Abstract
Dedifferentiation of chondrocyte greatly restricts its function and application, however, it is poorly understood except a small number of canonical markers. The non-cell-adhesive property endows polysaccharide hydrogel with the ability to maintain chondrocyte phenotype, which can serve as a platform to identify new molecular markers and therapeutic targets of chondrocyte dedifferentiation. In this study, the high-throughput RNA sequencing (RNA-seq) was first performed on articular chondrocytes at primary (P0) and passage 1 (P1) stages to explore the global alteration of gene expression along with chondrocyte dedifferentiation. Significantly, several potential marker genes, such as PFKFB3, KDM6B, had been identified via comparatively analyzing their expression in P0 and P1 chondrocytes as well as in 3D constructs (i.e. chondrocyte-laden alginate hydrogel and HA-MA hydrogel) at both mRNA and protein level. Besides, the changes in cellular morphology and enriched pathway of differentially expressed genes during chondrocyte dedifferentiation was studied in detail. This study developed the use of hydrogel as a platform to investigate chondrocyte dedifferentiation; the results provided new molecular markers and potential therapeutic targets of chondrocyte dedifferentiation.
Keywords: Chondrocyte dedifferentiation, Hydrogel, Gene expression, RNA sequencing
Graphical abstract
Highlights
-
•
Non-cell-adhesive hydrogel scaffold is first developed as a platform to study chondrocyte dedifferentiation.
-
•
The full spectrum of gene expression programs in chondrocyte dedifferentiation has been captured by RNA-seq.
-
•
Potential new markers of chondrocyte dedifferentiation, such as PFKFB3, KDM6B, have been identified.
1. Introduction
Articular Osteoarthritis (OA) is characterized by chondrocyte dedifferentiation and degradation of extracellular matrix (ECM) [1,2]. So far, there is no effective treatment to restore the damaged articular cartilage. In late stages of OA, surgery often is performed to replace or repair the damaged joint [3].
Autologous chondrocyte implantation (ACI) is one of promising treatment methods for OA, in which articular chondrocytes are extracted from autologous cartilage and expanded in vitro to obtain a sufficient number of chondrocytes [4]. However, in vitro chondrocyte expansion by multiple monolayer passaging often leads to dedifferentiation [[5], [6], [7]]. Moreover, cartilage tissue engineering is considered as a promising strategy for cartilage repair, yet, in which chondrocyte expansion is also an essential step [8]. The dedifferentiation of chondrocytes is a great concern for functional reconstruction of articular hyaline cartilage.
Chondrocyte dedifferentiation has been a troubling and unsolved problem in clinical applications and tissue engineering, in which chondrocytes undergo changes in morphologies and molecular markers, such as decreased expression of type II collagen (COL2), aggrecan (ACAN) as well as increased expression of type I collagen (COL1) [[5], [6], [7],9,10]. Although we have gained some understandings for the several classical markers, it is regrettable that the full spectrum of gene expression programs during this process has not been captured and confirmed.
Many scientists have strived hard to inhibit or slow down the process of chondrocyte dedifferentiation through various methods [[11], [12], [13], [14], [15]]. Among them, three-dimensional (3D) culture in hydrogels has exhibited great advantages to maintain the differentiated phenotype of chondrocytes [8,[16], [17], [18]]. Especially for polysaccharide hydrogels, their water-swollen, non-cell-adhesive structures can provide unique aqueous environments to mimic in vivo environments for chondrocyte growth [16,19]. Alginate that can form hydrogels with divalent cations (such as calcium, magnesium ions), has been an attractive material to facilitate proliferation of chondrocytes with native morphology [20]. Furthermore, hyaluronan (HA) is a crucial component of articular cartilage ECM [8,18]. HA-based hydrogel was also reported to support phenotype maintenance and amplification of chondrocytes in vitro [8,21].
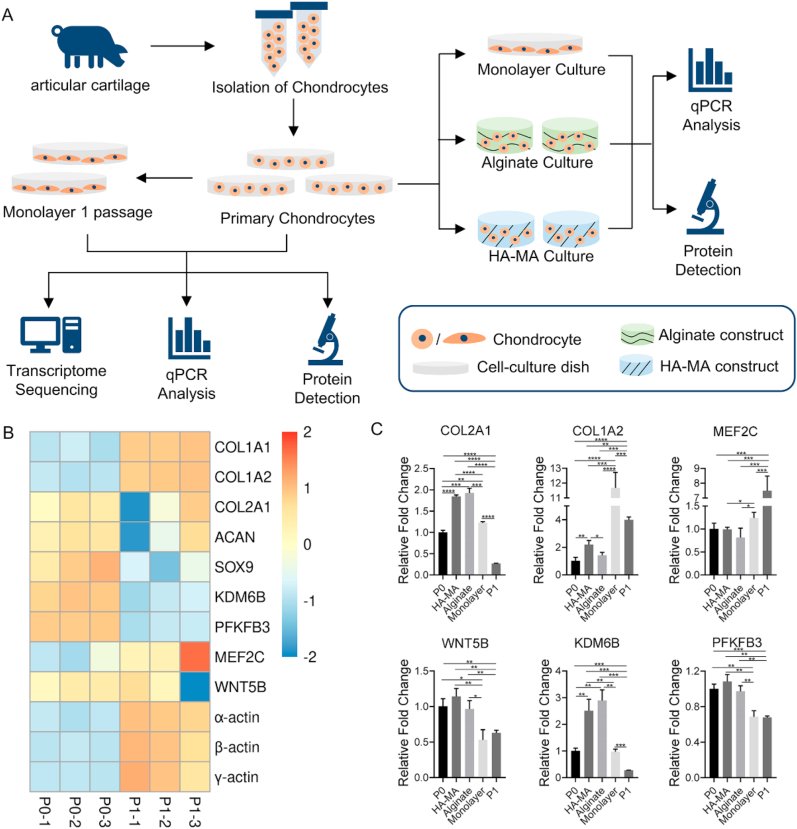
The inspiring functions of hydrogels in retaining chondrocyte phenotype motivated us to utilize them as a platform to identify new marker genes as well as therapeutic targets of chondrocyte dedifferentiation by combining high-throughput RNA sequencing (RNA-seq). In this work, we first studied the transcriptome profile changes induced by chondrocyte passaging from primary (P0) to passage 1 (P1) cells using RNA-seq. Comparative studies in cellular morphologies and gene expression were carried out on the chondrocytes cultured in monolayer and hydrogels (namely alginate gel and hyaluronan gel) at both RNA and protein levels.
2. Materials and methods
2.1. Isolation and culture of chondrocytes
Cartilage tissues were collected from the joints of 5-month-old pigs, and cut into small cubes according to the established protocol [19]. After five times of washing with the phosphate-buffered saline (PBS, pH 7.4) containing 10% (v/v) of penicillin-streptomycin mixture (Sigma), the cartilage cubes were digested for 14 h in Dulbecco's modified Eagle medium (DMEM, Gibco) containing type II collagenase (0.15%, Gibco) and 5% (v/v) fetal bovine serum (FBS, Gibco). The resultant cell suspension was filtered by a 70 μm cell-strainer and the primary chondrocytes were subsequently collected by centrifugation. The chondrocytes were resuspended and cultured in chondrocyte culture medium (CCM, DMEM containing 10% (v/v) FBS, 0.05 mg/mL vitamin C, 0.4 mM l-proline, 10 mM 4-(2-hydroxyethyl)-piperazine-1-ethanesulfonic acid, 0.1 mM nonessential amino acids (NEAA, Gibco, 11140050), 0.1 mg/mL streptomycin and 100 units/mL penicillin (Sigma).
2.2. Fabrication of gel-cell constructs
Primary chondrocytes were encapsulated in non-cell-adhesive hydrogels and cultured for 5 days (Fig. 1). Meanwhile, the primary chondrocytes were also cultivated on a petri-dish as control (2D monolayer culture).
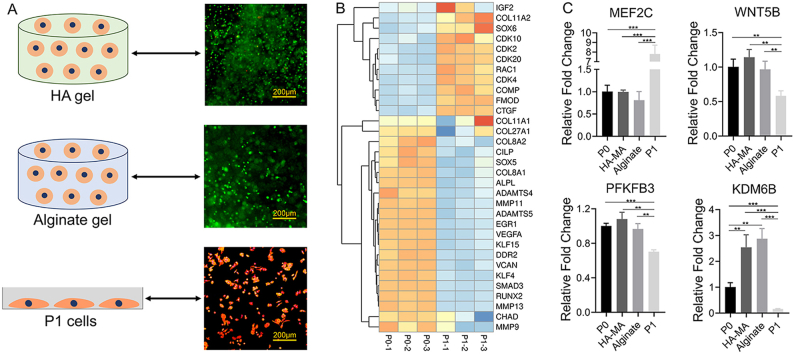
Fig. 1.
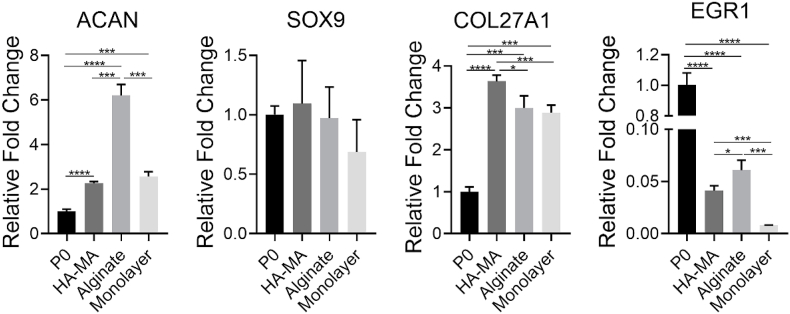
A) Schematic illustration of experimental procedures. B) Heatmap of typical DEGs associated with chondrocyte dedifferentiation. C) qPCR analysis for expression of representative genes in the chondrocytes at P0 and P1, in HA-MA gel and alginate gel constructs as well as by 5 days of monolayer culture, respectively; * indicated p < 0.05, ** indicated p < 0.005, *** indicated p < 0.0005, **** indicated p < 0.0001.
For hyaluronic acid-methacrylate (HA-MA) [22], 2 g of HA powder was thoroughly dissolved in 200 mL of PBS by stirring, 40 mL of N,N-dimethylformamide (Sigma-Aldrich) was slowly added, and the mixture was stirred for 2 h. Subsequently, 13.3 g of methacrylic anhydride (MA, Sigma-Aldrich) and 6.7 g of triethylamine (Sigma-Aldrich) was added, respectively, into the reaction solution. After 4 days of slowly stirring at room temperature, the mixture was precipitated in 2 L of ethanol, filtered, dialyzed for 5 days in deionized (DI) water, and finally lyophilized to obtain HA-MA foam. 107 of P0 chondrocytes were resuspended in 1 mL of HA-MA precursor (1%, g/mL) solution containing photo-initiator Irgacure 2959 (0.1%, g/mL, Ciba Specialty Chemicals), and 50 μL of the suspension was rapidly injected into a cylindrical mold (diameter: 5.2 mm) with a pipette. Subsequently, they were exposed to UV light (365 nm, 30 mW/cm2) for 10 min to gelation, and resultant HA-MA gel constructs were placed into CCM medium at 37 °C for 5 days.
