Abstract
The differentiation shift from osteogenesis to adipogenesis of bone marrow mesenchymal stem cells (BMSCs) characterizes many pathological bone loss conditions. Stromal cell-derived factor-1 (SDF1) is highly enriched in the bone marrow for C-X-C motif chemokine receptor 4 (CXCR4)-positive hematopoietic stem cell (HSC) homing and tumor bone metastasis. In this study, we displayed CXCR4 on the surface of exosomes derived from genetically engineered NIH-3T3 cells. CXCR4+ exosomes selectively accumulated in the bone marrow. Then, we fused CXCR4+ exosomes with liposomes carrying antagomir-188 to produce hybrid nanoparticles (NPs). The hybrid NPs specifically gathered in the bone marrow and released antagomir-188, which promoted osteogenesis and inhibited adipogenesis of BMSCs and thereby reversed age-related trabecular bone loss and decreased cortical bone porosity in mice. Taken together, this study presents a novel way to obtain bone-targeted exosomes via surface display of CXCR4 and a promising anabolic therapeutic approach for age-related bone loss.
Keywords: Exosomes, Bone targeting, miR-188, CXCR4, Osteoporosis
Graphical abstract
Highlights
-
•
Surface display of CXCR4 grants exosomes bone targeting properties.
-
•
Exosome-liposome hybrid nanoparticles carrying nucleic acid target bone.
-
•
Antagomir-188 loaded hybrid nanoparticles regulate MSC differentiation in aged mice.
1. Introduction
Bone marrow mesenchymal stem cells (BMSCs), the progenitors of bone marrow adipocytes and osteoblasts, constitute niches of hematopoiesis in the bone marrow [1]. Under various pathological conditions, BMSCs shift from osteogenesis to adipogenesis with impaired bone formation and increased adipocytes accumulation in the bone marrow [[2], [3], [4]]. To inhibit adipogenic differentiation and promote osteogenic differentiation and bone formation from BMSCs have been explored as a promising therapeutic option for bone loss diseases [[5], [6], [7], [8], [9]].
Multiple microRNAs (miRNAs/miRs) have been identified to regulate the differentiation fate of BMSCs [10,11]. MiR-188, which is remarkably increased with aging, significantly promotes adipogenesis and inhibits osteogenesis of BMSCs. Knockdown of miR-188 reversed the age-related BMSC adipogenic differentiation shift and ameliorated bone loss in aged mice [12].
RNA interference (RNAi) that regulates gene expression in a highly precise manner, could be theoretically used to target any pathogenic gene of interest [[13], [14], [15]]. To target mir-188 could serve as a treatment to promote bone formation. However, systemic administration of large doses of siRNA that need to stimulate bone formation could have adverse effects in non-skeletal tissues [16]. Several bone targeting delivery systems have been reported. Dioleoyl trimethylammonium propane (DOTAP)-based cationic liposomes attached to six repetitive sequences of aspartate, serine, serine ((AspSerSer)6) could deliver siRNAs specifically to bone-formation surfaces [17]. CH6 aptamer-functionalized lipid nanoparticles (LNPs) encapsulating osteogenic pleckstrin homology domain-containing family O member 1 (Plekho1) siRNA (CH6-LNPs-siRNA) nuclear acid aptamers could facilitate osteoblast selective uptake of Plekho1 siRNA [18]. Despite the effects, the toxicity of nanomaterials remains a challenge. For nanoparticles could result in both physical and chemical damages to cells [19]. Therefore, biologically friendly bone targeting delivery systems with high targeting and delivery efficiency and a wide source and easy acquisition are worth further exploring.
Exosomes (EXOs) are lipid bilayer extracellular vesicles with a diameter range of 40–160 nm [20]. As naturally derived nanocarriers, exosomes show intriguing potential in drug delivery, with low biotoxicity and high barrier penetrating capacity [21,22]. To improve their cell targeting properties, the surface display of signal regulatory molecules has been reported [23]. For example, CD47-expressing exosomes from genetically engineered fibroblasts could achieve targeted accumulation in tumors by binding to the signal regulatory protein alpha receptor (SIRPα) [24,25]. To overcome the encapsulating problems, an exosome-liposome hybrid delivery system was introduced by Lin et al. [26], which was capable of delivering large plasmids in vitro. The hybrid NPs could edit genes by the encapsulated CRISP/Cas 9 systems, indicating the excellent biocompatibility and medicinal potential of exosome-liposome hybrid NPs.
Stromal cell-derived factor 1 (SDF1), also known as C-X-C motif chemokine 12, acts as the ligand for C-X-C motif chemokine receptor 4 (CXCR4) and is predominantly expressed by BMSCs [27]. SDF1 constitutes niches of hematopoietic stem cells (HSCs) and regulates hematopoiesis [28]. The high levels of SDF1 in bone marrow recruits CXCR4+ peripheral HSC homing and promotes bone metastasis of several CXCR4+ tumor cells [29]. For example, breast cancers that highly express CXCR4 tend to seed in SDF1-rich environments like the bone marrow. Thus, blocking CXCR4 could either increase peripheral HSCs or block tumor bone metastasis [30,31]. Considering the critical role of the CXCR4-SDF1 axis in chemotaxis behavior, we assume that CXCR4-positive exosomes could be recruited to the bone marrow.
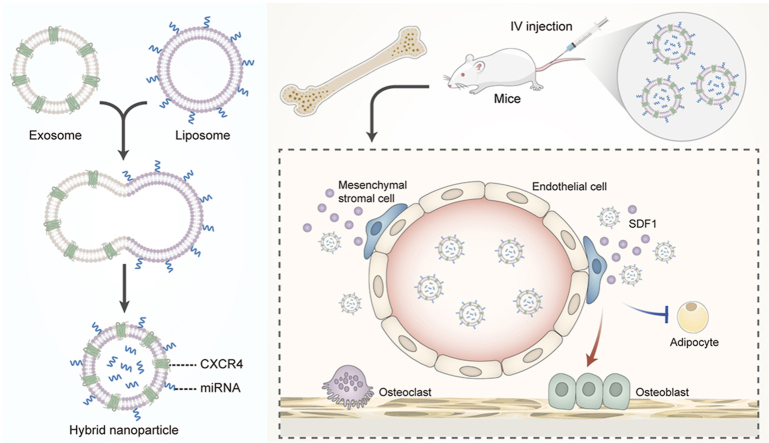
Based on the aforementioned evidence and our previous work on bone-targeting exosomes [32], we constructed genetically engineered NIH-3T3 cells that highly expressed CXCR4. We then collected the exosomes and demonstrated that the CXCR4 receptor was displayed on their surface, and fused them with liposomes carrying antagomir-188 to obtain hybrid NPs with both bone-targeting and anti-miR-188 capacities. We showed a specific bone gathering of the hybrid NPs, which significantly reversed age-related trabecular bone loss and decreased cortical bone porosity by inhibiting adipogenesis and promoting osteogenesis of BMSCs in aged mice. This study provides a novel strategy for constructing bone-targeted NPs for RNAi delivery as an anabolic therapy for aged bone loss (Fig. 1A).
Fig. 1.
