetic data over various time frames and locations (Khailany et al., 2020). They analyzed 95 complete genomes of SARS-CoV-2 submitted to various databases through April 2020 (Khailany et al., 2020).
Emameh et al. (2020) presented a data mining and computational study of SARS-CoV-2 isolates from oronasopharynx of Iranian patients to characterize the proteins in ORF1ab region of the genome. The polyproteins of SARS-CoV-2 are cleaved by virus-encoded cysteine proteinases comprising 16 nonstructural proteins (NSP's) including the expression of NSP1 to NSP11 by ORF1a and encoding NSP12 to NSP16 by ORF1b (Emameh et al., 2020).
The SARS-CoV-2 research projects and studies are very recent, and all publications are in 2020. In general, most research projects and studies related to SARS-CoV-2 can be divided into one of five distinct categories as summarized in the following table:
| Area or task | References |
|---|---|
| 1. History and evolutionary studies on the SARS-CoV-2 virus | (Andersen et al., 2020; Li et al., 2020; Li et al., 2020) |
| 2. Characterization studies: Studying the structural details of the virus with its genes and proteins | (Li et al., 2020; Yoshimoto, 2020; Emameh et al., 2020; Shah et al., 2020) |
| 3. Comparison of SARS-CoV-2 with other viruses in this family like SARS and MERS-CoV | (Li et al., 2020; Li et al., 2020; Tai et al., 2020; Prathiviraj et al., 2020) |
| 4. Studying the mutations in SARS-CoV-2 | (Center for Disease Control and Prevention (CDC), n.d.; Saha et al., 2020; Dorp et al., 2020; Khailany et al., 2020; Junejo et al., 2020) |
| 5. Studying the virus in connection with the COVID-19 disease (for example, for drugs, treatment, and vaccine) | (Center for Disease Control and Prevention (CDC), n.d.; Mirza and Froeyen, 2020; Tai et al., 2020; Emameh et al., 2020; Junejo et al., 2020) |
Note: Literature and publications related to the COVID-19 disease and pandemic (see Junejo et al., 2020; El Idrissi, 2020; Rothan and Byrareddy, 2020; Wilder-Smith et al., 2020; Anastassopoulou et al., 2020) are mainly interested in the disease outbreak data analysis, timeline and infection rate analysis, pandemic declining prediction, etc. (Junejo et al., 2020; El Idrissi, 2020; Rothan and Byrareddy, 2020; Wilder-Smith et al., 2020; Anastassopoulou et al., 2020).
3. Materials and methods
3.1. Genomic data and method
We collected 12 sets of complete genomic sequences of SARS-CoV-2 over the first seven months of 2020 based on the date of collection (or acquisition), and obtained from NCBI (NCBI, n.d.). Approximately, the 12 datasets S 1…S 12 are collected during the months Jan. 2020 through Sep. 2020 as shown in Table 3 .
Table 3.
Details of the twelve dataset of genome sequences.
| Dataset | Number of genomes | Collection date | Mean length (nt) | |
|---|---|---|---|---|
| S1 | S1-genomes-01-01-to-01-31 | ~100 | 1–31 January | 29,858 |
| S2 | S2-101-genomes-01-01-to-02-29 | ~100 | 1 Jan.–29 Feb. | 29,921 |
| S3 | S3-103-genomes-02-01-to-02-29 | ~100 | 1 Feb.–29 Feb. | 29,752 |
| S4 | S4-99-genomes-02-01-03-25-. | ~100 | 2 Feb.–25 March | 29,676 |
| S5 | S5-93-genomes-3-1-3-26 | ~100 | 1–26 March | 29,839 |
| S6 | S6-95-genomes-04-21-04-30 | ~100 | 21–30 April | 29,703 |
| S7 | S7_100-genomes-05-01-to-05-07 | ~100 | 1–7 May | 29,666 |
| S8 | S8-100-genomes-05-22-to-05-31 | ~100 | 22–31 May | 29,822 |
| S9 | S9-128-genomes-06-09-06-12 | ~100 | 9–12 June | 29,686 |
| S10 | S10-101-genomes-07-01-07-17 | ~100 | 1–17 July | 29,836 |
| S11 | S11-100-genomes-08-01-08-31 | ~100 | 1–31 August | 29,824 |
| S12 | S12-100-genomes-09-01-09-30 | ~100 | 1–30 September | 29,886 |
Each set, S 1…S 10, contains approximately 100 complete genome sequences. We focus on our study on the date of collection of each genome which is available for all data. The total number of sequences available in NCBI based on the month of collection for each month is shown in Table 7. We decided to choose the same number of sequences in each one of the 12 sets. As shown in Table 7, March and April were the peak months of data collection for this virus's genome. We discarded each genome with high variations or high unknown nucleotides.
Table 7.