For alginate-based constructs, 50 μL of chondrocyte suspension (density: 107/mL) in alginate solution (1%, g/mL) was injected into the same mold. Subsequently, 20 μL of CaCl2 solution was added. After 10 min at 37 °C, the gel-cell constructs were formed. These alginate constructs were then transferred into CCM medium and cultured at 37 °C for 5 days.
2.3. Swelling property of hydrogel
The swelling properties of acellular hydrogels were evaluated. The alginate gels and HA-MA gels were fabricated from 100 μL of alginate solution (1%, g/mL) and 100 μL of HA-MA solution (1%, g/mL), respectively, as described above. Subsequently, they were transferred to PBS solution, respectively, for 2 days of incubation at 37 °C. Then, they were weighted to obtain the wet weight (Ww), respectively, after wiping the water on the surfaces. The dry weight (Wd) was also measured after thorough drying in an oven. The equilibrium swelling ratio (Q) was calculated by following equation [23,24]:
| Q = Ww/Wd |
2.4. RNA extraction
P0 and P1 (sub-cultured on a petri-dish) cells and cell-laden constructs were collected, respectively, for isolation of total RNAs using TRI reagent (Sigma) according to the manufacturer's protocol. Briefly, P0 and P1 cells were harvested by trypsinization and then suspended into 1 mL of TRI reagent. For the cell-laden constructs, they were transferred into a 1.5 mL RNAase-free Eppendorf tube, respectively, followed by the addition of 1 mL of TRI reagent. After 10 times of pippeting, 200 μl chloroform was added into each tube. After violent shaking, the tubes were placed at room temperature for 5 min, and followed by centrifuging for 1 min at 4 °C. The total RNA in the supernatant was precipitated in isopropanol. The resultant RNA pellet was washed with 75% ethanol for 3 times, and then it was dissolved in RNAase-free water. The concentration of total RNAs was measured by a NanoDrop 2000c. Three biological replicates were carried out for each group.
2.5. Sequencing and data analysis
The extracted total RNA was sent to ANNOROAD Co., Ltd (Beijing, China) for library preparation, RNA sequencing (RNA-seq), and data analysis. RNA-seq data were submitted into the National Center for Biotechnology Information (NCBI) under accession number PRJNA683807. In our analysis, a gene whose count value was equal to 1 or more than 1, was thereupon identified to be expressed in this sample. The DEGs, namely differentially expressed genes, were determined by log2fold-change >1 and p-value <0.05. The Omicshare (http://www.omicshare.com/) was used to draw heatmap; DAVID (https://david.ncifcrf.gov/) was used for Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis.
2.6. qPCR
Gene expression of type I collagen A2 (COL1A2), type II collagen A1 (COL2A1), aggrecan (ACAN), sex-determining region Y-related high mobility group-box gene9 (SOX9), type XVII collagen A1 (COL27A1), myocyte enhancer factor 2C (MEF2C), Wnt family member 5B (WNT5B), early growth response 1 (EGR1), lysine demethylase 6B (KDM6B) and 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (PFKFB3) was determined, respectively. Briefly, 0.5 μg of total RNA was reverse-transcribed into cDNA using a PrimeScript RT Reagent Kit with gDNA eraser (TaKaRa). Then 2 μL of cDNA was taken out to amplify the target genes. Real-time quantitative PCR (qPCR) was carried out with IQ SYBR Green Supermix (Bio RAD) by using a real-time PCR Amplifier (Bio-RAD). GAPDH, one housekeeping gene, was used as internal reference for the expression of each gene. Using a 2∧(-ddCt) formula, the expression level of each target gene was calculated. The forward and reverse primers were listed Table 1.
Table 1.
qPCR primer sequences.
| Gene | F (forward) | R (reverse) |
|---|---|---|
| GAPDH | GCACAGTCAAGGCGGAGAAC | GCCTTCTCCATGGTCGTGAA |
| COL2A1 | GTGGAGCAGCAAGAGCAAGGA | CAGTGTTAGGAGCCAGGTTGTCAT |
| COL27A1 | TGTGAGCAGAAGATGGTGGATGGTA | CGGTCATGTTAAGGCAGTGGATGG |
| ACAN | AACTTCTTCGGGGTGGGTGG | TGTAAAGGGCTCCTCAGGCTC |
| COL1A2 | TGAAAACATCCCAGCCAAAAA | CGCATGAAGGCAAGTTGTGT |
| SOX9 | CCGTGCTGTGATTGGCTAGAGG | CGAATTGGAGAGGAGGAGGAGAAGA |
| EGR1 | CCTGACCGCAGAGTCTTTTCC | CCACAAGGTGTTGCCACTGT |
| PFKFB3 | AGACACGTACCCCGAGGAGTAC | GTGGCAGATAACCAGCACGTT |
| KDM6B | CCAGGCCATCAAGAGAACAAC | CACCAGGAACCCGTCAAGTAA |
| WNT5B | CAGACGTAGCCTGCAAATGC | GCTGTTGACCAGCTCCAACTT |
| MEF2C | GCCAGTCTCCATCCCAGTGT | GCACTTGGAGGTCGATGTGTT |
2.7. CMFDA staining
5-chloromethylfluorescein diacetate (CMFDA) staining was carried out to analyze cell morphology. For group P0, primary chondrocytes were extracted and cultured in the 24-well plates containing CCM for 2 h. For group P1, the chondrocytes were passage cultured for 3 days. Subsequently, the cells were incubated with 5 μM CMFDA working solution in DMEM for 45 min, respectively. The cells were carefully washed three times with PBS solution and observed with a fluorescence microscope (Nikon T1-FL), respectively. Meanwhile, bright-field photographs were also taken.
2.8. Live/dead assay
Live/dead staining was carried out to determine chondrocytes’ viability in hydrogel constructs. The viable cells were indicated with green fluorescence, while the dead cells exhibited red fluorescence. Briefly, the cell-laden constructs were collected and cut into thin slices using a surgical blade, and which were then incubated within 0.5 mL of PBS solution containing calcein-AM (2 μM) and propidium iodide (PI, 4 μM) at 37 °C. After 30 min of incubation, the slices were rinsed twice using PBS solution and observed under a fluorescence microscope (Nikon T1-FL).
2.9. F-actin and immunofluorescence staining
For P0 group, primary chondrocytes were placed in the 24-well plates containing CCM per well. After 2 h of culture, the supernatant was discard and the plates were washed by PBS; subsequently, 1 mL of 4% paraformaldehyde (PFA) was added for fixation. For group P1, the chondrocytes were fixed with 1 mL of PFA solution after discarding the supernatant and washing with PBS. P0 and P1 cells were pretreated with 0.5% Triton-100 for 5min, and then added with 100 nM TRITC-Phalloidin solution for 30min in darkness. Besides, 4’,6-diamidino-2-phenylindole (DAPI, 100 nM) was used for nuclei staining.
For immunofluorescence staining, native articular cartilage, alginate constructs, and HA-MA constructs, and plate-cultured cells were fixed with 1 mL of PFA solution for 24 h, respectively. The native cartilage and constructs were embedded in paraffin wax, and cut into 6 μm-thick sections with a microtome, respectively. The P0, monolayer-cultured, and P1 cells as well as these sections were incubated with primary antibody (2 μg/mL in PBS) for COL1 (Bioss, bs-7158R), COL2 (Bioss, bs-10589R), KDM6B (ABclonal, A12763), and PFKFB3 (ABclonal, A14764) at 4 °C overnight, respectively; subsequently they were incubated with anti-IgG (5 μg/mL in PBS, ABclonal, AS011, ABclonal, AS011) at room temperature for 1 h in the dark, respectively. After triple washing with PBS, they were counterstained with DAPI, and observed under a fluorescence microscope (Nikon T1-FL).
2.10. Statistical analysis
All the experimental data were conducted with at least three biological replicates in each group. Statistical significance between the results of two groups was analyzed with Student's t-test, and statistically significant difference was defined as p < 0.05. Results were given as mean ± standard deviation.
3. Results and discussion
We hypothesized that chondrocyte's mRNA expression must be changed along with their dedifferentiation induced by monolayer sub-culture. P0 cells underwent one monolayer passage to obtain P1 cells (early stage of dedifferentiation), and then RNA-seq was performed on P0 and P1 cells to explore the changes of gene expression at the transcriptome level in the early stage of chondrocyte dedifferentiation (Fig. 1). Transcriptome-wide analysis showed gene expression profile during chondrocyte dedifferentiation, however, a large amount of genes were differentially expressed. Due to the ability to maintain chondrocyte phenotype, hydrogel platform probably serves as an aid to identify reliable markers of chondrocyte dedifferentiation. P0 cells were cultured under three different conditions, namely 2D monolayer culture and 3D culture in alginate and HA-MA gel constructs, to explore the changes of gene expression in different culture conditions. Significantly, by comparing gene expression in P0, P1 and gel constructs, the canonical markers (namely COL2A1 and COL1A2) and potential markers of chondrocyte dedifferentiation, namely MEF2C, WNT5B, KDM6B, and PFKFB3, had been identified. Interestingly, the chondrocytes under monolayer culture (without passage) also exhibited dramatically elevated expression of COL1A2 and decreased expression of WNT5B, respectively, compared to P0.
3.1. Cell morphology and hydrogel properties
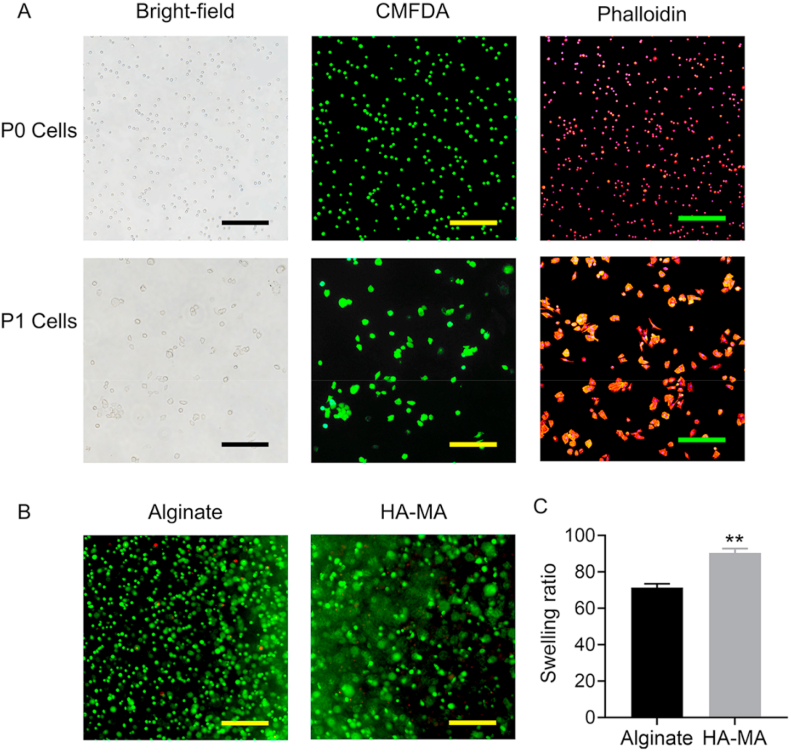
In 2D monolayer culture, the morphologies of most chondrocytes were changed under bright-field observation (Fig. 2A). To more clearly observe cellular morphology, CMFDA staining was carried out. Compared with P0 cells, P1 chondrocytes showed increased cellular size, and meanwhile some P1 cells exhibited irregular polygon shapes. To further verify the results, F-actin was stained with TRITC-phalloidin to illustrate cytoskeletal changes. The expression of F-actin in P1 chondrocytes was significantly increased compared to P0 chondrocytes at protein level (Fig. 2A). Interestingly, the round chondrocytes would not be detected after three passages in monolayer culture (data not shown), suggesting that the change in morphology of chondrocytes from small-round shape to size enlargement and then polygon shapes is a gradual process in company with chondrocyte dedifferentiation [25]. Taken together, the morphology change of chondrocytes and accompanying dedifferentiation is extremely affected by monolayer culture and passage. We have reason to believe that the dedifferentiation of chondrocytes must be related with drastic changes in global gene expression.