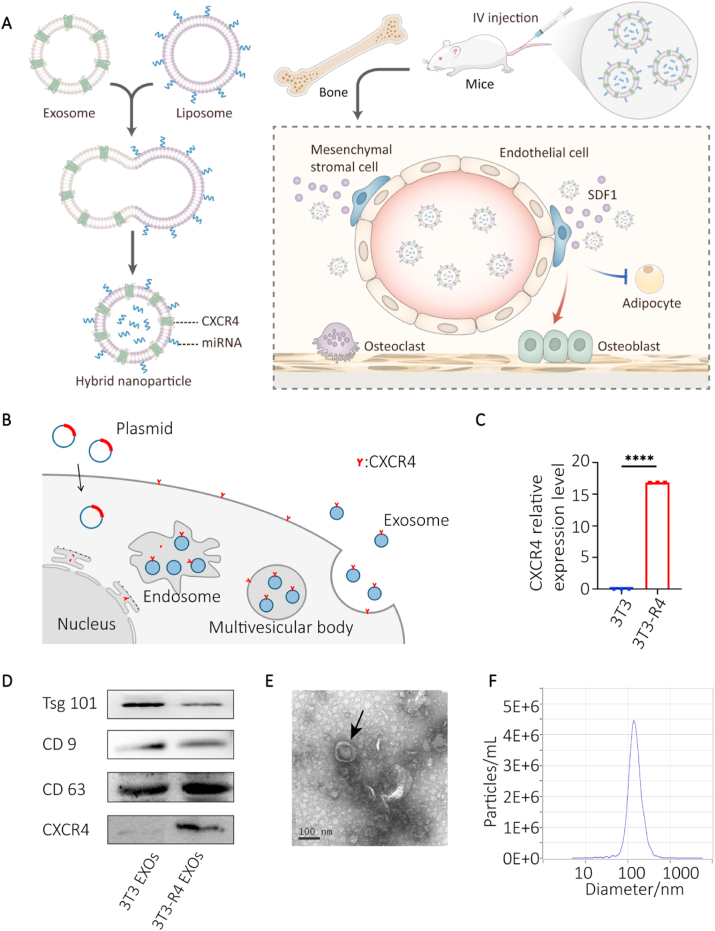
Characterization of CXCR4+ exosomes. A) Schematic illustration of exosome-guided miRNA blocking. B) Schematic illustration of CXCR4+ exosomes construction. C) CXCR4 expression levels in NIH-3T3 cells (3T3) and CXCR4-expressing NIH-3T3 cells (3T3-R4) determined by RT-qPCR (p < 0.0001). D) Western blotting results of exosome markers (Tsg101, CD9, and CD63) and CXCR4 in 3T3-R4 exosomes. E) TEM results showing the classical bilayer structure (black arrow) of CXCR4+ exosomes (scale bar = 100 nm). F) NTA results demonstrating the size distribution of CXCR4+ exosomes.
2. Materials and methods
2.1. Cell culture
NIH-3T3 cells were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China), and cultured in high glucose Dulbecco's Modified Eagle's Medium supplemented with 10% (v/v) fetal bovine serum (FBS) and 100 μg mL−1 penicillin-streptomycin (Gibco, Waltham, MA, USA) at 37 °C in a 5% CO2 humidified incubator. The cells were mycoplasma-free and passaged at 80–90% confluence.
2.2. Real-time quantitative polymerase chain reaction (RT-qPCR)
For CXCR4 transfection, total RNA from the cultured cells was extracted using a TRIzol regent kit (Invitrogen, Carlsbad, CA, USA), and normalized to an internal RNA reference control. Quantification was performed using a one-step RT-qPCR kit with a Roche PCR system (Basel, Switzerland). The primers used for this analysis were as follows: CXCR4, forward: 5′-GACTGGCATAGTCGGCAATGGA-3′ and reverse: 5′-CAAAGAGGAGGTCAGCCACTGA-3′; GAPDH, forward: 5′-CATCACTGCCACCCAGAAGACTG-3′ and reverse: 5′-ATGCCAGTGAGCTTCCCGTTCAG-3′; Sp7, forward: 5′-ACCAGGTCCAGGCAACAC-3′ and reverse: 5′-GCAAAGTCAGATGGGTAAGTAG-3′; Bglap, forward: 5′-TGAGGACCATCTTTCTGCTCACT-3′ and reverse: 5′-GGCATCTGTGAGGTCAGAGAGA-3′; Pparγ, forward: 5′-GGGATCAGCTCCGTGGATCT-3′ and reverse: 5′-TGCACTTTGGTACTCTTGAAGTT-3′; Fabp4, forward: 5′-AAATCACCGCAGACGACA-3′ and reverse: 5′-CACATTCCACCACCAGCT-3′; Mice were treated with hybrid NPs, antagomir-negative control (NC), and hybrid NPs + antagomir-188 for two weeks (n = 12). BMSCs were sorted by flow cytometry. The antibodies used were Sca-1 (108108; BioLegend, San Diego, CA, USA), CD29 (102206; BioLegend), CD45 (103132; BioLegend), and CD11b (101226; BioLegend). The primers used for this experiment were as follows: mir-188, forward: 5′-GCCGCCATCCCTTGCATG-3′ and reverse: 5′-CCAGTGCAGGGTCCGAGGTA-3′; U6, forward: 5′-CTCGCTTCGGCAGCACA-3′ and reverse: 5′-AACGCTTCACGAATTTGCGT-3′.
2.3. Exosome isolation
Exosome-depleted FBS was prepared from original FBS by ultracentrifugation at 110000×g for 70 min. When the cells reached nearly 75% confluence, they were treated with exosome-free medium for 24–48 h before the supernatant harvest. Non-adherent cells and debris in the culture medium were removed by centrifugation at 2000 rpm for 5 min, and organelle and diminutive debris were purged by high-speed centrifugation at 10,000×g for 60 min. The final supernatant was ultracentrifuged at 110,000×g for 70 min, resuspended in PBS buffer and the ultracentrifugation process was repeated once again. Exosomes were stored at −80 °C no more than 30 days before use or further detection.
2.4. Biophotonic imaging analysis
Six-week-old Balb/c male mice were purchased from Slack Laboratory Animal Corp. (Shanghai, China), and all animal experiments were approved by the Ethics Committee of Changhai Hospital. For the in vivo tracking process, exosomes with or without cyanine 5 (Cy5) labeling were intravenously injected through the tail vein of mice. For the tracking process of hybrid NPs, a Cy5-labeled miRNA analogue was used instead of the dye itself. We set a concentration gradient to determine the optimal ratio of exosomes and liposomes; 1:4 stands for 200 μg protein equivalent exosomes versus 4 mg of liposome in 1 mL PBS, 1:1 implies 200 μg protein equivalent exosomes versus 1 mg of liposome in 1 mL PBS, and 4:1 represents 800 μg protein equivalent exosomes versus 1 mg of liposome in 1 mL PBS. Animals were sacrificed 4 h after injection, and relevant organs were harvested. Finally, fluorescence imaging of the distribution was performed on a Quickview 3000 system (Bio-Real Sciences, Salzburg, Austria).