Total number of genome sequences available at NCBI based on the collection month starting from December 2019.
| Month of collection | No. of genome sequences |
|---|---|
| December/2019 | 22 |
| January | 399 |
| February | 555 |
| March | 6189 |
| April | 5157 |
| May | 2508 |
| June | 3005 |
| July | 4545 |
| August | 652 |
| September | 200 |
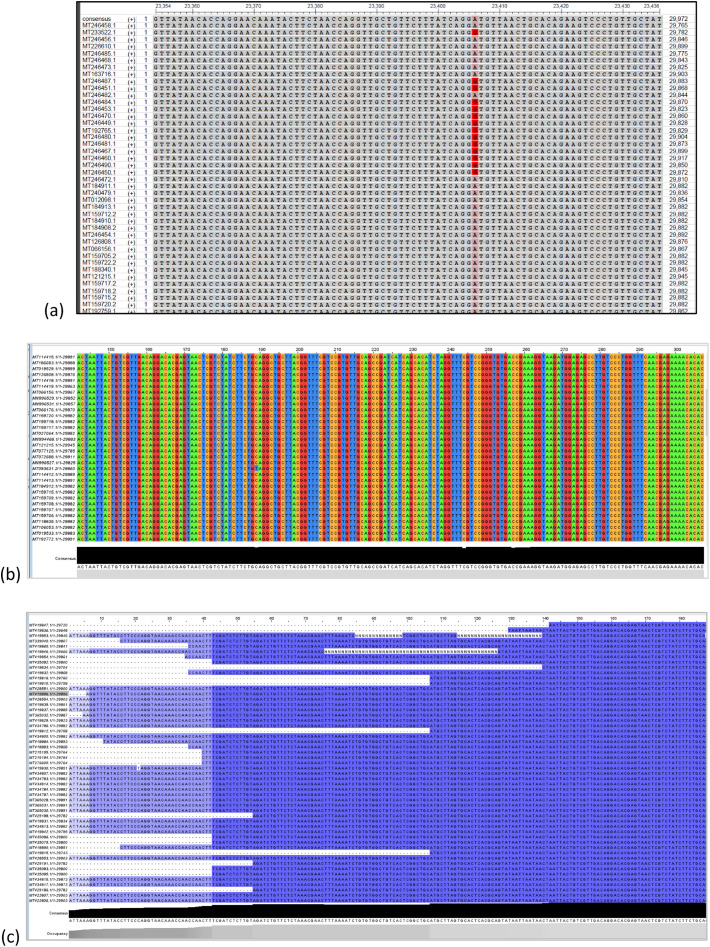
We ran each dataset for alignment in the Ebi Clustal Omega tool and used Jalview (Ebi Clustal Omega, n.d.; Waterhouse et al., n.d.; Rice et al., 2000) for alignment visualization. We used both Clustal Omega results and Jalview for extracting and determining the genetic variants in each dataset S i. Fig. 3 shows some of the sequence alignment results in various methods of visualization. In some cases, for the analysis, some variants were converted from nucleotide to amino acid levels. Mutations in each set S i were identified, extracted, and analyzed against all sequences in the other sets for the goal of finding significant mutations. Also, our analysis focused on determining whether or not a mutation is occurring randomly in the genome sequence. Since the virus continues to spread month after month throughout the world, we anticipate that it changes and mutates itself continuously over time starting January 2020.
Fig. 3.
Visualization of the genomic sequences showing the alignment and genetic variants.
(a) This shows 23406A>G using NCBI msa viewer (this is the dataset S5 (this is only partial view showing position 23354 to position 23463 with a sample of 30 genomes).
(b) Visualization of the multiple sequence alignment (using Jalview tool).
(c) S1c-100-genome (aligned with clustal msa)>>partial view with Jalview.
Note: Y is C or T; R is A or G; W is A or T.
Note: (a) is courtesy of the US National Center for Biotechnology Information NCBI. (b) and (c) are courtesy of Jalview tool (Waterhouse et al., n.d.): https://www.jalview.org/.
3.2. Comparison with other methods
Most of the related and similar work on mutations and genetic variations of SARS-CoV-2 focuses on extracting and determining mutations, studying and analyzing one or few specific mutations, or analyzing mutations within a specific coding region, e.g. Spike. This work, on the other hand, focuses on extracting most of the significant mutations and examining the important ones from various perspectives, for example, that do not occur randomly (see Section 4 Results). Moreover, while most of the other studies tend to focus on the differences in mutations based on geographic regions, our approach determines and analyzes the significant mutations based on time progression from January through September where the peak of the spread of the virus was during April 2020. This is the period that experienced the greatest number of genomes submitted/collected as reported in NCBI https://www.ncbi.nlm.nih.gov/sars-cov-2/. Therefore, the main contributions are in the Results section (Section 4). Koyama et al. (2020) studied the SARS-CoV genome and were able to identify more than 5700 distinct genome mutations. The most common was the synonymous 3037C>T (Koyama et al., 2020). This is also reported in this research in this paper with 41% in Table 4 and 47% in Table 5 for this 3037C>T mutation.
Table 4.