Fig. 2.
A) P0 and P1 chondrocyte observation under bright-field microscopy or fluorescence microscopy after staining with CMFDA and Phalloidin, respectively. In Phalloidin staining, the nucleus was counterstained with DAPI (blue). B) Live/Dead staining of chondrocytes encapsulated in alginate and HA-MA constructs. C) Equilibrium swelling ratio of alginate gel and HA-MA gel, respectively. Magnification was 100x. Scale bars were 200 μm ** indicated p < 0.005.
Alginate gels and HA-MA gels is widely used in tissue engineering [[26], [27], [28]], including cartilage tissue engineering. The swelling property and cytocompatibility of both alginate gels and HA-MA gels was first evaluated. As expected, the swelling ratio of alginate gels and HA-MA gels was up to 71.3% and 90.4%, respectively, and their high water contents were beneficial to maintenance of the chondrogenic phenotype. The results of live/dead assay indicated the high cell viability in both alginate gels and HA-MA gels; significantly, the round or oval morphology of the laden chondrocytes was also observed (Fig. 2B), which is crucial for sustaining normal chondrocyte function.
3.2. Overview of the transcriptome changes in P0 and P1 cells
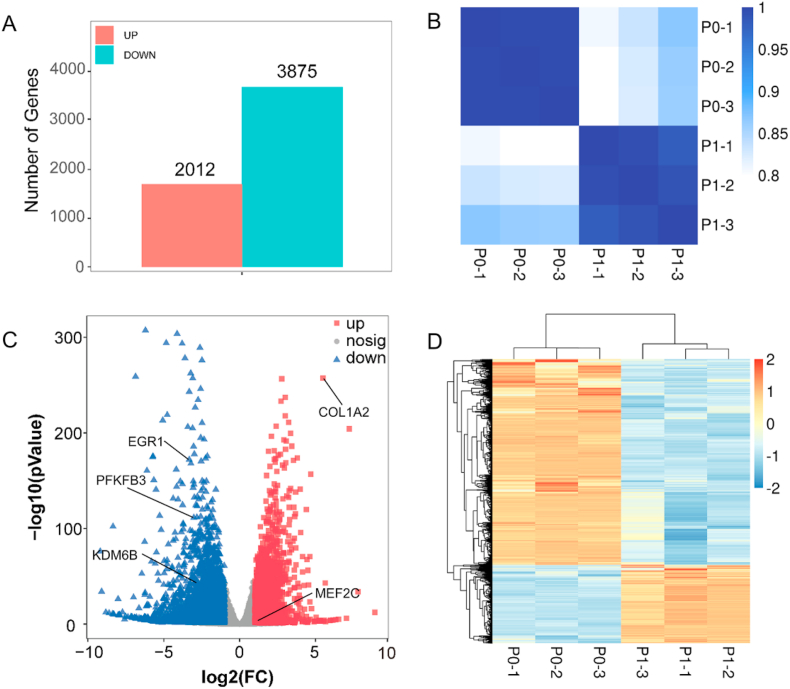
To explore global gene expression changes in early stage of dedifferentiation, RNA-seq based on next-generation sequencing technology was performed on P0 and P1 chondrocytes, respectively. 5887 DEGs were obtained between P0 and P1 cells. Compared with P0 cells, 2012 up-regulated genes and 3875 down-regulated genes were detected in P1, respectively (Fig. 3A). The number of down-regulated genes was obviously larger than that of up-regulated genes in P1 cells. The consistency among the samples was verified by performing Pearson correlation analysis. High similarity among three biological replicates of the P0 and P1 group, respectively, demonstrated that the sequencing results were consistent (Fig. 3B). The volcano plot was used to reflect the DEGs (Fig. 3C). Among these DEGs between P0 and P1, some cartilage-related genes had also been detected. MEF2C and COL1A2 was significantly up-regulated in P1 cells, but in the contrary, KDM6B, EGR1, and PFKFB3 was significantly down-regulated in P1 cells (Fig. 3C). The heatmap made by clustering analysis of genes visually revealed the difference in gene expression between P0 and P1 samples (Fig. 3D), and which further clearly showed that there were more down-regulated genes than up-regulated ones in P1 compared to P0 as illustrated in Fig. 3A.
Fig. 3.
A) Number of differentially expressed genes (DEGs), including up-regulated (UP) and down-regulated (DOWN) gens in P1 cells compared to P0 cells. B) Correlation analysis between P0 and P1 samples. C) Volcano plot of gene expression profile in P1 cells versus P0 cells; up, nosig, and down indicates up-regulated genes, non-significantly differentially expressed genes, and down-regulated genes, respectively. D) Heatmap of DEGs in P0 and P1 cells.
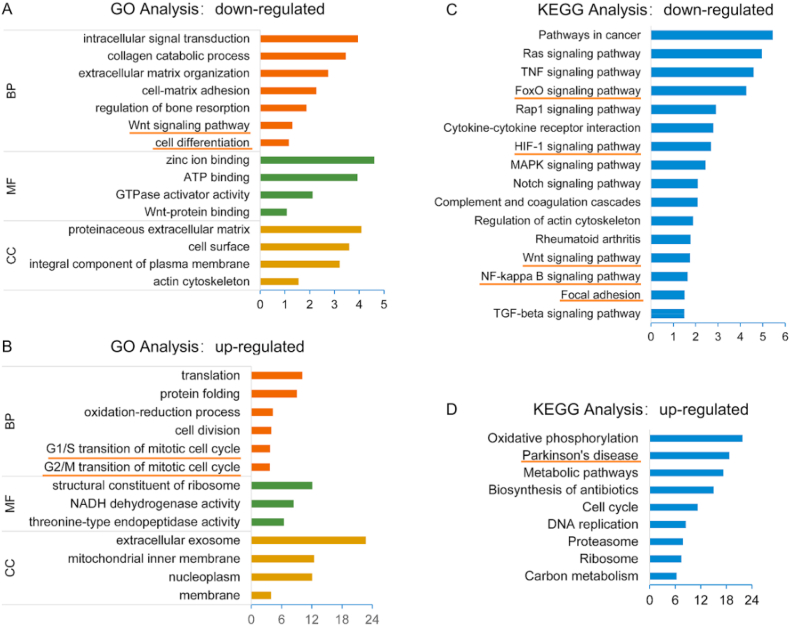
In order to assess the function of DEGs, GO analysis was carried out. The results showed that GO terms enriched in P1 down-regulated genes mainly included membrane, structural constituent of ribosome, protein folding and translation, and so on (Fig. 4A). Furthermore, the GO terms enriched in P1 up-regulated genes were significantly correlated with actin cytoskeleton, integral component of plasma membrane, extracellular matrix, cell-matrix adhesion, extracellular matrix organization as well as collagen catabolic process, etc. (Fig. 4B). These results showed that at the early stage of chondrocyte dedifferentiation, collagen metabolism was singularly raised, cell membranes and cytoskeleton was altered, and focal adhesion occurred.
Fig. 4.
A) Gene Ontology (GO) enrichment analysis of the down-regulated genes in P1 cells. B) GO enrichment analysis of the up-regulated genes in P1 cells. C) KEGG pathway analysis of down-regulated genes in P1 cells. D) KEGG pathway analysis of up-regulated genes in P1 cells.
In addition, we performed KEGG analysis for DEGs to analyze signaling pathways that should be involved in chondrocyte dedifferentiation. The top pathways enriched were shown in Fig. 4C and D. On the one hand, the up-regulated DEGs in P1 involved many pathways, such as carbon metabolism, proteasome, DNA replication, cell cycle, biosynthesis of antibiotics, metabolic pathways as well as oxidative phosphorylation (Fig. 4D). Among them, the oxidative phosphorylation and metabolic pathways were first found to be highly correlated with the early stage of chondrocyte dedifferentiation. The down-regulated pathways in P1 cells mainly included TGF-beta signaling pathway, NF-κB signaling pathway, Wnt signaling pathway, rheumatoid arthritis, TNF signaling pathway, regulation of actin cytoskeleton, notch signaling pathway, MAPK signaling pathway, HIF-1 signaling pathway, focal adhesion, cytokine-cytokine receptor interaction, Rap1 signaling pathway, etc. (Fig. 4C).
Among the pathways down-regulated in P1 cells, firstly, several pathways previously proven to associated with chondrocyte dedifferentiation were enriched, including TGF-beta signaling pathway, MAPK signaling pathway, regulation of actin cytoskeleton, FoxO signaling pathway, HIF-1 signaling pathway [15,[29], [30], [31], [32]] (Fig. 4C). Among them, the FoxO signaling pathway and HIF-1 signaling pathway are considered to be related to the maintenance of articular cartilage homeostasis [15]. Secondly, other dysregulated pathways considered to be linked to OA were also enriched, such as NF-κB signaling pathway, rheumatoid arthritis, Wnt signaling pathway, HIF-1 signaling pathway, FoxO signaling pathway, and TNF signaling pathway [15,[33], [34], [35]] (Fig. 4C). Especially, NF-κB signaling pathway has previously been identified to be involved in OA [33,[35], [36], [37]]. These results indicated that chondrocyte dedifferentiation might exhibit similar molecular process with OA. Some molecules that targeting and inhibiting Wnt signaling pathway were considered as promising treatment strategy for OA [38,39]. Moreover, the Wnt signaling pathway has also been shown to be associated with chondrocyte dedifferentiation [40,41]. Therefore, we suspected that these associated molecules probably play essential roles in preventing occurrence of chondrocyte dedifferentiation, which needs to be further explored in the future. On the other hand, previous studies proven that NF-κB signaling pathway was cross-linked with Wnt signaling pathway during inflammation [42,43]. Immune and inflammation-related signaling pathways are activated in the dedifferentiated chondrocytes, and which is similar to OA [33], further suggesting the close relationship between chondrocyte dedifferentiation and OA. Besides, we found some new pathways that were related to chondrocytes dedifferentiation, including Rap1 signaling pathway and Ras signaling pathway (Fig. 4C). Since these pathways have not been previously explored in chondrocytes, they are promising targets for further studies to disclose the mechanism as well as to achieve inhibition of chondrocyte dedifferentiation.