2.5. Liposome and hybrid NPs synthesis
1,2-dioleoyl-3-trimethylammonium-propane (890890; Avanti Polar Lipids, Alabaster, AL, USA) and cholesterol were dissolved in chloroform at a molar ratio of 1:1, and the vacuum rotatory evaporator was used for the concentration. The lipid layer was washed with RNase-free water and scattered by sonication for the extrusion assay. Exosomes (200 μg protein equivalent) and liposomes (100 μL of 10 mM concentration) were added into RNase-free water, filled to 1 mL and extruded 20 cycles through a 100 μm polycarbonate membrane (610005; Avanti Polar Lipids) in a mini extruder (610000; Avanti Polar Lipids) at 45 °C, according to the manufacturer's instructions.
2.6. Fluorescence resonance energy transfer (FRET) assay
Membrane fusion of exosomes and liposomes was demonstrated using the FRET process. Briefly, 1.5% of nitrobenzoxadiazole (NBD)-phosphatidylserine (PS; 810195l; Avanti Polar Lipids) and rhodamine (Rho)-phosphatidylethanolamine (PE; 810150; Avanti Polar Lipids) were added during liposome synthesis, then the liposomes and exosomes were extruded as described previously. When excited at a wavelength of 460 nm, the emission energy was transferred to adjacent Rho-PE through the FRET process, which was subsequently detected at a wavelength of 580 nm. On the other hand, excited NBD-PS without FRET emitted fluorescence at 534 nm. We measured the fluorescence value of 534 nm and 580 nm simultaneously at an excitation wavelength of 460 nm. The proportion of NBD fluorescence increased while the relative distance between these two dyestuffs was separated by the newly added exosome membrane ingredient.
2.7. Age-related osteoporosis mouse model
All animal studies were performed in a specific pathogen-free laboratory at Changhai Hospital (Shanghai, China). All procedures were carried out under the supervision of the Ethics Committee of Changhai Hospital. Eighteen-month-old male mice were purchased from Sino-British SIPPR/BK Lab Animals Ltd. (Shanghai, China) and randomly divided into three groups: hybrid NPs, antagomir, and hybrid NPs + antagomir. For the antagomir group, 100 μL PBS containing 2 μL antagomir (20 μM) was injected through the tail vein once a week for 8 weeks. For the hybrid NPs group, 100 μL of hybrid NPs without loaded cargoes was injected using the aforementioned procedure. For the hybrid NPs + antagomir group, 2 μL antagomir (20 μM) was encapsulated into the liposome before extrusion, and a total of 100 μL liquor was injected as describe previously.
2.8. Micro-CT analysis
Femora were fixed in 70% ethanol overnight and then 4% paraformaldehyde at 4 °C in EP tubes each. The scan was performed under conditions of 8 μm per pixel, a voltage of 80 kV, and a current of 124 μA. We used the corresponding software DataViewer, CTAn and CTVox for data analysis. At the distal femur, nearly 100 consecutive sections under the growth plate were reconstructed and observed. We uses bone volume fraction (BV/TV), trabecular bone pattern factor (Tb.Pf), trabecular number (Tb.N), trabecular separation (Tb.Sp) and trabecular thickness (Tb.Th) for trabecular description as well as cortical thickness (Ct.Th) and cortical porosity (Po) for cortical description. Micro-CT analysis was performed a Skyscan X-ray microtomography system (Skyscan 1275; Bruker, Germany).
2.9. ALP staining
BMSCs were cultured in 12-well plates at a density of 5 × 104 cells/well with osteogenic induction medium (MUBMX-90021; Cyagen, Sata Clara, CA, USA) for 2 weeks, and the culture medium was changed every 3 days. Alkaline phosphatase (ALP) staining was performed using an ALP color development kit (C3206; Beyotime Biotechnology, Shanghai, China) according to the manufacturer's instructions.
2.10. Oil Red O staining
BMSCs were treated with adipogenic induction medium (MUBMX-90031; Cyagen) for 2 weeks following the manufacturer's instructions. Oil Red O staining was performed to detect mature adipocytes (G1262; Solarbio, Beijing, China). Adipogenic markers were detected after 2 weeks of induction.
2.11. Western blotting
Cultured cells were lysed by 100 μL of RIPA buffer on ice for 20 min. Western blotting was conducted according to the standard protocols. Antibodies used for RUNX2 (ab92336; Abcam), FABP4 (ab92501; Abcam) and PPARγ (ab272718; Abcam) were incubated with samples overnight, according to manufacturer's instruction, secondary antibody (ab6721; Abcam) was applied before protein band visualizing.
2.12. Immunohistochemical staining
Immunohistochemical staining was performed as previously reported [12]. Briefly, femur sections were prepared after decalcification for 30 days. Antigen retrieval was carried out using sodium citrate antigen retrieval solution (C1032; Solarbio). Sections were incubated with the primary fatty acid-binding protein 4 (FABP4) antibody (ab92501; Abcam, Cambridge, UK) at 4 °C overnight, and the corresponding secondary antibody (ab6721; Abcam) for 90 min at room temperature. A horseradish peroxidase-3, 3′-Diaminobenzidine detection system (MK210; Takara Bio, Dalian, China) was used for visualization, followed by hematoxylin counterstaining (C1080; Solarbio).
2.13. Statistical analysis
All data are presented as means ± standard deviations, or direct values. Analysis was conducted using a one-way analysis of variance or the unpaired Student's t-test with or without Welch's correction, as appropriate. All statistical analysis were performed using GraphPad Prism 8 software (GraphPad Software, San Diego, CA, USA), and p-values < 0.05 were considered statistically significant.
3. Results and discussion
3.1. Characterization of CXCR4+ exosomes
Despite the advantages of exosomes, their application is limited by the technical challenges of producing sufficient quantities for in vivo administration [33]. In this study, NIH-3T3 cells were selected because they are highly reproductive and easily utilized for gene editing.
Surface modification of exosomes with the introduction of specific receptors or ligands is a potential strategy for adjusting the in vivo distribution properties of exosomes. The SDF1/CXCR4 axis plays a critical role in HSC trafficking and homing to the bone marrow [34,35], as well as hematopoietic niche maintenance [36]. Knockout of SDF1 or CXCR4 leads to impaired trafficking of HSCs from the periphery to the bone marrow [37]. Upregulated CXCR4 expression in HSCs enhances HSC homing [38]. As the “seed and soil” hypothesis suggests, many CXCR4+ tumor cells, such as breast cancer cells, use the axis to metastasize to the bone [39], thus, neutralizing the CXCR4-SDF1 axis prevents bone metastasis [40]. As a result, we hypothesized that the surface display of CXCR4 could grant exosomes bone targeting properties.