The most common genetic variants found in the 100 genomes of dataset S7 (s7_100-genomes-05-01-to-05-07) which includes genomes collected during 1–7 May.
| Results from S7 100-genomes-05-01-to-05-07 |
|---|
| — 100% occupancy and 100% identity started from position 142 |
| — last 100% occupancy 29,666 (last 100% identity 29,652 100%T) |
| — 241t>c 37% (i.e., T is 63% and mutation c is 37%) |
| — 1059c>t 36% |
| — 2416c>t 14% |
| — 2447g>a 3% |
| — 3037c>t 41% |
| — 4523g>a 13% |
| — MT434802: 5698c>t |
| — mt434786 6639:a>g |
| — 438551 8653 g>t |
| — 8782 c>t 17% |
| — 11083 g>t 6% |
| — 13265 a>t 3% |
| — 14408 t>c 37% |
| 14805 c>t 6% |
| 17747 c>t 15% |
| 17845 a>g 14% |
| 18060 c>t 14% |
| 23403 g>a 37% |
| 25563 t>g 47% |
| 26144g>a 8% |
| 27964 c>t 3% (including: mt429186, mt422807, and mt422806) |
| 28883 g>c 4% |
| 29540 g>a 10% |
| 29711g>t 7% |
Table 5.
The genetic variants in the set S8 (100 genomes).
| Results from S8 100-genomes-05-29-to-05-31 |
|---|
| — 100% occupancy and 100% identity started from position 50 with 100% C. |
| — last 100% occupancy 29,806 (also last 100% identity 29,806 100% A) |
| — 241 t>c 46% (i.e., T is 54% and mutation c is 46%) |
| — MT539162 490 t>a (this mutation is 4% >> in 4 sequences) |
| — MT535481 833 t>c (this mutation 3%) |
| — 1059 c>t 19% |
| — 2416c>t 14% |
| Mt536953 370 g>t |
| Mt539159 2243g>a |
| 3037 t>c 47% |
| 3177 c>t 4% (mt539163 is one of them). |
| 4084 c>t 7% |
| 6512 a>c |
| 8782 c>t 12% |
| 10129 a>y 18% ?? |
| MT534285: 11083 g>t (this 3%) |
| 12557 a>g 25% |
| 14408 t>c 46% |
| 14940 a>g 5% |
| 15771 t>y (or t>k) 15% |
| 17747 c>t 6% |
| 18877 c>t 10% |
| 23403 g>a 46% >>> see this in Table 3c |
| 24904 c>t 25% |
| 25563 g>t 33% |
| 25916 c>t 25% |
| 27359 a>g 25% |
| 27964 c>t 11% |
| 28144 t>c 12% |
| {near: 22 mutations in positions: 28878–28896} |
| 29360 t>k 7% |
4. Results
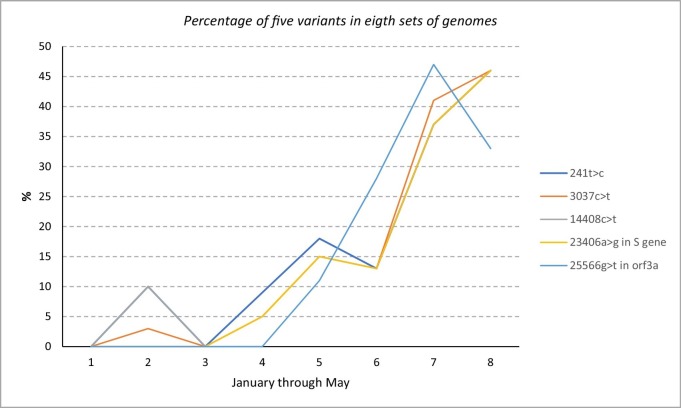
We identified and collected over 300 genetic variants (not all shown) from these genomes at the nucleotide level and we reported and discussed the important and significant ones. Table 4 contains the most commonly occurring genetic variants in the set S 7 which contains100 genomes collected during (the beginning of) the month of May. In Table 5, we see the most occurring and prevalent genetic variants in the set S 8 collected around the end (last three days) of May. Moreover, we collected 13 genetic variants common to almost all ten sets as shown in Table_6. The percentage of each one of the 13 variants in Table 6 are shown. We also identified 5 genetic variants that showed a clear trend of increase from January to May across eight datasets as shown in Fig. 4 . Table 8, Table 9 include the most frequent genetic variants in the August and September (S 11 and S 12 sets) genomes.
Table 6.