3.3. Analysis of DEGs in P0 and P1 cells
Fig. 1B showed the expression of chondrocyte-associated genes. The increased expression of COL1A1 and COL1A2 as well as decreased expression of COL2A1, ACAN and SOX9 indicated the dedifferentiation of chondrocytes at P1. This result of up-regulated COL1A1 and COL1A2 in P1 cells was similar to the clinical observation that the fibrocartilage was often formed after chondrocytes transplantation [44,45]. Global analysis for gene expression alterations at transcriptome-level might provide new targets to study and depress chondrocyte dedifferentiation. In addition, the PFKFB3 and KDM6B associated with OA were also differentially expressed. PFKFB3 is a critical stimulator of glycolysis, and which is down-regulated in OA [46]. Besides, PFKFB3 also plays roles in glycolytic metabolism and endoplasmic reticulum stress of OA chondrocytes [34]. KDM6B is involved in many cellular processes, such as proliferation, differentiation, apoptosis, and senescence [47]. Furthermore, KDM6B plays a key role in tissue development, cancer, neurodegenerative diseases, aging, and inflammatory diseases [47]. A recent study had shown that KDM6B is involved not only in maintaining chondrogenesis, but also in maintaining chondrocyte homeostasis and the development of OA [48]. In our results, the expression of PFKFB3 and KDM6B showed similar down-regulated trend as those in OA [48,49] (Fig. 1B). We assumed that PFKFB3 and KDM6B is participated in chondrocyte dedifferentiation and they will probably be new markers of chondrocyte dedifferentiation. In addition, MEF2C and WNT5B, considered to be associated with hypertrophy [50,51], was up-regulated and down-regulated in P1 cells, respectively. Besides, α-actin, β-actin, and γ-action were significantly up-regulated in P1 chondrocytes (Fig. 1B). Actin is the main component of cytoskeleton, and whose up-regulation suggested the reconstruction of cytoskeleton during chondrocyte dedifferentiation.
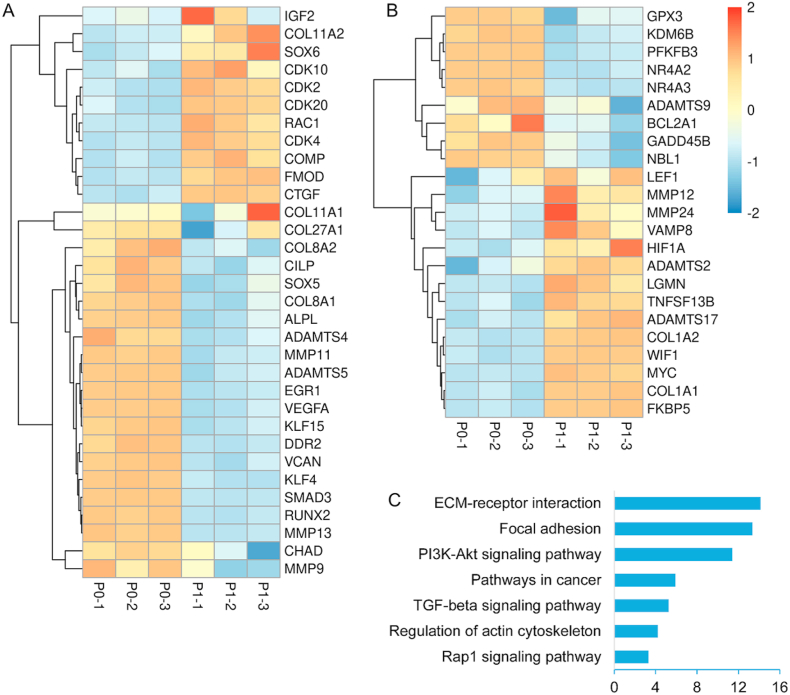
In addition to the genes mentioned above, other cartilage-related DEGs were also displayed in Fig. 5A. The up-regulated genes in P1 included collagen type XI alpha 1 (COL11A1), COL11A2, insulin like growth factor 2 (IGF2), fibromodulin (FMOD), cartilage oligomeric matrix protein (COMP), cellular communication network factor 2 (CTGF), SOX6, Rac Family Small GTPase 1 (RAC1), cyclin dependent kinase 4 (CDK4), CDK10, CDK2, and CDK20. The down-regulated genes in P1 included COL27A1, collagen type VIII alpha 1 (COL8A1) and COL8A2, EGR1, SOX5, SMAD family member 3 (SMAD3), versican (VCAN), cartilage intermediate layer protein (CILP), RUNX family transcription factor 2 (RUNX2), alkaline phosphatase, biomineralization associated (ALPL), vascular endothelial growth factor A (VEGFA), kruppel like factor 4 (KLF4), KLF15, a disintegrin metalloproteinase with thrombospondin motifs4 (ADAMTS4), ADAMTS5, MMP11, MMP13, MMP9, chondroadherin (CHAD), and discoidin domain receptor tyrosine kinase 2 (DDR2).
Fig. 5.
A) Heatmap of chondrocyte-related DEGs between P0 cells and P1 cells. B) Heatmap of OA-related DEGs between P0 cells and P1 cells. C) KEGG pathway analysis of chondrocyte-related DEGs between P0 and P1.
Among them, the genes of hyaline cartilage-specific ECM, such as COL2A1, were down-regulated during dedifferentiation, meanwhile the COL1A1 and COL1A2, associated with cartilage degradation were markedly up-regulated. This result indicated that chondrocytes could produce ECM in the early stage of dedifferentiation, but the composition of ECM changed significantly. Interestingly, the IGF2 that favors cell proliferation [52] was up-regulated in P1 cells; this phenomenon indicated that the changes in cellular phenotype, namely early dedifferentiation, might enhance cell proliferation. Furthermore, the cell cycle-related genes (such as CDK4 that regulates cell progression from G1 phase to S phase, CDK10 and CDK20 that are positively regulated during G2 or M phase) showed higher expression in P1 compared to P0 cells [[53], [54], [55]]. In addition, our data indicated that ADAMTS4, ADAMTS5, MMP11, and MMP13 (associated with matrix degradation) were dramatically down-regulated in P1 cells [19]. A previous study showed that mechanical stimulation can lead to the increase of matrix synthesis activity and upregulation of proteases [56]. Therefore, the down-regulation of ADAMTS4, ADAMTS5, ADAMTS9, MMP11 and MMP13 at P1 might be resulted from the loss of the dense environments in cartilage tissue. Moreover, the genes related to chondrogenesis such as SOX9 and SOX5 were down-regulated in P1 cells, while SOX6 was slightly up-regulated in P1 cells. A slight up-regulation of SOX6 in the early stages of chondrocyte dedifferentiation is also reported in previous studies [[57], [58], [59]]. EGR1 is engaged in the regulation of chondrocyte terminal differentiation and the catabolic response of pro-inflammatory cytokines [60]. To our knowledge, this is the first report on significant down-regulation of EGR1 in chondrocyte dedifferentiation. CTFG regulates matrix remodeling during bone development and plays an important role in regulating formation of cartilage as well as promotion of chondrocyte maturation and hypertrophic differentiation [61,62]. Compared with P0 cells, the higher expression of CTFG in P1 chondrocytes was consistent with great alterations of ECM in P1 cells. CHAD plays key roles not only in orchestrating crosstalk between chondrocytes and ECM, but also in regulating linkages between collagens and other ECM molecules [63]. Moreover, CHAD is implicated in ECM-receptor interaction and focal adhesion pathways [63]. Therefore, the down-regulation of CHAD in P1 cells should be an indicator for the changes in chondrocyte or ECM. CILP was another highly down-regulated gene in P1 cells, which was found to be present in the middle or intermediate zone of articular cartilage [64,65]. CILP plays a role in mediating growth factor signaling dynamics [63]. Further studies are needed to validate its functions during chondrocyte dedifferentiation. FMOD is small leucine-rich proteoglycan [66], and which likely plays a role in collagen fibrillogenesis [67]. Therefore, up-regulation of FMOD may aim to produce ECM in dedifferentiated chondrocytes (such as COL1). COMP is important in fibril formation and ECM assembly as well as in promoting attachment of chondrocytes to ECM [68,69]. Previous studies showed that COMP was down-regulated in fully dedifferentiated chondrocytes, however, interestingly, it was up-regulated in P1 chondrocytes. The exact mechanism under the phenomenon remains unclear. In limb chondrogenesis, VCAN is expressed in condensing mesenchyme, and then it is decreased expressed with cartilage formation [70]. Interestingly, the dramatic down-regulation of VCAN was observed in chondrocyte dedifferentiation.
The expressions of the genes associated with OA, were evaluated (Fig. 5B). The up-regulated genes in P1 cells included ADAMTS2, ADAMTS17, MMP12, MMP24, legumain (LGMN), MYC proto-oncogene (MYC), lymphoid enhancer binding factor 1 (LEF1), TNF superfamily member 13B (TNFSF13B), Wnt inhibitory factor 1 (WIF1), vesicle associated membrane protein 8 (VAMP8), COL1A2, and COL1A1; the down-regulated genes in P1 cells included NR4A2, NR4A3, ADAMTS9, KDM6B, PFKFB3, glutathione peroxidase 3 (GPX3), growth arrest and DNA damage inducible beta (GADD45B), FKBP prolyl isomerase 5 (FKBP5), hypoxia inducible factor 1 subunit alpha (HIF1A), neuroblastoma suppressor of tumorigenicity 1 (NBL1) as well as BCL2 related protein A1 (BCL2A1). Studies had shown that ADAMTS9 is associated with tumor suppression in many cancers [71,72]. Moreover, ADAMTS9 polymorphism could be used to predict the prognosis of OA [49]. Recent research showed that ADAMTS9 was a valuable biomarker to identify OA [34]. The decreased expression of ADAMTS9 in P1 was similar to that in OA chondrocytes versus healthy ones. GADD45B and GPX3 are involved in resistance to cellular stress [15]. Compare with normal cartilage, GADD45B and GPX3 was significantly down-regulated in OA [15]. The down-regulation of GADD45B and GPX3 was also observed in P1 cells. In addition, as mentioned above, PFKFB3 and KDM6B are generally considered to be down-regulated in OA. Similarly, we found for the first time that PFKFB3 and KDM6B was significantly related with chondrocyte dedifferentiation. Above results showed a similarity in expression of some genes between OA and the early dedifferentiated chondrocyte. Therefore, we believed early stage of chondrocyte dedifferentiation formed in monolayer culture might be serve as a simple pathological OA model for treatment studies. Besides, most of these genes have not been investigated in chondrocyte dedifferentiation, and their functions should be explored in future.
We performed KEGG pathway analysis for the cartilage-related DEGs. Fig. 5C showed the main pathways that were enriched, such as ECM-receptor interaction, focal adhesion, PI3K-Akt signaling pathway, and TGF-beta signaling pathway. ECM-receptor interaction was the most enriched pathway, which indicated that ECM (including COL1, COL2, ACAN, etc.) has undergone significant changes in the early stages of dedifferentiation. Actually, this result was also verified by significantly differentially expressed ECM genes, such as COL1, COL2. Focal adhesion symbolized the disorder of chondrocyte phenotype and the occurrence of dedifferentiation [73]. Previous studies proved that PI3K-Akt signaling pathway was highly enriched in OA and even considered as a potential therapeutic target for OA [74,75]. Interestingly, the PI3K-Akt signaling pathway was highly enriched, further confirming the similarity of dedifferentiated chondrocytes with OA at mRNA level.