To test this hypothesis, the CXCR4 gene (species: mouse, gene ID: 12767) was introduced into NIH-3T3 cell lines via the CMV-MCS-PGK-Puro lentiviral packaging system (Fig. 1B). The successful overexpression of CXCR4 on NIH-3T3 cells was validated by RT-qPCR (Fig. 1C) and immunofluorescence (Figure S1). Then, exosomes were isolated from the supernatant of cultured CXCR4+ NIH-3T3 cells. CXCR4 protein was also detected on CXCR4+ NIH-3T3 exosomes (CXCR4+ EXOs), compared to the negative control group using western blotting (Fig. 1D, bottom). Purified exosomes were characterized by morphological structure, size contribution and marker protein expression. A typical bilayer structure of CXCR4+ EXOs was observed by transmission electron microscopy (TEM) (Fig. 1E). Nanoparticle tracking analysis (NTA) showed a peak particle size distribution value at 142.6 nm (Figs. 1F), and 99.2% of particles were at that size, which confirmed the efficiency of the exosome isolation. The exosome protein markers Tsg101, CD9 and CD63 were detected by western blotting, which further validated the successful isolation of exosomes (Fig. 1D). Above all, we successfully constructed high CXCR4-expressing NIH-3T3 cells and demonstrated that the CXCR4 was expressed on the secreted exosomes.
3.2. In vivo distribution of CXCR4+ exosomes
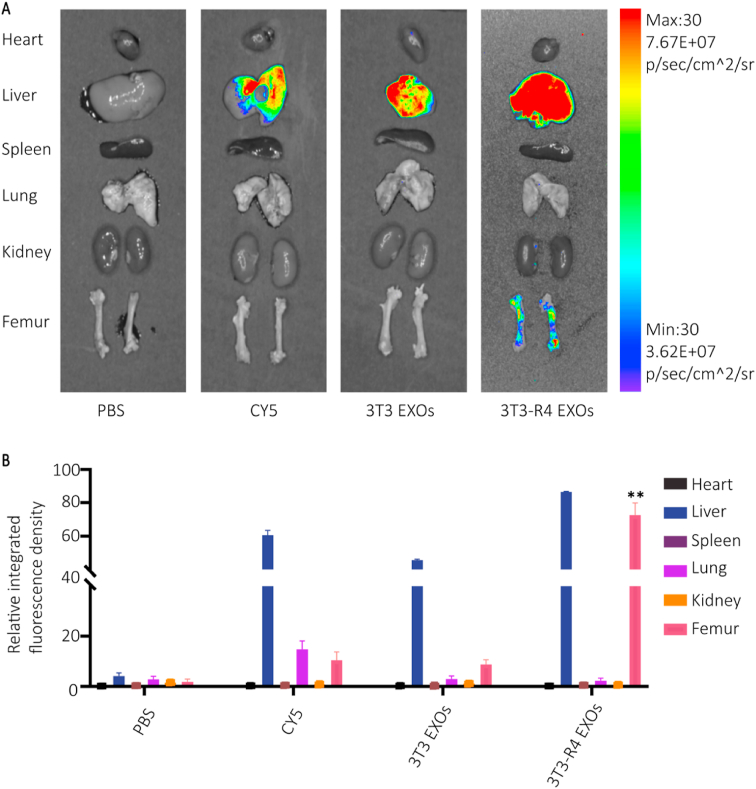
To test our hypothesis that CXCR4+ EXOs possess bone-targeting properties, we performed an in vivo tracing test. We used Cy5 fluorescent dye to label exosomes from both NIH-3T3 cells and CXCR4+ NIH-3T3 cells. Six-week-old male Balb/c mice were randomly separated into four groups: PBS, Cy5, EXOs and CXCR4+ EXOs. The fluorescence intensity of different organs from each group was measured using the Quickview 3000 system 4 h after tail intravenous injection (Fig. 2). We found a specific dye enrichment in the femora of the CXCR4+ EXOs group compared to the other groups, which indicated that CXCR4+ EXOs specifically targeted the bone marrow. To our knowledge, this is the first study to produce bone-targeted exosomes by surface displaying CXCR4 which may provide guidance for exosomes gathering in the bone marrow.
Fig. 2.
In vivo distribution of CXCR4+exosomes. A) Fluorescence of organs from mice sacrificed 4h after intravenous injection of PBS, equivalent Cy5 dissolved in PBS, Cy5 labeled CXCR4- exosomes and Cy5 labeled CXCR4+ exosomes. B) Fluorescence intensity quantitation in organs (**p < 0.01).
3.3. Characterization of hybrid nanoparticles
Exosomes are regarded as promising nanocarriers for nuclear acid cargoes because of their excellent penetrability, biostability and immunogenicity. However, inserting exogenous anionic nucleic acids into exosomes leads to low yields [41,42]. Several methods have been proposed to increase either the yield of exosomes or the loading efficiency of nuclear acid cargoes. In this study, we tested the loading efficiency of RNA cargoes into exosomes. The efficiency was measured using both incubation and electroporation protocols (9.58% and 33.66%, respectively) (Figure S2). Cationic lipoplexes have been reported to be promising for gene delivery [43]. Exosome-liposome hybrid NPs were designed to combine the advantages of both systems, and were applied for gene delivery in genetic editing systems in previous studies [44,45].
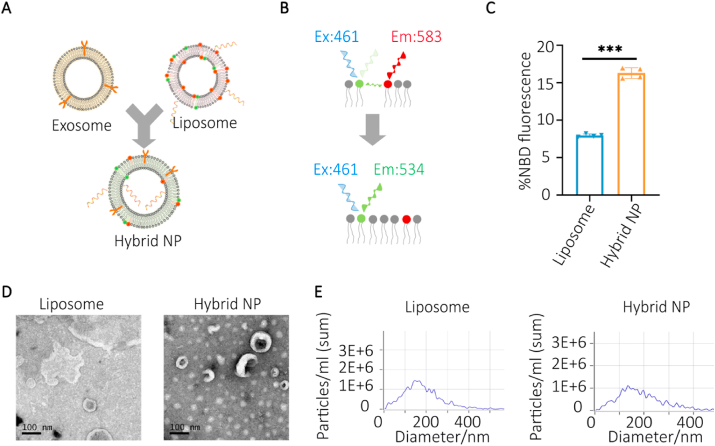
Here, we used the extrusion technique to design an exosome-liposome hybrid NP that is capable of loading RNA cargoes and targeting the bone marrow. The zeta-potential of the exosomes and liposomes were −12.4 to −15 mV and 39.7–40.6 mV, respectively and the value of the hybrid NPs was −14.2 to −18.8 mV after extrusion. The encapsulation efficiency reached to 99.54% and 97.63% in the lipoplexes and hybrid NPs group, respectively (Figure S2). To distinguish a membrane fusion from the physical mixture, a novel FRET assay was performed [46]. The results demonstrated that the membrane fusion ratio of NBD fluorescence increased from 7.98% to 16.30% after the extrusion procedure with statistical significance (p < 0.001) (Fig. 3A–C). We further characterized the liposomes and hybrid NPs by TEM (Fig. 3D) and NTA (Fig. 3E). TEM confirmed the typical lipid bilayer structure of the hybrid NPs with the peak diameters of lipoplexes and hybrid NPs being 159.7 nm and 135.7 nm, respectively.
Fig. 3.
Construction and description of hybrid NPs. A) Schematic description of exosome-liposome extrusion. B) Schematic illustration of FRET analysis. C) NBD ratio of the total fluorescence intensity indicates a membrane fusion process (***p < 0.001). n = 3 for each group). Representative TEM and NTA analyses are shown in D) and E) (scale bar = 100 nm).