Percentage of 13 genetic variants across ten sets of genomes. Each row represents on set of genomes (~about 100 genomes in each set). The 13 genetic variants are shown as column headers 1…13.
| 1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 241t>c | 1059c>t: 19% | 3037c>t | 8782C>T in ORF1ab gene | 11083g>t | 14408c>t | 17747c>t | 17858a>g | 18060c>t | 23406a>g in S gene | 25566g>t in orf3a | 28144T>C in ORF8 gene | 29095C>T | ||
| 1 | S1 | 0% | 0% | 0% | 35% | 6% | 0% | 0% | 0% | 6% | 0% | 0% | 35% | 18% |
| 2 | S2 | 232t>c: 10% | 1060c>t: 3% | 3% 3037t>c | 8798c>t: 39% | 1% 11069 | 10% 14407c>t | 6% 17749c>t | 10% 17836 | 0% | 0% | 0% | 28192t>c: 42% | 29143c>t: 23% |
| 3 | S3 | 0% | 0% | 0% | 8785c>t: 30% | 4% 11086 | 0% | 0% | 0% | 6%: 18063 | 0% | 0% | 28147t>c: 30% | 29098c>t: 9% |
| 4 | S4 | 9% | 1071: 2% | 3045: 5% | 8790: 29% | 11091t>g: 46% | 5% | 17755: 14% | 17868: 14% | 18068: 14% | 23414: 5% | 0% | 28155: 29% | 0% |
| 5 | S5 | 18%: 241c>t | 10% | 15% | 46% | 35% | 15% | 34% | 34% | 37% | 15% *1 | 11% | 46% (28147) | 0% |
| 6 | S6 | 13% | 32% 1059t>c | 13% t>c | 9% | 4% | 13% 14408t>c | 1% | 3% | 3% | 13% 23403 | 28% 25563t>g | 9% | 0% |
| 7 | S7 | 37% | 34% | 41% | 17% | 6% | 37% t>c | 15% | 14% | 14% | 37% g>a | 47% 25563t>g | 17% | 0% |
| 8 | S8 | 46% | 19% | 46% t>c | 12% | 3% | 46% t>c | 6% | 6% | 6% | 46% 23403g>a | 33% 25563g>t | 12% | 1% |
| 9 | S9 | 11.7% 241 t>c | 1163 a>t 10% | 8.60% | 7% | 0% (zero) | 14408t>c: 14% | 1.60% | 17858 1.6% | 1.60% | 23403: 8.6% | 25563: 48.44% | 4.70% | 0% |
| 10 | S10 | 19% | 11% | 17% t>c | 8% | 15% | 17% t>c | 4% | 4% | 4% | 23403: 17% | 32% | 6% | 11% |
Fig. 4.
Illustration of the percentage of five mutations in eight genomic sets (sets S1…S8) collected between 1 January and 31 May.
Table 8.
Top genetic variants in genome sets S11-August (a); and in S12-September (b).
| (a) | |
|---|---|
| 1059t>c | 42% |
| 25563t>g | 33% |
| 16260c>t | 23% |
| 28821c>a | 22% |
| 28881g>a | 19% |
| 28882g>a | 19% |
| 28883g>c | 19% |
| 27964c>t | 16% |
| 20268a>g | 15% |
| 28854c>t | 13% |
| 11498c>t | 11% |
| 21575c>t | 11% |
| 9115c>t | 10% |
| 19603a>g | 10% |
| 21304c>t | 10% |
| (b) | |
|---|---|
| 28881g>a | 41% |
| 28882g>a | 41% |
| 28883g>c | 41% |
| 25563g>t | 39% |
| 1163a>t | 27% |
| 7540t>c | 27% |
| 16647g>t | 27% |
| 1855c>t | 27% |
| 22992g>t | 27% |
| 23401g>a | 27% |
| 28854c>t | 19% |
| 20268a>g | 19% |
| 22162t>c | 17% |
| 27964c>t | 17% |
| 18486c>t | 16% |
| 13665c>t | 14% |
| 10319c>t | 11% |
Table 9.
The frequent genetic variants that found in common between S11-August and S12-September genome sequences.
| 16269c>t |
| 20268a>g |
| 21304c>t |
| 27964c>t |
| 28821c>a |
| 28854c>t |
| 28881g>a |
| 28882g>a |
| 28883g>c |
Of the genetic variants identified, the ORF1a region contained mostly synonymous mutations that retained the amino acid encoded by the codon, which is due to the degeneracy of the genetic code. The mutations found in this region were alterations of the third nucleotide in each respective codon triplet, which retained the amino acid of the reference genome in all but one mutation, which is due to the wobble hypothesis (Näsvall et al., 2007). The only mutation that did not was the mutation 11083 g>t, which was found in the region that coded for the NSP6 subunit. This mutation changed the TTG codon for Leucine to the TTT codon of Phenylalanine. Though this changes the amino acid, the hydrophobicity of the two amino acids are the same, with the only difference being the size of the side chain, which may add some steric hindrance and potentially cause the protein NSP6 to fold differently (The Center for Systems Science and Engineering (CSSE) at JHU and the Worldmeter, n.d.; Center for Disease Control and Prevention (CDC), n.d.; Biological Sciences, National Institutes of Health (US), 2007; Saha et al., 2020; Andersen et al., 2020; Waterhouse et al., n.d.; Petropoulos and Makridakis, 2020; Yoshimoto, 2020; Emameh et al., 2020). According to Table 1(c), this protein functions to generate autophagosomes which play an important part in the virulence of this virus, as autophagosomes designate cell parts for intracellular degradation by the lysosome of the cell. More research needs to be done about this specifically, however. For this mutation, we identified that it was most prevalent in the S 4 and S 5 genomes, which means it was most frequent around late February or early March (Table 1, Table 2, Table 3, Table 4, Table 5, Table 6). Through mid-June (S 9), the prevalence of this mutation quickly approached and maintained close to 0% of the genomes, then started to reemerge in S10, making a presence of 15%. This may have been due to the outbreaks in the United States, which began around this time as well, which could have caused this mutation to arise due to different environmental conditions as an effort of the virus to be effective in a new region.