3.4. qPCR validation of representative genes
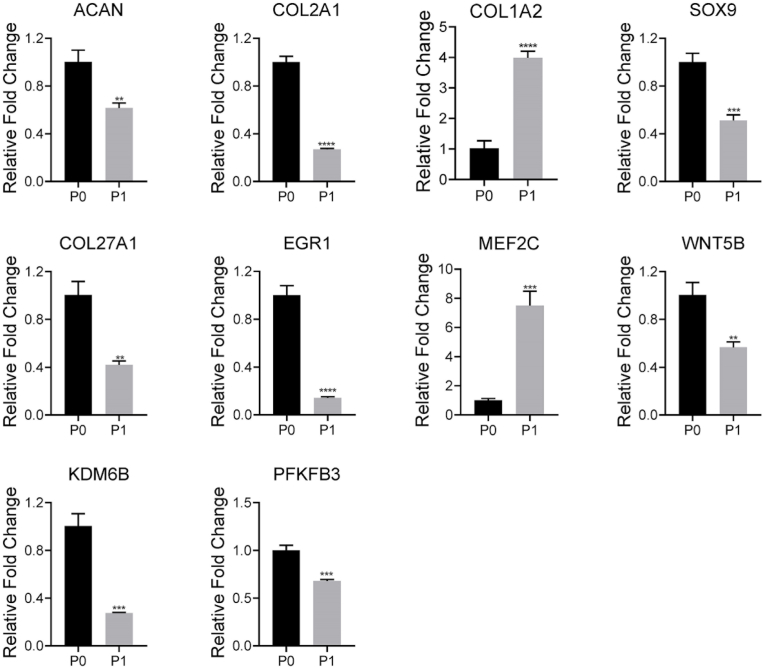
In order to verify the sequencing results, a series of representative genes, including COL1A2, COL2A1, SOX9, ACAN, EGR1, MEF2C, COL27A1, WNT5B, PFKBF3 and KDM6B, were selected for validation with qPCR (Figs. 1C and 6). We found that the relative expression levels of these genes obtained by qPCR were consistent with the sequencing results, confirming the accuracy of the sequencing data. Typically, the expression of COL2A1 was slightly increased and then severely decreased from P0 to monolayer culture and P1, respectively. Nevertheless, the dramatic increase in expression of COL1A2 was observed from P0 to monolayer culture, and as expected, which maintained high expression in P0. The similar significant changes were also detected in other potential markers.
Fig. 6.
qPCR analysis for representative gene expressions in P0 and P1 chondrocytes; * indicated p < 0.05, ** indicated p < 0.005, *** indicated p < 0.0005, and **** indicated p < 0.0001.
3.5. Immunofluorescence staining
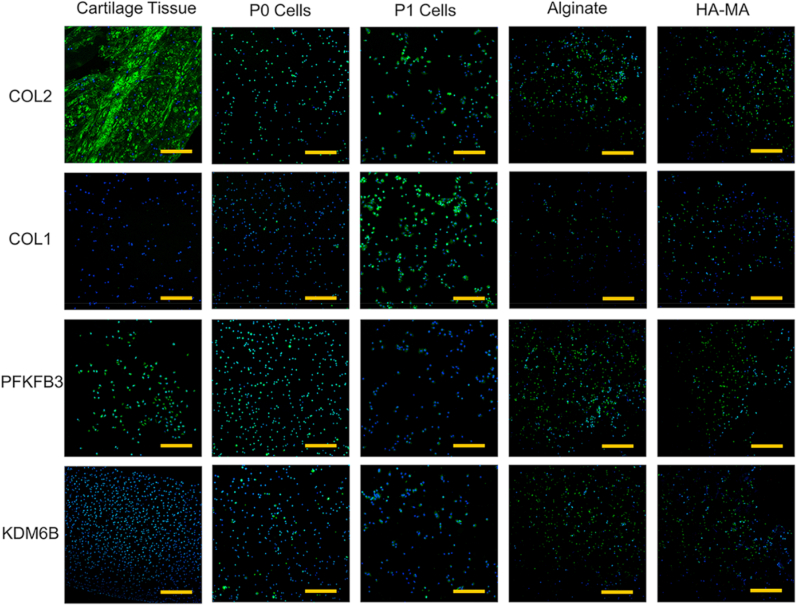
Alginate and HA-based hydrogels are considered as ideal scaffolds for maintaining chondrocyte phenotype [[76], [77], [78], [79]], and which were fabricated for determining the newly identified potential markers of chondrocyte dedifferentiation. As typical representatives of potential markers, KDM6B and PFKFB3 as well as classical chondrocyte markers (i.e. COL2, COL1) were evaluated via immune-fluorescent staining of native cartilage, P0 cells, P1 cells, chondrocytes-laden alginate and HA-MA constructs, respectively (Fig. 7). As expected, a dense staining of COL2 was observed in native cartilage tissues, meanwhile comparable COL2 staining was detected between P0 cells, alginate constructs, and HA-MA constructs. Interestingly, COL2 was significantly produced over 5 days of monolayer culture (Fig. S1). However, most P1 chondrocytes were negatively stained by COL2, which was in agreement with the results of qPCR showed in Fig. 6. This result suggested the occurrence of gradual dedifferentiation of those chondrocytes. COL1, an established marker of chondrocyte dedifferentiation, showed significantly positive staining in P1 chondrocytes than that in native cartilage tissue, P0, alginate and HA-MA constructs. At the same time, COL1 was also densely stained after 5 days of monolayer culture (Fig. S1), which also indicated the start of dedifferentiation during monolayer-culture of P0 chondrocytes. The increased COL1 expression and decreased COL2 expression from P0 to P1 demonstrated further dedifferentiation of chondrocytes after one passage of subculture. Besides, the negative staining of COL1 and positive staining of COL2 in HA-MA and alginate constructs indicated their capability to maintain the phenotype of chondrocytes.
Fig. 7.
Immunofluorescence staining of cartilage tissue, P0 cells, P1 cells, alginate constructs, and HA-MA constructs for COL2, COL1, PFKFB3, and KDM6B, respectively. The nucleus was counterstained with DAPI (blue). Magnification was 100x. Scale bars were 200 μm.
Next, the typical potential markers, namely KDM6B and PFKFB3, were subjected to immunofluorescence staining. Compared with P1 cells, both KDM6B and PFKFB3 have a higher expression in cartilage tissue, P0 cells, alginate constructs, and HA-MA constructs, respectively. These observations consisted with the results of qPCR (Fig. 1, Fig. 6). Our results showed that in the early stages of chondrocyte dedifferentiation (P1), PFKFB3 and KDM6B exhibited a dramatic down-regulation at the mRNA and protein levels. Interestingly, similar to observation for COL2, both PFKFB3 and KDM6B was densely stained after 5 days of monolayer culture (Fig. S1). PFKFB3 and KDM6B should be important elements for maintaining or indicating the normal phenotype of chondrocytes, so the roles of PFKFB3 and KDM6B in chondrocyte dedifferentiation should be further explored. In addition, these results also suggested the DEGs should be significant, and further studies should be carried out in future.
3.6. qPCR validation of potential marker genes
The chondrocytes cultured in alginate and HA-MA hydrogel were used as the control to identify the potential marker genes of chondrocyte dedifferentiation by qPCR (Figs. 1C and 8). Firstly, as a canonical marker gene of chondrocyte dedifferentiation, the COL1A2 was sharply up-regulated in monolayer culture, indicating the occurrence of dedifferentiation. Interestingly, the chondrocytes in hydrogel constructs also exhibited higher expression of COL1A2 than P0 cells, albeit significantly lower than that in monolayer culture. This phenomenon might be attributed to the changes of growth environments. Besides, we found that the COL1 was lower expressed in alginate constructs compared with HA-MA constructs. Compared to P0 cells, the chondrocytes cultured in hydrogel constructs expressed more COL2A1 and ACAN (classical chondrocyte markers). Meanwhile, as expected, much higher expression of COL2A1 was observed in hydrogel constructs than that in monolayer culture. Furthermore, intriguingly, ACAN was significantly higher expressed in alginate constructs than that in HA-MA constructs. Alginate constructs might provide a more favorable environment for producing cartilage ECM compared to HA-MA constructs. Interestingly, we found that the expression of COL2A1 was increased from P0 chondrocytes to the cells by 5 days of monolayer culture, but in contrast, it exhibited drastically decreased expression in P1 cells compared to those in monolayer culture. Hence, cell passaging could evidently enhance dedifferentiation of chondrocytes, and meanwhile, the comparison between P1 cells and the chondrocytes cultured in gel constructs should be more beneficial to identify potential markers of chondrocyte dedifferentiation. In addition, interestingly, some differences in the expression of marker genes were observed between the chondrocytes encapsulated in alginate constructs and HA-MA constructs (Figs. 1C and 8). In the future, it is significant to further explore the influence of hydrogels prepared from different materials on the maintenance of chondrocyte phenotype.
Fig. 8.
qPCR analysis for expression of representative genes in P0 and monolayer cultured chondrocytes as well as HA-MA gel and alginate gel constructs, respectively. * indicated p < 0.05, ** indicated p < 0.005, *** indicated p < 0.0005, and **** indicated p < 0.0001.
As a key regulator of chondrogenesis, SOX9 exhibited comparable expression for the chondrocytes cultured in P0, monolayer culture, and hydrogel constructs. Nevertheless, average expression of SOX9 is lower in 2D monolayer culture than 3D culture, and this trend became more obvious with sub-culture (Fig. 6). COL27A1 is one member of the family of fibrillar collagens, and is considered as a marker of cartilage development [80,81]. Similar to COL2A1, the up-regulated COL27A1 was also observed after culture through the three methods. EGR1 is engaged in the regulation of chondrocytes terminal differentiation and the catabolic response of pro-inflammatory cytokines [60]. Interestingly, it was dramatically down-regulated in the three culture conditions, especially for chondrocytes cultured in monolayer. The underlying mechanism of sharp down-regulation of EGR1 was not reported till now. EGR1 probably is an attractive target for investigating chondrocyte dedifferentiation. Significantly, we observed obvious up-regulation of MEF2C as well as down-regulation of WNT5B in the chondrocytes cultured in monolayer than those in gel constructs, and furthermore, the trends were enhanced in P1 chondrocytes (Fig. 1, Fig. 8).
KDM6B could regulate cartilage development and homeostasis [48]. Significantly, it was remarkably up-regulated in hydrogel constructs, exhibiting the similar tendency with ECM components, namely COL2A1, ACAN. Moreover, similar to COL2A1, the significant down-regulation of KDM6B was observed in P1 cells (Fig. 1C). Therefore, we speculated that KDM6B might be related to phenotype maintenance of chondrocytes, which should be investigated in further researches. In addition, the expression of PFKFB3 was maintained in hydrogel constructs. However, PFKFB3 was greatly down-regulated in monolayer culture, and this trend was enhanced after sub-culture (Fig. 1C). Taken together, above typical DEGs from RNA-seq, such as MEF2C, WNT5B, KDM6B and PFKFB3, were confirmed using the chondrocytes cultured in hydrogels, and which probably serve as potential markers of chondrocyte dedifferentiation.