3.4. Validation of bone targeting properties
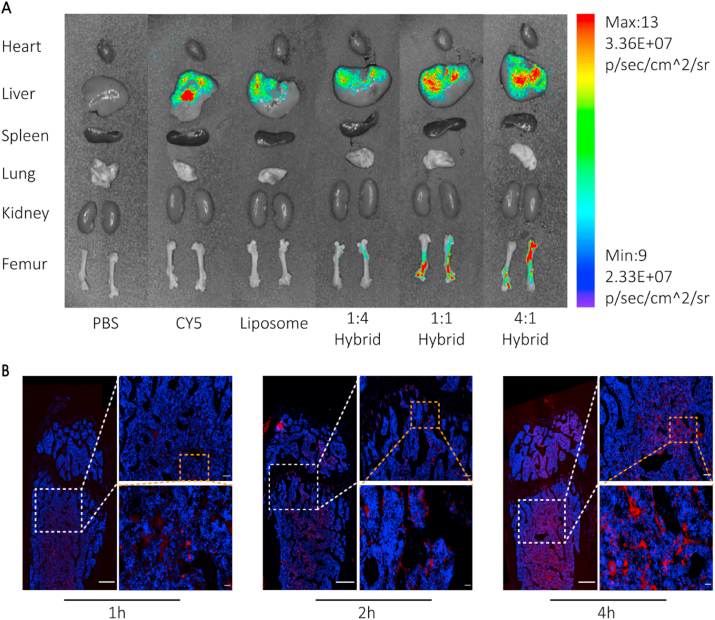
We next sought to explore the ratio of exosomes to liposomes. Mice were injected via the tail vein with PBS (control group, 100 μL), liposomes loaded with Cy5-labeled antagomir-188 (dissolved in PBS, 100 μL) and hybrid NPs loaded with Cy5-labeled antagomir-188 (equivalent microRNA encapsulated in corresponding vehicles, 100 μL). As shown in the tracing study, hybrid NPs with exosome/liposome ratios of 1:1 and 4:1 could gather in the bone marrow (Fig. 4A, Figure S3). The fluorescent results in different organs were consistent with the in vivo tracing results. In the heart, spleen, lung and kidney, fluorescence intensity varied at 1h and 2h, but little residual fluorescence was detected 4h after injection (Figure S4). Fluorescence intensity was maintained at a high level in the liver, but there was no significant difference between groups. In addition, a progressive increase in fluorescence intensity was observed in mouse femora, which indicated that fluorescence-labeled cargoes were captured in the bone marrow (Fig. 4B). The cytotoxicities of exosomes, liposomes, and hybrid NPs were then evaluated 48 h after intravenous administration. No obvious pathological changes were observed in the heart, liver, spleen, lung, and kidney, after 48 h (Figure S5) or 8 weeks (Figure S4) of administration. Liver and renal function remained stable 48 h after administration, which confirmed that the NPs had no short-term or long-term cytotoxicity in vivo. In summary, we combined the advantages of exosomes and liposomes and constructed biocompatible CXCR4+ hybrid NPs with bone-targeting properties as novel RNA nanocarriers.
Fig. 4.
In vivo distribution of CXCR4+hybrid NPs. A) Biophotonic images of the organ distribution 4h after intravenous injection of PBS, Cy5, Cy5 labeled liposomes and Cy5-labeled hybrid NPs, with various exosome-liposome ratios. B) Representative fluorescence microscopic images of the femur 1, 2, and 4 h after injection of Cy5-labeled CXCR4+ hybrid NPs (exosomes-liposomes ratio = 1:1; scale bars on the left, upper right, and bottom right represent 500 μm, 100 μm and 20 μm, respectively).
3.5. Validation of anabolic effects in an age-related bone loss model
MiRNAs are small, non-coding RNA segments that regulate gene expression after transcription. Elevated mir-188 was found in BMSCs from aged patients, and knockout of mir-188 inhibited adipogenic differentiation of senescent BMSCs. In this study, we loaded antagomir-188 into the hybrid NPs in order to decrease bone marrow mir-188 levels and rescued aging-related bone loss.
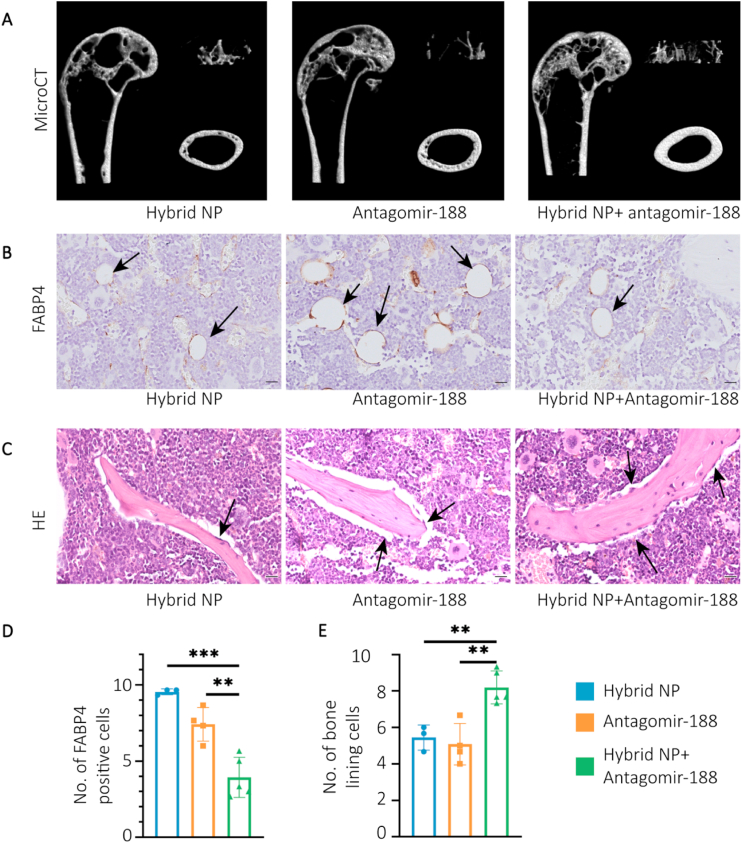
To further investigate the efficacy of hybrid NPs carrying antagomir-188, we tested the effects on an age-related osteoporosis mouse model in vivo. Hybrid NPs, antagomir-188 and antagomir-188-loaded hybrid NPs were weekly injected intravenously into 18-month-old male mice for 8 weeks. Mice treated with antagomir-loaded NPs showed a significantly higher bone mass reservation compared to vehicle and antagomir-188 groups (Fig. 5A). The relative expression levels of mir-188 were significantly lower in the antagomir and antagomir-loaded NPs groups (Figure S6). Micro-CT analysis showed that BV/TV and Tb.N were elevated in the hybrid NPs group compared to the control group, whereas Tb.Pf, Tb.Sp, were decreased after treatment with drug-loaded hybrid NPs (Figure S7A-D). However, no significant difference in Tb.Th was observed between the three groups of mice (Figure S7E). Notably, mice treated with the hybrid NPs loaded with antagomir-188 showed significantly higher Ct.Th (Figure S7F). Cortical bone porosity is used to reflect the bone formation capacity [47]. Po normally refers to channels for vessels that traverse the cortex and is the major predictor of fracture risk and bone mechanical strength [48]. Interestingly, the cortical bone porosity index Po.V and Po indicated a decreased cortical bone porosity (Figure S7G-H), which is commonly observed as a result of anabolic drugs, such as teriparatide [49,50]. The hybrid NPs loaded with antagomir-188 reduced cortical bone porosity and increased cortical bone quality.