The other subunit of the ORF1ab region, which is composed of NSP12a, NSP13, NSP14, NSP15a, and NSP16 also contained mutations. Of the four mutations studied, two of the mutations were synonymous mutations in the proteins NSP12a and NSP13. However, NSP13 also contained another mutation at nucleotide 17858 a>g. This mutation changed the codon from ATG, which codes for Methionine to GTG, coding for Valine. This mutation changes this codon from coding for a polar, hydrophilic amino acid to a nonpolar hydrophobic amino acid, which will cause this amino acid to be faced inwards rather than outwards, which could change the way the amino acid is folded, as well as its functionality. The last mutation in NSP13 identified was at nucleotide 18060 c>t. This changed the TCT codon of the reference to TTT, which consequentially changed the amino acid from Serine to Phenylalanine. This missense mutation is similar to the previous mutation discussed, as serine is a polar hydrophilic amino acid and Phenylalanine is a nonpolar hydrophobic amino acid. NSP13, according to Table 1(c), has a proposed functionality of a helicase, which has an enzymatic catalytic activity to unzip DNA for the virus to function. This mutation fluctuated between 0% and 14% through the month of February as indicated by the date for S1-S4 and peaked at 34% in March according to S5 and continued to decrease until S10. Interestingly, this missense mutation at nucleotide 17858 had almost the same trend as the other synonymous mutation in NSP13 at nucleotide 17747, as well as the missense mutation at nucleotide 18060. Because of such a similar trend between three independent mutations, this dismisses the potential for their trend to be random. Rather, a prediction from this data is that this may be due to an effort of the virus to change its code arbitrarily to cloak itself because this trend is near identical. Further research needs to be done over these mutations, as well as any other mutations found in NSP13.
In the second gene, ORF2 (S), which codes for the Spike protein, we studied a mutation at nucleotide position 23406 where we observed a transition mutation from a>g. The Spike protein acts as a ligand binding to cell receptors to initiate the attack of the virus on the cellular level, and a persistent mutation in this region may be indicative of the virus becoming more effective in human to human transmission. Furthermore, this protein is important for the body to develop antibodies as an immune response to this virus, as the Spike protein is the antigen that is used to make the antibody (Ebi Clustal Omega, n.d.; Waterhouse et al., n.d.; Petropoulos and Makridakis, 2020; Yoshimoto, 2020; Emameh et al., 2020). With this mutation specifically at nucleotide 23,406, it causes the codon to change from GGT to GAT, which changes the amino acid from Glycine to Aspartate. This missense mutation replaces the highly flexible, nonpolar amino acid Glycine with an acidic amino acid, which may change the folding of the Spike protein, as well as the function, as mentioned with the other missense mutations discussed previously. From S1 to S8 genomes, this transition missense mutation consistently increased in prevalence up to 46%. S9 and S10 saw dramatic decreases from this figure, but the mutation remained consistently prevalent in the genome, nonetheless. This mutation must have added functionality or had a favorable outcome for it to still be consistently prevalent in the genome. This particular mutation is interesting, as it occurs in the gene of an important protein, but also because the frequency of the mutation in the genomes studied never approached zero as the others did. Further research needs to be done on this mutation to make a conclusive determination of the function of this mutation.
The third gene, ORF3a, also contained a mutation that we studied extensively. This gene codes for its self-named protein, ORF3a, of 275 amino acids, Table 1. The particular mutation studied occurs at nucleotide 25566 where the transversion mutation of g>t occurs. This mutation changes the reference codon of AGA to ATA, which consequentially changes the amino acid encoded from Arginine to Isoleucine. This missense mutation changes the previously basic, highly polar, hydrophilic amino acid to a largely nonpolar and hydrophobic amino acid, which may lead to misfolded proteins with different functions. While the effect in this missense mutation is relatively similar to the others, this mutation is unique in that it consistently increases, and never truly peaks but rather fluctuates between 34% and 48% prevalence in the genomes, as indicated by the data. S1 through S3 does not exhibit this mutation at all in these genomes but quickly picks up over time in the data set of S5 through S7. From this, we can identify this as a relatively late arriving mutation, as it does not appear until March, and increases through the first week of May, and is still relatively highly prevalent through June up till the first week of July in the data sets of S8 to S10. However, as opposed to some of the other mutations that were early arriving, this mutation never experiences a drastic drop over time. This mutation needs to be monitored in future genomes, as it may experience a drop in frequency like the other mutations. The fact that this missense mutation is relatively high indicates that the functionality is not affected by the swap of the amino acids and can be used interchangeably. This mutation may need to be studied molecularly in future research to identify the effects of this mutation on protein folding and function.