4. Conclusion
Chondrocyte is a promising therapeutic cell for cartilage repair, however, it undergoes dedifferentiation during 2D expansion in vitro and clinical applications. Considering the great advantages of hydrogel in maintaining chondrocyte phenotype, in this study, alginate- and HA-based hydrogel was used as an aid to identify new marker genes and potential therapeutic targets of chondrocyte dedifferentiation by combining RNA-seq. We comprehensively analyzed the changes in gene expression profile during chondrocyte dedifferentiation. The differentially expressed genes and enriched pathways were presented, which serve as a database for further investigation of chondrocyte dedifferentiation. Significantly, several new marker genes, such as MEF2C, PFKFB3, EGR1, KDM6B, were identified with the assistance of hydrogel scaffolds. This work expanded the application of hydrogel in studying chondrocyte dedifferentiation, and also provided potential pathways and targets to prevent chondrocyte dedifferentiation.
CRediT authorship contribution statement
Yang Ling: Investigation, Methodology, Data curation, Writing – original draft. Weiyuan Zhang: Data curation, Writing – original draft. Peiyan Wang: Investigation. Wanhua Xie: Software, Data curation. Wei Yang: Investigation. Dong-An Wang: Conceptualization, Supervision, Writing – review & editing, Project administration. Changjiang Fan: Conceptualization, Supervision, Writing – review & editing, Project administration.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
The work is financially supported by the China Postdoctoral Science Foundation (Grant No. 2020M671993), Start-Up Grant for Professor (SGP 9380099 to D.-A.W.), City University of Hong Kong, and the National Natural Science Foundation of China, China (Grant No. 51973180 and 21604045).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2021.02.018.
Contributor Information
Dong-An Wang, Email: dwang229@cityu.edu.hk.
Changjiang Fan, Email: cjfan@qdu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Blagojevicy M., Jinksy C., Jefferyz A., Jordany K.P. Risk factors for onset of osteoarthritis of the knee in older adults: a systematic review and meta-analysis. Osteoarthritis Cartilage. 2010;18(1):24–33. doi: 10.1016/j.joca.2009.08.010. https://doi:10.1016/j.joca.2009.08.010 [DOI] [PubMed] [Google Scholar]
- 2.Kim C., Linsenmeyer K.D., Vlad S.C., Guermazi A., Felson D.T. Prevalence of radiographic and symptomatic hip osteoarthritis in an urban United States community: the framingham osteoarthritis study. Arthritis Rheum. 2014;66(11):3013–3017. doi: 10.1002/art.38795. https://doi: 10.1002/art.38795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Demoor M., Ollitrault D., Gomez-Leduc T., Bouyoucef M., Hervieu M., Fabre H., Lafont J., Denoix J.M., Audigie F., Mallein-Gerin F., Legendre F., Galera P. Cartilage tissue engineering: molecular control of chondrocyte differentiation for proper cartilage matrix reconstruction. Bba-Gen. Subjects. 2014;1840(8):2414–2440. doi: 10.1016/j.bbagen.2014.02.030. https://doi: 10.1016/j.bbagen.2014.02.030 [DOI] [PubMed] [Google Scholar]
- 4.Brittberg M., Lindahl A., Nilsson A., Ohlsson C., Isaksson O., Peterson L. Treatment of deep cartilage defects in the knee with autologous chondrocyte transplantation. N. Engl. J. Med. 1994;331(14):889–895. doi: 10.1056/NEJM199410063311401. https://doi: 10.1056/NEJM199410063311401 [DOI] [PubMed] [Google Scholar]
- 5.Benya P.D., Padilla S.R., Nimni M.E. Independent regulation of collagen types by chondrocytes during the loss of differentiated function in culture. Cell. 1978;15(4):1313–1321. doi: 10.1016/0092-8674(78)90056-9. https://doi: 10.1016/0092-8674(78)90056-9 [DOI] [PubMed] [Google Scholar]
- 6.Kang S.W., Yoo S.P., Kim B.S. Effect of chondrocyte passage number on histological aspects of tissue-engineered cartilage. Bio Med. Mater. Eng. 2007;17(5):269–276. https://doi: 10.1080/10731190701784823 [PubMed] [Google Scholar]
- 7.Schulze-Tanzil G., Souza P., Castrejon H.V., John T., Merker H.J., Scheid A., Shakibaei M. Redifferentiation of dedifferentiated human chondrocytes in high-density cultures. Cell Tissue Res. 2002;308(3):371–379. doi: 10.1007/s00441-002-0562-7. https://doi: 10.1007/s00441-002-0562-7 [DOI] [PubMed] [Google Scholar]
- 8.Balakrishnan B., Banerjee R. Biopolymer-based hydrogels for cartilage tissue engineering. Chem. Rev. 2011;111(8):4453–4474. doi: 10.1021/cr100123h. https://doi: 10.1021/cr100123h [DOI] [PubMed] [Google Scholar]
- 9.Mayne R., Vail M.S., Miller M.E.J. Changes in type of collagen synthesized as clones of chick chondrocytes grow and eventually lose division capacity. P. Natl. Acad. Sci. USA. 1976;73(5):1674–1678. doi: 10.1073/pnas.73.5.1674. https://doi: 10.1073/pnas.73.5.1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D Mark K.V., Gauss V., D Mark H.V., Müller P. Relationship between cell shape and type of collagen synthesised as chondrocytes lose their cartilage phenotype in culture. Nature. 1977;267(5611):531–532. doi: 10.1038/267531a0. https://doi: 10.1038/267531a0 [DOI] [PubMed] [Google Scholar]
- 11.Chua K.H., Aminuddin B.S., Fuzina N.H., Ruszymah B.H. Insulin-transferrin-selenium prevent human chondrocyte dedifferentiation and promote the formation of high quality tissue engineered human hyaline cartilage. Eur. Cell. Mater. 2005;9(9):58–67. doi: 10.22203/ecm.v009a08. https://doi: 10.22203/eCM.v009a08 [DOI] [PubMed] [Google Scholar]
- 12.Kuettner K., Pauli B., Gall G., Memoli V., Schenk R. Synthesis of cartilage matrix by mammalian chondrocytes in vitro: isolation, culture characteristics, and morphology. J. Cell Biol. 1982;93(3):743–750. doi: 10.1083/jcb.93.3.743. https://doi: 10.1083/jcb.93.3.743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mandl E.W., Veen S.W., Verhaar J.A., Osch G.J. Multiplication of human chondrocytes with low seeding densities accelerates cell yield without losing redifferentiation capacity. Tissue Eng. 2004;10(1–2):109–118. doi: 10.1089/107632704322791754. https://doi: 10.1089/107632704322791754 [DOI] [PubMed] [Google Scholar]
- 14.Elima K., Vuorio E. Expression of mRNAs for collagens and other matrix components in dedifferentiating and redifferentiating human chondrocytes in culture. FEBS Lett. 1990;258(2):195–198. doi: 10.1016/0014-5793(89)81651-5. https://doi: 10.1016/0014-5793(89)81651-5 [DOI] [PubMed] [Google Scholar]
- 15.Fisch K.M., Gamini R., Alvarez-Garcia O., Akagi R., Saito M., Muramatsu Y., Sasho T., Koziol J.A., Su A.I., Lotz M.K. Identification of transcription factors responsible for dysregulated networks in human osteoarthritis cartilage by global gene expression analysis. Osteoarthritis Cartilage. 2018;26(11):1531–1538. doi: 10.1016/j.joca.2018.07.012. https://doi: 10.1016/j.joca.2018.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L., Yu F., Zheng L., Wang R., Yan W., Wang Z., Xu J., Wu J., Shi D., Zhu L. Natural hydrogels for cartilage regeneration: modification, preparation and application. J. Orthop. Transl. 2019;17:26–41. doi: 10.1016/j.jot.2018.09.003. https://doi: 10.1016/j.jot.2018.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schindler O.S. Current concepts of articular cartilage repair. Acta Orthop. Belg. 2011;77(6):709. https://doi: 10.1016/j.mporth.2009.05.002 [PubMed] [Google Scholar]
- 18.Menzel E.J., Farr C. Hyaluronidase and its substrate hyaluronan: biochemistry, biological activities and therapeutic uses. Canc. Lett. 1998;131(1):3–11. doi: 10.1016/s0304-3835(98)00195-5. https://doi: 10.1016/s0304-3835(98)00195-5 [DOI] [PubMed] [Google Scholar]
- 19.Zhang W.Y., Xia Y.J., Ling Y., Yang W., Dong Z.X., Wang D.A., Fan C.J. A transcriptome sequencing study on genome-wide gene expression differences of 3D cultured chondrocytes in hydrogel scaffolds with different gel density. Macromol. Biosci. 2020;20(5) doi: 10.1002/mabi.202000028. https://doi: 10.1002/mabi.202000028 [DOI] [PubMed] [Google Scholar]
- 20.Bidarra S.J., Barrias C.C., Granja P.L. Injectable alginate hydrogels for cell delivery in tissue engineering. Acta Biomater. 2014;10(4):1646–1662. doi: 10.1016/j.actbio.2013.12.006. https://doi: doi:10.1016/j.actbio.2013.12.006 [DOI] [PubMed] [Google Scholar]
- 21.Miao Z., Lu Z., Wu H., Liu H., Li M., Lei D., Zheng L., Zhao J. Collagen, agarose, alginate, and matrigel hydrogels as cell substrates for culture of chondrocytes in vitro: a comparative study. J. Cell. Biochem. 2017;119(10):7924–7933. doi: 10.1002/jcb.26411. https://doi: 10.1002/jcb.26411 [DOI] [PubMed] [Google Scholar]
- 22.Fan C.J., Ling Y., Deng W., Xue J., Sun P., Wang D.A. A novel cell encapsulatable cryogel (CECG) with macro-porous structures and high permeability: a three-dimensional cell culture scaffold for enhanced cell adhesion and proliferation. Biomed. Mater. 2019;14(5) doi: 10.1088/1748-605X/ab2efd. https://doi: 10.1088/1748-605X/ab2efd [DOI] [PubMed] [Google Scholar]
- 23.Leach J.B., Bivens K.A., Patrick C.W., Schmidt C.E. Photocrosslinked hyaluronic acid hydrogels: natural, biodegradable tissue engineering scaffolds. Biotechnol. Bioeng. 2003;82(5):578–589. doi: 10.1002/bit.10605. https://doi: 10.1002/bit.10605 [DOI] [PubMed] [Google Scholar]
- 24.Fan C.J., Wang D.A. Effects of permeability and living space on cell fate and neo‐tissue development in hydrogel‐based scaffolds: a study with cartilaginous model. Macromol. Biosci. 2015;15(4):535–545. doi: 10.1002/mabi.201400453. https://doi: 10.1002/mabi.201400453 [DOI] [PubMed] [Google Scholar]