Fig. 5.
Hybrid NPs carrying antagomir-188 reverse aging-related bone loss. A) Representative micro-CT images of femora from the hybrid NPs, antagomir and hybrid NPs + antagomir groups. A sagittal section, a three-dimensional reconstruction of trabecular bone, and a horizontal section of cortical bone were presented in the left, upper right and bottom right, respectively. B) Representative images of FABP4 immunohistochemical staining of the distal femora, scale bar = 20 μm. C) Representative images of H&E staining of the distal femora. D) Quantification of FABP4 positive cells, scale bar = 20 μm. E) Quantification of bone lining cells. Statistic difference: *p < 0.05; **p < 0.01; ***p < 0.001.
3.6. Hybrid NPs promote osteogenic differentiation and inhibit adipogenic differentiation of BMSCs
We next explored osteogenesis and adipogenesis via hematoxylin and eosin (H&E) staining and the immunohistochemical (IHC) staining of fatty acid binding protein 4 (FABP4). Antagomir-188-loaded hybrid NPs decreased the number of marrow adipocytes and increased the number of bone-lining cells around the trabecular surface (Fig. 5B–E).
For in vitro studies, alkaline phosphatase (ALP) staining demonstrated that antagomir-188-loaded hybrid NPs increased ALP positive osteoblasts formation (Figure S8A, B). Osteogenic protein marker runt-related transcription factor 2 (RUNX2) was up-regulated by antagomir-188 treatment (Figure S8C). The osteoblast differentiation gene marker osterix (Sp7) and osteocalcin (Bglap) were significantly increased after antagomir-188 treatment (Figure S8D-E).
Antagomir-188-loaded NPs inhibited lipid drops formation in BMSCs after adipogenic induction demonstrated by the Oil Red O staining (Figure S9A, B). Western blotting results showed that antagomir-188 inhibited peroxisome proliferation-activated receptor gamma (PPARγ) and FABP4 (Figure S9C). QPCR results showed that antagomir-188 decreased Pparγ and Fabp4 expression levels (Figure S9D, E).
Taken together, the constructed bone targeting nanotherapeutics delivering antagomir-188 reversed age-related bone loss by promoting osteogenic differentiation and inhibiting adipogenic differentiation of BMSCs.
4. Conclusions
Surface display of signaling molecules on exosomes granted them specific in vivo behaviors including distribution and clearance etc. In this study, we displayed CXCR4 on the surface of exosomes which were fused with liposomes carrying antagomir-188. The CXCR4+ exosome-liposome hybrid NPs tended to accumulate in the bone marrow and release antagomir-188 which promotes osteogenic differentiation, inhibits adipogenic differentiation and reverses the age-related bone loss. Overall, this study provides an exosome-based bone-targeting RNAi delivery strategy as an anabolic therapy against pathological bone loss.
CRediT authorship contribution statement
Yan Hu: Conceptualization, Methodology, Investigation, Writing - original draft. Xiaoqun Li: Methodology, Visualization. Qin Zhang: Validation, Writing - original draft. Zhengrong Gu: Formal analysis, Software. Ying Luo: Methodology. Jiawei Guo: Investigation, Data curation. Xiuhui Wang: Investigation. Yingying Jing: Project administration, Data curation. Xiao Chen: Supervision, Funding acquisition, Writing - original draft. Jiacan Su: Supervision, Funding acquisition.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgement
This work was supported by grants from National Key R&D Program of China (No.2018YFC2001500) and National Natural Science Foundation of China (No.91749204, 81771491 to J. Su, and 81871099 to X. Chen).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.bioactmat.2021.02.014.
Contributor Information
Yingying Jing, Email: jingy4172@shu.edu.cn.
Xiao Chen, Email: sirchenxiao@126.com.
Jiacan Su, Email: drsujiacan@163.com.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Zhou B.O., Yue R., Murphy M.M., Peyer J.G., Morrison S.J. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell. 2014;15(2):154–168. doi: 10.1016/j.stem.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambrosi T.H., Scialdone A., Graja A., Gohlke S., Jank A.M., Bocian C., Woelk L., Fan H., Logan D.W., Schurmann A., Saraiva L.R., Schulz T.J. Adipocyte accumulation in the bone marrow during obesity and aging impairs stem cell-based hematopoietic and bone regeneration. Cell Stem Cell. 2017;20(6):771–784. doi: 10.1016/j.stem.2017.02.009. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zou W., Rohatgi N., Brestoff J.R., Li Y., Barve R.A., Tycksen E., Kim Y., Silva M.J., Teitelbaum S.L. Ablation of fat cells in adult mice induces massive bone gain. Cell Metabol. 2020;32(5):801–813. doi: 10.1016/j.cmet.2020.09.011. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu T., Sun J., Tan L., Yan Q., Li L., Chen L., Liu X., Bin S. Enhanced osteogenesis and therapy of osteoporosis using simvastatin loaded hybrid system. Bioactive materials. 2020;5(2):348–357. doi: 10.1016/j.bioactmat.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Metavarayuth K., Maturavongsadit P., Chen X., Sitasuwan P., Lu L., Su J., Wang Q. Nanotopographical cues mediate osteogenesis of stem cells on virus substrates through BMP-2 intermediate. Nano Lett. 2019;19(12):8372–8380. doi: 10.1021/acs.nanolett.9b02001. [DOI] [PubMed] [Google Scholar]