In the ORF8 gene, we studied mutations in this genome that yielded changes in the 121-amino-acid ORF8 protein. The mutation studied occurs at nucleotide 28144 where the transition mutation of t>c occurs. This mutation changes the codon from TTC to TTT, which is a synonymous mutation for the amino acid Phenylalanine. While this mutation may not affect protein structure or function, it may be a mutation prevalent because it helps cloak itself in the human body or may affect human to human transmission. As opposed to the trends for the other mutations, this mutation is more frequent in the earlier records of the genomes for the virus and continues to decrease. From S1 to S5, the genomes fluctuate between 29% and 46% in the frequency of this mutation. From the S6 genome set onwards, the frequency of this mutation sees a drastic drop, hovering between 4.7% and 17% in the data sets of S6 to S10. This is the reverse trend from the previous mutation studied in ORF3a, which had a dramatic increase in the frequency of its mutation and maintained that high frequency. Though this is a synonymous mutation, the virus may have been more effective in transmission without this mutation, or human cells were able to target this mutation more easily than the viruses that did not have this mutation. However, further research needs to be done to find conclusive evidence of this.
Of our findings, the most interesting results came from the mutations located in the coding region for the NSP13 protein including 17747 c>t, 17858 a>g, and 18060 c>t. Specifically, the frequency of these mutations over time were similar to one another. That is, these three mutations had similar frequencies of occurrence in each of the genome pools that we studied. This is interesting, as these mutations in the genome must be happening simultaneously and have similar changes on the NSP13 protein that served to be useful at one point (S5 genome had a frequency of 34–37% for the three mutations). According to (Dorp et al., 2020), recurrent non-synonymous mutations suggest the possible ongoing adaptation of the SARS-CoV-2 genome. As we found these mutations to be prevalent in genomes over time, with the trend being similar between the three mutations, this may be indicative of convergent evolution, as suggested by (Dorp et al., 2020). From an evolutionary standpoint, these three mutations (2 of which are non-synonymous) start with low frequency in the S1 genomes collected from January and remain low until they all reach 14% frequency in S4, the genomes collected between February and March of 2020. This mutation peaks in frequency in March, as indicated by the 34 – 37% frequency of the three mutations observed in the NSP13 region for the S5 genomes. Because of this increase in prevalence, we can infer that this mutation offered the virus adaptive value in human to human transmission, which may have increased its virulence. At the end of April (S6) we see a drastic drop of the frequency of this mutation, heavily favoring the wild-type nucleotide as indicated by the reference genome, as the frequency of the mutation was found at 1 – 3% in the S6 genomes. In S7 for the first week of May, the mutation frequency rose to 14–15% for the three mutations, then consistently decreased to 6%, 4.6%, and 1.6% for each of the three mutations for S8, S9, and S10, respectively. This steady decrease may represent a lack of need for this mutation, as it is not as prevalent in the last three genome sets. The drastic drop to 1–3% for the three NSP13 mutations in S6 may not be representative of the entire month of April, as these 100 genomes were collected in the last two days of April only. Further research may be needed to fill in the gaps in time for our data.
The evolution of these NPS13 mutations is particularly interesting because it affects the NSP13 protein coded by the ORF1ab. In fact, according to Mirza and Froeyen (2020), the gene coding for NSP13 is highly conserved and should be used to produce inhibitors and treatments for this virus, as this region does not have as much variation as other genes of this virus. Because we found three mutations that are found nearby in this region, two of which are non-synonymous, these results are interesting because this region is relatively conserved. Furthermore, the NSP13 region is conserved within the SARS virus, as a high degree of similarity was found between the two helicase proteins, as indicated in Mirza and Froeyen (2020), which results in a “strikingly conserved overall architecture” of NSP13 with its SARS counterpart. They found that this region has a higher similarity than the other two conserved regions, NSP12 and Mpro, with NSP13 having 99.83% similarity and the other two having 96.35% and 96.08% similarity to SARS. Thus, mutations found in the NSP13 region are very strange because this region is highly conserved, and three mutations were found in nearby areas according to our results. To make matters more interesting, Yuen et al. (2020) also discovers another function of NSP13 as an interferon antagonist, as it is used to “suppress primary interferon production and signaling.” This means that the cell cannot indicate to other cells that it contains a foreign invader to activate an immune response, which increases virulence and cell to cell transmission of this virus, as well as human to human transmission potentially.