- 25.Tan G.K., Dinnes D.L., Myers P.T., Cooper-White J.J. Effects of biomimetic surfaces and oxygen tension on redifferentiation of passaged human fibrochondrocytes in 2D and 3D cultures. Biomaterials. 2011;32(24):5600–5614. doi: 10.1016/j.biomaterials.2011.04.033. https://doi: 10.1016/j.biomaterials.2011.04.033 [DOI] [PubMed] [Google Scholar]
- 26.Su T., Zhang M., Zeng Q., Pan W., Huang Y., Qian Y., Dong W., Qi X., Shen J. Mussel-inspired agarose hydrogel scaffolds for skin tissue engineering. Bioact. Mater. 2021;6(3):579–588. doi: 10.1016/j.bioactmat.2020.09.004. https://doi: 10.1016/j.bioactmat.2020.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qi X., Su T., Zhang M., Tong X., Pan W., Zeng Q., Zhou Z., Shen L., He X., Shen J. Macroporous hydrogel scaffolds with tunable physicochemical properties for tissue engineering constructed using renewable polysaccharides. ACS Appl. Mater. Interfaces. 2020;12(11):13256–13264. doi: 10.1021/acsami.9b20794. https://doi: 10.1021/acsami.9b20794 [DOI] [PubMed] [Google Scholar]
- 28.Qi X., Su T., Zhang M., Tong X., Pan W., Zeng Q., Shen J. Sustainable, flexible and biocompatible hydrogels derived from microbial polysaccharides with tailorable structures for tissue engineering. Carbohydr. Polym. 2020;237:116160. doi: 10.1016/j.carbpol.2020.116160. https://doi: 10.1016/j.carbpol.2020.116160 [DOI] [PubMed] [Google Scholar]
- 29.Blaise R., Mahjoub M., Salvat C., Barbe U., Brou C., Corvol M., Savouret J., Rannou F., Berenbaum F., Bausero P. Involvement of the notch pathway in the regulation of matrix metalloproteinase 13 and the dedifferentiation of articular chondrocytes in murine cartilage. Arthritis Rheum. 2009;60(2):428–439. doi: 10.1002/art.24250. https://doi: 10.1002/art.24250 [DOI] [PubMed] [Google Scholar]
- 30.Li G., Song X., Li R., Sun L., Gong X., Chen C., Yang L. Zyxin-involved actin regulation is essential in the maintenance of vinculin focal adhesion and chondrocyte differentiation status. Cell proliferat. 2019;52(1) doi: 10.1111/cpr.12532. https://doi: 10.1111/cpr.12532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eo S., Kim D., Choi S., Kim H., Kim S. PEP-1-SIRT2 causes dedifferentiation and COX-2 expression via the MAPK pathways in rabbit articular chondrocytes. Exp. Cell Res. 2015;339(2):351–359. doi: 10.1016/j.yexcr.2015.09.001. https://doi: 10.1016/j.yexcr.2015.09.001 [DOI] [PubMed] [Google Scholar]
- 32.Ahn J., Kumar H., Cha B., Park S., Arai Y., Han I., Park S., Lee S. AIMP1 downregulation restores chondrogenic characteristics of dedifferentiated/degenerated chondrocytes by enhancing TGF-β signal. Cell Death Dis. 2016;7 doi: 10.1038/cddis.2016.17. https://doi: 10.1038/cddis.2016.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen C., Bao G.F., Xu G., Sun Y., Cui Z.M. Altered Wnt and NF-kappaB signaling in facet joint osteoarthritis: insights from RNA deep sequencing. Tohoku J. Exp. Med. 2018;245(1):69–77. doi: 10.1620/tjem.245.69. https://doi: 10.1620/tjem.245.69 [DOI] [PubMed] [Google Scholar]
- 34.Li Z., Zhang R., Yang X., Zhang D., Li B., Zhang D., Li Q., Xiong Y. Analysis of gene expression and methylation datasets identified ADAMTS9, FKBP5, and PFKBF3 as biomarkers for osteoarthritis. J. Cell. Physiol. 2019;234(6):8908–8917. doi: 10.1002/jcp.27557. https://doi: 10.1002/jcp.27557 [DOI] [PubMed] [Google Scholar]
- 35.Wu L., Huang X., Li L., Huang H., Xu R., Luyten W. Insights on biology and pathology of HIF-1α/-2α, TGFα/BMP, Wnt/β-catenin, and NF-κB pathways in osteoarthritis. Curr. Pharmaceut. Des. 2012;18(22):3293–3312. doi: 10.2174/1381612811209023293. https://doi: 10.2174/1381612811209023293 [DOI] [PubMed] [Google Scholar]
- 36.Corr M. Wnt/beta-catenin signaling in the pathogenesis of osteoarthritis. Nat. Clin. Pract. Rheumatol. 2008;4(10):550–556. doi: 10.1038/ncprheum0904. https://doi: 10.1038/ncprheum0904 [DOI] [PubMed] [Google Scholar]
- 37.Zhou Y., Wang T., Hamilton J.L., Chen D. Wnt/β-catenin signaling in osteoarthritis and in other forms of arthritis. Curr. Rheumatol. Rep. 2017;19(9):53. doi: 10.1007/s11926-017-0679-z. https://doi: 10.1007/s11926-017-0679-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blom A.B., Lent P.L., Kraan P.M., Berg W.B. To seek shelter from the Wnt in osteoarthritis? Wnt-signaling as a target for osteoarthritis therapy. Curr. Drug Targets. 2010;11(5):620–629. doi: 10.2174/138945010791011901. https://doi: 10.2174/138945010791011901 [DOI] [PubMed] [Google Scholar]
- 39.Akira T., Bisei O., Mikako I., Akio M., Tadahiro S., Naoki I., Kinji O., Liu C. Verapamil protects against cartilage degradation in osteoarthritis by inhibiting Wnt/β-catenin signaling. PloS One. 2014;9(3) doi: 10.1371/journal.pone.0092699. https://doi: 10.1371/journal.pone.0092699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Öztürk E., Despot-Slade E., Pichler M., Zenobi-Wong M. RhoA activation and nuclearization marks loss of chondrocyte phenotype in crosstalk with Wnt pathway. Exp. Cell Res. 2017;360(2):113–124. doi: 10.1016/j.yexcr.2017.08.033. https://doi: 10.1016/j.yexcr.2017.08.033 [DOI] [PubMed] [Google Scholar]
- 41.Luo S., Shi Q., Zha Z., Yao P., Lin H., Liu N., Wu H., Sun S. Inactivation of Wnt/β-catenin signaling in human adipose-derived stem cells is necessary for chondrogenic differentiation and maintenance. Biomed. Pharmacother. 2013;67(8):819–824. doi: 10.1016/j.biopha.2013.03.008. https://doi: 10.1016/j.biopha.2013.03.008 [DOI] [PubMed] [Google Scholar]
- 42.Ma B., Hottiger M.O. Crosstalk between Wnt/β-catenin and NF-κB signaling pathway during inflammation. Front. Immunol. 2016;7:378. doi: 10.3389/fimmu.2016.00378. https://doi: 10.3389/fimmu.2016.00378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma B., Fey M., Hottiger M.O. WNT/β-catenin signaling inhibits CBP-mediated RelA acetylation and expression of proinflammatory NF-κB target genes. J. Cell Sci. 2015;128(14):2430–2436. doi: 10.1242/jcs.168542. https://doi: 10.1242/jcs.168542 [DOI] [PubMed] [Google Scholar]
- 44.Demoor M., Ollitrault D., Gomez-Leduc T., Bouyoucef M., Hervieu M., Fabre H., Lafont J., Denoix J.M., Audigié F., Mallein-Gerin F., Legendre F., Galera P. Cartilage tissue engineering: molecular control of chondrocyte differentiation for proper cartilage matrix reconstruction. Bba-Gen. Subjects. 2014;1840(8):2414–2440. doi: 10.1016/j.bbagen.2014.02.030. https://doi: 10.1016/j.bbagen.2014.02.030 [DOI] [PubMed] [Google Scholar]
- 45.Shapiro F., Koide S., Glimcher M.J. Cell origin and differentiation in the repair of full-thickness defects of articular cartilage. J. Bone Joint Surg. Am. 1993;75(4):532–553. doi: 10.2106/00004623-199304000-00009. https://doi: 10.2106/00004623-199304000-00009 [DOI] [PubMed] [Google Scholar]
- 46.Qu J., Lu D., Guo H., Miao W., Wu G., Zhou M. PFKFB3 modulates glycolytic metabolism and alleviates endoplasmic reticulum stress in human osteoarthritis cartilage. Clin. Exp. Pharmacol. Physiol. 2016;43(3):312–318. doi: 10.1111/1440-1681.12537. https://doi: 10.1111/1440-1681.12537 [DOI] [PubMed] [Google Scholar]
- 47.Burchfield J.S., Li Q., Wang H.Y., Wang R.F. JMJD3 as an epigenetic regulator in development and disease. Int. J. Biochem. Cell Biol. 2015;67:148–157. doi: 10.1016/j.biocel.2015.07.006. https://doi: 10.1016/j.biocel.2015.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dai J., Yu D., Wang Y., Chen Y., Sun H., Zhang X., Zhu S., Pan Z., Heng B.C., Zhang S., Ouyang H. Kdm6b regulates cartilage development and homeostasis through anabolic metabolism. Ann. Rheum. Dis. 2017;76(7):1295–1303. doi: 10.1136/annrheumdis-2016-210407. https://doi: 10.1136/annrheumdis-2016-210407 [DOI] [PubMed] [Google Scholar]
- 49.Gok K., Cemeroglu O., Cakirbay H., Gunduz E., Acar M., Cetin E.N., Gunduz M., Demircan K. Relationship between cytosine-adenine repeat polymorphism of ADAMTS9 gene and clinical and radiologic severity of knee osteoarthritis. Int. J. Rheum. Dis. 2016;21(4):821–827. doi: 10.1111/1756-185X.12849. https://doi: 10.1111/1756-185X.12849 [DOI] [PubMed] [Google Scholar]
- 50.Bradley E.W., Drissi M.H. Wnt5b regulates mesenchymal cell aggregation and chondrocyte differentiation through the planar cell polarity pathway. J. Cell. Physiol. 2011;226(6):1683–1693. doi: 10.1002/jcp.22499. https://doi: 10.1002/jcp.22499 [DOI] [PubMed] [Google Scholar]
- 51.Diederichs S., Tonnier V., März M., Dreher S.I., Geisbüsch A., Richter W. Regulation of Wnt5A and Wnt11 during MSC in vitro chondrogenesis: Wnt inhibition lowers BMP and hedgehog activity, and reduces hypertrophy. Cell. Mol. Life Sci. 2019;76(19):3875–3889. doi: 10.1007/s00018-019-03099-0. https://doi:10.1007/s00018-019-03099-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Müller S., Lindemann S., Gigout A. Effects of sprifermin, IGF1, IGF2, BMP7, or CNP on bovine chondrocytes in monolayer and 3D Culture. J. Orthop. Res. 2020;38(3):653–662. doi: 10.1002/jor.24491. https://doi:10.1002/jor.24491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Q., Ma J., Lu Y., Zhang S., Huang J., Chen J., Bei J.X., Yang K., Wu G., Huang K., Chen J. CDK20 interacts with KEAP1 to activate NRF2 and promotes radiochemoresistance in lung cancer cells. Oncogene. 2017;36(37):5321–5330. doi: 10.1038/onc.2017.161. https://doi: 10.1038/onc.2017.161 [DOI] [PubMed] [Google Scholar]