- 6.Shang F., Yu Y., Liu S., Ming L., Zhang Y., Zhou Z., Zhao J., Jin Y. Advancing application of mesenchymal stem cell-based bone tissue regeneration. Bioactive materials. 2021;6(3):666–683. doi: 10.1016/j.bioactmat.2020.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X., Wang L., Huang B., Gu Y., Luo Y., Zhi X., Hu Y., Zhang H., Gu Z., Cui J., Cao L., Guo J., Wang Y., Zhou Q., Jiang H., Fang C., Weng W., Chen X., Chen X., Su J. Targeting actin-bundling protein L-plastin as an anabolic therapy for bone loss. Sci Adv. 2020;6(47):eabb7135. doi: 10.1126/sciadv.abb7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X., Zhi X., Wang J., Su J. RANKL signaling in bone marrow mesenchymal stem cells negatively regulates osteoblastic bone formation. Bone research. 2018;6:34. doi: 10.1038/s41413-018-0035-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L., Huang B., Chen X., Su J. New insight into unexpected bone formation by denosumab. Drug Discov. Today. 2020;20(S1359–6446):30340–30348. doi: 10.1016/j.drudis.2020.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Frith J.E., Kusuma G.D., Carthew J., Li F., Cloonan N., Gomez G.A., Cooper-White J.J. Mechanically-sensitive miRNAs bias human mesenchymal stem cell fate via mTOR signalling. Nat. Commun. 2018;9(1):257. doi: 10.1038/s41467-017-02486-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H., Li T., Fan J., Li T., Fan L., Wang S., Weng X., Han Q., Zhao R.C. miR-216a rescues dexamethasone suppression of osteogenesis, promotes osteoblast differentiation and enhances bone formation, by regulating c-Cbl-mediated PI3K/AKT pathway. Cell Death Differ. 2015;22(12):1935–1945. doi: 10.1038/cdd.2015.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li C.J., Cheng P., Liang M.K., Chen Y.S., Lu Q., Wang J.Y., Xia Z.Y., Zhou H.D., Cao X., Xie H., Liao E.Y., Luo X.H. MicroRNA-188 regulates age-related switch between osteoblast and adipocyte differentiation. J. Clin. Invest. 2015;125(4):1509–1522. doi: 10.1172/JCI77716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pecot C.V., Wu S.Y., Bellister S., Filant J., Rupaimoole R., Hisamatsu T., Bhattacharya R., Maharaj A., Azam S., Rodriguez-Aguayo C., Nagaraja A.S., Morelli M.P., Gharpure K.M., Waugh T.A., Gonzalez-Villasana V., Zand B., Dalton H.J., Kopetz S., Lopez-Berestein G., Ellis L.M., Sood A.K. Therapeutic silencing of KRAS using systemically delivered siRNAs. Mol. Canc. Therapeut. 2014;13(12):2876–2885. doi: 10.1158/1535-7163.MCT-14-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan T.L., Fellmann C., Lee C., Ritchie C.D., Thapar V., Lee L.C., Hsu D.J., Grace D., Carver J.O., Zuber J., Luo J., McCormick F., Lowe S.W. Development of siRNA payloads to target KRAS-mutant cancer. Canc. Discov. 2014;4(10):1182–1197. doi: 10.1158/2159-8290.CD-13-0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xue W., Dahlman J.E., Tammela T., Khan O.F., Sood S., Dave A., Cai W., Chirino L.M., Yang G.R., Bronson R., Crowley D.G., Sahay G., Schroeder A., Langer R., Anderson D.G., Jacks T. Small RNA combination therapy for lung cancer. P Nat Acad Sci USA. 2014;111(34):E3553–E3561. doi: 10.1073/pnas.1412686111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itaka K., Ohba S., Miyata K., Kawaguchi H., Nakamura K., Takato T., Chung U., Kataoka K. Bone regeneration by regulated in vivo gene transfer using biocompatible polyplex nanomicelles. Mol. Ther. : the journal of the American Society of Gene Therapy. 2007;15(9):1655–1662. doi: 10.1038/sj.mt.6300218. [DOI] [PubMed] [Google Scholar]
- 17.Zhang G., Guo B., Wu H., Tang T., Zhang B., Zheng L., He Y., Yang Z., Pan X., Chow H., To K., Li Y., Li D., Wang X., Wang Y., Lee K., Hou Z., Dong N., Li G., Leung K., Hung L., He F., Zhang L., Qin L. A delivery system targeting bone formation surfaces to facilitate RNAi-based anabolic therapy. Nat. Med. 2012;18(2):307–314. doi: 10.1038/nm.2617. [DOI] [PubMed] [Google Scholar]
- 18.Liang C., Guo B., Wu H., Shao N., Li D., Liu J., Dang L., Wang C., Li H., Li S., Lau W.K., Cao Y., Yang Z., Lu C., He X., Au D.W.T., Pan X., Zhang B., Lu C., Zhang H., Yue K., Qian A., Shang P., Xu J., Xiao L., Bian Z., Tan W., Liang Z., He F., Zhang L., Lu A., Zhang G. Aptamer-functionalized lipid nanoparticles targeting osteoblasts as a novel RNA interference-based bone anabolic strategy. Nat. Med. 2015;21(3):288–294. doi: 10.1038/nm.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Q., Yan J., Yang J., Li B. Nanomaterials promise better bone repair. Mater. Today. 2016;19(8):451–463. [Google Scholar]
- 20.Kalluri R., LeBleu V.S. The biology, function, and biomedical applications of exosomes. Science. 2020;367(6478) doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elsharkasy O.M., Nordin J.Z., Hagey D.W., de Jong O.G., Schiffelers R.M., Andaloussi S.E., Vader P. Extracellular vesicles as drug delivery systems: why and how? Adv. Drug Deliv. Rev. 2020;159:332–343. doi: 10.1016/j.addr.2020.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Perets N., Betzer O., Shapira R., Brenstein S., Angel A., Sadan T., Ashery U., Popovtzer R., Offen D. Golden exosomes selectively target brain pathologies in neurodegenerative and neurodevelopmental disorders. Nano Lett. 2019;19(6):3422–3431. doi: 10.1021/acs.nanolett.8b04148. [DOI] [PubMed] [Google Scholar]
- 23.Quinn Z., Mao W., Xia Y., John R., Wan Y. Conferring receptors on recipient cells with extracellular vesicles for targeted drug delivery. Bioactive materials. 2021;6(3):749–756. doi: 10.1016/j.bioactmat.2020.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nie W., Wu G., Zhang J., Huang L.L., Ding J., Jiang A., Zhang Y., Liu Y., Li J., Pu K., Xie H.Y. Responsive exosome nano-bioconjugates for synergistic cancer therapy. Angew Chem. Int. Ed. Engl. 2020;59(5):2018–2022. doi: 10.1002/anie.201912524. [DOI] [PubMed] [Google Scholar]
- 25.Kamerkar S., LeBleu V.S., Sugimoto H., Yang S., Ruivo C.F., Melo S.A., Lee J.J., Kalluri R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546(7659):498–503. doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin Y., Wu J., Gu W., Huang Y., Tong Z., Huang L., Tan J. Exosome-liposome hybrid nanoparticles deliver CRISPR/Cas9 system in MSCs. Adv. Sci. 2018;5(4) doi: 10.1002/advs.201700611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geminder H., Sagi-Assif O., Goldberg L., Meshel T., Rechavi G., Witz I.P., Ben-Baruch A. A possible role for CXCR4 and its ligand, the CXC chemokine stromal cell-derived factor-1, in the development of bone marrow metastases in neuroblastoma. J. Immunol. 2001;167(8):4747–4757. doi: 10.4049/jimmunol.167.8.4747. [DOI] [PubMed] [Google Scholar]
- 28.Zehentmeier S., Pereira J.P. Cell circuits and niches controlling B cell development. Immunol. Rev. 2019;289(1):142–157. doi: 10.1111/imr.12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smit M.J., Schlecht-Louf G., Neves M., den Bor J.V., Penela P., Siderius M., Bachelerie F., Mayor F.J. The CXCL12/CXCR4/ACKR3 Axis in the tumor microenvironment: signaling, crosstalk, and therapeutic targeting. Annu. Rev. Pharmacol. Toxicol. 2020;61:541–563. doi: 10.1146/annurev-pharmtox-010919-023340. [DOI] [PubMed] [Google Scholar]