The novel coronavirus has been notoriously contagious causing a global pandemic spreading to every corner of the world (The Center for Systems Science and Engineering (CSSE) at JHU and the Worldmeter, n.d.; Center for Disease Control and Prevention (CDC), n.d.; Biological Sciences, National Institutes of Health (US), 2007; Saha et al., 2020; Andersen et al., 2020; Li et al., 2020; Li et al., 2020; Mirza and Froeyen, 2020; Tai et al., 2020; Yuen et al., 2020). Thus, studies for mutations have happened regionally to see how these viruses have adapted to new environments, diverse climates, and new populations. However, these results need to be contextualized with time, as the virus can mutate and adapt to new conditions in a region that arise with time. Our results indicate that common mutations found to fluctuate over time, and analyzing these trends are important to frame the results of other research that study the variation in mutations between different regions. As we only have reliable genomic data for 6–7 months of this novel virus, more research needs to be conducted as to how these mutations persist over time in the future as well. Furthermore, these results need to be studied while factoring in different regions and should be compared between these regions. Our findings provide a platform for further studies to analyze such mutations that we have found, as well as other mutations from aggregate genomic data collected and compared with genomes collected from different periods, as opposed to just comparing two data sets of genomes from different areas.
The second most common mutation found by Koyama et al. is the 14408C>T (Koyama et al., 2020) which agrees with our findings as shown in Table 4, Table 5, Table 6 (as in Table 5 this mutation occurs 46% in S 8).
Finally, from studying the variations in the August (S11) and September (S12) genome sequences (200 sequences) we found that the majority of the mutations are c>t and g>t. These two types comprise more than 63% (Table 8, Table 9) of the most frequently occurring mutations in these genomes. The three most frequent mutations in the September S12 set (28881g>a, 28882g>a, and 28883g>c) are found to be major distinguishing points between two genome clusters with genome haplotype analysis in a recent study by Toshiki Takenouchi in Japan (Takenouchi et al., 2020). These three mutations (Takenouchi et al., 2020; Yin, 2020) belong to the nucleocapsid protein (N protein) which prepares the formation of the helical nucleocapsid during virion assembly (Yin, 2020; Zhao et al., 2005). More importantly, the N protein can trigger immune responses which may lead to progress towards developing a vaccine using this N protein and so these mutations will be considered and further studied for the vaccine development purpose (Takenouchi et al., 2020; Yin, 2020; Zhao et al., 2005).
5. Conclusions
We presented a study of analysis and comparison of genetic variants and mutations of SARS-CoV-2 genome. The study and analysis used almost 1000 (complete) genomes of sampled across the first seven months of 2020. We showed experimentally that certain mutations in the SARS-CoV-2 genome are not occurring randomly as it has been commonly believed. This finding is in agreement with other recently published research in this domain. Therefore, these results and outcomes validate other findings in this direction. Some mutations' ratios (frequency percentage) fluctuate over time to adapt the virus to various environmental factors, climate, and populations. One of the interesting findings in this paper is that the coding region, at the nucleotide level for NSP13 protein is relatively conserved compared with other protein regions in the ORF1ab gene which makes this protein a good candidate for developing drug targets and treatment for the COVID-19 disease. This result was already inferred and reported by other researchers and here we corroborated their result with our work with a different approach and another experimental setting with over almost one thousand (complete) genomes. This study includes close to 1000 genomes and was able to identify over 35 different mutations most of which are common to almost all genomes groups. We presented and discussed all these results and findings with tables of results and illustrating figures.
CRediT authorship contribution statement
Zaid Almubaid: Writing, Reviewing and editing, Data curation, Conceptualization
Hisham Al-Mubaid: Writing, Investigation, Methodology, Software.
Declaration of competing interest
No conflict of interest.