- 54.Zehra M., Mushtaq S., Ghulam Musharraf S., Ghani R., Ahmed N. Association of cyclin dependent kinase 10 and transcription factor 2 during human corneal epithelial wound healing in vitro model. Sci. Rep. 2019;9(1):11802. doi: 10.1038/s41598-019-48092-6. https://doi: 10.1038/s41598-019-48092-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lemmens B., Lindqvist A. DNA replication and mitotic entry: a brake model for cell cycle progression. J. Cell Biol. 2019;218(12):3892–3902. doi: 10.1083/jcb.201909032. https://doi: 10.1083/jcb.201909032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lomas C., Tang X.D., Chanalaris A., Saklatvala J., Vincent T.L. Cyclic mechanical load causes global translational arrest in articular chondrocytes: a process which is partially dependent upon PKR phosphorylation. Eur. Cell. Mater. 2011;22:78–89. doi: 10.22203/ecm.v022a14. https://doi: 10.22203/ecm.v022a14 [DOI] [PubMed] [Google Scholar]
- 57.Liu C., Lefebvre V. The transcription factors SOX9 and SOX5/SOX6 cooperate genome-wide through super-enhancers to drive chondrogenesis. Nucleic Acids Res. 2015;43(17):183–203. doi: 10.1093/nar/gkv688. https://doi: 10.1093/nar/gkv688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tan Z., Niu B., Tsang K., Melhado I., Ohba S., He X., Huang Y., Wang C., McMahon A., Jauch R., Chan D., Zhang M., Cheah K. Synergistic co-regulation and competition by a SOX9-GLI-FOXA phasic transcriptional network coordinate chondrocyte differentiation transitions. PLoS Genet. 2018;14(4) doi: 10.1371/journal.pgen.1007346. https://doi: 10.1371/journal.pgen [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park J., Yi S., Kim H., Kim S., Kim J., Park K. Construction of PLGA nanoparticles coated with polycistronic SOX5, SOX6, and SOX9 genes for chondrogenesis of human mesenchymal stem cells. ACS Appl. Mater. Interfaces. 2017;9(2):1361–1372. doi: 10.1021/acsami.6b15354. https://doi: 10.1021/acsami.6b15354 [DOI] [PubMed] [Google Scholar]
- 60.Florian D., Mohamed K. The role of early growth response 1 (EGR1) in brain plasticity and neuropsychiatric disorders. Front. Behav. Neurosci. 2017;11:35. doi: 10.3389/fnbeh.2017.00035. https://doi: 10.3389/fnbeh.2017.00035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nishida T., Nakanishi T., Asano M., Shimo T., Takigawa M. Effects of CTGF/Hcs24, a product of a hypertrophic chondrocyte-specific gene, on the proliferation and differentiation of chondrocytes in culture. J. Cell. Physiol. 2000;184(2):197–206. doi: 10.1002/1097-4652(200008)184:2<197::AID-JCP7>3.0.CO;2-R. https://doi: 10.1210/endo.141.1.7267 [DOI] [PubMed] [Google Scholar]
- 62.Frahs S.M., Reeck J.C., Yocham K.M., Frederiksen A., Fujimoto K., Scott C.M., Beard R.S., Brown R.J. Prechondrogenic ATDC5 cell attachment and differentiation on graphene foam; modulation by surface functionalization with fibronectin. ACS Appl. Mater. Interfaces. 2019;11(13):41906–41924. doi: 10.1021/acsami.9b14670. https://doi: 10.1021/acsami.9b14670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grogan S.P., Duffy S.F., Pauli C., Lotz M.K., D'Lima D.D. Gene expression profiles of the meniscus avascular phenotype: a guide for meniscus tissue engineering. J. Orthop. Res. 2018;36(7):1947–1958. doi: 10.1002/jor.23864. https://doi: 10.1002/jor.23864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lorenzo P., Bayliss M.T., Heinegard D. A novel cartilage protein (CILP) present in the mid-zone of human articular cartilage increases with age. J. Biol. Chem. 1998;273(36):23463–23468. doi: 10.1074/jbc.273.36.23463. https://doi: 10.1074/jbc.273.36.23463 [DOI] [PubMed] [Google Scholar]
- 65.Bernardo B.C., Belluoccio D., Rowley L., Little C.B., Hansen U., Bateman J.F. Cartilage intermediate layer protein 2 (CILP-2) is expressed in articular and meniscal cartilage and down-regulated in experimental osteoarthritis. J. Biol. Chem. 2011;286(43):37758–37767. doi: 10.1074/jbc.M111.248039. https://doi: 10.1074/jbc.M111.248039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wilda M., Bächner D., Just W., Geerkens C., Kraus P., Vogel W., Hameister H. A comparison of the expression pattern of five genes of the family of small leucine-rich proteoglycans during mouse development. J. Bone Miner. Res. 2000;15(11):2187–2196. doi: 10.1359/jbmr.2000.15.11.2187. https://doi: 10.1359/jbmr.2000.15.11.2187 [DOI] [PubMed] [Google Scholar]
- 67.Jepsen K.J., Wu F., Peragallo J.H., Paul J., Roberts L., Ezura Y., Oldberg A., Birk D.E., Chakravarti S. A syndrome of joint laxity and impaired tendon integrity in lumican- and fibromodulin-deficient mice. J. Biol. Chem. 2002;277(38):35532–35540. doi: 10.1074/jbc.M205398200. https://doi: 10.1074/jbc.M205398200 [DOI] [PubMed] [Google Scholar]
- 68.Rock M.J., Holden P., Horton W.A., Cohn D.H. Cartilage oligomeric matrix protein promotes cell attachment via two independent mechanisms involving CD47 and αVβ3 integrin. Mol. Cell. Biochem. 2010;338(1–2):215–224. doi: 10.1007/s11010-009-0355-3. https://doi: 10.1007/s11010-009-0355-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Södersten F., Hultenby K., Heinegård D., Johnston C., Ekman S. Immunolocalization of collagens (I and III) and cartilage oligomeric matrix protein in the normal and injured equine superficial digital flexor tendon. Connect. Tissue Res. 2013;54(1):62–69. doi: 10.3109/03008207.2012.734879. https://doi: 10.3109/03008207.2012.734879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shibata S., Fukada K., Imai H., Abe T., Yamashita Y. In situ hybridization and immunohistochemistry of versican, aggrecan and link protein, and histochemistry of hyaluronan in the developing mouse limb bud cartilage. J. Anat. 2003;203(4):425–432. doi: 10.1046/j.1469-7580.2003.00226.x. https://doi: 10.1046/j.1469-7580.2003.00226.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lo P.H., Lung H.L., Cheung A.K., Apte S.S., Chan K.W., Kwong F.M., Ko J.M., Cheng Y., Law S., Srivastava G. Extracellular protease ADAMTS9 suppresses esophageal and nasopharyngeal carcinoma tumor formation by inhibiting angiogenesis. Canc. Res. 2010;70(13):5567–5576. doi: 10.1158/0008-5472.CAN-09-4510. https://doi: 10.1158/0008-5472.CAN-09-4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Du W., Wang S., Zhou Q., Li X., Chu J., Chang Z., Tao Q., Ng E.K., Fang J., Sung J.J., Yu J. ADAMTS9 is a functional tumor suppressor through inhibiting AKT/mTOR pathway and associated with poor survival in gastric cancer. Oncogene. 2013;32(28):3319–3328. doi: 10.1038/onc.2012.359. https://doi: 10.1038/onc.2012.359 [DOI] [PubMed] [Google Scholar]
- 73.Shin H., Lee M.N., Choung J.S., Kim S., Choi B.H., Noh M., Shin J.H. Focal adhesion assembly induces phenotypic changes and dedifferentiation in chondrocytes. J. Cell. Physiol. 2016;231(8):1822–1831. doi: 10.1002/jcp.25290. https://doi: 10.1002/jcp.25290 [DOI] [PubMed] [Google Scholar]
- 74.Akasaki Y., Alvarez-Garcia O., Saito M., Caramés B., Iwamoto Y., Lotz M. FoxO transcription factors support oxidative stress resistance in human chondrocytes. Arthritis Rheum. 2014;66(12):3349–3358. doi: 10.1002/art.38868. https://doi: 10.1002/art.38868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu S., Cao C., Zhang Y., Liu G., Ren W., Ye Y., Sun T. PI3K/Akt inhibitor partly decreases TNF-α-induced activation of fibroblast-like synoviocytes in osteoarthritis. J. Orthop. Surg. Res. 2019;14(1):425. doi: 10.1186/s13018-019-1394-4. https://doi: 10.1186/s13018-019-1394-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mhanna R., Kashap A., Palazzolo G., Vallmajo-Martin Q., Zenobi-Wong M. Chondrocyte culture in three dimensional alginate sulfate hydrogels promotes proliferation while maintaining expression of chondrogenic markers. Tissue Eng. 2014;20(9–10):1454–1464. doi: 10.1089/ten.tea.2013.0544. https://doi: 10.1089/ten.TEA.2013.0544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Balakrishnan B., Joshi N., Jayakrishnan A., Banerjee R. Self-crosslinked oxidized alginate/gelatin hydrogel as injectable, adhesive biomimetic scaffolds for cartilage regeneration. Acta Biomater. 2014;10(8):3650–3663. doi: 10.1016/j.actbio.2014.04.031. https://doi: 10.1016/j.actbio.2014.04.031 [DOI] [PubMed] [Google Scholar]
- 78.Knudson C.B. Hyaluronan receptor-directed assembly of chondrocyte pericellular matrix. J. Cell Biol. 1993;120(3):825–834. doi: 10.1083/jcb.120.3.825. https://doi: 10.1083/jcb.120.3.825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu Y., Stoddart M.J., Wuertz-Kozak K., Grad S., Ferguson S.J. Hyaluronan supplementation as a mechanical regulator of cartilage tissue development under joint-kinematic-mimicking loading. J. R. Soc. Interface. 2017;14(133):20170255. doi: 10.1098/rsif.2017.0255. https://doi: 10.1098/rsif.2017.0255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Milan D.C., Arce G.S., Moaied M., Caturla M.J., Untiedt C. Identification, characterization and expression analysis of a new fibrillar collagen gene, COL27A1, Matrix. Biol. 2003;22(1):3–14. doi: 10.1016/s0945-053x(03)00007-6. https://doi: 10.1016/s0945-053x(03)00007-6 [DOI] [PubMed] [Google Scholar]
- 81.Jenkins E., Moss J.B., Pace J.M., Bridgewater L.C. The new collagen gene COL27A1 contains SOX9-responsive enhancer elements. Matrix Biol. 2005;24(3):177–184. doi: 10.1016/j.matbio.2005.02.004. https://doi: 10.1016/j.matbio.2005.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.