- 30.Conley-LaComb M.K., Semaan L., Singareddy R., Li Y., Heath E.I., Kim S., Cher M.L., Chinni S.R. Pharmacological targeting of CXCL12/CXCR4 signaling in prostate cancer bone metastasis. Mol. Canc. 2016;15(1):68. doi: 10.1186/s12943-016-0552-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muller A., Homey B., Soto H., Ge N., Catron D., Buchanan M.E., McClanahan T., Murphy E., Yuan W., Wagner S.N., Barrera J.L., Mohar A., Verastegui E., Zlotnik A. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410(6824):50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 32.Song H., Li X., Zhao Z., Qian J., Wang Y., Cui J., Weng W., Cao L., Chen X., Hu Y., Su J. Reversal of osteoporotic activity by endothelial cell-secreted bone targeting and biocompatible exosomes. Nano Lett. 2019;19(5):3040–3048. doi: 10.1021/acs.nanolett.9b00287. [DOI] [PubMed] [Google Scholar]
- 33.Stewart M.P., Sharei A., Ding X., Sahay G., Langer R., Jensen K.F. In vitro and ex vivo strategies for intracellular delivery. Nature. 2016;538(7624):183–192. doi: 10.1038/nature19764. [DOI] [PubMed] [Google Scholar]
- 34.Peled A., Petit I., Kollet O., Magid M., Ponomaryov T., Byk T., Nagler A., Ben-Hur H., Many A., Shultz L., Lider O., Alon R., Zipori D., Lapidot T. Dependence of human stem cell engraftment and repopulation of NOD/SCID mice on CXCR4. Science. 1999;283(5403):845–848. doi: 10.1126/science.283.5403.845. [DOI] [PubMed] [Google Scholar]
- 35.Morrison S.J., Scadden D.T. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505(7483):327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin D.K., Shido K., Kopp H.G., Petit I., Shmelkov S.V., Young L.M., Hooper A.T., Amano H., Avecilla S.T., Heissig B., Hattori K., Zhang F., Hicklin D.J., Wu Y., Zhu Z., Dunn A., Salari H., Werb Z., Hackett N.R., Crystal R.G., Lyden D., Rafii S. Cytokine-mediated deployment of SDF-1 induces revascularization through recruitment of CXCR4+ hemangiocytes. Nat. Med. 2006;12(5):557–567. doi: 10.1038/nm1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teicher B.A., Fricker S.P. CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin. Canc. Res. 2010;16,(11):2927–2931. doi: 10.1158/1078-0432.CCR-09-2329. [DOI] [PubMed] [Google Scholar]
- 38.Huang X., Guo B., Liu S., Wan J., Broxmeyer H.E. Neutralizing negative epigenetic regulation by HDAC5 enhances human haematopoietic stem cell homing and engraftment. Nat. Commun. 2018;9(1):2741. doi: 10.1038/s41467-018-05178-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Staller P., Sulitkova J., Lisztwan J., Moch H., Oakeley E.J., Krek W. Chemokine receptor CXCR4 downregulated by von Hippel-Lindau tumour suppressor pVHL. Nature. 2003;425(6955):307–311. doi: 10.1038/nature01874. [DOI] [PubMed] [Google Scholar]
- 40.Muller A., Homey B., Soto H., Ge N., Catron D., Buchanan M.E., McClanahan T., Murphy E., Yuan W., Wagner S.N., Barrera J.L., Mohar A., Verastegui E., Zlotnik A. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410(6824):50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 41.El-Andaloussi S., Lee Y., Lakhal-Littleton S., Li J., Seow Y., Gardiner C., Alvarez-Erviti L., Sargent I.L., Wood M.J. Exosome-mediated delivery of siRNA in vitro and in vivo. Nat. Protoc. 2012;7(12):2112–2126. doi: 10.1038/nprot.2012.131. [DOI] [PubMed] [Google Scholar]
- 42.Yang Z., Shi J., Xie J., Wang Y., Sun J., Liu T., Zhao Y., Zhao X., Wang X., Ma Y., Malkoc V., Chiang C., Deng W., Chen Y., Fu Y., Kwak K.J., Fan Y., Kang C., Yin C., Rhee J., Bertani P., Otero J., Lu W., Yun K., Lee A.S., Jiang W., Teng L., Kim B., Lee L.J. Large-scale generation of functional mRNA-encapsulating exosomes via cellular nanoporation. Nat Biomed Eng. 2020;4(1):69–83. doi: 10.1038/s41551-019-0485-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pedroso D.L.M., Simoes S., Pires P., Faneca H., Duzgunes N. Cationic lipid-DNA complexes in gene delivery: from biophysics to biological applications. Adv. Drug Deliv. Rev. 2001;47(2–3):277–294. doi: 10.1016/s0169-409x(01)00110-7. [DOI] [PubMed] [Google Scholar]
- 44.Zhang L., Wu S., Qin Y., Fan F., Zhang Z., Huang C., Ji W., Lu L., Wang C., Sun H., Leng X., Kong D., Zhu D. Targeted codelivery of an antigen and dual agonists by hybrid nanoparticles for enhanced cancer immunotherapy. Nano Lett. 2019;19(7):4237–4249. doi: 10.1021/acs.nanolett.9b00030. [DOI] [PubMed] [Google Scholar]
- 45.Lv Q., Cheng L., Lu Y., Zhang X., Wang Y., Deng J., Zhou J., Liu B., Liu J. Thermosensitive exosome-liposome hybrid nanoparticle-mediated chemoimmunotherapy for improved treatment of metastatic peritoneal cancer. Adv. Sci. 2020;7(18) doi: 10.1002/advs.202000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piffoux M., Silva A., Wilhelm C., Gazeau F., Tareste D. Modification of extracellular vesicles by fusion with liposomes for the design of personalized biogenic drug delivery systems. ACS Nano. 2018;12(7):6830–6842. doi: 10.1021/acsnano.8b02053. [DOI] [PubMed] [Google Scholar]
- 47.Bala Y., Zebaze R., Seeman E. Role of cortical bone in bone fragility. Curr. Opin. Rheumatol. 2015;27(4):406–413. doi: 10.1097/BOR.0000000000000183. [DOI] [PubMed] [Google Scholar]
- 48.Cooper D.M., Kawalilak C.E., Harrison K., Johnston B.D., Johnston J.D. Cortical bone porosity: what is it, why is it important, and how can we detect it? Curr. Osteoporos. Rep. 2016;14(5):187–198. doi: 10.1007/s11914-016-0319-y. [DOI] [PubMed] [Google Scholar]
- 49.Tsai J.N., Uihlein A.V., Burnett-Bowie S.M., Neer R.M., Derrico N.P., Lee H., Bouxsein M.L., Leder B.Z. Effects of two years of teriparatide, denosumab, or both on bone microarchitecture and strength (DATA-HRpQCT study) J. Clin. Endocrinol. Metabol. 2016;101(5):2023–2030. doi: 10.1210/jc.2016-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsai J.N., Nishiyama K.K., Lin D., Yuan A., Lee H., Bouxsein M.L., Leder B.Z. Effects of denosumab and teriparatide transitions on bone microarchitecture and estimated strength: the DATA-switch HR-pQCT study. J. Bone Miner. Res. 2017;32(10):2001–2009. doi: 10.1002/jbmr.3198. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.