Abbreviations
- CoV
corona virus
- COVID-19
coronavirus disease 2019
References
- Anastassopoulou C., Russo L., Tsakris A., Siettos C. Data-based analysis, modelling and forecasting of the COVID-19 outbreak. PLoS one. 2020;15(3):1–10. doi: 10.1371/journal.pone.0230405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen Kristian G., Rambaut Andrew, Lipkin W. Ian, Holmes Edward C., Garry Robert F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biological Sciences, National Institutes of Health (US) National Institutes of Health; Bethesda (MD): 2007. Curriculum Study. [Google Scholar]
- Center for Disease Control and Prevention (CDC) H5N1 genetic changes. https://www.cdc.gov/flu/avianflu/h5n1/inventory-qa.htm
- Dorp Lucyvan, Acmana Mislav, Richard Damien, Shaw Liam P., Ford Charlotte E. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infection, Genetics and Evolution. 2020;83 doi: 10.1016/j.meegid.2020.104351. (Sept. 2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Idrissi H.H. COVID-19: what you need to know. Gene Reports. 2020;20 doi: 10.1016/j.genrep.2020.100756. (September) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emameh R.Z., Nosrati R.H., Taheri R.A. Combination of biodata mining and computational modelling in identification and characterization of ORF1ab polyprotein of SARS-CoV-2 isolated from oronasopharynx of an Iranian patient. Biological procedures online. 2020;22(8) doi: 10.1186/s12575-020-00121-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebi Clustal Omega. https://www.ebi.ac.uk/Tools/msa/clustalo/
- The Center for Systems Science and Engineering (CSSE) at JHU and the Worldmeter. https://www.worldometers.info/coronavirus/worldwide-graphs/#case-timeline
- Junejo Y., Ozaslan M., Safdar M., Khailany R.A., Rehman S., Yousaf W., Khan M.A. Novel SARS-CoV-2/COVID-19: origin, pathogenesis, genes and genetic variations, immune responses and phylogenetic analysis. Gene Rep. 2020;20 doi: 10.1016/j.genrep.2020.100752. (September) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khailany R.A., Safdar M., Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020;19(100682):2020. doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Platt D., Parida L. Variant analysis of SARS-CoV-2 genomes. Bull. World Health Organ. 2020;98(7):495–504. doi: 10.2471/BLT.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zai J., Zhao Q., Nie Q., Li Y., Foley B.T., Chaillon A. Evolutionary history, potential intermediate animal host, and cross-species analyses of SARS-CoV-2. J. Med. Virol. 2020 doi: 10.1002/jmv.25731. (Advance online publication) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza Muhammad Usman, Froeyen Matheus. Structural elucidation of SARS-CoV-2 vital proteins: computational methods reveal potential drug candidates against main protease, Nsp12 polymerase and Nsp13 helicase. J. Pharm. Anal. 2020 doi: 10.1016/j.jpha.2020.04.008. (April 2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näsvall S.J., Chen P., Björk G.R. The wobble hypothesis revisited: uridine-5-oxyacetic acid is critical for reading of G-ending codons. RNA J. 2007;13(12):2151–2164. doi: 10.1261/rna.731007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI SARS-CoV-2 resources. https://www.ncbi.nlm.nih.gov/sars-cov-2/
- Petropoulos F., Makridakis S. Forecasting the novel coronavirus COVID-19. PLoS One. 2020;15(3) doi: 10.1371/journal.pone.0231236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prathiviraj R., Kiran G.S., Selvin Joseph. Phylogenomic proximity and comparative proteomic analysis of SARS-CoV-2. Gene Reports. 2020;20 doi: 10.1016/j.genrep.2020.100777. (September) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P., Longden I., Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16(6):276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020;109:102433–102442. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha P., Banerjee A.K., Tripathi P.P., Srivastava A.K., Ray U. A virus that has gone viral: amino acid mutation in S protein of Indian isolate of Coronavirus COVID-19 might impact receptor binding, and thus, infectivity. Biosci Rep. 2020;40(5) doi: 10.1042/BSR20201312. (29 May) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan R., Domingo-Calap P. Mechanisms of viral mutation. Cell Mol Life Sci. 2016;73(23):4433–4448. doi: 10.1007/s00018-016-2299-6. (December) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah A., Rashid F., Aziz A., Jan A., Suleman M. Genetic characterization of structural and open reading Fram-8 proteins of SARS-CoV-2 isolates from different countries. Gene Rep. 2020;21 doi: 10.1016/j.genrep.2020.100886. (December) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai Wanbo, He Lei, Zhang Xiujuan, Jing Pu, Voronin Denis, Jiang Shibo, Zhou Yusen, Lanying Du. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020;17:613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenouchi T., Iwasaki Y.W., Harada S. Clinical utility of SARS-CoV-2 whole genome sequencing in deciphering source of infection. J. Hospital Infections. 2020 doi: 10.1016/j.jhin.2020.10.014. (Oct. 24) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse, A.M., Procter, J.B., Martin, D.M.A, Clamp, M. and Barton, G. J. “Jalview Version 2 - a multiple sequence alignment editor and analysis workbench”. Bioinformatics 25 (9) 1189–1191 doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed]
- Wilder-Smith A., Chiew C.J., Lee V.J. Can we contain the COVID-19 outbreak with the same measures as for SARS? Lancet Infect. Dis. 2020;20:e102–e107. doi: 10.1016/S1473-3099(20)30129-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C. Genotyping coronavirus SARS-CoV-2: methods and implications. Genomics. 2020;112(5):3588–3596. doi: 10.1016/j.ygeno.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto Francis K. The proteins of severe acute respiratory syndrome coronavirus-2 (SARS CoV-2 or n-COV19), the cause of COVID-19. Protein J. 2020:1–19. doi: 10.1007/s10930-020-09901-4. (May 23) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen Chun-Kit, Lam Joy-Yan, Wong Wan-Man, Mak Long-Fung, Wang Xiaohui. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg. Microbes Infect. 2020;9(1):1418–1428. doi: 10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Cao J., Zhao L.-J., Qin Z.-L., Ke J.-S., Pan W., Ren H., Yu J.-G., Qi Z.-T. Immune responses against SARS-coronavirus nucleocapsid protein induced by DNA vaccine. Virology. 2005;331(1):128–135. doi: 10.1016/j.virol.2004.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]